Summary
Background
The CCR5 (R5) to CXCR4 (X4) coreceptor switch in natural HIV-1 infection is associated with faster progression to AIDS, but the mechanisms remain unclear. The difficulty in elucidating the evolutionary origin of the earliest X4 viruses limits our understanding of this phenomenon.
Methods
We tracked the evolution of the transmitted/founder (T/F) HIV-1 in RV217 participants identified in acute infection. The origin of the X4 viruses was elucidated by single genome amplification, deep sequencing and coreceptor assay. Mutations responsible for coreceptor switch were confirmed by mutagenesis. Viral susceptibility to neutralization was determined by neutralization assay. Virus CD4 subset preference was demonstrated by sequencing HIV-1 RNA in sorted CD4 subsets.
Findings
We demonstrated that the earliest X4 viruses evolved de novo from the T/F strains. Strong X4 usage can be conferred by a single mutation. The mutations responsible for coreceptor switch can confer escape to neutralization and drive the X4 variants to replicate mainly in the central memory (CM) and naïve CD4 subsets. Likely due to the smaller viral burst size of the CM and naïve subsets, the X4 variants existed at low frequency in plasma. The origin of the X4 viruses preceded accelerated CD4 decline. All except one X4 virus identified in the current study lost the conserved V3 N301 glycan site.
Interpretations
The findings demonstrate co-evolution of HIV-1 antigenicity, coreceptor usage and CD4 subset targeting which have implications for HIV-1 therapeutics and functional cure. The observations provide evidence that coreceptor switch can function as an evolutionary mechanism of immune evasion.
Funding
Institute of Human Virology, National Institutes of Health, Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Thai Red Cross AIDS Research Centre, Gilead Sciences, Merck, and ViiV Healthcare.
Keywords: HIV-1, Coreceptor switch, Immune evasion, CD4 subset, Pathogenesis
Research in context.
Evidence before this study
The correlation between HIV-1 coreceptor switch and faster progression to AIDS has been well demonstrated. However, the molecular mechanisms responsible for coreceptor switch remains poorly understood. We searched PubMed, without language restrictions from database inception to May 3, 2023, using the terms “HIV” and “coreceptor switch”. The search yielded 212 publications, including 32 reviews. While previous studies provided evidence that the earliest CXCR4 (X4) variants evolved de novo from the pre-existing CCR5 (R5) viruses, no studies have demonstrated the evolutionary pathway of coreceptor switch of the transmitted/founder (T/F) virus. Consequently, it remains a matter of debate whether the earliest X4 viruses originated de novo from the R5 tropic T/F virus, or were initially transmitted, archived, and re-emerged later when the immune system waned, as discussed in several recent reviews. The difficulty in elucidating the evolutionary origin of the earliest X4 variants also hampers our understanding of the biological mechanisms driving coreceptor switch, as well as the causal link between coreceptor switch and accelerated CD4 depletion. While a widely accepted hypothesis is that the loss of CCR5-expressing CD4+ T cells drives coreceptor switch, to our knowledge, no study has proven that the CCR5 positive CD4 subsets were depleted or decreased before the origin of the X4 virus in vivo. Emerging evidence showed that X4 viruses of both HIV-1 and HIV-2 are relatively resistant to V3 specific neutralizing antibodies (NAbs). These studies have raised the possibility that humoral responses could drive coreceptor switch. However, no studies have demonstrated how the X4 variants originated from the T/F virus and why they tend to be resistant to V3 NAbs.
Added value of this study
The RV217 cohort which has frequently collected samples from participants identified very early in acute HIV-1 infection provides a unique opportunity to better understand the phenomenon of coreceptor switch. By tracking the genetic and phenotypic evolution of the T/F HIV-1 in three RV217 participants, we visualized the evolutionary process of coreceptor switch of the T/F HIV-1 with high resolution. The findings demonstrated that the earliest (founder) X4 variants originated de novo from the R5 tropic T/F strains and had impaired ability in using CCR5. A single mutation is sufficient to confer strong X4 usage on the cognate T/F virus. The driver mutations responsible for coreceptor switch can confer escape to autologous neutralization or V3 specific broadly neutralizing antibodies (bNAbs). The driver mutations promoted the X4 viruses to replicate mainly in the central memory and naïve CD4 subsets, while the R5 viruses remained predominant in the effector memory (EM) and transitional memory (TM) CD4 subsets. The X4 variants existed at low frequency in plasma as compared to the co-existing R5 viruses, which is likely due to the smaller viral burst size of the naïve and CM CD4 subsets as compared to the more activated EM and TM subsets. Determination of the evolutionary origin of the X4 variants allowed us to demonstrate that an X4-using phenotype is a causal factor, rather than a consequence of accelerated CD4 depletion. Among the total of 21 participants studied, those harboring X4 viruses had significantly faster CD4 decline.
Implications of all the available evidence
The study demonstrates co-evolution of HIV-1 antigenicity, coreceptor usage, CD4 subset targeting, and possibly the reservoir landscape during natural HIV-1 infection, which have direct implications for HIV-1 therapeutics and functional cure approaches. The finding that immune escape mutation can alter HIV-1 coreceptor usage and CD4 subset preference emphasizes the need to maintain HIV-1 entry pathway stability during the intervention. Our observations, together with previous evidence that X4 variants are generally resistant to V3 specific NAbs, provide evidence that HIV-1 coreceptor switch can function as an evolutionary mechanism of immune evasion. During the co-evolution of HIV-1 and the humoral responses, certain variants may exploit genetic pathways to evade the humoral response at the cost of CCR5 usage. Mutations which can confer X4 usage could be selected for as compensatory mutations to maintain virus entry capacity. Characterization of the co-evolution of HIV-1 coreceptor usage and autologous monoclonal antibodies (mAbs) will be an important future direction. Given that the overlap between virus antigenicity epitopes and receptor binding sites is widely observed for viruses from diverse families, and that many viruses have the potential to use more than one receptor or coreceptor, we propose a conceptual model to explain the co-evolution of virus antigenicity and entry pathway termed “escape by shifting”. The central hypothesis is that for viruses with entry pathway flexibility, entry pathway alteration can function as an evolutionary mechanism of immune evasion to maintain viral entry capacity.
Introduction
Despite the demonstrated correlation between HIV-1 coreceptor switch and faster progression to AIDS, the underlying mechanisms have not been clearly elucidated. Several fundamental questions remain. First, it is unclear whether the earliest X4 viruses evolved de novo from the R5 tropic T/F viruses or were initially transmitted and archived. Second, the precise biological condition driving coreceptor switch is poorly understood. Third, the causal relation between coreceptor switch and accelerated CD4 depletion remains controversial.
Although it is generally considered that the earliest X4 viruses originated de novo from the pre-existing R5 viruses,1, 2, 3, 4 the evolutionary pathway of coreceptor switch of the T/F HIV-1 has not been documented. This limits our understanding of the fundamental aspects of this phenomenon, and consequently, the biological mechanisms remain controversial. It has been hypothesized that the depletion of the memory CD4+ T cells, the major target cells of the R5 HIV-1, could drive the emergence of the X4 viruses in late infection stages.1,5 However, studies on sequential viral isolates showed that the first isolation of the syncytium-inducing (SI) variants usually preceded the onset of CD4 depletion.6, 7, 8 Moreover, the target cells-based hypothesis could not explain why the R5 viruses tend to be predominant in plasma while the X4 viruses tend to exist at low frequency, even at late infection stages when the CD4 count was low, as observed in both subtype B and CRF01_AE HIV-1 infections.9, 10, 11, 12, 13 Another hypothesis is that X4 viruses could be better controlled by the immune system, in particular the neutralizing antibodies (NAbs) and tend to emerge later when the immune system waned.1,4 However, as NAbs with diverse specificities were identified, emerging evidence indicated that X4 variants from both HIV-1 and HIV-2 are more resistant to neutralization, especially to V3 specific NAbs.14, 15, 16, 17 Clearly, further efforts are needed to better understand the mechanisms underlying coreceptor switch, which has direct relevance to HIV-1 pathogenesis as well as therapeutic and cure strategies.
