Highlights
-
•
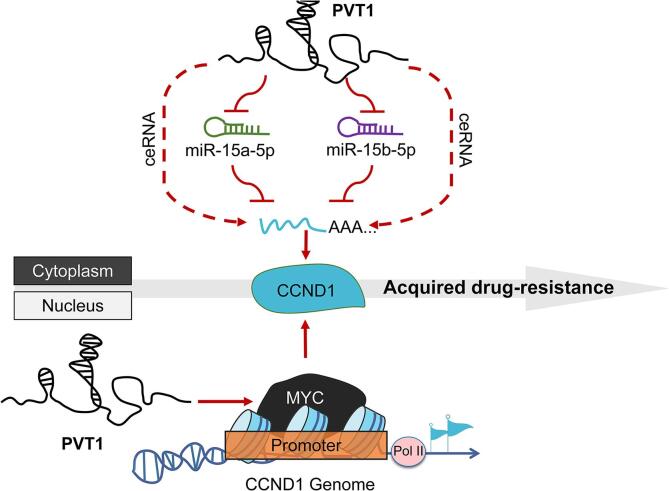
PVT1 mediated doxorubicin resistant of osteosarcoma cells through dual mechanisms of up-regulation of CCND1 expression: adsorption of miR-15a-5p and miR-15b-5p through endogenous competitive RNA mechanism to protect CCND1 mRNA from degradation, and activation of CCND1 transcription on the basis of ensuring the stability of MYC protein.
Keywords: Osteosarcoma, PVT1, CCND1, Acquired drug-resistance, MYC
Abstract
Background
Acquired drug-resistance is the major risk factor for poor prognosis and short-term survival in patients with osteosarcoma (OS). Accumulating evidence has revealed that long noncoding RNAs (lncRNAs), including plasmacytoma variant translocation 1 (PVT1), play potential regulatory roles in the malignant development of OS. Considering the subcellular distribution of PVT1 as both nuclear and cytoplasmic lncRNA, a thorough exploration of its extensive mechanisms becomes essential.
Methods
The GEO database was utilized for the acquisition of gene expression data, which was subsequently analyzed to fulfill the research objectives. The subcellular localization of PVT1 in OS cells was determined using fluorescence in situ hybridization (FISH) and quantitative real-time polymerase chain reaction (qRT-PCR). Additionally, the sensitivity of OS cells to doxorubicin was comprehensively evaluated through measurements of cell viability, site-specific proliferation capacity, and the relative expression abundance of multidrug resistance-related proteins (MRPs). In order to investigate the differential response of OS cells with varying levels of PVT1 expression to doxorubicin, pulmonary metastasis mice models were established for in vivo studies. Molecular interactions were further examined using the dual-luciferase assay and RNA immunoprecipitation assay (RIP) to analyze the binding sites of miR-15a-5p and miR-15b-5p on PVT1 and G1/S-specific cyclinD1 (CCND1) mRNA. Furthermore, the chromatin immunoprecipitation (ChIP) and dual-luciferase assay were employed to assess the transcriptional activation of the proto-oncogene c-myc (MYC) on the CCND1 promoter and identify the corresponding binding sites.
Results
In doxorubicin resistant OS cells, transcription levels of PVT1, MYC and CCND1 were significantly higher than those in original cells. In vivo experiments demonstrated that OS cells rich in PVT1 expression exhibited enhanced tumorigenicity and resistance to doxorubicin. In vitro experiments, it has been observed that overexpression of PVT1 in OS cells is accompanied by an upregulation of CCND1, thereby facilitating resistance to doxorubicin. Nonetheless, this PVT1-induced resistance can be effectively attenuated by the knockdown of CCND1. Mechanistically, PVT1 functions as a competitive endogenous RNA (ceRNA) that influences the expression of CCND1 by inhibiting the degradation function of miR-15a-5p and miR-15b-5p on CCND1 mRNA. Additionally, as a neighboring gene of MYC, PVT1 plays a role in maintaining MYC protein stability, which further enhances MYC-dependent CCND1 transcriptional activity.
Conclusion
The resistance of OS cells to doxorubicin is facilitated by PVT1, which enhances the expression of CCND1 through a dual mechanism. This findings offer a novel perspective for comprehending the intricate regulatory mechanisms of long non-coding RNA in influencing the expression of coding genes.
1. Introduction
Despite its low incidence, osteosarcoma (OS) is the most common malignancy of the skeletal system with poor prognosis [1], [2]. Acquired drug-resistance occurs in a large proportion of patients with OS and have shorter time intervals until metastasis, posing a great threat to the survival of patients [3].
Long noncoding RNA PVT1 (hereinafter referred to as PVT1) has been demonstrated to be associated with tumour proliferation, metastasis, and drug resistance, and was widely identified as an "oncogene" in various cancer types [4], [5], [6], [7], [8]. Studies have revealed that PVT1 is overexpressed in OS, suggesting its potential as an intervention target [9]. Currently, the promoting effect of PVT1 on the malignant progression of OS is mainly reported as a molecular "sponge" for a series of miRNAs, including miR-497, miR-183-5p, miR-484, miR-195 and miR-152 [10], as well as the direct binding effect with motif containing family 28 (TRIM28) to enhance the degradation of tumor suppressor complex 2 (TSC2) [11]. Notably, it is worth mentioning that PVT1 is a lncRNA, specifically an intergenic lncRNA (lincRNA), situated approximately 100 to 500 Kb 3-prime of myelocytomatosis oncogene (MYC, also referred to as c-myc) on chromosome 8 [12], and studies have indicated a concurrent upregulation of PVT1 in over 98 % of MYC-amplified cancers [13]. Given that the amplification of MYC is a major driver of high-grade tumors in childhood OS [14], our aim was to investigate the underlying mechanism in which PVT1 is implicated.
G1/S-specific cyclin-D1 (CCND1) functions as a regulatory component of the cyclin D1-CDK4 (DC) complex, which controls the cell cycle during the transition from G (1) to S phase [15], [16], [17], [18], [19], [20]. Furthermore, it was discovered that the breakdown of CCND1 via proteosome-dependent mechanisms has the ability to halt the advancement of cancerous cells during the G (1) phase [21]. Of note, a study investigating genes responsible for somatic cell copy number changes (SCNAs) in cancer revealed a significant overlap with well-known cancer genes, such as MYC, CCND1 in breast cancer, and PVT1 in various cancer types [22], suggesting their important roles in cancer promotion. Currently, numerous studies have demonstrated that the overexpression of CCND1 leads to chemotherapy resistance in various malignancies, including testicular cancer [23], breast cancer [24], prostate cancer [25] and OS [26], [27] etc. However, whether PVT1 is involved in CCND1-driven chemotherapy resistance in OS remains to be verified.
The current study showcased that PVT1 and CCND1 play a role in enhancing doxorubicin resistance in OS cells, and shed light on the molecular mechanisms through which PVT1 affects CCND1 expression. These findings hold significant implications in the quest to identify therapeutic targets for OS.
2. Material and methods
2.1. Tissue samples acquisition from patients
The cancer tissue samples for analysis (nN = 3) were obtained from the remaining needle biopsy samples from three male OS patients aged 12, 14, and 15 years who underwent puncture biopsy with cancer pathologic grades of IA, IIA, and III. The inclusion criteria were patients diagnosed with OS who had not previously received any therapeutic interventions. All patients signed informed consent, and obtained the ethical approval of the scientific research project of the Medical Ethics Committee of the Second Xiangya Hospital of Central South University and the approval number is (2022) No. (Research 023).
