Abstract
Sulfoquinovose (SQ, 6-deoxy-6-sulfoglucose) is a sulfosugar that is the anionic head group of plant, algal, and cyanobacterial sulfolipids: sulfoquinovosyl diacylglycerols. SQ is produced within photosynthetic tissues, forms a major terrestrial reservoir of biosulfur, and is an important species within the biogeochemical sulfur cycle. A major pathway for SQ breakdown is the sulfoglycolytic Embden–Meyerhof–Parnas pathway, which involves cleavage of the 6-carbon chain of the intermediate sulfofructose-1-phosphate (SFP) into dihydroxyacetone and sulfolactaldehyde, catalyzed by class I or II SFP aldolases. While the molecular basis of catalysis is understood for class I SFP aldolases, comparatively little is known about class II SFP aldolases. Here, we report the molecular architecture and biochemical basis of catalysis of two metal-dependent class II SFP aldolases from Hafnia paralvei and Yersinia aldovae. 3D X-ray structures of complexes with substrate SFP and product dihydroxyacetone phosphate reveal a dimer-of-dimers (tetrameric) assembly, the sulfonate-binding pocket, two metal-binding sites, and flexible loops that are implicated in catalysis. Both enzymes were metal-dependent and exhibited high KM values for SFP, consistent with their role in a unidirectional nutrient acquisition pathway. Bioinformatic analysis identified a range of sulfoglycolytic Embden–Meyerhof–Parnas gene clusters containing class I/II SFP aldolases. The class I and II SFP aldolases have mututally exclusive occurrence within Actinobacteria and Firmicutes phyla, respectively, while both classes of enzyme occur within Proteobacteria. This work emphasizes the importance of SQ as a nutrient for diverse bacterial phyla and the different chemical strategies they use to harvest carbon from this sulfosugar.
Keywords: sulfur cycle, structural biology, enzymes, bioinformatics, enzyme mechanism
Sulfoglycolysis is a catabolic process through which the sulfosugar sulfoquinovose (SQ) is broken down to smaller fragments that can enter central carbon metabolism (1, 2, 3). SQ is produced by phototrophs as part of the sulfolipid sulfoquinovosyl diacylglycerol and forms part of the photosynthetic thylakoid membranes of all plants and most cyanobacteria (4, 5). With an estimated 10-billion-tons synthesized annually, SQ comprises a major organosulfur reservoir that rivals that of the sulfur amino acids cysteine and methionine (6). Microbial sulfoglycolysis pathways therefore represent key links in the biogeochemical carbon and sulfur cycles.
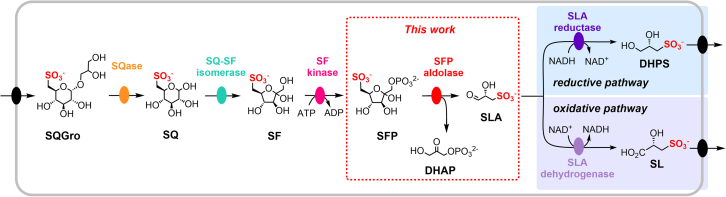
The first sulfoglycolysis pathway identified for the breakdown of SQ was a sulfoglycolytic Embden–Meyerhof–Parnas (sulfo-EMP) (7, 8) pathway in Escherichia coli that involves reactions and sulfonate intermediates that mirror the reactions and phosphorylated intermediates in the classical EMP glycolysis pathway (Fig. 1). Initially, SQ is released by the sulfoquinovosidase-catalyzed hydrolysis of sulfoquinovosyl diacylglycerol or its delipidated form, sulfoquinovosyl glycerol (9, 10). The sulfo-EMP pathway involves isomerization of SQ to sulfofructose (SF), by SQ-SF isomerase; phosphorylation of SF to SF-1-phosphate (SFP), by ATP-dependent SF kinase; and retroaldol cleavage of SFP to sulfolactaldehyde (SLA) and dihydroxyacetone phosphate (DHAP) by SFP aldolase (11). The fate of SLA depends on the specific organism, and it may be reduced to dihydroxypropane-1-sulfonate (DHPS) by NADH-dependent SLA reductase (12) or oxidized to sulfolactate (SL) by SLA dehydrogenase (13, 14). DHPS and SL are both excreted, where they can be biomineralized to inorganic sulfur by C3-organosulfonate–degrading bacteria (15, 16, 17).
Figure 1.
The sulfoglycolytic Embden−Meyerhof−Parnas (sulfo-EMP) pathway enables the catabolism of sulfoquinovosyl glycerol (SQGro) and sulfoquinovose (SQ) to 2,3-dihydroxypropanesulfonate (DHPS) (reductive pathway) or sulfolactate (SL) (oxidative pathway). Class I and class II 6-deoxy-6-sulfofructose-1-phosphate (SFP) aldolases catalyze the scission of the C3-C4 bond to give dihydroxyacetone phosphate (DHAP) and sulfolactaldehyde (SLA). SF, sulfofructose; SFP, SF-1-phosphate.
Within the sulfo-EMP pathway, the critical ‘lysis’ step to break the six-carbon chain into two three-carbon fragments is catalyzed by SFP aldolase. The SFP aldolase (YihT) encoded by the E. coli sulfo-EMP cluster is a class I aldolase, which catalyzes the reaction via a Schiff base intermediate formed with an active site lysine (11). Evidence for this mechanism was obtained through the determination of 3D structures of a SFP aldolase YihT with the active site lysine covalently attached to the acyclic SFP substrate and DHAP product, each as Schiff bases (11). Highlighting the functional segregation of the sulfo-EMP and EMP pathways, YihT is inactive on the structurally related glycolytic intermediate fructose bisphosphate (FBP), showing that YihT can discriminate between sulfoglycolytic and glycolytic/gluconeogenic intermediates (11). Subsequently, Zhang et al. identified sulfoglycolytic organisms that utilize the same sulfo-EMP pathway using nonhomologous enzymes and which use a class II aldolase (SqiA) (13). This enzyme was proposed to require divalent metal ions for activity (3). However, little is known of the metal ion dependence, structural basis of sulfonate binding, and organosulfonate selectivity of class II SFP aldolases.
Here, we identify class II SFP aldolases from Hafnia paralvei and Yersinia aldovae and report their biochemical and structural characterization. These enzymes exhibit metal-ion–dependent SFP aldolase activity. Crystal structures of the metalloenzymes in complex with bound ligands, SFP and DHAP, reveal that these SFP aldolases form homotetrameric complexes and illuminate the molecular details of substrate binding, sulfonate recognition, and metal interactions. We apply sequence similarity network (SSN) analysis to study the distribution of class I and II SFP aldolases, showing them to have mutually exclusive occurrence within gram-positive Actinobacteria and Firmicutes, respectively, and to co-occur within the gram-negative Proteobacteria.
Results
Identification and kinetic characterization of class II SFP aldolases
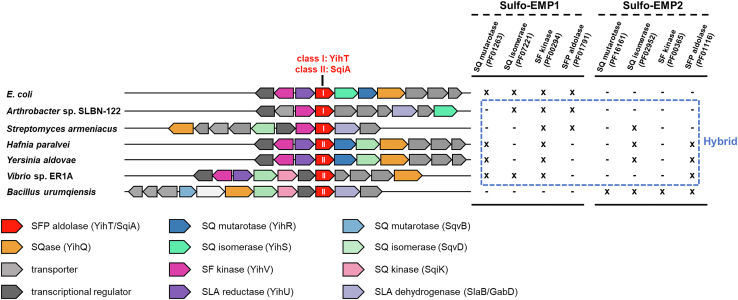
We conducted a survey of sulfoglycolytic pathways using a MultiGeneBLAST (18) search of the nonredundant protein set of the National Center for Biotechnology Information for gene clusters that contain sulfo-EMP genes but which lack homologs of the class I SFP aldolase from E. coli. Examination of the genome neighborhood led to identification of genes encoding class II aldolases that could potentially encode SFP aldolases. We selected H. paralvei and Y. aldovae for further study (Fig. 5).
Figure 5.
Representative proposed gene clusters for variant Embden−Meyerhof−Parnas sulfoglycolysis pathways.Escherichia coli is a representative sulfo-EMP1 pathway organism containing YihR-YihS-YihV-YihT; Bacillus urumqiensis is a representative sulfo-EMP2 pathway organism containing SqvB-SqvD-SqiK-SqiA. Boxed in blue are hybrid organisms containing various combinations of sulfo-EMP1/2 genes. sulfo-EMP, sulfoglycolytic Embden–Meyerhof–Parnas.
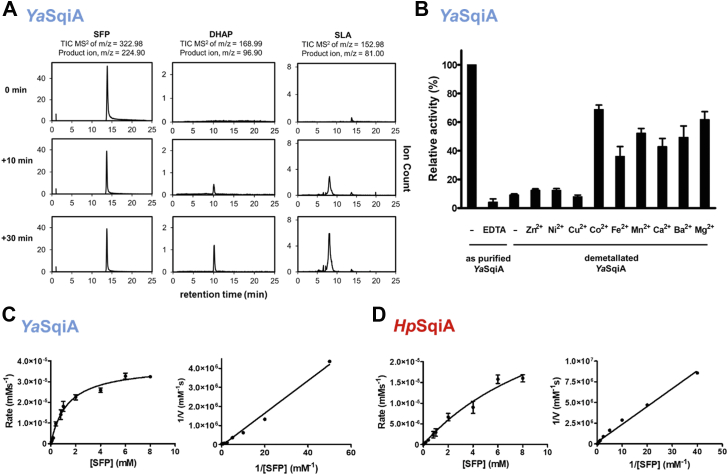
We synthesized genes for the putative SFP aldolases from H. paralvei (WP_234610022.1) and Y. aldovae (WP_042545622.1) and expressed and then purified the corresponding recombinant proteins (HpSqiA and YaSqiA, respectively) (Fig. S1). To establish whether these proteins are SFP aldolases, we incubated reaction mixtures containing YaSqiA or HpSqiA and SFP in Tris.HCl buffer containing MgCl2. Analysis of reaction mixtures by LC-MS using a triple quadrupole (QqQ) mass spectrometer in product ion mode revealed the time-dependent formation of SLA and DHAP (Figs. 2A and S2), demonstrating that YaSqiA and HpSqiA are SFP aldolase enzymes. These enzymes exhibited activity across the pH range 6.5 to 8.5, with optimum activity for YaSqiA at pH 8 and a flat pH profile for HpSqiA (Fig. S3).