The availability of longitudinal samples soon after HIV-1 acquisition in the RV217 cohort provides a unique opportunity to determine the mechanisms underlying coreceptor switch.18 By tracking the genetic and phenotypic evolution of the T/F HIV-1, we not only elucidated how the earliest X4 variants originated from the T/F viruses, but also identified the co-evolution of HIV-1 antigenicity, coreceptor usage and CD4 subset targeting underlying the phenomenon of coreceptor switch. We propose a conceptual model to explain the co-evolution of virus antigenicity and entry pathway termed “escape by shifting”. The central hypothesis is that for viruses with entry pathway flexibility, entry pathway alteration can function as an evolutionary mechanism of immune evasion to maintain virus entry capability.
Methods
Study participants
Study participants were from the RV217 and RV254 cohorts who were identified in acute HIV infection18,19 (Supplementary Table S1). Among the 28 participants identified in the RV217 Thailand cohort,18 21 participants who had pre-ART PBMC and plasma samples available up to at least two years from HIV-1 transmission were included in this study. All participants were infected by R5 tropic T/F viruses except for participant 40700 who was infected by an X4 tropic T/F virus as phenotypically determined in previous studies.20,21 Four phenotypically confirmed T/F env clones, including one R5 tropic and three X4 tropic, were identified in the RV254 cohort. All study participants were infected by CRF01_AE HIV-1 except for participants 40168 and 2548758 who were infected by CRF01_AE/B recombinant viruses (Supplementary Table S1).
Ethics
Written consent was provided by all participants. The study was approved by the local ethics review boards, the Walter Reed Army Institute of Research, and the institutional review boards of the University of Maryland School of Medicine (IRB approval number: HP-00089353).
Single genome amplification
HIV-1 RNA in plasma was extracted using the QIAamp Viral RNA Mini Kit (Qiagen). The 3’ half HIV-1 genome was amplified as described in appendix methods (Appendix p 3). The PCR amplicons were directly sequenced by the cycle sequencing and dye terminator methods.
Determination of coreceptor usage
Coreceptor usage of primary viral isolates or pseudoviruses were determined on NP-2 cells lines expressing CD4 together with either the CCR5 or CXCR4 coreceptors. To determine the coreceptor usage of primary viral isolates, NP-2 cell lines were seeded in a 48-well plate at a density of 2 × 105 cells per well one day before infection. The next day, the cells were infected with 200 μL undiluted viral isolates. After 6 h incubation at 37 °C, the cells were washed 3 times and cultured in fresh medium at 37 °C for 72 h. The p24 concentration in the culture supernatant was measured three days post infection (PerkinElmer). Virus infectivity was considered positive if the p24 concentration was at least three times higher than the background concentration in parental cell line. All infections were performed in duplicates.
To determine the coreceptor usage of the pseudovirus, NP-2 cells were seeded in a 96-well plate one day before infection at a density of 1 × 105 cells per well. On the next day, the cells were infected with approximately 200 TCID50 of each pseudovirus. After 6 h of incubation at 37 °C, the cells were washed twice with the culture medium and cultured at 37 °C for three days. At 72 h post infection, the infected cells were lysed, and the infectivity was determined by measuring the relative luciferase units (RLU) in the cell lysates using the Britelite plus system (PerkinElmer). Viral infectivity was considered positive if the RLU value was at least 5-fold higher than the background value in the parental cell line. All experiments were performed in triplicate. The NP-2 cell lines were kindly provided by Dr. Hiroo Hoshino and were not validated further.
Neutralization assay
The neutralization activity of plasma samples and monoclonal antibodies (mAbs) was measured by using a luciferase reporter system in TZM-bl cells. Plasma samples were heat inactivated at 56 °C for 45 min. The inactivated plasma was diluted at a 1:3 serial dilution starting from 1:20. The mAbs were diluted at a 1:3 serial dilution from a starting concentration of 25 μg/mL. The virus stocks were diluted to a concentration that achieved approximately 150,000 RLU in the TZM-bl cells (or at least 10 times above the background RLU of the cells control). The serial diluted plasma samples or mAbs were then incubated with the viruses for 1 h at 37 °C in duplicate before the TZM-bl cells were added. The 50% inhibitory dilution (ID50) was determined as the dilution at which the relative luminescence units (RLUs) were reduced by 50% in comparison to the RLUs in the virus control wells after subtraction of the background RLUs in cell control wells. The details for all monoclonal neutralizing antibodies (mAbs) were summarized in Supplementary Table S2. The mAbs were not validated further. The TZM-bl cell line was obtained from the HIV Reagent Program (ARP-8129) and was not validated further.
Statistical analysis
The neutralization sensitivity of different viral variants was compared using a two-sample t test. The rate of CD4 decline was determined using a linear mixed effect model (LME). The LME model was hierarchical in the sense that it estimated a population specific slope and intercept with time, as well as subject-specific slopes and intercepts. The longitudinal data contained CD4 data from the earliest available time point to the last available time point before ART initiation was used for the analysis.
Role of the funders
The founders had no roles in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
We determined virus entry tropism for 20 Thai participants in the RV217 cohort at approximately two years after HIV-1 transmission. Coreceptor assay using primary viral isolates identified X4 viruses in four participants (Supplementary Table S3). Of these four, participants 40094, 40436 and 40257 were initially infected by R5-tropic T/F viruses without evidence of superinfection (Supplementary Fig. S1). Thus, their X4 variants most likely evolved from the T/F strains. Participant 40100 was superinfected by an X4 variant at day 262 (Supplementary Fig. S1).
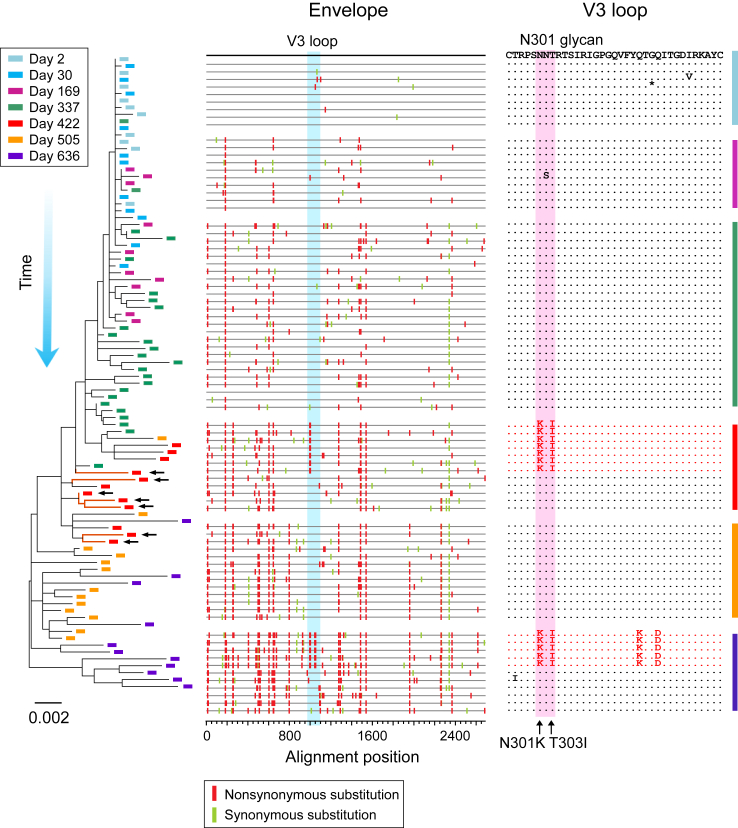
We tracked the evolution of the T/F viruses in participants 40094 and 40257. Both participants were infected by a single T/F virus as demonstrated by single genome amplification (SGA) and deep sequencing (Fig. 1 and Supplementary Figs. S2 and S3). This provided an ideal condition to determine the evolutionary origin of the earliest X4 viruses (Participant 40436 was infected by multiple T/F viruses. Due to rapid recombination between different T/F lineages, it was not possible to determine from which T/F lineage(s) the earliest X4 viruses originated. Thus, we did not focus on participant 40436 to study the evolutionary origin of the earliest X4 variants). In 40094, the earliest potential X4 variants, which carried two amino acid substitutions at the V3 N301 glycan site (N301K and T303I), appeared at day 422 (Fig. 1). In 40257, the earliest suspected X4 variants, which contained three mutations in V3 (S306R, Q313H and Q327R), emerged at day 264 (Supplementary Fig. S2). Coreceptor prediction using Geno2Phoeno22 indicated high likelihood of X4 usage of these V3 variants (FPR = 1.7 for 40094; FPR = 0.2% for 40257). In these variants, most of the substitutions outside of the V3 region were descended from the earlier viruses (Fig. 1 and Supplementary Fig. S2). These observations demonstrate that these earliest potential X4 variants evolved de novo from the T/F ancestor.
