Abstract
Macrophage polarization is a dynamic process determining the outcome of various physiological and pathological situations through inducing pro-inflammatory responses or resolving inflammation via exerting anti-inflammatory effects. The miRNAs are epigenetic regulators of different biologic pathways that target transcription factors and signaling molecules to promote macrophage phenotype transition and regulate immune responses. Modulating the macrophage activation, differentiation, and polarization by miRNAs is crucial for immune responses in response to microenvironmental signals and under various physiological and pathological conditions. In term of clinical significance, regulating macrophage polarization via miRNAs could be utilized for inflammation control. Also, understanding the role of miRNAs in macrophage polarization can provide insights into diagnostic strategies associated with dysregulated miRNAs and for developing macrophage-centered therapeutic methods. In this case, targeting miRNAs to further regulate of macrophage polarization may become an efficient strategy for treating immune-associated disorders. The current review investigated and categorized various miRNAs directly or indirectly involved in macrophage polarization by targeting different transcription factors and signaling pathways. In addition, prospects for regulating macrophage polarization via miRNA as a therapeutic choice that could be implicated in various pathological conditions, including cancer or inflammation-mediated injuries, were discussed.
Keywords: Macrophage polarization, miRNA, M1 classical phenotype, M2 alternative phenotype, Inflammation, Transcription factor
Abbreviation
- AAA
abdominal aortic aneurysm
- ABCA1
ATP-binding cassette transporter A1
- ADAR1
adenosine deaminase acting on double-stranded RNA 1
- AIS
acute ischemic stroke
- Akt
Ak strain transforming
- ALI
acute lung injury
- AMD
age-related macular degeneration
- AMPK
adenosine monophosphate kinase
- ARDS
acute respiratory distress syndrome
- Arg1
arginase 1
- ATF3
activating transcription factor 3
- α-SMA
alpha-smooth muscle active;
- BaP
Benzo-a-pyrene; Bcl6, B-cell lymphoma-6 protein
- BMDM
bone marrow-derived macrophage
- BMP-2
bone morphogenetic protein 2
- BPD
bronchopulmonary dysplasia
- Btk
Bruton's tyrosine kinase
- Cav-1
caveolin-1
- CBH
chronic brain hypo perfusion
- CDK
cyclin-dependent kinase
- CNS
central nervous system
- CNV
choroidal neovascularization
- COPD
chronic obstructive pulmonary disease
- CRC
colorectal cancer cell
- CREB-C/EBPβ
cyclic-AMP-response element-binding protein-CCAAT/enhancer-binding protein-beta
- CRLM
colorectal cancer liver metastasis
- CSE
cigarette smoke extract
- CSF-1R
colony-stimulating factor 1 receptor
- CVB3
coxsackievirus B3
- CYM
Cypermethrin;
- DBP
Dibutyl phthalate
- DSS
dextran sodium sulfate
- EAE
experimental autoimmune encephalomyelitis
- EMT
epithelial-mesenchymal transition
- EOC
epithelial ovarian cancer
- EREG
epiregulin
- ERK
extracellular signal-regulated kinase
- Ern1
endoplasmic reticulum to nucleus signaling 1
- ETV6
E26 transformation-specific variant 6 gene
- FOXO1
forkhead box transcription factor 1
- GM-CSF
granulocyte monocyte-colony stimulating factor
- GMP
granulocyte-monocyte progenitor
- GPCR
G-protein coupled receptor
- GSK3B
glycogen synthase kinase 3 beta
- HIF-1α
hypoxia-induced facor-1 alpha
- HNSCC
head and neck squamous cell carcinoma
- HMGB1
high mobility group box 1
- HPV
human papilloma virus
- HSC
hematopoietic stem cell
- HUVEC
human umbilical vein epithelial cell
- I/R
ischemia/reperfusion
- IBD
inflammatory bowel disease
- ICAM-1
intercellular adhesion molecule-1
- ICH
intracerebral hemorrhagic
- IFN-γ,
interferon-gamma
- IL,
interleukin
- iNOS
induced nitric oxide synthase
- IRAK
IL-1 receptor-associated kinase 1
- IRF
interferon regulatory factor
- JAK
Janus kinase
- JMJD1C
Jumonji domain containing 1C
- KLF
Kruppel-like factor
- KOA SF
knee osteoarthritis synovial fluid
- LPS
lipopolysaccharide;
- LRP6
low-density lipoprotein receptor-related protein 6
- LTBP1
latent transforming growth factor β binding protein 1
- MATN2
matrilin-2
- MAPK
