Abstract
Crimean-Congo Hemorrhagic Fever (CCHF) is a viral disease that can infect humans via contact with tick vectors or livestock reservoirs and can cause moderate to severe disease. The first human case of CCHF in Uganda was identified in 2013. To determine the geographic distribution of the CCHF virus (CCHFV), serosampling among herds of livestock was conducted in 28 Uganda districts in 2017. A geostatistical model of CCHF seroprevalence among livestock was developed to incorporate environmental and anthropogenic variables associated with elevated CCHF seroprevalence to predict CCHF seroprevalence on a map of Uganda and estimate the probability that CCHF seroprevalence exceeded 30% at each prediction location. Environmental and anthropogenic variables were also analyzed in separate models to determine the spatially varying drivers of prediction and determine which covariate class resulted in best prediction certainty. Covariates used in the full model included distance to the nearest croplands, average annual change in night-time light index, percent sand soil content, land surface temperature, and enhanced vegetation index. Elevated CCHF seroprevalence occurred in patches throughout the country, being highest in northern Uganda. Environmental covariates drove predicted seroprevalence in the full model more than anthropogenic covariates. Combination of environmental and anthropogenic variables resulted in the best prediction certainty. An understanding of the spatial distribution of CCHF across Uganda and the variables that drove predictions can be used to prioritize specific locations and activities to reduce the risk of future CCHF transmission.
Keywords: Crimean-Congo hemorrhagic fever, Geostatistical model, Seroprevalence prediction, Epidemiology, Livestock
Highlights
-
•
CCHF identified in Uganda in 2013.
-
•
Geostatistical model of CCHF seroprevalence among livestock in Uganda.
-
•
Predictions of seroprevalence made across country.
-
•
Environmental variables drove predictions more than anthropogenic ones.
1. Introduction
Crimean-Congo Hemorrhagic Fever (CCHF) is a zoonotic disease of humans and domesticated and wild animal species. Ixodid ticks, especially those from the genus Hyalomma, are considered the primary reservoirs of CCHF virus (CCHFV) and act as vectors to transmit the virus to susceptible individuals [1]. Humans can become infected with CCHFV through contact with animal body fluids and tissue, and seroprevalence studies have found livestock handlers, abattoir workers, butchers, and others who live or work in close contact with livestock or animal derived products to be at highest risk of infection [2,3]. Human-to-human transmission can also occur via contact with infectious blood or body fluids of an infected individual [4]. Animal hosts typically remain asymptomatic upon infection, but humans can develop severe symptoms such as high fever and uncontrolled bleeding. Human case fatality rates from past outbreaks have ranged from 3 to 30% [5].
The geographic distribution of CCHFV encompasses countries across the African, Asian, and European continents [1,6]. Habitat suitability for CCHFV varies by tick species, some viable CCHFV tick vectors being most common in wooded or forested areas while others thrive in arid climates. Land conversion for agricultural purposes has also been shown to create suitable tick habitat and result in increased CCHF prevalence [1,7,8]. Ticks generally prefer geographies with well-drained sandy soils and vegetation that provides protection from extreme weather [9]. CCHFV is maintained within tick populations and in an enzootic cycle between ticks and vertebrate hosts. Within tick populations, CCHFV survives both transovarially (to offspring) and transstadially (through life stages) [1]. Ticks can seek blood meals from mobile vertebrates for extended periods of time, leading to their potential dispersal across borders and continents and between susceptible populations [1,[10], [11], [12]].
The first ever human CCHF case in Uganda was reported in 2013, and subsequent cases have been identified throughout the country, most in Uganda's “Cattle Corridor” [13,14]. Recently, an outbreak of two CCHF cases was investigated in a refugee camp in western Uganda in April 2021 where sero-epidemiological investigations found evidence of antibodies against CCHFV in 71% of goats sampled within the camp [15].
Aside from recent outbreak investigations, the geographic distribution of CCHF in Uganda remains unknown. We conducted serosampling among livestock (cattle, sheep, goats) herds in 2017 and fit a geostatistical model to estimate the distribution of CCHF on a map of Uganda. This model was then divided into two partial models representing environmental and anthropogenic predictors of CCHF seroprevalence, and subsequent predictions from partial models were compared to determine possible spatial heterogeneity in drivers of CCHF seroprevalence.
2. Methods
2.1. Ethical statement
The Uganda Virus Research Institute (UVRI) in collaboration with the Viral Special Pathogens Branch at the Centers for Disease Control and Prevention received permission from the Uganda Ministry of Health to gather blood samples from livestock to prevent future morbidity and mortality from RVF. Human subjects approval for this study was granted from review by the UVRI Research Ethics Committee (UVRI REC: GC/127/16/03/551). Animal subjects work was conducted according to Uganda national guidelines and performed by officers from the Ministry of Agriculture, Animal Industries and Fisheries.
