Abstract
Malaria is a severe public health problem in several developing tropical and subtropical countries. Anopheles aquasalis is the primary coastal malaria vector in Central and South America and the Caribbean Islands, and it has the peculiar feature of living in water with large changes in salinity. Recent research has recognised An. aquasalis as an important model for studying the interactions of murine and human Plasmodium parasites. This study presents the complete genome of An. aquasalis and offers insights into its evolution and physiology. The genome is similar in size and gene content to other Neotropical anophelines, with 162 Mb and 12,446 protein-coding genes. There are 1387 single-copy orthologs at the Diptera level (eg. An. gambiae, An. darlingi and Drosophila melanogaster). An. aquasalis diverged from An. darlingi, the primary malaria vector in inland South America, nearly 20 million years ago. Proteins related to ion transport and metabolism belong to the most abundant gene families with 660 genes. We identified gene families relevant to osmosis control (e.g., aquaporins, vacuolar-ATPases, Na+/K+-ATPases, and carbonic anhydrases). Evolutionary analysis suggests that all osmotic regulation genes are under strong purifying selection. We also observed low copy number variation in insecticide resistance and immunity-related genes for all known classical pathways. The data provided by this study offers candidate genes for further studies of parasite-vector interactions and for studies on how anophelines of brackish water deal with the high fluctuation in water salinity. We also established data and insights supporting An. aquasalis as an emerging Neotropical malaria vector model for genetic and molecular studies.
Subject terms: Genome evolution, Entomology
Introduction
Malaria is a severe public health problem in several tropical and subtropical areas, mostly in countries in Africa, Asia, and America. It is a leading cause of death and disease in many developing countries, where young children and pregnant women are most affected1. Mosquitoes in the genus Anopheles are vectors of human malaria parasites, Plasmodium sp., which annually generate approximately 229 million cases that result in nearly half a million deaths worldwide1. Parasites are transmitted through the bite of a female mosquito of the Anopheles genus. However, although this genus comprises 400 species, only 41 recognised as vectors, nine of which are found in the Americas2. The biological characteristics, influenced by variations in the ability of Anopheles vectors to transmit the parasite (e.g., molecular components of the immune response, microbiota, and intestinal physiology), are well studied and have been characterised in established models such as Anopheles gambiae and Anopheles stephensi3–6.
Publication and availability of Anopheles genomes accelerated research that has enhanced our fundamental understanding of mosquito genetics, behaviour, physiology, and roles in transmission and can contribute to new strategies for combating malaria7. Although our knowledge that genome information will facilitate the development of innovative approaches to combat malaria and other mosquito-borne diseases, until today, only two Neotropical malaria vectors have had their genomes sequenced and are publicly available: Anopheles albimanus and Anopheles darlingi.
Anopheles aquasalis is among the species considered important in malaria transmission in the New World and is the primary coastal vector in Central and South America and the Caribbean Islands8,9. It is considered the primary Plasmodium vivax malaria vector from northeastern Venezuela to southern Brazil10,11, where its larvae develop in the brackish waters of mangroves. An. aquasalis is among the few anophelines capable of surviving in severe water salinity changes. Rising sea levels due to climate change have raised concerns regarding a greater risk of disease transmission in coastal regions12. However, few studies have addressed how anopheline larvae deal with saline stress. Physiological studies have suggested that morphological changes in the localisation of vacuolar-ATPases (V-ATPases) and K+/Na+ ATPase proteins are essential for osmotic regulation in An. albimanus13,14. Other molecular and transcriptome studies have also suggested that other genes, such as aquaporin and carbonic anhydrases, in conjunction with transcriptional modulation, are essential for mosquito survival in hyperosmotic or hypo-osmotic environments15–18.
Besides its peculiar ecological features as the primary coastal malaria vector, recent research has recognised An. aquasalis as an essential model for studying the interaction with human Plasmodium and murine parasites such as P. yoelii19–21. As such, it has been possible to identify and functionally characterise the role of molecular components relevant during the mosquito invasion by Plasmodium species19,22–24. Our understanding of the interactions between An. aquasalis and the malaria parasites are rapidly improving. Thus An. aquasalis is emerging as a model species for genetic and molecular studies.
However, despite its vectorial, ecological, and modelling importance, few genomic studies on An. aquasalis have been carried out to date. Genomic studies are fast and reliable methods for the genome exploration of medically necessary non-model insects25,26. Genomic and transcriptomic studies have provided a better understanding of the genetic characteristics of more than 18 anopheline species. They have established the composition of conserved regions of genes, the identification of highly divergent genes, the recognition of gene families, and the evolution of species-specific physiological or behavioural genetic variations24,27–29. Here, we present the analysis of the genome of An. aquasalis as a resource and platform for fundamental and translational studies. By identifying its protein-coding genes, we uncovered insights into genome evolution, structure, proteins relevant to osmotic regulation, insecticide resistance, and genes pertinent to vector-parasite and vector-host interactions, among others.
Results
Genome assembly and annotation of An. aquasalis
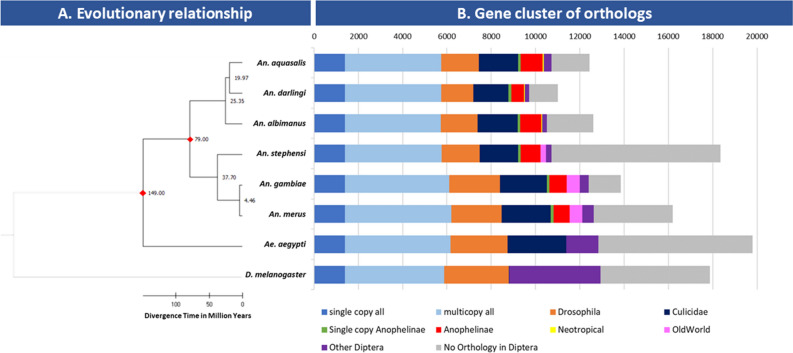
The genome sequencing generated ~ 123 million 100b paired-end reads (SRA SRX21970089). The assembled An. aquasalis genome (Accession GCA_002846955.1) spans 162,944 Mb, distributed on 16,504 scaffolds (N50 14,431). A total of 12,446 protein-coding genes were annotated in the genome. Benchmarking Universal Single-Copy Orthologues (BUSCO) analysis for genome [94.6% of complete single-copy genes, 0.2% complete and duplicated, 1.9% fragmented and 3.3% missing genes] and for annotation [92.1% of complete single-copy genes, 0.3% complete and duplicated, 2.2% fragmented and 5.4% missing genes] shows similar quality when compared to other mosquitoes (Additional file 1: Figure S1). The gene structure models generated with the MAKER program were evaluated according to the annotation edit distance (AED), with most gene structures supported by evidence, with 90% having a value of between 0 and 0.5 for AED (Additional file 1: Figure S2). Orthology analysis comparing An. aquasalis with An. darlingi, An. albimanus, An. gambiae, An. merus, An. stephensi, Aedes aegypti, and Drosophila melanogaster (Fig. 1B) identified 1387 single-copy orthologs (SCO) among the compared genomes; 4359 genes that are multicopy; 1766 genes exclusive to mosquitoes; 121 genes that are single-copy only in anophelines; 992 genes are present in at least one other Anophelinae, 55 genes that are exclusive to Neotropical anophelines; 345 genes with orthology to other Diptera and 1721 genes that presented no orthology at Diptera level. Based on the 1387 SCO, we reconstructed the evolutionary tree of An. aquasalis and other Neotropical Anophelinae and calculated the divergence times for each branch (Fig. 1A). The phylogenetic tree suggests that An. aquasalis diverged from An. darlingi ~ 19.97 million years ago (mya), while South American anophelines diverged from An. albimanus ~ 25.35 mya.
Figure 1.