2.2. Protein-double labeling and RNA FISH (IF/FISH)
On the slips of osteosarcoma tissues, double-labeling immunofluorescence (IF) for MYC and CCND1 was first performed, followed by FISH assay for PVT1. Briefly, tissue slips were fixed in 4 % PFA, permeabilized in 0.1 % Triton X-100, and blocked with 1 % BSA. After antigen retrieval, the slips were incubated with anti-MYC antibody (Abcam, Cambridge, MA, USA, diluted 1:100) at 37 °C for 1 h, followed by incubation with HRP-conjugated secondary antibody (Abcam, diluted 1:1000) in the dark for 1 h. Subsequently, the "color development" component from the Double Standard Trichromatic Multiplex Immunofluorescence Kit (AiFang Biological, Cat. AFIHC033, Hunan, China) was added for fluorescence presentation procedure (HRP catalyzes the addition of a tyramide fluorescent substrate to the reaction system, generating activated fluorescent substrate that covalently binds to tyrosine residues or other residues on the antigen, resulting in stable covalent binding of tyramide fluorescent substrate on the sample). Antigen retrieval was then performed again, and the same procedure was repeated with the addition of anti-CCND1 antibody (Abcam, diluted 1:100), HRP-conjugated secondary antibody (Abcam, diluted 1:1000), and the color development reagent from the kit. Next, FISH operation was performed on the slips with Cy7- conjugated probe for PVT1 as mentioned above. Finally, slips were cleaned with PBS to remove residual fluid and stained with DAPI for 10 min at 25 °C. The excitation wavelengths of fluorescence of various colors are as follows: blue (DAPI, EX: 480 nm), green (MYC, GFP, EX: 520 nm), red (CCND1, TYR-570, EX: 620 nm), purple (PVT1, Cy7, EX:780 nm). Images of the specimens were captured under a KFBio slide viewer (KFBio, Ningbo, Hangzhou, China).
2.3. Cell culture and transfection
The human OS cell lines MG63 and U2OS and the corresponding doxorubicin-resistant cells (abbreviated as MG63-Dox and U2OS-Dox) with accurate STR identification were presented as a gift from the Fenghui Biotechnology Co., LTD (Hunan, China). Cells were cultured in the way our lab always does [28]. The full-length sequence of PVT1 or the coding sequence of MYC was cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) to construct their overexpression vectors, designated as OE-PVT1 and OE-MYC, respectively. The shRNA constructs targeting PVT1 (sh-PVT1) and CCND1 (sh-CCND1), as well as the negative control (sh-NC), were provided by RiboBio (Guangzhou, China) for interference of their expression levels. Synthetic mimics of miR-15a-5p and miR-15b-5p, as well as the antagonists (GenePharma CO.,LT, Shanghai, China), were separately transfected into OS cell lines to establish cell models with either overexpression or knockdown of the the target miRNAs. The cells were first cultured in 6-well plates for 24 h prior to transfection. Subsequently, the plasmids were mixed with Lipofectamine 3000 (Invitrogen) and added to the culture medium for a duration of 48 h.
2.4. Cell viability assay
An MTT assay was applied to assess cell viability. Briefly, MG63, U2OS, MG63-Dox, or U2OS-Dox cells were seeded into 96-well plates, and then were treated with doxorubicin at different concentrations. The detailed operation steps were carried out in accordance with the manufacturer's instructions (Merck Chemicals, Shanghai, China), and the absorbance at 490 nm was measured by a spectrometer (Thermo, USA).
The resistance index (RI) is determined through the assessment of cellular resistance to doxorubicin. Initially, both the parental and resistant strains are cultivated in media containing varying concentrations of doxorubicin ranging from 0 to 1500 nM for 24 h. Subsequently, the viability of each group of cells is evaluated using the MTT cell viability assay. The RI is then calculated utilizing the subsequent formula: RI = IC50 (resistant cells) / IC50 (orignal cells). In this context, IC50 denotes the half maximal inhibitory concentration, which is the drug concentration that reduces cell viability by 50 %. A higher RI value signifies a more pronounced resistance of cells towards the drug.
2.5. Cell proliferation assay
In order to evaluate the impact of PVT1 on doxorubicin tolerance, the vector carrying PVT1 shRNA (sh-PVT1) and the empty control vector (sh-NC) were transfected into the parental osteosarcoma cell lines (MG63 and U2OS) and their doxorubicin-resistant cell lines (MG63-Dox and U2OS-Dox), respectively. Subsequently, cells were seeded in 6-well plates at a density of 1 × 105 cells per well and incubated for 14 days to observe clone formation. To evaluate the involvement of the PVT1-CCND1 axis in doxorubicin resistance, we conducted independent or combined interventions to manipulate PVT1 and CCND1 expression in the original OS cell lines. Subsequently, a uniform dosage of doxorubicin (0.4 μg/mL) was administered for 24 h as a pretreatment, followed by a colony formation assay to quantify the number of formed clones. Microscopy was used to capture images of colonies in each well, which were then analysed using ImageJ software V1.8.0 (National Institutes of Health, USA).
2.6. Real-time quantitative PCR (qRT–PCR)
Complementary DNA (cDNA) from various cell or tissue samples were amplified by real-time quantitative PCR with the SYBR Green Supermix (Takara, Japan) and specific primers for target genes. All reactions were performed in triplicate. Data were then normalized to GAPDH (for mRNA and lncRNA) or U6 small nuclear RNA (for miRNA) expression and calculated by the 2−ΔΔCt method. The following primers were designed for qRT-PCR amplification and sequencing: PVT1 forward: 5′-TGGAATGTAAGACCCCGACTCT-3′ and reverse: 5′-GATGGCTGTATGTGCCAAGGT-3′; MYC forward: 5′-TTTATAATGCGAGGGTCTGGA-3′ and reverse: 5′-GCATTCGACTCATCTCAGCA-3′; CCND1 forward: 5′-GGATGCTGGAGGTCTGCGA-3′ and reverse: 5′-AGAGGCCACGAACATGCAAG-3′.
2.7. Western blotting
Proteins were extracted and then separated through SDS-PAGE gel and electrotransferred onto polyvinylidene difluoride (PVDF) membranes. Next, the membranes were incubated with primary antibodies, including anti‐CCND1 (Abcam, 1:1000 dilution), anti‐ABCC1 (CST, Massachusetts, USA, 1:500 dilution), anti-ABCC2 (CST, 1:500 dilution), anti‐ABCC3 (CST, 1:500 dilution), anti‐Ki67 (Abcam, 1:100 dilution), anti‐ Caspase 9 (Abcam, 1:500 dilution), and anti‐β-actin (CST, 1:2000 dilution) at 4 °C overnight. Then, the blots were incubated with secondary antibodies (Abcam, 1:3000 dilution) at 25 °C for 1 h. Enhanced chemiluminescence (ECL) substrates (Millipore, Burlington, MA, USA) were used to expose the blots. Gray intensity analysis was performed using ImageJ software (NIH, Bethesda, MD, USA).
2.8. Animal study
Male BALB/c nude mice (6 ∼ 8-week-old, N = 5 per group) were from SJA Co., LTD (Hunan, China). For bioluminescent imaging, luciferase stable expressed MG63 cells were injected intravenously, and mice were injected with 200 mg/kg D-luciferin (Invitrogen) at 15 min post-inoculation. Briefly, mouse models with lung metastases were established by intravenously injecting stable transfected PVT1 overexpression (OE-PVT1) MG63 cells or negative control (OE-NC) cells (5 × 106 cells/0.2 mL RPMI1640). Subsequently, doxorubicin (5 mg/kg) was administered intraperitoneally once a week for a total of 3 weeks after two weeks of inoculation. Bioluminescent imaging was performed using the In Vivo Imaging System (Xenogen, Alameda, CA, USA). Following the designated observation period, euthanasia was conducted on the mice within each respective group, and the pulmonary tissue was extracted for subsequent analysis. Animal experiments in this study was approved by The Experimental Animal Welfare Ethical Review Consent of The Second Xiangya Hospital of Central South University (2022014).
2.9. TUNEL assay
Cell apoptosis in lung tissues was assessed using TUNEL assay kit (Roche® Life Science Products, Basel, Switzerland), and the detailed operation steps were carried out in accordance with the manufacturer's instructions. Briefly, paraffin slips of lung tissues were stained with mixed staining solution (50 μL TdT + 450 μL fluorescein labeled dUTP) at 37 °C for 1 h. Images were acquired through whole slide imaging (3D-HISTECH, Budapest, Hungary) for TUNEL+ cell analysis.
2.10. Fluorescence in situ hybridization (FISH)
The FITC-conjugated probe for PVT1 was designed and synthesized by RiboBio (Guangzhou, China). OS cells grown on slips were fixed with 1 % formaldehyde and permeabilized with 70 % ethanol at 4 °C overnight. Then the cells were incubated with the PVT1 probe in hybridization buffer at 37° C overnight, followed by mounted with Prolong Gold antifade reagent and DAPI (Invitrogen). Slivers were then scanned for each channel through whole slide imaging (3D-HISTECH) for analysis.
2.11. RNA immunoprecipitation (RIP)
RIP assay was conducted using the Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Briefly, immunoprecipitation was performed by the anti-AGO2 antibody (Abcam, 1:100 dilution) or normal mouse IgG (negative control), and then PVT1, miR-15a-5p, miR-15b-5p, as well as CCND1 were identified by qRT-PCR from the precipitated RNA products.