Figure 2.
Biochemical characterization of SFP aldolases.A, HPLC mass spectrometry (triple quadrupole, QqQ) chromatograms showing YaSqiA- (class II aldolase) catalyzed conversion of 6-deoxy-6-sulfofructose-1-phosphate (SFP) to 3-sulfolactaldehyde (SLA) and dihydroxyacetone phosphate (DHAP) at time (t) = 0, 10, 30 min. B, metal ion dependence of YaSqiA. As-purified protein was assessed for activity using SFP as substrate in the absence (−) or presence of EDTA. Demetalated YaSqiA was obtained by treatment with Chelex resin and supplemented with various divalent cations and assessed for activity with SFP as substrate. C and D, Michaelis–Menten and Lineweaver–Burk plots for reaction rates measured using SFP as substrate, for YaSqiA and HpSqiA, respectively. In all experiments, error bars represent SEM, n = 3.
Class II aldolase enzymes are metal-dependent (19). The metal dependency of both YaSqiA-His6 and HpSqiA-His6 was studied by addition of EDTA to the reaction mixtures containing 4.4 μM YaSqiA-His6 (8.8 μM HpSqiA-His6) as expressed from E. coli (i.e. containing endogenous metal) and 0.5 mM SFP. Addition of EDTA to reaction mixtures containing YaSqiA-His6 (Fig. 2B) or HpSqiA-His6 (data not shown) decreased the rate of DHAP production. These results suggest that both YaSqiA and HpSqiA enzymes are metalloenzymes.
To explore the metal-dependent activity of these SFP aldolases, purified YaSqiA-His6 and HpSqiA-His6 were passed over the metal-chelating resin Chelex to remove divalent metals, to give what we denote as demetalated proteins. Demetalated YaSqiA-His6 exhibited 10-fold lower activity than as-purified (but not demetalated) YaSqiA-His6 (Fig. 2B). Enzyme activity for demetalated YaSqiA-His6 was examined in the presence of 50 M M2+ in reaction mixtures containing 25 mM Tris.HCl, 25 mM NaCl, 0.5 mM SFP, and 4.4 M YaSqiA. Maximum activity was observed in the presence of Co2+ followed by Mg2+ > Mn2+ ≈ Ba2+ > Ca2+ > Zn2+ ≈ Ni2+ > Cu2+ > no metal. Demetalated HpSqiA-His6 was inactive and did not recover activity when incubated with different divalent metal ions, suggesting that demetalation caused permanent inactivation, possibly through destabilization of the protein. Enzymes lacking His6-tags were less stable: demetalated HpSqiA and YaSqiA lacking His6 tags had no activity, and addition of M2+ ions did not recover any activity in these proteins.
Given that the optimum activity for both enzymes was obtained using as-purified His6-tagged proteins, we used these enzymes to measure kinetic parameters using LCMS. Initial rate data was fit to the Michaelis–Menten equation to give kinetic parameters (Table 1). YaSqiA-His6 exhibited saturation and gave kcat = 0.085 s−1, KM = 1.33 ± 0.17 mM, and kcat/KM = 0.063 s−1 mM−1 (Fig. 2C) and HpSqiA-His6 did not exhibit saturation, preventing determination of kcat and KM values but still allowing determination of kcat/KM = 0.0052 s−1 mM−1 (Fig. 2D). We independently confirmed the LCMS rate measurements and obtained a commensurate estimate of kcat/KM (at [S] << KM) using a stopped assay in which NAD(P)H-dependent SLA dehydrogenase GabD was used to quantify SLA production (see Experimental procedures) (20). The specificity of both enzymes was examined with the structural analog fructose bisphosphate, a metabolic intermediate from glycolysis/gluconeogenesis. Neither enzyme catalyzed formation of any detectable product, and thus we conclude that both class II aldolase enzymes (YaSqiA and HpSqiA) are specific for SFP.
Table 1.
Michaelis–Menten kinetic parameters for YaSqiA- and HpSqiA-catalyzed formation of DHAP from SFP
| Parameter | YaSqiA | HpSqiA |
|---|---|---|
| Vmax (mM s−1) | (3.78 ± 0.16) × 10−5 | ---- |
| KM (mM) | 1.33 ± 0.17 | ---- |
| kcat (s−1) | 0.085 ± 0.003 | ---- |
| kcat/KM (s−1 mM−1) | 0.063 ± 0.008 | 0.0052 ± (0.17 × 10−3) |
| R2 | 0.977 | 0.991 |
Overall structure of class II SFP aldolases
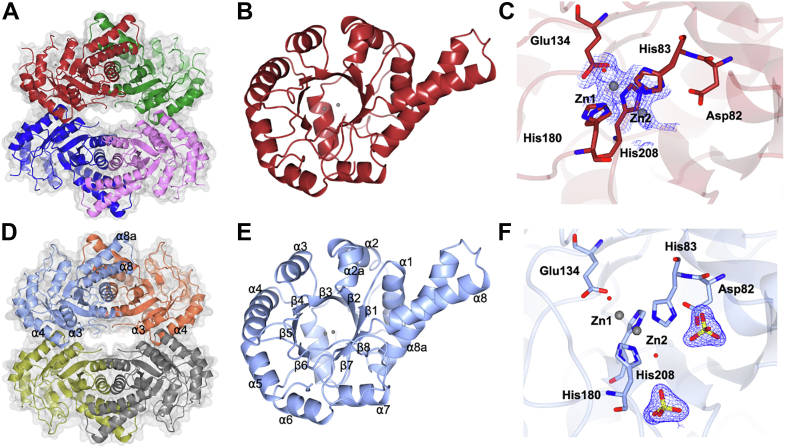
To elucidate the structural basis of catalysis, 3D X-ray crystal structures of YaSqiA and HpSqiA were obtained. HpSqiA crystallized in the I222 space group with a metal atom bound with a single chain in the asymmetric unit and diffracted to 1.7 Å resolution (Table S1). YaSqiA crystallized in the I2 space group with a single metal bound, also diffracting to 1.7 Å resolution. In both cases, analysis of symmetry-related molecules revealed a dimer-of-dimers assembly (Fig. 3, A and D). This was consistent with both proteins adopting tetramers in the solution state as shown by size-exclusion chromatography-multiangle laser light scattering analysis (Fig. S1). The four subunits in the SqiA homotetramer are arranged in a fashion distinct from the tetrameric oligomerization displayed by FBP aldolase from Mycobacterium tuberculosis (4DEL.pdb) or tagatose bisphosphate (TBP) aldolase from Thermus aquaticus (1RV8.pdb). The tetrameric solution state species for SFP aldolases observed here are also different from the homodimeric FBP aldolase assemblies reported for E. coli (1DOS.pdb) and Hafnia pylori (5UCK.pdb) (21) (Fig. S4).
Figure 3.
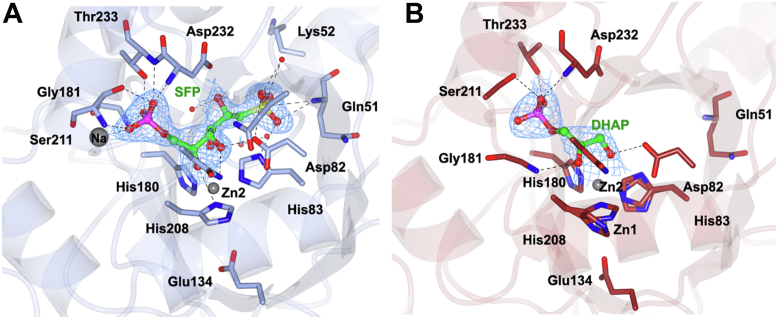
3D structures of metal-dependent class II SFP aldolases.A, crystal structure of HpSqiA•Zn showing tetrameric solution state assembly. B, overall fold of HpSqiA monomer showing dimerization domain comprised of helices α8 and α8a. C, active site of HpSqiA•Zn showing two mutually exclusive metal-binding sites and the coordinating residues. D, crystal structure of YaSqiA showing tetrameric assembly. E, overall fold of YaSqiA monomer. F, active site of YaSqiA•Zn•sulfate showing two mutually exclusive metal-binding sites and the location of bound sulfate anions. Electron density in blue corresponds to 2Fo − Fc map contoured at 1σ. SFP, sulfofructose-1-phosphate.
