Abstract
Dengue is a mosquito-borne disease caused by the dengue virus belonging to family flaviviridae and has grown to be a major global public health issue. Despite decades of effort, the global comeback of dengue is evidence of the inadequacy of present management techniques. Due to the loss of healthy lives and the depletion of scarce medical resources, dengue has a significant negative economic impact in underdeveloped countries. In recent years, research for tackling the incidences of dengue infection has increased. The structure of the viral genome has been deciphered with the non-structural protein, known as NS5 serving as a potential target. NS5 consisting of an MTase domain involved in RNA capping and an RdRp domain involved in viral replication. In the presented work, a series of new Oxindoline Carboxamide derivatives were designed and synthesized for inhibiting the viral RNA dependent RNA-polymerase (RdRp) activity of DENV. The novel compounds were put through tests including molecular docking and surface plasmon resonance (SPR) binding analysis to evaluate their affinity for the viral protein and their potential as novel inhibitors of the virus. From a total of 12 derivative compounds, four compounds OCA-10c, OCA-10f, OCA-10j & OCA-10i, were found to exhibit high affinity for NS5 RdRp, the KD values being 1.376 μM, 1.63 μM, 7.08 μM & 9.32 μM respectively. Overall, we report novel inhibitors of DENV RdRp activity with potential to be utilized against DENV for treating humans after further optimization.
Keywords: Dengue virus, Oxindoline carboxamide derivatives, RNA dependent RNA polymerase, Molecular docking, SPR binding analysis
1. Introduction
The dengue virus (DENV) is the most common human infectious arbovirus in the globe. Dengue fever (DF), has been known for more than 200 years and is typically marked by temperature, headache, eye pain, myalgia, arthralgia, and rash. Dengue hemorrhagic fever (DHF) is even more serious dengue virus illness marked by hemostasis problems and plasma leakage, and was not generally known until the 1950s [1]. Aedes aegypti (Yellow fever mosquito), the primary vector of dengue, has been expanding its range throughout Asia, Central America, South America, Africa, and the Pacific, causing dengue throughout tropical and subtropical areas [2].
Dengue is a public health issue because DHF can be deadly unless its accompanying plasma leakage is managed promptly, even though the majority of dengue cases produce a self-limited febrile sickness [3]. Patients who exhibit early illness symptoms should be immediately hospitalized for monitoring. Better patient outcomes can be achieved by more resource-intensive hospitalization policies. A more efficient and affordable approach for case management could be developed if the early determinants of dengue illness severity were thoroughly known and accordingly treated or prevented.
DENV belongs to the family Flaviviridae and the species Flavivirus. It is a single-stranded positive sense ribonucleic acid virus with 10,700 bases. Other arthropod-borne viruses like West Nile virus (WNV), tick-borne encephalitis virus, Zika virus, and yellow fever virus (YFV) are all members of the Flavivirus family. Each year, it affects 50–200 million people, endangering the lives of over 3.6 billion people who live in humid areas and result in 20,000 fatalities. The contemporary dynamics of globalization, climate change, travel, trade, socioeconomics, population, as well as viral evolution, are all blamed for the spread of dengue. Based on the variations in their virus structural and nonstructural proteins, there are four antigenically distinct DENV serotypes (DENV1, 2, 3, and 4). In terms of Open Reading Frame (ORF) amino acid sequences, these serotypes are roughly 70–80 % identical to each other [4]. One serotype infection can confer lifetime protection against that serotype, but heterologous infection causes serious dengue hemorrhagic fever owing to antibody-dependent enhancement [5]. The first vaccine was recently approved for use after decades of work, but it only offers limited cross protection for the four DENV serotypes [6]. Dengue and other flaviviral illnesses have not been authorized for treatment with antivirals [7].
The DENV genome encodes seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) in addition to three structural proteins (capsid protein C, membrane protein M, and envelope protein E) [8,9].
A multi-protein replication complex (RC) made up of the viral NS proteins and host cofactors is where flavivirus RNA replication takes place in host cells on endoplasmic reticulum-derived membranes [10,11]. The flavivirus RC's biggest and most conserved protein is called NS5, it has 900 amino acid residues. Its N-terminal region is an S-adenosyl-l-methionine (SAM)-dependent methyltransferase (MTase), which methylates the viral RNA genome cap (residues 1–265 in DENV3) [[12], [13], [14]]. The N-terminal region of NS5 was also suggested to have guanylyltransferase function [15]. The virus's genomic RNA is synthesized by its C-terminal RNA dependent RNA polymerase (RdRp) region (residues 267–900) [[16], [17], [18], [19]]. By modifying the interactions between MTase and RdRp, a possibly flexible linker region that links the two catalytic domains of NS5, RdRp activities and viral propagation could be controlled [[20], [21], [22]]. Serotype-dependent localization of NS5 to the nucleus of infected cells by DENV regulates host functions [23]. Following DENV infection, the RdRp uses a de novo start mechanism to synthesis viral RNA in the lack of a primer strand. In this mechanism, the viral RNA template's positive strand is translated into a complementary RNA strand with a negative polarity [16,17]. This duplex then acts as a template for the creation of extra positive polarity RNA strands that are either packed into virions or function as mRNA for protein translation.
In similarity with other polymerase families, DENV RdRp has a design resembling a right hand, with three subdomains referred to as "fingers," "palm," and "thumb" [19,22,24]. Seven conserved amino acid sequence motifs within these subdomains are crucial for binding RNA, NTPs, metal ions, and catalysis [25]. The apo-DENV RdRp structure was discovered to assume a "closed" pre-initiation state configuration, with a well-organized priming loop extending into a small RNA binding tunnel. In patterns F, G, and at the C-terminal end, disorganized peptide sequences were seen [19,22,24]
Because NS5 RNA dependent RNA polymerase is crucial for viral replication, it is a prime candidate for the development of inhibitors to address illnesses brought on by flaviviruses such DENV [[26], [27], [28]]. Only some of the DENV RdRp non-nucleoside inhibitors have been characterized despite several high-throughput screening efforts being carried out [[29], [30], [31]]. In the present study, OCA derivatives were tested for their capabilities to inhibit DENV NS5 RdRp activities which eventually disrupts the viral replication pathway of the Dengue virus and thereby inhibits its multiplication in the human body.
2. Experimental section
2.1. Materials and methods
All of the commercial reagents and anhydrous solvents were procured from Sigma Aldrich chemicals limited, India and used without further purification. The progresses of the reactions and purity of the compounds were monitored by thin layer chromatography (TLC) with F254 silica-gel precoated sheets using CH3OH/dichloromethane (0.5:9.5 and 0.7:9.3) as eluent. 1H NMR and 13C NMR spectra were recorded on Bruker and Varian spectrometer (400 MHz), CDCl3 and DMSO‑d6 were used as solvents, TMS was used as an internal standard. Mass spectral measurements were carried by ESI on Micro mass, Quattro LC using ESI + software with capillary voltage of 3.98 kV and ESI mode positive ion trap detector.
2.2. Experimental procedures
2.2.1. Synthesis of 5-(2-chloroacetyl) indolin-2-one (3)
Dropwise addition of chloroacetyl chloride (2) (3.56 mL, 45 mmol) to a stirred solution of aluminum chloride (24 g, 180 mmol) in 1,2-dicholroethane (160 mL) over a period of 5 min under a nitrogen atmosphere at 0 °C, and the reaction mixture was stirred at 0 °C for 30 min. Over a 20-min period, a solution of indoline-2-one (1) (4.0 g, 30 mmol) dissolved in 1,2-dichloroethane (50 mL) was added dropwise to the mixture. After complete addition, the reaction mixture was warmed to room temperature and stirred for 4 h. The reaction mixture was poured into ice-cold water (160 mL), whereupon the solid was precipitated. The obtained solid was filtered, washed with cold-water (30 mL) and dried under vacuum to obtain 5-(2-chloro acetyl)indolin-2-one (3) (3.5 g, 56 % yield) as an off-white solid.