Fig. 1.
The evolutionary pathway leading to coreceptor switch of the 40094 T/F virus. Longitudinal HIV-1 envelope (env) sequences of participant 40094 were obtained by SGA. The evolutionary pathway of the T/F virus is illustrated by using the phylogenetic tree (left) and highlighter plot (middle). Sequences from different time points (days since the first positive test for HIV-1 RNA) are color coded. The phylogenetic tree was constructed using the maximum likelihood method. The earliest X4 viruses are indicated by black arrows. In the highlighter plot, the black line on the top represents the T/F virus, and the red and green tics indicate non-synonymous and synonymous substitutions as compared to the T/F strain, respectively. The V3 loop is highlighted in blue. In the V3 amino acids alignment (right), the X4 variants carrying the driver mutations are shown in red while the R5 variants are shown in black. The position of the driver mutations (the N301 glycan site) is shaded in red, and the driver mutations are indicated by black arrows.
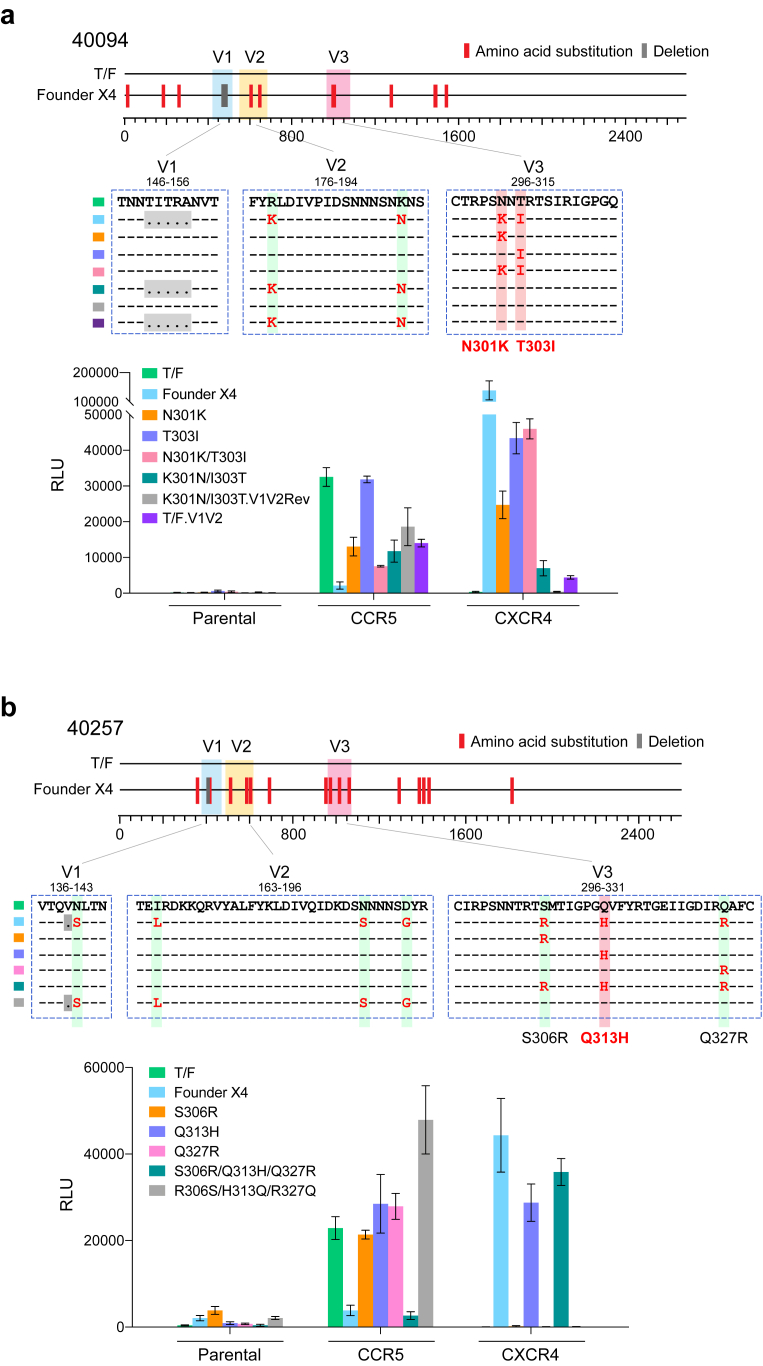
To phenotypically confirm whether these V3 variants were the earliest/founder X4 viruses, we constructed env clones which represented the consensus sequences of these variants. For both participants, the consensus sequences exhibited strong CXCR4 usage and significantly impaired CCR5 tropism in comparison to the cognate T/F virus (15.3-fold and 6.0-fold reduction in 40094 and 40257, respectively) (Fig. 2a–b). In 40094, the N301K/T303I mutations, which emerged together at day 422 conferred strong X4 usage on the T/F virus and compromised the efficiency in using R5 (4.3-fold reduction) (Fig. 2a). These two mutations were independently sufficient to confer strong X4 usage and the N301K mutation compromised CCR5 tropism (Fig. 2a). Reversion of these two mutations to the T/F sequence nearly abrogated X4 usage of the founder X4 virus (Fig. 2a). Further restoration of the V1/V2 regions to the T/F sequence completely abrogated the X4 usage (the “K301N/I303T.V1V2 Rev” mutant) (Fig. 2a). Introduction of the V1/V2 regions of the founder X4 virus into the T/F virus (The “T/F. V1V2” mutant) could confer weak X4 usage (Fig. 2a). In 40257, the three V3 mutations emerged together at day 264 conferred strong X4 usage and significantly compromised CCR5 tropism (8.6-fold reduction) (Fig. 2b). Among them, only the Q313H mutation in the V3 crown was able to confer X4 usage (Fig. 2b). Reversion of the V3 mutations to the T/F sequence completely abrogated the X4 usage (Fig. 2b). These results demonstrate that coreceptor switch of the T/F HIV-1 can be triggered by a single mutation.
Fig. 2.