The class II SFP aldolases adopt (α/β)8 TIM-barrel folds (Fig. 3, B and E) that are characteristic of other bacterial class II aldolases, including FBP and TBP aldolases. Pairwise DALI analysis identified the closest structural homologs: FBP aldolases from E. coli (PDB ID: 5VJD with a DALI z score of 34.2 and 1.9 Å rmsd and 25% sequence ID) (22), H. pylori (PDB ID: 5UCK with a DALI z score of 39.7 and 1.3 Å rmsd and 38% sequence ID) (21), and T. aquaticus (PDB ID: 1RV8, DALI z score of 39.5, 1.6 Å rmsd and 48% sequence ID) (23). Structure-based sequence alignment with these FBP aldolases shows high conservation of structural motifs, as well as catalytic and metal-binding residues, despite the low sequence identities in some cases (Fig. S5).
Class II FBP and TBP aldolases exhibit reciprocal interactions that are deemed essential for both their structures and catalysis (24, 25, 26). Likewise, at the dimer interfaces of HpSqiA and YaSqiA, the extended antiparallel helices α8 and α8a of one subunit make tight hydrophobic interactions with corresponding helices of partner subunit. The two monomers within the subunit dimer bury the small hydrophobic residues Val243, Leu256, and Leu260 and aromatic Phe239 and Tyr254 at the core of their dimerization motif. Two dimers then stack to give the final dimer-of-dimers through interactions of α3 and α4 helices from adjacent subunits (Fig. 3, A and D). Analysis of the YaSqiA and HpSqiA structures using the PISA server (27) shows the homotetrameric organization buries 9100 and 12,270 Å2 for HpSqiA and YaSqiA, respectively, as a result of intersubunit interactions for both proteins. This corresponds to 19 to 24% of the total tetramer surface, which is in the same range as seen for other tetrameric class II aldolases, despite the dimer-of-dimers arrangement of SqiA enzymes being distinct from tetrameric assemblies seen for TBP aldolase from T. aquaticus (1RV8.pdb) or FBP aldolase from M. tuberculosis (4DEL.pdb) (Figs. S4 and 6).
Figure 6.
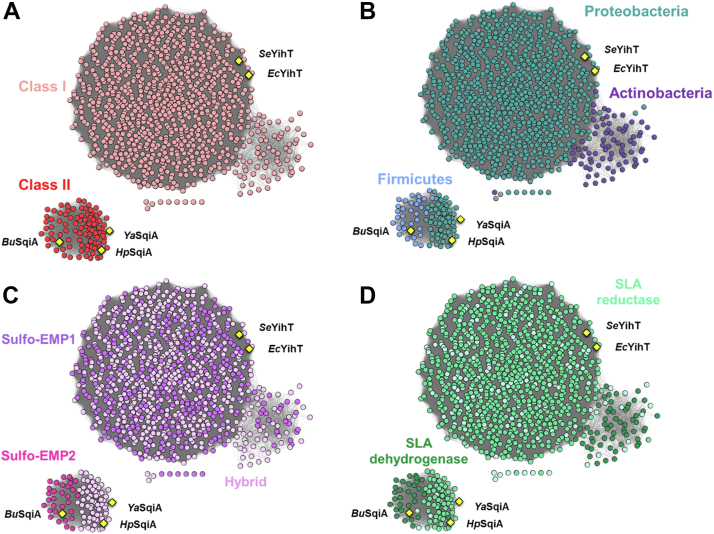
Sequence similarity networks of SFP aldolases at alignment score 90 (i.e., ≥ 52.5% similarity).A, the large cluster contains 1000 class I SFP aldolases and the small cluster contains 102 class II SFP aldolases. B, SSN colored based on phyla of bacteria containing SFP aldolase, (C) SSN colored based on genetic context of SFP aldolase gene within proposed sulfo-EMP1, sulfo-EMP2, or hybrid sulfo-EMP1/2 gene clusters, (D) SSN colored based on presence of SLA reductase (YihU) or SLA dehydrogenase (SlaB/GabD) in the proposed gene cluster. SFP, sulfofructose-1-phosphate; SLA, sulfolactaldehyde; SSN, sequence similarity network; sulfo-EMP, sulfoglycolytic Embden–Meyerhof–Parnas.
Both SFP aldolases crystallize with two bound divalent metal ions, apparently recruited during recombinant expression. Anomalous difference electron density maps identified two mutually exclusive metal sites at different occupancies (Fig. S7A). In order to uncover the identity of the bound metal, an emission spectrum for HpSqiA crystal was recorded using a beam with the energy of 12.66 keV (selenium absorption K edge). On the basis of X-ray fluorescence scans of the crystals used for data collection, zinc was modeled in as the divalent metal. The electron density for residues 138 to 151 and an active site histidine containing β6-α8 loop (residues 181–186) were missing in the structure of ligand-free HpSqiA, suggesting that these may be flexible loops involved in substrate binding and catalysis as seen for other class II aldolases (21, 23, 28). A ligand-free structure of HpSqiA was obtained that contained two metal atoms: Zn2+ at site Zn1 was modeled at occupancy of 0.8 and Zn2+ at site Zn2 at a lower occupancy of 0.2. Zn2+ at site Zn1 is buried at the core of the β-barrel and binds to three histidines: His83 (Nε2 at 2.1 Å), His180 (Nε2 at 2.2 Å), and His208 (Nδ1 at 2.2 Å); a glutamate, Glu134 at 2.0 Å, and a water (2.6 Å). Zn2+ at site Zn2 resides in an alternate, surface-accessible site 3.4 Å away from Zn1. The modeling of this second zinc center required rotation of imidazole side chains of His83 and His208 to engage the metal. Zn2+ at Zn2 is bound by His83 (Nε2 at 1.9 Å), His208 (Nδ1 at 2.8 Å), and three waters each at 3.4 Å. His 180 within flexible β6-α8 loop does not bind metal at this site. The presence of zinc ion at the buried Zn1 site precludes binding at the Zn2 site and requires conformational transitions and side chain rotations of active site histidines, His83 and His208 about the Cβ-Cγ bonds. Similar metal-binding interactions and side chain rotations of corresponding His110 and His226 were previously seen in E. coli FBPA (1DOS.pdb) where two divalent zinc ions were present in mutually exclusive sites 3.2 Å apart from each other and Zn1/Zn2 were modeled at occupancies of 0.6/0.4 (22).
The YaSqiA structure revealed the metal-binding interactions at these two sites and also density for two bound sulfate ions at the active site along with displacement of active site β6-α8 loop containing His180 (Figs. 3C and S6). Within the YaSqiA•Zn2+•sulfate structure, Zn2+ at site Zn1 (buried at the core of the β-barrel) was modeled at 0.3 occupancy, with Zn2+ bound to three histidines: Nε2 of His83 at a distance of 2.7 Å, His180 (Nε2 at 2.5 Å), and His208 (Nδ1 at 2.6 Å); a glutamate, Glu134 (3.4 Å); and a water molecule (2.6 Å). Zn2+ at site Zn2 (located 1.9 Å from Zn1) was modeled at higher 0.5 occupancy and sits at a relatively surface-exposed site in a similar position to the two metal ions bound in E. coli FBP aldolase (PDB ID: 5VJD) (Fig. S7B). The zinc ion binds three histidines: His83 (Nε2 at 2.1 Å), His180 (Nε2 2.0 Å), His208 (Nδ1 1.9 Å); and a water (2.6 Å). The two sulfate ions, originating from the crystallization buffer, are positioned 8.5 Å apart and occupy opposite sides of the Zn2 center. One sulfate occupies the same position as phosphate seen in 5UCK.pdb (Fig. S7C) (21), with the sulfate oxygens engaged in H-bonding interactions with side chains of Ser211 (at 2.6 Å) and Thr233 (2.7 Å), Nδ1 of His180 (3.1 Å), and backbone amides of Gly181, Gly210, and Asp232 and a sodium cation. The second sulfate H-bonds to active site His83 (2.8 Å); the backbone amides of Gln51 (2.9 Å) and Lys52 (3.0 Å); and four water molecules.
SFP–aldolase complex and in-crystal cleavage to give DHAP
Soaking of a crystal of YaSqiA with the SFP substrate gave diffraction data to a resolution of 2.0, and the anomalous map revealed the location of the bound metal (Table S2). The Zn2+ metal cofactor occupies the Zn2 site at full occupancy and is braced by His83 (2.1 Å), His180 (2.0 Å), and His208 (2.0 Å), the substrate C2 carbonyl (2.4 Å), and a water molecule (2.3 Å) (Fig. 4A). Next to site Zn2, contiguous density corresponding to an open-chain sugar molecule was observed and SFP was modeled at an occupancy of 0.9 and 0.8 in the two subunits in the asymmetric unit. The hexose sugar makes extensive interactions with the active site residues: C2-carbonyl interacts with backbone amide of Gly209 (3.5 Å) and Zn2+ at site Zn2, the C3-hydroxyl H-bonds to Asp82 (2.5 Å) and Asn230 (3.0 Å), and the C5-hydroxy binds Asp232 (2.4 Å) and a water molecule (2.5). The phosphate-binding pocket includes H-bonding interactions with the side chains of Ser211 (at 2.7 Å) and Thr233 (2.6 Å), as well as backbone amides of Gly181, Gly210, and Asp232, and includes a sodium cation. Collectively, these interactions position SFP near the Zn2 site, poised for C3-C4 retro-aldol cleavage to give DHAP and SLA. The structure of the YaSqiA•Zn2+•SFP complex reveals details of sulfonate recognition: one of the sulfonate oxygens of SFP binds backbone amide of Gln51 (3.0 Å) and side chain Nδ of His83 (2.8 Å), a second oxygen makes H-bonding interactions with backbone amide of Lys52 and a water molecule (both at 3.1 Å), and the third sulfonate oxygen binds two water molecules in the sulfonate pocket (at 2.6 and 2.8 Å).