1H NMR (400 MHz, DMSO‑d6): δ 10.8 (s, 1H), 7.88 (d, J = 8.2 Hz, 1H), 7.83 (s, 1H), 6.92 (d, J = 8.2 Hz, 1H), 5.08 (s, 2H), 3.57 (s, 2H).
2.2.2. Synthesis of 2-oxoindoline-5-carboxylic acid (4)
A suspension of 5-(2-chloro acetyl)indolin-2-one (3) (3.5 g, 16.7 mmol) in pyridine (35 mL) was stirred at 90 °C for 3 h. The reaction mixture was cooled to room temperature, obtained solid was filtered, washed with ethanol (20 mL) and dried under vacuum to obtain pyridine derivative. The product was dissolved in 2.5 N NaOH solution (35 mL) and stirred the reaction mixture at 75 °C for 3 h. The mixture was cooled to room temperature and acidified to pH 2 with 1 N HCl (40 mL), whereupon the solid was precipitated. The obtained solid was filtered, washed with water (50 mL) and dried under vacuum to obtain 2-oxoindoline-5-carboxylic acid (4) (2.5 g, 84 % yield) as a brown solid.
1H NMR (400 MHz, DMSO‑d6): δ 12.5 (brs, 1H), 10.6 (s, 1H), 7.94–7.68 (m, 2H), 6.88 (d, J = 8.7 Hz, 1H), 3.53 (s, 2H).
Preparation of Intermediate-5.
2.2.3. Synthesis of tert-butyl 2,4-difluorobenzylcarbamate (12)
Triethylamine (14.6 mL, 104.7 mmol) and Boc anhydride (9.6 mL, 41.8 mmol) were added to a stirred solution of (2,4-difluorophenyl)methanamine) (11) (5.0 g, 34.9 mmol) in anhydrous CH2Cl2 (20 mL) under Ar atmosphere at 0 °C. After complete addition, the reaction mixture was stirred at room temperature for 2 h. The reaction mixture was diluted with water (100 mL) and extracted with dichloromethane (2 × 100 mL). The combined organic layers were washed with brine (50 mL), dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure to obtain the crude product. The product was purified by silica gel chromatography (1–40 % EtOAc in hexanes). The fractions containing the pure product were combined and concentrated under reduced pressure to obtain tert-butyl 2,4-difluorobenzylcarbamate (12) (7.0 g, 82 % yield) as colorless oil.
1H NMR (400 MHz, DMSO‑d6): δ 7.41 (t, J = 4.0 Hz, 1H), 7.33 (q, J = 8.0 Hz, 1H), 7.22–7.17 (m, 1H), 7.09–7.05 (m, 1H), 4.13 (d, J = 4.0 Hz, 2H), 1.38 (s, 9H).
2.2.4. Synthesis of tert-butyl 2,4-difluorobenzyl (methyl)carbamate (13)
After cooling a suspension of 60 % NaH (4.60 g, 115.1 mmol) in anhydrous THF (50 mL) to 0 °C, tert-butyl 2,4-difluorobenzylcarbamate (12) (7.00 g, 28.7 mmol) dissolved in THF (50 mL) was added dropwise over a period of 10 min under a nitrogen atmosphere and stirred for 30 min. Iodomethane (7.1 mL, 115.1 mmol) was added dropwise over a period of 5 min to the above mixture and stirred at 40 °C for 2 h. The reaction mixture was cooled to 10 °C, quenched with aq. ammonium chloride solution (100 mL) and extracted with EtOAc (2 × 100 mL). The combined organic layers were washed with brine (50 mL), dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure to obtain the crude product. The product was purified silica gel chromatography (1–20 % EtOAc in hexanes). The fractions containing the pure product were combined and concentrated under reduced pressure to obtain tert-butyl 2,4-difluorobenzyl (methyl)carbamate (13) (4.5 g, 61 % yield) as colorless oil.
1H NMR (400 MHz, DMSO‑d6): δ 7.30–7.23 (m, 2H), 7.10 (t, J = 8.0 Hz, 1H), 4.39 (s, 2H), 2.78 (s, 3H), 1.41–1.36 (m, 9H).
2.2.5. Synthesis of 1-(2,4-difluorophenyl)-N-methyl methanamine hydrochloride (5)
TMS-Cl (6.6 mL, 52.5 mmol) was added dropwise over 10 min at 0 °C to a stirred solution of tert-butyl 2,4-difluorobenzyl (methyl)carbamate (13) (4.5 g, 17.5 mmol) in 2,2,2-trifluoroethanol (30 mL) under Ar atmosphere. After complete addition, the reaction mixture was warmed to room temperature and stirred for 2 h. Excess solvents were removed under reduced pressure to obtain 1-(2,4-difluorophenyl)-N-methylmethanamine hydrochloride (5) (2.5 g, 74 % yield) as an off-white solid.
1H NMR (400 MHz, DMSO‑d6): δ 9.47 (s, 2H), 7.80–7.74 (m, 1H), 7.40–7.34 (m, 1H), 7.23–7.18 (m, 1H), 4.13 (s, 2H), 2.51 (s, 3H).
2.2.6. Synthesis of N-(2,4-difluorobenzyl)-N-methyl-2-oxoindoline-5-carboxamide (6)
At 0 °C, DIPEA (5.89 mL, 33.8 mmol), EDC•HCl (3.22 g, 16.8 mmol), and HOBt (2.27 g, 16.8 mmol) were added to a stirred solution of 2-oxoindoline-5-carboxylic acid (4) (2.00 g, 11.2 mmol) in anhydrous CH3CN (20 mL) under Ar and stirred the reaction mixture at 0 °C for 15 min. To the above mixture, 1-(2,4-difluorophenyl)-N-methylmethanamine hydrochloride (5) (2.38 g, 12.3 mmol) was added and stirred the reaction mixture at room temperature for 16 h. Excess acetonitrile was removed under reduced pressure. The crude reaction mixture was diluted with water (100 mL) and extracted with dichloromethane (2 × 100 mL). The combined organic layer was washed with brine (100 mL), dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure to obtain the crude product. The product was purified by silica-gel chromatography (1–10 % CH3OH in CH2Cl2). The fractions containing the pure product were combined and concentrated under reduced pressure to obtain N-(2,4-difluorobenzyl)-N-methyl-2-oxoindoline-5-carboxamide (6) (2.5 g, 70 % yield) as an off-white solid.
1H NMR (400 MHz, DMSO‑d6): δ 10.5 (s, 1H), 7.40–7.38 (brs, 1H), 7.30–7.23 (m, 3H), 7.13–7.08 (m, 1H), 6.85 (d, J = 8.0 Hz, 1H), 4.63 (s, 2H), 3.51 (s, 2H), 2.88 (s, 3H).
2.2.7. Synthesis of (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (8)
N-(2,4-difluorobenzyl)-N-methyl-2-oxoindoline-5-carboxamide (6) (2.0 g, 6.32 mmol) was dissolved in ethanol (20 mL) and agitated for 5 min before pyrrolidine (1.57 mL, 18.9 mmol) was added. The reaction was carried out at room temperature. To the above mixture thiazole-2-carbaldehyde (7) (1.07 g, 9.48 mmol) was added and stirred at 50 °C for 2 h. The reaction mixture was cooled to room temperature, whereupon the solid was precipitated. The obtained solid was filtered, washed with ethanol (25 mL), and dried under vacuum to obtain (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (8) (1.5 g, 58 % yield) as a light-yellow solid.
1H NMR (400 MHz, DMSO‑d6): δ 10.9 (s, 1H), 9.31 (s, 1H), 8.22–8.14 (m, 2H), 7.69 (s, 1H), 7.46–7.14 (m, 4H), 6.94 (d, J = 8.0 Hz, 1H), 4.69 (s, 2H), 2.96 (s, 3H); MS (ESI + APCI): m/z = 412 [M + H]+. IR (KBr): υ (cm−1) 3159, 3121, 3082, 1709, 1621, 1500, 1393, 1316, 1257, 1094, 1060, 840, 754, 636, 608.