mitogen-activated protein kinase
- MCP-1
monocyte chemoattractant protein-1
- MDM2
murine double minute 2
- MEC
mammary epithelial cell
- MEK
mitogen activated protein kinase kinase
- MET-1
mammary epithelial tumor cell-1
- METTL3
methyltransferase-like 3
- MI
myocardial infarction
- miR
microRNA
- MKP1
mitogen-activated protein kinase phosphatase 1
- MMP
matrix metalloproteinase
- MRC1
mannose receptor C-type 1
- MS
multiple sclerosis
- MSC
mesenchymal stem cell
- MyD88
myeloid differentiation primary response 88
- NF-κB
nuclear factor-kappa B
- NGF
neuronal growth factor
- NLRP3
NOD-like receptor P3; NO, nitric oxide;
- NSCLC
non-small cell lung cancer
- OLR1
ox-LDL receptor 1
- OSCC
oral squamous cell carcinoma
- ox-LDL,
oxidized-low density lipoprotein
- PAI-2
plasminogen activator inhibitor 2
- PBMC
peripheral blood mononuclear cell
- PDAC
pancreatic ductal adenocarcinoma
- PDCD4
programmed cell death protein 4
- PI3K
phosphoinositide 3-kinase
- Pknox1
PBX/Knotted 1 Homebox 1
- PLCB3
1-phosphatidylinositol-4, 5-bisphosphate phosphodiesterase beta-3
- PM
particular matter; PPAR, peroxisome proliferator-activated receptor
- PRKCD
protein kinase C delta type
- PTGDS
prostaglandin D2 synthase
- PTGS
prostaglandin-endoperoxide synthase 2
- PTPRD
protein tyrosine phosphatase receptor type D
- PTPRO
protein tyrosine phosphatase receptor type O
- PTEN
phosphatase and TENsin homolog deleted on chromosome 10
- ROCK2
Rho associated coiled-coil containing protein kinase 2
- ROS
reactive oxygen species
- RSA
recurrent spontaneous abortion
- SCI
spinal cord injury
- SCIRI
spinal cord ischemia-reperfusion injury
- SFTSV
severe fever with thrombocytopenia syndrome virus
- SHIP1
Src homology-2 domain-containing inositol 5-phosphatase 1
- SIRPβ1
signal-regulatory protein beta1
- SIRT1
sirtuin1
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- STZ
streptozotocin
- T2D
type-2 diabetes
- TACE
TNF-α converting enzyme
- TAM
tumor-associated macrophage
- TBL1X
transducing (beta)-like 1X-linked
- TGF-β,
transforming growth factor-beta
- Th1
type 1 helper T lymphocyte
- TLR
toll-like receptor
- TMEM229B
protein transmembrane 229B
- TNBC
triple negative breast cancer cell
- TNFAIP3
tumor necrosis factor alpha-induced protein 3
- TNF-α,
tumor necrosis factor-alpha
- Trib1
tribbles homolog 1
- TRAF
TNF receptor-associated factor
- TRIF
TIR-domain-containing adapter-inducing interferon-beta
- TSC2
tuberous sclerosis complex 2
- UCB-MSC
umbilical cord blood-derived mesenchymal stem cell
- UTR
untranslated region
- UVRAG
UV radiation resistance-associated gene protein
- VEGF
vascular endothelial growth factor
- VSMC
vascular smooth muscle cell
1. Introduction Macrophage
1.1. Biogenesis, characteristics, and polarization
Monocytes originate from bone marrow-derived myeloid precursors that include a small proportion of peripheral blood leukocytes (4–10 %) in a healthy human. Due to their short halflife, these cells migrate into tissues and differentiate into long-lived macrophages [1,2]. For years, circulatory monocytes were believed to be the only source of differentiated macrophages. However further studies proved that most tissue-resident macrophages are monocyte-independent cells with self-renewal potential and originate from the yolk sac during embryonic development. A unique combination of embryonic and monocyte-derived cells forms the unique macrophage composition in each tissue [3,4]. Macrophages are involved in a wide variety of processes. Eliminating pathogens via phagocytosis, modulating adaptive immune responses through processing and presenting antigens, secreting different signaling proteins, and playing as scavengers to clear dead cells and cellular debris [[3], [4], [5]]. Macrophages are highly heterogeneous cell populations with various inflammatory and anti-inflammatory functions [6]. Their flexible phenotype results from a dynamic process known as macrophage polarization and is regulated in response to signals and microenvironmental stimulants. To clear the paradigm about opposite macrophage polarization phenotypes, these cells are classified into M1 and M2 polarization states [[7], [8], [9], [10]].
