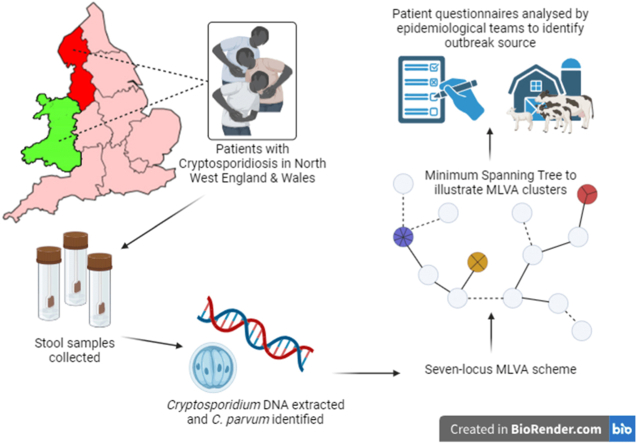
Abstract
The protozoan Cryptosporidium parvum is an important cause of gastroenteritis in humans and livestock, and cryptosporidiosis outbreaks are common. However, a multi-locus genotyping scheme is not widely adopted. We describe the further development and application of a seven-locus multi-locus variable number of tandem repeats analysis (MLVA) scheme. From 28th March to 31st July 2022, confirmed C. parvum stools (n = 213) from cryptosporidiosis patients (cases) in Wales (n = 95) and the north west of England (n = 118) were tested by MLVA. Typability (defined as alleles identified at all seven loci in a sample) was 81.2% and discriminatory power estimated by Hunter Gaston Discriminatory Index was 0.99. A MLVA profile was constructed from the alleles, expressed in chromosomal order. Profiles were defined as simple (single allele at each locus) or mixed (more than one allele at any locus). A total of 161 MLVA profiles were identified; 13 were mixed, an additional 38 simple profiles contained null records, and 110 were complete simple profiles. A minimum spanning tree was constructed of simple MLVA profiles and those identical at all seven loci defined genetic clusters of cases (here, null records were considered as an allele); 77 cases formed 25 clusters, ranging from two to nine (mode = two) cases. The largest cluster, following epidemiological investigation, signalled a newly-identified outbreak. Two other cases with mixed profiles that contained the outbreak alleles were included in the outbreak investigation. In another epidemiologically-identified outbreak of six initial cases, MLVA detected two additional cases. In a third, small outbreak of three cases, identical MLVA profiles strengthened the microbiological evidence. Review of the performance characteristics of the individual loci and of the seven-locus scheme suggested that two loci might be candidates for review, but a larger dataset over a wider geographical area and longer timeframe will help inform decision-making about the scheme by user laboratories and stakeholders (such as public health agencies). This MLVA scheme is straightforward in use, fast and cheap compared to sequence-based methods, identifies mixed infections, provides an important tool for C. parvum surveillance, and can enhance outbreak investigations and public health action.
Keywords: Cryptosporidium parvum, MLVA, Subtyping, Outbreak, Cluster, Multi-locus
Graphical abstract
Highlights
-
•
The performance of seven VNTR loci and the MLVA scheme was evaluated.
-
•
The typability of 213 Cryptosporidium parvum isolates was 81.2%.
-
•
A new outbreak was identified by MLVA.
-
•
MLVA strengthened the microbiological evidence in outbreak investigations.
1. Introduction
Protozoan parasites of the genus Cryptosporidium, especially Cryptosporidium parvum and Cryptosporidium hominis, are notable causes of acute gastroenteritis (cryptosporidiosis) worldwide (Davies and Chalmers, 2009; Checkley et al., 2015; Feng et al., 2018). Symptoms include profuse watery diarrhoea, abdominal cramps, vomiting and/or nausea, low-grade fever and loss of appetite, and can be prolonged, with a mean duration in one UK study of 12.7 days (Hunter et al., 2004). Infection with this parasite is a particular risk for young children, the elderly and the immunocompromised. People with profound T-cell deficiencies can develop protracted, severe diarrhoea, with complications including sclerosing cholangitis and rarely, biliary cirrhosis and pancreatitis (Hunter and Nichols, 2002). Cryptosporidium spp. are one of the leading causes of diarrhoea in young and malnourished children and present an increased risk of death in those under 24 months of age (Kotloff et al., 2013; Sow et al., 2016; Levine et al., 2020; Hossain et al., 2023).
Genotyping Cryptosporidium spp. from patient specimens (usually stools), and further subtyping is important for the epidemiological surveillance of the parasite, inferring linkage and for the investigation of outbreaks (Ryan et al., 2021). In England and Wales, Cryptosporidium-positive stools are submitted to the Cryptosporidium Reference Unit (CRU) for species identification by PCR-based methods (Chalmers et al., 2019; Robinson et al., 2020). Further subtyping is then undertaken (if required) by sequencing part of the hyper-variable 60 kDa glycoprotein (gp60) gene (Strong et al., 2000; Chalmers et al., 2019). Most C. parvum and C. hominis infections and outbreaks in England and Wales have been caused by a few common gp60 subtypes (Chalmers et al., 2019), but a multilocus scheme would be more suitable for investigating the potential variation within the genome arising from recombination during the sexual stage of the parasitic life-cycle, resulting in heterogeneity (Widmer and Lee, 2010; Morris et al., 2019). The choice of markers would be different for each species due to their genetic variances (Robinson and Chalmers, 2012).
Cryptosporidium parvum has a wider host range than C. hominis and infects humans, primarily but not exclusively through zoonotic transmission (Chalmers and Giles, 2010) either by direct animal-person contact (especially young livestock) or indirectly through fomites or the consumption of contaminated drinking water or food (EFSA Panel on Biological Hazards (BIOHAZ) et al., 2018). A multi-locus variable number of tandem repeats (VNTR) analysis (MLVA) scheme was prioritised, developed and validated for C. parvum on a range of samples from gp60 subtype families IIa, IId and IIc by Robinson et al. (2022). The applicability of the scheme for other Cryptosporidium spp., especially C. hominis, was addressed in that paper. The scheme utilises fragment sizing of seven loci for rapid discrimination through generation of MLVA profiles that can then be mapped with minimum spanning trees to infer linkage. During validation on a large number of patient specimens, a high proportion (79%) of MLVA profiles were unique, suggesting genetic clusters might indicate unrecognised outbreaks (Robinson et al., 2022). Further work has been done to investigate the use of MLVA for the epidemiological surveillance of C. parvum to identify otherwise-missed epidemiological links, during the spring when C. parvum is most prevalent in the human population; here, we describe the identification of VNTR alleles using the SeqStudio Genetic Analyser (Applied Biosystems) for fragment sizing, the performance of the loci and the laboratory findings. The epidemiological analysis is described elsewhere (Chandra et al., in preparation).