2.2.8. General procedure for synthesis of (E)-N-(2,4-difluorobenzyl)-1-alkyl-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA 10a-l)
To a stirred solution of (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene) indoline-5-carboxamide (8) (100 mg, 0.24 mmol) in anhydrous DMF (2.0 mL), potassium carbonate (100.7 mg, 0.729 mmol) was added at room temperature and stirred for 5 min. To the above mixture was added Alkyl halide (9a-l, 0.36 mmol) and stirred the reaction mixture at room temperature for 16 h. The mixture was diluted with water (50 mL) and extracted with EtOAc (2 × 50 mL). The combined organic layer was washed with brine (50 mL), dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure. The crude residue was purified by silica gel chromatography (1–10 % CH3OH in dichloromethane) to obtain (E)-N-(2,4-difluorobenzyl)-1-alkyl-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA 10a-l, 59–77 % yields) derivatives.
2.2.9. (E)-N-(2,4-difluorobenzyl)-N,1-dimethyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10a)
Yellow solid, Rf (0.4), C22H17F2N3O2S, Yield: 60 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.37 (s, 1H), 8.16–8.10 (m, 2H), 7.79 (s, 1H), 7.54–7.47 (m, 2H), 7.29 (bs, 1H), 7.14–7.12 (m, 2H), 4.71 (s, 2H), 3.25 (s, 3H), 2.96 (s, 3H). 13C NMR (100 MHz, CDCl3): 177.9, 168.9, 163.7, 161.7, 162.5, 161.2, 145.6, 131.4, 129.5, 127.4, 126.9, 124.8, 123.9, 120.8, 120.2, 111.84, 111.6, 108.1, 103.9, 44.5, 38.1, 26.3. UPLC-MS (ES + APCI): 426 [M+H] +.
2.2.10. (E)-N-(2,4-difluorobenzyl)-1-ethyl-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10b)
Yellow solid, Rf (0.4), C23H19F2N3O2S, Yield: 66 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.39 (s, 1H), 8.19–8.11 (m, 2H), 7.79 (s, 1H), 7.54 (s, 1H), 7.46 (s, 1H), 7.29 (bs, 1H), 7.21 (d, J = 8.0 Hz, 1H), 7.15 (bs, 1H), 4.70 (s, 2H), 3.85 (q, J = 6.8 Hz, 2H), 2.96 (s, 3H), 1.19 (t, J = 7.2 Hz, 3H). 13C NMR (100 MHz, CDCl3): 171.7, 168.9, 163.7, 162.5, 161.2, 145.6, 131.4, 129.5, 127.4, 126.9, 124.8, 123.9, 120.8, 120.2, 111.84, 111.81, 111.6, 108.1, 103.9, 44.5, 38.1, 35.1, 12.9. UPLC-MS (ES + APCI): 440 [M+H] +.
2.2.11. (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-1-propyl-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10c)
Yellow solid, Rf (0.4), C24H21F2N3O2S, Yield: 66 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.39 (s, 1H), 8.29–8.19 (m, 2H), 7.79 (s, 1H), 7.53 (s, 1H), 7.45 (s, 1H), 7.31 (s, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.14 (bs, 1H), 4.71 (s, 2H), 3.75 (t, J = 6.8 Hz, 2H), 2.97 (s, 3H), 1.68–1.62 (m, 2H), 0.90 (t, J = 7.6 Hz, 3H). 13C NMR (100 MHz, CDCl3): 171.7, 168.9, 163.7, 162.5, 161.2, 145.6, 131.4, 129.5, 127.4, 126.9, 124.8, 123.9, 120.8, 120.2, 111.81, 111.84, 111.6, 108.1, 103.9, 44.5, 41.9, 38.1, 21.0, 11.4. IR (KBr): υ (cm−1) 3119, 3067, 2962, 2919, 1703, 1625, 1457, 1396, 1344, 1210, 954, 728, 600.3. UPLC-MS (ES + APCI): 454 [M+H]+.
2.2.12. (E)-1-Allyl-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10d)
Yellow solid, Rf (0.4), C24H19F2N3O2S, Yield: 69 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.40 (s, 1H), 8.20–8.17 (m, 2H), 7.82 (s, 1H), 7.52 (s, 1H), 7.46 (s, 1H), 7.42 (bs, 1H), 7.14 (bs, 1H), 7.09 (d, J = 8.0 Hz, 1H), 5.92–5.85 (m, 1H), 5.19–5.15 (m, 2H), 4.71 (s, 2H), 4.44 (d, J = 4.0 Hz, 2H), 2.96 (s, 3H). 13C NMR (100 MHz, CDCl3): 171.6, 168.6, 163.7, 162.4, 161.2, 145.7, 145.2, 131.0, 129.7, 127.2, 126.9, 125.0, 124.1, 120.8, 120.1, 117.7, 111.84, 111.81, 111.6, 108.7, 103.9, 44.4, 42.6, 38.0. UPLC-MS (ES + APCI): 452 [M+H] +.
2.2.13. (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-1-(prop-2-yn-1-yl)-3-(thiazol-2-ylmethylene) indoline-5-carboxamide (OCA-10e)
Yellow solid, Rf (0.4), C24H17F2N3O2S, Yield: 64 %. 1H NMR (400 MHz, CDCl3): δ 9.52 (s, 1H), 8.10–8.06 (bs, 1H), 7.79 (s, 1H), 7.65 (d, J = 3.2 Hz, 1H), 7.60 (bs, 1H), 7.52–7.49 (bs, 1H), 7.10 (d, J = 8.4 Hz, 1H), 6.88–6.71 (m, 2H), 4.81 (s, 2H), 4.66 (d, J = 2.4 Hz, 2H), 3.07 (s, 3H), 2.26 (t, J = 2.4 Hz, 1H). 13C NMR (100 MHz, CDCl3): 171.6, 168.0, 163.7, 162.3, 161.3, 145.8, 144.0, 131.3, 130.2, 126.8, 125.4, 124.2, 120.9, 120.1, 111.83, 111.81, 111.6, 108.9, 103.9, 76.7, 72.7, 44.5, 38.1, 29.6. UPLC-MS (ES + APCI): 450 [M+H] +.
2.2.14. (E)-1-Benzyl-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10f)
Yellow solid, Rf (0.4), C28H21F2N3O2S, Yield: 74 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.41 (s, 1H), 8.25–8.21 (m, 2H), 7.88 (s, 1H), 7.55–7.45 (m, 2H), 7.35–7.28 (m, 6H), 7.13 (bs, 1H), 7.07 (d, J = 8.0 Hz, 1H), 5.07 (s, 2H), 4.70 (s, 2H), 2.97 (s, 3H). 13C NMR (100 MHz, CDCl3): 171.9, 169.0, 163.7, 162.4, 161.2, 145.7, 145.1, 135.5, 131.3, 129.8, 128.9 (2C), 127.8, 127.3 (2C), 127.1, 126.8, 125.2, 124.1, 121.0, 120.2, 111.8, 111.6, 108.8, 103.9, 44.4, 44.0, 38.0. UPLC-MS (ES + APCI): 502 [M+H]+.
