Abstract
The rate, progress, and limits of trichloroethylene (TCE) degradation by Ralstonia eutropha AEK301/pYK3021 whole cells were examined in the absence of aromatic induction. At TCE concentrations up to 800 μM, degradation rates were sustained until TCE was no longer detectable. The Ks and Vmax for TCE degradation by AEK301/pYK3021 whole cells were determined to be 630 μM and 22.6 nmol/min/mg of total protein, respectively. The sustained linear rates of TCE degradation by AEK301/pYK3021 up to a concentration of 800 μM TCE suggest that solvent effects are limited during the degradation of TCE and that this construct is little affected by the formation of toxic intermediates at the TCE levels and assay duration tested. TCE degradation by this strain is subject to carbon catabolite repression.
Like many other chlorinated hydrocarbons, trichloroethylene (TCE) has become an important environmental pollutant because of its toxic properties and widespread occurrence in groundwater. TCE is the most commonly reported volatile organic pollutant of groundwater in the United States (20). While there are no reports of bacterial growth on TCE as a sole carbon and energy source, cometabolic oxidation of TCE by nonspecific catabolic oxygenases has been described for several types of microorganisms (3) and TCE is perhaps the best-studied compound subject to aerobic cometabolism.
The application of bacteria for the aerobic bioremediation of TCE has been proposed and investigated for a wide variety of microorganisms. The most critical factors in consideration for such studies are the specific activity of the cells for TCE and the possible formation of toxic intermediates. For example, in wild-type Pseudomonas putida and P. putida F1, in which TCE oxidation is mediated by toluene dioxygenase, observed inhibition of growth has been attributed to covalent modification of cellular molecules through reactive products of TCE degradation (9, 23). For the wild type and F1, the rate of TCE removal declines rapidly in batch cultures when TCE is supplied at initial concentrations greater than 10 or 80 μM, respectively. Furthermore, it has been shown that growth substrates added to induce the catabolic genes involved in oxidation of TCE can be competitive inhibitors of TCE conversion (4).
Most studies on the kinetics of aerobic TCE degradation have been done with methane- and toluene-utilizing mixed and pure cultures (5, 9, 12, 14, 16–18, 22). Limited data on the kinetics of aerobic TCE degradation by phenol-induced monooxygenases and on the possible toxic effects of TCE oxidation metabolites are available.
Ralstonia eutropha (formerly Alcaligenes eutrophus) JMP134 is able to degrade TCE by an inducible, chromosomally encoded phenol hydroxylase (7, 11). The isolation of the chromosomally encoded phenol hydroxylase genes, through complementation of a mutant deficient in phenol degradation with a JMP134 genomic cosmid library, has been reported previously (10). The subcloning of restriction fragments from this cosmid resulted in a recombinant plasmid (pYK3021) conferring phenol hydroxylase activity and TCE degradation in the absence of phenol induction. Preliminary studies using this construct have shown a high capacity for TCE removal in the absence of aromatic induction with limited sensitivity to TCE-mediated toxicity (11).
The purpose of the work presented here was to determine the whole-cell kinetics of TCE degradation by suspended batch cultures of R. eutropha containing the recombinant plasmid pYK3021. Ks and Vmax were estimated for comparison to previously published values for these parameters. Our objective was to establish the feasibility of stable constitutive expression for application to the remediation of TCE contamination. Constitutive expression allows TCE degradation to occur in the absence of induction by the normal aromatic substrate. This obviates the biochemical problems related to competitive inhibition at the active site and regulatory problems which would prohibit the introduction of aromatic inducers during in situ remediation. The degree of TCE degradation was determined during growth on a variety of noninducing carbon sources in an effort to maximize TCE removal.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, growth conditions, and chemicals.