2. Materials and methods
The materials and methods described here refer to the validated 7-locus MLVA scheme (Robinson et al., 2022), and further details, the size and sequence of each repeat unit, along with figures showing sequence alignments of example alleles, were included in Supplementary file 1 of that paper; an updated version is available as “MLVA assay protocol” on our website at https://phw.nhs.wales/services-and-teams/cryptosporidium-reference-unit/.
2.1. MLVA scheme
The following loci were examined and results expressed in chromosomal order: cgd1_470_1429 (cgd1); cgd4_2350_796 (cgd4); cgd5_10_310 (MSF); cgd5_4490_2941 (cgd5); cgd6_4290_9811 (cgd6); cgd8_4440_NC_506 (cgd8); cgd8_4840_6355 (MM19) (Pérez-Cordón et al., 2016; Robinson et al., 2022).
2.2. Specimens, DNA extraction and PCR
All C. parvum specimens identified between 28th March 2022 and 31st July 2022 from diagnostic microbiology laboratories in Wales and the northwest of England were subtyped by MLVA. To encourage continued referral of all Cryptosporidium-positive stools to the CRU, laboratories in these regions were informed in writing of the study prior to its initiation. DNA was extracted using the QIAamp Fast DNA Stool Kit (Qiagen, Hilden, Germany) and C. parvum was confirmed by real-time PCR (Robinson et al., 2020).
Two multiplex PCRs (a 4-plex: cgd1, cgd4, cgd8 and MM19; and a 3-plex: MSF, cgd5 and cgd6) for the seven VNTR markers were performed with the Type-it Microsatellite PCR kit (Qiagen). PCR amplicons were diluted 1 in 10 with HiDi Formamide (Thermo Fisher Scientific, Paisley, UK) and 2 μl added to a master mix comprised of 12 μl HiDi Formamide and 0.5 μl GeneScan 600 Liz dye Size Standard 2.0 (Thermo Fisher Scientific) per sample. Amplicons were sized on a SeqStudio Genetic Analyser (Thermo Fisher Scientific) using the following settings: (i) size standard: GS600_LIZ (60–400); (ii) dye set: G5 (D2-33); (iii) run module 1: FragAnalysis.
2.3. Variable number of tandem repeats analysis
Raw.fsa files were imported into BioNumerics software (version 7.6, Applied Maths), which allows the loci with longer repeat units (e.g. MSF is a 12 bp repeat and cgd4 a 15 bp repeat) to be analysed readily, and also allows for simultaneous visualisation of alleles in individual or multiple channels. The MLVA management function was used construct bins to capture peaks and determine VNTR alleles by the calculated number of repeats. The bin ranges were initially created from the validation panel of 259 specimens described previously using the mean band sizes for peaks from the same allele ± 0.7 bp (Robinson et al., 2022). New alleles were confirmed by sequencing.
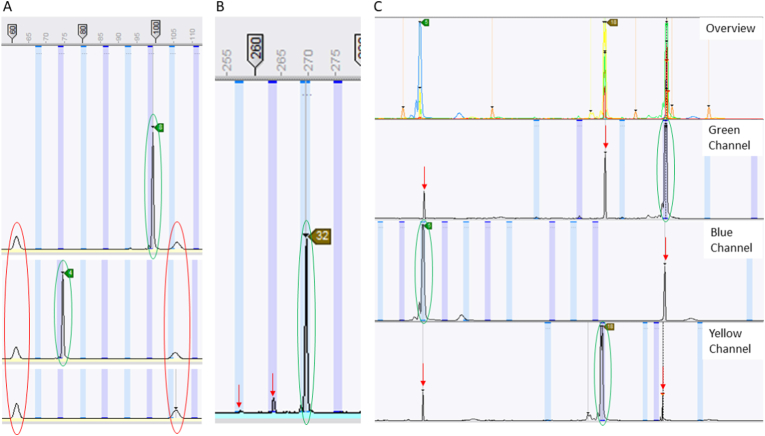
Care was taken to identify and report only true peaks, determined as follows: (i) the peak must sit within a confirmed allele size bin (Fig. 1A–C); (ii) the peak must be tall and narrow (Fig. 1A–C); (iii) the peak must not be artefactual fluorescence from this or another channel (artefactual peaks are typically broader than true peaks and usually appear in every sample on the run, or in the same position in multiple loci) (Fig. 1A); (iv) where multiple peaks are present the weaker peak(s) should be > 25% of the height of the largest peak to be considered true (Fig. 1B); (v) the peak must not be bleed-through from a true peak from another locus/channel (observed when all channels are overlaid and visualised simultaneously) (Fig. 1C); and (vi) the peak height should be ≥ 150 relative fluorescence units (RFU).
Fig. 1.
Illustration of true, artefactual and bleed-through peaks from the SeqStudio Genetic Analyser in BioNumerics software. A True peaks (circled in green) (with peak heights of ≥ 150 RFU) as well as short and wide artefactual peaks (circled in red), which can be observed at the same number of base pairs across different samples, including an NTC (displayed in the bottom image). B A true peak (circled in green) as well as short and narrow stutter peaks (indicated by red arrows). C Bleed through of strong peaks that occur in the same position across other channels. In the green channel, the true peak (highlighted in green) bleeds through to the blue and yellow channels (indicated by red arrows). In the blue channel, the true peak (circled in green) also bleeds through to the green and yellow channels (highlighted with red arrows). In the yellow channel, the true peak (circled in green) also bleeds through to the green channel (indicated by a red arrow).
