Abstract
The sense of taste is an important sentinel governing what should or should not be ingested by an animal, with high pH sensation playing a critical role in food selection. Here we explore the molecular identities of taste receptors detecting the basic pH of food using Drosophila melanogaster as a model. We identify a chloride channel named alkaliphile (Alka), which is both necessary and sufficient for aversive taste responses to basic food. Alka forms a high-pH-gated chloride channel and is specifically expressed in a subset of gustatory receptor neurons (GRNs). Optogenetic activation of alka-expressing GRNs is sufficient to suppress attractive feeding responses to sucrose. Conversely, inactivation of these GRNs causes severe impairments in the aversion to high pH. Altogether, our discovery of Alka as an alkaline taste receptor lays the groundwork for future research on alkaline taste sensation in other animals.
Introduction
The sense of taste, which lies at the interface between the interior and the exterior of the body, ensures that food of nutritional value is consumed, whereas potentially noxious substances are rejected1,2. Acids and bases are opposite chemical substances that are broadly present in food sources3. While it is generally accepted that animals use sour taste to assess the acidity, or low pH, of food4–6, whether animals have a taste modality to sense the basicity, or high pH, of food is a long-standing open question. Given that acid, or low pH, has a sour taste, it would be logical to hypothesize that base, or high pH, also produces a gustatory sensation.
Previous studies in humans and animal models provide initial clues to the existence of alkaline taste sensations. In the 1940s, psychophysical studies conducted on human participants reported that the tip portion of the tongue, which is enriched with taste buds, exhibits a higher sensitivity to sodium hydroxide (NaOH) than the mid-dorsal part of the tongue with few taste buds, implying that basic solutions may have taste qualities7. Furthermore, electrophysiological recordings of the taste nerves in cats document that a subpopulation of the chorda tympani nerves, which relay taste input from the oral cavity to the brain, can be activated by high pH but not by other stimuli such as temperature8, indicating that cats interpret the stimulation by high-pH solutions as a sense of taste rather than as an irritating noxious chemical. Moreover, insects such as beetles show robust avoidance of alkaline environments associated with unfavorable habitats and food sources9. The beetle’s high-pH sensitivity is mediated by pH receptor cells in the beetle’s taste organ, which display increased firing activities proportional to basic pH10. Collectively, these earlier studies imply but do not resolve whether the high-pH sensation is a discrete taste modality. Since these early studies, little mechanistic research had been done to unravel the molecular and cellular underpinnings of alkaline taste sensation. In particular, the molecular identities of taste receptors and taste receptor cells orchestrating alkaline taste sensation had not yet been established in animals.
Like mammals, the fruit fly, Drosophila melanogaster, employs different classes of taste receptors to detect sugars11–15, salts16–18, acids6,19–21, bitter substances22,23, and other chemicals24–29. Given that flies have such a remarkable capability to detect a wide range of substances through taste, we inferred that flies are also able to sense the alkalinity of food. Indeed, we isolated a vital fly gene dubbed alkaliphile (alka) that regulates gustatory responses to strong alkalinity. Our molecular genetic studies revealed that alka is both necessary and sufficient to avoid highly basic food. We performed extensive electrophysiological assays and discovered that the Alka protein forms a chloride (Cl−) channel, which is specifically activated by hydroxide (OH−). Moreover, we found that alka is expressed in a subset of GRNs in the peripheral taste organ. At the sensory cell level, alka-expressing GRNs are both necessary and sufficient for the rejection of strongly alkaline food. In summary, our work establishes Alka as the long-sought-after taste receptor responsible for sensing the basic pH of food.
Results
Requirement for alka to avoid highly basic food
We aimed to decipher the molecular nature of alkaline taste sensation by leveraging the fruit fly, Drosophila melanogaster, a well-established model organism for taste perception24,30. We chose the strong base NaOH to alter food pH because it is completely dissociated into Na+ and OH− ions once dissolved, therefore enabling us to obtain a wide range of basic pH with relatively low concentrations of NaOH. Furthermore, side effects arising from sodium taste can be genetically removed from flies. We found that wild-type flies, when given a choice between neutral food comprising 2 mM glucose alone (pH 7) and basic food containing the same concentration of glucose plus 100 mM NaOH (pH 13), predominantly preferred neutral over basic food (Extended Data Fig. 1a). Thus, our behavioral data indicate that wild-type flies are repelled by basic food. We surmise that flies detect food basicity through specific taste receptors, which are very likely to be transmembrane receptors or ion channels.
To identify the alkaline taste sensor, we employed the two-choice feeding assay to screen a plethora of mutants, which are defective for a large variety of receptors or ion channels whose physiological functions were either established or unclear (Extended Data Fig. 1a). These include the gustatory receptors (GRs)11,31, the ionotropic glutamate receptors (IRs)32, transient-receptor-potential (TRP) cation channels33, otopetrin channels5,6,20,34, and putative receptors or channels with unknown functions. According to our screen, the majority of mutant animals rejected alkaline food, but one mutant (CG12344MI11416), carrying a transposon insertion in the uncharacterized gene CG12344, displayed significantly reduced aversion to basic food (Extended Data Fig. 1a). The physiological function of CG12344 was unknown, yet bioinformatic analyses indicated that the protein encoded by CG12344 is distantly related to glycine receptors (GlyRs)35 and belongs to the ligand-gated chloride channel (LGCC) family in Drosophila36,37.
The fly genome encodes 12 LGCC members, which exhibit pronounced divergence with regard to ligand activation, expression pattern, and cellular function38–47. Among the fly LGCC family, many of the LGCCs are orphan channels because their activating ligands and physiological functions remain largely unclear. Consequently, we asked whether the other LGCC family members in Drosophila were involved in the detection of basic food. We used either null mutants or GRN-specific knockdown to determine if any of the LGCC genes were required for the taste response to alkaline food. Specifically, we used the poxn-Gal4 line, which drives expression in the entire population of GRNs6,48, to specifically knock down most of the LGCC genes in the labellum, a primary taste organ of the fly. Our two-choice feeding assays revealed that selective knockdown of CG12344 but not the other LGCC genes led to significant defects in the avoidance of basic food (pH 13) (Extended Data Fig. 1b). In conclusion, we propose that CG12344 is an excellent candidate alkaline-taste receptor. Since CG12344 mutant flies showed enhanced acceptance of basic foods compared to wild-type flies, we dubbed the CG12344 gene alkaliphile (alka).
To determine the function of alka in detecting basic food, we successfully generated a null alka1 allele using CRISPR/Cas9-mediated genome editing. In the alka1 mutant, the DNA sequences encoding all four transmembrane segments (TM1-TM4) were ablated, leading to severe disruption of any potential ion channel activities (Extended Data Fig. 2a-c). As demonstrated by our two-choice feeding assays, wild-type animals exhibited increasing aversion toward moderately to highly alkaline food (Fig. 1a). In contrast, alka1 mutant flies displayed profound defects in alkaline food aversion, even showing a strong preference for food mixed with low levels of NaOH (for example, 10 mM) (Fig. 1a). We deduced that this might be caused by the inherent attraction to low concentrations of sodium (Na+)16 from NaOH when the aversion to OH− is eliminated in alka1 mutant flies. To test this hypothesis, we generated alka1;Ir76b1 double-mutant flies lacking both alka and Ir76b, the latter of which is required for salt taste sensation16. Indeed, we found that alka1;Ir76b1 double-mutant flies also showed severe impairments in avoiding alkaline food but no longer preferred moderately basic food (Fig. 1a), indicating that the preference for basic food in the alka1 mutant is due to low Na+. Further, to exclude any potential effects arising from Na+ and osmolarity on our two-choice feeding assays, we allowed the flies to choose between basic food containing 0.1–100 mM NaOH (pH 10–13) and neutral food comprising the same concentrations of NaCl (0.1–100 mM; pH 7) so that Na+ and osmolarity in both food options were balanced. Consistent with the above results, alka1 flies showed significantly reduced aversion to basic food (Fig. 1b); however, alka1 mutants were not more attracted to NaOH than to NaCl, because the sodium levels for both food options are identical (Fig. 1b). Furthermore, the mutant phenotype of alka1 flies was fully restored to normal by expressing a wild-type alka transgene in the alka1 mutant background (alka1;alka-Gal4/UAS-alka) (Fig. 1a, 1b). In summary, our two-choice feeding assays demonstrate that alka is selectively required for sensing high pH instead of Na+ or osmolarity.
Fig. 1. Behavioral and electrophysiological taste responses to alkaline foods depend on alka1.
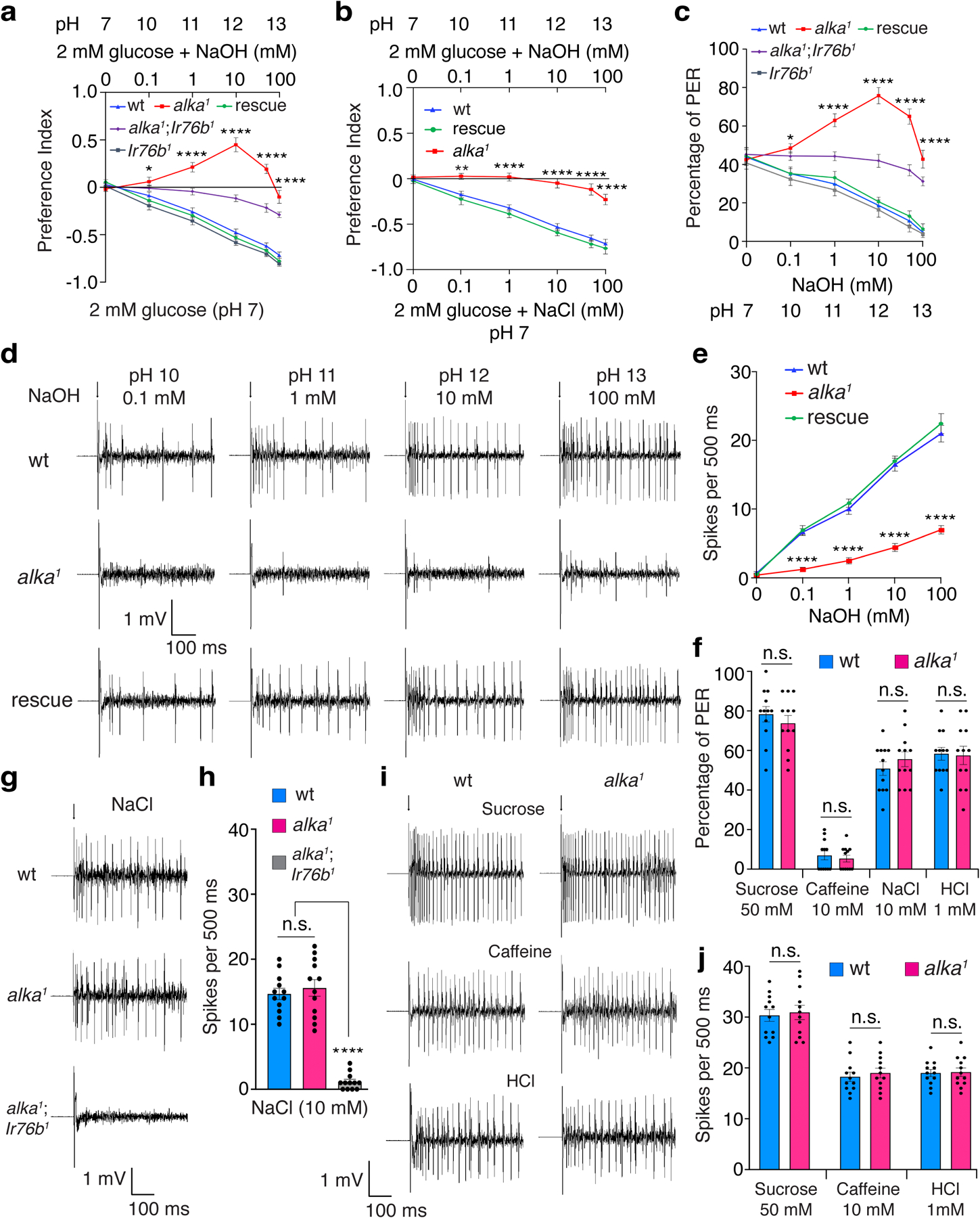
(a) Discrimination of neutral (pH 7) food comprising 2 mM glucose from basic foods (pH 10–13) containing 2 mM glucose plus 0.1–100 mM NaOH by wild-type (wt), alka1 mutant, Ir76b1 mutant, alka1;Ir76b1 double mutant, and rescue (alka1;alka-Gal4/UAS-alka) flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, *p=0.045, ****p<0.0001. (b) Feeding preference of neutral (pH 7) foods containing 2 mM glucose plus 0.1–100 mM NaCl versus basic foods (pH 10–13) comprising 2 mM glucose plus 0.1–100 mM NaOH among wt, alka1 mutant, and rescue (alka1;alka-Gal4/UAS-alka) flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, **p=0.0037, ****p<0.0001. (c) PERs to foods containing 30 mM sucrose with various concentrations of NaOH among wt, alka1 mutant, Ir76b1 mutant, alka1;Ir76b1 double mutant, and rescue (alka1;alka-Gal4/UAS-alka) flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, *p=0.014, ****p<0.0001. (d) Representative tip recording traces showing spikes elicited by S6 sensilla in wt, alka1 mutant, and rescue (alka1;alka-Gal4/UAS-alka) flies in response to alkaline pH stimuli. (e) Statistical analyses of the frequencies of spikes collected in d. n=15 flies. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests. ****p<0.0001. (f) PERs to sucrose (50 mM, p=0.418), caffeine (10 mM, p=0.583), NaCl (10 mM, p=0.359), and HCl (1 mM, p=0.884) in wt and alka1 flies. n=12 trials. Mean ± SEM, unpaired two-tailed Student’s t-tests. (g, h) Representative spikes (g) and quantification of the frequencies of spikes (h) evoked by NaCl at L4 sensilla among wt, alka1, and alka1;Ir76b1 flies. n=12 flies. Mean ± SEM, one-way ANOVA with Tukey’s post-hoc tests, p=0.763 (wt vs. alka1), ****p<0.0001. (i, j) Representative spikes (i) and quantification of the frequencies of spikes (j) elicited by sucrose (50 mM, p=0.753) at L4 sensilla, caffeine (10 mM, p=0.581) at S6 sensilla, and HCl (1 mM, p=0.881) at L6 sensilla for wt and alka1 flies. n=12 flies. Mean ± SEM, unpaired two-tailed Student’s t-tests. Arrows indicate the onset of taste stimuli (d, g). n.s., not statistically significant.
To directly assess the fly’s taste response to alkaline food, we employed proboscis-extension-reflex (PER) assays to directly assess the fly’s taste response to basic food49. Wild-type flies extended their proboscises to feed if their proboscises were transiently touched with a probe containing an appealing food. Wild-type flies showed about 45% PERs toward 30 mM sucrose, but the PER percentage declined as the food became more basic (Fig. 1c). In contrast, alka1 mutants displayed severe defects in rejecting alkaline food. Similar to our previous results, alka1 mutants showed dramatically increased PERs to moderately basic food. This abnormal PER phenotype was abolished in alka1;Ir76b1 double mutants (Fig. 1c), suggesting that the attraction to moderately basic food is due to low concentrations of salt. Additionally, the abnormality of the alka1 mutant’s PERs to basic food was fully rescued by expressing a wild-type alka transgene (Fig. 1c). Collectively, our behavioral and genetic analyses indicate that alka is required for perceiving basic food.
alka is necessary for GRNs to sense high pH
To address whether alka is functionally required for GRNs to sense high pH, we performed tip recording, a robust electrophysiological technique used to interrogate the neuronal activity of GRNs6,16. There are three groups of taste sensilla that house GRNs in the fly proboscis, including the large (L), intermediate (I), and small (S) types6,50,51. To determine whether these three types of sensilla differentially respond to high pH, we carried out an electrophysiological survey of their responses to 10 mM NaOH. Our tip recordings demonstrated that among the L-, I-, and S-type sensilla, S-type sensilla produced the most robust responses to NaOH (Extended Data Fig. 3a). Therefore, we focused our electrophysiological analyses on S-type sensilla. After being stimulated with a series of NaOH concentrations (0.1–100 mM), S-type sensilla, such as S6 in wild-type flies, fired a train of action potentials. As the NaOH concentration rose, the spike intensity increased remarkably (Fig. 1d, 1e). In keeping with our behavioral data, the frequency of spikes produced by the alka1 mutant was significantly lower than wild type. This defect was fully restored in rescue animals (alka1;alka-Gal4/UAS-alka) (Fig. 1d, 1e).
To ascertain that the action potentials detected at S6 sensilla were mainly evoked by OH−, we examined the responses of S6 sensilla to low salt (0.1–100 mM NaCl). We only detected weak responses to low salt, and the spike frequencies were comparable between wild-type and alka1 mutant flies (Extended Data Fig. 3b, 3c). As such, this indicates that the spike responses of S6 sensilla evoked by NaOH are predominantly contributed by OH− instead of Na+ ions.
To determine whether the loss of alka affects other taste modalities, including sweet, bitter, salty, and sour tastes, we examined the PERs to sucrose, caffeine, sodium chloride (NaCl), and hydrochloric acid (HCl) in alka1 mutants, with wild-type flies as controls. According to our PER assays, alka1 mutant and wild-type flies showed no significant difference in the percentage of PER to sucrose, caffeine, salt, or acid (Fig. 1f). In accordance with the PER assays, our tip recording analyses demonstrated that wild-type and alka1 flies displayed comparable gustatory responses to salt. As controls, the spikes evoked by NaCl were eliminated in the alka1;Ir76b1 double mutant (Fig. 1g, 1h). Furthermore, wild-type and alka1 flies also exhibited similar taste responses to sucrose, caffeine, and acid (Fig. 1i, 1j). In summary, alka is responsible for the gustatory sensation of high pH rather than other taste substances tested.