2.12. Luciferase assay
For analysis of targeted degradation of RNAs (PVT1 or CCND1 mRNA) by miR-15a-5p and miR-15b-5p, the sequences of PVT1 and 3′- UTR clones of CCND1, including wild-type (WT) and mutant (Mut) clones, were synthesized by and purchased from Sangon Biotech (Shanghai, China). WT-PVT1/Mut-PVT1 and the 3′- UTRs of WT-CCND1/Mut-CCND1 were individually cloned into pGL4 luciferase reporter plasmids (Promega, Wisconsin, USA). Then, HEK293T cells were co-transfected with them and miR-15a-5p or miR-15b-5p mimics or negative control (mimics-NC). 48 h after transfection, the cells were subjected to a dual luciferase reporter assay according to the manufacturer’s instructions (Promega). The relative luciferase activity was normalized to Renilla luciferase activity.
For analysis of transcriptional regulation of CCND1 by MYC, the expression vector of MYC and the pGL4 luciferase reporter vector that carrying the CCND1 promoter with the WT binding sites or the Mut sites were co-transferred into HEK293 cells. Luciferase activity was detected and analyzed with the same procedures as before.
2.13. Chromatin immunoprecipitation (ChIP) assay
Cell samples were lysed and ultrasonically broken to produce 200 ∼ 500 bp chromatin fragments. Subsequently, the fragments were precipitated following the kit manufacturer's instructions (CST, Cat. No.9003) using the anti-MYC ChIP grade antibody (CST, Cat. No. 3400, 1:100 dilution). qRT-PCR was conducted to amplify the promoter fragments of CCND1 in ChIP products. Obtained data were finally displayed in percentage of the Input (% Input).
2.14. Statistical analysis
Data are expressed as the mean ± standard deviation (SD). Comparisons between two groups were performed using Student’s t test. Comparisons among three or more groups were conducted with one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. Bivariate correlation analysis and Wilcoxon signed-rank test were performed to analyze the correlation between two different indicators. The results were statistically analysed using GraphPad Prism 9.0 (San Diego, CA, USA). Differences were considered statistically significant when the P value was less than 0.05.
3. Results
3.1. Advanced progression of OS is related to the elevated expression of PVT1, MYC and CCND1
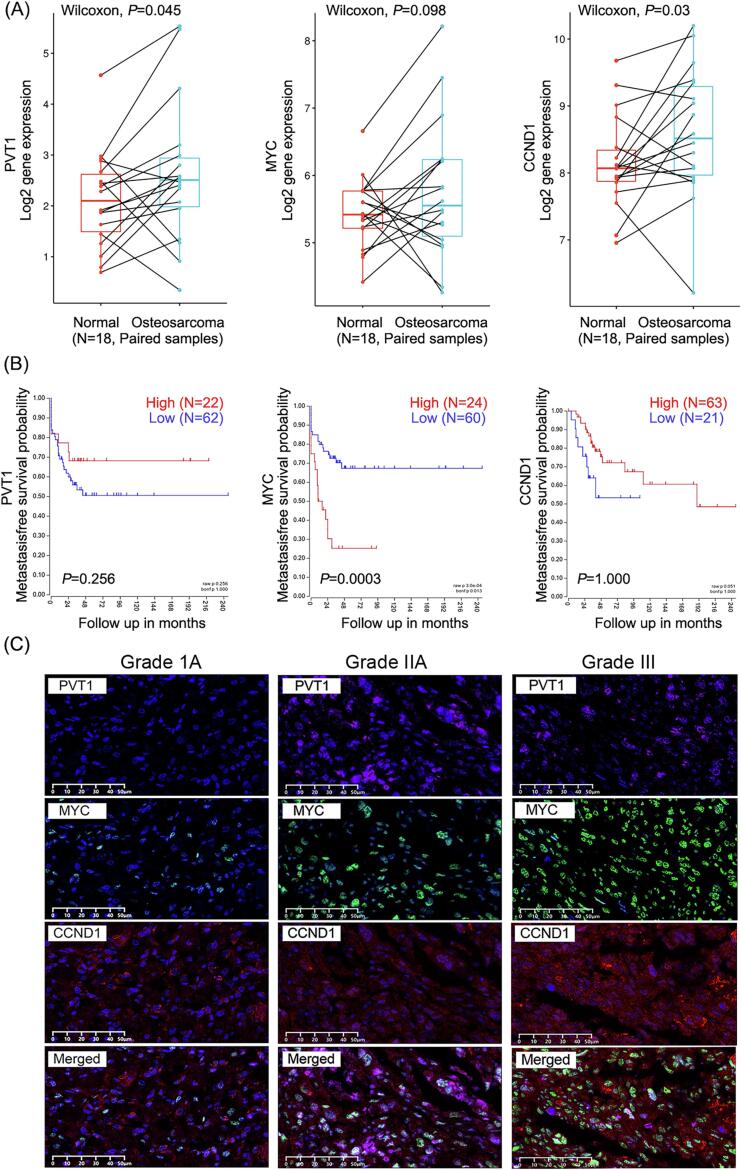
The analysis findings obtained from a GEO database consisting of 18 paired samples (cancerous tissue matched 1:1 with adjacent osteogenic tissue, GSE99671) demonstrated a significant increase in the transcription levels of PVT1 and CCND1 in cancerous tissues compared to osteogenic tissues (PVT1, P = 0.045; CCND1, P = 0.03; MYC, P = 0.098) (Fig. 1A). Additionally, another gene expression array database comprising cancer tissues from 84 OS patients (GSE42352) revealed that patients with high MYC expression experienced notably shorter durations of metastasis-free survival in comparison to those with low MYC expression (P = 0.0003). However, this association was not observed for PVT1 and CCND1 (P = 0.256, P = 1.000, respectively) (Fig. 1B). These findings suggest a potential association between PVT1 and CCND1 as indicators for the occurrence of OS, whereas MYC may serve as a prognostic indicator for poor outcomes. To further investigate this, we examined the expression levels of PVT1 (RNA), MYC (protein), and CCND1 (protein) in three biopsy samples obtained from patients with varying grades of OS, and the results demonstrated an upward trend in the expression levels of PVT1, MYC, and CCND1 with advanced cancer grade (Fig. 1C), indicating a possible correlation between the upregulated expression of these genes and the advanced progression of OS.
Fig. 1.
Advanced progression of OS is related to the elevated expression of PVT1, MYC and CCND1. (A) Relative mRNA levels of PVT1, MYC and CCDN1 in 18 paired samples of cancer tissues and adjacent tissues from patients with OS. Wilcoxon test, P<0.05: statistically significant. (B) Metastasis-free survival probability of 84 OS patients with high or low expression of PVT1, MYC and CCDN1 via Kaplan-Meier method. Log-rank test, P<0.05: statistically significant. (C) Detection of PVT1, MYC and CCDN1 in caner tissues via IF/FISH assay.
3.2. PVT1 mediates doxorubicin resistance in OS cells
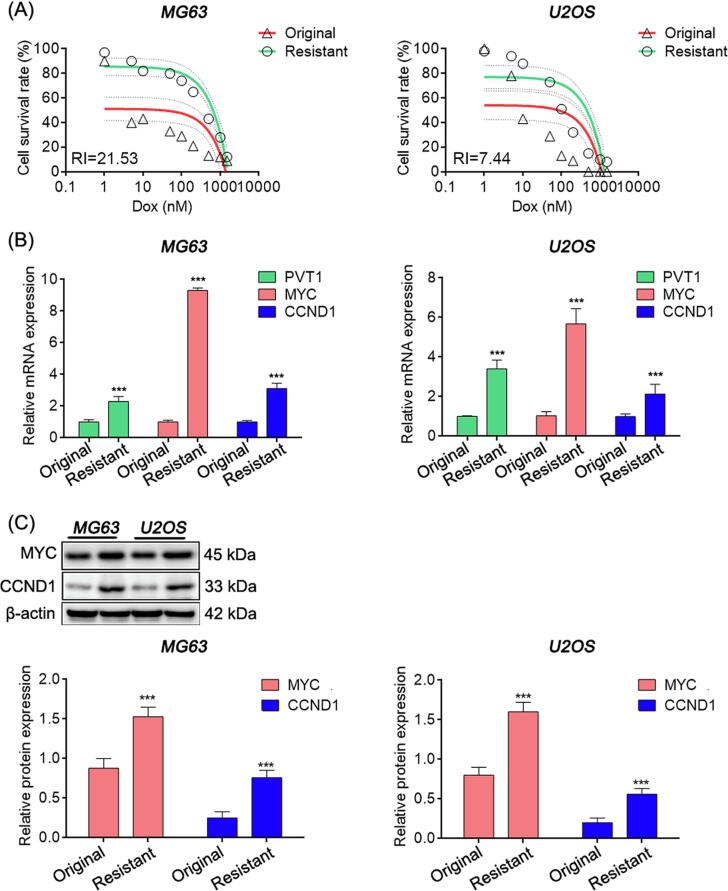
The occurrence of acquired drug resistance is the primary reason for recurrence and unfavorable prognosis in patients with OS. Therefore, we aimed to investigate whether the high expression levels of PVT1, MYC, and CCND1 could contribute to chemotherapy resistance. Our findings demonstrated that the mRNA expressions of PVT1, MYC, and CCND1 were markedly upregulated in doxorubicin-resistant OS cell lines (MG63 and U2OS), with resistance indices (RIs) of 21.53 and 7.44, respectively, which were significantly higher than those in the original cells. Moreover, the protein levels of MYC and CCND1 were also elevated (Fig. 2A-C).