2.2. Livestock sampling data
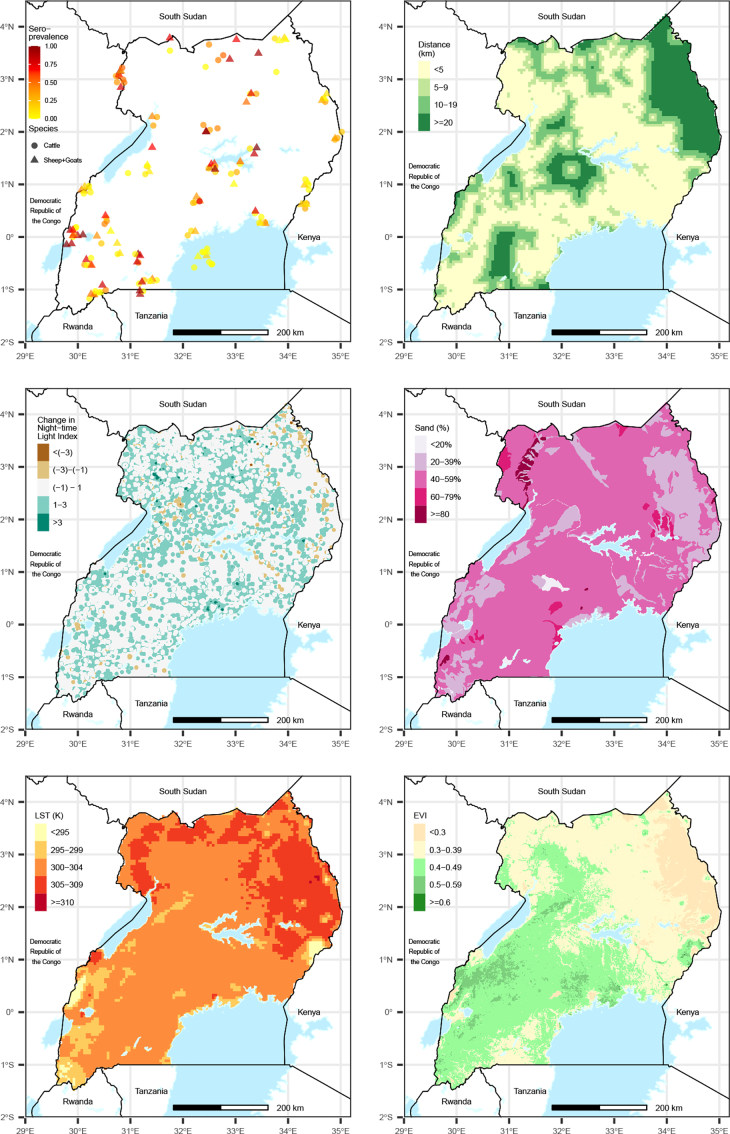
Blood samples were collected from 3181 animals selected from herds at 112 sites in 28 Uganda districts between February–August 2017 (Fig. 1). Approximately half of animals sampled were cattle (54.4%), while 34.3% were goats and 11.3% were sheep. Sampling locations targeted the various geographies within the country and border districts where virus importation could occur. For each herd represented, a random sample of animals was selected from which to collect a blood sample for serological testing.
Fig. 1.
Observed herd-level seroprevalence of Crimean-Congo Hemorrhagic Fever (CCHF) among Ugandan livestock in 2017 and spatial distributions of predictor variables.
2.3. Spatial predictor data
Given the complex transmission cycle of CCHF between ticks, humans, and vertebrates, we evaluated the relationship between CCHF seroprevalence and a wide range of variables to identify those with the strongest predictive ability. Covariate raster data were obtained on a 1km2 grid of Uganda and values were extracted at sampling locations for analysis. Covariate data unavailable at a resolution of 1km2 were resampled using bilinear interpolation to match this resolution. We assumed that the average lifespan of livestock in Uganda was 8 years (personal communication, FAO Uganda), therefore annual values of each variable were averaged over the 8 years prior to sampling (2009–2016).
Potential model covariates were classified into the following domains describing unique ecological processes associated with CCHF transmission: bioclimatic, geologic, land cover, and population density of humans and animals. Variables within each domain were evaluated to identify those with the best fit to sampling data. We sought to represent each domain within the final model to encompass each ecological system associated with CCHF transmission. Evaluated covariates under each domain are listed in Supplemental Table 1.