Determination of driver mutations responsible for coreceptor switch. In participants 40094 (a) and 40257 (b), the sequence of the founder X4 virus is shown as the consensus sequence of the earliest variants carrying the driver mutations. Amino acid substitutions and deletions in comparison to the T/F strain are indicated by red and gray tics, respectively. The V1, V2 and V3 regions are color coded. Part of the V1, V2 and V3 regions spanning the genetic substitutions are shown. The numbers on the top show the HXB2 locations of the start and the end of each fragment. The driver mutations responsible for coreceptor switch are shaded in red. Different genetic mutants are color coded. In panel (a), the “K301N/I303T.V1V2” mutant had both the N301 glycan site and the V1/V2 regions restored to the T/F sequence. Coreceptor usage of the T/F virus, the founder X4 virus, as well as each mutant was determined in NP2 cell lines. All experiments were performed in triplicate and the error bar shows the standard deviation (SD).
To gain better insight into the origin of the founder X4 viruses, we performed deep sequencing to track the evolution of the variants carrying the driver mutations. Because plasma samples from the earliest time points (day 2 for 40094 and day 4 for 40257) were not available for the current study, the earliest plasma samples used for deep sequencing were from day 16 and day 14 (around the time of peak viremia) for 40094 and 40257, respectively. In both participants, these mutations could be detected as random mutations at low frequency during acute infection (Supplementary Figs. S4 and S5). Similar to the majority of variants detected at acute infection, the variants carrying the V3 mutations only differed from the T/F virus by a single base initially. Tracking their evolution showed that they were evolving over time, as additional mutations appeared, some of which became detectable by SGA at subsequent time points (Supplementary Figs. S4 and S5). However, the driver mutations persisted at low frequency until they were strongly selected for within a short period of time. This “deep evolutionary pathway” confirmed that the founder X4 viruses evolved from the T/F virus in a stepwise manner. It also indicated that coreceptor switch of the T/F virus was driven by strong positive selection favoring these V3 mutations.
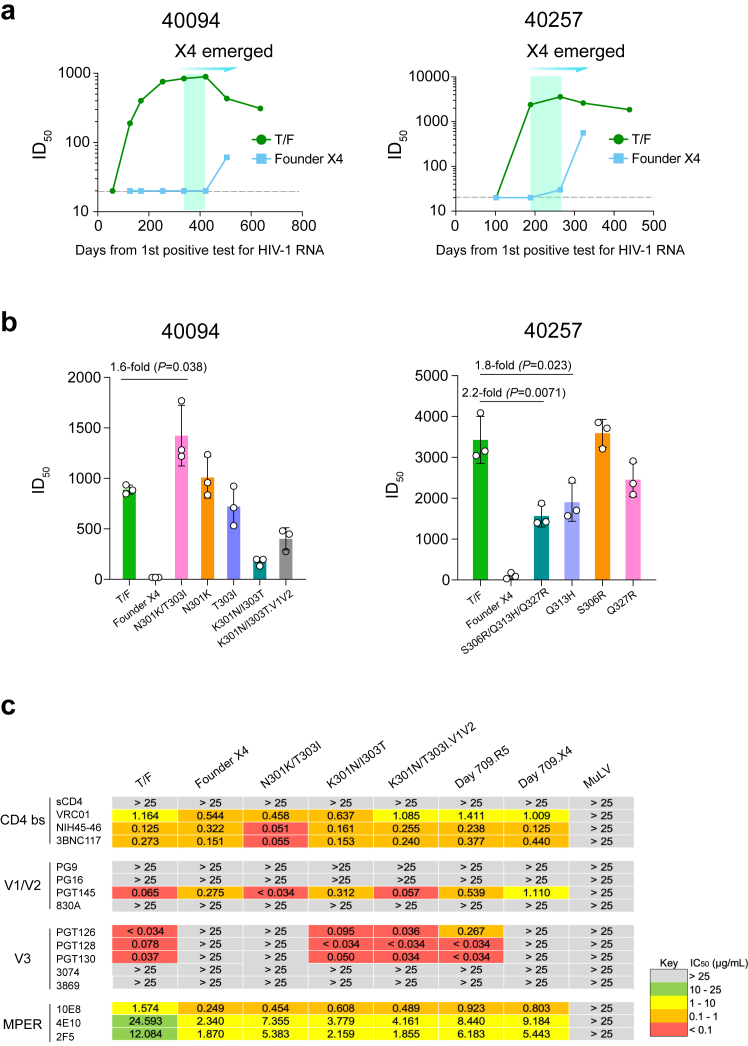
Previous studies showed that X4 viruses are more resistant to V3 specific NAbs.14,15 Investigation of the kinetics of autologous neutralization showed that in both participants, the V3 mutations were strongly selected for when the autologous neutralization activity targeting the T/F virus was increasing to the peak (Fig. 3a). Determination of the neutralization sensitivity of the founder X4 viruses demonstrated that they were immune escape variants resistant to autologous neutralization before and during the time when they were strongly selected in vivo, while they can be neutralized by plasma samples from later time points (Fig. 3a–b). We then determined the impact of the V3 mutations on neutralization susceptibility. The N301K/T303I mutations at the N301 glycan sites, when introduced into the 40094 T/F virus, could increase the susceptibility to autologous plasma [1.6-fold, P = 0.038; two-sample t test] (Fig. 3b). However, as previously reported, this effect is likely due to the exposure of other epitopes when the N301 glycan was removed.23 Restoration of the N301 glycan by the K301N/I303T mutations could partially restore the neutralization susceptibility, indicating the existence of N301 glycan dependent NAbs in 40094 (Fig. 3b). Reversion of both the N301 glycan site and the V1/V2 regions to the T/F sequence (the K301N/I303T.V1V2 Rev mutant), which fully restored the CCR5 phenotype (Fig. 2a), can further restore the neutralization sensitivity of the founder X4 virus (Fig. 3b). Introduction of the V1/V2 regions of the founder X4 virus into the T/F virus led to a 1.9-fold reduction in neutralization sensitivity (data not shown. The neutralization assay for the T/F. V1V2 mutant was not repeated due to the availability of the plasma samples). In 40257, the S306R/Q313H/Q327R mutations significantly decreased the susceptibility to autologous plasma [2.2-fold, P = 0.0071; two-sample t test] (Fig. 3b). Among them, only the Q313H mutation could significantly reduce the neutralization susceptibility [1.8-fold, P = 0.023; two-sample t test] (Fig. 3b). We also observed that the S306R/Q313H/Q327R mutations impaired the infectivity of the 40257 T/F virus (84-fold reduction in infectivity), indicating potential fitness cost incurred by these three mutations (Supplementary Table S4). The infectivity of the S306R/Q313H/Q327R mutant could be restored when the V1/V2 mutations from the founder X4 virus were introduced (Supplementary Table S4), which suggested compensatory effect of the V1/V2 mutations during coreceptor switch as previously observed.24 The V1/V2 mutations did not confer further immune escape to autologous plasma (data not shown). The reason why the S306R/Q313H/Q327R mutations were selected together in vivo in participant 40257 remains to be determined.
Fig. 3.
Kinetics of autologous neutralization activity and the impact of driver mutations on neutralization susceptibility. (a) Kinetics of autologous neutralization activity against the T/F virus and the founder X4 strain. The time frame when the earliest (founder) X4 virus became detectable in plasma by SGA is highlighted in green. (b) Neutralization susceptibility of the founder X4 viruses to autologous plasma and the impact of V3 mutations on neutralization susceptibility. For each participant, plasma samples with the highest neutralization activity against the autologous T/F virus were used for the neutralization assay (day 422 for 40094; day 264 for 40257). The mean titer of three independent neutralization assays is shown and the error bar represents the SD. The statistical significance was determined by using a two-sample t test. P value < 0.05 is considered to be statistically significant. (c) Neutralization susceptibility of the 40094 T/F virus, the founder X4 virus, as well as different genetic variants to a panel of bNAbs. The IC50 (μg/mL) of each bNAb is shown. The murine leukemia virus (MuLV) was used as the negative control.