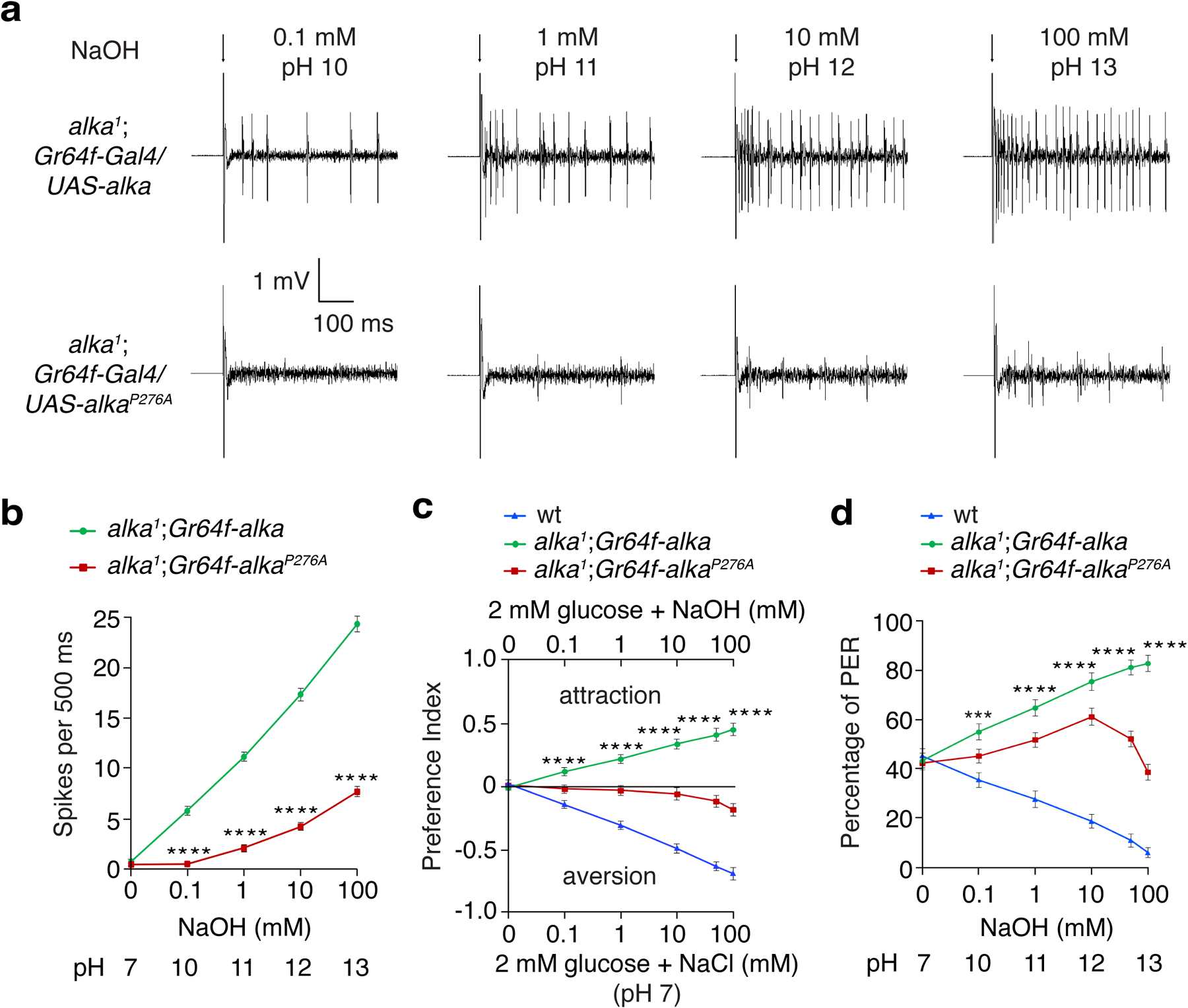
Moreover, we were curious to know whether alka was also required to detect high pH contributed by weak bases in addition to strong bases such as NaOH. To test this idea, we chose sodium carbonate (Na2CO3)52 because it is a weak nonvolatile base that yields little interference from the olfactory system, enabling us to specifically determine its effect on taste. Furthermore, Na2CO3 is broadly present in the natural ecosystem, where many animals such as insects live and eat. Excessive carbonate consumption can imbalance pH homeostasis, consequently leading to alkalosis of the animal’s body fluid53. Therefore, Na2CO3 is an ethologically and ecologically relevant taste substance frequently encountered by animals during food foraging and feeding. Our two-choice feeding and PER assays revealed that alka1 flies showed significantly decreased aversions to basic foods containing Na2CO3 (Fig. 2a, 2b). Similar to NaOH, alka1 mutants exhibited preferences for foods containing low concentrations of Na2CO3. This was due to the remaining attractive response to low salt because no attraction to Na2CO3 was observed in alka1;Ir76b1 double-mutant flies (Fig. 2a, 2b). In support of this result, our tip-recording analyses demonstrated that there was a significant decrease in the number of action potentials evoked by Na2CO3 in alka1 mutants compared to wild-type controls (Fig. 2c, 2d). These behavioral and electrophysiological abnormalities were fully rescued in alka1;alka-Gal4/UAS-alka flies (Fig. 2a–d). Collectively, we propose that alka is required to detect high pH produced by either strong or weak bases.
Fig. 2. alka regulates the aversive feeding and physiological responses to foods containing sodium carbonate (Na2CO3).
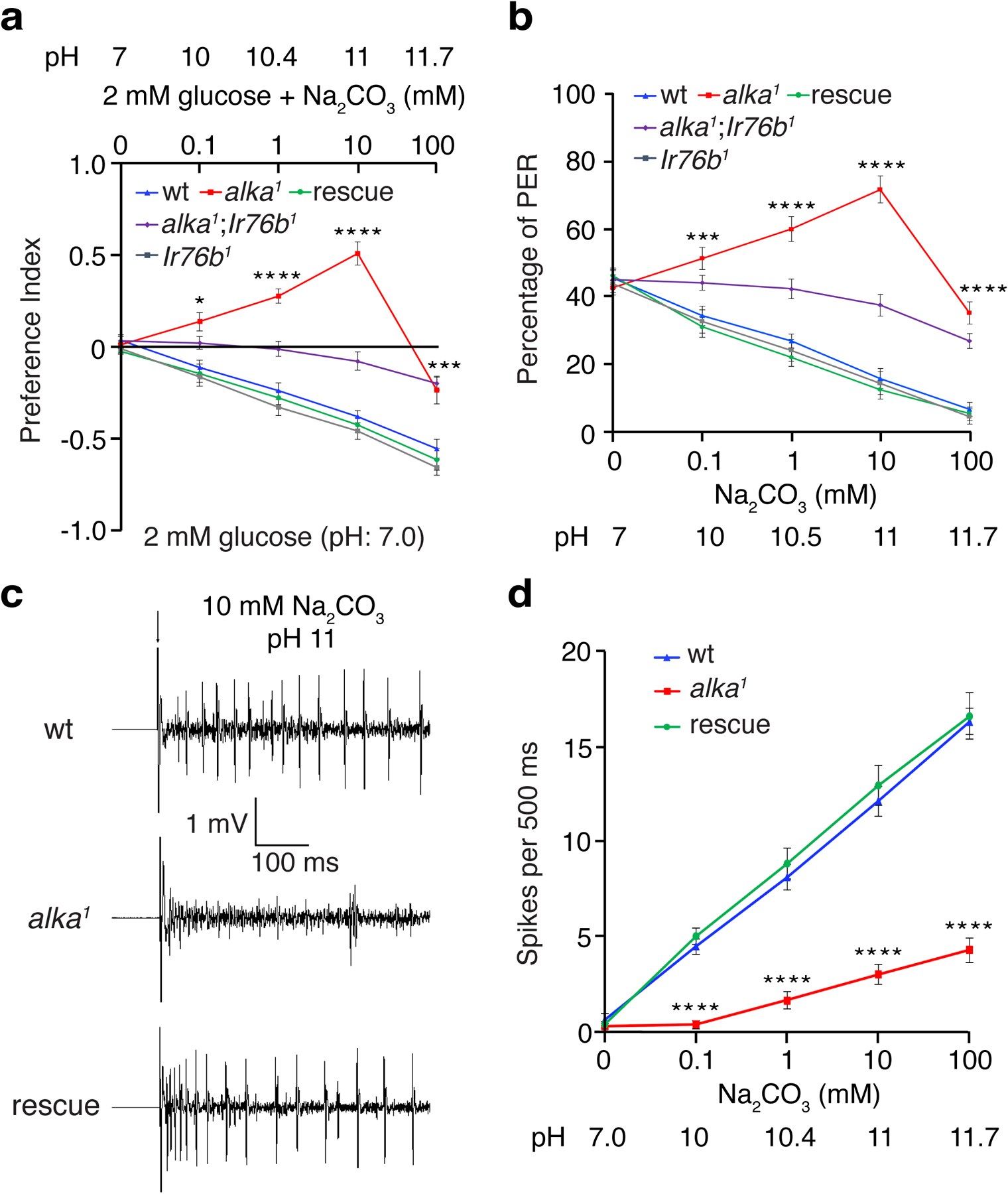
(a) Feeding choices of wild-type (wt), alka1 mutant, Ir76b1 mutant, alka1;Ir76b1 double mutant, and rescue (alka1;alka-Gal4/UAS-alka) flies when presented with neutral food containing 2 mM glucose and basic foods composed of 2 mM glucose mixed with varying concentrations of Na2CO3 (0.1 mM, pH 10; 1 mM, pH 10.4; 10 mM, pH 11; and 100 mM, pH 11.7). n=11 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, *p=0.014, ***p=0.001, ****p<0.0001. (b) PERs to liquid foods containing 30 mM sucrose and different concentrations of Na2CO3 among wt, alka1 mutant, Ir76b1 mutant, alka1;Ir76b1 double mutant, and rescue flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, ***p=0.0003, ****p<0.0001. (c) Representative action potentials evoked by 10 mM Na2CO3 (pH 11) at S6 sensilla for wt, alka1 mutant, and rescue flies. The arrow indicates stimulus onset. (d) Statistical analyses of action potentials produced by S6 sensilla in response to the indicated concentrations of Na2CO3 among wt, alka1 mutant, and rescue flies. n=12 flies. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001.
alka is expressed in a subset of GRNs in the fly labellum
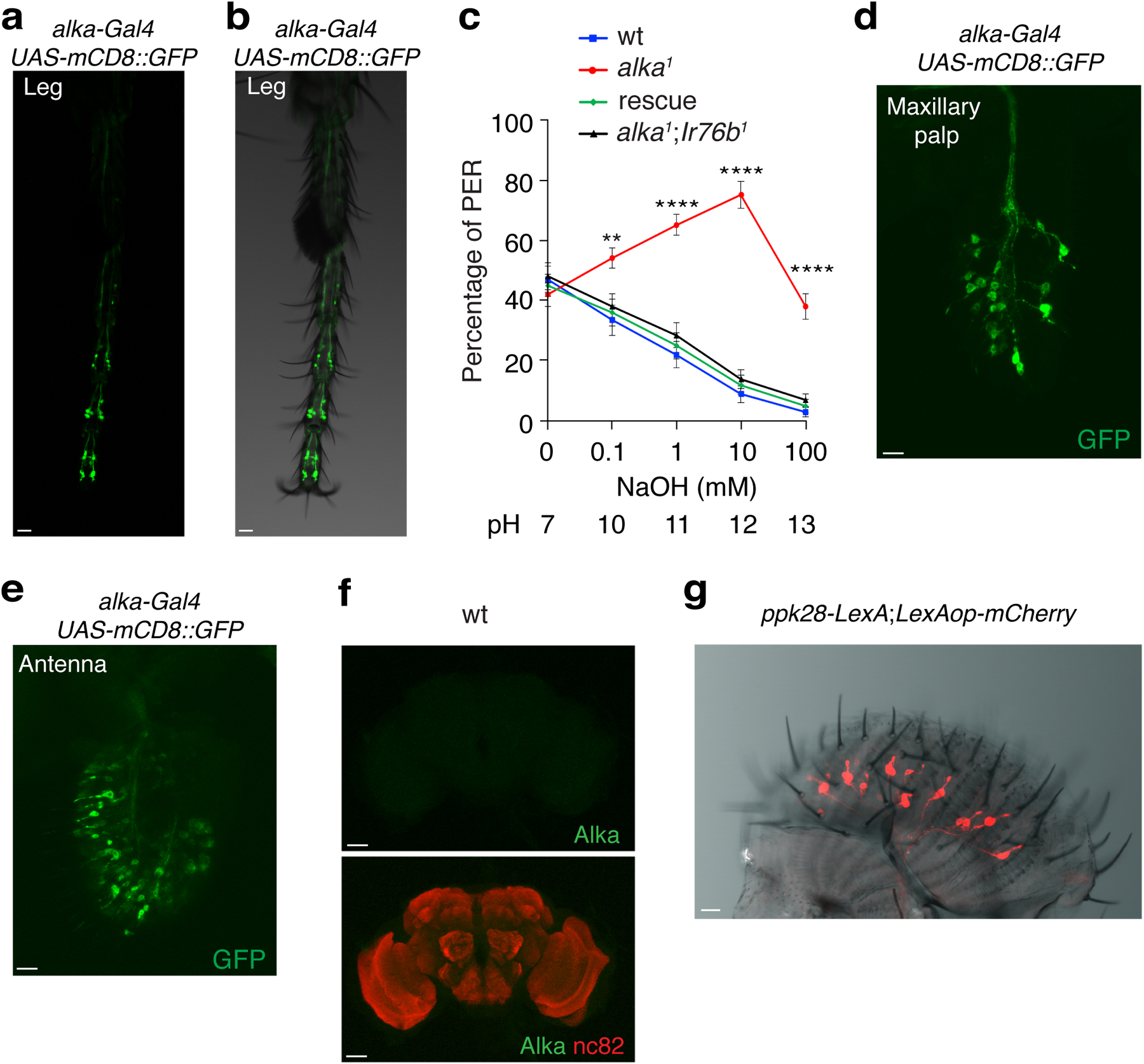
To examine the expression pattern of alka in the labellum, the fly’s primary taste organ, we conducted immunohistochemical assays using Alka antibodies that we generated in this study. We observed a group of immunoreactive GRNs present in the wild type (Fig. 3a) but not in the alka1 mutant (Fig. 3b), affirming the specificity of the Alka antibody. Moreover, we generated a promoter Gal4 line for alka and examined the expression of the alka-Gal4 using a green fluorescent protein (GFP) reporter54. We found that the alka-Gal4 was specifically expressed in a subset of GRNs, a bipolar-neuron type with characteristic structures including dendrite, soma, and axon (Fig. 3c). In agreement with our tip recordings, the dendrites of alka-expressing GRNs mostly innervated S-type sensilla (Fig. 3d). Our double-labeling of alka-Gal4;UAS-mCD8::GFP flies revealed that nearly all the alka-Gal4 GRNs colocalized with those marked by Alka antibodies (Fig. 3e–g), indicating that the alka-Gal4 mostly reflected the endogenous expression of alka in the fly labellum. Further, we found that only a small percentage of GRNs specified by the alka-Gal4 (21.6%) overlapped with Gr66a-expressing (bitter) GRNs (Fig. 3h)55, while showing little colocalization with Gr64f-expressing (sweet) GRNs (Fig. 3i)15,56 or Ir76b-expressing GRNs involved in the taste sensations of low concentrations of salt (Fig. 3j)16,18 and other substances57. Furthermore, the alka-expressing GRNs exhibited no obvious overlap with other types of GRNs, such as ppk28-expressing GRNs responsible for detecting water (Fig. 3k)58,59 or ppk23-expressing GRNs involved in sensing high concentrations of calcium as well as pheromones (Fig. 3l)28,60. Additionally, the axons of alkaline GRNs projected to the subesophageal zone (SEZ), a brain region involved in processing taste signals (Fig. 3m). In addition to the fly labellum, we found that alka-Gal4 was also expressed in chemosensory neurons at the tarsus, the last segment of the fly leg (Extended Data Fig. 4a, 4b). To determine whether the alka-expressing tarsal neurons contributed to the detection of high pH, we used high pH solutions to selectively stimulate the tarsal segment rather than the proboscis of the fly while performing PER assays. In keeping with our previous PERs collected on the fly proboscis, alka1 mutant flies exhibited significant deficits in rejecting high pH. This defect was restored to normal in alka1;alka-Gal4/UAS-alka rescue flies (Extended Data Fig. 4c), suggesting that alka was also required at the tarsi to sense high pH from the external environment. Additionally, alka-Gal4 expression was detected in the peripheral olfactory organs such as the antenna and maxillary palp (Extended Data Fig. 4d, 4e). Notably, we failed to observe any alka-Gal4 expression in the whole fly brain except the axonal projections of chemosensory neurons from the labellum, the antenna, and the maxillary palp (Fig. 3m). Consistently, we observed little, if any, Alka expression in the central brain of the wild-type adult fly (Extended Data Fig. 4f). This distinct expression pattern of alka further substantiated the notion that Alka mainly functioned as a chemoreceptor instead of a neurotransmitter receptor like GlyR, as the latter is typically expressed broadly in the brain61.
Fig. 3. Expression pattern of alka in the fly labellum.