Fig. 2.
PVT1, MYC and CCND1 were high expressed in doxorubicin resistance OS cells. (A) Survival rate curves of MG63 and U2OS cells treated with different doxorubicin concentrations via MTT assay. (B) Relative mRNA expression in OS cells detected by qRT-PCR. (C) Relative protein expression detected by western blot. In (B) and (C), student’s t test, *: P < 0.05; **: P < 0.01; ***: P < 0.005.
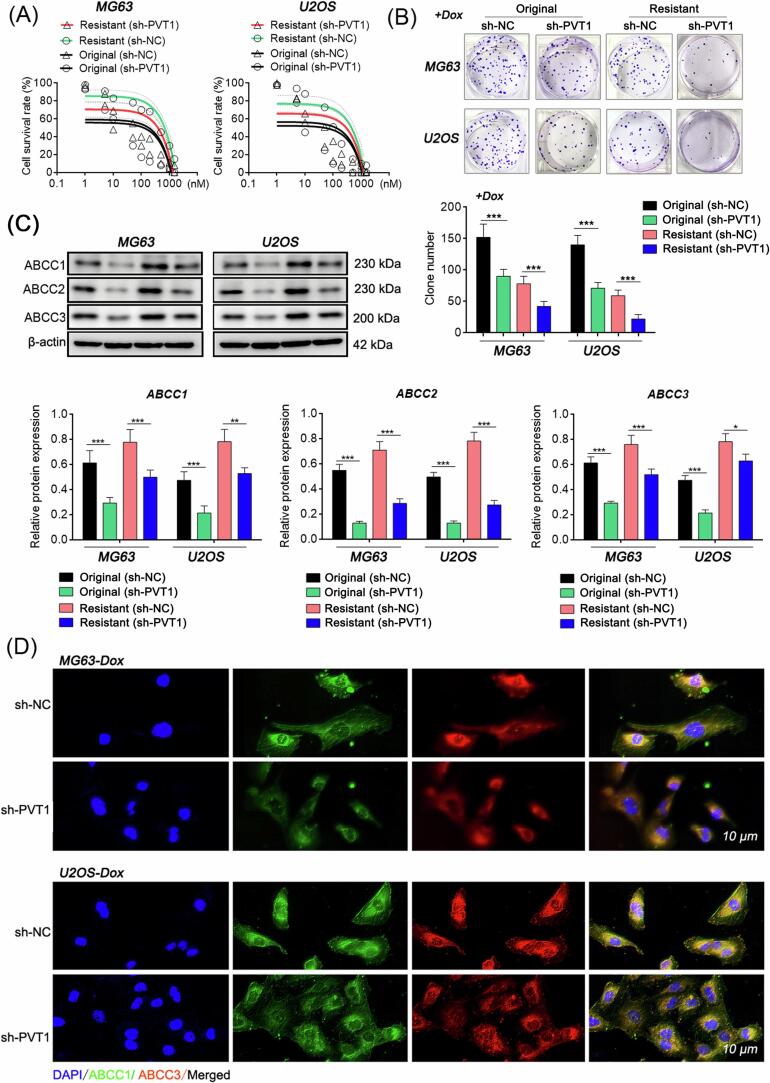
Next, shRNA technology was used to interfere with the expression of PVT1 in both parental and resistant OS cell lines. The impact of PVT1 expression levels on the sensitivity of OS cells to doxorubicin was evaluated by measuring changes in cell viability when exposed to different concentrations of doxorubicin (ranging from 0 to 1500 nM) in the culture medium for 24 h. The results revealed that interference with PVT1 in the resistant cell strains significantly restored cellular sensitivity to doxorubicin, as evidenced by a greater decrease in cell viability (Fig. 3A). Additionally, when PVT1 expression was disrupted in cells and subsequently treated with equimolar doses of doxorubicin (0.4 μg/mL), the site-specific proliferative capacity (Fig. 3B) and expression levels of multidrug resistance-related proteins ABCC1, ABCC2, and ABCC3 (Fig. 3C-D) were significantly reduced. These data suggest that downregulation of PVT1 enhances the sensitivity of OS cells to doxorubicin in OS cells.
Fig. 3.
Knockdown of PVT1 enhances doxorubicin susceptibility of OS cells. (A) Survival rate curves of different concentrations of doxorubicin-treated groups measured by the MTT assay. Doxorubicin concentration range: 0–1500 nM; duration: 24 h. (B) Site-specific proliferation of cancer cells detected by plate cloning method. (C) Relative protein expression detected by western blot. (D) ABCC1 and ABCC3 expression levels in cells detected by double-labelling immunofluorescence. In (A), (B), and (C), student’s t test, *: P < 0.05; **: P < 0.01; ***: P < 0.005.
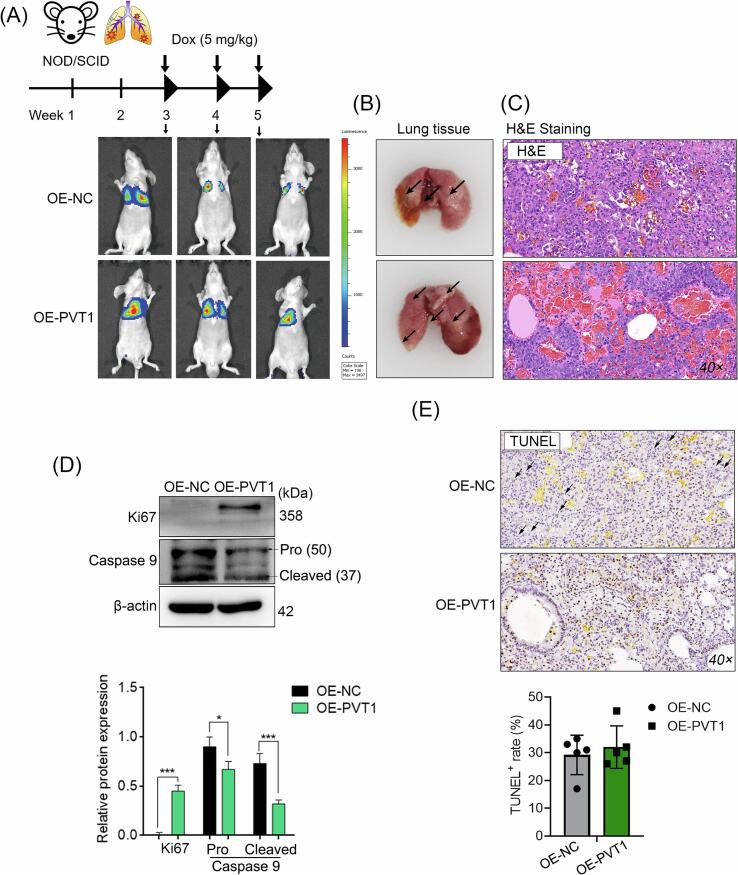
We employed intravenous injection of MG63 cells via the tail vein to establish a murine model of pulmonary metastasis of OS, taking into account the strong tendency of OS to undergo distant metastasis, specifically in the lungs. Results showed that cells overexpressing PVT1 in the murine OS model with lung metastasis exhibited increased resistance to doxorubicin treatment compared to the negative control cells (Fig. 4A). This was characterized by a higher number of lung nodules, denser cell nuclei within the tumor tissue, higher expression of the proliferation marker protein Ki67, but lower levels of the apoptosis-related cysteine peptidase (Caspase 9) (Fig. 4B-D). In addition, the TUNEL assay was employed to assess the extent of nuclear genomic damage caused by doxorubicin treatment in tumor tissues. No significant difference was observed in the proportion of TUNEL+ cells between the two groups. However, distinct nuclear dissolution was observed in the tumor tissues of the OE-NC group, as indicated by the black arrows, suggesting a potentially higher degree of cell death and indicating a relatively better therapeutic effect of doxorubicin in the tumor tissues of the OE-NC group (Fig. 4E).