Genetic substitution at the N301 glycan site is a common route to evade V3 glycan NAbs.23,25,26 However, mutations at the N301 glycan can confound plasma neutralization due to the exposure of other epitopes.23 To better understand the impact of the N301K/T303I mutations on neutralization susceptibility, we performed neutralization assays using a panel of broadly neutralizing antibodies (bNAbs) (Fig. 3c). The 40094 T/F viruses can be potently neutralized by bNAbs targeting the CD4 binding site, the V2 bNAb PGT145, and the V3 glycan bNAbs PGT126, PGT128 and PGT130 (Fig. 3c). However, the founder X4 virus was completely resistant to all V3 glycan bNAbs and had reduced susceptibility to PGT145 (Fig. 3c). Mutations N301K/T303I, when introduced into the 40094 T/F virus, conferred complete escape to all V3 glycan bNAbs (Fig. 3c). Reversion of the N301K/T303I mutations to the T/F sequence fully restored the susceptibility to all V3 glycan bNAbs (Fig. 3c). Further reversion of the V1/V2 regions restored the sensitivity to PGT145 (Fig. 3c). These results demonstrate that the N301K/T303I mutations, which are responsible for coreceptor switch in 40094, are also the exact mutations conferring immune escape to V3 bNAbs. Notably, the N301K/T303I mutant had increased sensitivity to PGT145 and the CD4 binding site bNAbs (Fig. 3c), which could be due to the exposure of the corresponding epitopes and might explain its increased sensitivity to autologous plasma (Fig. 3b).
Neither 40094 nor 40257 developed neutralization breadth. This limited the possibility to accurately predict the neutralization specificity using the fingerprinting assay. For participant 40436 who underwent coreceptor switch and developed neutralization breadth, fingerprinting assay predicted the exist of PGT128-like NAbs in plasma.27 Indeed, the X4-using variants isolated on day 671 from 40436 contained substitutions at the N301 glycan site as in 40094, a typical mutational route to evade PGT128 family NAbs26 (Supplementary Fig. S1). This prompted us to determine the origin of the N301 glycan mutations in 40436 and their role in coreceptor switch. Sequence analysis of longitudinal plasma samples from all available time points identified that the earliest mutations at the N301 glycan site (N302K/T303I) appeared at day 504 (Supplementary Figs. S6 and S7). The two variants carrying the N302K/T303I mutations were predicted to be X4-using by Geno2Pheno (FPR = 0.7). Recombination analysis showed that all sequences obtained at day 504, including these two predicted X4 variants, were recombinants between different T/F lineages (Supplementary Fig. S7a), and the two predicted X4 variants were mainly derived by recombination between T/F1 and T/F2 (Supplementary Fig. S7a). Phenotypic characterization of one variant demonstrated that it had strong X4 usage, impaired CCR5 usage, and was completely resistant to PGT128 neutralization (Supplementary Fig. S7b) (We did not examine the other variant because the sequence had a large deletion). Restoration of the N301 glycan site by reversion mutations K302N/I303T restored both the CCR5 phenotype and the sensitively to PGT128 of the founder X4 virus (Supplementary Fig. S7b). These data demonstrate that, similar as in participant 40094, the mutations at the N301 glycan site in 40436 are responsible for both coreceptor switch and immune escape to V3 bNAb.
We further investigated the neutralization sensitivity for another six phenotypically confirmed X4 viruses identified in the RV217 and RV254 cohorts. While most of the R5 tropic CRF01_AE T/F viruses in the RV217 Thailand cohort can be neutralized by V3 bNAbs PGT126, PGT128 and PGT130 as shown in our recent study,20 all X4 viruses identified were fully resistant to bNAbs of this class (Supplementary Fig. S8). These data demonstrated that the resistant phenotype to V3 bNAbs is specific for the X4 tropic viruses, instead of due to an overall neutralization resistance of the CRF01_AE HIV-1.
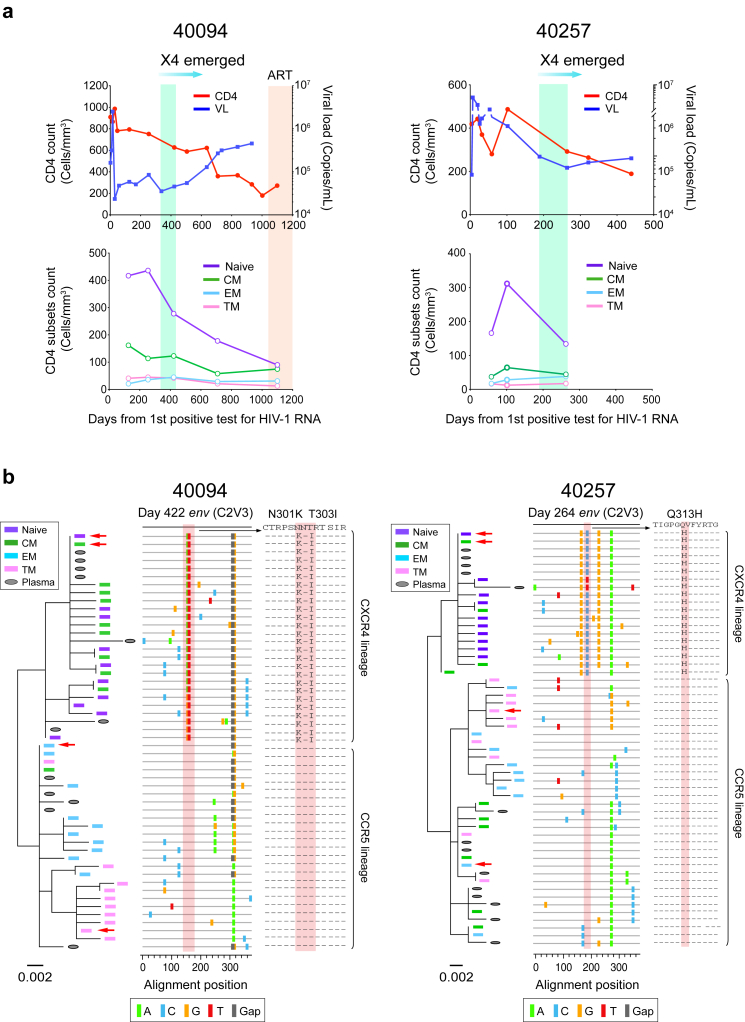
In participants 40094, 40257 and 40436, the origin of the founder X4 virus preceded an accelerated CD4 decline (Fig. 4a and Supplementary Fig. S9). In participants 40094 and 40257, the origin of the founder X4 virus was also followed by a continuous increase of the plasma viral load (VL) (Fig. 4a). Further analysis of the CD4 subset dynamics in 40094 showed that the naïve and central memory (CM) CD4 subsets declined faster than effector memory (EM) and transitional memory (TM) CD4 subsets (Fig. 4a).
Fig. 4.
CD4 and viral load dynamics before and after coreceptor switch and diverged CD4 subset targeting of the R5 and X4 HIV-1. (a) Dynamics of CD4 count, plasma viral load (VL) and different CD4 subsets in participants 40094 and 40257. The time frame when the founder X4 viruses emerged is highlighted in green. In 40094, the time after ART initiation is highlighted in red. (b) Phylogenetic trees and highlighter plots showing the evolutionary relationship between cell associated HIV-1 RNA and the plasma viruses. The cell associated HIV-1 RNA in each CD4 subset were sequenced by next-generation sequencing using the Illumina MiSeq. Ten unique sequences with the highest frequency in each CD4 subset are shown. Sequences from different CD4 subsets are color coded. The red arrows indicate the most frequent sequence in each subset. In the highlighter plot, the driver mutations responsible for coreceptor switch are shaded in red.