2.4. Statistical analysis
Initial data analysis explored the relationship between logit-transformed CCHF seroprevalence with each covariate in a binomial generalized linear model (GLM) to identify those with positive or negative relationships with logit-CCHF seroprevalence. Covariates associated with CCHF seroprevalence in a non-spatial GLM (α = 0.05) were then analyzed using a Generalized Linear Geostatistical Model (GLGM) to determine whether the relationship remained after accounting for the spatial structure of the sampling data. We fit the GLGM using Model-based Geostatistics [16,17] in the R package PrevMap [18]. A multivariate GLGM model was developed in a step-wise process, first retaining the covariate resulting in the best model fit, and subsequently adding the next variable resulting in the best model fit until each ecological domain was represented and the best fitting model was identified. Covariates included in the model were tested for collinearity, and a correlation coefficient of 0.6 was used as a cutoff, in which case the variable resulting in the better model fit was retained. Model overfitting was evaluated using a correlation matrix of the parameter maximum likelihood estimates, and we used a cutoff value of 0.6 to determine the presence of linear dependence between any covariates. The best fitting model was identified as the one which reduced uncertainty most in the cumulative output probabilities of exceeding 30% CCHF seroprevalence in each prediction location, as described in Giorgi et al. [19]. The final model included six covariates: animal species (SPECIES), distance to cropland (DIST CROP), average annual change in a night-time light index (NTL CHANGE), percent sand soil content (% SAND), land surface temperature (LST), mean annual enhanced vegetation index (EVI), which is a satellite-derived index measure of vegetation “greenness” [12,[20], [21], [22], [23]]. Because sheep and goats are typically maintained under similar grazing and movement patterns, they were combined for analysis. The model of the probability that a given sampled animal had positive antibodies for CCHF was implemented as follows:
where p(x) represents CCHF seroprevalence in location x, β represents the effects of each covariate at a given prediction location, S(x) represents a spatial random effect following a Gaussian process with mean zero and variance σ2 and a Matern correlation function. Model covariates were considered statistically significant using α = 0.05. The “nugget effect” Ui represents the unstructured random variation in the outcome and is included to capture the effects of unmeasured explanatory variables unassociated with spatial structure in the data. The shape parameter of the Matern correlation function was specified as k = 1, based on the profile likelihood for the shape parameter in a logit-transformed linear Gaussian model. Covariance parameters were determined using a semi-variogram of model residuals.
The final binomial GLGM was fit using Markov Chain Maximum Likelihood (MCML), using 200,000 simulations to calculate final model coefficients and covariance parameters with PrevMap [18] in R 1.3.1075 version [24]. Model parameters were then fit to covariate values on a 1km2 resolution grid of Uganda to predict CCHF seroprevalence on a map of Uganda. Maps were created separately for sheep and goats compared to cattle given differences in seroprevalence and animal management practices, and were generated using the ggplot2 package in R 1.3.1075 version [24,25]. Prediction uncertainty was quantified by generating a map of the probability that CCHF seroprevalence exceeded 30% in each prediction location. We used 30% as a cutoff because overall seroprevalence was approximately 30%, therefore allowing us to determine the probability that seroprevalence was above average. Locations with a high (>0.95), moderately high (0.7), moderately low (0.3), or low (<0.05) probability are those at which the model reduces error sufficiently to determine with relative confidence whether CCHF truly exceeds 30%. Conversely, locations with a probability 0.3–0.7 are those at which there is inadequate sampling and/or covariate data to confidently determine whether seroprevalence exceeds 30%.
The full model was divided into two partial models; one including only environmental covariates (percent sand soil content, land surface temperature, EVI) and the other one including only anthropogenic covariates (distance to croplands, average annual change in night-time light index). Both partial models (environmental and anthropogenic) were fit using MCML as previously outlined, and individual maps of predicted seroprevalence and probability that seroprevalence exceeded 30% were also generated. An intercept-only model was also run to estimate CCHF seroprevalence without using covariate information and used to estimate CCHF seroprevalence on a map of Uganda. Because it was necessary to make predictions separately for each species category (cattle, sheep/goats), we averaged the predictions to get a single prediction of livestock seroprevalence for each model, which required the assumption of equal distribution of livestock species across the country. We used two methods to compare the models as outlined by Giorgi et al. [19]. First, we used the root-mean-integrated-square-error (RMISE) criterion to determine which partial model resulted in greater similarity in predictions to full model; the model with a RMISE value closer to zero is that which produces predictions closer to those from the full model. Second, we calculated a certainty index for each model to determine that which produced exceedance probabilities further from 0.5; a probability of 0.5 indicates complete uncertainty in determining whether CCHF seroprevalence exceeds 30% at a given prediction location. Certainty index values further from zero result from models that produce exceedance probabilities closer to 0 or 1, while values closer to zero result from models that produce exceedance probabilities closer to 0.5. Using RMISE and the certainty index in combination identified which covariate class (environmental or anthropogenic) drives predictions in the full model, and which improves certainty in predictions.
2.5. Model validation
We simulated 1000 semi-variograms using Monte Carlo iterations given the full model to determine the validity and fit of the spatial correlation structure to the data. A 95% interval defining the variability of the model simulations was used to evaluate whether the model accurately fit the data. After simulation, the fitted semi-variogram from the observed data fell within the 95% interval of variograms estimated from the simulated data, and it was determined that our specified correlation structure adequately fit the spatial structure in our data to proceed with prediction [17].
3. Results
3.1. CCHF seroprevalence model and prediction map
Overall CCHF seroprevalence was 31.4%, being 16.9% among cattle, 48.7% among goats, and 49.2% among sheep. There were 84 herds of cattle sampled, among whom CCHF seroprevalence ranged from 0 to 75%, and there were 75 herds of sheep and goats sampled among whom CCHF seroprevalence ranged from 0 to 100%.