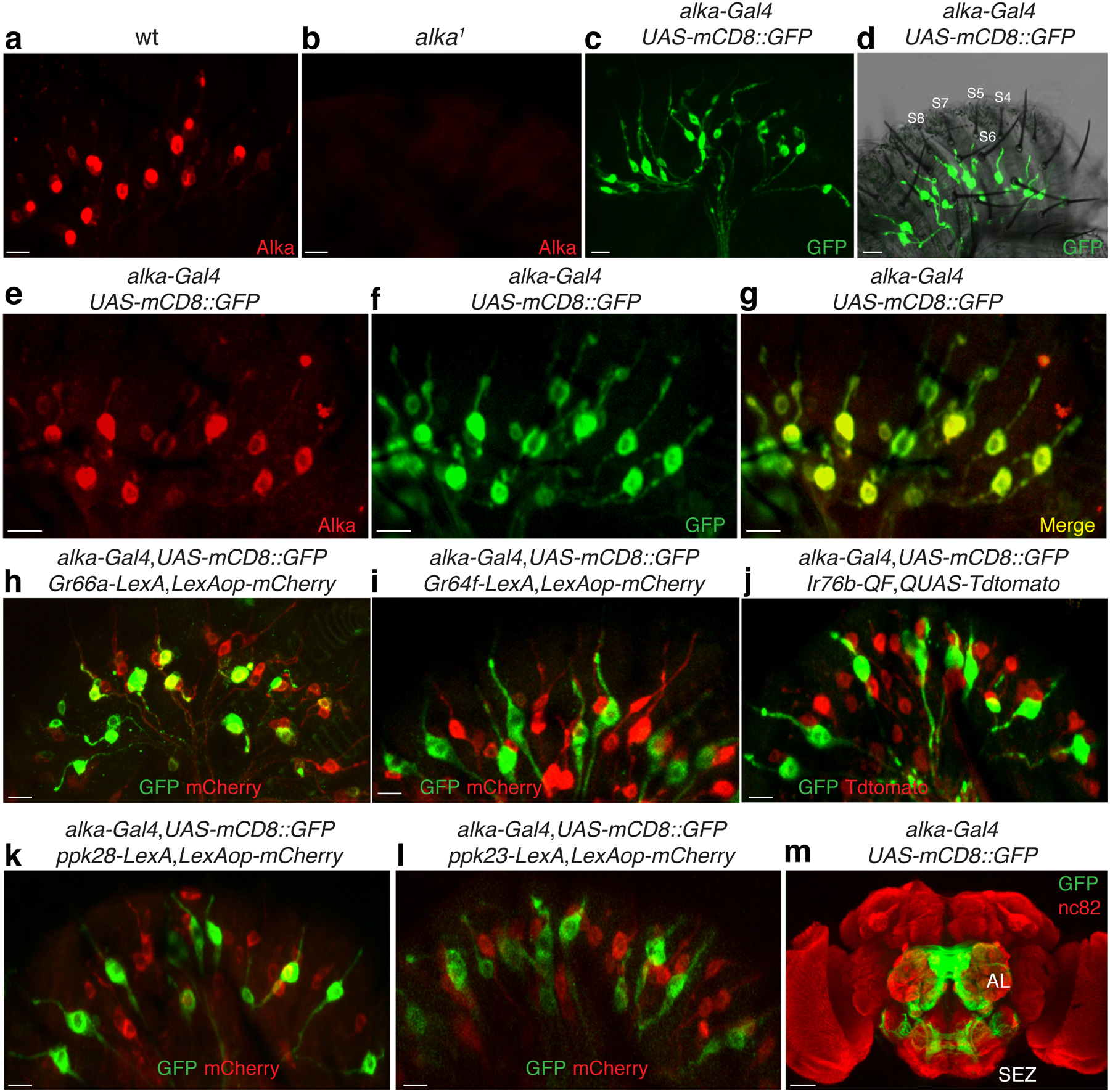
(a, b) Immunoreactivities of the Alka antibody in wild-type (wt) (a) and alka1 mutant (b) labella. (c) GRNs detected by the alka-Gal4;UAS-mCD8::GFP reporter. (d) alka-expressing GRNs predominately innervate the S-type sensilla. Several S-type sensilla (S4-S8) are marked. (e-g) Double-labeling of the alka-Gal4;UAS-mCD8::GFP fly with (e) anti-GFP and (f) anti-Alka. The (g) merged image shows that anti-GFP and anti-Alka signals mostly colocalize. (h-l) Localization pattern of alkaline GRNs relative to (h) Gr66a-, (i) Gr64f-, (j) Ir76b-, (k) ppk28-, and (l) ppk23-expressing GRNs. (m) The projection pattern of alka-Gal4;UAS-mCD8::GFP in the fly brain. Subesophageal zone (SEZ), antennal lobe (AL). Scale bars: 10 μm (a-l), 50 μm (m).
It has been established that there are four GRNs embedded underneath each of the 12 S-type taste hairs51. Consistent with the literature55, we found that most S-type sensilla (S1-S12) housed one GRN expressing Gr66a. Moreover, most S-type sensila included one alka-expressing GRN. A portion of the S-type taste hairs contained ppk23-expressing GRNs and Gr64f-expressing GRNs. Additionally, we observed few ppk28-expressing GRNs innervating the S-type taste sensilla; instead, most of them innervated the L-type sensilla (Extended Data Figure 4g). In keeping with our anatomical results, no obvious water-induced neuronal spikes were detected at the S-type sensilla in wild-type or ppk28 mutant flies, indicating that ppk28 is not functionally required at the S-type sensilla. Our finding is consistent with the established role of ppk28 in attractive water taste sensation at the L-type sensilla58,59.
Alka forms a high-pH-activated Cl− channel
At this stage, our findings implied that Alka was likely a direct sensor for high pH. Moreover, protein sequence analysis revealed that the fly Alka protein showed a low level of homology (30% identity) to vertebrate GlyRs, such as glycine receptor alpha1 (GlyRa1), in zebrafish, mice, and humans (Extended Data Fig. 5). Since GlyRs are Cl− channels35,62, we deduced that Alka might form a high-pH-activated ion channel. To test this hypothesis, we used whole-cell patch-clamp recording6 to interrogate the potential channel function of Alka in heterologous HEK293 cells. We observed that Alka proteins were expressed at the surface of HEK293 cells (Extended Data Fig. 6a), making them suitable for whole-cell patch-clamp analyses. To stimulate the HEK293 cells, we used a micropipette to locally apply basic isotonic solutions to the patched cells (Extended Data Fig. 6b). Of primary importance, Alka-expressing HEK293 cells generated robust inward currents when stimulated by basic isotonic solutions (pH 12). In contrast, few currents were detected in control cells without Alka expression (Fig. 4a, 4b). To ascertain what ions contribute to the inward conductance, we replaced the ions in the external bath and internal pipette solutions. Under isotonic conditions, when CsCl was used as the internal solution and N-methyl-D-glucamine (NMDG)-Cl or NMDG-gluconate as the external solution (NMDG and gluconate ions are membrane-impermeable)6, we still detected comparable levels of inward currents (Fig. 4a, 4b). However, when we further replaced the internal Cl− with membrane-impermeable methanesulfonate (MeSO4−), we observed no obvious currents (Fig. 4a, 4b). Therefore, our ion replacements suggested that the high-pH-evoked inward currents were primarily due to Cl− efflux instead of Na+ influx through the Alka channel. To further establish that Alka encodes a Cl− ion channel, we analyzed differential activations of the Alka channel by high pH (pH 12) under a series of intracellular chloride concentrations. Increasing the intracellular chloride concentration potentiated inward currents considerably (Fig. 4c, 4d) and positively shifted the reversal potential (Erev) (Fig. 4e, 4f) in a manner closely following the prediction of Nernst equation for a chloride-selective ion channel35. The current/voltage (I/V) relationship curve of the Alka channel was nearly linear, indicating that its conductance was mostly independent of voltage gating (Fig. 4e). Further, we used bi-ionic conditions to analyze the Alka channel’s permeability for Cl− relative to other anions such as F−, Br−, and I−. We found that the Alka channel was essentially conductive to all halide ions with a slight preference for I− and Br− over Cl− (Extended Data Fig. 6c-e). Furthermore, we found that Alka-expressing HEK293 cells failed to produce any obvious currents when stimulated by isotonic acidic solutions (pH 2.0–6.0) (Extended Data Fig. 6f, 6g). Moreover, no obvious currents were detected when the Alka-expressing HEK293 cells were stimulated by glycine (Extended Data Fig. 6h, 6i) or gamma-aminobutyric acid (GABA) (Extended Data Fig. 6j, 6k) within the physiological concentration range (less than 1 mM)63. Collectively, our whole-cell patch-clamp analyses indicate that Alka forms a high-pH-activated Cl− channel.
Fig. 4. Alka forms a high-pH-activated Cl− channel in HEK293 cells.
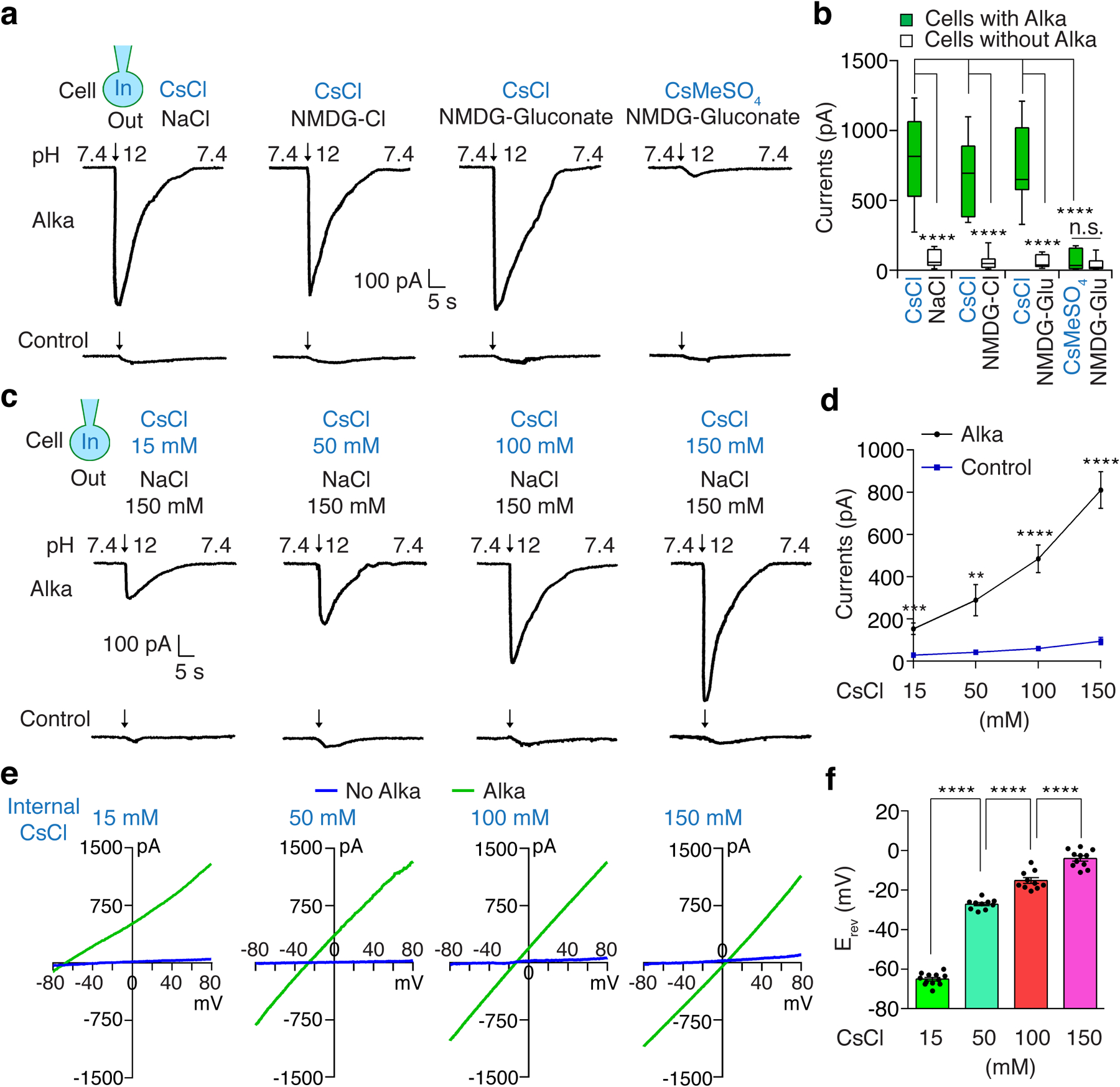
(a) Representative whole-cell currents evoked by the alkaline pH (pH 12) in HEK293 cells with or without Alka expression under different bath (out)/pipette (in) solutions (150 mM each): NaCl/CsCl, NMDG-Cl/CsCl, NMDG-gluconate/CsCl, and NMDG-gluconate /CsMeSO4. (b) Statistical analyses of inward currents from a. Cells expressing Alka: n=12 for NaCl/CsCl, NMDG-Cl/CsCl, or NMDG-gluconate/CsCl; n=11 for NMDG-gluconate/CsMeSO4. Cells without Alka expression: n=12 for NMDG-Cl/CsCl; n=11 for any other groups. One-way ANOVA with Tukey’s post-hoc tests, p=0.996 (NMDG-gluconate/CsMeSO4: Alka vs. control), ****p<0.0001, n.s., not statistically significant. (c) Whole-cell currents evoked by basic isotonic solutions (pH 12) in HEK293 cells with or without Alka expression. The pipette solutions contained various levels of CsCl (15–150 mM). Cs-methanesulfonate was used to maintain the osmolarity of pipette solutions; 150 mM NMDG-Cl served as the bath solution. Arrows indicate the onset of basic-pH stimuli. (d) Statistical analyses of currents in c. n=11 cells. Mean ± SEM, unpaired two-tailed Student’s t-tests, **p=0.0034, ***p=0.0001, ****p<0.0001. (e) Current/voltage (I/V) relationships of Alka-expressing cells and control cells without Alka from experiments in c, with voltage ramps from −80 to + 80 mV. (f) Reversal potentials of the Alka conductances with varying intracellular Cl− concentrations: 15 mM CsCl (n=12 cells); 50 mM CsCl (n=10 cells); 100 mM CsCl (n=10 cells); and 150 mM CsCl (n=11 cells). The bath solution contained 150 mM NMDG-Cl. Mean ± SEM, one-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001. Cells were clamped at −70 mV (a-d).
Topologically resembling the glycine-gated Cl− channels64, the Alka channel complex is predicted to be a pentamer with five transmembrane segments 2 (TM2) helices lining the ion pore (Fig. 5a, 5b). We focused on an evolutionarily conserved proline residue P276 located in the TM2 segment (Fig. 5a–c), given that proline residues play important roles in shaping the conformation of transmembrane helices65. To determine the function of P276, we mutated it to alanine (A). Like wild-type Alka, AlkaP276A was readily detected at the cell surface of HEK293 cells (Extended Data Fig. 6a). For HEK293 cells expressing wild-type Alka, the amplitude of inward current significantly elevated as the pH of the basic isotonic solution increased from 10 to 13. However, few currents were detectable in cells expressing the AlkaP276A mutant (Fig. 5d–f), demonstrating that P276 is critical for Cl− permeation across the channel. In summary, we propose Alka as a high-pH-gated Cl− channel. This raises the question of how fly GRNs sense high pH through Alka. Typically, the intracellular Cl− concentration in the fly taste receptor neurons is higher than in the extracellular lymph66. Thus, we inferred that the opening of Alka channels by OH− ions led to massive Cl− efflux from the GRN, thereby depolarizing the GRN (Fig. 5g).
Fig. 5. The P276 residue is essential for the conductance of Alka.
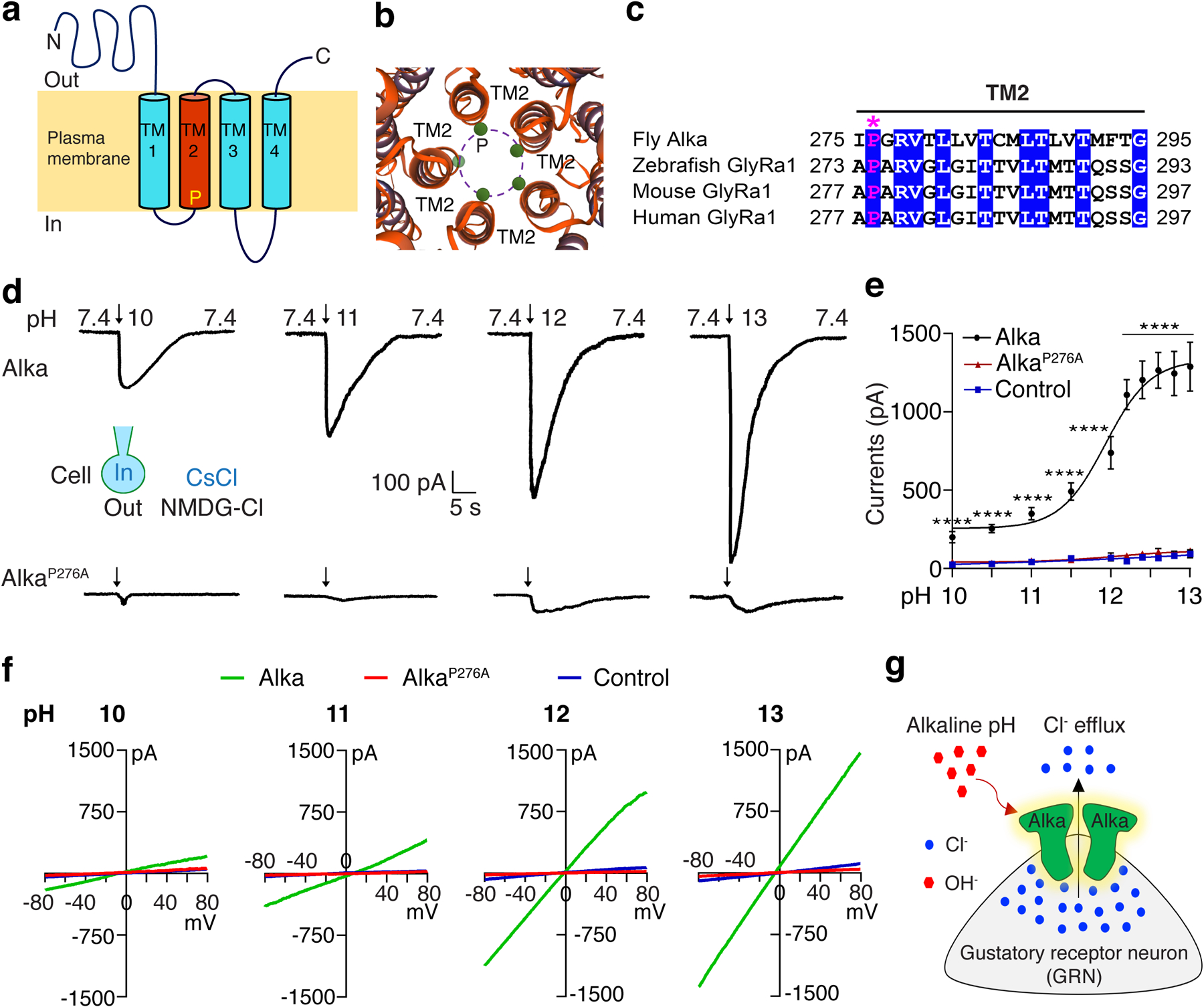
(a) The predicted topology of Alka, with a conserved proline residue (P) in the second transmembrane segment (TM2). (b) Alka is predicted to form a pentameric Cl− channel with five TM2 helices lining the ion pore. The green circles depict the pore-lining proline residues. (c) Multi-sequence alignment of TM2 for fly Alka and GlyRa1 from zebrafish, mice, and humans. The asterisk (*) marks the highly conserved proline residue. (d) Representative whole-cell currents detected in Alka- or AlkaP276A-expressing cells in response to basic-pH stimuli (pH 10–13). Bath/pipette solutions were NMDG-Cl/CsCl (150 mM each). (e) Dose-response curve of inward currents evoked by varying basic-pH solutions. Cells were held at −70 mV. The pH50 for Alka is 11.9. Alka: n=11 cells (pH 12, 12.2, 12.4, 12.6,12.8, and 13); n=12 cells (pH 10, 10.5, and 11.5); n=14 cells (pH 11). AlkaP276A: n=11 cells for each group. Control: n=11 cells (pH 10.5, 11.5, 12.2, 12.4, 12.6, 12.8, and 13); n=12 cells (pH 10); n=13 cells (pH 11); n=14 cells (pH 12). Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests. ****p<0.0001. (f) Current/voltage (I/V) relationships of Alka-expressing cells, AlkaP276A-expressing cells, and control cells without Alka or AlkaP276A expression in response to basic-pH stimuli (pH 10–13). Voltage ramps from −80 to +80 mV were applied during the peak current. Bath/pipette solutions were NMDG-Cl/CsCl (150 mM each). (g) A proposed model illustrating that Alka channel activation by hydroxide (OH−) results in GRN depolarization. The intracellular concentration of Cl− of a fly GRN is higher than on the extracellular side. When the animal is exposed to alkaline food, the external OH− opens Alka Cl− channels, presumably through deprotonation, which in turn leads to the flow of Cl− from the cytosol to the outside of the GRN. The Cl− efflux results in GRN depolarization, which subsequently causes the GRN to fire action potentials.