Evolutionary and orthology analysis of Neotropical anophelines. (A) The species phylogeny tree was inferred based on 1387 single-copy orthologs at Diptera level and rooted on Drosophila melanogaster. Divergence times were calculated using two fixed calibration constraints (red diamonds). (B) Bars show the orthology of annotated genes in each taxon. Single copy all: single copy orthologs present in all taxa; multicopy all: multicopy orthologs present in all taxa; Drosophila: orthologs present in Drosophila melanogaster and at least one mosquito; Culicidae: orthologs present in Ae. aegypti and at least one anopheline; Single-copy Anophelines: single-copy orthologs present in all anophelines only; Anophelinae: orthologs present in more than one anopheline (but not all); Neotropical: orthologs present only in Neotropical anophelines; Old World: orthologs present only in African and Asian anophelines; Other: genes with orthology with other Diptera; No Orthology: genes that presented no orthology at Diptera level.
Gene cluster analysis (Additional file 1: Figure S3) of orthologous genes suggests that 3154 genes are specific to An. aquasalis when compared to An. darlingi, An. albimanus and An. gambiae. A cluster of 7012 orthologs are present in all four anophelines, while 331 orthologs are present only in Neotropical anophelines. The mean transcript size was 4059,96 bp, while coding sequences presented a mean length of 420.88 bp (Additional file 1: Figure S4). In all, 35,352 introns were identified in the genome of An. aquasalis with an average size of 666.45 bp. Finally, the average gene size was predicted to be 3508.37 bp (Additional file 1: Table S1).
Repetitive element analysis suggests that ~ 6.3% of An. aquasalis genome is composed of such elements (Table 1). The most abundant classes were Interspaced Repeats (3.1%) and simple repeats (2.4%). Transposable elements accounted for 0.62% of the genome, and the most representative families were Jockey, Gypsy, Bel/Pao and Mariner/TC1 (Additional file: Figure S5).
Table 1.
Repetitive Element identification in the An. aquasalis genome.
| Repetitive element class | Subclass | Number of elements | Length (BP) | % seq |
|---|---|---|---|---|
| Transposable elements | ||||
| SINEs | Alu | 33 | 6075 | 0.004% |
| Others SINE | 36 | 5908 | 0.004% | |
| Others LINE | 5996 | 490,975 | 0.301% | |
| LINES | LINE1 | 73 | 4118 | 0.003% |
| LINE2 | 136 | 26,889 | 0.017% | |
| L3/CR1 | 117 | 17,933 | 0.011% | |
| LTR | Others LTR | 2385 | 457,214 | 0.281% |
| ERV Class I | 6 | 383 | 0.000% | |
| ERV Class II | 24 | 1457 | 0.001% | |
| Repetitive DNA elements | 10,094 | 1,388,204 | 0.852% | |
| Unclassified | 25,547 | 2,742,549 | 1.683% | |
| Interspased repeats | 5,085,017 | 3.121% | ||
| RNA small | 63 | 4446 | 0.003% | |
| Simple repeats | 121,702 | 3,976,717 | 2.441% | |
| Low complexity repeats | 9266 | 415,557 | 0.255% | |
| Total | 44,510 | 10,231,168 | 6.279% | |
Functional prediction of encoded genes from the An. aquasalis genome
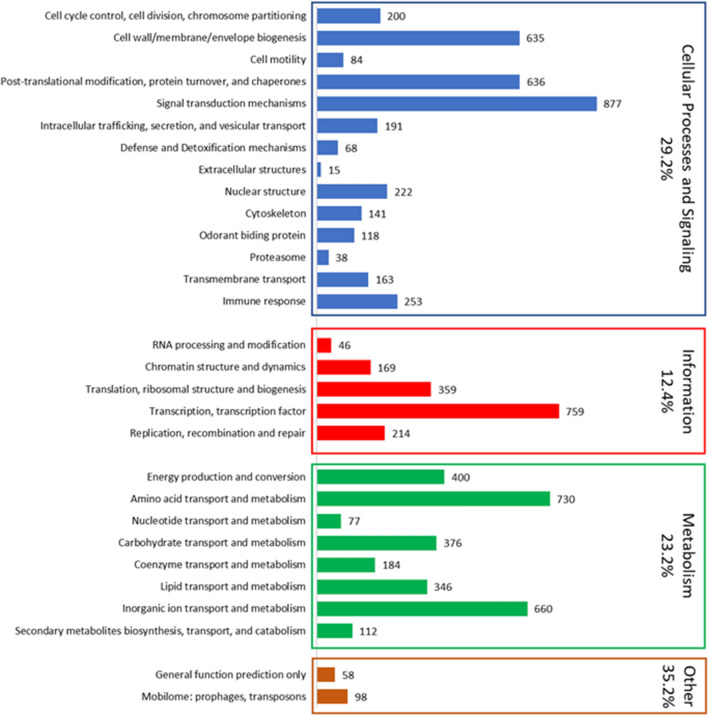
The putative functions were inferred for 65.9% (8208) of the predicted protein-coding genes (Fig. 2; Additional file 2). From the genes with identified ontologies and putative functions, our analysis indicated that most terms corresponded to the category of cellular processes and signalling (29.2%), followed by metabolism with 23.2%, and information storage and processes (12.4%) (Fig. 2). Looking at the more specific classification, the most abundant classes of genes belonged to signal transduction mechanisms (877 genes); transcription and transcription factors (759 genes); amino acid transport and metabolism (730 genes); inorganic ion transport and metabolism (660 genes); post-translational modification, protein turnover, and chaperones (636 genes) and cell wall/membrane/envelope biogenesis (635 genes). Altogether, the genes in these six functional classes encompass 51.7% of all genes with ascertained putative function. As expected, most annotated genes have unknown functions (4242 genes: 34.1%), though 98 genes were classified into the mobilome functional class (transposons and prophages).
Figure 2.
Summary of annotated genes in the An. aquasalis genome. Bars represent the number of genes annotated in each functional class. Colours represent major functional groups, and the percentage of genes in each category is represented. The percentage of genes belonging to the category of other genes (orange) includes genes with unknown functions (4217 genes not represented by a bar in the figure due to scalability).
The analysis of the composition of domains with the InterproScan tool recognized among the main families of most representative proteins those that are composed of the domains zinc finger C2H2-type (IPR013087) with 301 proteins, the zinc finger, RING/FYVE/PHD-type (IPR013083) with 196 proteins and zinc finger, RING-type (IPR001841) with 101 proteins. Domains related to catalytic processes, such as protein kinase domains (IPR000719) with 212 proteins and serine proteases and trypsin domain (IPR001254) with 200 proteins, also had considerable representation. Other well-represented domains were those related to cell recognition processes, DNA repair, which is part of cell surface receptors and the immune response, such as immunoglobulin-like domain (IPR007110) with 168 proteins, leucine-rich repeat, typical subtype (IPR003591) with 122 proteins, leucine-rich repeat (IPR001611), RNA recognition motif domain (IPR000504) with 120 proteins and fibronectin type III (IPR003961) with 64 proteins. Finally, some groups of domains related to the structure of the cuticle were abundant, such as insect cuticle protein (IPR000618) with 97 proteins and the chitin-binding domain (IPR002557) with 96 proteins (Additional file 1: Table S3). In the next section, some groups of interest are discussed.
Osmoregulation, ion metabolism and transport
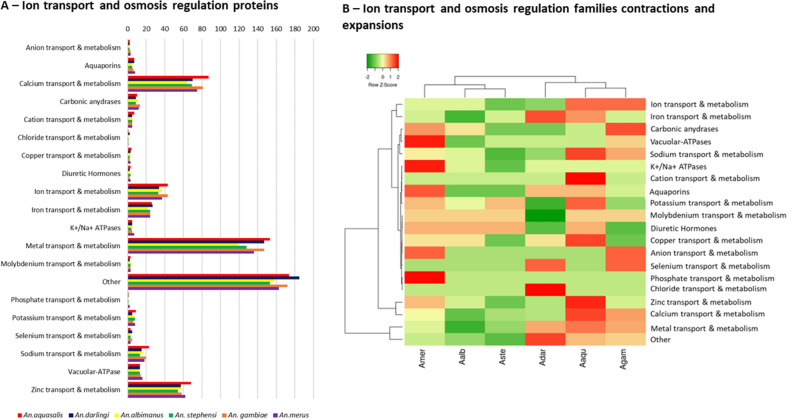
An. aquasalis larvae live in brackish waters, and osmoregulation is a crucial process for these anophelines, involving ion transport and metabolism genes, water permeability, and tissue modifications. We found 685 proteins related to ion transport and metabolism in An. aquasalis, which is higher than other anophelines, particularly Neotropical anophelines (Fig. 3A; Additional File 3—Dataset 2). The An. aquasalis genome presented a higher number of Cation transport proteins, zinc, copper, sodium and calcium transport and metabolism proteins (Fig. 3A; Additional File 3—Dataset 2).