Output from the full model showed that the odds of CCHF seropositivity among sheep and goats was 2.16 times the odds among cattle (95% CI: 1.68–2.64). An inverse association was found between CCHF seroprevalence and distance in kilometers to the nearest croplands, where seroprevalence increased as distance decreased (Beta: −0.03; p = 0.01) Seropositivity tended to increase in locations that had an average annual increase in night-time light indices, however, this relationship was not statistically significant based on a 95% confidence limit (Beta: 0.06, p = 0.22). CCHF seroprevalence was higher in locations that had higher average land surface temperature (p < 0.01) and in drier environments that had lower EVI values (p = 0.03). Seroprevalence tended to be higher in locations with higher proportions of sand soil content, but this relationship was not considered statistically significant (p = 0.07) (Table 1). Compared to the full model, parameter estimates from the partial models remained relatively stable in magnitude and direction of association and are shown in Supplemental Tables 2–3.
Table 1.
Generalized Linear Geostatistical Model parameter estimates.
| Parameter | Coefficient | Standard Error | 0.025 CI |
0.975 CI |
P-value |
|---|---|---|---|---|---|
| Intercept | −66.80 | 23.90 | −113.64 | −19.96 | <0.01 |
| Sheep/Goat Species (Ref: Cattle) | 2.16 | 0.25 | 1.68 | 2.64 | <0.01 |
| Change in Night-time Light Index | 0.06 | 0.05 | −0.04 | 0.16 | 0.22 |
| Distance to Croplands (km) | −0.03 | 0.01 | −0.05 | −0.01 | 0.01 |
| Sand (% Soil Content) | 2.48 | 1.37 | −0.20 | 5.16 | 0.07 |
| Land Surface Temperature | 0.22 | 0.08 | 0.06 | 0.37 | <0.01 |
| EVI | −4.56 | 2.14 | −8.75 | −0.36 | 0.03 |
| log(sigma^2)† | −0.34 | 0.45 | −1.22 | 0.54 | – |
| log(phi)†† | 1.65 | 0.57 | −2.77 | −0.54 | – |
| log(tau^2) ††† | 0.3 | 0.98 | −1.62 | 2.22 | – |
Abbreviations: km = kilometers.
Variance of the Gaussian process.
Scale of the spatial correlation.
Variance of the nugget effect.
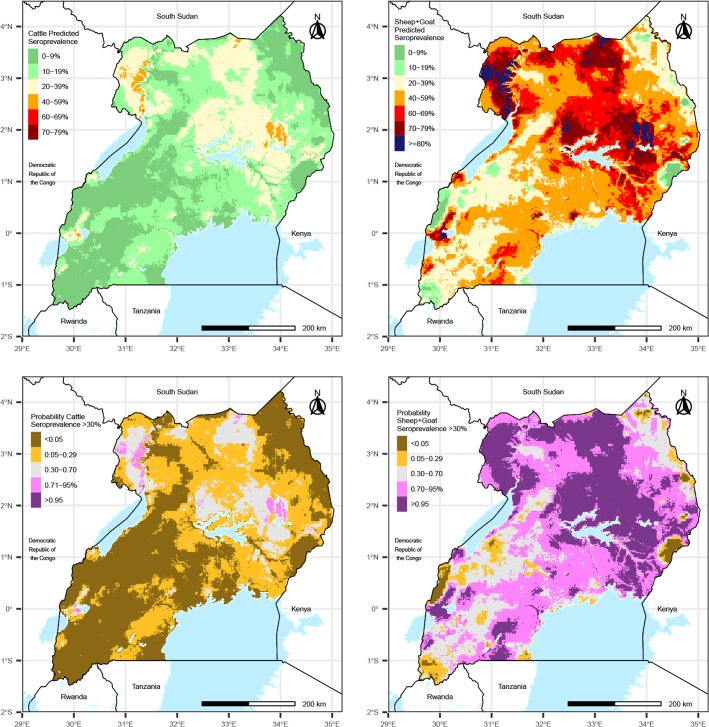
Predicted CCHF seroprevalence among sheep and goats ranged from 0 to 93% and was considerably higher throughout Uganda compared to predicted seroprevalence among cattle, which ranged from 0 to 64%. Predictions were highest in northern Uganda, especially near the northwestern Nile River and in locations with very sandy soil (Fig. 1, Fig. 2). Patches of locations with high predictions also occurred surrounding Lake Kwania and Lake Kyoga, near the border of South Sudan, along the border of Kenya, and around the Queen Elizabeth National Park in Southwestern Uganda. Comparing the model-predicted CCHF seroprevalence to the observed herd seroprevalence showed overall strong correlation between the values, where the greatest differences in predicted and observed seroprevalence occurred in locations where the sample size was low (Supplemental Fig. 1). Maps of the probability that CCHF seroprevalence exceeds 30% show stark differences between cattle and sheep and goats. Among cattle, <1% of the country had a probability >0.95 that CCHF seroprevalence exceeded 30%, and only 1% of the country had a probability >0.7 that CCHF seroprevalence exceeded 30%. In contrast, among sheep and goats, 29% of the country had a probability >0.95 that CCHF seroprevalence exceeded 30%, and 63% of the country had a probability >0.7 that CCHF seroprevalence exceeded 30% (Fig. 2).