Figure 4.
3D crystal structures of SFP aldolase complexes with substrate SFP and in-crystal retroaldol cleavage of SFP to DHAP.A, close-up view of YaSqiA•Zn•SFP complex showing active site interactions with the substrate SFP. B, close-up view of HpSqiA•Zn•DHAP active site showing interactions with the C3 product DHAP after cleavage. Electron density in blue mesh corresponds to 2Fo − Fc map contoured at 0.9σ. DHAP, dihydroxyacetone phosphate; SFP, sulfofructose-1-phosphate.
The location and interactions of SFP are largely conserved with those observed for FBP in the class II FBP aldolase from E. coli (5UD3.pdb). However, notable differences exist in the binding of the 6-phosphate and the 6-sulfonate groups (Fig. S8). First, the 6-sulfonate of SFP projects into a different pocket to interact with the backbone amides of Gln51/Lys52 and side chain of active site His83. Compared to this, the oxyanion interactions of 6-phosphate of fructose bisphosphate observed previously in class II FBP aldolases are conserved and extensive. For example, FBP complexes of class II FBP aldolases from H. pylori (E142A variant, 5UCP.pdb) and E. coli FBPA (5UD3.pdb), one of 6-phosphate oxygen binds three water molecules, while the other two oxygens are stabilized by Arg259 (2.9 Å) and Ser49 (2.4 Å) within the protomer chain, while Arg280 (2.7 Å) protrudes into the substrate-binding pocket from a partner protomer. A second difference is that in SFP aldolases, any such reciprocal subunit interactions in 6-sulfonate pocket are not possible because Arg280 is replaced by Ala257. A third difference involves the 1-phosphate of SFP/FBP aldolases: E. coli FBPA contains a cationic Lys184 that projects into the substrate-binding site and binds the 1-phosphate group of FBP (5UD3.pdb), while SqiA lacks this cationic lysine and this role is fulfilled by a sodium cation, which is bound by backbone carbonyls of active site residues Gly209, Ser211, Ala179, and Gly181 and may be important for loop closure (see below).
A longer soak of HpSqiA crystals at saturating concentration of SFP enabled observation of the product DHAP, arising from cleavage of SFP by the enzyme in the crystalline state (Table S2). DHAP was modeled into the electron density at an occupancy of 0.7 (Fig. 4B), but no density was observed for the other product, the C3 sulfonate SLA. The interactions of the phosphate group of DHAP are identical to those observed for SFP discussed above. Zn2+ at site Zn2 was modeled at occupancy of 0.7 and binds the DHAP carbonyl and C3 hydroxyl at distances of 2.7 and 2.4 Å. Asp82 makes H-bonding interactions with the C3 hydroxyl (2.6 A). Zn2+ at site Zn1 was modeled at occupancy 0.3 and is bound by histidines, His83, His180, and His208, and Glu134 as seen for the SFP complex.
The class II SFP aldolases possess two flexible loops, β6-α8 (residues 179–190) that contains the active site His180 and the accompanying β5-α7 loop (residues 141–153). These loops undergo conformational transitions initiating lid closure over the active site upon ligand binding, as seen previously for class II FBP aldolases (21, 23, 28). In the ligand bound YaSqiA•Zn•SFP and HpSqiA•Zn•DHAP complexes, the previously disordered active site β6-α8 loop (stabilized through aforementioned interactions with the Na+ cation and triose/hexose sugars) closes over the active site. However, part of the other surface loop remains disordered with electron density for residues 142 to 149 not observed in our structures (Fig. S9).
Bioinformatics analysis of sulfo-EMP gene clusters in representative organisms
BLAST search of the UniProt database using E. coli class I SFP aldolase YihT and the class II SFP aldolase YaSqiA, respectively, retrieved representative class I (YihT, belonging to PFAM protein family PF01791) and class II (SqiA, belonging to PFAM protein family PF01116) SFP aldolases for a wide range of bacteria. The retrieved lists were inspected and curated to exclude those sequences that were not located within sulfo-EMP gene groupings. This yielded a list of 1000 class I YihT homologs and 102 class II SqiA homologs.
To define which variant sulfoglycolytic pathways the YihT/SqiA homologs occur within, we used the combined list of 1102 YihT/SqiA sequences to retrieve the genome neighborhood diagrams within a ±10 open reading frame window of the yihT/sqiA genes using the Enzyme Function Initiative web tools (https://efi.igb.illinois.edu/efi-gnt/) (29). Representative examples of putative sulfoglycolytic gene clusters are shown in Figure 5. The first genome neighborhood diagram (GND) shows the sulfo-EMP pathway in E. coli and is comprised of SQ mutarotase (PF01263), SQ isomerase (PF07221), SF kinase (PF00294), and class I SF aldolase (PF01791), with this combination of PFAM members denoted sulfo-EMP1. The last GND is that of Bacillus urumqiensis, and this encodes a nonhomologous series of functionally equivalent enzymes, namely SQ mutarotase (PF16161), SQ isomerase (PF02952), SF kinase (PF00365), and class II SF aldolase (PF01116), with this combination of PFAM members denoted sulfo-EMP2, as proposed by Liu et al. (13). Figure 5 shows that there are a range of hybrid gene clusters that contain various combinations of sulfo-EMP1 and sulfo-EMP2 PFAM members and can include either class I (PF01791) or class II (PF01116) SFP aldolases.
The combined list of 1102 YihT/SqiA sequences were used to generate a series of SSNs (30) at various minimum alignment scores (AS) (Fig. S10). At AS 50 and 80, two clusters are evident, with little separation of Actinobacteria and Proteobacteria. At AS = 90, Actinobacteria mostly segregate from Proteobacteria into a subcluster, with limited formation of singletons. At AS = 100, segregation of most Actinobacteria is almost complete; however, some Actinobacteria still cluster with Proteobacteria, with an increased number of singletons. We chose the compromise YihT/SqiA SSN with AS = 90 for further analysis (Fig. 6A). Figure 6B shows the SSN colored for PFAM PF01791 (YihT) or PF01116 (SqiA) and reveals that the chosen AS results in clean segregation of class I and class II SFP aldolases.
To perform a comprehensive analysis of the genome neighborhoods of each gene encoding YihT/SqiA SFP aldolases, we used the Enzyme Function Initiative Genome Neighborhood Tool to retrieve the gene neighbor accession codes from the GNDs described above. An all-by-all pairwise BLAST was conducted of the neighbors and the output used to generate a SSN of neighbors (SSNN) (31). A minimum alignment score was selected that resulted in clustering of the neighbors into isofunctional groups, as assessed by protein family (PFAM) number distribution (Fig. S11). We then colored the YihT/SqiA SSN based on the occurrence of various sulfoglycolytic pathways. Figure 6C shows that the sulfo-EMP1 PFAM combination is present exclusively within Proteobacteria and Actinobacteria, while the sulfo-EMP2 PFAM combination is found only within Firmicutes. Hybrid sulfo-EMP1/2 protein combinations were distributed across Proteobacteria and Actinobacteria (Fig. 6C). Coloring of the YihT/SqiA SSN based on the occurrence of SLA reductase or dehydrogenase shows that SLA dehydrogenase (which results in the production of DHPS) associations with Firmicutes and most Actinobacteria, while SLA reductase (which results in SL) associates with Proteobacteria (Fig. 6D).
Phylogenetic analysis of SFP and FBP aldolases in assorted organisms
To explore the phylogenic relationships of class I and II SFP and FBP aldolases, we selected a subset of taxonomically diverse YihT and SqiA homologs and supplemented these with FBP aldolases selected from eukaryotes, bacteria, and archaea. The class I FBP aldolases included a subset of archaeal enzymes that have been classified as type IA (32). Class II FBP aldolases have been classified into types IIA and IIB on the basis of sequence (33). The rectangular phylogenetic tree reveals good segregation of class I and II aldolases. It also shows that class I and II SFP aldolases cluster into groups that are separate from the class I and II FBP aldolases (Fig. S12).
Discussion
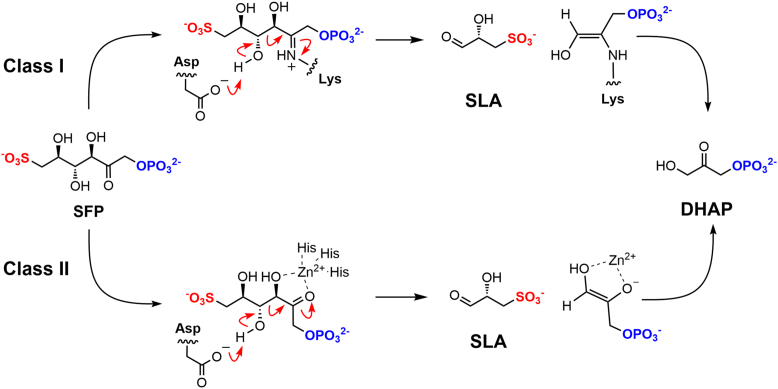
Aldolases are classified into two mechanistically distinct groups. The class I aldolases operate through a multistep mechanism involving a covalent Schiff base intermediate formed with a conserved active site lysine, while class II aldolases are metalloenzymes containing divalent cations as cofactors that act as Lewis acid catalysts. Experimentally determined 3D X-ray structures of Salmonella enterica class I SFP aldolase in complex with SFP or the triose phosphate product DHAP revealed Schiff-base intermediates defining the retroaldol reaction coordinate (Fig. 7, upper pathway) (13). The 3D structures of the class II SFP aldolases of YaSqiA and HpSqiA show a Zn2+ in the active site cleft and identified the binding modes of substrate SFP and product DHAP. The structural data is consistent with a mechanism involving the active site Zn2+ coordinating with the substrate and promoting the formation of a metal enolate intermediate (Fig. 7, lower pathway). Enzyme activity was considerably attenuated in the presence of the chelator EDTA, consistent with a role in catalysis for Zn2+.