1.1.1. Classical-activated or M1 phenotype
The classically activated M1 macrophage is primarily induced by type 1 helper T lymphocyte (Th1)-secreted cytokines, including IFN-γ and tumor necrosis factor-alpha (TNF-α). Also, granulocyte monocyte-colony stimulating factor (GM-CSF), lipopolysaccharide (LPS), and other toll-like receptors (TLR) ligands are among the M1-inducers [8,11]. In addition, previous studies showed that oxidized-low density lipoprotein (Ox-LDL), high mobility group box 1 (HMGB1), and caveolin-1 (Cav-1) are involved in the macrophage phenotypic shift to the classical phenotype [[12], [13], [14]]. M1 macrophages, which highly express CD80, CD86, MHC-II, and TLR4 molecules, secrete various pro-inflammatory cytokines and chemokines, including interleukin (IL)-1β, IL-6, IL-12, IL-23, TNF-α, CCL2, CCL5, CXCL1-3, CXCL5, and CXCL8-10, and produce massive amounts of reactive oxygen species (ROS) to support Th1-oriented responses against invading pathogens [15,16]. LPS/TLR4 is considered the main pathway for M1 polarization that activates nuclear factor-kappa B (NF-κB) and interferon regulatory factor 3 (IRF3) and induces pro-inflammatory cytokines including IL-6 and TNF-α [2,17]. Further studies revealed that LPS/TLR4-dependent classical macrophage activation occurres due to signal transducer and activator of transcription 1 (STAT1) dimerization and activation through a myeloid differentiation primary response 88 (MyD88)-associated pattern [18]. Janus kinase (JAK)-STAT is considered an essential pathway for classical polarization, and IRF5 plays a crucial role in promoting IL-12, IL-23, and TNF-α secretion [19]. Bruton's tyrosine kinase (Btk) was also identified in LPS-induced M1 polarization and Btk absence, which is explicitly associated with M2 polarization [20]. The P2Y(2)R, a G-protein coupled receptor (GPCR) involved in nitric oxide (NO) production [21], the suppressor of cytokine signaling 3 (SOCS3) that activates the NF-κB/phosphoinositide 3-kinase (PI3K) pathway [22], and activating A that downregulates IL-10 expression [23] are among other molecules involved in classical polarization.
1.1.2. Alternative-activated or M2 phenotype
Macrophage activation via Th2-associated factors leads to M2 or alternative-activated polarization involved in injury healing, dead cell clearance, vascularization, and tumor promotion or invasion. Based on the specific inducers, M2 macrophages are categorized into M2a, M2b, M2c, and M2d subsets [24,25]. IL-4 induces the M2a subset, IL-13, fungal, and helminth infections and highly expresses CD206, the decoy IL-1 receptor, and the IL-1 receptor antagonist. IL-1 receptor ligands, immune complexes, TLR agonists, and LPS induce the M2b subset that produces various cytokines such as TNF-α, IL-1β, IL-6, and IL-10. The M2c subset is polarized by IL-10, transforming growth factor-beta (TGF-β), and glucocorticoids and exerts intense anti-inflammatory activities against apoptotic cells by releasing high amounts of IL-10 and TGF-β. Also, the M2d subset is induced by TLR agonists via adenosine receptors [26,27]. Collectively, the M2 macrophages are characterized by highly expressed CD163, scavenger, mannose, and galactose receptors, upregulated levels of CXCR1, CXCR2, and CCR2, elevated production of IL-10, vascular endothelial growth factor (VEGF), matrix metalloproteinases (MMPs), ornithine, and polyamines, and low levels of inflammatory factors such as IL-12, CD86, MHC-II, and induced nitric oxide synthase (iNOS) [15,16,28]. The IL-4 and IL-13 cytokines activate macrophages through STAT6 [29,30]. Also, other transcriptional factors, including IRF4, peroxisome proliferator-activated receptor gamma (PPARγ), and Kruppel-like factor 4 (KLF4), were suggested to promote alternative polarization [[31], [32], [33]].