2.2.15. (E)-1-(4-(tert-butyl)benzyl)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10g)
Yellow solid, Rf (0.5), C32H29F2N3O2S, Yield: 77 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.40 (s, 1H), 8.24–8.21 (m, 2H), 7.87 (s, 1H), 7.51–7.46 (m, 2H), 7.36–7.34 (m, 2H), 7.29–7.26 (m, 3H), 7.14 (bs, 1H), 7.11 (d, J = 8.0 Hz, 1H), 4.91 (s, 2H), 4.70 (s, 2H), 2.94 (s, 3H), 1.12 (s, 9H). 13C NMR (100 MHz, CDCl3): 171.6, 169.0, 163.7, 162.4, 161.2, 150.8, 145.7, 145.2, 132.5, 131.4, 129.7, 127.2, 127.1 (2C), 126.8, 125.8 (2C), 125.1, 124.0, 120.9, 120.1, 111.8, 111.6, 108.9, 103.9, 44.4, 43.7, 38.0, 34.6, 31.3 (3C). UPLC-MS (ES + APCI): 558 [M+H] +.
2.2.16. (E)-1-(cyclopropylmethyl)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10h)
Yellow solid, Rf (0.5), C25H21F2N3O2S, Yield: 67 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.40 (s, 1H), 8.20–8.17 (m, 2H), 7.80 (s, 1H), 7.53–7.47 (m, 2H), 7.35 (bs, 1H), 7.27 (d, J = 8.0 Hz, 1H), 7.18 (s, 1H), 4.71 (s, 2H), 3.70 (d, J = 6.8 Hz, 2H), 2.97 (s, 3H), 1.29–1.19 (m, 1H), 0.49–0.47 (m, 2H), 0.38–0.37 (m, 2H). 13C NMR (100 MHz, CDCl3): 171.6, 168.9, 163.7, 162.5, 161.2145.7, 131.4, 129.5, 127.5, 127.0, 124.8, 123.9, 120.8, 120.1, 111.86, 111.82, 111.6, 108.3, 103.9, 44.6, 38.1, 32.0, 9.04, 4.09 (2C). UPLC-MS (ES + APCI): 466 [M+H] +.
2.2.17. (E)-1-(cyclobutylmethyl)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene) indoline-5-carboxamide (OCA-10i)
Yellow solid, Rf (0.4), C26H23F2N3O2S, Yield: 69 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.38 (s, 1H), 8.19–8.18 (m, 2H), 7.79 (s, 1H), 7.52–7.46 (m, 2H), 7.35 (bs, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.14 (bs, 1H), 4.72 (s, 2H), 3.84 (d, J = 7.2 Hz, 2H), 2.96 (s, 3H), 2.74–2.67 (m, 1H), 1.97–1.93 (m, 2H), 1.86–1.78 (m, 4H). 13C NMR (100 MHz, CDCl3): 171.6, 169.1, 163.7, 162.5, 161.2145.8, 145.6, 131.3, 129.5, 127.4, 126.9, 124.8, 123.9, 120.8, 120.2, 111.8, 111.6, 108.3, 103.9, 45.4, 44.5, 38.0, 34.1, 26.3 (2C), 18.3. UPLC-MS (ES + APCI): 480 [M+H] +.
2.2.18. (E)-N-(2,4-difluorobenzyl)-1-(2-(4-methoxyphenyl)-2-oxoethyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10j)
Yellow solid, Rf (0.3), C30H23F2N3O4S, Yield: 59 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.42 (s, 1H), 8.25–8.21 (m, 2H), 8.10 (s, 1H), 8.08 (s, 1H), 7.85 (s, 1H), 7.50–7.47 (m, 2H), 7.35 (s, 1H), 7.14–7.08 (m, 4H), 5.44 (s, 2H), 4.72 (s, 2H), 3.88 (s, 3H), 2.98 (s, 3H). 13C NMR (100 MHz, CDCl3): 190.4, 171.5, 169.0, 164.3, 163.7, 162.3, 161.2, 145.7, 145.2, 131.3, 130.6 (3C), 129.9, 127.5, 127.0, 125.2, 124.1, 120.9, 120.1, 114.2 (2C), 111.9, 111.7, 108.4, 103.9, 55.6, 46.4, 44.4, 38.1. UPLC-MS (ES + APCI): 560 [M+H] +.
2.2.19. (E)-N-(2,4-difluorobenzyl)-1-(2-(4-fluorophenyl)-2-oxoethyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10k)
Yellow solid, Rf (0.3), C29H20F3N3O3S, Yield: 75 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.43 (s, 1H), 8.22–8.19 (m, 4H), 7.85 (s, 1H), 7.48–7.43 (m, 4H), 7.31 (s, 1H), 7.18 (s, 1H), 7.14 (d, J = 8.0 Hz, 1H), 5.51 (s, 2H), 4.71 (s, 2H), 2.89 (s, 3H). 13C NMR (100 MHz, CDCl3): 190.6, 171.5, 169.0, 167.7, 165.1, 162.3, 145.8, 144.9, 131.2 (2C), 131.1, 131.0, 130.1 (2C), 127.0, 126.8, 125.5, 124.3, 120.8, 120.1, 116.5, 116.2, 111.8, 111.9, 108.3, 104.0, 46.6, 44.5, 38.1. UPLC-MS (ES + APCI): 548 [M+H] +.
2.2.20. (E)-N-(2,4-difluorobenzyl)-N-methyl-1-(2-(2-nitrophenyl)-2-oxoethyl)-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA-10l)
Yellow solid, Rf (0.3), C29H20F2N4O5S, Yield: 72 %. 1H NMR (400 MHz, DMSO‑d6): δ 9.44 (s, 1H), 8.81 (s,1H), 8.58–8.53 (m, 2H), 8.31–8.22 (m, 2H), 7.92 (t, J = 7.6Hz, 1H), 7.87 (s, 1H), 7.51–7.48 (m, 2H), 7.31 (bs, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.14 (bs, 1H), 5.65 (s, 2H), 4.72 (s, 2H), 2.98 (s, 3H). 13C NMR (100 MHz, CDCl3): 190.5, 171.4, 168.9, 163.6, 162.2, 161.3148.6, 145.8, 144.5, 135.6, 133.9, 131.5, 131.2, 130.4, 128.4, 127.0, 126.5, 125.6, 124.4, 123.2, 120.9, 120.2, 111.8, 111.6, 108.1, 103.9, 46.9, 44.4, 38.1. UPLC-MS (ES + APCI): 575 [M+H] +.
2.3. Cloning, expression and purification of the viral NS5 in E. Coli
The entire NS5 viral protein sequence (272–900 AA) of DENV was downloaded from the UniProt and then cloned into the expression clone pET21a without any tags and to pET28a with the N terminus 6-His tag. Both expression clones were used for transformation of the expression host BL21 (DE and Rosetta.2 (DE3) of E. Coli. NiNTA column chromatography method was applied to isolate the RdRp protein. It was demonstrated that the protein without a tag is more soluble than the protein with a tag in the BL21 (DE3) system. SDS-PAGE and Coomassie blue staining were used to determine the purity of the RdRp protein, which may be used for research on the enzymatic activity of the RdRp protein and SPR testing to determine how the RdRp protein interacts with test compounds [33,34].
2.4. Characterization of DENV NS5 by SPR on BIAcore T-200 platform
A high-performance device for real-time bimolecular interaction research, the BIAcoreTM T200 from GE Healthcare uses Surface Plasmon Resonance (SPR) technology. It is used to test the binding affinity interaction of all the compounds for DENV NS5. Real-time, label-free detection of bimolecular interactions is possible with Surface Plasmon Resonance (SPR). Polarized light striking an electrically conducting surface at the boundary of two media causes SPR. This produces Plasmon waves, which are waves of electron charge density that reduce the intensity of reflected light in proportion to the mass on a sensor surface at a certain angle known as the resonance angle [35]. CM5 chip immobilization of DENV NS5 via analyte couples with more than 10,000 RU. The binding interactions of all substances to DENV NS5 were assessed by producing the stocks as 30 mM in 100 % DMSO (referred to as the analyte) as well as PBS with 5 % DMSO as a running buffer. The experiment involved the compound's (the analyte's) association with DENV NS5(Ligand) at a 50 μM concentration for 60 s, subsequently followed by its dissociation for 60 s and further regeneration for 120 s using running buffer [36].