Fig. 4.
PVT1 mediates doxorubicin resistance in OS cells in vivo. Murine model of pulmonary metastasis of OS was established through the injection of MG63 cells. (A) Representative photos of pulmonary metastasis in nude mouse models of OS in different groups. (B) Lung tissues obtained from mice in each group after the observation period. (C) Micropathology examination by H&E staining. (D) Protein levels of Ki67 and Caspase 9 were assessed in mixture of intra-group samples from different groups using western blotting (N = 5). (E) Cellular apoptosis within the tissues detected via the TUNEL assay. In (D), student’s t test, *: P < 0.05; **: P < 0.01; ***: P < 0.005. Black arrow: region of significant nuclear dissolution in the tissue.
Taken together, the aforementioned data provide evidence of the role played by PVT1 in the in vivo development of doxorubicin tolerance.
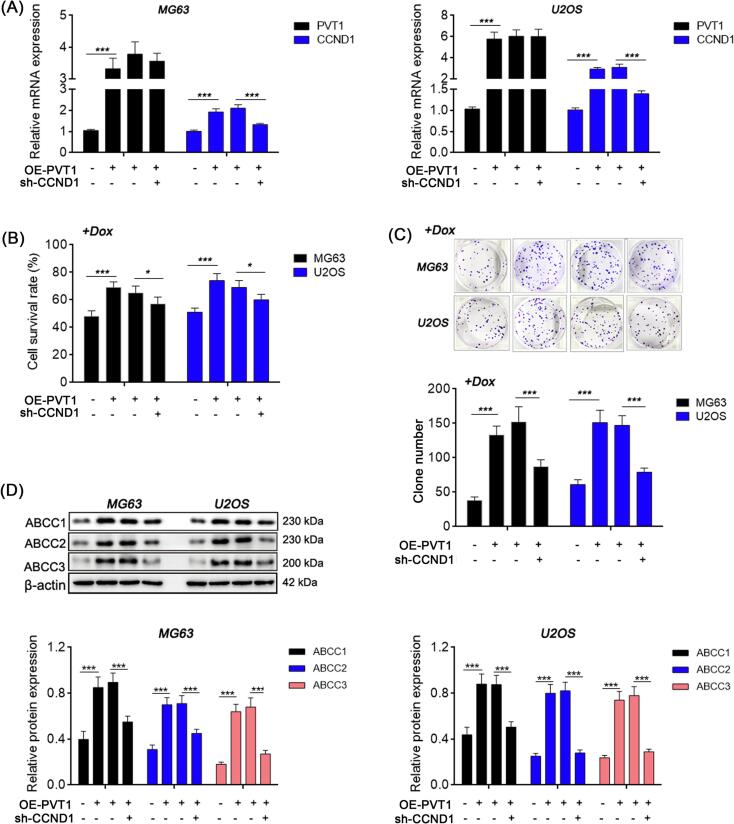
3.3. CCND1 knockdown alleviates doxorubicin resistance caused by PVT1
CCND1 was then knocked down in the PVT1-overexpressed OS cells, followed by treatment with doxorubicin (0.4 μg/mL, 24 h), to evaluate the impact of CCND1 on PVT1-induced doxorubicin resistance (Fig. 5A). Compared with the blank load transfection group, the cell viability, colony forming ability and expression levels of multi-drug resistance related proteins (ABCC1, ABCC2, and ABCC3) were significantly reduced in the CCND1 knockdown group (Fig. 5B-D). The data presented indicates that inhibition of CCND1 may potentially enhance the sensitivity of OS cells with upregulated PVT1 expression to doxorubicin.
Fig. 5.
CCND1 knockdown alleviates doxorubicin resistance caused by PVT1. (A) Relative mRNA expression in OS cells detected by qRT-PCR. (B) Cell survival rate detected by MTT method. (C) Site-specific proliferation detected by plate cloning method. (D) Relative protein expression detected by western blot. One-way ANOVA. *: P < 0.05; **: P < 0.01; ***: P < 0.005.
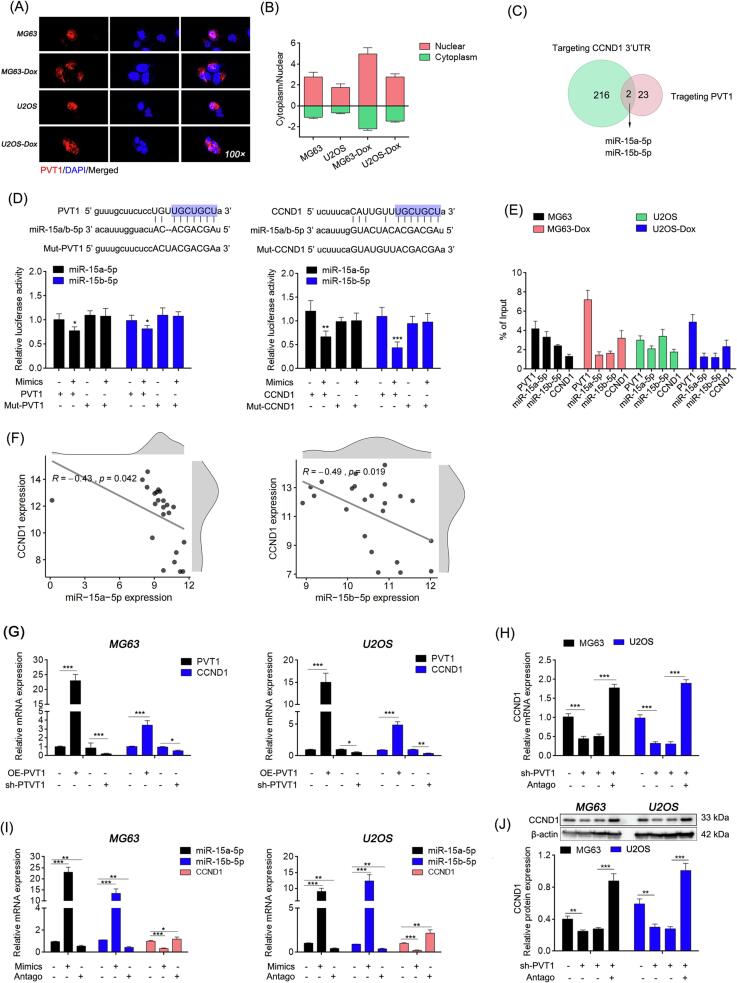
3.4. PVT1 acted as molecular sponge of miR-15a-5p and miR-15b-5p to up-regulate CCND1 expression levels
PVT1 is an lncRNA primarily localized in the nucleus and subsequently in the cytoplasm of OS cells (Fig. 6A), and it is upregulated in OS cells resistant to doxorubicin (Fig. 6B). Various studies have suggested that cytoplasmic PVT1 acts as a molecular “sponge” for microRNAs (miRNAs). The StarBase database (https://starbase.sysu.edu.cn/) indicates that there are 216 miRNAs that could potentially modulate the expression of CCND1. Among these, 23 miRNAs can be adsorbed by PVT1, and only miR-15a-5p and miR-15b-5p (referred to as miR-15a/b) have the potential to interact with both (Fig. 6C). Intriguingly, luciferase assays confirmed that PVT1 and the 3’-UTR region of CCND1 share the same binding site (5'-UGCUGCU-3') for miR-15a/b (Fig. 6D). Furthermore, PVT1, miR-15a/b and CCND1 3’-UTR were simultaneously detected in the RNA precipitation products of AGO2, a key component of the RISCs complex responsible for target RNA degradation [29] (Fig. 6E). These findings provide further support for the ceRNA role of PVT1 and CCND1 mRNA.
Fig. 6.