Alka is sufficient to form an alkaline taste receptor in vivo
To determine whether the Alka channel is sufficient to sense basic food in vivo, we misexpressed Alka or AlkaP276A at sweet GRNs using the Gr64f-Gal4 as a driver14,15 in an alka1 mutant background, as Gr64f-expressing GRNs were separate from alka-expressing GRNs (Fig. 3i). We chose one of the L-type sensilla, L7, to conduct tip-recording assays because the L7 sensilla showed quite weak responses to the stimuli of high pH (Extended Data Fig. 3a) as well as salt16. We found that flies misexpressing wild-type Alka in sweet GRNs acquired a robust sensitivity to high pH (Fig. 6a, 6b). In contrast, sweet GRNs misexpressing AlkaP276A produced few action potentials in response to high-pH stimuli (Fig. 6a, 6b). Further, the control flies such as alka1;Gr64f-Gal4 or alka1;UAS-alka failed to produce obvious spikes in response to high-pH stimuli (Extended Data Fig. 7a, 7b). Furthermore, L7 sensilla misexpressing wild-type Alka or AlkaP276A yielded comparable responses to sucrose (50 mM) (Extended Data Fig. 7c, 7d), indicating that the failure of sweet GRNs misexpressing AlkaP276A to evoke action potentials responding to high-pH stimuli was not due to the dysfunction of these GRNs. Next, we allowed these flies to choose between basic foods consisting of 0.1–100 mM NaOH and neutral foods containing the same concentration of NaCl. Of great interest, selectively misexpressing Alka in Gr64f-expressing GRNs enabled the flies to show a progressively increasing preference for basic foods as the food pH rose from 10 to 13, whereas AlkaP276A-misexpressing flies, like alka1 mutant flies (Fig. 1b), displayed profound defects in avoiding alkaline food (Fig. 6c). Furthermore, our PER assays revealed that Alka-misexpressing flies also exhibited significantly increased PERs to alkaline food when compared to AlkaP276A-misexpressing flies (Fig. 6d). Altogether, we propose that Alka is sufficient to serve as a taste sensor for high pH in vivo.
Fig. 6. Alka is sufficient to be an alkaline taste sensor in vivo.
(a) Representative spikes evoked by basic pH in sweet GRNs (L7 sensilla) misexpressing Alka or AlkaP276A driven by the Gr64f-Gal4 in the alka1 mutant background. (b) Quantification of the frequencies of spikes detected in a. n=15 flies. Mean ± SEM, unpaired two-tailed Student’s t-tests, ****p<0.0001. (c) Two-choice feeding assays showing the feeding responses to neutral versus alkaline foods among alka1;Gr64f-Gal4/UAS-alka (alka1;Gr64f-alka), alka1;Gr64f-Gal4/UAS-alkaP276A (alka1;Gr64f-alkaP276A), and wild-type (wt) flies . n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001. (d) PERs to 30 mM sucrose at different alkaline pH for alka1;Gr64f-Gal4/UAS-alka (alka1;Gr64f-alka), alka1;Gr64f-Gal4/UAS-alkaP276A (alka1;Gr64f-alkaP276A), and wt flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, ***p=0.0004, ****p<0.0001. Arrows indicate onset of stimuli (a).
alka-expressing GRNs regulate feeding responses
To exclusively manipulate the activities of alka-expressing GRNs, we employed the Orco-Gal80 (refs.67,68) line to repress the alka-Gal4 expression in olfactory organs (Extended Data Fig. 8a, 8b) while leaving its expression in GRNs intact (Extended Data Fig. 8c, 8d). Next, we selectively expressed a red-shifted channelrhodopsin CsChrimson69 in alka-positive GRNs. We then acutely activated alka-positive GRNs with red light, to which flies are almost blind, greatly minimizing the visual interference. Control flies readily accepted an offered drop of sucrose (500 mM), even under very strong red light (Extended Data Video 1 and Extended Data Video 1 caption). Similarly, both alka-Gal4;UAS-CsChrimson and alka-Gal4,UAS-CsChrimson;Orco-Gal80 flies also preferred sucrose in the absence of red light (Fig. 7a); however, upon stimulation with a moderate level of red light, the same flies instantly retracted their proboscis to reject sucrose (Fig. 7b, Extended Data Video 2, and Extended Data Video 2 caption). These flies showed substantially enhanced avoidance of sucrose as the red-light intensity increased (Fig. 7c, 7d). Thus, our results implied that alkaline taste negatively influences sweet taste, presumably through cross-modal inhibition. Furthermore, we selectively inactivated alka-expressing GRNs using a neuronal tetanus toxin (TNT)70 and subsequently examined their feeding responses to alkaline food. Similar to the alka1 mutant, suppressing the activity of alka-expressing-GRNs caused severe defects in aversion to basic foods (Fig. 7e, 7f). Like alka1 mutants, flies with suppressed alkaline GRNs preferred basic foods. We deduced that this was due to the attractive response to low concentrations of Na+ from NaOH when the aversion to OH− was abolished. In summary, our studies establish that alka and alka-expressing GRNs are both necessary and sufficient to detect highly basic food.
Fig. 7. Effects on feeding responses due to activating or suppressing alka-expressing GRNs.
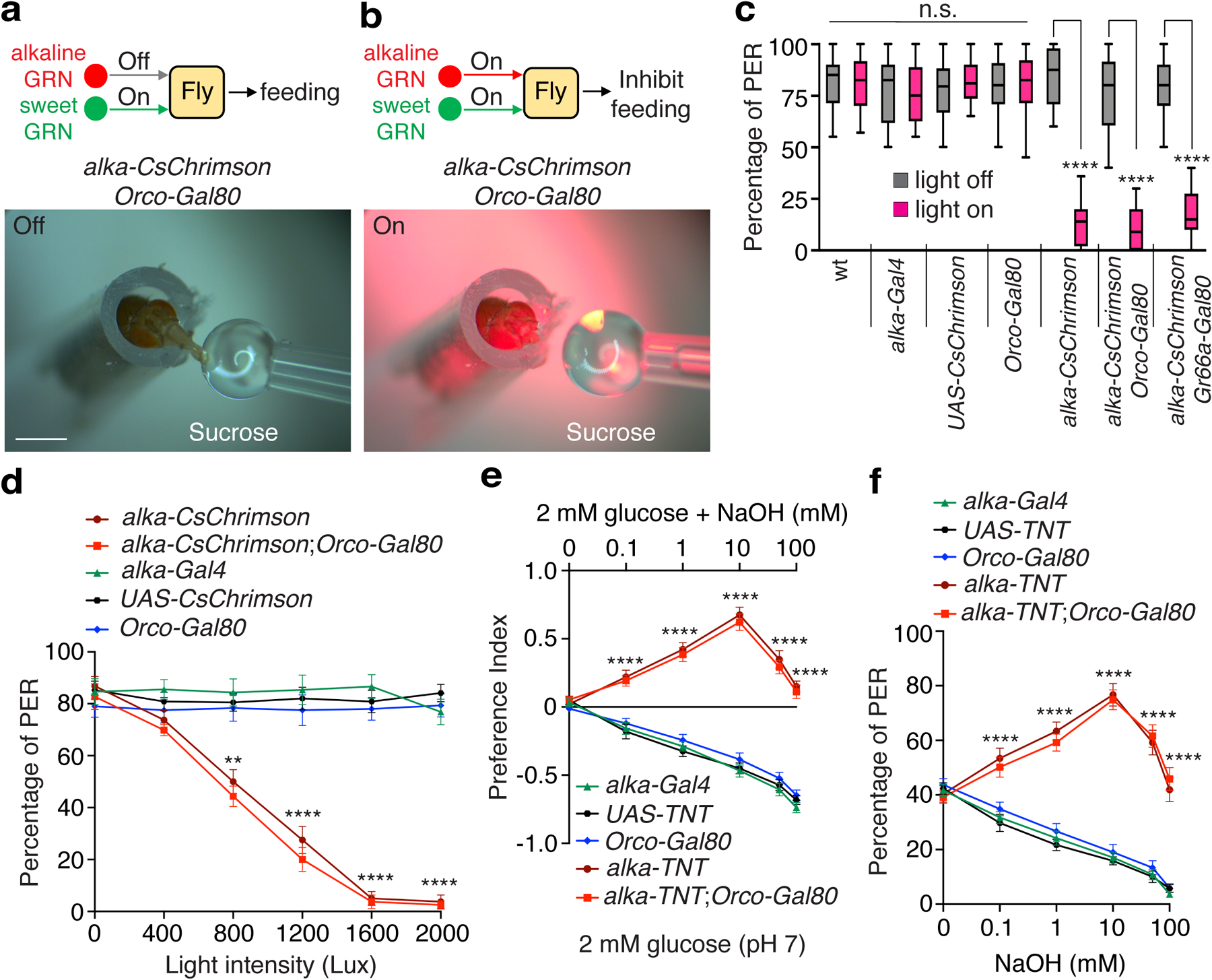
(a, b) Representative images showing feeding responses to 500 mM sucrose without (a) or with (b) red-light stimulation (1200 Lux) of alka-CsChrimson;Orco-Gal80 (alka-Gal4,UAS-CsChrimson;Orco-Gal80) flies. Scale bars: 1 mm. (c) Statistical analyses of PER percentages with or without red-light stimulation (1200 Lux) for alka-CsChrimson (alka-Gal4,UAS-CsChrimson), alka-CsChrimson;Orco-Gal80, alka-CsChrimson;Gr66a-Gal80 (alka-Gal4,UAS-CsChrimson;Gr66a-LexA,LexAop-Gal80), and indicated control flies. n=12 trials. Mean ± SEM, one-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001. (d) Relationship between PER percentages and red-light intensity among alka-CsChrimson, alka-CsChrimson;Orco-Gal80, and indicated control flies. n=10 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, **p=0.0012, ****p<0.0001. (e) Feeding responses to neutral versus alkaline foods among alka-TNT (alka-Gal4,UAS-TNT), alka-TNT;Orco-Gal80 (alka-Gal4,UAS-TNT;Orco-Gal80), and indicated control flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001. (f) PERs to 30 mM sucrose mixed with different NaOH concentrations for alka-TNT (alka-Gal4,UAS-TNT), alka-TNT;Orco-Gal80 (alka-Gal4,UAS-TNT;Orco-Gal80), and control flies. n=12 trials. Mean ± SEM, two-way ANOVA with Tukey’s post-hoc tests, ****p<0.000
Our cell biological studies revealed that alkaline GRNs show a small fraction of overlap with bitter GRNs marked by Gr66a (Fig. 3h). This raised a question as to whether alkaline taste and bitter taste are distinct from each other. The double-labeling approach with two fluorescent reporters has limitations in discerning the relative localizations among different GRN markers. For example, two fluorescent-reporter transgenes, when combined in one animal, may affect each other’s GRN morphology, number, or even innervation pattern. To overcome these problems and better evaluate the relative localization between Gr66a- and alka-expressing GRNs in the fly labellum, we employed the Gal80-based intersectional genetic approach71. Typically, the transcriptional repressor Gal80 potently inhibits the activity of Gal4, a transcriptional factor used to drive the expression of a reporter in the UAS/Gal4 system72. As a result, Gal80 effectively represses the GFP-reporter expression driven by Gal4 only when Gal80 and Gal4 are present in the same cell. To ascertain whether alka-expressing GRNs are distinct from Gr66a-expressing GRNs, we examined the expression of alka-Gal4/UAS-GFP in the presence of Gr66a-Gal80. When the Gr66a-Gal4;UAS-GFP (Gr66a-GFP) reporter was combined with the Gal80 alone, as no Gal80 was expressed, Gr66a-positive GRNs were easily detected by the Gr66a-GFP reporter (Extended Data Fig. 9a). In contrast, after expressing Gal80 repressors in Gr66a-positive GRNs using the Gr66a-Gal80 driver, we failed to observe any obvious GRNs, indicating that expression of Gr66a-GFP reporter was successfully shut down by Gr66a-Gal80 (Extended Data Fig. 9b). Of great interest, we found that even in the presence of Gr66a-Gal80, most of the alka-expressing GRNs were still readily detected based on the GFP expression. Moreover, the remaining alka-expressing GRNs mostly innervated S-type sensilla such as S6 (Extended Data Fig. 9c). In support of our double-labeling assays, our intersectional genetic studies demonstrated that alka-expressing GRNs were largely separate from Gr66a-expressing GRNs in the fly labellum.
Next, we used the optogenetic approach to selectively activate the subgroup of alkaline GRNs with little overlap with bitter GRNs. We found that the alka-CsChrimson;Gr66a-Gal80 (alka-Gal4,UAS-CsChrimson;Gr66a-lexA,LexAop-Gal80) flies immediately rejected sucrose as soon as the alka-expressing GRNs were activated by light (Fig. 7c). This suggested that this subset of alkaline GRNs was sufficient to repress sucrose feeding when acutely activated. Furthermore, we employed Gr66a-Gal80 to inactivate bitter GRNs when we silenced alka-expressing alkaline GRNs with the neuronal toxin TNT. We used PER assays to examine the resultant fly’s taste response to NaOH, sucrose, and bitter substances such as caffeine. We found that in the absence of overlapping bitter GRNs, the remaining alkaline GRNs were sufficient to avoid basic food (Extended Data Fig. 9c). Moreover, silencing the alkaline GRNs with TNT had no statistically significant effect on the PERs to sucrose or caffeine stimuli (Extended Data Fig. 9c). Meanwhile, silencing bitter GRNs failed to cause defects in aversion to alkaline food (Extended Data Fig. 9c). As controls, inactivating ppk23-expressing or ppk28-expressing GRNs with TNT yielded little effect on the feeding responses to basic food (Extended Data Fig. 9c). Altogether, our work establishes that alkaline taste is sensed by the Alka channel and alka-expressing GRNs in Drosophila.
Discussion
Since most organisms’ optimal physiological activities and enzymatic reactions can occur only in a narrow pH range (around 7.4), excessively high pH can disrupt acid-base balance and lead to alkalosis of the body, a life-threatening condition53,73. There are many places where organisms can encounter high-pH conditions in their ecosystem such as in the food and water they may consume. Moreover, many naturally occurring toxins, including alkaloids and aqueous ammonia, are quite basic. Ethological research has documented well-defined behavioral responses to basic pH in a large variety of species, such as nematodes74,75, insects9, fish76, and mammals7,8,77.
The impact of alkaline pH on fly physiology has been well documented across a variety of studies. It has been reported that the fly’s overall body pH exhibits a dynamic change over the course of development: the body pH starts as approximately neutral at the larval stage and gradually becomes more acidic as the animal advances to the pupal and adult stages78. In addition, there is remarkable variation in the luminal pH at different regions of the fly midgut, with its posterior segment more alkaline79. As a result, alkaline pH sensation is strongly implicated in fly health and longevity. Flies fed moderately alkaline diets display notably reduced lifespan and survivability80. Furthermore, chronic exposure to a highly alkaline environment impairs development, shortens lifespan, and causes lethality81. Consequently, female flies robustly avoid alkaline substrates when selecting a location to deposit eggs81. Taken together, alkaline pH sensation serves as an essential self-protection strategy that enables flies and other animal species to effectively avoid toxic environments during food foraging and habitat selection. We propose that alkaline taste dramatically increases the fly’s evolutionary fitness by enhancing its survival, growth, and reproduction.
Alkaloids taste very bitter and are poorly soluble, meaning their strong bitterness can confound the investigation of the taste component solely contributed by high pH. In addition, ammonia is highly volatile and can interfere with the contact-dependent taste sensation by strongly activating the olfactory system. To avoid these limitations, we chose NaOH and Na2CO3 in our feeding assays because these two basic substances are simple and ecologically relevant. Our molecular genetic study demonstrates that flies have the capability to avoid highly basic food mainly through their gustatory system. Based on our findings, the fly shows specific and robust taste responses to basic food, suggesting that it is a well-suited model organism to explore alkaline taste sensation. We provide several lines of evidence supporting that Alka is a taste sensor specifically tuned to high pH. First, alka is expressed in a subset of GRNs in the fly labellum, which functions similarly to the mammalian tongue. The alka-expressing GRNs are both necessary and sufficient to detect basic food. Second, alka1 mutant flies show remarkably decreased aversion to basic food. Third, using in vivo electrophysiological analyses, we found that the S-type sensilla of alka1 flies exhibit considerably reduced responses to high pH but maintain normal responses to bitter compounds such as caffeine. Last, misexpressing alka in sweet GRNs attracts the flies to the otherwise aversive basic food. In summary, our work establishes that Alka is a bona fide taste receptor dedicated to sensing food basicity in Drosophila.
Over the past 20 years, the fly model has made enormous contributions to the discovery of various classes of ionotropic taste receptors24. These include GRs11,31,82, IRs16,32,83,84, otopetrin (Otop) channels6,20, TRP channels33,85–87, and the degenerin/epithelial Na+ channels (DEG/ENaC) or pickpocket (Ppk) channels58,59. Notably, Alka shows remarkable differences from previously identified taste receptors because it is an anion Cl− channel, whereas the IRs, Otops, TRPs, and Ppks are cation channels. Therefore, we believe our identification of the Cl− channel Alka as an alkaline taste receptor is substantial and innovative, adding another class of receptors to the diverse taste receptor repertoire in Drosophila.
The combination of optogenetic and intersectional genetic approaches enables us to selectively manipulate the activity of alka-expressing GRNs at will. We demonstrate that acutely activating alka-expressing GRNs alone is sufficient to trigger aversive taste responses, supporting the conclusion that alka-expressing GRNs are both necessary and sufficient for alkaline taste sensation. Like bitter taste, alkaline taste can suppress sweet taste responses. Although the neuronal mechanism underlying this cross-modal inhibition is currently not clear, our assay serves as a robust behavioral paradigm allowing us to screen for the higher-order neurons mediating the integration between alkaline taste and sweet taste.