-
2.
microRNA (miRNA)
In 1993, the miRNA was first described in Caenorhabditis elegans as non-coding, small, 19–25 nucleotide, endogenous RNA with an evolutionarily conserved sequence [5,34]. Following transcription, miRNAs are processed by Drosha and Dicer enzymes, and their abnormal expression is associated with various disorders [[35], [36], [37]]. The miRNAs control gene silencing in the post-transcription phase and primarily bind to the 3′- or 5′-untranslated region (UTR) of target transcripts to suppress translation and degrade mRNA and influence gene expression, cellular function and various pathophysiological processes, e.g., cell proliferation, metabolism, apoptosis, and organ development [[38], [39], [40]]. Based on bioinformatics predictions, approximately 33 % of protein-coding genes are regulated post-transcriptionally via miRNAs [34].
2. miRNA and macrophage polarization
Macrophages are dynamic cells with phenotypic transition abilities and a microenvironment milieu-based activation potential [41]. Multiple lines of evidence support associations between miRNAs deregulation and inadequate or overloaded inflammatory responses [42]. Monocyte-derived macrophages originate from hematopoietic stem cells (HSC), and miRNAs are critical regulators of HSC's renewal and fate [43]. As increased activation of PU.1 is in favor of granulocyte-monocyte progenitor (GMP) differentiation to monocyte linguae, this transcription factor induces the expression of multiple miRNAs, including miR-146a, miR-342, miR-338, and miR-15, which are involved in various steps of myeloid cell differentiation and control monocyte and macrophage maturation. For instance, miR-146 overexpression negatively regulates innate immune responses and is enough to induce stem cell maturation into monocyte during hematopoiesis [[44], [45], [46]].
Zhang et al. evaluated the miRNA expression profile during macrophage polarization in murine bone marrow-derived macrophages (BMDM). The results of microarray analysis showed that mir-109 expression was altered between M1 and M2 phenotypes. Also, the expression of miR-181a, miR-155-5p, miR-204-5p, and miR-451 was upregulated in classically activated macrophages compared to the alternative form, while a downregulation was reported in the miR-125-5p, miR-146a-3p, miR-143-3p, and miR-145-5p expressions [47]. Also, Graff et al. conducted a comprehensive study using TaqMan low-density array human miRNA assays to evaluate different miRNA expressions in various M1, M2a, M2b, and M2c subsets. They reported specifically expressed miR-125a-3p and miR-26a in the M1 and miR-222, miR-132, miR-29b, miR-27a, and miR-193b in the M2b subset [48].
In another study, Cobos Jimenez and colleagues analyzed the miRNA profile in peripheral blood mononuclear cell (PBMC)-derived polarized monocytes and reported upregulation or downregulation in the expression of 303 different miRNAs. In this case, miR-125a-5p, miR-125b-5p, miR-181a-5p, and miR-193b-3p were significantly upregulated in M1-polarized macrophages and dysregulated in M2 macrophages. In addition, upregulation in miR-146a-5p, miR-145-5p, miR-29b-3p, and miR-193a-5p and downregulation in miR-629-5p were observed only in classic phenotype. Also, elevated miR-502-3p and miR-500a-5p and reduced miR-181-5p expressions were observed in the M2a-polarized macrophage, while increased expressions in miR-22-3p, miR-21-5p, and miR-146b-5p and decreased levels in miR-200a-3p and miR-339-3p were reported in the M2c phenotype [49].