To better understand the biological mechanisms, we sequenced cell associated HIV-1 RNA in each CD4 subset. While the R5 viruses were predominant in the EM and TM subsets, the emerging X4 viruses were enriched in the CM and naïve subsets (Fig. 4b). The analysis also revealed distinct cellular origins of the R5 and X4 viruses in plasma. While the R5 viruses mainly originated from the EM, TM and CM subsets, the X4 viruses mainly originated from the naïve and the CM subsets (Fig. 4b). In both participants, the most frequent sequence in the CM subset was X4 tropic, indicating a replication advantage of X4 viruses in the CM cells (Fig. 4b). Consistent with the genetic analysis, the appearance of the founder X4 variants coincided with an emerging cell associated RNA in the naïve cells, which was undetectable earlier, and a substantially increased amount of cell associated RNA (20.5-fold) in the CM cells (Supplementary Fig. S10). We did not measure the dynamic changes of the cell associated RNA in 40257 due to sample availability, but it was also undetectable in the naïve cells before coreceptor switch (Supplementary Fig. S10). Quantification of CCR5 and CXCR4 expression suggested that the compartmentalization was most likely due to the differential expression of the two coreceptors as previously reported28 (Supplementary Fig. S11). It is important to note that there was no depletion of the TM and EM subsets before coreceptor switch in either participant (Fig. 4a). Thus, the origin of the founder X4 virus could not be explained by the depletion of the target cells of the R5 virus.
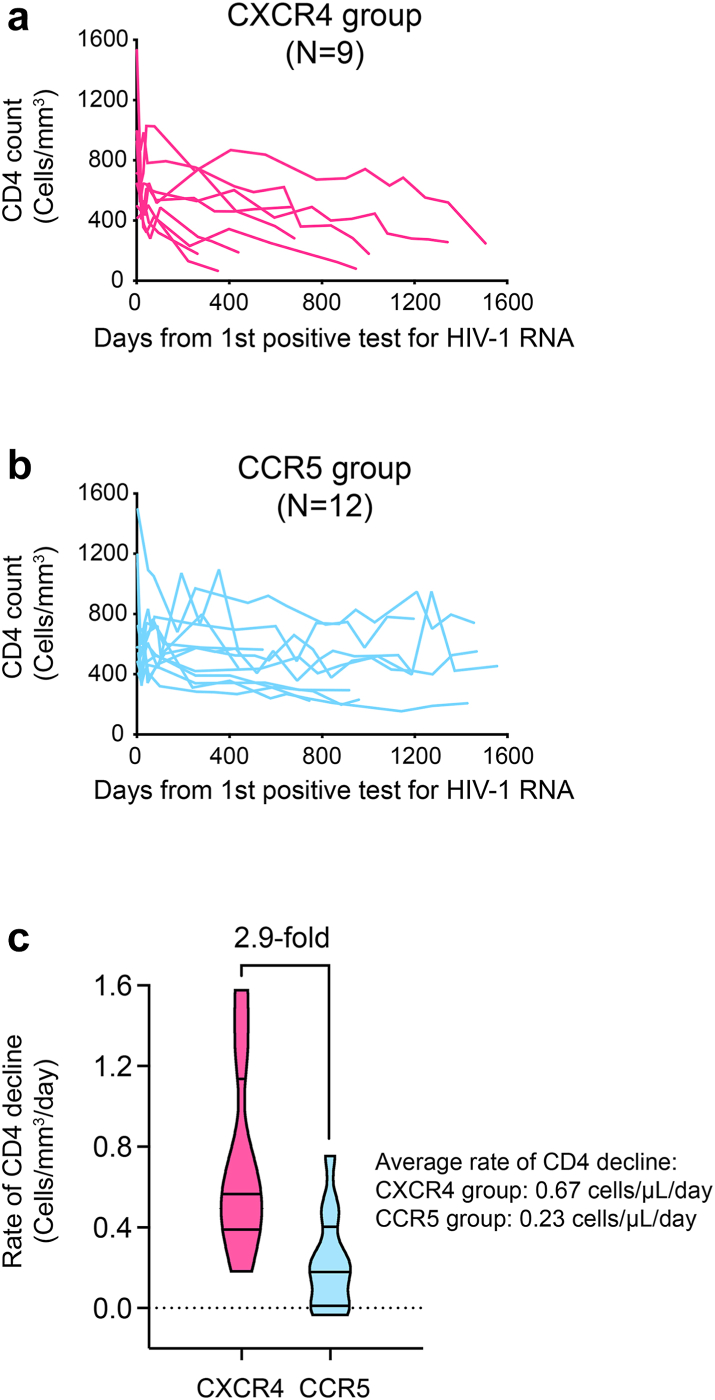
To better understand the distinct CD4 subset preferences between R5 and X4 viruses, we quantified cell associated RNA for another eleven participants for whom the primary viral isolates showed R5 tropism. Cell associated RNA was unexpectedly detected in naïve CD4+ T cells in four participants (Supplementary Fig. S12a). Sequencing of the viruses in naïve cells identified X4 variants in participants 40250 and 40168 (Supplementary Fig. S12b). While we were unable to amplify the env region from naïve cells for participants 40577 and 40503, deep sequencing of the plasma identified low frequency X4 variants in 40577 (Supplementary Fig. S12b). This prompted us to deep sequence the plasma for other participants for whom the PBMC samples were not sufficient to perform cell associated RNA assay. A low frequency of X4 viruses was detected in 40123 (Supplementary Fig. S12b). Notably, all X4 variants identified lost the conserved N301 glycan site as in 40049 and 40436 (Supplementary Fig. S12b). Among the total of 21 participants, those harboring X4 viruses showed 2.9-fold faster CD4+ T cell decline (Fig. 5 and Supplementary Fig. S13). The average rate of CD4+ T cell decline was 0.23 cells/μL/day in the R5 group (95% confidence interval: 0.10–0.35) and 0.67 cells/μL/day in the X4 group (95% confidence interval: 0.38–0.96). Of note, participant 40700, who was infected by an X4 tropic T/F virus, had the fastest rate of CD4 depletion (Supplementary Fig. S13). These results demonstrate distinct CD4 subset preferences of the R5 and X4 HIV-1. The data also demonstrate that an X4-using phenotype is a causal factor of accelerated CD4 depletion.
Fig. 5.
Significantly faster CD4 decline in participants harboring X4 viruses. (a) CD4 dynamics of participants harboring X4 viruses. (b) CD4 dynamics of participants without the evidence of harboring X4 viruses. (c) The rate of CD4 decline of the R5 and X4 groups were determined by a linear mixed effect model. The average rate of CD4 decline for each group was shown.
Discussion
Identification of the co-evolution of HIV-1 antigenicity, coreceptor usage and CD4 subset preference advances our understanding of the immunopathogenesis of HIV-1. It is generally considered that X4 viruses are more virulent because they can infect a broader range of CD4+ T cells, including the naïve subset which is important for CD4 homeostasis and renewal.2 Our study provided further insights into the pathogenesis of X4 tropic HIV-1. While the X4 viruses had a replication advantage in the naïve and the CM subsets, they could not outcompete the R5 viruses in the EM and TM subsets. The accelerated CD4 decline upon coreceptor switch was mainly due to the loss of the naïve and the CM cells. These observations together provide direct evidence that an altered CD4 subset targeting toward the CM and naïve cells could be a key factor in driving the loss of CD4 homeostasis. Several mechanisms could be involved. First, productive infection can kill the cells directly.29 Second, considering the relatively quiescent or resting status of the naïve and CM cells, it is reasonable to hypothesize that a higher proportion of the cells could be abortively infected than those infected productively. Abortive infections can induce pyroptosis, which could in turn enhance immune activation and inflammation.30 Third, the migration of the X4 viruses into the naïve and the CM cells would increase the virus burden in lymph nodes due to the expression of the homing receptor CCR7 on these cells.28 While the exact mechanisms remain to be determined, these observations indicate the importance of CD4 subset preference in influencing the CD4 virulence of HIV-1. Indeed, preferential sparing of the CM CD4 subset is one potential mechanism for the nonprogressive nature of simian immunodeficiency viruses (SIVs) infection in their natural hosts.31 In people with HIV infection, a lower infection level of CM cells is a feature of long-term nonprogressors.32 In light of this, it will be necessary to determine the heterogenicity of CD4 subset targeting by the R5 HIV-1, as well as its association with pathogenesis, immune activation and reservoir landscape. The low frequency of X4 viruses in plasma is likely due to the smaller viral burst size of the naïve and CM cells as compared to the more activated TM and EM cells. This raises the concern that X4 viruses could be underestimated in people with HIV infection. Direct characterization of the viruses in the naïve and CM cells where the X4 viruses are enriched may enhance the sensitivity to detect the X4 variants.