The membrane potential of a living cell is established and maintained by the flows of different cation and anion species, such as Na+, K+, and Cl−, across the plasma membrane. While extensive studies have focused on the roles played by cations and cation channels in the regulation of membrane excitability, the functional importance of the anion Cl− and Cl− channels had been overlooked. In recent years, Cl− has emerged as an essential player in electrical signal transduction, and great progress has been made toward the molecular identification of various types of Cl− channels such as the acid-sensitive Cl− channel PAC/TMEM20688,89. Our work demonstrates that Alka functions as a distinct Cl− channel because it is strongly activated by highly basic pH (pH50, 11.9). In contrast to other LGCCs such as pHCl44, Alka is insensitive to slightly basic or acidic pH. Thus, Alka is well suited to act as an external sensor to detect high pH in the natural ecosystem, which can become noxiously basic due to excessive carbonation or nitration.
Protein sequence analyses suggest that Alka is distantly related to glycine-gated Cl− channels in vertebrates. Nevertheless, we found that Alka functions as a taste receptor rather than acting as a glycine or GABA receptor, as Alka is not activated by physiological concentrations of glycine or GABA (Extended Data Fig. 6h-k). In support of this notion, Alka is selectively expressed in the chemosensory organs. The functional divergence of Alka from canonical GlyRs is reminiscent of the evolution of the IR family in Drosophila32, which adopted chemosensory functions as olfactory or gustatory receptors in the periphery rather than as glutamate neurotransmitter receptors in the brain. In summary, by discovering that Alka is a high-pH-activated Cl− channel in taste receptor cells, our work provides important insights into the highly diversified nature of Cl− channels in terms of gating and function.
Upon further interrogation of the channel properties of Alka using patch-clamp experiments, we conclude that Alka mainly conducts Cl−. This creates a logical quandary, as mature neurons in the brain usually experience an intracellular-facing Cl− gradient, and the Cl− influx in mature neurons is hyperpolarizing instead of depolarizing. However, the answer lies in the unusual distribution of the Cl− gradient across the fly GRN. From the perspective of comparative physiology, the extracellular receptor lymph of the fly GRN is analogous to the saliva bathing taste receptor cells in humans, which contains lower levels of Cl− than does blood plasma90. Furthermore, like mammalian olfactory sensory neurons91 and airway epithelial cells92, the Cl− concentration of the extracellular receptor lymph appears to be even lower than in the cytosol of the taste receptor neuron in flies66. Thus, Alka perfectly fits the unusual ionic milieu of fly GRNs. Based on our model, in response to OH− stimuli, Alka is induced to adopt an open conformation, which leads to the flow of intracellular Cl− out of the GRN. The Cl− efflux depolarizes the GRN and results in the production of action potentials, thereby enabling the animals to sense alkaline food. In summary, our work highlights the important roles played by Cl− and Cl− channels in regulating taste transduction.
In conclusion, we propose that Alka, which forms a previously unknown hydroxide-gated Cl− channel, is a taste receptor responsible for detecting alkaline food. As far as we know, Alka represents an important Cl− channel identified in the animal kingdom, which is dedicated to sensing highly basic pH. Furthermore, by showing that basic pH has a discrete taste in Drosophila, our work resolves a long-standing debate as to whether alkaline taste really exists. Moreover, we have demonstrated that Alka functions as a Cl− channel that is potently activated by highly alkaline pH. Therefore, our molecular discovery of Alka as a taste receptor of alkaline pH advances our understanding of the roles and modes of activation of Cl− channels. Finally, given that detecting basic pH is crucial for food selection across many different species, our work provides the conceptual basis for investigating the neuronal mechanisms underlying alkaline taste sensations in other animals.
Methods
1. Flies
All fly strains were raised on a standard cornmeal and sucrose medium. The flies used in this study are as follows: wild type (w1118) (ref.6), alka1 (generated in this study), Gr66aEx83 (ref.55), Gr33a1 (ref.82), Ir76b1 (ref.16), Ir25a1 (ref.32), trpl302 (ref.85), trpA11 (ref.87), otopla1 (ref.6), tmc1 (ref.49), CG12344MI11416 (RRID_BDSC_56318), ort1 (RRID_BDSC_1133)42, RdlMD-RR (RRID_BDSC_1675)37, UAS-Grd RNAi (RRID_BDSC_29589)40, UAS-HisCl1 RNAi (RRID_BDSC_28013)93, UAS-GluCla RNAi (RRID_BDSC_53356)41, UAS-pHCl-1 RNAi (RRID_BDSC_38909)44, UAS-pHCl-2 RNAi (RRID_BDSC_26003)45, UAS-Lcch3 RNAi (RRID_BDSC_50668)37, UAS-CG6927 RNAi (RRID_BDSC_25895), UAS-CG7589 RNAi (RRID_BDSC_27090), UAS-CG8916 RNAi (RRID_BDSC_39031), UAS-CG12344 RNAi (RRID_BDSC_26250), UAS-alka (generated in this study), UAS-alkaP276A (generated in this study), UAS-mCD8::GFP (RRID_BDSC_5137)54, alka-Gal4 (generated in this study), Gr66a-Gal4 (RRID_BDSC_57670)55, Gr64f-LexA56,94, Gr66a-LexA94,95, Ir76b-QF16, ppk23-LexA94, ppk28-LexA94, QUAS-mtdTomato (RRID_BDSC_30005)96,Orco-Gal80 (RRID_BDSC_80559)68, LexAop-Gal80 (RRID_BDSC_32215), and LexAop-mCherry (RRID_BDSC_52271)97.
2. Antibodies and molecular genetics
The detailed information for antibodies, transgenic fly lines, and molecular cloning are available in the Supplementary Information.
3. Immunocytochemistry
The fly proboscis and brain tissues were dissected in ice-cold phosphate-buffered saline (PBS) and fixed with fresh 4% paraformaldehyde for 20 minutes at room temperature. We washed the samples at room temperature with PBS four times for 30 minutes each. We incubated them with primary antibodies in 1x PBS containing 0.3% Triton™ X-100 (PBST) for 48 hours at 4 °C. In this study, we used the following primary antibodies: rabbit anti-GFP (1:200 dilution, Thermo Fisher, cat. no. A-11122, RRID:AB_221569); mouse anti-GFP (1:200 dilution, Thermo Fisher, cat. no. A11120, RRID:AB_221568); mouse anti-nc82 (1:50 dilution, DSHB, cat. no. nc82, RRID:AB_528108); mouse anti-mCherry (1:50 dilution, DSHB, cat. no. DSHB-mCherry-3A11, RRID:AB_2617430); mouse anti-Myc (1:100 dilution, Cell Center Services, University of Pennsylvania, cat. no. 3207); and rabbit anti-Alka polyclonal antibody (1:200 dilution, generated in this study).
Following incubation with primary antibodies, we rewashed the samples with 1x PBST four times for 30 minutes each and incubated them in secondary antibodies for 24 hours at 4 °C. We used the following secondary antibodies: goat anti-rabbit AlexaFluor 488 (1:200 dilution, Jackson Immuno Research, cat. no. 111–545-003, RRID:AB_2338046); goat anti-rabbit AlexaFluor 594 (1:200 dilution, Jackson Immuno Research, cat. no. 111–585-003, RRID:AB_2338059); goat anti-mouse AlexaFluor 488 (1:200 dilution, Jackson Immuno Research, cat. no. 115–545-003, RRID:AB_2338840); and donkey anti-mouse AlexaFluor 594 (1:200 dilution, Jackson Immuno Research, cat. no. 715–585-150, RRID:AB_2340854). After overnight incubation, we washed the samples in 1x PBST four times for 30 minutes each and incubated them in VECTASHIELD antifade mounting media (Vectorlabs, cat. no. H-1200) overnight. After incubation, we mounted the samples on transparent glass slides (Electron Microscopy Sciences, cat. no. 71870–10) with thin coverslips (Electron Microscopy Sciences, cat. no. 72200–11).
For the immunostaining of cultured HEK293 cells, the HEK293 cells transfected with pCS2+MT-Myc-alka or pCS2+MT-Myc-alkaP276A were cultured on a coverslip (Fisher Scientific, cat. no. 12542BP) for 24–48 hours. After washing the coverslip with 1x PBS, we fixed the cells using fresh 4% paraformaldehyde for 20 minutes. We washed the residual paraformaldehyde away from the coverslip with 1x PBST. We incubated the cells with mouse anti-Myc monoclonal antibodies (1:100 dilution, Cell Center Services, University of Pennsylvania, cat. no. 3207) overnight at 4 °C. Next, we washed the coverslip with 1x PBST three times for 30 minutes each and added goat anti-mouse AlexaFluor 488 antibodies (1:200 dilution, Jackson Immuno Research, cat. no. 115–545-003, RRID:AB_2338840). We incubated the coverslip with secondary antibodies at 4 °C overnight. After washing the coverslip with 1x PBST, we mounted the cells with the VECTASHIELD antifade mounting media.
We imaged and analyzed the stained tissues or cells using Zeiss LSM 710 confocal microscopy (Cell and Developmental Biology Microscopy Core, University of Pennsylvania).
4. Cell culture and transient transfection
We cultured and transfected human embryonic kidney 293T (HEK293T) cells as described previously6. Briefly, HEK293T cells were cultured at 37 °C with 5% CO2 in the Opti-MEM medium supplemented with 10% fetal bovine serum. We placed the cells in 35-mm Petri dishes before transfection. When the cell density reached about 70% confluency, we transfected the pCS2+MT-alka or pCS2+MT-alkaP276A construct with the pcDNA3-GFP reporter plasmid (molar ratio: 5:1) into HEK293T cells using the Lipofectamine 3000 transfection reagent. Cells transfected only with the pcDNA3-GFP reporter plasmid served as controls. We plated the cells onto glass coverslips (6 mm) for patch-clamp recordings 2–3 days after transfection.
5. Whole-cell patch-clamp electrophysiology
We used whole-cell patch-clamp recordings to examine the channel properties of Alka or AlkaP276A. To perform whole-cell recordings, we used a MultiClamp 700B amplifier and an Axon Digidata 1440A digitizer (Molecular Devices). The bath solution contained 150 mM NaCl and 10 mM HEPES (pH 7.4 with HCl, 320 mOsm/kg). The pipette solution consisted of 150 mM CsCl, 2 mM Mg-ATP, and 10 mM HEPES (pH 7.4 with CsOH, 320 mOsm/kg). Using the P-1000 micropipette puller (Sutter Instrument Co.) we made the patch pipettes (6–10 MΩ) from borosilicate glass capillaries. Then we used a Microforge MF- 900 (Narishige) to polish the tips of the glass pipettes. We used Clampex10.5 software to record the electrophysiological signals.
We chose the GFP-positive cells randomly for recordings. After forming the whole-cell configuration, positive pressure was applied from a nitrogen tank using a Picospritzer III (Parker) to a stimulating glass pipette (3 μm in diameter) containing a stimulus such as alkaline or acidic buffer, glycine, or GABA. The stimulating solution was delivered to the patched cell at a very close range (5–10 μm from the cell). For solutions used to stimulate the HEK293 cells, the alkalization or acidification of the NMDG-Cl buffer was adjusted with CsOH or HCl, respectively. The glycine- or GABA-containing solution was prepared by diluting a 1 M glycine or 100 mM GABA stock to the indicated glycine or GABA concentration with the bath solutions (150 mM NaCl and 10 mM HEPES, pH 7.4). The osmolarities of high-pH, low-pH, glycine, or GABA solutions were adjusted to 320 mOsm/kg. The currents were recorded at a holding potential of −70 mV or ramped from −80 mV to +80 mV at room temperature. We analyzed the data with Clampfit 10.5, and only gigaohm-sealed cells were selected for subsequent data analysis.
For ion replacement experiments, 150 mM NaCl was replaced by isosmotic bath solutions, including 150 mM NMDG-Cl or 150 mM NMDG-gluconate. The pipette solution contained 150 mM CsCl or 150 mM Cs-methanesulfonate, 2 mM Mg-ATP, and 10 mM HEPES (pH 7.4 with CsOH, 320 mOsm/kg). To test the relative permeability, NaCl was replaced with equimolar NaF, NaBr, or NaI. The pipette solution contained (in mM): 150 CsCl, 2 Mg-ATP, and 10 HEPES (pH 7.4, 320 mOsm/kg). To calculate the relative permeability of a particular anion versus Cl−, we used the Goldman-Hodgkin-Katz equation Er(X) - Er(Cl) = (RT/zF)ln(Px[X]o/ PCl[Cl]o). To determine the effects of intracellular Cl− concentrations on the conductance of Alka, we used the following pipette solutions (in mM): (1) 15 CsCl, 135 Cs-methanesulfonate; (2) 50 CsCl, 100 Cs-methanesulfonate; (3) 100 CsCl, 50 Cs-methanesulfonate; and (4) 150 CsCl. All pipette solutions also received 2 mM Mg-ATP and 10 mM HEPES (pH 7.4 with CsOH, 320 mOsm/kg).
6. Two-choice feeding assays
We assayed food preference between two different food options using the well-established two-choice feeding assay. two- to four-day-old flies were wet-starved for 24 hours. After starvation, we placed flies in an evenly partitioned Petri dish containing two types of food. One side contained 2 mM glucose and the experimental taste chemical (for example, NaOH) in 1% agarose, and the other side consisted of 2 mM glucose in 1% agarose. To determine food preference, we mixed each food with either blue dye (Fujifilm Wako Chemicals, cat. no. 027–12842) or red dye (MiliporeSigma, cat. no. S9012–5G). We adjusted the red and blue dye concentration for all genotypes tested to eliminate the flies’ potential bias to either dye. After 90 min of feeding in dark conditions, flies were examined under light microscopy, and the color of food showing through their abdomens was used as an indicator of which food they ate. The preference index (PI) was calculated by the following equation:
A PI of zero indicates no preference, and a PI ranging from +1 to −1 indicates that one food is more attractive than the other. In each trial (n), approximately 70 flies were tested. To eliminate experimental bias, we encoded fly genotypes while setting up the assay, so that a separate researcher who counted the assay would be blind to genotype and experimental condition.
7. PER and optogenetics assays
To conduct the PER assay, we wet-starved two- to four-day-old flies for 6–12 hours. To restrict contact with the stimulus solely to the proboscis, we wedged each fly into the barrel of a 10-μl pipette tip. The narrow tip end was trimmed back to expose the fly’s head and proboscis. The rest of the fly, such as the legs, wings, and thorax, remained inside the tip. We then arranged the fly with its proboscis facing up and used a standard dissection microscope (AmScope, cat. no. SM-1TSZ-144) to visualize the PER. We applied a single drop (2–5 μl) of test solutions to the proboscis and immediately recorded any PERs. To perform optogenetics, using the same fly preparation, we stimulated the fly proboscis with red LED light (AmScope, cat. no. LED-RGB-60W) and immediately assessed the PER. The representative videos of PERs in the presence or absence of red light stimuli were captured by a Basler ace USB camera (Basler, acA2000–165uc). In each trial (n), 10 or more flies were assayed, and the PER percentage was calculated by the percentage of the total number of flies that responded to the given stimulus.
8. Tip recordings
We performed tip recordings according to our published work6. Specifically, we used the P-1000 micropipette puller to make a long-tapered reference glass pipette. The sharp end of this pipette was impaled into the thorax of flies, whose ages ranged from 2 to 4 days old. Next, we carefully maneuvered the reference pipette through the fly neck to reach the distal end of the proboscis. The reference pipette solution (pH 7.2) consisted of (in mM) 130 NaCl, 2 CaCl2, 2 KCl, 40 sucrose, and 10 HEPES. We stimulated taste sensilla with a micropipette filled with specific taste solutions. In the recording pipette, KCl (1 mM) was used as an electrolyte for NaOH, Na2CO3, NaCl, caffeine, quinine, or HCl, whereas tricholine citrate (30 mM) served as an electrolyte for sucrose. The recording system included a computer-controlled taste probe, a head stage, and a data acquisition controller (Syntech). We used Autospike software (Syntech) to record the electrical signals generated by the GRNs. The taste spikes were sampled at a rate of 12,000 and filtered at 1kHz.
9. Quantification and statistical analysis
We used two-way analysis of variance (ANOVA) with Tukey’s post hoc test to analyze interactions between two independent variables such as genotype and concentration, one-way ANOVA with Tukey’s post hoc test to analyze the differences among more than two experimental samples, and an unpaired two-tailed Student’s t-test for two experimental samples. We followed LaMorte’s power calculation and our published work16,6,49 to determine the sample sizes and replicates. A p value less than 0.05 (p< 0.05) was considered statistically significant. For other experiments such as immunohistochemistry and gel electrophoresis, we independently repeated them at least three times.
Regarding the box-and-whisker plots (Fig. 4b, 7c) presented in our paper, the center line indicates a median value, whereas the first and third quartiles are presented as the bottom and top boundary lines. The whiskers represent maxima and minima for each dataset.
Extended Data
Extended Data Fig. 1. Screening for receptors or ion channels required for the feeding responses to alkaline food.