Figure 3.
Ion metabolism and transport and osmosis proteins in An. aquasalis. (A) shows the number of genes related to ion metabolism and transport found in An. aquasalis in comparison to other anophelines. (B) Heatmap of expansions (red) and contractions (green) of gene families in anophelines.
Studies of the tolerance of An. albimanus to saltwater also suggests the importance of V-type ATPase (V-ATPase), carbonic anhydrase, and K+/Na+ ATPase proteins in osmoregulation13–17, and studies of other insects have shown the role of aquaporins in this process. Orthology searches suggest that An. aquasalis has a similar, but slightly lower, number of osmoregulation genes (Figs. 3B and 4) compared to other anophelines, especially An. merus. Heatmap analysis on the number of genes in osmoregulation and ion transport/metabolism (Fig. 3B) shows that An. aquasalis has a significantly higher number of cation transporters (7 genes while the mean is 5.3 ± 0.8 for other anophelines) and zinc transport/metabolism (68 genes while the mean is 59.5 ± 4.9 for other anophelines), while An. merus has a significantly higher number of phosphate transport/metabolism (two copies instead of one for all other anophelines) and canonical osmoregulation genes such as vacuolar ATPsaes (16 genes while the mean is 13.5 ± 1.4) and K+/Na+ ATPases (7 genes while the mean is 5.1 ± 1.0).
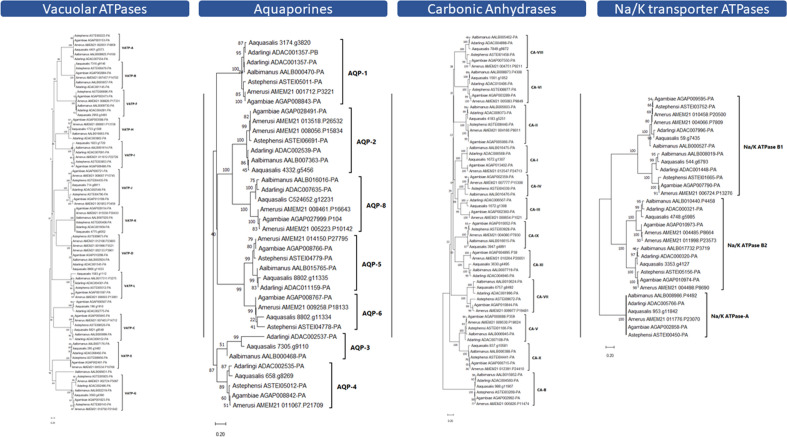
Figure 4.
Evolutionary relationships of the osmotic regulation genes. The evolutionary relationships of An. aquasalis (Aaq); An. darlingi (Adar); An. albimanus (Aalb) An. merus (Amer) and An. gambiae (Agam) osmotic regulation genes. (A) The Vacuolar ATPases proteins; (B) Aquaporins; (C) Na/K ATPases and; (D) Carbonic Anhydrase. Trees were inferred by neighbour-joining, using amino acid sequences, and bootstrapped (10,000 replicates).
Evolutionary analyses and codon-based tests (Fig. 4; Additional file 3—Dataset 3) suggest that almost all osmotic regulation genes evolve via strong purifying selection. The significantly higher number of An. merus V-ATPases seem to originate from multiple duplication events in the V-ATPase D family (Fig. 4), while the increase in the K+/Na+ ATPases of An. merus originates from two duplications in the B1 and B2 families. Purifying selection was observed even between gene subfamilies (e.g., all seven aquaporin gene subfamilies), except for vacuolar ATPases, in which evolutionary analysis suggests purifying selection within subfamilies but not between subfamilies (e.g., VATP-F and VATP-G).
Immune response genes
Identifying genes involved in the mosquito responses against Plasmodium sp. Is particularly interesting for understanding parasite-vector interactions. Hence, genes of the immune response system in An. aquasalis were discussed in depth in a companion paper30, and here we will briefly present the most important findings and insights. We identified 278 immune-related proteins, divided into 24 families or signalling pathways groups, in the An. aquasalis genome. All genes from the classic signalling pathways; Janus kinase (JAK)/signal transducer and activator of transcription (STAT), immune deficiency (Imd), Toll and Jun N-terminal kinase (JNK)—were identified with one-to-one orthologs for An. darlingi. Cascade modulators (e.g., serine proteases) accounted for 25.64% of identified immune response genes. Signalling pathways genes correspond to 13.46%. Other abundant families in the immune response group are Fibrinogen-Related Proteins (FREPs) with 29 genes (9.29%), autophagy process with 20 genes (6.41%), leucine-rich repeats (20; 6.41%) and C-type lectins (13; 4.16%). In general, An. aquasalis has a similar number of immune response genes to other Neotropical anophelines (An. darlingi 294; An. albimanus 304); but a significantly lower number of genes when compared to An. gambiae (410).
Chemosensory system
Anophelines use a series of chemosensory proteins to perceive the environment they are in, such as identifying hosts and oviposition sites. Chemosensory genes are classified as chemosensory proteins (CSPs) and odorant binding proteins (OBPs). They can be divided into three families of significant importance: odorant receptors (ORS), gustatory receptors (GRs), and ionotropic receptors (IRs). Our analysis found 44 ORS, 32 GRs, and 15 IRS in the An. aquasalis genome is a lower number than all anophelines studied so far, including An. darlingi, with 57 ORS and 56 GRs. Despite the low conservation of OBP amino acid sequences, we identified six conserved cysteine residues characteristic of this gene family. All An. aquasalis OBPs were also classified into Classic, Atypical, and Plus-C subfamilies according to their homology to An. gambiae sequences and phylogeny (Additional file 1: Figures S6, S7 and Table S4). Motif analysis identified eight conserved motifs in OBPs, with conserved cysteine residues in four (Additional file 1: Figure S8).
Insecticide resistance and detoxification
In all, 133 genes were identified related to metabolic detoxification, 73 (54%) from the P450s family (Table 2), 25 glutathione-S-transferases (GST), and 36 carboxylesterases (Additional file 1: Table S5 and Table S6). Evolutionary analyses with An. gambiae P450 genes (Additional file 1: Figure S9, S10 and S11) allowed us to classify An. aquasalis P450 into four classic clans: CYP2 (8 genes), CYP3 (32 genes), CYP4 (25 genes), and mitochondrial CYP (8 genes). These numbers were similar to the genes found in the genome of An. darlingi; however, An. albimanus and An. gambiae presented a greater number of genes.
Table 2.
An. aquasalis cytochrome P450 gene subfamilies compared with An. darlingi, An. albimanus and An. gambiae.
| An. aquasalis | An. darlingi | An. albimanus | An. gambiae | |
|---|---|---|---|---|
| CYP2 | 8 | 10 | 10 | 11 |
| CYP3 | 32 | 32 | 35 | 39 |
| CYP4 | 25 | 22 | 36 | 45 |
| Mitochondrial CYP | 8 | 8 | 8 | 9 |
| Total | 73 | 72 | 89 | 104 |
Evolutionary analyses (Additional file 1: Figure S9, S10 and S11) identified three An. aquasalis genes orthologous to An. gambiae genes related to insecticide resistance: 1921.g1856 is orthologous to AGAP002862 (CYP6AA1); 1805.g1684 related to AGAP008213 (CYP6M3)—both from the CYP3 family; and C559148.g12387, which is orthologous to AGAP001861-PA (CYP4H14)—from the CYP4 family. We also identified two losses of genes in the CYP2 family: the orthologous gene of CYP350 (AGAP005660, AALB015553, and ADAC003150) and the orthologous gene of CYP11179 (AGAP003065, AALB015657, ADAC007012). Finally, the An. gambiae mitochondrial CYP12F2 (AGAP008020-PA) and CYP12F3 (AGAP008019-PA) genes were not found in the genome of An. aquasalis.