PVT1 acted as molecular sponge of miR-15a-5p/ miR-15b-5p to up-regulate CCND1 expression levels. (A) Subcellular localization of PVT1 via FISH assay. (B) PVT1 expression abundance in cytoplasm and nucleus detected by qRT-PCR after nucleo-plasmic mRNA separation. (C) The number and intersection of miRNAs predicted targeting PVT1 and CCND1 3′-UTR (data obtained from the StarBase database). (D) Binding sites analysis via dual-luciferase assay. (E) Binding relationship between PVT1, miR-15a and miR-15b-5p, and CCND1 mRNA analyzed by AGO2 RIP. (F) Correlation analysis between miR-15a-5p, miR-15b-5p and CCND1 (GSE database). (G-I) Relative mRNA expression levels detected by qRT-PCR. (J) Relative protein expression levels detected by western blot. In (D, G-J), One-way ANOVA, *: P < 0.05; **: P < 0.01; ***: P < 0.005. In (F), Bivariate correlation analysis.
Moreover, analysis based on miRNA and mRNA expression in paraffin-archived human OS specimens (GSE39058) revealed a significant negative correlation between miR-15a/b and CCND1 (R = -0.43, P = 0.042 and R = -0.49, P = 0.019, respectively) (Fig. 6F). In OS cells, the expression of CCND1 can be influenced by PVT1 or miR-15a/b (Fig. 6G, I). Furthermore, interfering with PVT1 to release miR-15a/b also led to decreased CCND1 expression, but this effect could be restored when miR-15a/b was subsequently antagonized (Fig. 6H, J). Data presented here demonstrated that PVT1 competes with CCND1 mRNA to bind miR15a/b to regulate the expression level of CCND1 in OS cells.
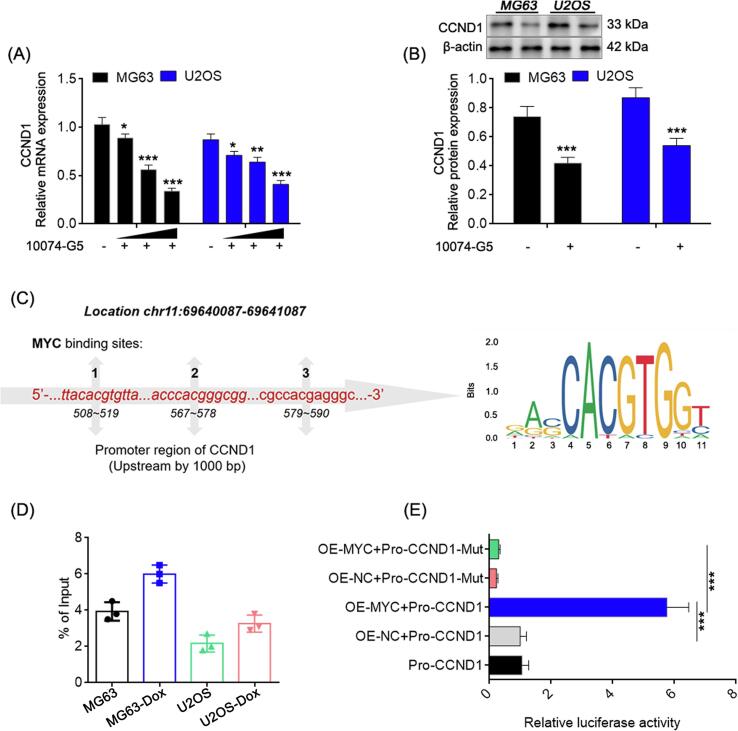
3.5. PVT1 guarantees the expression of MYC to promote its transcriptional activity against CCND1
MYC is a carcinogenic transcription factor that drives the development of OS [30], [14], and the recognition site (5′-CAC[GA]TG-3′) of which is highly frequent in the promoter region of CCND1, making making our attempt to explore whether intranuclear PVT1 is capable to regulate CCND1 transcription through MYC.
Significantly reduced mRNA and protein expression levels of CCND1 were observed in OS cells treated with MYC inhibitor (10074-G5, 10 μM, 24 h) (Fig. 7A-B). The primary binding site of MYC was identified as the chromatin region located approximately 500–600 bp upstream of the transcription start site (TSS) of the CCND1 gene (Fig. 7C). Moreover, the binding abundance of MYC in this region was found to be higher in resistant cells compared to original cells (Fig. 7D). Additionally, the dual-luciferase assay demonstrated a significant enhancement in the transcriptional activity of the CCND1 promoter when MYC was overexpressed (Fig. 7E), indicating that MYC functions as a transcriptional activator for CCND1.
Fig. 7.
MYC promotes transcriptional activity of CCND1. Relative mRNA and protein expression detected by qRT-PCR (A) and western blot (B). (C) Binding sites of MYC in the promoter region of CCND1. (D) Relative abundance of CCND1 promotor fragments amplified by qRT-PCR from immunoprecipitate products by MYC ChIP assay (% of Input > 1, defined as high strength binding). (E) CCND1 promoter activity detection with wild type (WT) or mutation (Mut) of MYC binding sites via dual-luciferase. In (A) and (E), one-way ANOVA. In (B) and (D), student’s t test, *: P < 0.05; **: P < 0.01; ***: P < 0.005.
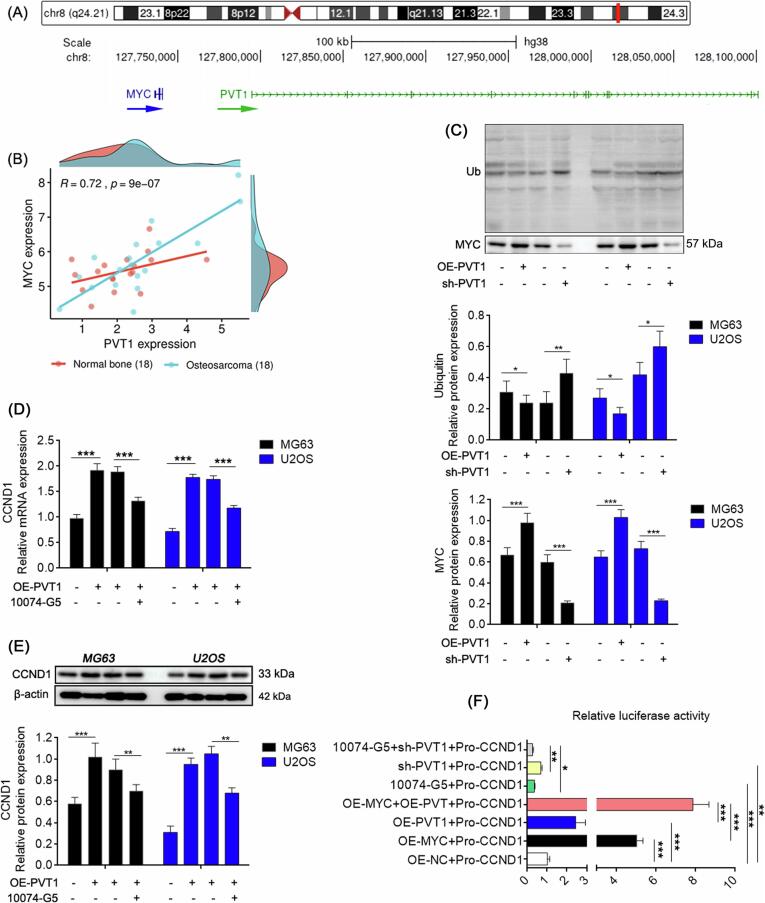
PVT1, located adjacent to MYC (Fig. 8A), exhibits a significant positive correlation with MYC expression in cancerous tissues of patients with OS, as evidenced by data from the GEO database (GSE99671) (Fig. 8B). Experiments conducted on OS cell lines indicate that upregulation of PVT1 prevents MYC degradation through the ubiquitination pathway, whereas knockdown of PVT1 has the opposite effect (Fig. 8C). Furthermore, the impact of PVT1 upregulation on CCND1 expression is partially counteracted by MYC inhibition (Fig. 8D-E), suggesting that PVT1 regulates CCND1 expression through MYC. Additionally, simultaneous overexpression of PVT1 and MYC significantly enhances CCND1 transcription compared to overexpression of either alone, and interference with both PVT1 and MYC results in a more pronounced reduction in CCND1 transcription (Fig. 8F).
Fig. 8.