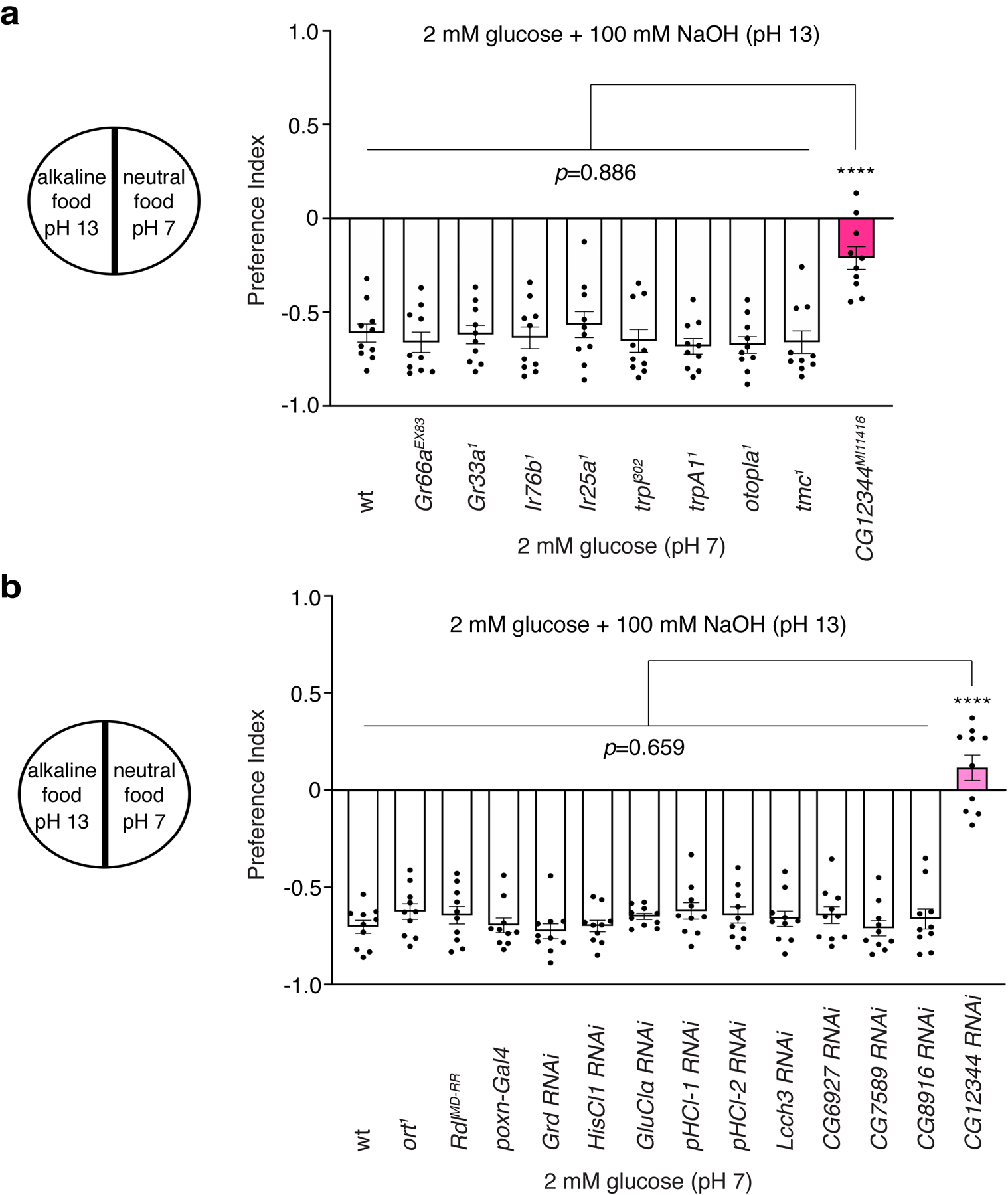
(a) We tested a wide array of receptor and ion channel candidates, a representative sample of which is shown in the bar graph. These include the gustatory receptor (Gr) family, such as Gr66a and Gr33a; the ionotropic glutamate receptor (Ir) family, such as Ir76b and Ir25a; the transient-receptor-potential (trp) ion channel family, such as trpl and trpA1; the otopetrin family, such as otopla; the transmembrane channel-like (tmc); and genes with unknown functions such as CG12344. n=10 trials. (b) Feeding responses to neutral versus alkaline foods among control flies and mutant flies of the fly LGCC family. n=10 trials. Mean ± SEM, one-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001.
Extended Data Fig. 2. Generating the alka1 null mutant flies.
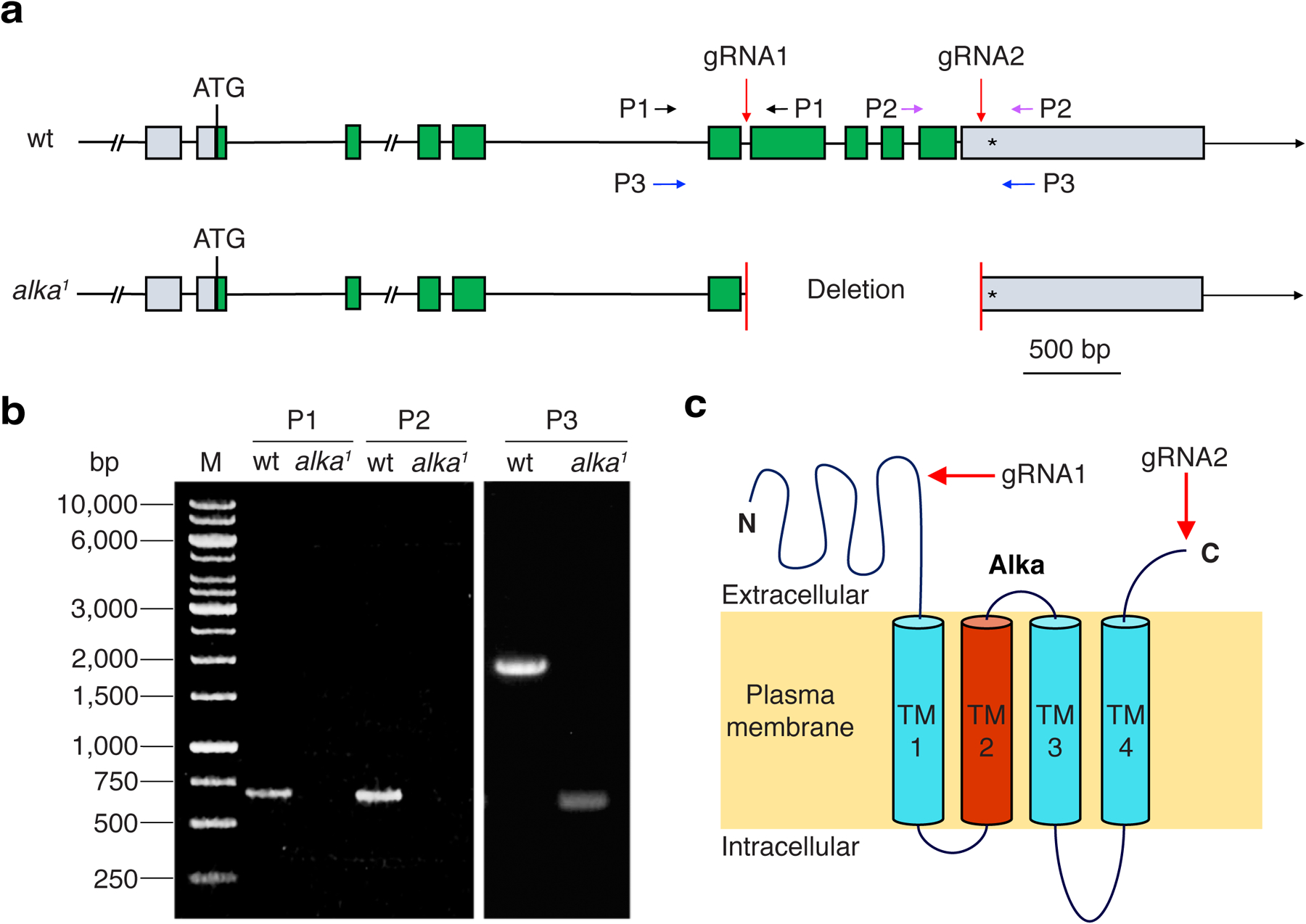
(a) Genomic composition of the wild-type and mutant alka genes, including the translational start (ATG) and stop (*) codons, exons, and introns. The red arrows indicate the guide RNA (gRNA) target sites (gRNA1 and gRNA2). To screen for the alka1 mutant, we designed three sets of primers, P1, P2, and P3, which flank the gRNA targeting sites. (b) PCR analyses of genomic DNA with the P1, P2, and P3 primers for wild-type and alka1 mutant. (c) The predicted topology of the Alka protein comprising four transmembrane segments. The TM2 domain (Red) is predicted to line the channel pore. Both the N- and C-terminal ends of the Alka protein reside on the extracellular side. The red arrows show the ablated protein regions in the alka1 mutant.
Extended Data Fig. 3. Electrophysiological responses to NaOH and NaCl.
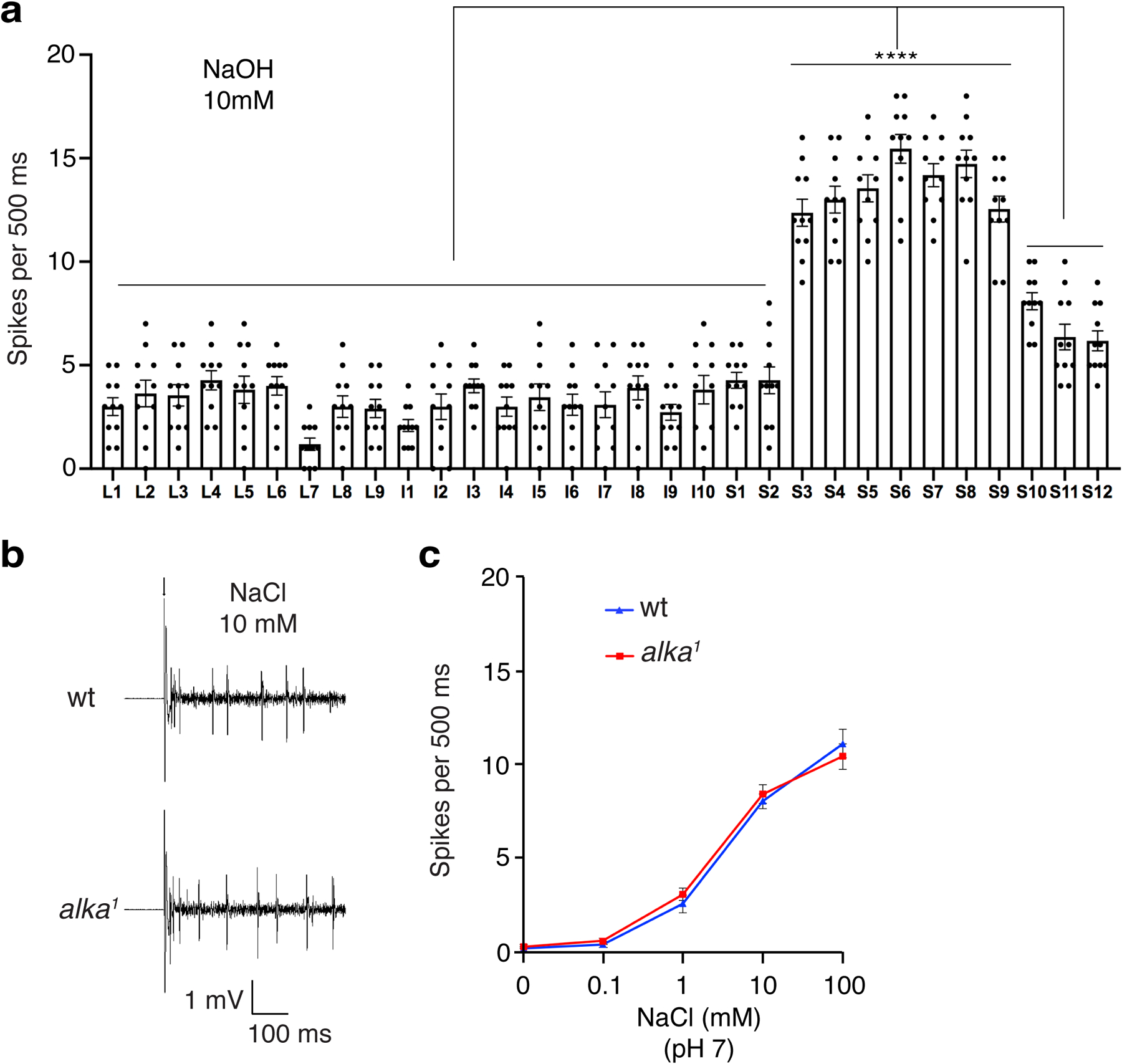
(a) Statistical analyses of the frequencies of spikes produced by L-, I-, and S-type sensilla responding to 10 mM NaOH in wild-type (wt) flies. One-way ANOVA with Tukey’s post-hoc tests, Mean ± SEM, n=11 flies, ****p<0.0001. (b) Representative spikes evoked by 10 mM NaCl (pH 7) at S6 sensilla for wt and alka1 mutant. The arrow indicates the stimulus onset. (c) Statistical analyses of the frequencies of spikes produced by S6 sensilla in response to different concentrations of NaCl for wt and alka1 mutant flies. n=10 flies. Mean ± SEM, unpaired two-tailed Student’s t-tests.
Extended Data Fig. 4. alka is required to detect alkaline food through the legs.
(a, b) Expression of alka-gal4;UAS-mCD8::GFP at the tarsal segment of the fly foreleg. (c) PERs to alkaline solutions containing 30 mM sucrose and various concentrations of NaOH among wild-type (wt), alka1, alka1;Ir761, and rescue flies. n=12 trial. Two-way ANOVA with Tukey’s post-hoctests, Mean ± SEM, **p=0.0081, ****p<0.0001. (d) Expression of alka-gal4;UAS-mCD8::GFP in the maxillary palp. (e) Expression of alka-gal4;UAS-mCD8::GFP in the antenna. (f) No obvious anti-Alka signals were detected in the wt adult brain. (g) Relative localization pattern between ppk28-expressing GRNs and taste sensilla. Scale bars: 10 μm (a, b, d, e, and g), 50 μm (f).
Extended Data Fig. 5. Multisequence alignment of the full protein sequences among fly Alka and the glycine receptor alpha 1 (GlyRa1) from zebrafish, mice, or humans.
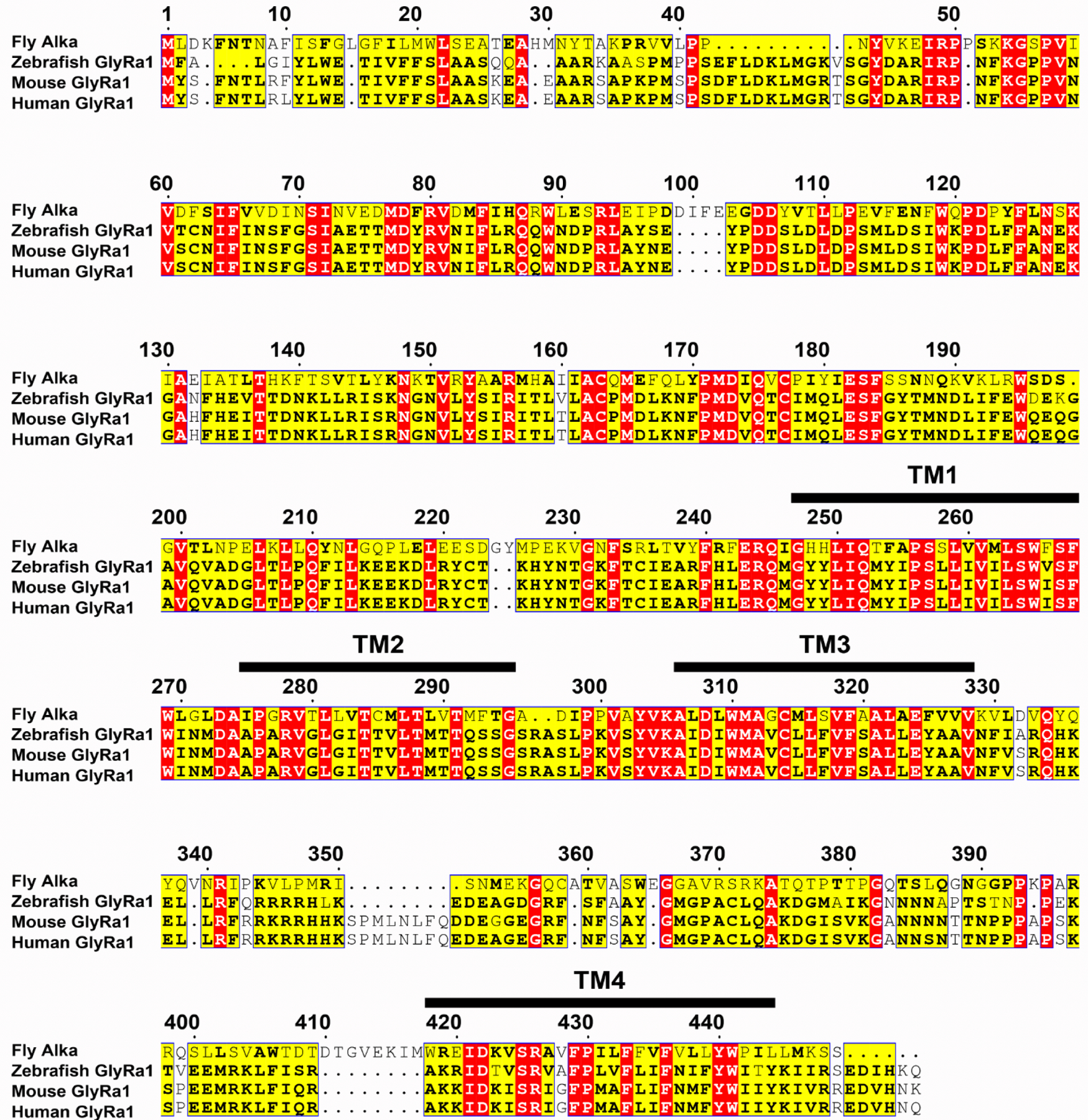
The protein sequence identities between Alka and GlyRa1 in other species are as follows: zebrafish, 30% identity; mouse, 30% identity; human, 30% identity. Identical amino acid residues are labeled in red, whereas similar amino acid residues among at least three species are labeled in yellow and highlighted in bold. The four transmembrane regions (TM1-TM4) are denoted by black bars above their amino acid sequences.
Extended Data Fig. 6. Ion selectivity of Alka and conductance of Alka in response to acidic pH, glycine, or GABA stimuli.