R. eutropha AEK301 was derived from R. eutropha JMP134. The plasmid pJP4 was cured to create strain AEO106 (8). AEO106 was subjected to rifampin selection to produce the Rifr strain AEK101 (11). Random Tn5 mutagenesis was used to obtain strain AEK301, deficient in phenol metabolism (10). The recombinant plasmid pYK3021 contains an 8.6-kb XhoI-BamHI fragment in the pMMB67EH vector, confers TCE degradation in the absence of phenol induction, and confers resistance to carbenicillin when placed in AEK301 (10). Cultures of R. eutropha AEK301 with and without pYK3021 were maintained at 30°C on minimal salts medium (MMO) (21) supplemented with 20 mM sodium citrate or on tryptone-yeast extract-glucose medium (TNB) (19). Where indicated, MMO was supplemented with benzoate (2.5 mM), citrate (20 mM), sodium citrate (20 mM), gluconate (20 mM), lactate (20 mM), malate (20 mM), or 0.3% Casamino Acids. Concentrated stock solutions of each carbon source were prepared and adjusted to a pH of 7.0 with NaOH where necessary. When required, 50 μg of carbenicillin, 150 μg of rifampin, or 100 μg of kanamycin per ml was added to the growth medium. Yeast extract, tryptone, and agar were purchased from Difco. Other medium additives, bovine serum albumin, and chromatography-quality n-pentane were all purchased from Sigma. Chromatography-quality TCE and 1,2-dibromoethane (EDB) were purchased from Aldrich. Teflon-butyl septa and reactor vials were purchased from Fisher Scientific.
Analytical methods.
TCE was measured by gas chromatography (GC) analysis with a Hewlett-Packard model 5890 gas chromatograph equipped with a 25-m cross-linked methyl silicone gum capillary column (Hewlett-Packard) and an electron capture detection system. Peak integrations were obtained with a Hewlett-Packard model 3390A integrator. The following operating conditions were used: injector temperature, 150°C; detector temperature, 250°C; column temperature, 40 to 100°C at 20°C/min; and helium carrier gas flow, 6 ml/min. Under these conditions TCE and EDB (internal standard) in pentane extracts had retention times of 2.2 and 2.9 min, respectively.
Standard TCE degradation kinetics assay.
AEK301/pYK3021 was grown in MMO containing 10 mM sodium citrate and appropriate levels of kanamycin and carbenicillin at 30°C with shaking at 180 rpm to mid-log phase at an optical density of 0.6 to 0.8 at 425 nm. Cells were harvested by centrifugation at 8,000 × g for 10 min. Cell pellets were then suspended in fresh MMO containing 10 mM sodium citrate at an optical density of 1.0 at 425 nm. The cultures were then returned to 30°C and shaking at 180 rpm. After 1 h, 2-ml samples were dispensed into 20-ml glass vials, and the vials were crimp-sealed with Teflon-butyl septa. The appropriate volume of an 8 mM TCE stock was added to each vial by injection through the septum with a gastight syringe (Hamilton, Reno, Nev.). The vials were inverted to minimize TCE loss and returned to 30°C and shaking at 180 rpm. All assays were performed in triplicate.
At the appropriate time, the reactions were stopped by the addition of 2 ml of n-pentane containing 1 ppm of EDB. EDB was added as an internal standard to correct for GC sampling imprecision. The vials were placed at room temperature on a shaker platform for 15 min and then centrifuged at 4,000 × g for 10 min to aid in the separation of the organic phase. Following centrifugation, approximately 0.5 ml was transferred with a gastight syringe to a Teflon-butyl septum-sealed vial. A 1-μl sample was then removed and analyzed by GC for TCE concentrations. Control samples of sterile medium gave TCE recoveries of 95 to 97% under these conditions. The GC data represent an average of two or more injections per assay. TCE stocks of 8 mM were prepared by completely filling a 20-ml glass vial (containing eight 3-mm-diameter glass beads to facilitate mixing) with sterile water. Once each vial had been crimp-sealed with a Teflon-butyl septum with no trapped air, the appropriate volume of pure TCE was added by injection through the septum and then allowed to dissolve completely overnight at room temperature with constant mixing.