PVT1 is necessary for MYC transcription and protein stability. (A) Genome locations of PVT1 and MYC on chromosome 8. (B) Expression correlation between PVT1 and MYC in cancer tissues and adjacent tissues of patients with OS (GEO database). (C) The ubiquitination level of MYC protein. Relative mRNA and protein expression detected by qRT-PCR (D) and western blot (E). (F) CCND1 promoter activity detection in cells with PVT1 and MYC co-expression or co-silence via dual-luciferase. In (B), Bivariate correlation analysis. In (C-F), One-way ANOVA. *: P < 0.05; **: P < 0.01; ***: P < 0.005.
In summary, both PVT1 and MYC contribute to CCND1 transcription, with MYC serving as a direct transcriptional activator of CCND1, while PVT1 guarantees the protein stability of MYC.
4. Discussion
OS, the most common type of bone cancer in children and adolescents, frequently encounters treatment failure attributed to the development of chemoresistance, for which the underlying molecular mechanisms remains elusive [28]. In this study, we aimed to elucidate the mechanism of PVT1 in acquired resistance to OS and proposed a novel molecular target: CCND1. It is well known that CCND1 regulates the cell cycle and its impact on chemotherapy resistance in various malignant tumors has been studied [23], [24], [25]. This study confirmed that the upregulation of CCND1 induced by PVT1 is one of the reasons for doxorubicin resistance in OS cells. Mechanistically, PVT1 and CCND1 3′-UTR share the same miR-15a/b binding sites and mutually regulate their expression in OS cells. Additionally, PVT1 facilitates the stabilization of MYC protein, thereby promoting the transcriptional activation of CCND1 (Fig. 9).
Fig. 9.
Schematic diagram of the molecular regulatory mechanism elaborated in this study.
LncRNAs play regulatory roles in disease progression through a variety of mechanisms, which vary based on their distinct subcellular localization [31]. In the case of PVT1 in OS cells, we observed a higher expression tendency in the nucleus compared to the cytoplasm, consistent with observations in numerous other cancer cells based on data from the LncALTAs database (https://lncatlas.crg.eu/). This finding suggests that PVT1 may possess diverse biological functions.
LncRNAs present in the cytoplasm have the potential to function as molecular sponges for specific miRNAs by virtue of their free bases, which can exhibit complete or partial complementarity with miRNAs, thus preventing the degradation of miRNA's target mRNA [32], [3]. In this study, we discovered that miR-15a/b play a crucial role in mediating the impact of PVT1 on CCND1 expression. Specifically, PVT1 acts as a molecular decoy for miR-15a/b, thereby impeding their ability to degrade CCND1 mRNA.
The high similarity of miR-15a and miR-15b suggests that both clusters likely regulate a similar set of target genes and may have overlapping functions. Previous studies have shown that miR-15a/b played inhibitory roles in a variety of cancers such as childhood neuroblastoma [33], colorectal cancer [34], cervical cancer [35] and chronic lymphocytic leukemia [36], supporting the tumor suppressor identity of miR-15a/b. Specifically, the miR-15a/b cluster has been extensively shown to play a central role in B-cell oncogenesis, as over two-thirds of B-cell chronic lymphocytic leukemia cases exhibit deletion of the miR-15a/b locus at 13q14 [37]. But to our knowledge, the roles of miR-15a/b in OS remain unclear. In this study, our data provide evidence for a negative correlation between miR-15a/b expression and CCND1 in OS. Furthermore, we identified a shared miR-15a/b binding site between PVT1 and CCND1 3′-UTR, suggesting that functional loss of miR-15a/b in OS may contribute to PVT1- or CCND1-dependent cell cycle dysregulation. Consequently, this dysregulation may lead to uncontrolled malignant proliferation and resistance to chemotherapy.
LncRNAs in the nucleus may participate in the regulation of transcription, splicing, and mRNA nuclear export processes [38], [39]. It is widely recognized that normal cells have evolved diverse mechanisms to control the levels of MYC, aiming to hinder genomic instability and tumorigenesis. Nevertheless, these mechanisms may encounter disruption in cancer cells [40]. Thus, keeping MYC in check is an important part of avoiding carcinogenic transformation [41]. Through genomic analysis of MYC on chromosome 8, there is a "gene desert" was found approximately 1444 Mb upstream of the MYC genome that contains various non-coding RNAs, including PVT1 which is the closest to MYC. Studies have shown that increased expression of PVT1 is required for high MYC protein levels in 8q24-amplified human cancer cells, and removal of PVT1 from MYC-driven cancer cells decreases their tumorigenic potential [13]. Therefore, PVT1 is considered a promising therapeutic target for MYC-driven tumors.
Currently, the impact of PVT1 on the expression or biological functioning of MYC has been identified in different ways, including its role as a intergenetic super enhancer lncRNA (SE lncRNA) [42], or as an enhancer competitor in cis [43], or as a protein stability protector [44], among others. In this study, our findings in OS cells suggest that PVT1 plays a beneficial role in maintaining MYC protein levels. Specifically, knocking down PVT1 leads to increased degradation of MYC through ubiquitylation. Given that targeted degradation by the ubiquitin-proteosome system is one of the main ways in which MYC levels are controlled in cells [40], inhibiting PVT1 could be a feasible approach to combat the OS process driven by MYC.
Collectively, the present investigation centered on elucidating the pathways through which PVT1 and CCND1 contribute to the development of doxorubicin resistance, thereby offering novel therapeutic targets against acquired resistance in OS.
5. Ethics Statement
The patient tissue samples involved in this study obtained informed consent from all the patients and the scientific research project was approved by the Medical Ethics Committee of the Second Xiangya Hospital of Central South University with number is (2022) No. (Research 023). The animal ethics in this study have been approved by the Experimental Animal Welfare Ethical Review Consent of The Second Xiangya Hospital of Central South University (2022014).
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China with Grant No. 82273263.
CRediT authorship contribution statement
Zi Li: Conceptualization, Data curation, Validation, Writing – original draft. Jia-Ming Tian: Data curation, Methodology, Writing – original draft. Yi Chu: Formal analysis, Software, Supervision, Visualization. Hong-Yi Zhu: Formal analysis, Software, Supervision. Jun-Jie Wang: Investigation, Data curation. Jun Huang: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Mirabello L., Troisi R.J., Savage S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer-Am. Cancer Soc. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe N., Puri A., Gelderblom H. Osteosarcoma: evolution of treatment paradigms. Sarcoma. 2013;2013:1–7. doi: 10.1155/2013/203531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J.-Y., Yang Y., Ma Y., Wang F., Xue A., Zhu J., Yang H., Chen Q.i., Chen M., Ye L., Wu H., Zhang Q. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed. Pharmacother. 2020;121 doi: 10.1016/j.biopha.2019.109627. [DOI] [PubMed] [Google Scholar]
- 4.Xu M., Wang Y., Weng W., Wei P., Qi P., Zhang Q., et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin. Cancer Res. 2017;23(8):2071–2080. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]
- 5.Du P., Hu C., Qin Y., Zhao J., Patel R., Fu Y., Zhu M., Zhang W., Huang G. LncRNA PVT1 Mediates Antiapoptosis and 5-Fluorouracil Resistance via Increasing Bcl2 Expression in Gastric Cancer. J. Oncol. 2019;2019:1–10. doi: 10.1155/2019/9325407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., Du H., Bao L., Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol. Med. 2018;15(3):238–250. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., Li C., Mudd A., Gu X. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Biosci. Biotech. Bioch. 2017;81(12):2301–2306. doi: 10.1080/09168451.2017.1387048. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J., Du P., Cui P., Qin Y., Hu C., Wu J., Zhou Z., Zhang W., Qin L., Huang G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37(30):4094–4109. doi: 10.1038/s41388-018-0250-z. [DOI] [PubMed] [Google Scholar]