Figure 7.
Proposed mechanism for class I and class II SFP aldolases. SFP, sulfofructose-1-phosphate.
The presence of two Zn2+ ions in the structures of YaSqiA and HpSqiA is similar to the bimetallic structures observed with E. coli class II FBP aldolase (22, 34). Within the E. coli enzyme, Zn2+ in the surface accessible site (Zn2) plays an unambiguous role in catalysis. However, the role of the second Zn2+ at site Zn1 is ambiguous, and it has been suggested to play a structural or activating role (34). As for the structures obtained for SqiA enzymes described here, metal binding to the E. coli protein is mutually exclusive (22), with rotation of the orientation of coordinating histidine residues required to allow metal binding at the catalytic Zn2 site.
Compared to class II FBP aldolases, the kcat/KM values for SFP as substrate for both bacterial SFP aldolases are significantly lower, arising from both lower kcat and a KM value in the millimolar range, whereas both class I and class II FBP aldolases such as from rabbit muscle (35), Staphylococcus aureus (35), E. coli (25), and Giardia lamblia (36) have KM values for FBP in the low micromolar range. Notably, the class I SFP aldolase YihT from E. coli also has a KM value for SFP in the millimolar range (11). The high KM values for SFP may reflect the roles of both enzymes in a unidirectional sulfoglycolysis pathway where flux occurs only in the catabolic direction, allowing flux through the pathway to scale with substrate concentration. On the other hand, FBP aldolases operate within the EMP pathway, which can operate in both glycolytic and gluconeogenic directions, and contains branchpoints that lead to the pentose phosphate pathway and cell wall biogenesis. A lower KM value for FBP for FBP aldolases will result in saturation as substrate concentrations rise, causing accumulation of substrate and allowing metabolite overflow and partitioning of carbon intermediates into other pathways.
The two SFP aldolases studied exhibit unexpectedly low kcat values. Structural analysis reveals incomplete 0.5 occupancy in the catalytic Zn1 site and the other Zn2 site. Possibly, recombinant expression within E. coli results in incomplete loading of Zn2+, which may benefit from an endogenous loading system in the original host organisms. We were unable to fully reconstitute activity by addition of Zn2+ after removal of bound metal using a chelating resin and observed that Co2+ was better able to restore activity, although activity did not return to levels seen with endogenous metal loading. The assignment of the native metal to class II aldolases and the study of their metal ion dependence is a subject of some complexity. Galkin et al. (36) showed that FBP aldolase from Giardia lambia containing Zn2+ could be only partially inactivated with EDTA and extensive dialysis and that activity could not be restored by addition of Zn2+. Complicating the analysis further, high concentrations of Zn2+ inhibited enzyme catalysis (36). In the case of the class II FBP aldolase of T. aquaticus, EDTA caused inactivation, and Co2+ but not Zn2+ could restore activity (37). We propose that incomplete loading of our enzymes with Zn2+ occurs during expression, and an inability to reconstitute Zn2+ into demetalated enzyme by inclusion into buffer may contribute to the low kcat value. Additionally, while we assign Zn2+ as the metal present in both metal-binding sites in the 3D structures determined by X-ray crystallography, the metal(s) present natively within the host organisms are unknown.
Our phylogenetic analysis reveals that broadly, class I SFP aldolases are more abundant in sequenced bacterial genomes than class II SFP aldolases. Class I SFP aldolases occur in gram-negative Proteobacteria and gram-positive Actinobacteria, while class II SFP aldolases are present in gram-negative Proteobacteria and gram-positive Firmicutes. It has been proposed that the sulfo-EMP1 and sulfo-EMP2 pathways, which use distinct isozymes for SQ degradation, arose by convergent evolution (13). The mutually exclusive distribution of the two pathways in Firmicutes and Actinobacteria provides some support for this hypothesis in these two phyla. A related but alternative hypothesis is that the isozyme pairs for each biochemical step arose independently by convergent evolution (presumably by neofunctionalization of glycolytic enzymes) and were incorporated into functional sulfo-EMP pathways in a stepwise manner. The existence of hybrid pathways that contain mosaic combinations of proteins drawn from these sulfo-EMP1/2 PFAM groupings distributed across all three phyla provide support for this alternative view.
Sulfoglycolysis is an important pathway in the recycling of SQ that allows bacteria to access three of the six carbons in this sugar. This work shows the structural basis of class II aldolase cleavage of SFP, including substrate- and product-bound complexes that provide insights into the chemical mechanism of catalysis. We reveal that two class II SFP aldolases share striking structural similarities with class II FBP aldolases, including a bimetallic metalloenzyme analogous to FBP aldolase of E. coli. Thus, evolution has yielded class I and class II SFP aldolases for cleavage of SFP in sulfoglycolysis pathways mimicking the existence of class I and class II FBP aldolases in glycolysis/gluconeogenesis. This work provides a deeper molecular and sequence-based understanding of bacterial degradation of the prevalent organosulfur sugar SQ.
Experimental procedures
General
SFP was synthesized as previously described (38). HPLC–LC–MS/MS was achieved using a Vanquish Horizon UHPLC system (Thermo Fisher Scientific), HPLC system fitted with a ZIC-HILIC column (5 μm, 150 × 2.1 mm; Merck) connected to a TSQ Altis triple quadrupole mass spectrometer (Thermo fisher Scientific). Fructose bisphosphate was purchased from Sigma.
Cloning, expression, and purification of recombinant enzymes
The gene sequence coding for target enzymes (HpSqiA and YaSqiA) were synthesized (GenScript) with codon optimization for expression in E. coli cloned into pET28a(+) with N-terminal His6-tag (Table S3).
The plasmids containing the genes for each enzyme were used to transform E. coli BL21(DE3)-competent cells for gene expression. Starter cultures were grown in LB medium (5 ml) containing 30 μg ml−1 kanamycin for 18 h at 37 °C with shaking at 220 r.p.m. One liter volume cultures were inoculated with the starter culture (5 ml) and incubated at 37 °C with shaking at 220 r.p.m. until an A600 of 0.6 to 0.8 was reached. Gene expression was induced by the addition of IPTG (0.5–1 mM) and shaking was continued overnight at 16 °C with shaking at 200 r.p.m. The cells were then harvested by centrifugation at 5000g for 20 min and resuspended in 50 mM Tris buffer pH 7.5, containing 300 mM NaCl and 30 mM imidazole. Cells were disrupted by ultrasonication for 3 × 5 min, 30 s on, 30 s off cycles, and the suspension was centrifuged at 50,000g for 30 min to yield a clear lysate. The N-terminal His6-tagged protein was purified using immobilized-metal affinity chromatography using nickel nitrilotriacetic acid (Ni-NTA) column, followed by size-exclusion chromatography (Fig. S1). For immobilized-metal affinity chromatography, the lysate was loaded onto a pre-equilibrated Ni-NTA column, followed by washing with a load buffer (50 mM Tris, 300 mM NaCl, 30 mM imidazole pH 7.5). The bound protein was eluted using a linear gradient with buffer containing 300 mM imidazole. Protein fractions were pooled, concentrated, and loaded onto a HiLoad 16/600 Superdex 200 gel filtration column pre-equilibrated with 50 mM Tris, 300 mM NaCl pH 7.5 buffer. The protein was concentrated using a Vivaspin centrifugal concentration with 10 kDa molecular weight cut-off to a final concentration of 14 to 16 mg ml−1 for crystallization experiments.
For His6-tag cleavage, solutions of purified HpSqiA and YaSqiA at concentrations of 2 mg/ml was prepared in 25 mM Tris buffer pH 8 containing 150 mM NaCl. In order to optimize conditions for cleavage of His6-tag using thrombin, test reactions were attempted with 0.5 to 2.5 U of thrombin per mg of protein at room temperature, with gentle shaking at 100 rpm. The reaction was monitored over a period of 16 h by analyzing the progress of the cleavage by SDS-PAGE. Cleavage at the thrombin site was complete within 6 h using 0.1 U or within 3 h using 0.5 U of thrombin per mg of protein at room temperature. No nonspecific cleavage or over-digestion were observed when using thrombin under the conditions employed. Preparative cleavage of the His6-tag using thrombin was achieved by treating 4 mg of HpSqiA and YaSqiA (2 mg/ml in 25 mM Tris buffer pH 8 containing 150 mM NaCl) with 0.5 U thrombin for 4 h at room temperature and shaking at 150 rpm. The cleaved protein was passed through an Ni-NTA column to separate it from the cleaved His-tag peptide and the remaining uncleaved protein. Fractions were pooled and passed through a HiLoad 16/600 Superdex 75 size-exclusion column to separate SqiA from thrombin. The cleaved protein (lacking the His6-tag) recovered after gel filtration was concentrated to 5 mg/ml and used for kinetics experiments.