No-headspace assay.
Cultures were incubated and prepared as described above. Following the 1-h preincubation, approximately 2 ml of each cell suspension and a glass bead (3-mm diameter) were placed in a 2-ml vial crimp-sealed with a Teflon-butyl septum and with no trapped air. The glass beads were added to facilitate thorough mixing of the contents. The appropriate volume of an 8 mM TCE stock was added by injection through the septum of each vial with a gastight syringe, and the vials were incubated at 30°C with constant mixing. At 5-min intervals, the reactions were stopped by the transfer of 0.5-ml aliquots of TCE-cell suspensions into other sealed vials containing 0.5 ml of n-pentane and 1 ppm of EDB. These vials were placed at room temperature for extraction as described above, and 1 μl of the organic phase was analyzed by GC for TCE concentrations.
Protein determinations.
Cell suspensions were solubilized by adding 0.2 volume of 5 M NaOH and heating at 85°C for 10 min. Following the addition of 0.2 volume of 4 M HCl, the total protein concentrations were determined by the Lowry assay (13). Bovine serum albumin which had been treated with NaOH, heat, and HCl in parallel was used as a standard in these assays.
Calculations and equations.
The doubling time of batch cultures, in hours, was determined during the logarithmic phase of growth by measuring the optical density of the culture at 425 nm at two separate time points. Standard logarithmic calculation of doubling time was then applied.
The air-water partitioning behavior of TCE was expressed with a modified equilibrium-partitioning-in-closed-systems (EPICS) equation, developed for predicting the partitioning of volatile C1 and C2 chlorinated hydrocarbons, with a dimensionless Henry’s law constant which has been adapted for studies conducted at different temperatures (6). The total moles of a volatile solute added to a sealed reactor vial will be partitioned at equilibrium according to the equation moles = Cg [(Vw/Hc) + Vg], where Cg is the concentration (micromolar) of TCE in the gas phase or headspace, Vw is the volume of the aqueous phase in the reactor, Vg is the volume of the headspace in the reactor, and Hc is the dimensionless Henry’s law constant for TCE, which was previously determined to be 0.492 at 30°C. The Ks, which is the Michaelis constant for cellular kinetics and is analogous to Km for enzymatic reactions, and Vmax were determined from the axis intercepts from a Lineweaver-Burk double-reciprocal plot.
RESULTS AND DISCUSSION
Effects of substrates on TCE removal by AEK301/pYK3021.
While TCE degradation by AEK301/pYK3021 occurred in the absence of phenol induction, the apparent rate of TCE cometabolism varied depending on the carbon and energy source provided (10, 11). To further characterize this observation and enhance rates of TCE removal by AEK301/pYK3021, the degree of TCE oxidation was examined with a variety of noninducing carbon sources. AEK301/pYK3021 was grown to mid-log phase in MMO supplemented with different single substrates or in TNB, an enriched medium, and subjected to a standard TCE degradation assay at an initial concentration of 40 μM TCE. After 2 h the reactions were stopped by the addition of pentane, and the concentrations of remaining TCE were determined. While measurable amounts of TCE were removed from all reaction mixtures within 2 h, the degree of removal varied among substrates. MMO containing citrate, sodium citrate, or gluconate provided the highest TCE removal rates. MMO containing malate provided relatively poor TCE removal, as did TNB (Table 1). The doubling time for AEK301/pYK3021 in MMO supplemented with these different substrates or in TNB ranged from 1.2 h in TNB to 2.6 h in MMO supplemented with citrate (Table 1). MMO containing citrate, sodium citrate, or gluconate had the slowest doubling times (2.6, 2.5, and 2.4 h, respectively) yet provided the greatest TCE removal of the carbon sources tested. Conversely, TNB had the fastest doubling time (1.2 h) while providing relatively poor TCE removal. No significant effect on doubling time was observed in the presence of 40 μM TCE (Table 1). Catabolite repression of phenol hydroxylase catabolic genes has been demonstrated, for example, in P. putida H (15) and R. eutropha (1). In P. putida H, repression is mediated by inhibition of a phenol hydroxylase-specific transcriptional activator and subsequent reduction in transcription of the phenol hydroxylase catabolic genes (15). Repression was observed with the addition of glucose, succinate, lactate, or acetate and was least affected by the addition of pyruvate or citrate (15). In our test system, the addition of citrate or sodium citrate appeared to be the least repressive. No significant difference between the addition of 10 mM sodium citrate or that of 20 mM sodium citrate was noted (data not shown). Therefore, 10 mM sodium citrate was selected as the carbon source for all subsequent TCE degradation assays.