- 9.Song J., Wu X., Liu F., Li M., Sun Y., Wang Y., Wang C., Zhu K., Jia X., Wang B., Ma X. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem. Biophys. Res. Commun. 2017;490(2):217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 10.Wu T., Ji Z., Lin H., Wei B.o., Xie G., Ji G., Fu S., Huang W., Liu H. Noncoding RNA PVT1 in osteosarcoma: The roles of lncRNA PVT1 and circPVT1. Cell Death Discov. 2022;8(1) doi: 10.1038/s41420-022-01192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsang S.V., Rainusso N., Liu M., Nomura M., Patel T.D., Nakahata K., Kim H.R., Huang S., Rajapakshe K., Coarfa C., Man T.-K., Rao P.H., Yustein J.T. LncRNA PVT-1 promotes osteosarcoma cancer stem-like properties through direct interaction with TRIM28 and TSC2 ubiquitination. Oncogene. 2022;41(50):5373–5384. doi: 10.1038/s41388-022-02538-w. [DOI] [PubMed] [Google Scholar]
- 12.Graham M., Adams J.M. Chromosome 8 breakpoint far 3' of the c-myc oncogene in a Burkitt's lymphoma 2;8 variant translocation is equivalent to the murine pvt-1 locus. EMBO J. 1986;5(11):2845–2851. doi: 10.1002/j.1460-2075.1986.tb04578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng Y.-Y., Moriarity B.S., Gong W., Akiyama R., Tiwari A., Kawakami H., Ronning P., Reuland B., Guenther K., Beadnell T.C., Essig J., Otto G.M., O’Sullivan M.G., Largaespada D.A., Schwertfeger K.L., Marahrens Y., Kawakami Y., Bagchi A. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Noon S., Ijaz J., Coorens T.H., Amary F., Ye H., Strobl A., et al. MYC amplifications are common events in childhood osteosarcoma. J. Pathol. Clin. Res. 2021;7(5):425–431. doi: 10.1002/cjp2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lew D.J., Dulic V., Reed S.I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66(6):1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Y., Connolly T., Futcher B., Beach D. Human D-type cyclin. Cell. 1991;65(4):691–699. doi: 10.1016/0092-8674(91)90100-d. [DOI] [PubMed] [Google Scholar]
- 17.Meyerson M., Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell Biol. 1994;14(3):2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates S., Bonetta L., MacAllan D., Parry D., Holder A., Dickson C., et al. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene. 1994;9(1):71–79. [PubMed] [Google Scholar]
- 19.Santra M.K., Wajapeyee N., Green M.R. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459(7247):722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simoneschi D., Rona G., Zhou N., Jeong Y.-T., Jiang S., Milletti G., Arbini A.A., O’Sullivan A., Wang A.A., Nithikasem S., Keegan S., Siu Y., Cianfanelli V., Maiani E., Nazio F., Cecconi F., Boccalatte F., Fenyö D., Jones D.R., Busino L., Pagano M. CRL4(AMBRA1) is a master regulator of D-type cyclins. Nature. 2021;592(7856):789–793. doi: 10.1038/s41586-021-03445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng L.H., Zhang H., Hayward L., Takemura H., Shao R.G., Pommier Y. Tetrandrine induces early G1 arrest in human colon carcinoma cells by down-regulating the activity and inducing the degradation of G1-S-specific cyclin-dependent kinases and by inducing p53 and p21Cip1. Cancer Res. 2004;64(24):9086–9092. doi: 10.1158/0008-5472.CAN-04-0313. [DOI] [PubMed] [Google Scholar]
- 22.Zhou W., Zhao Z., Wang R., Han Y., Wang C., Yang F., Han Y.a., Liang H., Qi L., Wang C., Guo Z., Gu Y. Identification of driver copy number alterations in diverse cancer types and application in drug repositioning. Mol. Oncol. 2017;11(10):1459–1474. doi: 10.1002/1878-0261.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noel E.E., Yeste-Velasco M., Mao X., Perry J., Kudahetti S.C., Li N.F., Sharp S., Chaplin T., Xue L., McIntyre A., Shan L., Powles T., Oliver R.T.D., Young B.D., Shipley J., Berney D.M., Joel S.P., Lu Y.-J. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am. J. Pathol. 2010;176(6):2607–2615. doi: 10.2353/ajpath.2010.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bostner J., Ahnstrom W.M., Fornander T., Skoog L., Nordenskjold B., Stal O. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene. 2007;26(49):6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q., Wu X., Wen C., Wang H., Wang H., Liu H., Peng J. Corrigendum to “Toosendanin induces caspase-dependent apoptosis through the p38 MAPK pathway in human gastric cancer cells” [Biochem. Biophys. Res. Commun. 505(1) (2018) 261-266] Biochem. Biophys. Res. Commun. 2020;532(3):496. doi: 10.1016/j.bbrc.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Cai C.K., Zhao G.Y., Tian L.Y., Liu L., Yan K., Ma Y.L., et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol. Rep. 2012;28(5):1764–1770. doi: 10.3892/or.2012.1995. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C., Zhao Y., Zeng B. Enhanced chemosensitivity by simultaneously inhibiting cell cycle progression and promoting apoptosis of drug-resistant osteosarcoma MG63/DXR cells by targeting Cyclin D1 and Bcl-2. Cancer Biomark. 2012;12(4):155–167. doi: 10.3233/CBM-130305. [DOI] [PubMed] [Google Scholar]
- 28.Huang J., Ni J., Liu K., Yu Y., Xie M., Kang R., et al. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72(1):230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 29.Yi T. Identifying RISC Components Using Ago2 Immunoprecipitation and Mass Spectrometry. Methods Mol. Biol. 2018;1720:149–159. doi: 10.1007/978-1-4939-7540-2_11. [DOI] [PubMed] [Google Scholar]
- 30.Czarnecka A.M., Synoradzki K., Firlej W., Bartnik E., Sobczuk P., Fiedorowicz M., Grieb P., Rutkowski P. Molecular Biology of Osteosarcoma. Cancers (basel) 2020;12(8):2130. doi: 10.3390/cancers12082130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chava S., Reynolds C.P., Pathania A.S., Gorantla S., Poluektova L.Y., Coulter D.W., Gupta S.C., Pandey M.K., Challagundla K.B. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol. Oncol. 2020;14(1):180–196. doi: 10.1002/1878-0261.12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji D., Zhan T., Li M., Yao Y., Jia J., Yi H., Qiao M., Xia J., Zhang Z., Ding H., Song C., Han Y., Gu J. Enhancement of Sensitivity to Chemo/Radiation Therapy by Using miR-15b against DCLK1 in Colorectal Cancer. Stem Cell Rep. 2018;11(6):1506–1522. doi: 10.1016/j.stemcr.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y., Zhang X., Wang L., Zhu X., Xia Z., Xu L., Xu J. FENDRR suppresses cervical cancer proliferation and invasion by targeting miR-15a/b-5p and regulating TUBA1A expression. Cancer Cell Int. 2020;20(1) doi: 10.1186/s12935-020-01223-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutter K., Rulicke T., Szabo T.G., Andersen L., Villunger A., Herzog S. The miR-15a/16-1 and miR-15b/16-2 clusters regulate early B cell development by limiting IL-7 receptor expression. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.967914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lovat F., Fassan M., Gasparini P., Rizzotto L., Cascione L., Pizzi M., Vicentini C., Balatti V., Palmieri D., Costinean S., Croce C.M. miR-15b/16-2 deletion promotes B-cell malignancies. PNAS. 2015;112(37):11636–11641. doi: 10.1073/pnas.1514954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bridges M.C., Daulagala A.C., Kourtidis A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021;220(2) doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z.-H., Chen T.-Q., Zeng Z.-C., Wang D., Han C., Sun Y.-M., Huang W., Sun L.-Y., Fang K.e., Chen Y.-Q., Luo X.-Q., Wang W.-T. Nuclear export of chimeric mRNAs depends on an lncRNA-triggered autoregulatory loop in blood malignancies. Cell Death Dis. 2020;11(7) doi: 10.1038/s41419-020-02795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farrell A.S., Sears R.C. MYC Degradation. Cold Spring Harb. Perspect. Med. 2014;4(3):a014365. doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felsher D.W., Bishop J.M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. PNAS. 1999;96(7):3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shigeyasu K., Toden S., Ozawa T., Matsuyama T., Nagasaka T., Ishikawa T., Sahoo D., Ghosh P., Uetake H., Fujiwara T., Goel A. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol. Cancer. 2020;19(1) doi: 10.1186/s12943-020-01277-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho S.W., Xu J., Sun R., Mumbach M.R., Carter A.C., Chen Y.G., Yost K.E., Kim J., He J., Nevins S.A., Chin S.-F., Caldas C., Liu S.J., Horlbeck M.A., Lim D.A., Weissman J.S., Curtis C., Chang H.Y. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell. 2018;173(6):1398–1412.e22. doi: 10.1016/j.cell.2018.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng Y.-Y., Bagchi A. The PVT1-MYC duet in cancer. Mol. Cell. Oncol. 2015;2(2):e974467. doi: 10.4161/23723556.2014.974467. [DOI] [PMC free article] [PubMed] [Google Scholar]