Fig. 2.
Predicted CCHF seroprevalence among livestock across Uganda in the sampling year 2017 (top) and probability that CCHF seroprevalence exceeds 30% (bottom) among cattle (left) and sheep and goats (right).
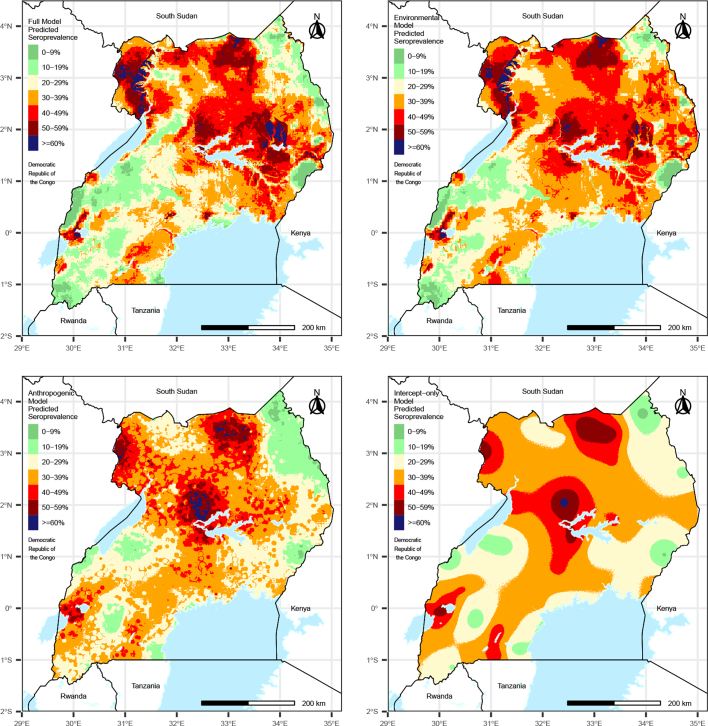
3.2. Environmental vs. anthropogenic drivers of transmission
The species-averaged prediction maps resulting from each of the partial models show varying spatial trends in predictions and suggest that predictions in the full model were driven by environmental variables more than anthropogenic variables, given that the predictions from the environmental partial model are more similar to those from full model compared to the anthropogenic and intercept-only models (Fig. 3). This was confirmed by comparison of RMISE values across the partial models, where the lowest RMISE values resulted from the environmental partial model, followed by the anthropogenic partial model, and then the intercept-only model, suggesting that predictions in the environmental model were most similar to the predictions from the full model (Table 2). RMISE values for the anthropogenic model were only marginally better than the intercept-only model, suggesting that these variables had little influence on the predictions in the full model. Certainty indices for each model showed that the full model resulted in the greatest certainty in predictions (values further from 0), followed by the environmental model, the anthropogenic model, and then the intercept-only model (Table 2). Therefore, we conclude that both environmental and anthropogenic variables should be accounted for in predicting CCHF seroprevalence, although environmental variables were more important for improving model fit and reducing error.
Fig. 3.
Species-averaged predicted CCHF seroprevalence among livestock across Uganda in 2017 resulting from the full model (top left), and partial environmental (top right) and anthropogenic (bottom left) sub models, and an intercept-only model (bottom right) that ignored covariate information.
Table 2.
Comparison of prediction accuracy and certainty of the full, partial (anthropogenic and environmental), and intercept models using RMISE and certainty index.
| Intercept-only Model | Environmental Model | Anthropogenic Model | Full Model | |
|---|---|---|---|---|
| RMISE: Cattle Predictions | 40.94 | 18.66 | 39.00 | – |
| RMISE: Sheep/Goat Predictions | 73.87 | 36.06 | 70.49 | – |
| Certainty Index: Cattle Exceedance probability | −33,723.35 | −36,917.2 | −35,447.23 | −41,926.64 |
| Certainty Index: Sheep/Goat Exceedance probability | −22,160.49 | −33,249.7 | −23,670.64 | −34,518.36 |
RMISE values closer to zero indicate better prediction accuracy (as compared to the full model). Certainty index values more distant from zero suggest greater certainty in determining exceedance of 30% seroprevalence.
Abbreviations: RMISE = Root mean integrated square error.
4. Discussion
Using a geostatistical model of CCHF seroprevalence among livestock herds in Uganda, we estimate CCHF livestock seroprevalence throughout Uganda among cattle, sheep, and goats and the probability that seroprevalence exceeds the country-wide average of approximately 30%. Variables resulting in the model with the best statistical fit to the sampling data included distance to croplands, average annual change in night-time light indices, percent sand soil composition, land surface temperature, and EVI. By dividing the full model into partial models comprised of anthropogenic and environmental variables, we found that environmental variables drove seroprevalence predictions more than anthropogenic variables and resulted in greater certainty in determining whether CCHF seroprevalence exceeded 30%.