Regarding glutathione-S-transferases, the genes were classified into seven classes: Delta, Epsilon, Omega, Sigma, Theta, Zeta, and Unclassified (Additional file 1: Table S5). The number of genes of the main classes Delta and Epsilon remained stable among the mosquitoes. Cholinesterases (CCEs) were classified into eight subfamilies: α-esterase, β-esterase, juvenile hormone esterase, acetylcholinesterase, gliotactin, glutactin, neurotactin and neuroligin (Additional file 1: Table S6). Among the identified CCEs, 36% belong to the α-esterase subfamily, the subfamily with the highest number of CCEs among anophelines (30–36%).
Discussion
The sequencing and annotation of anopheline genomes allowed the identification of these mosquitoes’ structural, functional, and evolutionary differences. This is a task that started with An. gambiae at the beginning of this century and has so far included more than 15 species29,31,32. Five species of anophelines are responsible for most of the malaria transmission in the Americas. Two of them, An. darlingi and An. albimanus have already been sequenced. Here, we present the genome of the primary coastal malaria vector in Central and South America and the Caribbean Islands, An. aquasalis. Besides its importance as a vector of human malaria parasites, An. aquasalis has a remarkable ecological feature since its larvae grow in the brackish water of mangroves. Hence, its genomic sequences could reveal genes related to interaction with Plasmodium sp. and unique adaptations that allow it to deal with salted water, in contrast to other anophelines.
Anophelines have genomes ranging from 134.7 Mb (An. darlingi) to 375.8 Mb (An. sinensis)—with a median size of 224.3 Mb±50.3 Mb29. The number of genes ranges from 10,457 (An. darlingi) to 16,149 (An. melas), with a mean of 13,162 genes ± 138029. Our results suggest that the size of the genome (162.9 Mb) and number of protein-coding genes (12,446) of An. aquasalis is relatively similar to other anophelines. We found a core of 1387 single-copy genes with orthologs in all Neotropical anophelines, An. gambiae, An. merus, An. stephensi and D. melanogaster and other Diptera. Based on these single-copy orthologs, we could reconstruct the evolutionary history of An. aquasalis and estimate its divergence time from other anophelines. Our data suggest that An. aquasalis diverged from An. darlingi approximately 20 mya33,34. The work of Martinez-Villegas33 was the first to estimate the divergence of An. aquasalis from other species using mitochondrial DNA data and found the divergence of An. aquasalis from An. darlingi to be ~ 39 mya. The results presented here are the first to calculate the divergence times of An. aquasalis to other anophelines based on a set of over a thousand nuclear genes and suggests not only that An. aquasalis is more closely related to An. darlingi than to An. albimanus, but also that both species have a much earlier divergence time than proposed by Martinez-Villegas.
The An. aquasalis repetitive elements and mobilome occupied of 6.28% of the genome, higher than that found in the American anophelines An. albimanus and An. darlingi ranges from 2.4 to 2.9% of the total genomic DNA. In general, repetitive elements in anophelines range from 0.13% in An. koliensis to 20% in An. gambiae34,35. However, the revision of repetitive elements in genomes assembled with long-read technologies has shown that the proportion of repetitive DNA is underestimated in many available genomes. Recent studies also found significant variations of repetitive elements in anopheline populations, such as that observed in An. darlingi, with repetitive elements varying from 2.9 to 5.6% of the genome36,37. Also, studies on the role of the mobilome in anophelines have shown the role of these DNA sequences in the modulation of immune response genes and detoxification genes38. All in all, the characterisation of repetitive elements and TE in anophelines has become a necessary process to understand how they are formed within the genome of these mosquitoes and as a possible explanation for the rapid response that some species have to unfavourable environmental factors and to the response to pathogens of medical interest34,37,38.
Insects have developed several mechanisms to live in saline environments, from regulation of ion transport and metabolism genes to morphological modifications of the rectum. We found 600 + proteins related to ion transport and metabolism, and our data suggest that An. aquasalis has a higher number of such proteins than other Neotropical anophelines and An. gambiae. Among the most abundant gene families related to ion transport, we identified many cation channels, zinc and copper transport and metabolism, and calcium binding, and transport proteins. An. albimanus and An. merus are other anophelines with tolerance to saline water, and both presented a lower number of such genes. Physiological studies have demonstrated that in the larvae of An. albimanus, when exposed to gradual changes in saline water, specialised non-dorsal anterior rectal (non-DAR) cells undergo changes in the localisation of V-type ATPases and K+/Na+ ATPases proteins, allowing the production of super osmotic urine and disrupting the ion reabsorption system in non-DAR cells13,14. Other studies of mosquitoes have also suggested the importance of carbonic anhydrase (CA) and aquaporins to osmoregulation in saline environments15,17. We found that An. aquasalis has a slightly higher number of such genes than An. albimanus. Interestingly, An. merus has a significantly higher number of canonical osmoregulation genes, especially in V-ATPases and K+/Na+ ATPases, with recent duplication events on these genes. It is possible that An. aquasalis and An. merus have different strategies to deal with saline environments, and further functional studies comparing both insects could reveal relevant adaptations for mosquito survivability in saline waters. We also hypothesised if the ability of An. aquasalis to live in saline water could be due to amino acid changes in such proteins (positive selection). However, as expected, our analysis suggests that all osmotic regulation genes are under strong purifying selection within and among orthologous groups in each protein family (the exception is the V-type ATPases in which each ortholog group seems to be evolving independently). Transcriptome studies in An. merus (another anopheline with high tolerance to saline environments)16 revealed several changes in gene expression upon salinity stress, raising a few candidates for further functional studies. Therefore, further transcriptome studies on An. aquasalis may reveal significant differences in gene expression and indicate candidates for functional studies.
An. aquasalis is one of the major malaria vectors in the New World, and studies in parasite-vector and vector-host interactions have been the focus of many researchers. For the Plasmodium to infect the anopheline, the parasite must overcome the insect’s immune system. The An. aquasalis genome revealed all genes from classical immune pathways in a one-to-one ortholog with An. darlingi. However, our study suggests that An. aquasalis (as the Neotropical An. darlingi and An. albimanus) has fewer immune-related genes than Old World anophelines.
In general, the family groups related to the control processes of reactive oxygen species (ROS) production and components regulating the expression of effectors of the immune response or signalling pathways were well conserved, with groupings of orthologs 1:1 for the four compared species. They are functionally relevant genes for the maintenance of the homeostasis of the organism and, in the case of signalling pathways such as the Toll pathway, they help in embryonic development processes and are constitutive for these insects39–41. On the other hand, the marked differences in the number of copies were in groups related to the recognition of molecular patterns of microorganisms, with sharp differences in the FREP and MLD proteins, especially with species-specific expansions in An. gambiae. A phenomenon that is possibly induced by the microbiota of each species or by the metabolic or sensory needs of each organism, as in the case of MLD proteins42,43.
Other families, such as PGRP or GNBP, had few differences in the number of copies, with losses mainly in American anophelines. These proteins activate signalling pathways, and some have been found as regulatory factors for other members of the same family44,45. The regulatory role of both families allowed a few variations to be maintained during evolution in American anophelines and An. gambiae. In addition, American mosquitoes suffered copy losses in cascade modulation proteins, mainly in serine proteases with CLIP domains, the most abundant family, and with gene expansions in An. gambiae46. These gene families activate signalling pathways and produce melanisation components through proteolytic cascades after recognising a pathogen46,47. It is recognised that specific sets of these proteins are organised for specific physiological and immune processes, sometimes with a redundant function that synergises to increase the intensity of the response48,49. In this sense, it is speculated how exposure to specific pathogens has shaped the set of serine proteases, serpins, and sometimes prophenoloxidase proteins in each species of mosquito41,46.