2.5. Computational section
Structural retrieval and preparation: The protein data bank database (www.rcsb.org) of Research Collaboratory for Structural Bioinformatics (RCSB) was used to download the molecular structure of APO DENV RdRp of serotype 2 (PDB ID: 6IZY). After the retrieval of the structure, AutoDock tools were utilized for generating the viral protein of DENV [37]. Water molecules, native ligands and non-interacting ions were all removed from the crystal structure of the target protein. The protein was supplied of all the missing hydrogens to reduce the pressure on the crystal structure of the protein and also for its efficient use while performing docking simulations in AutoDock programme. The chimera graphical user interface provided by UCSF was utilized to generate the final structure of protein after its structural reduction processes involving addition of H-atoms, combining non-polar hydrogen atoms with carbon atoms, and Gasteiger charge calculations.
ACD/ChemSketch Freeware: ChemSketch is a free sketching programme provided by Advanced Chemistry Development (ACD), Inc. This programme makes it possible to sketch chemical structure including organics, organometallics, polymers and Markush structures. Apart from its various features, it has the capacity to identify structures having less than 50 atoms and three rings, calculations of molecular parameters like those involving molecular weight, density and molar refractivity are also possible, even cleaning, predicting logP and visualization of 2D and 3D structures [38].
2.6. Molecular docking of the OCA derivatives against NS5 RdRp
The molecular docking analysis was performed for all the OCA derivatives against the NS5 RdRp using AutoDock Vina, one of the quickest and most popular open-source molecular docking software [37]. The target protein docking site was discovered to be at coordinates X: 56.859, Y: 4.648, and Z: 75.768. The docking was carried out by placing a grid box at the center of the best binding pose in the RdRp protein structure, which has 32 X 32 X 32 dimensions and a grid spacing of 1.0. The target protein docking site was discovered to be at coordinates X: 56.859, Y: 4.648, and Z: 75.768. The contacts between the active sites in the target and ligand conformations, as well as the kind of interaction and bond lengths, were identified using Discovery Studio Visualizer [39].
3. Result & discussion
A series of Oxindoline Carboxamide derivative compounds have been synthesized as a class of novel inhibitors against the RdRp activity of DENV to inhibit viral replication and propagation. The series OCA 10a-l was synthesized and tested for the presented work.
The synthetic approaches adopted to obtain (E)-N-(2,4-difluorobenzyl)-1-alkyl-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA 10a-l) derivatives were described in Scheme 1. 5-(2-chloroacetyl)indolin-2-one (3) synthesized by treating commercially available indolin-2-one (1) with chloroacetyl chloride (2) in presence of aluminum chloride at 0 °C to room temperature for 4 h [40]. 2-oxoindoline-5-carboxylic acid (4) was obtained by treating the compound (3) with pyridine at 70 °C for 4 h followed by sodium hydroxide at 70 °C for 4 h [41]. N-(2,4-difluorobenzyl)-N-methyl-2-oxoindoline-5-carboxamide (6) was obtained by treating 2-oxoindoline-5-carboxylic acid (4) with 1-(2,4-difluorophenyl)-N-methylmethanamine hydrochloride (5) using coupling reagent EDC•HCl, HOBt and DIPEA at room temperature for 16 h. (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (8) was prepared by treating 2-thiazolecaboxaldehyde with pyrrolidine in ethanol at 50 °C for 2 h [42,43]. The final (E)-N-(2,4-difluorobenzyl)-1-alkyl-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carboxamide (OCA 10a-l) derivatives were synthesized by treating (E)-N-(2,4-difluorobenzyl)-N-methyl-2-oxo-3-(thiazol-2-ylmethylene)indoline-5-carbox amide (8) with different amines (9a-l) and K2CO3 in DMF at room temperature for 16 h. All of these targets were purified by flash column chromatography and characterized by spectroscopic methods (1H NMR, 13C NMR) and mass spectrometry.
Scheme 1.
Reagents and conditions: (a) Chloroacetyl chloride, AlCl3, 1,2 DCE, 0 °C to rt, 4 h (b) i) Pyridine, 70 °C, 4 h ii) NaOH, 70 °C, 4 h (c) 1-(2,4-difluorophenyl)-N-methylmethanamine hydrochloride, EDC•HCl, HOBt, DIPEA, CH3CN, 0 °C to rt, 16 h d) 2-thiazolecaboxaldehyde, pyrrolidine, ethanol, 50 °C, 2 h e) Alkyl halide, K2CO3, DMF, rt, 16 h f) Boc2O, Et3N, CH2Cl2, rt, 2 h g) NaH, CH3I, THF, 0 °C–40 °C, 2 h h) TMS-Cl, TFE, 0 °C to rt, 2 h.
OCA analogues: The derivatives of oxindole carboxamide derivatives (OCA) with yields (59 %–77 %)
are presented below.
3.1. Docking
In the presented work, docking analysis was conducted to get more insights into the binding affinity and intermolecular interactions of Oxindoline carboxamide derivatives with RNA dependent RNA polymerase. S-adenosyl-l-homocysteine (SAH) PDB ID-6KR2 was utilized as a reference compound to validate molecular docking, this inhibitor SAH complexed with the viral protein RdRp of NS5 was used for screening of oxindoline carboxamide derivatives.
We docked 12 oxindoline carboxyamide compounds with DENV NS5 RdRp. The molecular docking findings are shown in Table 1 and comprise docking binding energy, interacting amino acid residues in the protein binding area, the kind of interaction, and the structure of the ligands (Fig. 1, Fig. 2, Fig. 3, Fig. 4). SAH displayed the highest affinity for the DENV NS5 RdRp active site residues, with a binding energy of 7.5 kcal/mol.
Table 1.
Results of Molecular Docking analysis using AutoDock Vina.