Biophysical characterization of SFP aldolases
SEC MALLS analysis
Experiments were conducted on a system comprising a Wyatt HELEOS-II multi-angle light scattering detector and a Wyatt rEX refractive index detector linked to a Shimadzu HPLC system (SPD-20A UV detector, LC20-AD isocratic pump system, DGU-20A3 degasser, and SIL-20A autosampler). Work was conducted at room temperature (20 ± 2 °C). Solvent was 0.2 μm filtered before use and a further 0.1 μm filter was present in the flow path. The column was equilibrated with at least two column volumes of buffer (50 mM NaPi, 300 mM NaCl, pH7.4) before use and flow was continued at the working flow rate until a stable baseline was obtained for UV, light scattering, and refractive index detectors. Sample injection volume was 100 μl HpSqiA or YaSqiA at 2 to 5 mg/ml in 50 mM Tris buffer, 300 mM NaCl pH 7.5; Shimadzu LabSolutions software (www.shimadzu.com) was used to control the HPLC and Astra V software (https://www.wyatt.com/products/software/astra.html) for the HELEOS-II and rEX detectors. The Astra data collection was 1 min shorter than the LC solutions run to maintain synchronization. Blank buffer injections were used as appropriate to check for carry-over between sample runs. Data were analyzed using the Astra V software. MWs were estimated using the Zimm fit method with degree 1. A value of 0.158 was used for protein refractive index increment (dn/dc).
Kinetic analysis of SFP aldolases
Product formation for class II aldolases (YaSqiA and HpSqiA) was measured using HPLC-ESI-MS/MS analysis. This used a triple quadrupole mass spectrometer (Agilent 6460 QQQ) coupled with Agilent 1260 Infinity Series LC system. The column was ZIC-HILIC column (5 μm, 50 × 2.1 mm; Merck). HPLC conditions were from 90% B to 40% B over 15 min; then 40% B for 5 min; back to 90% in 2 min (solvent A: 20 mM ammonium acetate in 1% acetonitrile; solvent B: 100% acetonitrile); flow rate, 0.30 ml/min; injection volume, 2 μl. The mass spectrometer was operated in negative ionization mode. Quantification was done using MS/MS multiple reaction monitoring mode of Agilent Mass Hunter Quantitative Analysis software (https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software/data-analysis/quantitative-analysis) and normalized using α-MeSQ as internal standard. The sensitivity for each multiple reaction monitoring-MS/MS transition was optimized for each analyte before analysis. The retention times and ESI-MS/MS fragmentation patterns of analytes were:
SLA retention time, 7.98 min; SLA ESI-MS/MS m/z of [M-H]- 153; product ions 81, 71;
α-MeSQ (internal standard) retention time, 8.57 min; α-MeSQ ESI-MS/MS m/z of [M-H]- 257; product ions 224.9, 164.9, 81;
DHAP retention time, 10.01 min; DHAP ESI-MS/MS m/z of [M-H]- 169; product ions 96.9, 78.9;
SFP retention time, 13.72 min; SFP ESI-MS/MS m/z of [M-H]- 323; product ions 224.9, 206.9, 152.9.
A calibration curve for response to DHAP was constructed using varying concentrations of DHAP and a constant concentration of internal standard, α-MeSQ.
pH profile of YaSqiA and HpSqiA
Assays for YaSqiA were conducted using 100 μl reaction mixtures containing 25 mM Tris.HCl buffer (pH 6.5–8.5), 25 mM NaCl, 5 mM MgCl2, 0.5 mM SFP, and 0.015 mg/ml YaSqiA. Assays for HpSqiA were conducted using 100 μl reaction mixtures containing 25 mM Tris.HCl buffer (pH 6.5–8.5), 25 mM NaCl, 5 mM MgCl2, 0.5 mM SFP, 0.5% bovine serum albumin (BSA), and 0.030 mg/ml HpSqiA. Reactions were initiated by addition of enzyme and incubating at 30 °C for 2 h. After 2 h, reactions were quenched by heating the reaction mixture at 80 °C for 4 min. The quenched reaction mixture (20 μl) was mixed with 30 μl of internal standard (i.e., final concentration 0.025 mM α-MeSQ) and analyzed by HPLC-ESI-MS-MS.
The relative enzyme activity was calculated by measuring the ion count for DHAP (area under the curve), divided by the ion count for the internal standard (0.025 mM α-MeSQ). The pH profile showed YaSqiA has the highest activity at pH 8.0. The pH profile for HpSqiA gave a similar rate for pH 6.5 to 8.5. Thus, Tris.HCl buffer solution (pH 8.0) was used for kinetic analysis of both enzymes.
Linearity of reaction rates
To demonstrate linearity of reaction rates, enzyme assays for YaSqiA were conducted in 100 μl reaction mixtures containing 25 mM Tris.HCl buffer solution (pH 8.0), 5 mM MgCl2, 25 mM NaCl, 0.5 mM SFP, and 0.015 mg/ml YaSqiA. Reactions were initiated by addition of enzyme and incubated for 1 h at 30 °C. At 10, 20, 30 min time intervals, 20 μl of sample were removed and quenched by heating at 80 °C for 4 min. The quenched reaction mixtures were mixed with 30 μl of internal standard (i.e., 0.025 mM α-MeSQ) and analyzed by MS-MS.
Similarly, enzyme assays for HpSqiA were conducted in 100 μl reaction mixtures containing 25 mM Tris.HCl buffer solution (pH 8.0), 5 mM MgCl2, 25 mM NaCl, 0.5 mM SFP, 0.5% BSA, and 0.030 mg/ml HpSqiA. Reactions were initiated by addition of enzyme and incubated for 1 h at 30 °C. At 10, 20, 30 min time intervals, 20 μl of sample were removed and quenched by heating at 80 °C for 4 min. The quenched reaction mixtures were mixed with 30 μl of internal standard (i.e., 0.025 mM α-MeSQ) and analyzed by MS-MS.
Michaelis–Menten kinetics
YaSqiA assays were conducted in 100 μl reaction mixture containing 25 mM Tris.HCl buffer solution (pH 8.0), 5 mM MgCl2, 25 mM NaCl, and 0.015 mg/ml YaSqiA. SFP concentration varied from 0 to 8 mM. Reactions were initiated by the addition of enzyme and incubating for 10 min at 30 °C. After 10 min, the reaction was quenched by heating at 80 °C for 4 min. The quenched reaction mixtures were mixed with 30 μl of internal standard (i.e., 0.025 mM α-MeSQ) and analyzed by MS-MS.
HpSqiA assays were conducted in 100 μl reaction mixture containing 25 mM Tris.HCl buffer solution (pH 8.0), 5 mM MgCl2, 25 mM NaCl, 0.5% BSA, and 0.030 mg/ml HpSqiA. SFP concentration varied from 0 to 8 mM. Reactions were initiated by the addition of enzyme and incubating for 10 min at 30 °C. After 10 min, the reaction was quenched by heating at 80 °C for 4 min. The quenched reaction mixtures were mixed with 30 μl of internal standard (i.e., 0.025 mM α-MeSQ) and analyzed by MS-MS.
Comparison of assays using HPLC-ESI-MS analysis with a coupled assay using SLA dehydrogenase
Reaction rates measured using HPLC-ESI-MS/MS were compared with those using a coupled assay in which SLA production was coupled to NAD+ consumption catalyzed by SLA dehydrogenase GabD from Rhizobium leguminosarum (20). For YaSqiA, assays were conducted in triplicate in 100 μl reaction mixtures containing 25 mM Tris.HCl buffer solution (pH 8.0), 5 mM MgCl2, 25 mM NaCl, 0.4 mM SFP (approx KM/3), and 0.015 mg/ml YaSqiA. Reactions were initiated by the addition of enzyme and incubating for 10 min at 30 °C. After 10 min, the reaction was quenched by heating at 80 °C for 4 min. The quenched reaction mixture was incubated with 0.1% BSA, 1.5 mM NAD+, and 200 nM RlGabD, and the progress of reaction was monitored using a UV/vis spectrophotometer at 340 nm. Once the absorbance had stabilized, the change in absorbance was used to calculate the concentration of SLA and the reaction rate. The extinction coefficient used for NADH was 6363 M−1 cm−1. kcat/KM was estimated using rate/([S][E]). At [SFP] = 0.4 mM, the rate measured by HPLC-ESI-MS-MS was (0.99 ± 0.13) × 10−5 and by coupled assay was (1.01 ± 0.12) × 10−5 mM s−1. A full Michaelis–Menten analysis using the HPLC-ESI-MS-MS assay gave kcat/KM = 0.063 ± 0.008; using the coupled assay, the estimate kcat/KM = 0.056 ± 0.006 s−1 mM−1. Errors are SEM.
For HpSqiA, assays were conducted in triplicate in 100 μl reaction mixtures containing 25 mM Tris.HCl buffer solution (pH 8.0), 5 mM MgCl2, 25 mM NaCl, 2 mM SFP (<< KM), and 0.015 mg/ml YaSqiA. Reactions were initiated by addition of enzyme and incubating for 10 min at 30 °C. After 10 min, the reaction was quenched by heating at 80 °C for 4 min. The quenched reaction mixture was incubated with 0.1% BSA, 1.5 mM NAD+, and 200 nM RlGabD. The progress of reaction mixture was monitored by absorbance at 340 nm until the reaction had stabilized. The data was analyzed as described for YaSqiA. At [SFP] = 2 mM, the rate measured by HPLC-ESI-MS-MS was (0.76 ± 0.17) × 10−5 and by coupled assay was (1.10 ± 0.13) × 10−5 mM s−1. A full Michaelis–Menten analysis using the HPLC-ESI-MS-MS assay gave kcat/KM = 0.0052 ± 0.00017; using the coupled assay, the estimate kcat/KM = 0.0062 ± 0.0002 s−1 mM−1.