A MLVA profile was compiled for each sample by listing the calculated number of repeats at each locus in chromosomal order (cgd1, cgd4, MSF, cgd5, cgd6, cgd8, MM19), and expressed as shown in the following example: 4-14-5-7-27-32-15.
Samples that contained one true peak at each locus were described as having simple profiles, and those with > 1 true peak at any locus were defined as mixed profiles. Mixed MLVA profiles were recorded with a forward slash between the alleles observed at each locus, with the lowest copy number displayed first regardless of which was stronger, for example cgd4 here has two alleles: 4-13/14-5-7-27-32-15. Hypothetical simple profiles were not created from mixed profiles.
If no true peaks were visible, or if peaks were < 150 RFU, the DNA was concentrated by vacuum desiccation at 45 °C for 45 min (Concentrator Plus, Eppendorf), reconstituted in 20 μl nuclease free water and re-tested.
If no allele was identified either initially or on re-test, then a null symbol (Ø) was recorded. Patterns in the occurrence of null records were explored. To investigate sequence variation within the locus including polymorphisms in the primer binding sites, we looked for samples in our collection that had been genome sequenced, and applied MLVA PCRs to identify those with null records.
2.4. Quality control
PCR-positive (C. parvum DNA) and no template controls (nuclease-free water) were included in every PCR batch. The C. parvum DNA also acted as a reference sample of two known MLVA profiles: 4-14-5-7-27-32-15 and 6-13-3-6-23-15-4. A GeneScan 600 Liz size standard (v2.0, Thermo Fisher Scientific) was included to ensure accurate allele sizing. In addition to the PCR-positive DNA samples, two C. parvum DNA samples were repeated as quality control and produced the same results each time.
2.5. Data analysis: Scheme and marker performance
Initially, only samples with one or more alleles at all 7 loci were defined as typable. Following an investigation of null records at the cgd1 locus, re-analysis of the data as a 6-locus scheme without cgd1 was also explored. Typability was calculated for each individual locus as well as for the 6- and 7-locus schemes.
Samples with complex profiles were described but excluded from statistical and graphical analyses where their inclusion would have necessitated the construction of hypothetical profiles.
The variability of the VNTRs was investigated using the Hunter Gaston Diversity Index (HGDI) calculated for each locus individually, and for the complete schemes, for all non-outbreak (presumably unrelated) samples (Hunter and Gaston, 1988). For outbreaks, alleles from a single representative of each profile were included. Null alleles were excluded. Alleles from samples with mixed profiles were included in the calculation of HGDI for each locus but were excluded for the complete schemes.
To compare allelic variation over time, the present study and the historical case control study dataset in the validation study by Robinson et al. (2022) was investigated by plotting frequency distribution. The case-control study dataset was used as the sampling frame was similar to the present study.
To investigate the efficiency of the typing scheme, the maximum number of MLVA profiles that were generated from the best combination of each of one through to seven loci were inferred from Accurate Marker Choice for Accession Identification and Discrimination (AMaCAID) analysis using Model 1 in R (version 4.2.1) (Caroli et al., 2011). AMaCAID graphs were constructed to show the maximum number of MLVA profiles that were generated from the dataset when the optimal combination of loci was used. A single representative of each profile was used; samples with null records and mixed alleles were excluded from the dataset prior to this analysis. The analysis was re-run for a 6-locus scheme that excluded cgd1, thus including those samples previously excluded due to null records at this locus.
2.6. Data analysis: MLVA profiles and clusters
Clusters of cases with the same MLVA profile were identified with a macro in Microsoft Excel. Null records were regarded as an allele (Ø) when identifying clusters. Genetic clusters were defined as two or more cases with an identical MLVA profile. To display all simple profiles and arising clusters, a minimum spanning tree (MST) was constructed using BioNumerics software (version 7.6, Applied Maths). Samples were assigned as non-cluster, cluster or outbreak, depending on the epidemiological data obtained. Unique case identifiers were shared with the epidemiologists at the UKHSA Field Service North West for case specimens referred from laboratories in the north west of England and Public Health Wales’ Communicable Disease Surveillance Centre for those from Wales for further investigation with routinely collected epidemiological/exposure data, as described elsewhere (Chandra et al., in preparation).
3. Results
A total of 213 PCR-confirmed C. parvum specimens, from 213 cryptosporidiosis cases, were tested during the study, 95 from Wales and 118 from northwest England.
3.1. MLVA scheme and marker performance
Overall typability where all seven loci had identifiable alleles was 173/213 (81.2%).
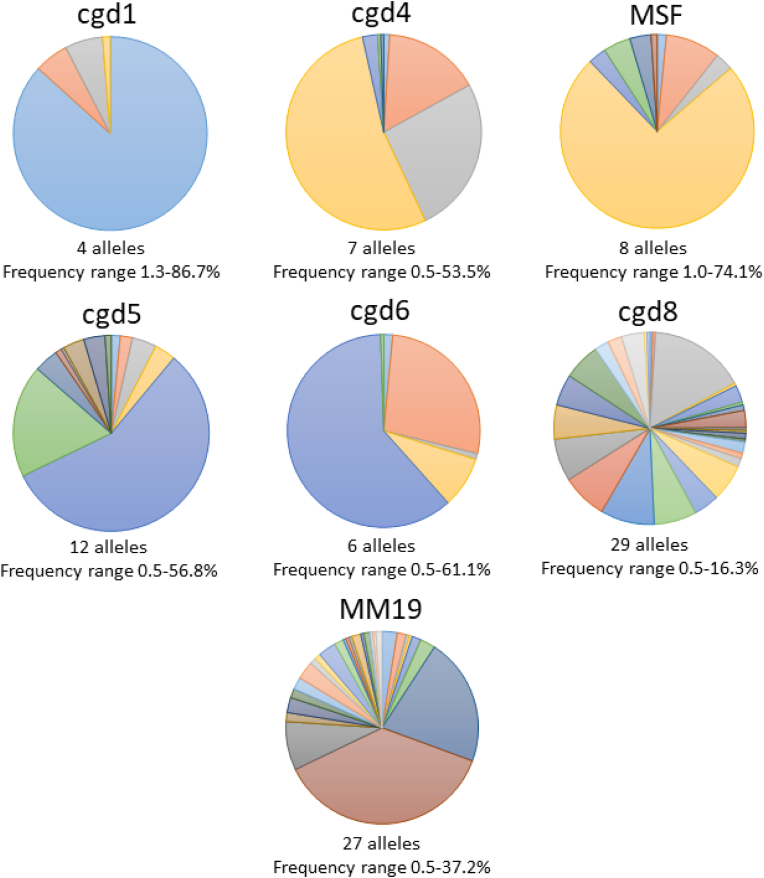
Further to those identified previously (Robinson et al., 2022), 19 new alleles were confirmed by Sanger sequencing and new bins were generated with the confirmed peak sizes. The most variable locus was cgd8 with 29 alleles and the least variable was cgd1 with four alleles (Table 1, Fig. 2).