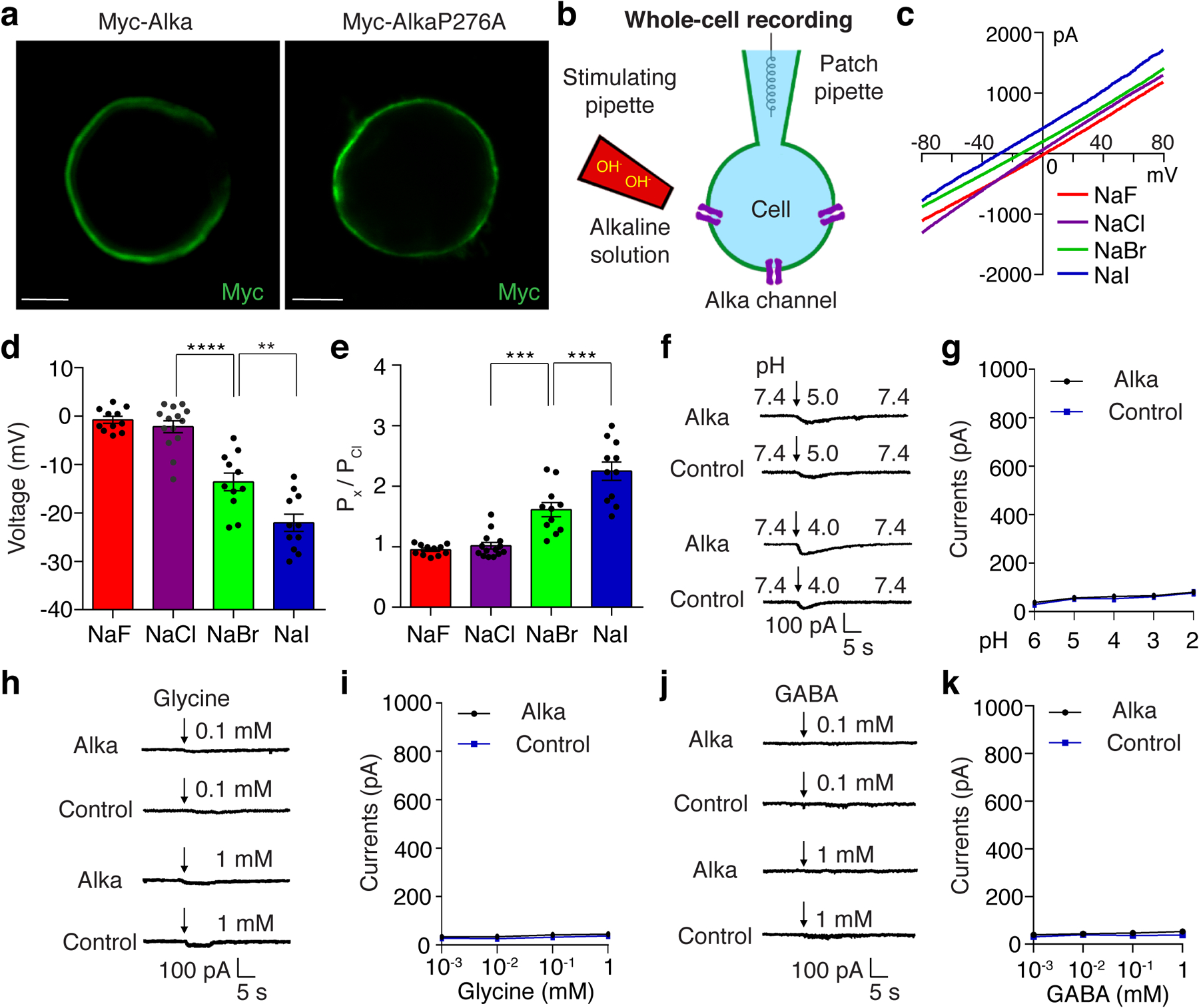
(a) Localization of Alka in HEK293 cells expressing Alka that is N-terminally fused with a Myc tag. Scale bar: 5 μm. (b) Configuration of the whole-cell patch-clamp recording setup. We used a stimulating pipette (red) to locally apply high-pH solutions to the cells, and a patch pipette (blue) to carry out whole-cell recordings. (c) Current-voltage (I-V) relationships of Alka-expressed HEK293 cells, which were elicited by voltage ramps from −80 mV to +80 mV. The bath solution contained 150 mM NaF, NaCl, NaBr, or NaI, and the intracellular solution contained 150 mM CsCl. (d) Statistical analyses of reversal potentials from experiments in c. n=11 cells (NaF, NaBr, or NaI); n=14 cells (NaCl). One-way ANOVA with Tukey’s post-hoc tests, Mean ± SEM, **p=0.0012, ****p<0.0001. (e) Relative anion permeability of Alka. n=11 cells (NaF, NaBr, and NaI); n=14 cells (NaCl). One-way ANOVA with Tukey’s post-hoc tests, Mean ± SEM, ***p=0.0003. (f, g) Currents from Alka-expressing cells and control cells without Alka expression responding to the stimuli of acidic isosmotic solutions. n=11 cells. Unpaired two-tailed Student’s t-tests. (h-k) Currents evoked by Alka-expressing cells and control cells without Alka expression in response to the stimuli of glycine (0.001–1 mM) (h, i) or GABA (0.001–1 mM) (j, k). n=11 cells. Unpaired two-tailed Student’s t-tests. Mean ± SEM. Cells were clamped at −70 mV. Arrows indicate the onset of stimulus.
Extended Data Fig. 7. Spikes elicited by NaOH in control flies and spikes evoked by sucrose in flies misexpressing Alka or AlkaP276A at the sweet GRNs.
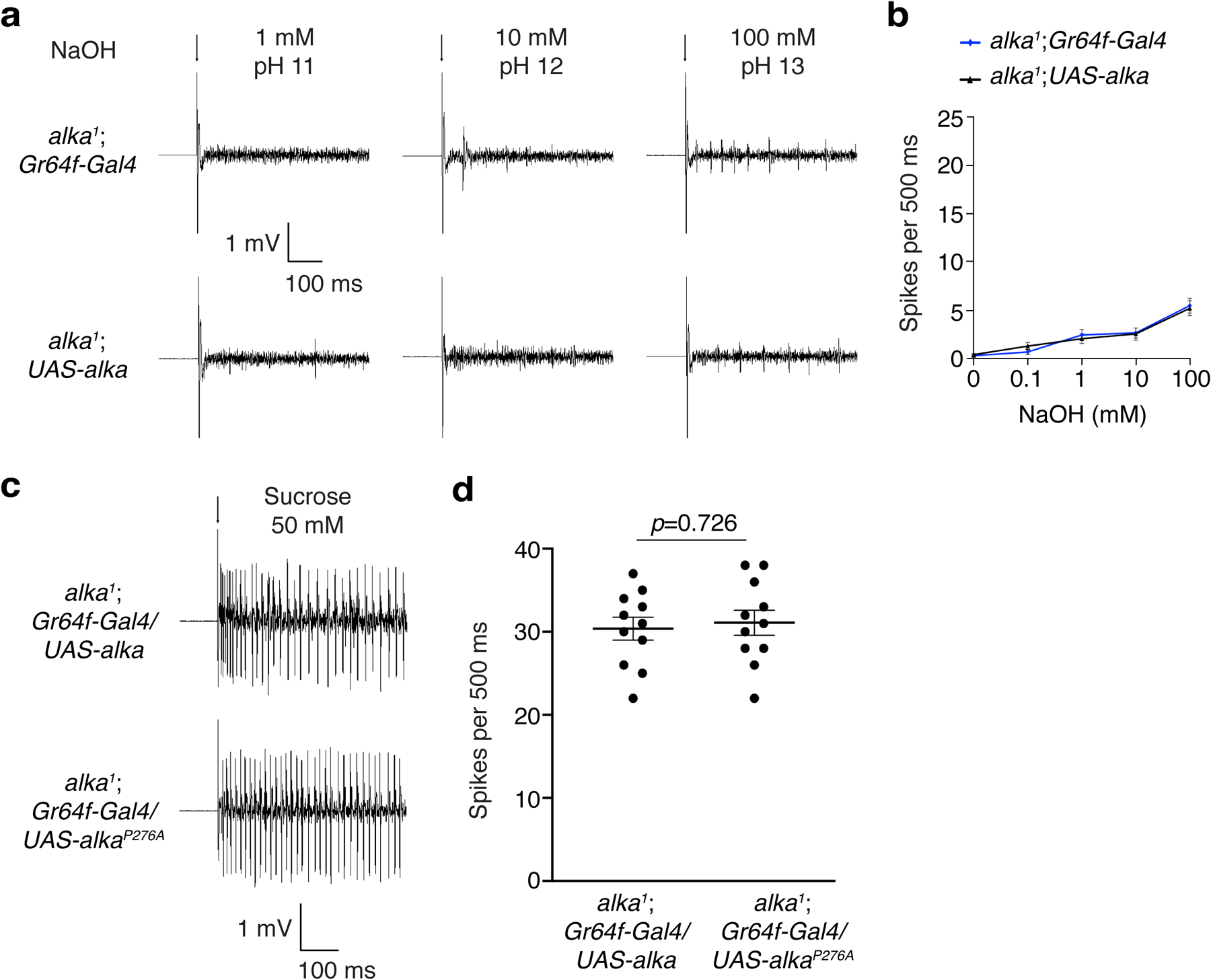
(a) Spikes evoked by L7 sensilla responding to high-pH stimuli in alka1;Gr64f-Gal4 or alka1;UAS-alka flies. (b) Statistical analyses of the spike frequencies for alka1;UAS-alka and alka1;Gr64f-Gal4 flies. n=11 flies, unpaired two-tailed Student’s t-tests. (c) Spikes produced by L7 sensilla responding to 50 mM sucrose in alka1;Gr64f-Gal4/UAS-alka or alka1;Gr64f-Gal4/UAS-alkaP276A flies. (d) Statistical analyses of the spike frequencies for alka1;Gr64f-Gal4/UAS-alka and alka1;Gr64f-Gal4/UAS-alkaP276A flies. n=11 flies, unpaired two-tailed Student’s t-tests. Mean ± SEM. Arrows indicate stimulus onset.
Extended Data Fig. 8. Expression of alka-Gal4 in combination with Orco-Gal80 in the fly labellum and brain.
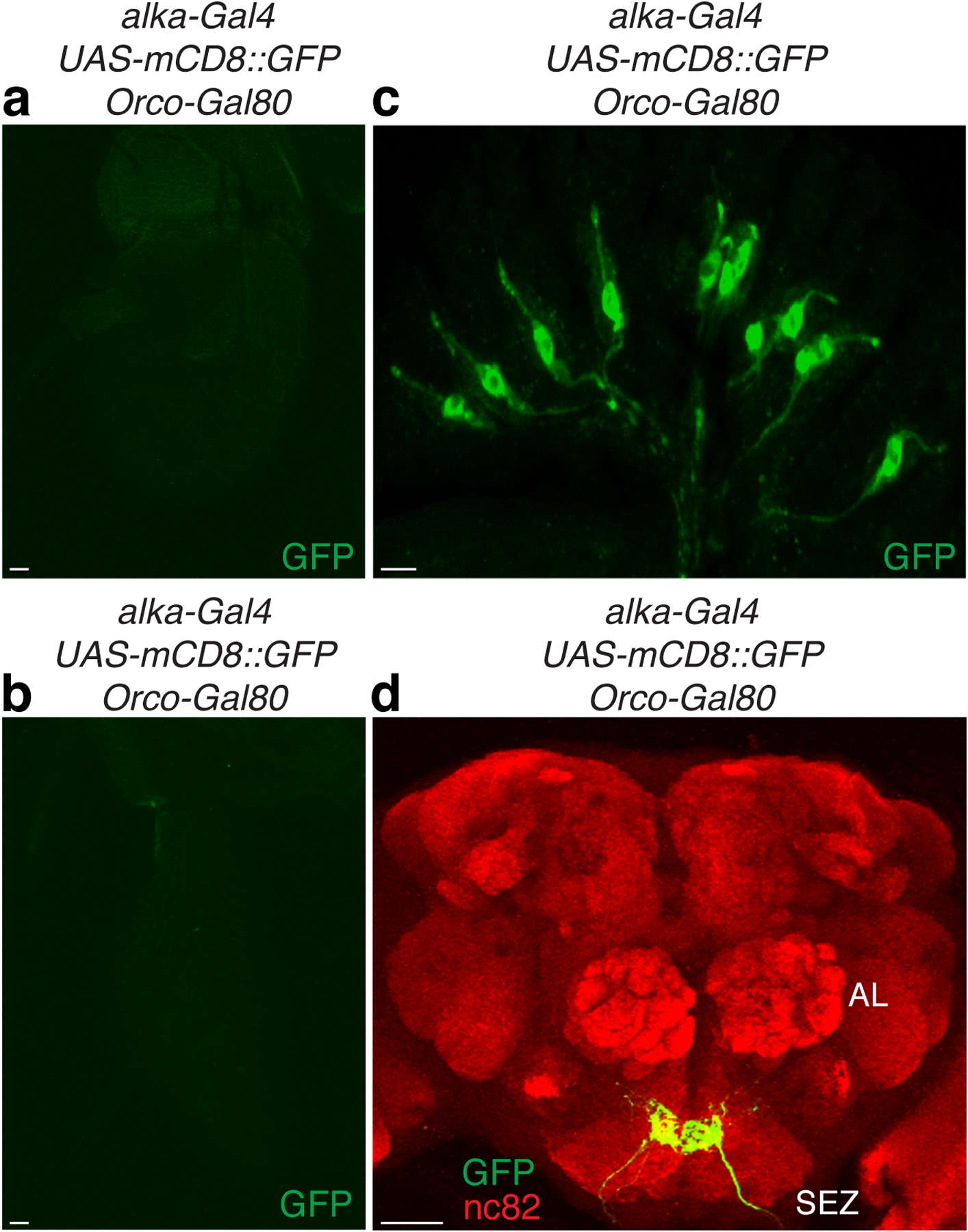
(a, b) Expression of alka-Gal4,UAS-mCD8::GFP;Orco-Gal80 in the antenna (a) and the maxillary palp (b). (c) Expression of alka-Gal4,UAS-mCD8::GFP;Orco-Gal80 in the labellum. (d) GRN projections in the brain of the alka-Gal4,UAS-mCD8::GFP;Orco-Gal80 fly. SEZ, subesophageal zone; AL, antennal lobe. Scale bars: 10 μm (a-c), 50 μm (d).
Extended Data Fig. 9. alka-expressing GRNs are selectively required to sense alkaline food.
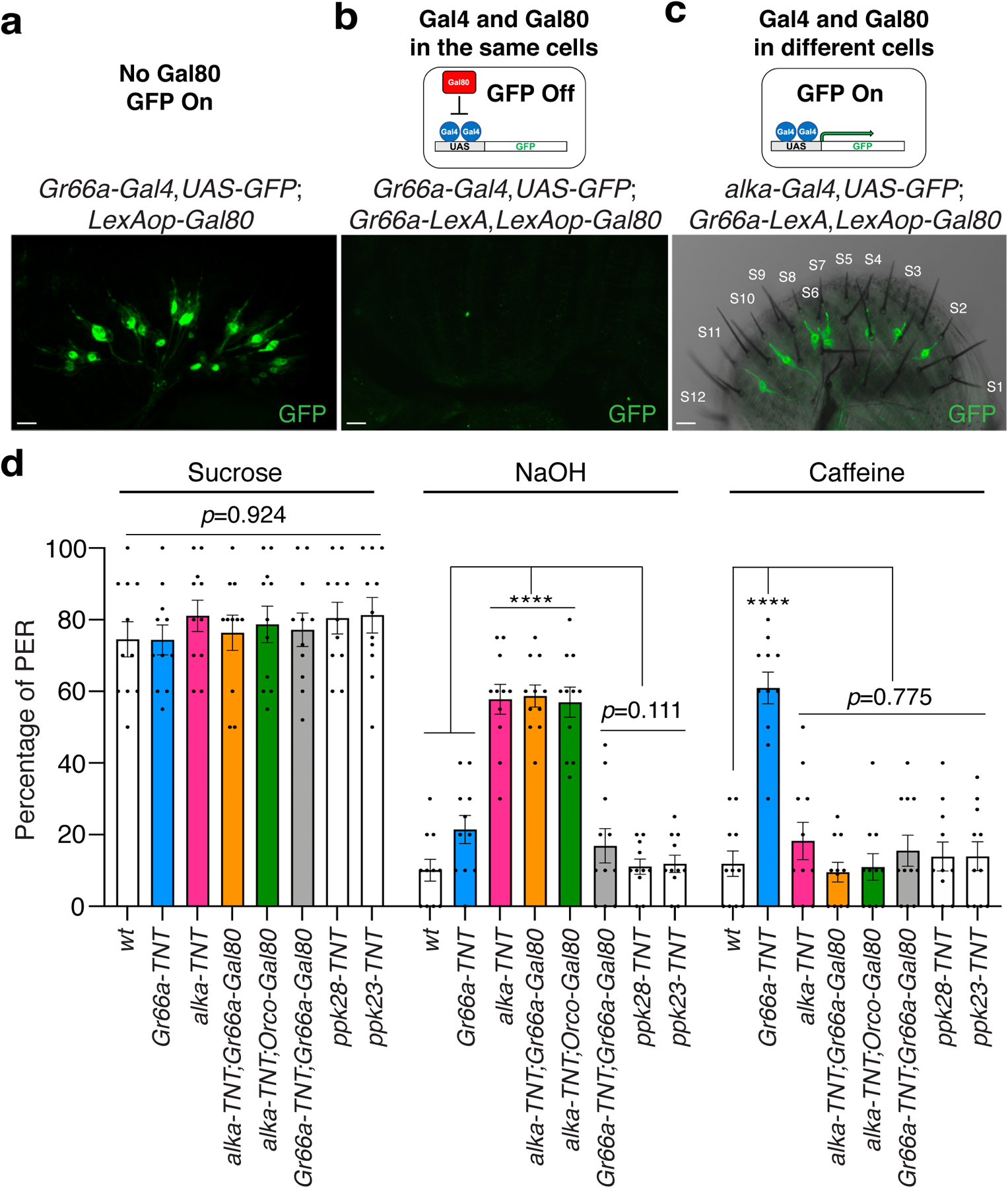
(a, b) GFP expression in the labellum of Gr66a-Gal4,UAS-GFP;LexAop-Gal80 (a) or Gr66a-Gal4,UAS-GFP;Gr66a-lexA,LexAop-Gal80 (b). Scale bar: 10 μm. (c) Relative localization between the alka-expressing GRNs and S-type taste sensilla in the alka-Gal4,UAS-GFP;Gr66a-lexA,LexAop-Gal80 fly labellum. Scale bar: 10 μm. (d) PERs to sweet food (50 mM sucrose), alkaline food (10 mM NaOH mixed with 30 mM sucrose), and bitter food (10 mM caffeine mixed with 30 mM sucrose) for alka-TNT (alka-Gal4,UAS-TNT), alka-TNT;Orco-Gal80 (alka-Gal4,UAS-TNT;Orco-Gal80), alka-TNT;Gr66a-Gal80 (alka-Gal4,UAS-TNT;Gr66a-lexA,LexAop-Gal80), Gr66a-TNT (Gr66a-Gal4,UAS-TNT), Gr66a-TNT;Gr66a-Gal80 (Gr66a-Gal4,UAS-TNT;Gr66a-lexA,LexAop-Gal80), ppk23-TNT (ppk23-Gal4,UAS-TNT), ppk28-TNT (ppk28-Gal4,UAS-TNT), and wild-type (wt) flies. n=11 trials, Mean ± SEM, one-way ANOVA with Tukey’s post-hoc tests, ****p<0.0001.
Extended Data Movie 1.
The alka-Gal4 control fly showing persistent sucrose (500mM) feeding in the presence and absence of an intense red-light stimulus (2000 Lux).
Extended Data Movie 2.
The alka-Gal4,UAS-CsChrimson;Orco-Gal80 fly exhibiting normal feeding of sucrose (500 mM) in the absence of red light but a cessation of feeding in response to a moderate red-light stimulus (1200 Lux).