Substrate specificity of YaSqiA and HpSqiA versus FBP
Specificity of YaSqiA was tested using 100 μl reaction mixtures containing 25 mM Tris.HCl buffer (pH 8.0), 25 mM NaCl, 5 mM MgCl2, 0.5 mM FBP with varied concentration of enzyme (i.e., 0.015 and 0.030 mg/ml). Specificity of HpSqiA was tested using 100 μl reaction mixtures containing 25 mM Tris.HCl buffer (pH 8.0), 25 mM NaCl, 5 mM MgCl2, 0.5% BSA, 0.5 mM FBP with varied concentration of enzymes (i.e., 0.015 and 0.030 mg/ml). Reactions were initiated by the addition of enzyme and incubating at 30 °C for 2 h. After 2 h, reactions were quenched by heating the reaction mixture at 80 °C for 4 min. The quenched reaction mixtures were mixed with 30 μl of internal standard (i.e., 0.025 mM α-MeSQ) and analyzed by MS-MS.
Protein crystallization
Initial screening and optimized crystallization conditions
Initial screening was performed using commercially available INDEX (Hampton Research), PACT premier, and CSSI/II (Molecular Dimensions) screens in 96-well sitting drop trays. Further optimization was carried out in a 48-well sitting drop or 24-well hanging-drop format to obtain optimal crystals for X-ray diffraction.
A crystal of HpSqiA•Zn was grown using a 14 mg ml−1 protein solution in 50 mM Tris buffer pH 7.5 containing 300 mM NaCl in a drop with 0.15 μl protein:0.15 μl mother liquor, the latter comprising 0.2 M sodium malonate dibasic monohydrate and 20% w/v PEG 3350.
A crystal of the YaSqiA•Zn(SO42−) complex was grown with YaSqiA at 16 mg ml−1 in 50 mM NaPi, 300 mM NaCl buffer pH 7.4, using a drop containing 0.6 μl protein:0.5 μl mother liquor, the latter comprising 1.5 M ammonium sulfate, 0.1 M NaCl, 0.1 M Bis-Tris pH 6.5.
A crystal of the HpSqiA•Zn•DHAP complex grew from 14 mg ml−1 enzyme in 50 mM Tris buffer pH 7.4 in a drop with 0.15 μl protein:0.15 μl mother liquor soaked with SFP for 2 min, with the reservoir solution containing 25% PEG 1500 w/v, 0.1 M SPG (succinic acid, propionate, glycine) buffer pH 7.
A crystal of the YaSqiA•Zn•SFP complex grew from 16 mg ml−1 enzyme in 50 mM NaPi buffer pH 7.4 soaked with SFP for 30 s in a drop with 0.1 μl protein:0.2 μl mother liquor, with the reservoir solution containing 1.5 M ammonium sulfate, 0.1 M Bis-Tris pH 5.5, 1% PEG 3350 w/v.
Crystals were harvested into liquid nitrogen, using nylon CryoLoops (Hampton Research) using mother liquor without any cryoprotectants. Data were collected at Diamond light source, on beamlines I04 (HpSqiA•Zn) to 1.7 Å, I04 YaSqiA•Zn(SO42−) to 1.7 Å, I03 HpSqiA•Zn•DHAP to 2.7 Å, and I03 (YaSqiA•Zn•SFP to 2.0 Å).
Data collection, processing, and refinement
The data were processed and integrated using XDS (39) and scaled using SCALA (40) included in the Xia2 processing system (41). Data collection and refinement statistics are given in Tables S1 and S2. The structures were solved using MOLREP (42), using the 5UCK.pdb file as initial search model. The structure was built and refined using iterative cycles using Coot (43) and REFMAC (44), the latter employing local NCS restraints. Following building and refinement of the protein and water molecules, clear residual density was observed in the omit maps for metal and ligands. The coordinate and refinement library files for ligands (DHAP, SFP) were prepared using ACEDRG (45). For HpSqiA•Zn structure, DHPS was soaked onto the crystal as substrate analog but density for DHPS was not observed in the structure. SFP was modeled at occupancy of 0.8 to 0.9 in YaSqiA•Zn•SFP structure and DHAP was modeled in at one occupancy in HpSqiA•Zn•DHAP; all other ligands/ions were modeled at occupancy of 0.7. The coordinate files and structure factors have been deposited in the Protein DataBank (PDB) with accession numbers: 8Q57 (YaSqiA•Zn•SO42−), 8Q58 (HpSqiA•Zn), 8Q59 (YaSqiA•Zn•SFP), and 8Q5A (HpSqiA•Zn•DHAP).
Structure-based analyses
All structure figures were generated using ccp4mg. The protein interactions, surfaces, and assemblies (PISA) server (46) was used to deduce the dimerization interface and buried surface area. Crystal packing interactions were analyzed using PISA (27). Structural comparisons and structure-based sequence alignments were conducted using a Dali search of the Protein Data Bank (47).
Bioinformatic analysis
Sequence alignments and phylogenetic analysis
Alignment and phylogenetic reconstructions were performed using the function "build" of ETE3 v3.0.0b32 (46) as implemented on the GenomeNet (https://www.genome.jp/tools/ete/). The phylogenetic tree of both FBP and SFP aldolases were constructed using MEGA (Molecular Evolutionary Genetics Analysis) software and formatted using iTOL (Interactive Tree of Life, https://itol.embl.de/) (48). Numbers at branches indicate the bootstrap proportion for 100 replicates.
SSN analysis
The protein sequence of class I aldolase from E. coli was used as query for a BLAST search of the Uniprot database, which retrieved 1000 sequences with more than 45% similarity. Similarly, class II aldolase from Y. aldovae was used as query for a BLAST search of the Uniprot database, which retrieved 100 sequences with more than 55% similarity (for a complete list, see Excel spreadsheet in SI (SI_aldolases.xlsx). These sequences were used to generate SSN by using the enzyme function initiative-enzyme similarity tool (49) with different alignment score, i.e., AS > 50 (corresponds to ≥ 41.3 percentage identity), AS > 80 (corresponds to ≥47.7 percentage identity), AS > 90 (corresponds to ≥52.5 percentage identity), and 100 (corresponds to ≥70 percentage identity). The SSN generated from the alignment score 90 was used to generate GNDs with ORF ± 10 neighbors using the Enzyme Function Initiative-Genome Neighborhood Tool (https://efi.igb.illinois.edu/efi-gnt/). A script was used to extract the accession codes of the retrieved neighbors. The aldolase neighbors were used to generate SSNN. The resulted network (SSNN) was plotted using Cytoscape v3.8 (50) with minimum alignment score of 50.
The nodes of aldolases were manually colored according to the occurrences of sulfo-EMP1 key degrading proteins (YihS, YihV) and sulfo EMP2 key degrading proteins (SqvD, SqiK). Initially, the EFI tool was used to generate a cluster number for each unique isofunctional cluster within the aldolases SSNN. The cluster number of neighborhood genes from the SSNN for YihS, YihV, SqvD, and SqiK were mapped onto aldolases SSN (alignment score at 90).
Data availability
Supporting information includes Figs. S1–S12, Tables S1–S3, and list of aldolase accession codes used for SSN analysis (Excel spreadsheet).
X-ray crystallography data is available at the Protein Databank archive (https://www.rcsb.org/) under accession codes 8Q58, 8Q57, 8Q5A, and 8Q59.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge Dr Johan P. Turkenburg and Sam Hart for assistance with X-ray data collection; the staff of the Diamond Light Source (U.K.) for provision of IO3 and IO4 beamline facilities (proposal numbers mx-18598 and mx-24948). We also acknowledge Dr Andrew Leech at the University of York Bioscience Technology Facility for assistance with SEC-MALLS analysis, and Dr Janice Mui for assistance with bioinformatics.
Author contributions
M. S., G. J. D., and S. J. W. conceptualization; M. S., A. K., E. D. G.-B., G. J. D., and S. J. W. writing–original draft; M. S., A. K., N. M. S., and J. P. L. investigation; M. S. and A. K. formal analysis; M. S. data curation; R. E. resources; E. D. G.-B., G. J. D., and S. J. W. funding acquisition.
Funding and additional information
This work was supported by the Australian Research Council (DP210100233, DP210100235), the Biotechnology and Biological Sciences Research Council (BB/W003805/1), the UKRI Future Leader Fellowship Program (MR/T040742/1), and the Royal Society for the Ken Murray Research Professorship to G. J. D. E. D. G.-B. acknowledges support from The Walter and Eliza Hall Institute of Medical Research, National Health and Medical Research Council of Australia (NHMRC) project grant GNT2000517, the Australian Cancer Research Fund, and the Brian M. Davis Charitable Foundation Centenary Fellowship. A. K. is supported by a Norma Hilda Schuster Scholarship.
Reviewed by members of the JBC Editorial Board. Edited by Chris Whitfield
Contributor Information
Gideon J. Davies, Email: gideon.davies@york.ac.uk.
Spencer J. Williams, Email: sjwill@unimelb.edu.au.