Table 1.
Variability of the VNTRs in 213 Cryptosporidium parvum samples.
| Locus | Range of calculated no. of repeats (Median) | No. of alleles | No. of samples with null records | No. of samples with identified alleles (% typability) | Hunter-Gaston discrimination index |
|---|---|---|---|---|---|
| cgd1 | 4–7 (4) | 4 | 40 | 173 (81.2) | 0.242 |
| cgd4 | 11–17 (14) | 7 | 4 | 209 (98.1) | 0.622 |
| MSF | 2–10 (5) | 8 | 1 | 212 (99.5) | 0.439 |
| cgd5 | 3–18 (7) | 12 | 1 | 212 (99.5) | 0.638 |
| cgd6 | 13–31 (23) | 6 | 1 | 212 (99.5) | 0.547 |
| cgd8 | 4–54 (26) | 29 | 2 | 211 (99.1) | 0.933 |
| MM19 | 4–40 (16) | 27 | 6 | 207 (97.2) | 0.807 |
Fig. 2.
Pie graphs illustrating the frequency with which alleles were detected at individual loci, with each segment representing the proportional fraction each allele contributed to all those at each locus. The number of alleles and range of allele frequency (%) for each locus is shown. The colours are used simply to make the segments clear within each pie.
Discriminatory power, determined by the HGDI, varied by locus (Table 1). HGDI is a function of the number of alleles detected at each locus and their frequency of detection. For example, although more alleles were detected in MSF than either cgd4 or cgd6, the HGDI was lower as one allele accounted for 74% of the total in MSF (Fig. 2).
The frequency distribution and allelic variation (Fig. 2) was similar to that observed in the case control study dataset in the validation study indicating little change over the different time periods (Supplementary file S1).
Fifty-six (26.2%) samples were desiccated as they initially had null record(s) at one or more loci. Sixteen of these samples became fully typable, leaving 40 samples that retained null records at one or more loci; 33 samples had null records in 1 locus, three at 2 loci, three at 3 loci and one at all 7 loci (this one was therefore negative by MLVA). All 40 samples that were not fully typable had a null record at the cgd1 locus. Null records occurred at a much lower frequency at other loci; six at MM19, four at cgd4, and two at cgd8.
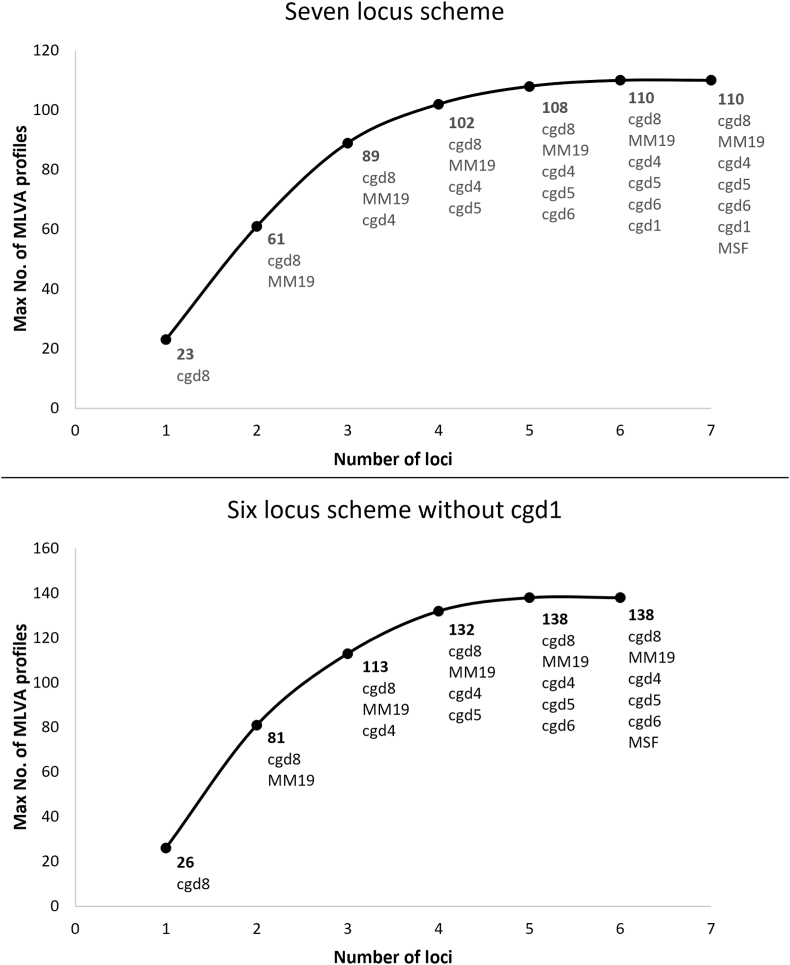
When cgd1 was excluded from analysis of the MLVA scheme, an additional 33 samples were typable and 28 more complete MLVA profiles were identified; the overall typability improved to 96.7% and the discriminatory power was 0.995 (Table 2). Due to the increased number of typable samples included in the dataset when cgd1 was removed prior to analysis, AMaCAID analysis showed that the maximum number of profiles generated from these data was greater at each number of combined loci with the remaining 6 loci, than when all 7 loci were included (Fig. 3).
Table 2.