On the other spectrum, vector-host interactions, mosquitoes rely on a repertoire of chemosensory proteins to identify their hosts. Many studies have demonstrated that CSPs and OBPs present rapid evolution, sometimes limiting the identification of orthologs even in close species. The 16 Anopheles genomes manuscript suggested that most anophelines have ~ 60 OR copies, while all anophelines of the gambiae complex gained ~ 10 OR copies29. On the other hand, copy numbers of GRs and IRs have remained stable in all species studies so far. Our data suggest that An. aquasalis has a much lower number of OR and GR genes than other anophelines (44 and 32, respectively). As rapidly evolving genes, it is expected that the identification of OBPs is underestimated in genomes, which could be the reason for our findings. Neafsey and colleagues29 tried to find a correlation between OBP copy number variation (CNV) and host preference. However, transcriptome studies suggest that such differences are more likely due to functional divergence and regulation of gene expression50,51.
The increased resistance to insecticides in insect vectors of diseases is of significant concern for public health programs. Metabolic insecticide resistance is mediated by multi-copy gene families, such as cytochrome P450, glutathione S-transferases (GSTs), and carboxyl/cholinesterases. Despite its large numbers, several studies have shown the conservation of these gene families. We found 72 P450 in all relevant clades (CYP2, CYP3, CYP4, and mitochondrial CYP), similar to Neotropical anophelines. The same is true for GSTs and carboxyl/cholinesterases. In most cases, we found a 1:1 ortholog to An. gambiae, including orthologs with genes related to insecticide resistance, such as CYP6AA1, CYP4H14 (resistance to pyrethroids), CYP6M2 (resistance to carbamates), CYP6M3 (resistance to organochlorines)52,53 and GSTE2, GSTE4, and GSTE6 (resistance do DDT, organochlorines and pyrethroids, respectively)54,55. We also observed a few gene duplications and losses, and recent studies have suggested that CNV has a relevant role in the rise of pyrethroid resistance56,57. We have no reports on the increase in insecticide resistance in An. aquasalis and only a few studies have addressed this issue58,59. Identifying orthologs of genes related to major insecticide resistance is relevant for future studies on insecticide metabolism and evaluating insecticide resistance in natural populations.
Conclusions
The data presented here brings new insights for An. aquasalis biology, Neotropical anopheline evolutionary relationships, and general anopheline evolution. Despite being the primary coastal malaria vector in Central and South America and the Caribbean Islands, the physiology of An. aquasalis still needs to be better understood. Recent research has elevated An. aquasalis as a significant model of vector-parasite interaction19–21, and the identification of immunity and digestion-related genes are essential for future research. Moreover, An. aquasalis is among the few anophelines capable of surviving drastic changes in water salinity, and with climate change and increased potential for saltwater invasion and salinisation of inland waters, studying the physiology of saltwater anophelines may be of great significance.
Materials and methods
Anopheles aquasalis mosquito sampling and sequencing
The mosquito sample used for this work came from the colony established in 1995 by the Laboratory of Medical Entomology at FIOCRUZ-MG. Genomic DNA was purified from a single adult female using a Qiagen DNeasy Kit for blood and tissues. The library was prepared using the Nextera DNA sample preparation kit (Epicentre Biotechnologies, Madison, WI), with an amplification step (5 cycles) as outlined in the Nextera protocol61. The fragment size distribution was analysed utilising a 2100 Bioanalyzer with a 7500 DNA assay kit (Agilent Technologies, Santa Clara, CA). Fragments of ~ 600 bp long were carried out for sequencing. The library was sequenced on one lane of an Illumina HiSeq2000 instrument to generate 50 base paired-end reads. Sequencing was performed by The Vincent J. Coates Genomics Sequencing Laboratory (GSL) at the University of California, Berkeley60. Sequences were assembled de novo using Velvet v1.2.1061 with a k-mer size of 41, according to the scripts and parameters suggested by the Velvet Manual (https://www.ebi.ac.uk/~zerbino/velvet/Manual.pdf) and in-house protocols from the UC Davis Vector Genetics Laboratory61. The assembled genome is available under the accession number GCA_002846955.1.
Databases and sequences used to predict the genome of An. aquasalis
As part of the evidence implemented for gene prediction, cDNAs generated from the transcriptome of24 were downloaded from the GEO database with accession number GSE124997 on the NCBI website. In addition, proteins from four species of anophelines deposited in the VEuPathDB database were downloaded: An. albimanus (Anopheles-albimanus-STECLA_PEPTIDES_AalbS2.6), An. darlingi (Anopheles-darlingi-Coari_PEPTIDES_AdarC3.8.fa), An. sinensis (Anopheles-sinensis-China_PEPTIDES_AsinC2.2.fa) and An. gambiae (Anopheles-gambiae-PEST_PEPTIDES_AgamP4.12.fa)62.
Annotation and prediction of repetitive elements in the genome of An. aquasalis
Simple repeats and complex repetitive elements in the genome of An. aquasalis were predicted using three approaches in the following order: 1st—we identified repeat elements using RepeatModeler (Version 2.0.4)63 with standard parameters; 2nd—we identified additional repeat elements using BLASTn against a database of repetitive elements from An. gambiae, An darlingi, An. albimanus and An. stephensi downloaded from VEuPathDB website64. 3rd—The repetitive elements identified with RepeatModeler and BLASTn were concatenated to build a database that was used to predict and mask repeat elements with RepeatMasker (Version 4.1.5)64,65. The options for searching the elements of interest were –nolow to mask only the interspaced sequences. –norna to not mask the smallRNA genes. The file with the program's predictions was obtained in gff format, with the –gff option66. The prediction results from all three steps were compared to build the final library of An. aquasalis repetitive elements.
Structural prediction of the genome of Anopheles aquasalis
For structural prediction, a total of 60,752 proteins from five anopheline species are available in VEuPathDB and An. aquasalis transcripts24 were used as a template for gene annotation. Initial prediction of genes was performed with the MAKER program v. 2.31.1 available online on the Galaxy server67 in two rounds to create a draft of An. aquasalis putative genes. Using the option to infer gene predictions directly from all Expressed Sequenced Tags (from An. aquasalis), gene predictions were inferred directly from all protein alignments (database created), and pre-identified repeat elements from an external GFF file (repeat masker output). The gff output from MAKER, along with the unique scaffolds from the An. aquasalis genome was used to perform ab initio training with the annotator AUGUSTUS v. 3.3.3 in two rounds to create a draft of An. aquasalis putative genes. The MAKER archive and An. aquasalis ESTs were then used to train the AUGUSTUS program68. After training, a second round in MAKER was performed, using the same files as the first round with the addition of the gene models trained by AUGUSTUS. After this second round of MAKER, the gff output was used for AUGUSTUS second training, and then finally the final annotation by AUGUSTUS.and models generated by the training file were used for final annotation with AUGUSTUS using ESTs as evidence of transcription.
Evaluation of An. aquasalis genome and annotation quality
The quality of the genome assembly and gene predictions were evaluated using the BUSCO v5.4.6, selecting the Diptera lineage and “genome assemblies” (for genome quality) or “protein” (for annotations) options on the Galaxy Australia server67,69. Additionally, the result obtained by the MAKER with the AED index was evaluated and established how much of the information used as evidence directly aligns with An. aquasalis’ genome and this agreement between evidence and prediction must be equal to 90% of the generated alignments. The AED cumulative fraction of the annotations graph was generated using the AED_cdf_generator.pl script70,71.