Our CCHF serological sampling results differed greatly by livestock species, where approximately 17% of cattle were CCHFV-seropositive, and almost 50% of sheep and goats were seropositive. A global review of CCHF seropositivity among domestic and wild animals similarly found that seroprevalence tended to be higher among sheep (23.85%) and goats (28.07%) compared to cattle (19.33%), however, the magnitude of the difference across species was notable [26]. Differences in animal handling practices could be a potential reason for higher levels of seroprevalence among smaller ungulates compared to large ungulates. For example, grazing patterns may vary by livestock species, where sheep and goats may be tethered or paddocked within smaller areas, whereas cattle may roam across larger areas or migrate annually according to rain patterns, which could lead to different levels of contact with ticks carrying CCHFV [12]. Similarly, treatment of livestock with insecticide products to prevent tick infestation may vary by animal species based on perceived economic value or access to insecticide treatments [27]. Given that seroprevalence was considerably lower among cattle, additional information should seek to determine whether cattle are prioritized for treatment with insecticides compared to smaller ungulates. If cattle tend to be prioritized for insect control measures, future public health efforts to reduce the risk of human infections with CCHFV could target improved hygiene and vector control among smaller ungulates that may have higher likelihood of being infected with CCHFV.
We found a statistically significant association between CCHF seroprevalence and proximity to agricultural land. These results are intuitive given that agricultural activities often occur in the same locations where animal husbandry is practiced, suggesting that proximity to croplands could act as a proxy indicator for high levels of livestock activity and congregation such as was found by a study in Greece where introduction of livestock and land conversion for agricultural purposes resulted in an increase in CCHF outbreaks [28]. Previous studies have also shown that abandoned agricultural lands were more conducive to ticks compared to actively cultivated lands, suggesting the potential for increased tick activity in areas that farmers allow their livestock to roam outside of actively cultivated fields [29]. We considered croplands as from MODIS land cover classes 12 and 14, which are defined as raster pixels where at least 60% of area is cultivated cropland (class 12) and mosaics of small-scale cultivation 40–60% with natural tree, shrub, or herbaceous vegetation (class 14). While such environments provide suitable habitat for ticks, agriculture is also commonly associated with management of livestock, shepherding, and pastoral activities, which can create opportunities for increased contact between humans, livestock, and ticks.
Measures of night-time light obtained from satellite imagery can be used to measure shifts in human population density, and increases in average annual night-time light measures could indicate increases in development and human population sizes [30]. While changes in average annual night-time light index values did not result in a statistically significant relationship in the full model, inclusion of this variable resulted in an improved model fit and could therefore represent some underlying relationship between changes in human population development and CCHF transmission activity. However, the mechanism of this relationship is unclear, as increased human presence has been shown to impact many ecological conditions that may make importation and transmission of infectious diseases more likely [31,32]. We analyzed data on human and livestock population density and changes in human population density and found these variables to be weaker predictors of CCHF seroprevalence compared to the night-time light index. Importation of CCHFV through livestock trade has been reported extensively and could be an explanation for high levels of CCHF seroprevalence in Uganda, especially given that CCHF only recently emerged in Uganda [5]. Previous research on livestock movement in Uganda has identified locations in the country with high levels of animal movement and congregation such as abattoirs, animal markets, border crossings, and communal grazing areas, however, analysis of proximity to these locations showed weak relationships with CCHF seroprevalence among livestock sampled in this study [12]. Future research would benefit from data on the origin and number of animals imported into districts of Uganda.
Stratification of environmental and anthropogenic variables into partial models showed that environmental variables (EVI, LST, and sand soil composition) both drove the predicted seroprevalence in the full model more than anthropogenic variables and resulted in greater certainty in predictions, but the best predictive certainty resulted in the combination of environmental and anthropogenic variables. According to these results, disease prevention or surveillance strategies such as vector control may be most effective when carried out in warmer and dryer areas with sandy soil conducive to ticks that is in close proximity to agricultural areas, such as those highlighted in the maps in Fig. 2. However, while environmental variables in our model drove predictions more than anthropogenic variables, predictors of CCHF seroprevalence could have been missed and therefore we cannot assume that anthropogenic variables do not play an important role in the ecology of CCHF in Uganda. As previously discussed, we were unable to include information on livestock movement, trade, and importation, which could play an important role in CCHFV ecology in Uganda given that is first identified in Uganda relatively recently in 2013. Likewise, measurement error in covariates evaluated in our model selection process could result in exclusion of variables that are important to the true transmission process of CCHF. Therefore care should be taken in the interpretation in the drivers of CCHF seroprevalence given that important considerations could be missing from this analysis or covariates we included could be correlated with external variables not evaluated here.