TABLE 1.
Effects of carbon sources on growth of and TCE degradation by AEK301/pYK3021
| Medium and carbon source | Concn of carbon source | Concn of TCE remaining (μM)a | % of TCE removed | Doubling time (h/generation)
|
|
|---|---|---|---|---|---|
| Without TCE | With 40 μM TCEc | ||||
| MMO | |||||
| Benzoate | 2.5 mM | 23.4 | 41.5 | 2.0 | 2.1 |
| Casamino Acids | 0.30% | 12.3 | 69.3 | 1.8 | 1.8 |
| Citrate | 20 mM | 1.2 | 97.0 | 2.6 | 2.5 |
| Gluconate | 20 mM | 6.5 | 83.8 | 2.4 | 2.5 |
| Lactate | 20 mM | 23.0 | 42.5 | 1.7 | 1.8 |
| Malate | 20 mM | 37.4 | 6.5 | 1.6 | 1.7 |
| Sodium citrate | 20 mM | NDb | >99.9 | 2.5 | 2.4 |
| TNB | 38.5 | 3.8 | 1.2 | 1.2 | |
After 2 h of incubation at 30°C; initial concentration, 40 μM TCE.
ND, not detected.
TCE was added by injection into sealed cultures during early exponential phase.
No-headspace assay.
Although TCE is highly volatile, for convenience it is often reported as an aqueous concentration in closed systems containing both air and water phases. This could be considered misleading when reporting kinetic data. To determine the effects, if any, of TCE volatility and phase partitioning on the measured kinetic parameters, a no-headspace assay was developed for comparison. Degradation of TCE by AEK301/pYK3021 is an aerobic process and is dependent on available oxygen. In such a closed system with no headspace to replenish the consumed dissolved oxygen, it is likely that oxygen would become a limiting factor. Accordingly, the degradation of TCE in a no-headspace assay was examined at a low initial concentration of TCE. Rate measurements were made immediately following the induction lag and were found to be consistent for several sampling intervals. After this short period of stable measurements, the rates in the no-headspace assay tended to diminish significantly. This probably indicates the eventual development of limiting oxygen levels.
The rate of TCE degradation in the no-headspace assay was determined to be 0.54 nmol/min/mg of total protein at an initial TCE concentration of 22 μM. This value was compared to rates observed at 16 and 80 μM TCE (headspace assay, uncorrected) (Table 2) and was found to be comparable to that observed in the standard assay at 16 μM, even though the aqueous concentrations were calculated to be different. Although the aqueous concentration of TCE in the no-headspace assay was similar to that in the standard assay at 80 μM, the rate of TCE degradation was significantly lower. These results suggest that the redistribution of TCE from the gas phase into the aqueous phase occurred faster than the degradation and was not a limiting factor in the well-mixed standard assay. Similar observations have been reported by Folsom et al. (5).
TABLE 2.