Our findings, together with previously available evidence, supports that HIV-1 coreceptor switch can function as an evolutionary mechanism of immune evasion. This is based on multiple lines of evidence. First, the mutations responsible for coreceptor switch were strongly selected for at the peak of autologous neutralizing activity and conferred escape to neutralization. Second, both the N301 glycan site and V3 crown are common targets for V3 specific NAbs.33 The footprints of these NAbs overlap with the CCR5 binding region.14 Third, the founder X4 viruses were immune escape variants with impaired CCR5 tropism. Fourth, phenotypically confirmed X4 viruses from different HIV-1 subtypes, including those investigated here and in previous studies, are in general resistant to V3 NAbs.14,15 These findings together support the following scenario underlying the phenomenon of coreceptor switch: During the co-evolution of HIV-1 and host immune responses, certain variants exploit genetic pathways to evade autologous neutralization at the cost of CCR5 usage. In this case, mutation(s) which can confer CXCR4 usage will compensate for the impairment of CCR5-mediated entry, and thus be selected for in the quasispecies to maintain virus entry capacity.
Indeed, the overlap between virus antigenic epitopes and receptor binding sites is widely observed for viruses from diverse families, and it has become increasingly apparent that many viruses have the potential to use more than one receptor or coreceptor (i.e., have entry pathway flexibility).34 Potential co-evolution of virus antigenicity and receptor specificity has been observed for other viruses in vitro, such as the foot-and-mouth disease virus,35,36 although evidence from natural viral infections remains lacking. These lines of evidence prompted us to propose a conceptual model termed “escape by shifting” (Supplementary Fig. S14). The central hypothesis is that for viruses with entry pathway flexibility, entry tropism alteration can function as an evolutionary mechanism of immune evasion in vivo. We coin the term “receptor tropism space” to describe the repertoire of receptors that could be utilized by a virus. Under the immune pressure, a virus explores its “receptor tropism space” while exploring the sequence space and fitness landscape. During this process, certain variants can achieve immune escape by altering the receptor they use. Regarding the molecular mechanism, we hypothesize two possible mutational pathways. First, an immune escape mutation itself could confer novel receptor usage (mutations at the N301 glycan could be an example). Second, when an immune escape mutation compromises the usage of the original receptor, mutations which can confer novel receptor usage could be selected as compensatory mutations to restore viral entry capacity. Exploring the escape by shifting scenario in different viral infections may provide insights into viral pathogenesis and therapeutic and prevention strategies. It may also shed light on how viruses gain zoonotic potential in natural reservoir species.
The limitation of the study is relatively small number of participants infected by a single clade. Longitudinal phenotype characterization of more participants is required to understand whether the evolutionary dynamics of coreceptor switch we observed here is generalizable to more participants infected by different HIV subtypes. Existing evidence indicate that the co-evolution of HIV-1 antigenicity, coreceptor usage and CD4 subset targeting is not a specific phenomenon for CRF01_AE HIV-1. First, the N301 glycan site, which is essential for V3 glycan NAbs neutralization, is also an important determinant for coreceptor usage for different HIV-1 subtypes.37, 38, 39 A previous study showed that mutations at the N301 glycan site were enriched in the X4 viruses from subtype A, B, C, D and CRF01_AE.39 Second, resistance to V3 NAbs was reported for diverse HIV-1 subtypes as well as for HIV-2.14,15,17 Third, a recent study on subtype B HIV-1 showed that the X4 variants were predominant in the CM and naïve CD4 subsets in the reservoir in people on ART.40 Thus, characterization of the co-evolution of HIV-1 antigenicity, entry pathway and the reservoir landscape for participants infected by different HIV-1 subtypes will be an important future direction which has implications for therapeutics and functional cure. The study also emphasizes the need to better understand the impact of immune escape mutations on viral pathogenesis beyond immune escape itself.
Contributors
H.S., V.R.P., and M.H.M. designed the study. M.H.M., M.Z., L.W., E.S-B., M.B., A.M.O., F.D-M., S.S. and H.S. performed the experiments. M.H.M., E.S-B., M.B., A.M.O., S.T., and H.S. were involved in viral sequencing and genetic analysis. D.K. and L.F. modeled the CD4 dynamics. Y.T. contributed to the design and data analysis of the flow cytometry and cell sorting experiments. N.C. provided the protocol of cell associated HIV-1 RNA assay and contributed to discussions of the experiments and data. N.P., J.A., D.H., S.V., N.L.M., L.A.E., and M.L.R. were involved in recruiting of the clinical cohorts, analysis of the clinical data and supplied clinical samples. H.S. wrote the manuscript with the input from all authors. H.S. and M.H.M accessed and verified all the data. All authors read the manuscript and approved the submission.
Data sharing statement
The GenBank accession numbers of the sequences used in the current study, including newly generated sequences and sequences submitted previously were summarized in Supplementary Table S5. The deep sequencing data were deposited in the Sequence Read Archive (SRA) under the project ID 913037 and 914717. All sequencing data are accessible prior to publication.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
The authors thank the study participants of the RV217 and RV254/SEARCH 010 cohorts. We thank Sebastian Molnar for technical assistance. This study was supported by the Institute of Human Virology, University of Maryland School of Medicine. Part of the study was supported by cooperative agreements between the Henry M. Jackson Foundation for the Advancement of Military Medicine, and the U.S. Department of Defense (DOD). RV254/SEARCH010 is supported by cooperative agreements (W81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, and the United States Army Medical Research and Development Command (USAMRDC) and by an intramural grant from the Thai Red Cross AIDS Research Centre and, in part, by the Division of AIDS, the National Institute of Allergy and Infectious Diseases, National Institutes of Health (AAI21058-001-01000). Antiretroviral therapy for RV254/SEARCH 010 participants was supported by the Thai Government Pharmaceutical Organization, Gilead Sciences, Merck and ViiV Healthcare. M.H.M. and H.S. were supported by the NIH grant R21AI147893.
Disclaimer: The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army, the Department of Defense, the National Institutes of Health, the Department of Health and Human Services, or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. The investigators have adhered to the policies for protection of human subjects as prescribed in AR-70-25.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104867.