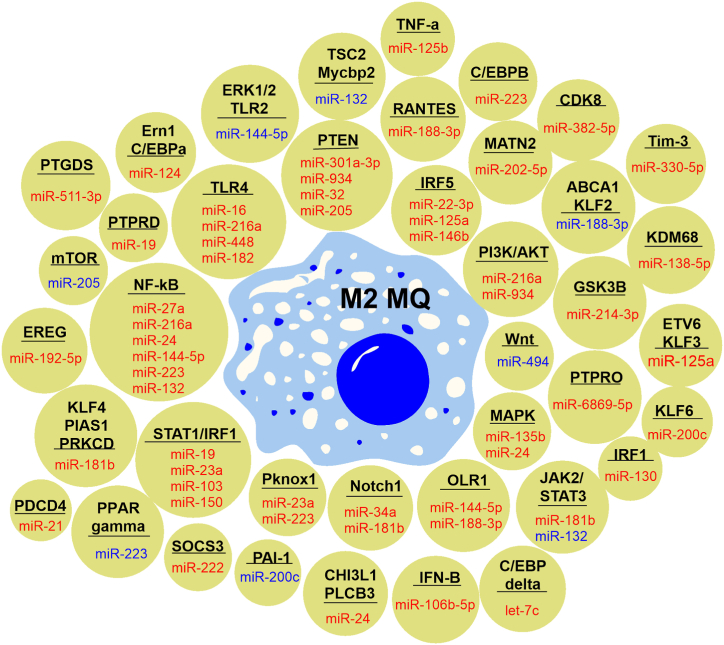
The miRNA-mediated macrophage polarization is a fully-conserved process controlled by specific transcription factors that promote different polarization patterns and maintain their balance [48,50,51]. Here, we categorized and summarized the prominent miRNAs involved in macrophage polarization in Table .1. Also, miRNA and identified targets were illustrated in Fig. 1 and Fig. 2 as regulators of M1 and M2 phenotypes, respectively.
Table 1.
Summary of main miRNAs involved in macrophage polarization.
| Type of miRNA | Target | Effects on M1/M2 polarization | Consequences of macrophage polarization | Ref. |
|---|---|---|---|---|
| miR-21 | PDCD4, PTEN/Akt/STAT6, and CSF-1R | M2 ↑ |
|
[[52], [53], [54], [55], [56], [57], [58], [59], [60]] |
| PI3K, NFκB | M1 ↑ | – | [61] | |
| miR-34a | KLF4 | M1 ↑ |
|
[[62], [63], [64]] |
| Notch1 and NLRP3 | M2 ↑ |
|
[[65], [66], [67]] | |
| miR-124 | STAT3, TACE, PU.1, and Ern1 | M2 ↑ |
|
[[68], [69], [70], [71]] |
| miR-125a-5p | A20 | M1 ↑ |
|
[48] |
| KLF13, IRF5, and ETV6 | M2 ↑ |
|
[[72], [73], [74], [75]] | |
| miR-125b | A20 and IRF4 | M1 ↑ |
|
[[76], [77], [78]] |
| miR-146a | Notch1, PPARγ, SIRT1, TRAF6, and STAT1 | M1 ↑ |
|
[[79], [80], [81]] |
| STAT1 | M2 ↑ |
|
[82] | |
| miR-146b | IRF5 and TLR4 | M2 ↑ |
|
[[83], [84], [85]] |
| miR-155 | SOCS1, SHIP1, Bcl6, C/EBPβ, and TGF-β/Smad | M1 ↑ |
|
[[86], [87], [88], [89], [90], [91], [92], [93]] |
| miR-9 | ERK1/2,AMPK, SIRT1, and PPARδ | M1 ↑ |
|
[[94], [95], [96], [97], [98]] |
| miR-181b | Notch1, KLF4, and PRKCD | M2 ↑ |
|
[99,100] |
| miR-200c | KLF6 | M2 ↑ |
|
[101,102] |
| GM-CSF | M1 ↑ |
|
[103,104] | |
| miR-223 | STAT3, Pknox, HIF-1α, and C/EBPβ | M2 ↑ |
|
[[105], [106], [107], [108], [109], [110]] |
| miR-511 | PTGDS, CCL2, LTBP1, ROCK2, TLR4, and C/EBPα | M2 ↑ | _ | [[111], [112], [113]] |
| Let-7c | C/EBPδ | M2 ↑ |
|
[114] |
| Le-7f | A20 | M1 ↑ | _ | [115] |
| miR-98 | Trib1 | M1 ↑ | Tumor invasion ↓ | [116] |
Abbreviation Akt; Ak strain transforming, ARDS; acute respiratory distress syndrome, C/EBPα; cyclic-AMP-response element-binding protein-CCAAT/enhancer-binding protein-alpha, CNS; central nervous system, COPD; chronic obstructive pulmonary disease, CSF-1R; colony-stimulating factor 1 receptor, CVB3; coxsackievirus B3, EAE; experimental autoimmune encephalomyelitis, EMT; epithelial-mesenchymal transition, Ern1; endoplasmic reticulum to nucleus signaling 1, ETV6; E26 transformation-specific variant 6, GM-CSF; granulocyte monocyte-colony stimulating factor, HIF-1α; hypoxia-induced facor-1 alpha, IBD; inflammatory bowel disease, KLF; Kruppel-like factor; LTBP1; latent transforming growth factor β binding protein 1, NLRP3; NOD-like receptor P3, NSCLS; non-small cell lung