Comparison of the standard TCE degradation assay with a no-headspace TCE degradation assay
| Assay method | Total amt of TCE (nmol/vial) | Concn of TCE (μM)
|
TCE degradation
|
||
|---|---|---|---|---|---|
| Uncorrected | Aqueousa | Relative rate (nmol/min)b | Adjusted rate (nmol/min/mg)c | ||
| 90% headspace | 32 | 16 | 4 | 0.14 | 0.57 |
| 90% headspace | 160 | 80 | 17 | 0.72 | 2.26 |
| No headspace | 40 | 22 | 22 | 0.11 | 0.54 |
Determined with a dimensionless Henry’s law constant adjusted for the assay temperature and accounting for the reactor volume and sample volume, according to the equation in Materials and Methods.
Raw rate obtained from the assay.
Rate adjusted for the amount of protein.
Time course of TCE degradation by AEK301/pYK3021.
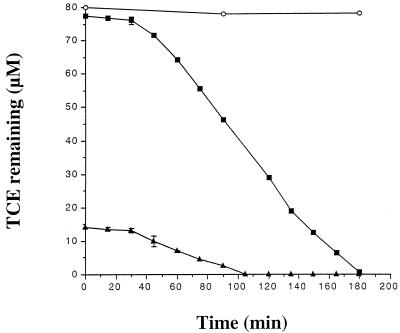
In previous studies of AEK301/pYK3021, TCE degradation was monitored over a period of many hours or even days (10, 11). For the purpose of determining the whole-cell kinetics of TCE degradation by AEK301/pYK3021, TCE degradation was instead monitored for 3 h at 15-min intervals at two different initial concentrations of TCE (16 and 80 μM). The negative control (AEK301 alone) was unable to degrade detectable amounts of TCE. For each initial TCE concentration tested, an initial lag period of approximately 40 min was observed prior to the onset of TCE degradation by AEK301/pYK3021 (Fig. 1). Following the initial lag, the rate of TCE degradation by AEK301/pYK3021 at an initial concentration of 80 μM TCE was sustained and remained essentially constant for a period of almost 2 h, until TCE was no longer detectable.
FIG. 1.
Degradation of TCE by AEK301/pYK3021 at an initial concentration of 16 μM TCE (▴) and AEK301/pYK3021 at an initial concentration of 80 μM TCE (■). The negative control, AEK301 alone (○), was used at an initial concentration of 80 μM TCE. Cultures were grown in MMO supplemented with 10 mM sodium citrate to mid-log phase, harvested by centrifugation, and suspended in fresh medium to an optical density of 1.0 at 425 nm. After 1 h at 30°C, 2-ml samples of each strain were then distributed into vials and the vials were sealed. Reactions were initiated by the injection of TCE through the septum. Samples were collected in duplicate every 15 min for a total of 3 h. Each data point represents the average of two or more samples, and error bars are provided where visible.
The lag period is likely the result of TCE-mediated induction of the structural gene operon and has not been observed in the phenol-induced wild type, JMP134 (11). Sequence analysis (GenBank accession no. AF065891) of the cloned region of pYK3021 indicates that the gene for a putative transport protein (2) is encoded and regulated separately from the phenol hydroxylase structural gene operon. If TCE degradation requires the induction and expression of both transport and hydroxylase functions, we hypothesize that the wild-type transport gene is not influenced by TCE-mediated induction, while the structural gene operon is. The cloning of the transport region has likely placed expression of the putative transport protein under the control of a plasmid-contained constitutive promoter. This would explain the absence of TCE-mediated induction in the wild-type strain and the lag phase in the pYK3021 construct.
Whole-cell kinetics of TCE degradation by AEK301/pYK3021.