Supplementary Material
Acknowledgments
We thank Samuel Chan and Peter Nguyen for their technical contributions to this study. We also thank the Bloomington Drosophila Research Center for fly stocks and the Drosophila Genome Research Center for DNA clones. We appreciate the labs of Drs. Danielle R. Reed, Mehmet Hakan Ozdener, and Ichiro Matsumoto at the Monell Chemical Senses Center for sharing equipment and facilities. Our work was supported by the National Institute on Deafness and other Communication Disorders grants R01 DC018592 (Y.V.Z.) and R01 DC007864 (C.M.), the Ambrose Monell Foundation (Y.V.Z.), and the National Key Research and Development Program of China Project 2018YFA0108001 (Z.-Q.T.).
Footnotes
Competing interests
The authors involved in this study declare no competing interests.
Data availability
All relevant data have been presented in this manuscript and its supplementary information. Any other related information is available upon request from Yali V. Zhang. In addition, we deposited the raw confocal videos for double-labeling experiments, including Fig. 3h (https://doi.org/10.6084/m9.figshare.22029284), Fig. 3i (https://doi.org/10.6084/m9.figshare.22029131), Fig. 3k (https://doi.org/10.6084/m9.figshare.22029488), and Fig. 3l (https://doi.org/10.6084/m9.figshare.22029350), in Figshare, a publicly accessible repository.
References
- 1.Yarmolinsky DA, Zuker CS & Ryba NJ Common sense about taste: from mammals to insects. Cell 139, 234–244, doi: 10.1016/j.cell.2009.10.001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liman ER, Zhang YV & Montell C Peripheral coding of taste. Neuron 81, 984–1000, doi: 10.1016/j.neuron.2014.02.022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiwull-Schone H, Kiwull P, Manz F & Kalhoff H Food composition and acid-base balance: alimentary alkali depletion and acid load in herbivores. J Nutr 138, 431S–434S, doi: 10.1093/jn/138.2.431S (2008). [DOI] [PubMed] [Google Scholar]
- 4.Huang AL et al. The cells and logic for mammalian sour taste detection. Nature 442, 934–938, doi: 10.1038/nature05084 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu YH et al. An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050, doi: 10.1126/science.aao3264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi T, Mack JO, Lee CM & Zhang YV Molecular and cellular basis of acid taste sensation in Drosophila. Nat Commun 12, 3730, doi: 10.1038/s41467-021-23490-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloehn NW & Brogden WJ The alkaline taste; a comparison of absolute thresholds for sodium hydroxide on the tip and mid-dorsal surfaces of the tongue. Am J Psychol 61, 90–93 (1948). [PubMed] [Google Scholar]
- 8.Liljestrand G & Zotterman Y The alkaline taste. Acta Physiol Scand 35, 380–389, doi: 10.1111/j.1748-1716.1955.tb01294.x (1956). [DOI] [PubMed] [Google Scholar]
- 9.Paje F, Mossakowski D pH-preferences and habitat selection in carabid beetles. Oecologia 64, 6 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Milius M et al. A new method for electrophysiological identification of antennal pH receptor cells in ground beetles: the example of Pterostichus aethiops (Panzer, 1796) (Coleoptera, Carabidae). J Insect Physiol 52, 960–967, doi: 10.1016/j.jinsphys.2006.06.003 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Clyne PJ, Warr CG & Carlson JR Candidate taste receptors in Drosophila. Science 287, 1830–1834, doi: 10.1126/science.287.5459.1830 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Dahanukar A, Foster K, van der Goes van Naters WM & Carlson JR A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci 4, 1182–1186, doi: 10.1038/nn765 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Singhvi A, Kong P & Scott K Taste representations in the Drosophila brain. Cell 117, 981–991, doi: 10.1016/j.cell.2004.06.011 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Slone J, Daniels J & Amrein H Sugar receptors in Drosophila. Curr Biol 17, 1809–1816, doi: 10.1016/j.cub.2007.09.027 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y, Moon SJ & Montell C A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci U S A 104, 14110–14115, doi: 10.1073/pnas.0702421104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YV, Ni J & Montell C The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338, doi: 10.1126/science.1234133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaeger AH et al. A complex peripheral code for salt taste in Drosophila. Elife 7, doi: 10.7554/eLife.37167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dweck HKM, Talross GJS, Luo Y, Ebrahim SAM & Carlson JR Ir56b is an atypical ionotropic receptor that underlies appetitive salt response in Drosophila. Curr Biol 32, 1776–1787 e1774, doi: 10.1016/j.cub.2022.02.063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rimal S et al. Mechanism of Acetic Acid Gustatory Repulsion in Drosophila. Cell Rep 26, 1432–1442 e1434, doi: 10.1016/j.celrep.2019.01.042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganguly A et al. Requirement for an Otopetrin-like protein for acid taste in Drosophila. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2110641118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Alcaniz JA et al. An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nat Commun 9, 4252, doi: 10.1038/s41467-018-06453-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shim J et al. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat Commun 6, 8867, doi: 10.1038/ncomms9867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung HY et al. Heterogeneity in the Drosophila gustatory receptor complexes that detect aversive compounds. Nat Commun 8, 1484, doi: 10.1038/s41467-017-01639-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montell C Drosophila sensory receptors-a set of molecular Swiss Army Knives. Genetics 217, 1–34, doi: 10.1093/genetics/iyaa011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn JE, Chen Y & Amrein H Molecular basis of fatty acid taste in Drosophila. Elife 6, doi: 10.7554/eLife.30115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown EB et al. Ir56d-dependent fatty acid responses in Drosophila uncover taste discrimination between different classes of fatty acids. Elife 10, doi: 10.7554/eLife.67878 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo R et al. Molecular basis and homeostatic regulation of Zinc taste. Protein Cell 13, 462–469, doi: 10.1007/s13238-021-00845-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Poudel S, Kim Y, Thakur D & Montell C Calcium Taste Avoidance in Drosophila. Neuron 97, 67–74 e64, doi: 10.1016/j.neuron.2017.11.038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisotsky Z, Medina A, Freeman E & Dahanukar A Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nat Neurosci 14, 1534–1541, doi: 10.1038/nn.2944 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Freeman EG & Dahanukar A Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol 34, 140–148, doi: 10.1016/j.conb.2015.06.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott K et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673, doi: 10.1016/s0092-8674(01)00263-x (2001). [DOI] [PubMed] [Google Scholar]
- 32.Benton R, Vannice KS, Gomez-Diaz C & Vosshall LB Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162, doi: 10.1016/j.cell.2008.12.001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatachalam K & Montell C TRP channels. Annu Rev Biochem 76, 387–417, doi: 10.1146/annurev.biochem.75.103004.142819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J et al. Sour Sensing from the Tongue to the Brain. Cell 179, 392–402 e315, doi: 10.1016/j.cell.2019.08.031 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Lynch JW Molecular structure and function of the glycine receptor chloride channel. Physiol Rev 84, 1051–1095, doi: 10.1152/physrev.00042.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Remnant EJ et al. Evolution, Expression, and Function of Nonneuronal Ligand-Gated Chloride Channels in Drosophila melanogaster. G3 (Bethesda) 6, 2003–2012, doi: 10.1534/g3.116.029546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knipple DC, The SDM ligand-gated chloride channel gene family of Drosophila melanogaster. Pesticide Biochemistry and Physiology 97, 140–148 (2010). [Google Scholar]
- 38.Ffrench-Constant RH, Mortlock DP, Shaffer CD, MacIntyre RJ & Roush RT Molecular cloning and transformation of cyclodiene resistance in Drosophila: an invertebrate gamma-aminobutyric acid subtype A receptor locus. Proc Natl Acad Sci U S A 88, 7209–7213, doi: 10.1073/pnas.88.16.7209 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson JE, Soderlund DM & Knipple DC Characterization of a putative gamma-aminobutyric acid (GABA) receptor beta subunit gene from Drosophila melanogaster. Biochem Biophys Res Commun 193, 474–482, doi: 10.1006/bbrc.1993.1648 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Harvey RJ et al. Sequence of a Drosophila ligand-gated ion-channel polypeptide with an unusual amino-terminal extracellular domain. J Neurochem 62, 2480–2483, doi: 10.1046/j.1471-4159.1994.62062480.x (1994). [DOI] [PubMed] [Google Scholar]
- 41.Cully DF, Paress PS, Liu KK, Schaeffer JM & Arena JP Identification of a Drosophila melanogaster glutamate-gated chloride channel sensitive to the antiparasitic agent avermectin. J Biol Chem 271, 20187–20191, doi: 10.1074/jbc.271.33.20187 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Gengs C et al. The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA). J Biol Chem 277, 42113–42120, doi: 10.1074/jbc.M207133200 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Gisselmann G, Pusch H, Hovemann BT & Hatt H Two cDNAs coding for histamine-gated ion channels in D. melanogaster. Nat Neurosci 5, 11–12, doi: 10.1038/nn787 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Schnizler K et al. A novel chloride channel in Drosophila melanogaster is inhibited by protons. J Biol Chem 280, 16254–16262, doi: 10.1074/jbc.M411759200 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Feingold D, Starc T, O’Donnell MJ, Nilson L & Dent JA The orphan pentameric ligand-gated ion channel pHCl-2 is gated by pH and regulates fluid secretion in Drosophila Malpighian tubules. J Exp Biol 219, 2629–2638, doi: 10.1242/jeb.141069 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Redhai S et al. An intestinal zinc sensor regulates food intake and developmental growth. Nature 580, 263–268, doi: 10.1038/s41586-020-2111-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frenkel L et al. Organization of Circadian Behavior Relies on Glycinergic Transmission. Cell Rep 19, 72–85, doi: 10.1016/j.celrep.2017.03.034 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Dambly-Chaudiere C et al. The paired box gene pox neuro: a determinant of poly-innervated sense organs in Drosophila. Cell 69, 159–172 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Zhang YV, Aikin TJ, Li Z & Montell C The Basis of Food Texture Sensation in Drosophila. Neuron 91, 863–877, doi: 10.1016/j.neuron.2016.07.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stocker RF The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 275, 3–26, doi: 10.1007/BF00305372 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Shanbhag SR, Park SK, Pikielny CW & Steinbrecht RA Gustatory organs of Drosophila melanogaster: fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res 304, 423–437, doi: 10.1007/s004410100388 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Dusek M, Chapuis G, Meyer M & Petricek V Sodium carbonate revisited. Acta Crystallogr B 59, 337–352, doi: 10.1107/s0108768103009017 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Khanna A & Kurtzman NA Metabolic alkalosis. Respir Care 46, 354–365 (2001). [PubMed] [Google Scholar]
- 54.Lee T & Luo L Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Moon SJ, Kottgen M, Jiao Y, Xu H & Montell C A taste receptor required for the caffeine response in vivo. Curr Biol 16, 1812–1817, doi: 10.1016/j.cub.2006.07.024 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Fujii S et al. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol 25, 621–627, doi: 10.1016/j.cub.2014.12.058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganguly A et al. A Molecular and Cellular Context-Dependent Role for Ir76b in Detection of Amino Acid Taste. Cell Rep 18, 737–750, doi: 10.1016/j.celrep.2016.12.071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cameron P, Hiroi M, Ngai J & Scott K The molecular basis for water taste in Drosophila. Nature 465, 91–95, doi: 10.1038/nature09011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Z, Wang Q & Wang Z The amiloride-sensitive epithelial Na+ channel PPK28 is essential for drosophila gustatory water reception. J Neurosci 30, 6247–6252, doi: 10.1523/JNEUROSCI.0627-10.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thistle R, Cameron P, Ghorayshi A, Dennison L & Scott K Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149, 1140–1151, doi: 10.1016/j.cell.2012.03.045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betz H Glycine receptors: heterogeneous and widespread in the mammalian brain. Trends Neurosci 14, 458–461, doi: 10.1016/0166-2236(91)90045-v (1991). [DOI] [PubMed] [Google Scholar]
- 62.Duran C, Thompson CH, Xiao Q & Hartzell HC Chloride channels: often enigmatic, rarely predictable. Annu Rev Physiol 72, 95–121, doi: 10.1146/annurev-physiol-021909-135811 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Germann AL et al. Activation and modulation of recombinant glycine and GABAA receptors by 4-halogenated analogues of propofol. Br J Pharmacol 173, 3110–3120, doi: 10.1111/bph.13566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X, Chen H, Michelsen K, Schneider S & Shaffer PL Crystal structure of human glycine receptor-alpha3 bound to antagonist strychnine. Nature 526, 277–280, doi: 10.1038/nature14972 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Schmidt T, Situ AJ & Ulmer TS Structural and thermodynamic basis of proline-induced transmembrane complex stabilization. Sci Rep 6, 29809, doi: 10.1038/srep29809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gödde J, Krefting ER Ions in the receptor lymph of the labellar taste hairs of the fly Protophormia terraenovae. J Insect Physiol 35, 107–111 (1989). [Google Scholar]
- 67.Larsson MC et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714, doi: 10.1016/j.neuron.2004.08.019 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Eliason J, Afify A, Potter C & Matsumura I A GAL80 Collection To Inhibit GAL4 Transgenes in Drosophila Olfactory Sensory Neurons. G3 (Bethesda) 8, 3661–3668, doi: 10.1534/g3.118.200569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klapoetke NC et al. Independent optical excitation of distinct neural populations. Nat Methods 11, 338–346, doi: 10.1038/nmeth.2836 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Umezaki Y, Yasuyama K, Nakagoshi H & Tomioka K Blocking synaptic transmission with tetanus toxin light chain reveals modes of neurotransmission in the PDF-positive circadian clock neurons of Drosophila melanogaster. J Insect Physiol 57, 1290–1299, doi: 10.1016/j.jinsphys.2011.06.004 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Venken KJ, Simpson JH & Bellen HJ Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72, 202–230, doi: 10.1016/j.neuron.2011.09.021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brand AH & Perrimon N Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 73.Adrogue HE & Adrogue HJ Acid-base physiology. Respir Care 46, 328–341 (2001). [PubMed] [Google Scholar]
- 74.Murayama T, Takayama J, Fujiwara M & Maruyama IN Environmental alkalinity sensing mediated by the transmembrane guanylyl cyclase GCY-14 in C. elegans. Curr Biol 23, 1007–1012, doi: 10.1016/j.cub.2013.04.052 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Li G, Liu J, Liu J & Xu XZ TMC-1 Mediates Alkaline Sensation in C. elegans through Nociceptive Neurons. Neuron 91, 146–154, doi: 10.1016/j.neuron.2016.05.023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye X & Randall DJ The effect of water pH on swimming performance in rainbow trout (Salmo gairdneri, Richardson). Fish Physiol Biochem 9, 15–21, doi: 10.1007/BF01987607 (1991). [DOI] [PubMed] [Google Scholar]
- 77.St John SJ & Boughter JD Jr. Orosensory responsiveness to and preference for hydroxide-containing salts in mice. Chem Senses 34, 487–498, doi: 10.1093/chemse/bjp023 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Massie HR, Williams TR & Colacicco JR Changes in pH with age in Drosophila and the influence of buffers on longevity. Mech Ageing Dev 16, 221–231, doi: 10.1016/0047-6374(81)90098-1 (1981). [DOI] [PubMed] [Google Scholar]
- 79.Shanbhag S & Tripathi S Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J Exp Biol 212, 1731–1744, doi: 10.1242/jeb.029306 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Deshpande SA et al. Acidic Food pH Increases Palatability and Consumption and Extends Drosophila Lifespan. J Nutr 145, 2789–2796, doi: 10.3945/jn.115.222380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu W et al. Symbiotic bacteria attenuate Drosophila oviposition repellence to alkaline through acidification. Insect Sci 28, 403–414, doi: 10.1111/1744-7917.12857 (2021). [DOI] [PubMed] [Google Scholar]
- 82.Moon SJ, Lee Y, Jiao Y & Montell C A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol 19, 1623–1627, doi: 10.1016/j.cub.2009.07.061 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koh TW et al. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865, doi: 10.1016/j.neuron.2014.07.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Min S, Ai M, Shin SA & Suh GS Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A 110, E1321–1329, doi: 10.1073/pnas.1215680110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang YV, Raghuwanshi RP, Shen WL & Montell C Food experience-induced taste desensitization modulated by the Drosophila TRPL channel. Nat Neurosci 16, 1468–1476, doi: 10.1038/nn.3513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang K et al. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464, 597–600, doi: 10.1038/nature08848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim SH et al. Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc Natl Acad Sci U S A 107, 8440–8445, doi: 10.1073/pnas.1001425107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ullrich F et al. Identification of TMEM206 proteins as pore of PAORAC/ASOR acid-sensitive chloride channels. Elife 8, doi: 10.7554/eLife.49187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang J et al. PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science 364, 395–399, doi: 10.1126/science.aav9739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nonaka T & Wong DTW Saliva Diagnostics. Annu Rev Anal Chem (Palo Alto Calif) 15, 107–121, doi: 10.1146/annurev-anchem-061020-123959 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dibattista M, Pifferi S, Boccaccio A, Menini A & Reisert J The long tale of the calcium activated Cl(−) channels in olfactory transduction. Channels (Austin) 11, 399–414, doi: 10.1080/19336950.2017.1307489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang W et al. Increased intracellular Cl(−) concentration improves airway epithelial migration by activating the RhoA/ROCK Pathway. Theranostics 10, 8528–8540, doi: 10.7150/thno.46002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng Y et al. Identification of two novel Drosophila melanogaster histamine-gated chloride channel subunits expressed in the eye. J Biol Chem 277, 2000–2005, doi: 10.1074/jbc.M107635200 (2002). [DOI] [PubMed] [Google Scholar]
- 94.Li Q & Montell C Mechanism for food texture preference based on grittiness. Curr Biol 31, 1850–1861 e1856, doi: 10.1016/j.cub.2021.02.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chu B, Chui V, Mann K & Gordon MD Presynaptic gain control drives sweet and bitter taste integration in Drosophila. Curr Biol 24, 1978–1984, doi: 10.1016/j.cub.2014.07.020 (2014). [DOI] [PubMed] [Google Scholar]
- 96.Potter CJ, Tasic B, Russler EV, Liang L & Luo L The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548, doi: 10.1016/j.cell.2010.02.025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shearin HK, Macdonald IS, Spector LP & Stowers RS Hexameric GFP and mCherry reporters for the Drosophila GAL4, Q, and LexA transcription systems. Genetics 196, 951–960, doi: 10.1534/genetics.113.161141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data have been presented in this manuscript and its supplementary information. Any other related information is available upon request from Yali V. Zhang. In addition, we deposited the raw confocal videos for double-labeling experiments, including Fig. 3h (https://doi.org/10.6084/m9.figshare.22029284), Fig. 3i (https://doi.org/10.6084/m9.figshare.22029131), Fig. 3k (https://doi.org/10.6084/m9.figshare.22029488), and Fig. 3l (https://doi.org/10.6084/m9.figshare.22029350), in Figshare, a publicly accessible repository.