Supporting information
3
References
- 1.Benson A.A., Shibuya I. Sulfocarbohydrate metabolism. Fed. Proc. 1961;20:79. [Google Scholar]
- 2.Snow A.J.D., Burchill L., Sharma M., Davies G.J., Williams S.J. Sulfoglycolysis: catabolic pathways for metabolism of sulfoquinovose. Chem. Soc. Rev. 2021;50:13628–13645. doi: 10.1039/d1cs00846c. [DOI] [PubMed] [Google Scholar]
- 3.Wei Y., Tong Y., Zhang Y. New mechanisms for bacterial degradation of sulfoquinovose. Biosci. Rep. 2022;42 doi: 10.1042/BSR20220314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson A.A., Daniel H., Wiser R. A sulfolipid in plants. Proc. Natl. Acad. Sci. U. S. A. 1959;45:1582–1587. doi: 10.1073/pnas.45.11.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goddard-Borger E.D., Williams S.J. Sulfoquinovose in the biosphere: occurrence, metabolism and functions. Biochem. J. 2017;474:827–849. doi: 10.1042/BCJ20160508. [DOI] [PubMed] [Google Scholar]
- 6.Harwood J.L., Nicholls R.G. The plant sulpholipid - a major component of the sulphur cycle. Biochem. Soc. Trans. 1979;7:440–447. doi: 10.1042/bst0070440. [DOI] [PubMed] [Google Scholar]
- 7.Denger K., Weiss M., Felux A.K., Schneider A., Mayer C., Spiteller D., et al. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature. 2014;507:114–117. doi: 10.1038/nature12947. [DOI] [PubMed] [Google Scholar]
- 8.Burrichter A., Denger K., Franchini P., Huhn T., Müller N., Spiteller D., et al. Anaerobic degradation of the plant sugar sulfoquinovose concomitant with H2S production: Escherichia coli K-12 and Desulfovibrio sp. Strain DF1 as Co-culture Model. Front. Microbiol. 2018;9:2792. doi: 10.3389/fmicb.2018.02792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speciale G., Jin Y., Davies G.J., Williams S.J., Goddard-Borger E.D. YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids. Nat. Chem. Biol. 2016;12:215–217. doi: 10.1038/nchembio.2023. [DOI] [PubMed] [Google Scholar]
- 10.Abayakoon P., Jin Y., Lingford J.P., Petricevic M., John A., Ryan E., et al. Structural and biochemical insights into the function and evolution of sulfoquinovosidases. ACS Cent. Sci. 2018;4:1266–1273. doi: 10.1021/acscentsci.8b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma M., Abayakoon P., Epa R., Jin Y., Lingford J.P., Shimada T., et al. Molecular basis of sulfosugar selectivity in sulfoglycolysis. ACS Cent. Sci. 2021;7:476–487. doi: 10.1021/acscentsci.0c01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma M., Abayakoon P., Lingford J.P., Epa R., John A., Jin Y., et al. Dynamic structural changes accompany the production of dihydroxypropanesulfonate by sulfolactaldehyde reductase. ACS Catal. 2020;10:2826–2836. [Google Scholar]
- 13.Liu J., Wei Y., Ma K., An J., Liu X., Liu Y., et al. Mechanistically diverse pathways for sulfoquinovose degradation in bacteria. ACS Catal. 2021;11:14740–14750. [Google Scholar]
- 14.Kaur A., van der Peet P.L., Mui J.W., Herisse M., Pidot S., Williams S.J. Genome sequences of Arthrobacter spp. that use a modified sulfoglycolytic Embden-Meyerhof-Parnas pathway. Arch. Microbiol. 2022;204:193. doi: 10.1007/s00203-022-02803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook A.M., Denger K., Smits T.H. Dissimilation of C3-sulfonates. Arch. Microbiol. 2006;185:83–90. doi: 10.1007/s00203-005-0069-1. [DOI] [PubMed] [Google Scholar]
- 16.Wei Y., Zhang Y. Glycyl radical enzymes and sulfonate metabolism in the Microbiome. Annu. Rev. Biochem. 2021;90:817–846. doi: 10.1146/annurev-biochem-080120-024103. [DOI] [PubMed] [Google Scholar]
- 17.Mayer J., Huhn T., Habeck M., Denger K., Hollemeyer K., Cook A.M. 2,3-Dihydroxypropane-1-sulfonate degraded by Cupriavidus pinatubonensis JMP134: purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology. 2010;156:1556–1564. doi: 10.1099/mic.0.037580-0. [DOI] [PubMed] [Google Scholar]
- 18.Medema M.H., Takano E., Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobes R.D., Simpson R.T., Vallee R.L., Rutter W.J. A functional role of metal ions in a class II aldolase. Biochemistry. 1969;8:585–588. doi: 10.1021/bi00830a018. [DOI] [PubMed] [Google Scholar]
- 20.Li J., Sharma M., Meek R.W., Alhifthi A., Armstrong Z., Madiedo Soler N., et al. Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum. Chem. Sci. 2023;14:11429–11440. doi: 10.1039/d3sc01594g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacques B., Coinçon M., Sygusch J. Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases. J. Biol. Chem. 2018;293:7737–7753. doi: 10.1074/jbc.RA117.001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blom N.S., Tétreault S., Coulombe R., Sygusch J. Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase. Nat. Struct. Biol. 1996;3:856–862. doi: 10.1038/nsb1096-856. [DOI] [PubMed] [Google Scholar]
- 23.Izard T., Sygusch J. Induced fit movements and metal cofactor selectivity of class II aldolases: structure of Thermus aquaticus fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 2004;279:11825–11833. doi: 10.1074/jbc.M311375200. [DOI] [PubMed] [Google Scholar]
- 24.Hall D.R., Leonard G.A., Reed C.D., Watt C.I., Berry A., Hunter W.N. The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity. J. Mol. Biol. 1999;287:383–394. doi: 10.1006/jmbi.1999.2609. [DOI] [PubMed] [Google Scholar]
- 25.Plater A.R., Zgiby S.M., Thomson G.J., Qamar S., Wharton C.W., Berry A. Conserved residues in the mechanism of the E. coli Class II FBP-aldolase. J. Mol. Biol. 1999;285:843–855. doi: 10.1006/jmbi.1998.2376. [DOI] [PubMed] [Google Scholar]
- 26.Zgiby S.M., Thomson G.J., Qamar S., Berry A. Exploring substrate binding and discrimination in fructose 1, 6-bisphosphate and tagatose 1,6-bisphosphate aldolases. Eur. J. Biochem. 2000;267:1858–1868. doi: 10.1046/j.1432-1327.2000.01191.x. [DOI] [PubMed] [Google Scholar]
- 27.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43:W314–W319. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pegan S.D., Rukseree K., Capodagli G.C., Baker E.A., Krasnykh O., Franzblau S.G., et al. Active site loop Dynamics of a class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis. Biochemistry. 2013;52:912–925. doi: 10.1021/bi300928u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberg N., Zallot R., Gerlt J.A. EFI-EST, EFI-GNT, and EFI-CGFP: enzyme function initiative (EFI) web resource for genomic enzymology tools. J. Mol. Biol. 2023;435 doi: 10.1016/j.jmb.2023.168018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atkinson H.J., Morris J.H., Ferrin T.E., Babbitt P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zallot R., Oberg N., Gerlt J.A. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry. 2019;58:4169–4182. doi: 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirovich D.B., Da'dara A.A., Skelly P.J. Multifunctional fructose 1,6-bisphosphate aldolase as a therapeutic target. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.719678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaumann M., Pelzer-Reith B., Martin W.F., Schnarrenberger C. Multiple recruitment of class-I aldolase to chloroplasts and eubacterial origin of eukaryotic class-II aldolases revealed by cDNAs from Euglena gracilis. Curr. Genet. 1997;31:430–438. doi: 10.1007/s002940050226. [DOI] [PubMed] [Google Scholar]
- 34.Cooper S.J., Leonard G.A., McSweeney S.M., Thompson A.W., Naismith J.H., Qamar S., et al. The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold. Structure. 1996;4:1303–1315. doi: 10.1016/s0969-2126(96)00138-4. [DOI] [PubMed] [Google Scholar]
- 35.Callens M., Kuntz D.A., Opperdoes F.R. Kinetic properties of fructose bisphosphate aldolase from Trypanosoma brucei compared to aldolase from rabbit muscle and Staphylococcus aureus. Mol. Biochem. Parasitol. 1991;47:1–9. doi: 10.1016/0166-6851(91)90142-s. [DOI] [PubMed] [Google Scholar]
- 36.Galkin A., Kulakova L., Melamud E., Li L., Wu C., Mariano P., et al. Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia. J. Biol. Chem. 2007;282:4859–4867. doi: 10.1074/jbc.M609534200. [DOI] [PubMed] [Google Scholar]
- 37.De Montigny C., Sygusch J. Functional characterization of an extreme thermophilic class II fructose-1,6-bisphosphate aldolase. Eur. J. Biochem. 1996;241:243–248. doi: 10.1111/j.1432-1033.1996.0243t.x. [DOI] [PubMed] [Google Scholar]
- 38.Abayakoon P., Epa R., Petricevic M., Bengt C., Mui J.W.Y., van der Peet P.L., et al. Comprehensive synthesis of substrates, intermediates and products of the sulfoglycolytic Embden-Meyerhoff-Parnas pathway. J. Org. Chem. 2019;84:2901–2910. doi: 10.1021/acs.joc.9b00055. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Xds. Acta Crystallogr. D: Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D: Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 41.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 2010;43:186–190. [Google Scholar]
- 42.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 43.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Long F., Nicholls R.A., Emsley P., Grazulis S., Merkys A., Vaitkus A., et al. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr. D: Biol. Crystallogr. 2017;73:112–122. doi: 10.1107/S2059798317000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlt J.A., Bouvier J.T., Davidson D.B., Imker H.J., Sadkhin B., Slater D.R., et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software Environment for integrated Models of Biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3
Data Availability Statement
Supporting information includes Figs. S1–S12, Tables S1–S3, and list of aldolase accession codes used for SSN analysis (Excel spreadsheet).
X-ray crystallography data is available at the Protein Databank archive (https://www.rcsb.org/) under accession codes 8Q58, 8Q57, 8Q5A, and 8Q59.