Appendix A. Supplementary data
References
- 1.Regoes R.R., Bonhoeffer S. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 2005;13(6):269–277. doi: 10.1016/j.tim.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Swanstrom R., Coffin J. HIV-1 pathogenesis: the virus. Cold Spring Harb Perspect Med. 2012;2(12) doi: 10.1101/cshperspect.a007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunnik E.M., Swenson L.C., Edo-Matas D., et al. Detection of inferred CCR5- and CXCR4-using HIV-1 variants and evolutionary intermediates using ultra-deep pyrosequencing. PLoS Pathog. 2011;7(6) doi: 10.1371/journal.ppat.1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuitemaker H., van 't Wout A.B., Lusso P. Clinical significance of HIV-1 coreceptor usage. J Transl Med. 2011;9(Suppl 1):S5. doi: 10.1186/1479-5876-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberger A.D., Perelson A.S. Persistence and emergence of X4 virus in HIV infection. Math Biosci Eng. 2011;8(2):605–626. doi: 10.3934/mbe.2011.8.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koot M., Keet I.P., Vos A.H., et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118(9):681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 7.Connor R.I., Mohri H., Cao Y., Ho D.D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67(4):1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor R.I., Ho D.D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68(7):4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irlbeck D.M., Amrine-Madsen H., Kitrinos K.M., Labranche C.C., Demarest J.F. Chemokine (C-C motif) receptor 5-using envelopes predominate in dual/mixed-tropic HIV from the plasma of drug-naive individuals. AIDS. 2008;22(12):1425–1431. doi: 10.1097/QAD.0b013e32830184ba. [DOI] [PubMed] [Google Scholar]
- 10.Verhofstede C., Vandekerckhove L., Eygen V.V., et al. CXCR4-using HIV type 1 variants are more commonly found in peripheral blood mononuclear cell DNA than in plasma RNA. J Acquir Immune Defic Syndr. 2009;50(2):126–136. doi: 10.1097/QAI.0b013e31819118fa. [DOI] [PubMed] [Google Scholar]
- 11.Song H., Ou W., Feng Y., et al. Disparate impact on CD4 T cell count by two distinct HIV-1 phylogenetic clusters from the same clade. Proc Natl Acad Sci U S A. 2019;116(1):239–244. doi: 10.1073/pnas.1814714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou S., Bednar M.M., Sturdevant C.B., Hauser B.M., Swanstrom R. Deep sequencing of the HIV-1 env Gene Reveals Discrete X4 Lineages and Linkage Disequilibrium between X4 and R5 Viruses in the V1/V2 and V3 Variable Regions. J Virol. 2016;90(16):7142–7158. doi: 10.1128/JVI.00441-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda Y., Takemura T., Chikata T., et al. Existence of replication-competent minor variants with different coreceptor usage in plasma from HIV-1-Infected individuals. J Virol. 2020;94(12) doi: 10.1128/JVI.00193-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sok D., Pauthner M., Briney B., et al. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity. 2016;45(1):31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin N., Gonzalez O.A., Registre L., et al. Humoral immune pressure selects for HIV-1 CXC-chemokine receptor 4-using variants. eBioMedicine. 2016;8:237–247. doi: 10.1016/j.ebiom.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polonis V.R., de Souza M.S., Darden J.M., et al. Human immunodeficiency virus type 1 primary isolate neutralization resistance is associated with the syncytium-inducing phenotype and lower CD4 cell counts in subtype CRF01_AE-infected patients. J Virol. 2003;77(15):8570–8576. doi: 10.1128/JVI.77.15.8570-8576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcelino J.M., Borrego P., Nilsson C., et al. Resistance to antibody neutralization in HIV-2 infection occurs in late stage disease and is associated with X4 tropism. AIDS. 2012;26(18):2275–2284. doi: 10.1097/QAD.0b013e328359a89d. [DOI] [PubMed] [Google Scholar]
- 18.Robb M.L., Eller L.A., Kibuuka H., et al. Prospective study of acute HIV-1 infection in adults in East Africa and Thailand. N Engl J Med. 2016;374(22):2120–2130. doi: 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ananworanich J., Schuetz A., Vandergeeten C., et al. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuriakose Gift S., Wieczorek L., Sanders-Buell E., et al. Evolution of antibody responses in HIV-1 CRF01_AE acute infection: founder envelope V1V2 impacts the timing and magnitude of autologous neutralizing antibodies. J Virol. 2023;97(2) doi: 10.1128/jvi.01635-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song H.S.-B.E., Mann B., Engeman E., et al. The 10th IAS Conference on HIV Science; 2019. Identification and characterization of a CXCR4-tropic transmitted/founder HIV-1 in an acute CRF01_AE infection. [Google Scholar]
- 22.Lengauer T., Sander O., Sierra S., Thielen A., Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25(12):1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 23.Longo N.S., Sutton M.S., Shiakolas A.R., et al. Multiple antibody lineages in one donor target the glycan-V3 supersite of the HIV-1 envelope glycoprotein and display a preference for quaternary binding. J Virol. 2016;90(23):10574–10586. doi: 10.1128/JVI.01012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastore C., Nedellec R., Ramos A., Pontow S., Ratner L., Mosier D.E. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J Virol. 2006;80(2):750–758. doi: 10.1128/JVI.80.2.750-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonsignori M., Zhou T., Sheng Z., et al. Maturation pathway from Germline to Broad HIV-1 neutralizer of a CD4-mimic antibody. Cell. 2016;165(2):449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krumm S.A., Mohammed H., Le K.M., et al. Mechanisms of escape from the PGT128 family of anti-HIV broadly neutralizing antibodies. Retrovirology. 2016;13:8. doi: 10.1186/s12977-016-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Townsley S.M., Donofrio G.C., Jian N., et al. B cell engagement with HIV-1 founder virus envelope predicts development of broadly neutralizing antibodies. Cell Host Microbe. 2021;29(4):564–578.e9. doi: 10.1016/j.chom.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sallusto F., Lenig D., Forster R., Lipp M., Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 29.Zagury D., Bernard J., Leonard R., et al. Long-term cultures of HTLV-III--infected T cells: a model of cytopathology of T-cell depletion in AIDS. Science. 1986;231(4740):850–853. doi: 10.1126/science.2418502. [DOI] [PubMed] [Google Scholar]
- 30.Doitsh G., Galloway N.L., Geng X., et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paiardini M., Cervasi B., Reyes-Aviles E., et al. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nat Med. 2011;17(7):830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Descours B., Avettand-Fenoel V., Blanc C., et al. Immune responses driven by protective human leukocyte antigen alleles from long-term nonprogressors are associated with low HIV reservoir in central memory CD4 T cells. Clin Infect Dis. 2012;54(10):1495–1503. doi: 10.1093/cid/cis188. [DOI] [PubMed] [Google Scholar]
- 33.Overbaugh J., Morris L. The antibody response against HIV-1. Cold Spring Harb Perspect Med. 2012;2(1):a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howley P.M., Knipe D.M. Vol. 1. 2021. Fields virology. [Google Scholar]
- 35.Baranowski E., Ruiz-Jarabo C.M., Domingo E. Evolution of cell recognition by viruses. Science. 2001;292(5519):1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- 36.Martinez M.A., Verdaguer N., Mateu M.G., Domingo E. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc Natl Acad Sci U S A. 1997;94(13):6798–6802. doi: 10.1073/pnas.94.13.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogert R.A., Lee M.K., Ross W., Buckler-White A., Martin M.A., Cho M.W. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J Virol. 2001;75(13):5998–6006. doi: 10.1128/JVI.75.13.5998-6006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollakis G., Kang S., Kliphuis A., Chalaby M.I., Goudsmit J., Paxton W.A. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J Biol Chem. 2001;276(16):13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]
- 39.Joshi A., Cox E.K., Sedano M.J., et al. HIV-1 subtype CRF01_AE and B differ in utilization of low levels of CCR5, Maraviroc susceptibility and potential N-glycosylation sites. Virology. 2017;512:222–233. doi: 10.1016/j.virol.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roche M., Tumpach C., Symons J., et al. CXCR4-Using HIV strains predominate in naive and central memory CD4(+) T cells in people living with HIV on antiretroviral therapy: implications for how latency is Established and maintained. J Virol. 2020;94(6) doi: 10.1128/JVI.01736-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.