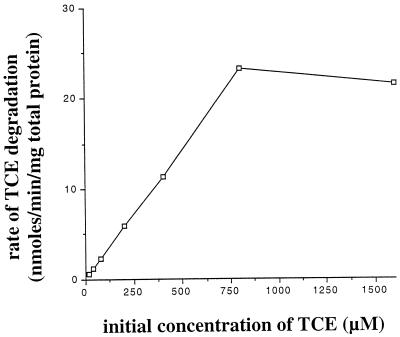
To accurately measure initial rates of TCE degradation by whole cells of AEK301/pYK3021, 5-min assays (beginning 50 min following the addition of TCE to the reactor vials) were conducted at various concentrations of TCE. For seven different initial TCE concentrations ranging from 16 to 1,600 μM, TCE degradation rates and total protein concentrations were measured. These results, summarized in Table 3, represent the averages for at least three different samples. The velocity or rate of degradation for each concentration was determined, and these values were plotted as a function of the initial TCE concentration (Fig. 2). The rate of TCE degradation by AEK301/pYK3021 was essentially linear from 16 to 800 μM TCE. The rate observed at 1,600 μM TCE was similar to that observed at 800 μM TCE, indicating that the saturation or inhibition of the catabolic enzymes occurred somewhere between these two concentrations.
TABLE 3.
Whole-cell kinetics of TCE degradation by AEK301/pYK3021
| Initial concn of TCE (μM) | Amt of TCE consumed (nmol/min) | Amt of total protein (mg/ml) | Rate of TCE degradation (nmol/min/mg of total protein) |
|---|---|---|---|
| 16 | 0.144 | 0.253 ± 0.006 | 0.57 |
| 40 | 0.299 | 0.238 ± 0.026 | 1.26 |
| 80 | 0.724 | 0.321 ± 0.016 | 2.26 |
| 200 | 2.010 | 0.342 ± 0.013 | 5.88 |
| 400 | 3.355 | 0.297 ± 0.010 | 11.30 |
| 800 | 7.240 | 0.312 ± 0.008 | 23.21 |
| 1,600 | 5.890 | 0.273 ± 0.004 | 21.58 |
FIG. 2.
Whole-cell kinetics of TCE degradation by AEK301/pYK3021. The initial rates of TCE degradation were determined as described in Materials and Methods. Rates are plotted as a function of initial TCE concentration.
Kinetics parameters were estimated by transforming the data (Table 3) to produce a double-reciprocal plot. The apparent cellular Ks and Vmax were then estimated to be 630 μM and 22.6 nmol/min/mg of total protein, respectively.
In R. eutropha AEK301/pYK3021, the rate of TCE degradation continued to increase at concentrations that are known to inhibit other enzyme systems (22). The highest observed rate of TCE degradation by P. putida F1 was 1.8 nmol/min/mg of total protein at 80 μM TCE, and the rate dropped rapidly at concentrations higher than 300 μM TCE (22). At 320 μM TCE, TCE degradation by P. putida F1 no longer occurred. In methane-induced Methylosinus trichosporium OB3b, toxicity was apparent at a concentration of 70 μM TCE, and cell suspensions were not able to degrade higher concentrations of TCE (17). In P. putida F1 and M. trichosporium the rates of TCE degradation were sustained only 20 and 60 min, respectively (22). Following an initial burst of TCE degradation, the rates declined rapidly, and even at initial concentrations of 15 μM TCE added to induced cultures of P. putida F1, significant quantities of TCE remained in reactor vials after 6 h of incubation (21). These studies suggest that TCE may be toxic to P. putida F1 and M. trichosporium through the formation of toxic intermediates (23).
Post-lag degradation rates were sustained until TCE was no longer detectable. This consistency of TCE degradation by AEK301/pYK3021 suggests that the process in this construct is less affected by the formation of toxic intermediates at the levels tested than in P. putida F1 or M. trichosporium. The linear rate of TCE degradation by AEK301/pYK3021 up to a concentration of 800 μM TCE suggests that general TCE effects are limited as well.
AEK301/pYK3021 is an ideal candidate for in situ remediation studies on the basis of the following attributes. (i) It is able to degrade significant quantities of TCE at relatively high and sustained rates in the absence of aromatic induction, (ii) its sensitivity to TCE-mediated toxicity and metabolite toxicity is limited, and (iii) high concentrations of TCE appear to be well tolerated by the system. Bench-scale studies involving a continuous culture and substrate are in progress.