Performance of the 7-locus and 6-locus MLVA schemes in 213 Cryptosporidium parvum samples.
| MLVA scheme | No. of samples with identified alleles (% typability) | Hunter-Gaston discrimination index | No. of complete, simple profiles |
|---|---|---|---|
| 7-locus | 173 (81.2) | 0.994 | 110 |
| 6-locus (without cgd1) | 206 (96.7) | 0.995 | 138 |
Fig. 3.
AMaCAID analysis of the maximum number of genotypes discriminated by optimal marker combinations for the 7-locus and 6-locus schemes. The graphs show the number of MLVA profiles generated by sequential inclusion of optimal loci within each dataset with cgd1 (top) and without cgd1 (bottom). A single representative of each profile was used; samples with null records and mixed alleles were excluded from the dataset prior to this analysis.
AMaCAID analysis further revealed that, in terms of discrimination, MSF appeared to be the least useful locus in both the 7- and 6-locus schemes for this data set (Fig. 3). Omitting MSF still achieved the maximum number of MLVA profiles whereas omitting other optimally combined loci reduced the maximum number (Fig. 3).
3.2. Exploration of cgd1
As all 40 samples containing null records included nulls at cgd1, we further investigated this locus to determine the cause. The null records may have indicated any or all of: a low concentration of DNA template; a truly absent or null allele; or variation in the primer sequence sites reducing amplification efficiency.
We first explored the Ct-values in the real-time PCRs that identified C. parvum. For the 40 samples that generated null records at cgd1, the mean Ct-value was 32.18 (range 29.22–38.16, SD 3.15), significantly higher than the mean Ct-value of 30.29 from the 173 typable samples (range 24.44–35.73, SD 3.05) (t-value = 2.72, P = 0.007), indicating that some of the null records may have been from samples with a low concentration of DNA template. The samples were from a variety of gp60 subtype families; IIa (n = 11), IIc (n = 6), IId (n = 19), IIe (n = 1) and three were not typable at gp60. To explore variation in primer sites we analysed whole genome sequence data from five other C. parvum samples in our collection (UKP97, UKP99, UKP127, UKP128 and UKP129 from BioProject PRJEB15112) for which MLVA PCR and fragment sizing generated null records at cgd1. The sequence alignment revealed mismatches in the cgd1 PCR primer sites potentially explaining the non-amplification at this locus (Supplementary file S2), and indicating that null records generated with this primer set should be treated with caution and not interpreted as true null alleles.
An alternative primer set, designed for allele confirmation by sequencing (Robinson et al., 2022), achieved amplification of the microsatellite region from 21/40 previously negative samples. Two samples had alleles of four repeats (both with weak alleles at other loci), 13 samples had five repeats and six had six repeats. These sequences confirmed the mismatches in the MLVA primer regions seen in the genome sequence data, and revealed further sequence variation and minisatellite repeats outside of the targeted microsatellite region. Additionally, apart from the two weak samples with four repeats, unexpected heterogeneity was seen in the upstream sequences. The samples with five repeats all had an additional 27 bp minisatellite (two or three repeats) in the upstream sequence. Samples with six repeats had a 3 bp microsatellite coding for glycine (six repeats).
3.3. MLVA profiles and clusters
A total of 161 MLVA profiles were identified from the 7-locus scheme (see Supplementary file S3); 13 (8.1%) of these were mixed profiles of which twelve were mixed at cgd8 (the most variable locus, Table 1), five at cgd4, four at MM19, two at cgd5, one at cgd6. None were mixed at cgd1 or MSF. Thirty-eight simple profiles contained null records, and 110 were complete simple profiles.
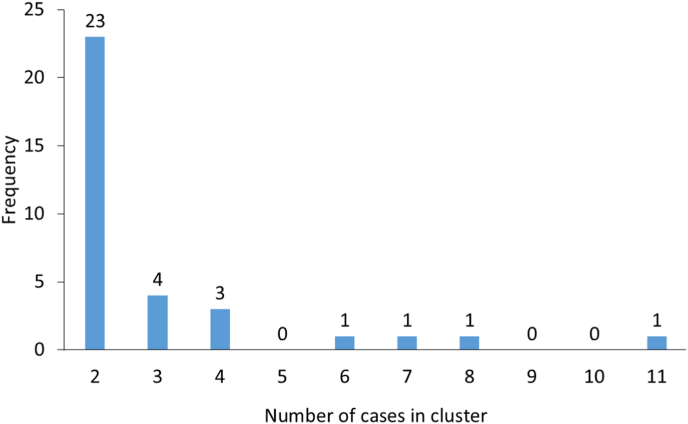
When all case samples were analysed, including those with mixed profiles, 92 out of 213 (43.2%) were part of 34 MLVA clusters (range 2–11 cases); the majority (23 clusters) were 2-case clusters (Fig. 4). A total of 56 cases were part of the 11 (32.4%) MLVA clusters that comprised three or more cases.
Fig. 4.
Size distribution of MLVA clusters with simple or mixed MLVA profiles, among 92/213 Cryptosporidium parvum cases. The numbers above the columns are the number of clusters with n cases. The MLVA profiles and notes on the public health significance of clusters are provided in Supplementary file S3.
To generate an MST, only the 148 simple profiles were included; 122 out of 148 (82.4%) profiles were unique (unlinked by MLVA) within this dataset, although there was one additional profile containing null records at all loci which was not shown in the MST. There were 77 out of 148 (52.0%) cases in 25 clusters, ranging from 2 to 9 (mode = 2) cases (Fig. 5). There were 10 out of 25 (40.0%) clusters that comprised 3 or more cases. Running the analysis without cgd1 resulted in no additional clusters in the MST but did increase the number of cases in three of the existing clusters by a single case each (data not shown). Omitting MSF from the analysis (as suggested by the AMaCAID analysis in Fig. 3) did not change the clustering in the MST (data not shown).
Fig. 5.
Minimum spanning tree of MLVA of 147 simple profiles, displaying non-cluster cases, outbreak cluster cases, and other clusters of cases. Each MLVA profile is indicated by one node or branch tip displayed as circles and connected by branches. The branch line style indicates the number of loci that differ between MLVA profiles: thick solid line, 1 difference; thin solid line, 2 differences; dashed line, 3 differences; no line, ≥ 4 differences. The branch length is not indicative of genetic distance. The wedges in the circles indicate the number of samples with that MLVA profile.