Functional prediction of the genome of An. aquasalis
Protein sequences originating from the An. aquasalis genome gene model was used for functional prediction, gene ontology (GO) assignments, and functional descriptions of the An. aquasalis genome was generated through the Pannzer program pipeline, selecting these annotations using ppv > 0.572. Regarding protein domains and some gene ontologies (GO), these were annotated and searched for with the InterproScan tool from the Galaxy Europe server, selecting the annotations as an e-score of 0.000173. The REVIGO tool (http://revigo.irb.hr/) was used to identify some GO terms not identified by functional annotation programs and to reduce the redundancy of these terms. Finally, a homology search was done using the Blastp program against the proteins of Drosophila melanogaster (Berkeley strain) (UP000000803), Culex quinquefasciatus (Southern House mosquito strain JHB) (UP000002320), Aedes aegypti (Yellow fever mosquito strain LVP_AGWV) (UP000008820), An. darlingi (UP000000673), An. albimanus (New World malaria mosquito strain STECLA/ALB19) (UP000069272) and An. gambiae (African malaria PEST strain) (UP000007062), choosing the sequences with a percentage identity > 50% and with an e-value of 0.000574. The data generated by each tool were concatenated in Excel and the online Google Colaboratory tool and classified according to significance to determine the most accurate protein function. Basically, we established an order of priority for functional prediction with priority to Pannzer results followed by InterproScan, REVIGO, and Blastp (which means that we only assigned protein functionality based on Blastp in those cases that we had no results for Pannzer, InterproScan or REVIGO). To classify the terms of the genetic ontology obtained in the functional prediction, the R GO.db package (version 3.13.0) was used, using the option “GOANCESTOR”. The functional prediction was complemented with orthology searches in OrthoDB (v11) at the Diptera level75.
Orthology analysis
Orthology assignments were retrieved from OrthoDB (v11)75 Diptera-level orthologous groups (116 species) for the species detailed in Dataset S1. An. aquasalis protein-coding genes were mapped to OrthoDB (v11) at the Diptera level using D. melanogaster as an anchor. Mapping was also performed for An. gambiae, An. darlingi, An. albimanus, An. stephensi, An. merus, A. aegypti and D. melanogaster. Each species was then merged to create the final orthologous groups, including all mapped An. aquasalis proteins. Single copy genes were then identified for all species to build a dataset of genes for phylogenomics. The presence, absence, and copy-numbers of orthologs were also assessed to partition genes from each Dipteran species into the categories shown in the bar chart (Fig. 1A) and classified as: (1) single copy orthologs present in all taxa; (2) multicopy orthologs present in all taxa; (3) orthologs present in D. melanogaster and at least one mosquito; (4) orthologs present in all mosquitoes; (5) orthologs present in other Diptera; (6) single-copy orthologs present in all anophelines; (7) orthologs present in more than one anopheline (but not all); (8) orthologs present only in Neotropical anophelines; (9) Orthologs present only in old-world anophelins; (11) proteins with no orthology at Diptera level. A Venn diagram was created with the VennDiagram package for R76.
Phylogenomics and gene evolutionary analysis
All evolutionary analyses were conducted in MEGA 1177. For the main evolutionary analysis of An. aquasalis, a total of 1387 single-copy orthologous proteins from An. aquasalis, An. darlingi, An. albimanus, An. gambiae, An. merus, An. stephensi, Ae. aegypti and D. melanogaster (outgroup) were used. Proteins were aligned with MUSCLE and the phylogenetic tree constructed by Maximum Likelihood (with 10,000 bootstrap replications, JTT model, Uniform Rates and Complete Deletion). Divergence times were inferred by the RelTime method78,79 specifying T. castaneum as the outgroup and using three fixed calibration constraints based on data available at http://www.timetree.org/80, being: D. melanogaster to An. gambiae (241 mya); Ae. aegypti to An. gambiae (149 mya) and; An. darlingi to An. gambiae (79 mya).
Protein-coding sequences were translated to amino acids for the evolutionary analysis and substitution rates of osmoregulation proteins and aligned using Muscle v581. Phylogenetic trees were constructed using amino acid sequences, using neighbour-joining (for orthology analysis)82 with 10,000 replicates and a complete deletion option77,78. Purifying and positive selection hypotheses were tested via synonymous and nonsynonymous substitutions per site (dS and dN, respectively) in MEGA11. P values less than 0.05 were considered significant at the 5% level within and among orthologous groups for each gene family. The variance of the difference was computed using the analytical method. Analyses were conducted using the Nei-Gojobori method83. For other simpler evolutionary analyses, phylogenetic trees were constructed using amino acid sequences aligned by Muscle, using neighbour-joining (for orthology analysis)82 with 10,000 replicates and a complete deletion option.
Heatmap analysis of the number of ion transport/metabolism and osmoregulation genes
The number of orthologous genes was retrieved from Additional File 3—Dataset 2 and clustered in family functions. The Heatmap was constructed using the Heatmapper software84 clustered by average linkage and Euclidean distance measurement method, applying clustering to rows and columns.
Supplementary Information
Acknowledgements
This manuscript is a part of the PhD thesis developed by RMA and CCPS and supervised by PFPP, WMM, APD and LBK (PPGMT-UEA/FMT-HVD). PFPP, NFCS, WMM, and MVGL are productivity fellows of the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). This research is part of the Institutos Nacionais de Ciencia e Tecnologia—Entomologia Molecular (INCT-EM).
Author contributions
P.F.P.P., L.B.K., N.F.C.S., M.V.G.L. and W.M.M. designed the research; L.M.V., P.F.P.P., L.B.K. and N.F.C.S. sequenced and assembled the genome; C.C.P.S., R.M.A., A.C.B., G.M.A.D., I.B.S., A.P.D., L.M.V. and L.B.K. performed the bioinformatics. C.C.P.S., R.M.A., L.B.K. and P.F.P.P. performed the data analysis. R.A.S. established and reared the genomic lineage; C.C.P.S., R.M.A., L.B.K. and P.F.P.P. wrote the manuscript. P.F.P.P. and L.B.K. participated in the study design, coordination, and writing of the final version of the manuscript. All authors read and approved the final version of the manuscript.
Funding
The following Brazilian agencies partially funded this study supporting the PhD and post-doctoral students with fellowships: Fundação e Instituto Oswaldo Cruz (FIOCRUZ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa de Minas Gerais e Amazonas (FAPEMIG and FAPEAM-PROESTADO). The funders had no role in study design, data collection, analysis, or manuscript preparation.
Data availability
The Anopheles aquasalis genome is deposited in GeneBank under accession GCA_002846955.1 (https://www.ncbi.nlm.nih.gov/data-hub/genome/GCA_002846955.1/). Annotated proteins are under submission to GeneBank and can be downloaded as Additional File 4 (CDS) and Additional File 5 (Proteins).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rodrigo Maciel Alencar and Cesar Camilo Prado Sepulveda.
Contributor Information
Paulo Filemon Paolucci Pimenta, Email: pfppimenta@gmail.com.
Leonardo Barbosa Koerich, Email: lbkoerich@ufmg.br.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-47830-1.
References
- 1.WHO. World Malaria Report. Vol. WHO/HTM/GM, World Health Organization. 238 (2022).
- 2.Manguin, S. Anopheles mosquitoes—New insights into malaria vectors. Anopheles mosquitoes—New insights into malaria vectors. (2013).