| Compound | Structure | Docking energy (kcal/ml | Amino acid Interactions |
|---|---|---|---|
| SAH | 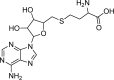 |
−7.5 | SerA:331, ThrA:336, ArgA:329, AsnA340, GlyA: 392, TyrA:337, CysA:439, AspA:394, AlaA:203, SerA:201, GlyA:200, GlyA:267, ThrA:270-Van der Waals; GlyA:332, SerA:391 Conventional hydrogen bond; AspA:393, LysA:199 – Attractive charges, AspA: 269 – Pi-Sulfur. |
| OCA-10a |  |
−9.1 | HisA:528, IleA:527, IleA:204, SerA:331, AsnA:340, TyrA:337, ValA:133, PheA:129, PheA:216, ValA:334, GlnA:333- Van der Waals; LysA:132- Conventional Hydrogen Bond; ThrA:336- Pi-Sigma; GlyA:332-Pi-Pi Stacked; AlaA:137, LeuA:139, AlaA:138- Pi-Alkyl. |
| OCA-10b |  |
−8.6 | SerA:526, IleA:204, TyrA:337, AsnA:340, GlyA:332, ValA:133, PheA:129, PheA:216, GlnA:333- Van der Waals; LysA:132- Conventional Hydrogen Bond; AlaA:137- Halogen (Fluorine); ThrA:336-Pi-Sigma; HisA:528, IleA:527; LeuA:139; ValA:334; AlaA:138- Pi-Alkyl. |
| OCA-10c | 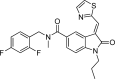 |
−10.2 | HisA:528, IleA:527, AsnA:340, GlyA:332, SerA:331, ValA:133, PheA:129, PheA:216, GlnA:333-Van der Waals; LysA:132- Conventional Hydrogen Bond; ThrA:336- Pi-Sigma; TyrA:337-Pi-Cation; IleA:204, LysA:187, AlaA:137, LeuA:139, AlaA:138, ValA:334- Pi-Alkyl. |
| OCA-10d |  |
−8.8 | HisA:528, IleA:204, GlyA:332, ThrA:336, PheA:216, ValA:133, GlnA:333- Van der Waals; ValA:334-Pi-Donor Hydrogen Bond; PheA:129-Halogen (Fluorine); TyrA:337, LysA:132-Pi-Cation; IleA:527, AlaA:137, LeuA:139, AlaA:138-Pi-Alkyl. |
| OCA-10e | 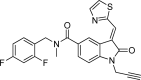 |
−8.9 | SerA:526, GlyA:332, IleA:204, PheA:216, ValA:133, GlnA:333-Van der Waals; LysA:132-Pi-Cation; TyrA:337-Pi-Anion; ThrA:336-Pi-Pi Stacked; PheA:129-Halogen (Fluorine); ValA:334, HisA:528-Pi- Donor Hydrogen Bond; LeuA:139, AlaA:137, AlaA:138, IleA:527-Pi-Alkyl. |
| OCA-10f |  |
−10.1 | AsnA:223, AsnA:340, AspA:393, GlyA:392, ThrA:336, IleA:204, HisA:528, GlyA:332, GlnA:333, PheA:129-Van der Waals; LysA:132-Conventional Hydrogen Bond; IleA:527, ValA:334- Pi-Sigma; TyrA:337, PheA:216- Pi-Pi T-Shaped; ArgA:212-Halogen (fluorine); LeuA:139, AlaA:138-Pi-Alkyl. |
| OCA-10g |  |
−9.8 | AlaA:203, SerA:526, ThrA:336, GlyA:332, GlnA:333, PheA:129-Van der Waals; AsnA:223, LysA:132-Conventional Hydrogen Bond; IleA:527, IleA:204, ValA:334-Pi-Sigma; TyrA:337, PheA:216- Pi-Pi Stacked; AlaA:137-Pi-Donor Hydrogen Bond; ArgA:212-Halogen (Fluorine); LysA:187, AlaA:138- Pi-Alkyl. |
| OCA-10h |  |
−9.3 | SerA:526, LysA:187, IleA:527, IleA:204, AsnA:340, TyrA:337, GlyA:332, GlnA:333, ValA:133-Van der Waals; ThrA:336-Pi-Sigma; AlaA:138-Pi-Donor Hydrogen Bond; LysA:132-Pi-Cation; AlaA:137, PheA:129-Halogen (Fluorine); HisA:528, LeuA:139, ValA:334-Pi-Alkyl. |
| OCA-10i |  |
−9.7 | TrpA:525, AalA137, GlnA:333, ValA:334, TyrA:337, SerA:201, LysA:199, GlyA:267, ThrA:336, SerA:331, LeuA:139-Van der Waals; SerA:526, TrpA:268, LysA:187-Pi- Donor Hydrogen Bond; IleA:527, IleA:204-Pi-Sigma; HisA:528, GlyA:332, AsnA:340-Conventional Hydrogen Bond; GlyA:200, AspA:269, AspA:393-Halogen (Fluorine); ArgA:188, AlaA:208-Pi-Alkyl. |
| OCA-10j |  |
−8.7 | GlyA:472, IleA:527, SerA:526, IleA:204, AsnA:340, TyrA:337, GlyA:332, ValA:133, PheA:129, PheA:216, ValA:334, GlyA:333-Van der Waals; HisA:528, LysA:132- Conventional Hydrogen Bond; LysA:187-Pi- Cation; ThrA:336-Pi-Sigma, AlaA:137-Halogen (Fluorine); LeuA:139, AlaA:138-Pi-Alkyl. |
| OCA-10k |  |
−9.7 | IleA:204, GlyA:332, GluA:238, GlyA:267, SerA:201, HisA:528-Van der Waals; GlnA:333, TrpA:268, IleA:527-Pi-Donor Hydrogen Bond; TyrA:337- Pi-Pi stacked; ThrA:336-Pi-Sigma; SerA:526, AspA:393, AspA:269, GlyA:200-Halogen (Fluorine); AsnA:340-Conventional Hydrogen Bond; LeuA:139, AlaA:137-Pi-Alkyl. |
| OCA-10l |  |
−9.2 | GlyA:332, AlaA:203, HisA:528, SerA:526, LysA:187, LysA:132, AsnA:223, GlnA:333, ThrA:336, AsnA:340, GlyA:392 – Van der Waals; AspA:393 – Carbon hydrogen bond; AspA:269, SerA:331, TyrA:337 – Carbon hydrogen bond; LeuA:139, IleA:527 – Pi-Alkyl; IleA:204 – Pi-Sigma; GluA:238 – Halogen (Fluorine). |
Fig. 1.
Molecular docking interaction of OCA-10c with DENV NS5 RdRp.
Fig. 2.
Molecular docking interaction of OCA-10f with DENV NS5 RdRp.
Fig. 3.
Molecular docking interaction of OCA-10g with DENV NS5 RdRp.
Fig. 4.
Molecular docking interaction of OCA-10i with DENV NS5 RdRp.
The Non-structural protein NS5 plays a major role in viral infection through its multiple enzymatic functions. It has C-terminal RdRp activity and N-terminal MTase activity, necessary for viral amplification and RNA capping of the DENV genome respectively. The MTase activity prevents viral mRNA from being degraded by 5′-exoribonucleases while also making sure that eukaryotic translation initiation factor can recognize the mRNA. There are three subdomains known as fingers (273–315, 416–496, and 543–600), palm (497–542, 601–705), and thumb (706–900) that together make up the RdRp domain. Two nuclear localization sequences that are recognized by cellular components, located in the amino acid sequence between residues 320 and 405, enable protein transport to the nucleus.
We observed the reference inhibitor SAH making a hydrogen bond with GlyA:332 and SerA:201, additional interactions observed included a Pi-Sulfur bond with the amino acid AspA:269. From the compounds subjected for docking with the DENV NS5 RdRp, we observed OCA-10c to be showing the best fit (dock score of −10.2 kcal/mol) with the active site of RdRp viral protein (Fig. 1). The OCA-10f, 10g, and 10i have highest energy values of −10.1, −9.8, and −9.7 kcal/mol, respectively after compound 10c (Fig. 2, Fig. 3, Fig. 4). All the oxindoline carboxamide derivatives, however, were subjected for further experimental analysis due to observed interactions that were crucial for inhibition of viral RdRp activity, having the dock scores in the range of −7.5 to −10.2 kcal/mol (Table 1).