REFERENCES
- 1.Ampe F, Léonard D, Lindley N D. Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl Environ Microbiol. 1998;64:1–6. doi: 10.1128/aem.64.1.1-6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne A M, Kukor J J, Olsen R H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;154:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 3.Ensley B D. Biochemical diversity of trichloroethylene metabolism. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- 4.Folsom B R, Chapman P J. Performance characterization of a model bioreactor for the biodegradation of trichloroethylene by Pseudomonas cepacia G4. Appl Environ Microbiol. 1991;57:1602–1608. doi: 10.1128/aem.57.6.1602-1608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folsom B R, Chapman P J, Pritchard P H. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interactions between substrates. Appl Environ Microbiol. 1990;56:1279–1285. doi: 10.1128/aem.56.5.1279-1285.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gossett J M. Measurement of Henry’s law constants for C1 and C2 chlorinated hydrocarbons. Environ Sci Technol. 1987;21:202–208. [Google Scholar]
- 7.Harker A R, Kim Y. Trichloroethylene degradation by two independent aromatic-degrading pathways in Alcaligenes eutrophus JMP134. Appl Environ Microbiol. 1990;56:1179–1181. doi: 10.1128/aem.56.4.1179-1181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harker A R, Olsen R H, Seidler R J. Phenoxyacetic acid degradation by the 2,4-dichlorophenoxyacetic acid (TFD) pathway of plasmid pJP4: mapping and characterization of the TFD regulatory gene, tfdR. J Bacteriol. 1989;171:314–320. doi: 10.1128/jb.171.1.314-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heald S, Jenkins R O. Trichloroethylene removal and oxidation toxicity mediated by toluene dioxygenase of Pseudomonas putida. Appl Environ Microbiol. 1994;60:4634–4637. doi: 10.1128/aem.60.12.4634-4637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Ayoubi P, Harker A R. Constitutive expression of the cloned phenol hydroxylase gene(s) from Alcaligenes eutrophus JMP134 and concomitant trichloroethylene oxidation. Appl Environ Microbiol. 1996;62:3227–3233. doi: 10.1128/aem.62.9.3227-3233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y. A recombinant approach to the isolation and characterization of a primary degrader of trichloroethylene. Ph.D. dissertation. Stillwater: Oklahoma State University; 1993. [Google Scholar]
- 12.Landa A S, Sipkema E M, Weijma J, Beenackers A A C M, Dolfing J, Janssen D B. Cometabolic degradation of trichloroethylene by Pseudomonas cepacia G4 in a chemostat with toluene as the primary substrate. Appl Environ Microbiol. 1994;60:3368–3374. doi: 10.1128/aem.60.9.3368-3374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 14.Mars A E, Houwing J, Dolfing J, Janssen D B. Degradation of toluene and trichloroethylene by Burkholderia cepacia G4 in growth-limited fed-batch culture. Appl Environ Microbiol. 1996;62:886–891. doi: 10.1128/aem.62.3.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman L M, Wackett L P. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J Bacteriol. 1997;179:90–96. doi: 10.1128/jb.179.1.90-96.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oldenhuis R, Oedzes J Y, van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oldenhuis R, Vink R L J M, Janssen D B, Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989;55:2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen R H, Hansen J. Evolution and utility of a Pseudomonas aeruginosa drug resistance factor. J Bacteriol. 1976;125:837–844. doi: 10.1128/jb.125.3.837-844.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajagopal R. Conceptual design for a groundwater quality monitoring strategy. Environ Prof. 1986;8:244–264. [Google Scholar]
- 21.Stanier R Y, Palleroni N, Doudorff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 22.Wackett L P, Gibson D T. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wackett L P, Householder S R. Toxicity of trichloroethylene to Pseudomonas putida F1 is mediated by toluene dioxygenase. Appl Environ Microbiol. 1989;55:2723–2725. doi: 10.1128/aem.55.10.2723-2725.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]