- 3.Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, et al. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science (1979) 2009;326(5949):147–150. doi: 10.1126/science.1175241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins WE, Sullivan JS, Williams A, Nace D, Williams T, Galland GG, et al. Aotus nancymaae as a potential model for the testing of anti-sporozoite and liver stage vaccines against Plasmodium falciparum. Am. J. Trop. Med. Hyg. 2006;74(3):422–424. doi: 10.4269/ajtmh.2006.74.422. [DOI] [PubMed] [Google Scholar]
- 5.Eldering M, Bompard A, Miura K, Stone W, Morlais I, Cohuet A, et al. Comparative assessment of An. gambiae and An. stephensi mosquitoes to determine transmission-reducing activity of antibodies against P. falciparum sexual stage antigens. Parasit. Vectors. 2017;10(1):1–10. doi: 10.1186/s13071-017-2414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Looker M, Taylor-Robinson AW. A protocol for a highly consistent, high level production <i>in Vivo</i> of <i>Plasmodium falciparum</i> Oocysts and Sporozoites. Adv. Biosci. Biotechnol. 2014;05(13):985–993. doi: 10.4236/abb.2014.513112. [DOI] [Google Scholar]
- 7.Neafsey DE, Christophides GK, Collins FH, Emrich SJ, Fontaine MC, Gelbart W, et al. The evolution of the Anopheles 16 genomes project. G3 Genes Genomes Genet. 2013;3(7):1191–1194. doi: 10.1534/g3.113.006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Póvoa MM, Conn JE, Schlichting CD, Amaral JCOF, Segura MNO, da Silva ANM, et al. Malaria vectors, epidemiology, and the re-emergence of Anopheles darlingi in Belém, Pará, Brazil. J. Med. Entomol. 2003;40(4):379–386. doi: 10.1603/0022-2585-40.4.379. [DOI] [PubMed] [Google Scholar]
- 9.Sinka ME, Rubio-Palis Y, Manguin S, Patil A, Temperley W, Gething P, et al. The dominant Anopheles vectors of human malaria in the Americas: Occurrence data. Parasit. Vectors. 2011;4:210–211. doi: 10.1186/1756-3305-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deane LM, Causey LM, Deane MP. Notas sobre a distribuição e a biologia dos anofelinos das regiões nordestina e amazônica do Brasil. Memórias do Instituto Evandro Chagas Parasitologia. 1948;1(4):827–965. [Google Scholar]
- 11.Berti J, Zimmerman R, Amarista J. Spatial and temportal distribution of anopheline larvae in two malarious areas in Sucre state, Venezuela. Mem. Inst. Oswaldo Cruz. 1993;88(3):353–362. doi: 10.1590/S0074-02761993000300003. [DOI] [PubMed] [Google Scholar]
- 12.Ramasamy R, Surendran SN. Possible impact of rising sea levels on vector-borne infectious diseases. BMC Infect. Dis. 2011;11(1):18. doi: 10.1186/1471-2334-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith KE, Raymond SL, Valenti ML, Smith PJS, Linser PJ. Physiological and pharmacological characterizations of the larval Anopheles albimanus rectum support a change in protein distribution and/or function in varying salinities. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010;157(1):55–62. doi: 10.1016/j.cbpa.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith KE, VanEkeris LA, Okech BA, Harvey WR, Linser PJ. Larval anopheline mosquito recta exhibit a dramatic change in localization patterns of ion transport proteins in response to shifting salinity: A comparison between anopheline and culicine larvae. J. Exp. Biol. 2008;211(19):3067–3076. doi: 10.1242/jeb.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon D, van Ekeris L, Linser P. Characterization of carbonic anhydrase 9 in the alimentary canal of aedes aegypti and its relationship to homologous mosquito carbonic anhydrases. Int. J. Environ. Res. Public Health. 2017;14(2):213. doi: 10.3390/ijerph14020213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uyhelji HA, Cheng C, Besansky NJ. Transcriptomic differences between euryhaline and stenohaline malaria vector sibling species in response to salinity stress. Mol. Ecol. 2016;25(10):2210–2225. doi: 10.1111/mec.13609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misyura L, Grieco Guardian E, Durant AC, Donini A. A comparison of aquaporin expression in mosquito larvae (Aedes aegypti) that develop in hypo-osmotic freshwater and iso-osmotic brackish water. PLoS One. 2020;15(8):e0234892. doi: 10.1371/journal.pone.0234892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith HA, White BJ, Kundert P, Cheng C, Romero-Severson J, Andolfatto P, et al. Genome-wide QTL mapping of saltwater tolerance in sibling species of Anopheles (malaria vector) mosquitoes. Heredity (Edinb) 2015;115(5):471–479. doi: 10.1038/hdy.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahia AC, Kubota MS, Tempone AJ, Pinheiro WD, Tadei WP, Secundino NFC, et al. Anopheles aquasalis infected by Plasmodium vivax displays unique gene expression profiles when compared to other malaria vectors and plasmodia. PLoS One. 2010;5(3):e795. doi: 10.1371/journal.pone.0009795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias-Lopes G, Borges-Veloso A, Saboia-Vahia L, Domont GB, Britto C, Cuervo P, et al. Expression of active trypsin-like serine peptidases in the midgut of sugar-feeding female Anopheles aquasalis. Parasit. Vectors. 2015;8(1):1–10. doi: 10.1186/s13071-015-0908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orfano AS, Duarte APM, Molina-Cruz A, Pimenta PF, Barillas-Mury C. Plasmodium yoelii nigeriensis (N67) is a robust animal model to study malaria transmission by South American anopheline mosquitoes. PLoS One. 2016;11(12):1–15. doi: 10.1371/journal.pone.0167178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahia AC, Kubota MS, Tempone AJ, Araújo HRC, Guedes BAM, Orfanó AS, et al. The JAK-STAT pathway controls plasmodium vivax load in early stages of Anopheles aquasalis infection. PLoS Negl. Trop. Dis. 2011;5(11):e1317. doi: 10.1371/journal.pntd.0001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahia AC, Oliveira JHM, Kubota MS, Araújo HRC, Lima JBP, Ríos-Velásquez CM, et al. The role of reactive oxygen species in Anopheles aquasalis response to plasmodium vivax infection. PLoS One. 2013;8(2):1–10. doi: 10.1371/journal.pone.0057014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santana RAG, Oliveira MC, Cabral I, Junior RCAS, De Sousa DRT, Ferreira L, et al. Anopheles aquasalis transcriptome reveals autophagic responses to Plasmodium vivax midgut invasion. Parasit. Vectors. 2019;12(1):1–14. doi: 10.1186/s13071-019-3506-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterhouse RM. A maturing understanding of the composition of the insect gene repertoire. Curr. Opin. Insect. Sci. 2015;7:15–23. doi: 10.1016/j.cois.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Oppenheim SJ, Baker RH, Simon S, Desalle R. We can’t all be supermodels: The value of comparative transcriptomics to the study of non-model insects. Insect. Mol. Biol. 2015;24(2):139–154. doi: 10.1111/imb.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Peery A, Hall AB, Sharma A, Chen XG, Waterhouse RM, et al. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol. 2014;15(9):459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padrón, A., Molina-cruz, A., Quinones, M., Ribeiro, J. M. C., Ramphul, U. In depth annotation of the Anopheles gambiae mosquito midgut transcriptome. In: Depth annotation of the Anopheles gambiae mosquito midgut transcriptome. (2014) [DOI] [PMC free article] [PubMed]
- 29.Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science (1979) 2015;347(6217):1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prado Sepulveda CC, Alencar RM, Santana RA, Belém de Souza I, D’Elia GMA, Godoy RSM, et al. Evolution and assembly of Anopheles aquasalis’s immune genes: primary malaria vector of coastal Central and South America and the Caribbean Islands. Open Biol. 2023;13(7):230061. doi: 10.1098/rsob.230061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holt RA, Mani Subramanian G, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science (1979) 2002;298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 32.Lau YL, Lee WC, Chen J, Zhong Z, Jian J, Amir A, et al. Draft genomes of Anopheles cracens and Anopheles maculatus: Comparison of simian malaria and human malaria vectors in peninsular Malaysia. PLoS One. 2016;11(6):1–24. doi: 10.1371/journal.pone.0157893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Villegas L, Assis-Geraldo J, Koerich LB, Collier Id TC, Lee Y, Main BJ, et al. Characterization of the complete mitogenome of Anopheles aquasalis, and phylogenetic divergences among Anopheles from diverse geographic zones. PLoS One. 2019 doi: 10.1371/journal.pone.0219523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Melo ES, Wallau GL. Mosquito genomes are frequently invaded by transposable elements through horizontal transfer. PLoS Genet. 2020;16(11):1–26. doi: 10.1371/journal.pgen.1008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science (1979) 2015;347(6217):1–20. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marinotti O, Cerqueira GC, De Almeida LGP, Ferro MIT, Da Silva Loreto EL, Zaha A, et al. The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res. 2013;41(15):7387–7400. doi: 10.1093/nar/gkt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diesel JF, Ortiz MF, Marinotti O, Vasconcelos ATR, Loreto ELS. A re-annotation of the Anopheles darlingi mobilome. Genet. Mol. Biol. 2019;42(1):125–131. doi: 10.1590/1678-4685-gmb-2017-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargas-Chavez C, Longo Pendy NM, Nsango SE, Aguilera L, Ayala D, González J. Transposable element variants and their potential adaptive impact in urban populations of the malaria vector Anopheles coluzzii. Genome Res. 2022;32(1):189–202. doi: 10.1101/gr.275761.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mapalo MA, Arakawa K, Baker CM, Persson DK, Mirano-Bascos D, Giribet G. The unique antimicrobial recognition and signaling pathways in tardigrades with a comparison across ecdysozoa. G3 Genes Genomes Genet. 2020;10(3):1137–1148. doi: 10.1534/g3.119.400734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer WJ, Jiggins FM. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol. Biol. Evol. 2015;32(8):2111–2129. doi: 10.1093/molbev/msv093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316(5832):1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mancini MV, Damiani C, Accoti A, Tallarita M, Nunzi E, Cappelli A, et al. Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 2018;18(1):1–10. doi: 10.1186/s12866-018-1266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, Sharma S, Maurya RK, De TD, Thomas T, Lata S, et al. Salivary glands harbor more diverse microbial communities than gut in Anopheles culicifacies. Parasit Vectors. 2014;7(1):1–7. doi: 10.1186/1756-3305-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gendrin M, Turlure F, Rodgers FH, Cohuet A, Morlais I, Christophides GK. The peptidoglycan recognition proteins PGRPLA and PGRPLB regulate anopheles immunity to bacteria and affect infection by plasmodium. J. Innate Immun. 2017;9(4):333–342. doi: 10.1159/000452797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meister S, Agianian B, Turlure F, Relógio A, Morlais I, Kafatos FC, et al. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5(8):e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao X, Gulati M, Jiang H. Serine protease-related proteins in the malaria mosquito, Anopheles gambiae. Insect. Biochem. Mol. Biol. 2017;176(5):139–148. doi: 10.1016/j.ibmb.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar A, Srivastava P, Sirisena PDNN, Dubey SK, Kumar R, Shrinet J, et al. Mosquito innate immunity. Insects. 2018;9(3):95. doi: 10.3390/insects9030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volz J, Osta MA, Kafatos FC, Müller HM. The roles of two clip domain serine protease in innate immune responses of the malaria vector Anopheles gambiae. J. Biol. Chem. 2005;280(48):40161–40168. doi: 10.1074/jbc.M506191200. [DOI] [PubMed] [Google Scholar]
- 49.Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8(9):1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 50.Rinker DC, Zhou X, Pitts RJ, Rokas A, Zwiebel LJ, Neafsey DE, et al. Antennal transcriptome profiles of anopheline mosquitoes reveal human host olfactory specialization in Anopheles gambiae. BMC Genom. 2013;14(1):1–15. doi: 10.1186/1471-2164-14-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinker DC, Pitts RJ, Zhou X, Suh E, Rokas A, Zwiebel LJ. Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 2013;110(20):8260–8625. doi: 10.1073/pnas.1302562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kouamo MFM, Ibrahim SS, Hearn J, Riveron JM, Kusimo M, Tchouakui M, et al. Genome-wide transcriptional analysis and functional validation linked a cluster of epsilon glutathione s-transferases with insecticide resistance in the major malaria vector anopheles funestus across Africa. Genes (Basel) 2021;12(4):561. doi: 10.3390/genes12040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atoyebi SM, Tchigossou GM, Akoton R, Riveron JM, Irving H, Weedall G, et al. Investigating the molecular basis of multiple insecticide resistance in a major malaria vector Anopheles funestus (sensu stricto) from Akaka-Remo, Ogun State, Nigeria. Parasit. Vectors. 2020;13(1):1–14. doi: 10.1186/s13071-020-04296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edi CV, Djogbénou L, Jenkins AM, Regna K, Muskavitch MAT, Poupardin R, et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10(3):e1004236. doi: 10.1371/journal.pgen.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou D, Liu X, Sun Y, Ma L, Shen B, Zhu C. Genomic analysis of detoxification supergene families in the Mosquito Anopheles sinensis. PLoS One. 2015;10(11):e0143387. doi: 10.1371/journal.pone.0143387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weetman D, Djogbenou LS, Lucas E. Copy number variation (CNV) and insecticide resistance in mosquitoes: Evolving knowledge or an evolving problem? Curr. Opin. Insect Sci. 2018;1(27):82–88. doi: 10.1016/j.cois.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lucas ER, Miles A, Harding NJ, Clarkson CS, Lawniczak MKN, Kwiatkowski DP, et al. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 2019;29(8):1250–1261. doi: 10.1101/gr.245795.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molina D, Figueroa LE. Resistencia metabólica a insecticidas organofosforados en Anopheles aquasalis Curry 1932, municipio Libertador, estado Sucre, Venezuela. Biomédica. 2009;29(4):604–615. doi: 10.7705/biomedica.v29i4.138. [DOI] [PubMed] [Google Scholar]
- 59.Floch H, Fauran P. Susceptibility of culex fatigans and anopheles aquasalis to chlorinated hydrocarbon insecticides in French Guiana. Bull. World Health Organ. 1958;18(4):667–673. [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Villegas L, Assis-Geraldo J, Koerich LB, Collier TC, Lee Y, Main BJ, et al. Characterization of the complete mitogenome of Anopheles aquasalis, and phylogenetic divergences among Anopheles from diverse geographic zones. PLoS One. 2019;14(9):1–22. doi: 10.1371/journal.pone.0219523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Emrich S, Collins F, et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43(D1):D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. 2020;117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amos B, Aurrecoechea C, Barba M, Barreto A, Basenko EY, Bażant W, et al. VEuPathDB: The eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2022;50(D1):D898–D911. doi: 10.1093/nar/gkab929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smit, A., Hubley, R. & Green, P. http://www.repeatmasker.org. RepeatMasker Open-4.0. (2015)
- 66.Permal E, Flutre T, Quesneville H. Mobile genetic elements: Protocols and genomic applications. Methods Mol. Biol. 2012;859:5–7. [Google Scholar]
- 67.Afgan E, Baker D, Batut B, Van Den Beek M, Bouvier D, Ech M, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stanke M, Morgenstern B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33(2):465–467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: User guide. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 70.Campbell MS, Law MY, Holt C, Stein JC, Moghe GD, Hufnagel DE, et al. MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 2014;164(2):513–524. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yandell M, Ence D. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012;13(5):329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 72.Törönen P, Medlar A, Holm L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018;46(W1):W84–W88. doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madden, T. The BLAST sequence analysis tool. BLAST Seq. Anal. Tool. 1–17 (2013).
- 75.Zdobnov EM, Kuznetsov D, Tegenfeldt F, Manni M, Berkeley M, Kriventseva E. v. OrthoDB in 2020: Evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2021;49(D1):D389–D393. doi: 10.1093/nar/gkaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maintainer C, Boutros P. Title generate high-resolution venn and euler plots. BMC Inform. 2022;12:1–7. [Google Scholar]
- 77.Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamura K, Tao Q, Kumar S. Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol. Biol. Evol. 2018;35(7):1770–1782. doi: 10.1093/molbev/msy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA. 2012;109(47):19333–19338. doi: 10.1073/pnas.1213199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34(7):1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 81.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 83.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 84.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Anopheles aquasalis genome is deposited in GeneBank under accession GCA_002846955.1 (https://www.ncbi.nlm.nih.gov/data-hub/genome/GCA_002846955.1/). Annotated proteins are under submission to GeneBank and can be downloaded as Additional File 4 (CDS) and Additional File 5 (Proteins).