3.2. SPR binding analysis outcomes
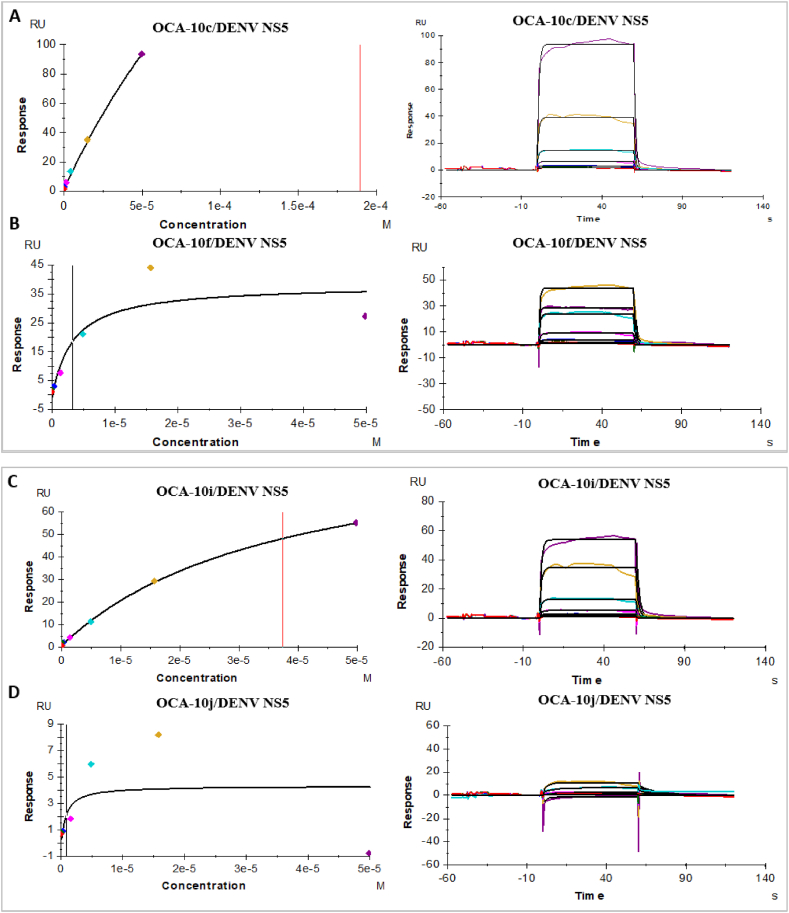
The following study is used to evaluate the binding affinity interactions of small compounds against DENV NS5 by SPR analysis on BIAcore-T200 platform. For SPR binding analysis all the analytes were prepared in PBS supplemented with 5 % of DMSO. All the analytes were made to a stock concentration of 50 μM for studying the interaction of the OCA derivatives with the bacterially overexpressed and purified NS5 of the virus. The relative response levels (RU) of the derivatives were studied and their Dissociation equilibrium constant (KD) value were determined with the aid of steady state kinetics that utilized 7 concentrations in the range of 50 μM–0.0502 μM in half log fold dilution (Table 2). The KD (Dissociation Equilibrium Constant) value of interactions as observed at various response levels is summarized in Table 3. For the reduction of background signal of the flow cell, solvent correction was conducted. The KD values for all the Oxindoline Carboxamide Derivatives (OCA) have been estimated using the used range of concentration through this analysis with OCA-10c and OCA-10f [Fig. 5 (A–B)] showing the highest affinity for RdRp followed by OCA-10j and OA-10i [Fig. 5 (C – D)].
Table 2.
Range of concentration of analyte used in the SPR binding analysis.
| S. No | Concentration of analytes (in half log fold dilution) |
|---|---|
| 1 | 50 μM |
| 2 | 15.82 μM |
| 3 | 5.00 μM |
| 4 | 1.584 μM |
| 5 | 0.501 μM |
| 6 | 0.158 μM |
| 7 | 0.0502 μM |
Table 3.
Observed binding interaction (KD) with the highlighted compounds at different response level.
| S. No. | Compound | Binding (in KD values) |
|---|---|---|
| 1 | OCA-10a | NA |
| 2 | OCA-10b | NA |
| 3 | OCA-10c | 1.376 μM |
| 4 | OCA-10d | NA |
| 5 | OCA-10e | NA |
| 6 | OCA-10f | 1.63 μM |
| 7 | OCA-10g | NA |
| 8 | OCA-10h | NA |
| 9 | OCA-10i | 9.31 μM |
| 10 | OCA-10j | 7.08 μM |
| 11 | OCA-10k | NA |
| 12 | OCA-10l | NA |
Fig. 5.
(A – D) SPR analysis results of interaction of the compounds OCA-10c, OCA-10f, OCA-10i, OCA-10j with DENV NS5, the KD values of the compounds are (Fig. A) OCA-10c - 1.376 μM, (Fig. B) OCA - 10f-1.63 μM, (Fig. C) OCA - 10i-9.31 μM and (Fig. D) OCA-10j - 7.08 μM.
Millions of instances of the dengue virus are reported globally, making it one of the most widely dispersed viruses. Any serotype of dengue infection may result in a variety of symptoms, usually infection with different serotypes may even be severe and fatal due to antibody-dependent enhancement [5]. The NS5 RdRp, which catalyses the production of the viral RNA causing the viral dengue infection, is found in the non-structural protein NS5. RdRp was found capable of synthesizing the viral genome of dengue. After the entry of the virus and protein translation, de-novo synthesis of RNA occurs for generating a negative polarity RNA from positive viral RNA template, this template is further used to produce more positive RNA strands that later undergo translation or get packed to generate the virions [28].
In research, a number of commercially available drug-like compounds were examined to see how effective they would be against the DENV NS5 protein. A total of 18 compounds were to show promise, and had great potential against the NS5 protein. They were found harmless to Huh-7 cell lines at concentrations less than 50 μM. Compared to the reference inhibitor NITD107, crystallized in the GDD motif region, Garcia-Ariza et al. found the compound 7sd binding to the GDD motif region of RdRp and specifically targeting serotypes 2 and 3, as well as showing that it had conserved interactions with the amino acids crucial for polymerase activity of RdRp [32].
4. Conclusion
In our research, we designed and synthesized novel compounds and prepared derivatives to test their potential in inhibiting the viral replication of DENV. We used molecular docking to find the compound with best docking score and then also performed SPR binding analysis on all the compounds to test the binding affinity of the derivatives towards the target protein. Considering the docking study and SPR analysis as well as the observed KD values, we report a total of 4 compounds OCA-10c, OCA-10f, OCA-10i, and OCA-10j as the most promissory for future research and human use after further optimization and analysis.
Consent for publication
We authorize to the publication of the article without any conflict.
Data availability statement
All data generated or analysed during this study are included in this published article.
CRediT authorship contribution statement
Chandra Prakash Koraboina: Investigation, Methodology, Writing – original draft. Parameswari Akshinthala: Data curation, Software. Naresh Kumar Katari: Visualization, Writing – review & editing. Ravi Adarasandi: Formal analysis, Validation. Sreekantha Babu Jonnalagadda: Funding acquisition, Resources. Rambabu Gundla: Conceptualization, Project administration, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to the GITAM Deemed to be University Hyderabad for providing the SEED Grant (Ref: F.No 2022/0174) to execute the above work and Department of Science and Technology (DST-SERB-ECR/2016/000288) India for providing financial assistance.
Contributor Information
Naresh Kumar Katari, Email: nkatari@gitam.edu.
Rambabu Gundla, Email: rgundla@gitam.edu.