This analysis had several limitations. First, selection of sampling locations may have been biased toward high-animal traffic areas, which could be more conducive to transmission of CCHFV. This model represented a single time point of seroprevalence sampling and was unable to account for temporal dynamics of CCHF transmission or the vast complexity of the CCHF transmission cycle, which includes numerous species tick and animals with differing feeding habits, geographic ranges, and environmental niches. This was the first of several planned longitudinal sampling efforts planned to be carried out among livestock herds in these sampling locations, and sampling results at additional time points will be reported separately. While we could not account for all possible modes of CCHFV survival or transmission, variables retained in the model after the model selection process represent conditions optimal for tick survival and viral transmission among susceptible animal hosts, and adequately reduced error in the model to confidently determine in most prediction locations whether CCHF seroprevalence greater or <30%.
5. Conclusion
This analysis provides predictions of CCHF seroprevalence in 2017 using serosampling data from livestock across 28 districts in Uganda. Predicted CCHF-seroprevalence occurred in patches throughout the country; the highest predicted seroprevalence occurring in the northern half of the country. Covariates resulting in the best statistical fit to sampling data included distance to croplands, average annual change in night-time light index, percent sand soil content, land surface temperature, and EVI. By separating the full model into partial models of anthropogenic and environmental covariates, we found that environmental covariates drove predictions in the full model and resulted in greater certainty in predictions compared to anthropogenic covariates. These results can guide future public health prevention efforts to specific locations where there may be higher likelihood of CCHF circulation, especially near agricultural areas that may attract ticks and result in higher levels of contact between ticks, livestock, and humans.
6. Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or any institutions with which the authors are affiliated.
Declaration of Competing Interest
Spatial prediction of Crimean Congo hemorrhagic fever virus seroprevalence among livestock in Uganda.
The authors report no conflicts of interest in publishing this research manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2023.100576.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- 1.Hoogstraal H. Review Article1: the epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa23. J. Med. Entomol. 1979 May 22;15(4):307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- 2.Akuffo R., Brandful J.A.M., Zayed A., Adjei A., Watany N., Fahmy N.T., et al. Crimean-Congo hemorrhagic fever virus in livestock ticks and animal handler seroprevalence at an abattoir in Ghana. BMC Infect. Dis. 2016 Jul 8;16(1):324. doi: 10.1186/s12879-016-1660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zohaib A., Saqib M., Athar M.A., Hussain M.H., Sial A. Ur R., Tayyab M.H., et al. Crimean-Congo hemorrhagic fever virus in humans and livestock, Pakistan, 2015–2017. Emerg. Infect. Dis. 2020 Apr;26(4):773–777. doi: 10.3201/eid2604.191154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celikbas A.K., Dokuzoğuz B., Baykam N., Gok S.E., Eroğlu M.N., Midilli K., et al. Crimean-Congo hemorrhagic fever among health care workers, Turkey. Emerg. Infect. Dis. 2014 Mar;20(3):477–479. doi: 10.3201/eid2003.131353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spengler J.R., Bergeron É., Spiropoulou C.F. Crimean-Congo hemorrhagic fever and expansion from endemic regions. Curr. Opin. Virol. 2019 Feb 1;34:70–78. doi: 10.1016/j.coviro.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ergönül Ö. Crimean-Congo haemorrhagic fever. Lancet Infect. Dis. 2006 Apr 1;6(4):203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estrada-Peña A., Vatansever Z., Gargili A., Ergönul Ö. The trend towards habitat fragmentation is the key factor driving the spread of Crimean-Congo haemorrhagic fever. Epidemiol. Infect. 2010 Aug;138(8):1194–1203. doi: 10.1017/S0950268809991026. [DOI] [PubMed] [Google Scholar]
- 8.Jameson L.J., Ramadani N., Medlock J.M. Possible drivers of Crimean-Congo hemorrhagic fever virus transmission in Kosova. Vector-Borne Zoonotic Dis. 2012 Sep 1;12(9):753–757. doi: 10.1089/vbz.2011.0773. [DOI] [PubMed] [Google Scholar]
- 9.Burtis J.C., Yavitt J.B., Fahey T.J., Ostfeld R.S. Ticks as soil-dwelling arthropods: an intersection between disease and soil ecology. J. Med. Entomol. 2019;56(6):1555–1564. doi: 10.1093/jme/tjz116. [DOI] [PubMed] [Google Scholar]