The largest cluster of nine cases in the MST (Fig. 5), with the simple MLVA profile 4-12-5-7-27-37-16, was further investigated by the local Health Protection Team. It was discovered to indicate a newly identified outbreak that had not been recognised during routine surveillance but was subsequently found linked to an open farm. Although the construction of hypothetical profiles was avoided and mixed profiles were excluded from the MST, analysis of the distribution of common alleles was useful, especially in this outbreak. Of two cases with mixed MLVA profiles that included the outbreak alleles, one with the profile 4-12/13-5-7-27-31/37-16 was from a case that had visited the open farm (Supplementary file S3).
Two MLVA clusters signalled outbreaks that were already under investigation by the local Health Protection Teams. In the cluster identified as part of outbreak A, also linked to an open farm, six cases had the MLVA profile 4-12-5-7-27-28-16. Two of the cases had not previously been identified as part of the outbreak and MLVA enabled their inclusion in the investigation. One further case that displayed in the MST as a non-cluster case had the MLVA profile 4-14-5-7-27-30-16, which differed from the main outbreak at two loci, cgd4 and cgd8, but had also visited the farm indicating that another MLVA profile might be connected to the outbreak. The finding of various profiles among cases linked to farm premises may occur due to the variety of animals present and/or duration of outbreaks, or the case may have acquired their infection elsewhere.
In outbreak B the identification of a single MLVA profile 4-14-5-7-27-31-17 in all three cases was important as it strengthened the microbiological evidence for the common exposure considered by the Health Protection Team as the source of the outbreak.
The three outbreaks, the investigation of the other 22 clusters in the MST, and the epidemiological/exposure data available for all cases shown in Fig. 5 and those with mixed infections are described in more detail by Chandra et al., in preparation.
4. Discussion
Here we describe the application of MLVA in real-time to a large number of C. parvum specimens, encompassing all those identified from Wales and the northwest of England, an area with a combined population of over 10.5 million, in late spring/early summer 2022. As such they represented the range of C. parvum diversity (including IIa, IIc, IId and IIe gp60 subtype families) in clinical cases in the area and study period. We have assessed the performance of the MLVA scheme and individual markers and investigated whether samples with identical MLVA profiles from a similar space and time are indicative of outbreaks during the spring peak period. The epidemiology and exposure data of the cases, their MLVA profiles and clustering are explored further elsewhere (Chandra et al., in preparation). This MLVA scheme was previously applied successfully to assist in the investigation of an outbreak associated with an on-farm milk vending machine, linked human cases and the cattle herd (Gopfert et al., 2022).
We have further adapted and developed the method from the validation study (Robinson et al., 2022), first by adoption of fragment sizing in-house on the SeqStudio (Thermo Fisher) instrument. This reduced the turnaround times and allowed us greater management of data and verification of alleles. Secondly, including the use of BioNumerics software (version 7.6, Applied Maths, Belgium) to produce a database of fragment sizing data for identification of number of repeats (alleles) at seven loci, and generation of MSTs for the analysis of case clusters and infer genetic linkage. The proportion of unique simple profiles (82.4%) among all samples, and the distribution of alleles, was similar to the relevant part of the validation study (79.4%) (Robinson et al., 2022), indicating little genetic drift at these loci over time. The use of BioNumerics software provided many benefits to the analysis, including timely identification of new alleles through the automatic assignment of VNTRs with bin placements, and the intuitive manipulation of MST to identify clusters. Unfortunately, BioNumerics will be phased out on December 31st, 2024 as bioMérieux state they are unable to support the ever-growing demands of sequencing (https://www.bionumerics.com/). As BioNumerics has been very fruitful for our research we hope that the software will be reincarnated in the future; however, we expect this is unlikely and alternative software should be explored for the same functionalities.
The MLVA scheme performed well with good typability of the 7-locus scheme of 81.2%, similar to that found during the validation study (85.3%) (Robinson et al., 2022), achieved through the addition of DNA concentration of initially untypable samples. During this study we defined a peak height threshold of ≥ 150 RFU for allele acceptance, and those not reaching threshold were desiccated and retested. Since the study and after reassessing the RFU values before and after desiccation for 1000 samples, we have reduced the threshold to ≥ 50 RFU as the same allele was observed after desiccation. This new threshold will be reflected in the transfer document on our website, https://phw.nhs.wales/services-and-teams/cryptosporidium-reference-unit/.
Most of the loss of typability was at cgd1 and omitting this locus from the scheme improved both typeability and discriminatory power estimated by HGDI. The most likely underlying cause was mismatches in the primer sequences at cgd1 which may have produced poor amplification resulting in no or weak peaks. Although the re-design of the primers for cgd1 would seem to be the solution, this would prove difficult within the requirements of fragment sizing (Nadon et al., 2013; Chalmers et al., 2017), due to the amount of sequence variation and the presence of additional mini- and microsatellites in some isolates.
AMaCAID analysis revealed the best combinations of loci and the “added value” of including certain loci to maximise the number of unique MLVA profiles that could be generated. This implied that while the inclusion of cgd1 in the 7-locus scheme generated two additional unique MLVA profiles, the increased typeability from excluding cgd1 resulted in an additional 28 unique MLVA profiles. Removal of cgd1 from the MST analysis increased three clusters by a single case each, but the epidemiological analysis will reveal whether or not this was helpful (Chandra et al., in preparation). AMaCAID analysis also implied there was nothing to gain from including MSF, also borne out by no change in the clusters identified in the MST following its exclusion. While it might be tempting to conclude that the 6-locus scheme performed better than the 7-locus scheme, higher typeability, fewer null alleles, a higher number of maximum MLVA profiles and higher HGDI-discrimination, this would be premature. The value of continuing to include these loci in the MLVA scheme needs to be established by the analysis of a larger data set over a wider geographical area, both of the UK and other countries, and a longer period of time. This work is underway and will help inform subsequent review of the scheme by its users.