References
- 1.Hammon W.M., Rudnick A., Sather G.E. Viruses associated with epidemic hemorrhagic fevers of the Philippines and Thailand. Science (New York, N.Y.). 1960;131(3407):1102–1103. doi: 10.1126/science.131.3407.1102. [DOI] [PubMed] [Google Scholar]
- 2.Leta S., Beyene T.J., De Clercq E.M., Amenu K., Kraemer M.U.G., Revie C.W. Global risk mapping for major diseases transmitted by Aedes aegypti and Aedes albopictus. Int. J. Infect. Dis. : Int. J. Infect. Dis. 2018;67:25–35. doi: 10.1016/j.ijid.2017.11.026. official publication of the International Society for Infectious Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke D.S., Nisalak A., Johnson D.E., Scott R.M. A prospective study of dengue infections in Bangkok. Am. J. Trop. Med. Hyg. 1988;38(1):172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 4.Kuno G., Chang G.J., Tsuchiya K.R., Karabatsos N., Cropp C.B. Phylogeny of the genus flavivirus. J. Virol. 1998;72(1):73–83. doi: 10.1128/JVI.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murugesan A., Manoharan M. 2020. Dengue Virus, Emerging and Reemerging Viral Pathogens; pp. 281–359. [DOI] [Google Scholar]
- 6.Capeding M.R., Tran N.H., Hadinegoro S.R.S., Ismail H.I.H.M., Chotpitayasunondh T., Chua M.N., Luong C.Q., Rusmil K., Wirawan D.N., Nallusamy R., Pitisuttithum P., Thisyakorn U., Yoon I.K., van der Vliet D., Langevin E., Laot T., Hutagalung Y., Frago C., Boaz M., Wartel T.A. CYD14 Study Group, Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet (London, England) 2014;384(9951):1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 7.Lim S.P., Wang Q.Y., Noble C.G., Chen Y.L., Dong H., Zou B., Yokokawa F., Nilar S., Smith P., Beer D., Lescar J., Shi P.Y. Ten years of dengue drug discovery: progress and prospects. Antivir. Res. 2013;100(2):500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 9.Rice C.M., Lenches E.M., Eddy S.R., Shin S.J., Sheets R.L., Strauss J.H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science (New York, N.Y.). 1985;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 10.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6(5):363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salonen A., Ahola T., Kääriäinen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. The EMBO journal. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y., Soh T.S., Lim S.P., Chung K.Y., Swaminathan K., Vasudevan S.G., Shi P.Y., Lescar J., Luo D. Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5) Proc. Natl. Acad. Sci. U.S.A. 2015;112(48):14834–14839. doi: 10.1073/pnas.1514978112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Issur M., Geiss B.J., Bougie I., Picard-Jean F., Despins S., Mayette J., Hobdey S.E., Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA (New York, N.Y.). 2009;15(12):2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 17.Nomaguchi M., Teramoto T., Yu L., Markoff L., Padmanabhan R. Requirements for West Nile virus (–)-and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 2004;279(13):12141–12151. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- 18.Malet H., Egloff M.P., Selisko B., Butcher R.E., Wright P.J., Roberts M., Gruez A., Sulzenbacher G., Vonrhein C., Bricogne G., Mackenzie J.M., Khromykh A.A., Davidson A.D., Canard B. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 2007;282(14):10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 19.Yap T.L., Xu T., Chen Y.L., Malet H., Egloff M.P., Canard B., Vasudevan S.G., Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007;81(9):4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim S.P., Koh J.H., Seh C.C., Liew C.W., Davidson A.D., Chua L.S., Chandrasekaran R., Cornvik T.C., Shi P.Y., Lescar J. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J. Biol. Chem. 2013;288(43):31105–31114. doi: 10.1074/jbc.M113.508606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potisopon S., Priet S., Collet A., Decroly E., Canard B., Selisko B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014;42(18):11642–11656. doi: 10.1093/nar/gku666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y., Soh T.S., Zheng J., Chan K.W., Phoo W.W., Lee C.C., Tay M.Y., Swaminathan K., Cornvik T.C., Lim S.P., Shi P.Y., Lescar J., Vasudevan S.G., Luo D. A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 2015;11(3) doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannemann H., Sung P.Y., Chiu H.C., Yousuf A., Bird J., Lim S.P., Davidson A.D. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J. Biol. Chem. 2013;288(31):22621–22635. doi: 10.1074/jbc.M113.481382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble C.G., Lim S.P., Chen Y.L., Liew C.W., Yap L., Lescar J., Shi P.Y. Conformational flexibility of the Dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 2013;87(9):5291–5295. doi: 10.1128/JVI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caillet-Saguy C., Lim S.P., Shi P.Y., Lescar J., Bressanelli S. Polymerases of hepatitis C viruses and flaviviruses: structural and mechanistic insights and drug development. Antivir. Res. 2014;105:8–16. doi: 10.1016/j.antiviral.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Keller T.H., Chen Y.L., Knox J.E., Lim S.P., Ma N.L., Patel S.J., Sampath A., Wang Q.Y., Yin Z., Vasudevan S.G. vol. 277. Novartis Foundation symposium; 2006. pp. 102–253. (Finding New Medicines for Flaviviral Targets). [DOI] [PubMed] [Google Scholar]
- 27.Noble C.G., Shi P.Y. Structural biology of dengue virus enzymes: towards rational design of therapeutics. Antivir. Res. 2012;96(2):115–126. doi: 10.1016/j.antiviral.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Lim S.P., Noble C.G., Shi P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Yin Z., Chen Y.L., Schul W., Wang Q.Y., Gu F., Duraiswamy J.…Shi P.Y. An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. USA. 2009;106(48):20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Z., Chen Y.L., Kondreddi R.R., Chan W.L., Wang G., Ng R.H.…Keller T.H. N-sulfonylanthranilic acid derivatives as allosteric inhibitors of dengue viral RNA-dependent RNA polymerase. J. Med. Chem. 2009;52(24):7934–7937. doi: 10.1021/jm901044z. [DOI] [PubMed] [Google Scholar]
- 31.Niyomrattanakit P., Chen Y.L., Dong H., Yin Z., Qing M., Glickman J.F., Lin K., Mueller D., Voshol H., Lim J.Y., Nilar S., Keller T.H., Shi P.Y. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 2010;84(11):5678–5686. doi: 10.1128/JVI.02451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-Ariza L.L., Rocha-Roa C., Padilla-Sanabria L., Castaño-Osorio J.C. Virtual screening of drug-like compounds as potential inhibitors of the dengue virus NS5 protein. Front. Chem. 2022;10 doi: 10.3389/fchem.2022.637266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y.-H., Wu W.-y., Guo S.-X., He S.-J., Tang X.-D., Wu X.-Y., Nandakumar K.S., Zou M., Li L., Chen X.-G., Liu S.-W., Yao X.-G. [1,2,4]Triazolo[1,5-a]pyrimidine derivative (Mol-5) is a new NS5-RdRp inhibitor of DENV2 proliferation and DENV2-induced inflammation. Acta Pharmacol. Sin. 2020;41(5):706–718. doi: 10.1038/s41401-019-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian W., Xue J.-X., Xu J., Li F., Zhou G.-F., Wang F., Luo R.-H., Liu J., Zheng Y.-T., Zhou G.-C. Design, synthesis, discovery and SAR of the fused tricyclic derivatives of indoline and imidazolidinone against DENV replication and infection. Bioorg. Chem. 2022;120 doi: 10.1016/j.bioorg.2022.105639. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson R. SPR for molecular interaction analysis: a review of emerging application areas. J. Mol. Recogn. 2004;17(3):151–161. doi: 10.1002/jmr.660. [DOI] [PubMed] [Google Scholar]
- 36.Maddipati V.C., Mittal L., Kaur J., Rawat Y., Koraboina C.P., Bhattacharyya S., Asthana S., Gundla R. Discovery of non-nucleoside oxindole derivatives as potent inhibitors against dengue RNA-dependent RNA polymerase. Bioorg. Chem. 2023;131 doi: 10.1016/j.bioorg.2022.106277. [DOI] [PubMed] [Google Scholar]
- 37.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock 4 and AutoDockTools 4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu W., Chen L. The application of computer softwares in Chemistry teaching. International Journal of Education and Management Engineering. 2012;12:73–77. doi: 10.5815/ijeme.2012.12.12. [DOI] [Google Scholar]
- 39.Studio D. 2008. Discovery Studio. Accelrys [2.1] [Google Scholar]
- 40.Study of 5-Endro-Trig Radical Cyclisations, Application towards New Synthesis of Lycorine, Synthesis of New DNA Intercalating-Alkylating Agents, Analogues to Duocarmycins by Dauge. Delfine from null.; 2005. [Google Scholar]
- 41.Preparation of 3-Heteroarylmethylidene-2-Indoline Protein Kinase Inhibitors for Use against Cancer and Other Disorders by McMahon. Gerlad etal from U.S.; 26 Nov 2002. [Google Scholar]
- 42.Jui-Yi H., Kai-Cheng H., Ching S., Ching-Hsuan C., Tiny Eight L., Tzu-Ying S., Hui-Ju T., Shih-Chung Y., Chai-Ron Y., Wei-Jan H. Design synthesis, Biological evaluation of indolin-2-one derivatives as novel cyclin- dependent protein kinase 8 (CDK8) inhibitors. Biomed. Pharmacother. 2023;159:114258. doi: 10.1016/j.biopha.2023.114258. [DOI] [PubMed] [Google Scholar]
- 43.Taohua P., Maofei H., Lulu D., Jiang L., Yanhua F., Xiaojiang H., Shuzhen M. Design, synthesis, evaluation of the COX-2 inhibitory activities of new 1,3-dihydro-2H indolin-2-one derivatives. Molecules. 2023;28(12):4668. doi: 10.3390/molecules28124668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.