- 10.Jameson L.J., Morgan P.J., Medlock J.M., Watola G., Vaux A.G.C. Importation of Hyalomma marginatum, vector of Crimean-Congo haemorrhagic fever virus, into the United Kingdom by migratory birds. Ticks Tick-Borne Dis. 2012 Apr 1;3(2):95–99. doi: 10.1016/j.ttbdis.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Palomar A.M., Portillo A., Santibáñez P., Mazuelas D., Arizaga J., Crespo A., et al. Crimean-Congo hemorrhagic fever virus in ticks from migratory birds, Morocco1. Emerg. Infect. Dis. 2013 Feb;19(2):260–263. doi: 10.3201/eid1902.121193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medley A.M., Gasanani J., Nyolimati C.A., McIntyre E., Ward S., Okuyo B., et al. Preventing the cross-border spread of zoonotic diseases: multisectoral community engagement to characterize animal mobility—Uganda, 2020. Zoonoses Public Health. 2021;68(7):747–759. doi: 10.1111/zph.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balinandi S., Patel K., Ojwang J., Kyondo J., Mulei S., Tumusiime A., et al. Investigation of an isolated case of human Crimean–Congo hemorrhagic fever in Central Uganda, 2015. Int. J. Infect. Dis. 2018;68:88–93. doi: 10.1016/j.ijid.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kizito S., Okello P.E., Kwesiga B., Nyakarahuka L., Balinandi S., Mulei S., et al. Notes from the field: Crimean-Congo hemorrhagic fever outbreak—Central Uganda, august–september 2017. Morb. Mortal. Wkly Rep. 2018;67(22):646. doi: 10.15585/mmwr.mm6722a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyakarahuka L., Whitmer S., Kyondo J., Mulei S., Cossaboom C.M., Telford C.T., et al. Crimean-Congo hemorrhagic fever outbreak in refugee settlement during COVID-19 pandemic, Uganda, April 2021. Emerg. Infect. Dis. 2022;28(11):2326–2329. doi: 10.3201/eid2811.220365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diggle P.J., Tawn J.A., Moyeed R.A. Model-based geostatistics. J. R. Stat. Soc.: Ser. C: Appl. Stat. 1998;47(3):299–350. [Google Scholar]
- 17.Diggle P.J., Giorgi E. CRC Press; 2019. Model-Based Geostatistics for Global Public Health: Methods and Applications. [Google Scholar]
- 18.Giorgi E., Diggle P.J. PrevMap: an R package for prevalence mapping. J. Stat. Softw. 2017;78(8):1–29. [Google Scholar]
- 19.Giorgi E., Fronterrè C., Macharia P.M., Alegana V.A., Snow R.W., Diggle P.J. Model building and assessment of the impact of covariates for disease prevalence mapping in low-resource settings: to explain and to predict. J. R. Soc. Interface. 2021;18(179):20210104. doi: 10.1098/rsif.2021.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedl M., Sulla-Menashe D. 006. 2015. MCD12C1 MODIS/Terra+ Aqua Land Cover Type Yearly L3 Global 0.05 Deg CMG V006, NASA EOSDIS Land Processes DAAC. Accessed 2019-12-11 from. [DOI] [Google Scholar]
- 21.Nachtergaele F., van Velthuizen H., Verelst L., Batjes N.H., Dijkshoorn K., van Engelen V.W.P., et al. Proceedings of the 19th World Congress of Soil Science, Soil Solutions for a Changing World, Brisbane, Australia, 1–6 August 2010. 2010. The harmonized world soil database; pp. 34–37. [Google Scholar]
- 22.McNally A., Arsenault K., Kumar S., Shukla S., Peterson P., Wang S., et al. A land data assimilation system for sub-Saharan Africa food and water security applications. Sci. Data. 2017;4(1):1–19. doi: 10.1038/sdata.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didan K. NASA EOSDIS Land Process DAAC. 10(1) 2015. MOD13Q1 MODIS/Terra vegetation indices 16-day L3 global 250m SIN grid V006. [Google Scholar]
- 24.Team R.C. 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 25.Wickham H., Chang W. ggplot2: An Implementation of the Grammar of Graphics. R Package Version 07. https://CRANR-ProjOrgpackageGgplot2
- 26.Spengler J.R., Bergeron É., Rollin P.E. Seroepidemiological studies of Crimean-Congo hemorrhagic fever virus in domestic and wild animals. PLoS Negl. Trop. Dis. 2016;10(1) doi: 10.1371/journal.pntd.0004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Castillo P., Rist C., Rabinovich R., Chaccour C. Insecticide-treated livestock: a potential one health approach to malaria control in Africa. Trends Parasitol. 2022;38(2):112–123. doi: 10.1016/j.pt.2021.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Sargianou M., Panos G., Tsatsaris A., Gogos C., Papa A. Crimean-Congo hemorrhagic fever: seroprevalence and risk factors among humans in Achaia, western Greece. Int. J. Infect. Dis. 2013;17(12):e1160–e1165. doi: 10.1016/j.ijid.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 29.Mierzejewska E.J., Alsarraf M., Behnke J.M., Bajer A. The effect of changes in agricultural practices on the density of Dermacentor reticulatus ticks. Vet. Parasitol. 2015;211(3–4):259–265. doi: 10.1016/j.vetpar.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Bharti N., Tatem A.J., Ferrari M.J., Grais R.F., Djibo A., Grenfell B.T. Explaining seasonal fluctuations of measles in Niger using nighttime lights imagery. Science. 2011;334(6061):1424–1427. doi: 10.1126/science.1210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindblade K.A., Walker E.D., Onapa A.W., Katungu J., Wilson M.L. Land use change alters malaria transmission parameters by modifying temperature in a highland area of Uganda. Tropical Med. Int. Health. 2000;5(4):263–274. doi: 10.1046/j.1365-3156.2000.00551.x. [DOI] [PubMed] [Google Scholar]
- 32.Patz J.A., Daszak P., Tabor G.M., Aguirre A.A., Pearl M., Epstein J., et al. Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 2004;112(10):1092–1098. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.