Using the 7-locus scheme, we have shown that the microbiological evidence during known outbreaks can be strengthened by providing a genetic link between case specimens, and that additional cases can also be identified through genetic cluster detection. Importantly, previously unrecognised outbreaks can be identified that might have been missed in routine epidemiological surveillance. MLVA is already accepted for use in epidemiological surveillance and investigation of outbreaks caused by other pathogens, mainly bacteria (Keim et al., 2000; Malorny et al., 2008; Wuyts et al., 2013;Strydom et al., 2019; Kabała et al., 2021).
Mixed profiles were identified in this study. This concurs with evidence from other analyses, such as genome sequencing and re-analysis of gp60 chromatograms, that mixed C. parvum infections occur (Grinberg and Widmer, 2016; Morris et al., 2019; Dettwiler et al., 2022). There is no standardisation for interpreting mixed MLVA profiles for most pathogens, but their inclusion is useful in practice for potentially linking cases to outbreaks, as we discovered in the newly identified outbreak. It may therefore be important to develop a probabilistic approach to formalise how to include and interpret isolates with mixed profiles within putative clusters.
There were some limitations to our study. It is possible that some profiles and subsequently clusters were mis-classified due the presence of null records, and some alleles were indeed identified by further analysis of cgd1. It is possible that clusters sharing profiles at seven loci and those sharing six of those seven plus a null record at cgd1 are potentially related (or not). Again, a probabilistic framework will help decide how to approach these; we propose a cautionary approach for treating null records, and incorporation of epidemiological data may provide clarification. The study was also limited in time, covering a 4-month period, although this did span the spring peak in C. parvum cases. The study focused on a limited geographical area of Wales and northwest England; clusters of cases or outbreaks involving cases beyond the boundaries may have been missed, there may be different MLVA profiles elsewhere, and the applicability of the scheme to other countries is under investigation. Epidemiological analysis is underway to further refine our current definition of a MLVA cluster (two or more cases with an identical MLVA profile), for future practical application and incorporation in public health practice, taking into account temporal factors for example. The work described here was to evaluate the performance of the markers and scheme for identification of C. parvum clusters and outbreaks. We have not mapped all these cases to gp60 subtypes, but where that analysis was done it showed IIa, IIc, IId and IIe subtype families were included.
MLVA offers a fast turnaround time, which is crucial for outbreak investigations. From specimen reception to the MLVA profile can be two to three working days, and is a seamless addition to genotyping workflows, using DNA already extracted for species identification. All C. parvum specimens from Wales and northwest England will continue to be analysed for MLVA profile identification. The intention is to extend the scheme to specimens submitted from diagnostic laboratories in all regions, supported by automation of cluster identification, mechanisms for cluster reporting, and to provide context to systematic capture of MLVA profiles in surveillance systems. A PubMLST database (PubMLST.org) is being developed to allow MLVA profiles to be compared between users of the scheme and provide high-level international surveillance of predominant and emerging subtypes.
5. Conclusions
This MLVA scheme has the potential to provide benefits for public health investigations. Known outbreaks were characterised and additional cases (not initially recognised as epidemiologically-linked) were identified, strengthening evidence in ongoing outbreak investigations. Importantly, a previously unrecognised outbreak was identified. The scheme provides a valuable tool for the identification and control of C. parvum outbreaks, through interventions to limit transmission from the source and the prevention of further cases.
Funding
This work was supported by the National Institute for Health and Care Research (NIHR) Health Protection Research Unit in Gastrointestinal Infections, a partnership between the UK Health Security Agency, the University of Liverpool and the University of Warwick. The views expressed are those of the authors and not necessarily those of the NIHR, the UK Health Security Agency or the Department of Health and Social Care.
Ethical approval
Not applicable.
Data availability
The data supporting the conclusions of this article are included within the article and its supplementary files, and in Robinson et al. (2022). The laboratory and analytical methods have been made freely available on our website at: https://phw.nhs.wales/services-and-teams/cryptosporidium-reference-unit/.
CRediT authorship contribution statement
Harriet Risby: Data curation, Formal Analysis, Investigation, Methodology, Visualisation, Writing - original draft. Guy Robinson: Formal analysis, Investigation, Methodology, Supervision, Visualisation, Writing - review & editing. Nastassya Chandra: Investigation, Methodology, Writing - review & editing. Grace King: Investigation, Writing - review & editing. Roberto Vivancos: Funding acquisition, Methodology, Supervision, Writing - review & editing. Robert Smith: Funding acquisition, Methodology, Resources. Daniel Thomas: Funding acquisition, Methodology, Resources. Andrew Fox: Investigation, Writing - review & editing. Noel McCarthy: Conceptualisation, Methodology, Writing - review & editing. Rachel M. Chalmers: Conceptualisation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing - review & editing. All authors read and approved the final manuscript.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Rahma Mohammed, Cryptosporidium Reference Unit, for specimen preparation and DNA extraction, Kristin Elwin, Cryptosporidium Reference Unit, for Cryptosporidium species identification and helpful comments on the manuscript, Arthur Morris, Cardiff University, for Cryptosporidium parvum genome analysis, diagnostic microbiology laboratories for continuing to send Cryptosporidium-positive stools for genotyping, Health Protection Teams and Environmental Health Departments for following up cases. The Graphical Abstract was created with www.BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crpvbd.2023.100151.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Caroli S., Santoni S., Ronfort J. AMaCAID: A useful tool for accurate marker choice for accession identification and discrimination. Mol. Ecol. Res. 2011;11:733–738. doi: 10.1111/j.1755-0998.2011.02993.x. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M., Giles M. Zoonotic cryptosporidiosis in the UK - challenges for control. J. Appl. Microbiol. 2010;109:1487–1497. doi: 10.1111/j.1365-2672.2010.04764.x. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M., Robinson G., Elwin K., Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites Vectors. 2019;12:95. doi: 10.1186/s13071-019-3354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R.M., Robinson G., Hotchkiss E., Alexander C., May S., Gilray J., et al. Suitability of loci for multiple-locus variable-number of tandem-repeats analysis of Cryptosporidium parvum for inter-laboratory surveillance and outbreak investigations. Parasitology. 2017;144:37–47. doi: 10.1017/S0031182015001766. [DOI] [PubMed] [Google Scholar]
- Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A.P., Chalmers R.M. Cryptosporidiosis. BMJ. 2009;339:b4168. doi: 10.1136/bmj.b4168. [DOI] [PubMed] [Google Scholar]
- Dettwiler I., Troell K., Robinson G., Chalmers R.M., Basso W., Rentería-Solís Z.M., et al. TIDE analysis of Cryptosporidium infections by gp60 typing reveals obscured mixed infections. J. Infect. Dis. 2022;225:686–695. doi: 10.1093/infdis/jiab417. [DOI] [PubMed] [Google Scholar]
- EFSA Panel on Biological Hazards (BIOHAZ) Koutsoumanis K., Allende A., Alvarez‐Ordóñez A., Bolton D., Bover‐Cid S., et al. Public health risks associated with food‐borne parasites. EFSA J. 2018;16:5495. doi: 10.2903/j.efsa.2018.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Ryan U.M., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Gopfert A., Chalmers R.M., Whittingham S., Wilson L., Van Hove M., Ferraro C.F., et al. An outbreak of Cryptosporidium parvum linked to pasteurised milk from a vending machine in England: A descriptive study, March 2021. Epidemiol. Infect. 2022;150:e185. doi: 10.1017/S0950268822001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg A., Widmer G. Cryptosporidium within-host genetic diversity: Systematic bibliographical search and narrative overview. Int. J. Parasitol. 2016;46:465–471. doi: 10.1016/j.ijpara.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Hossain M.J., Powell H., Sow S.O., Omore R., Roose A., Jones J.C.M., et al. Clinical and epidemiologic features of Cryptosporidium-associated diarrheal disease among young children living in sub-Saharan Africa: The vaccine impact on diarrhea in Africa (VIDA) study. Clin. Infect. Dis. 2023;76(Suppl. 1):S97–S105. doi: 10.1093/cid/ciad044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpsonʼs index of diversity. J. Clin. Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.R., Hughes S., Woodhouse S., Syed Q., Verlander N.Q., Chalmers R.M., et al. Sporadic cryptosporidiosis case-control study with genotyping. Emerg. Infect. Dis. 2004;10:1241. doi: 10.3201/eid1007.030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.R., Nichols G. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin. Microbiol. Rev. 2002;15:145–154. doi: 10.1128/CMR.15.1.145-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabała M., Gofron Z., Aptekorz M., Sacha K., Harmanus C., Kuijper E., Martirosian G. Clostridioides difficile ribotype 027 (RT027) outbreak investigation due to the emergence of rifampicin resistance using multilocus variable-number tandem repeat analysis (MLVA) Infect. Drug Resist. 2021;17:3247–3254. doi: 10.2147/IDR.S324745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P., Price L.B., Klevytska A.M., Smith K.L., Schupp J.M., Okinaka R., et al. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Levine M.M., Nasrin D., Acácio S., Bassat Q., Powell H., Tennant S.M., et al. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: Analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Global Health. 2020;8:e204–e214. doi: 10.1016/S2214-109X(19)30541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malorny B., Junker E., Helmuth R. Multi-locus variable-number tandem repeat analysis for outbreak studies of Salmonella enterica serotype enteritidis. BMC Microbiol. 2008;8:84. doi: 10.1186/1471-2180-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A., Robinson G., Swain M.T., Chalmers R.M. Direct sequencing of Cryptosporidium in stool samples for public health. Front. Public Health. 2019;7:360. doi: 10.3389/fpubh.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon C.A., Trees E., Ng L.K., Møller Nielsen E., Reimer A., Maxwell N., et al. Development and application of MLVA methods as a tool for inter-laboratory surveillance. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.35.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cordón G., Robinson G., Nader J., Chalmers R.M. Discovery of new variable number tandem repeat loci in multiple Cryptosporidium parvum genomes for the surveillance and investigation of outbreaks of cryptosporidiosis. Exp. Parasitol. 2016;169:119–128. doi: 10.1016/j.exppara.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Robinson G., Chalmers R.M. Assessment of polymorphic genetic markers for multi-locus typing of Cryptosporidium parvum and Cryptosporidium hominis. Exp. Parasitol. 2012;132:200–215. doi: 10.1016/j.exppara.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Robinson G., Elwin K., Chalmers R.M. Cryptosporidium diagnostic assays: Molecular detection. Methods Mol. Biol. 2020;2052:11–22. doi: 10.1007/978-1-4939-9748-0_2. [DOI] [PubMed] [Google Scholar]
- Robinson G., Pérez-Cordón G., Hamilton C., Katzer F., Connelly L., Alexander C.L., Chalmers R.M. Validation of a multilocus genotyping scheme for subtyping Cryptosporidium parvum for epidemiological purposes. Food Waterborne Parasitol. 2022;27 doi: 10.1016/j.fawpar.2022.e00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan U.M., Feng Y., Fayer R., Xiao L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia - a 50 year perspective (1971–2021) Int. J. Parasitol. 2021;51:1099–1119. doi: 10.1016/j.ijpara.2021.08.007. [DOI] [PubMed] [Google Scholar]
- Sow S.O., Muhsen K., Nasrin D., Blackwelder W.C., Wu Y., Farag T.H., et al. The burden of Cryptosporidium diarrheal disease among children < 24 months of age in moderate/high mortality regions of sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS) PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong W.B., Gut J., Nelson R.G. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15-and 45-kilodalton zoite surface antigen products. Infect. Immun. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strydom H., Wang J., Paine S., Dyet K., Cullen K., Wright J. Evaluating sub-typing methods for pathogenic Yersinia enterocolitica to support outbreak investigations in New Zealand. Epidemiol. Infect. 2019;147:e186. doi: 10.1017/S0950268819000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer G., Lee Y. Comparison of single-and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl. Environ. Microbiol. 2010;76:6639–6644. doi: 10.1128/AEM.01268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts V., Mattheus W., De Laminne de Bex G., Wildemauwe C., Roosens N.H., Marchal K., et al. MLVA as a tool for public health surveillance of human Salmonella typhimurium: Prospective study in Belgium and evaluation of MLVA loci stability. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its supplementary files, and in Robinson et al. (2022). The laboratory and analytical methods have been made freely available on our website at: https://phw.nhs.wales/services-and-teams/cryptosporidium-reference-unit/.