cancer, PDCD4; programmed cell death protein 4, Pknox; PBX/Knotted 1 Homebox 1, PPARγ; peroxisome proliferator-activated receptor, PRKCD; protein kinase C delta type, PTGDS; prostaglandin D2 synthase, PTEN; phosphatase and TENsin homolog deleted on chromosome 10, ROCK2; Rho associated coiled-coil containing protein kinase 2, SCI; spinal cord injury, SCIRI; spinal cord ischemia-reperfusion injury, SFTSV; severe fever with thrombocytopenia syndrome virus, SIRT1; sirtuin1, STAT1; signal transducer and activator of transcription 1, STZ; streptozotocin, TACE; TNF-α converting enzyme, TRAF6; TNF receptor-associated factor 6, Trib1; tribbles homolog 1.
Fig. 1.
Schematic representations of miRNAs and identified targets that promote M1 macrophage polarization. The miRNAs highlighted in red suppress their targets to promote M1 phenotype, while miRNAs in blue positively influence targets to induce the M1 subset. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Schematic representations of miRNAs and identified targets that promote M2 macrophage polarization. The miRNAs highlighted in red suppress their targets to promote M2 phenotype, while miRNAs in blue positively influence targets to induce the M2 subset. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.1. miR-21
Due to its involvement in pathophysiological conditions, including solid tumors, cardiac injuries, and inflammation, miR-21 is considered one of the most critical mammalian miRNAs [117]. Recent studies showed that miR-21 promotes M2 macrophage polarization, significantly inhibits apoptosis, and induces chemoresistance in ovarian cancer cells, while miR-21 inhibitors reverse the mentioned effects [52]. In addition, mesenchymal stem cells (MSC) and KEL endometrial cancer cells under hypoxic conditions secrete miR-21-enriched exosomes that induce M2 polarization and promote lung cancer [53,54]. The programmed cell death protein 4 (PDCD4) is a tumor suppressor that activates NF-κB and inhibits IL-10 expression as a pro-inflammatory protein. Recent studies reported that miR-21 negatively regulated PDCD4 and was upregulated in different cancers as an oncomir [55,56]. Sheedy and colleagues reported that miR-21 suppressed PDCD4 expression in a MyD88-or TIR-domain-containing adapter-inducing interferon-beta (TRIF)-dependent manner. Also, in pre-miR-21-transfected RAW264.7 cells, suppressed NFκB signaling and elevated IL-10 production were observed following LPS stimulation [57]. In addition, miR-21 was upregulated in IL-1β-treated MSCs, and miR-21-enriched exosomes derived from treated MSCs effectively promoted M2 polarization both in vivo and in vitro. PDCD4 was the target gene for miR-21, and significant amelioration in symptoms and an increased survival rate were observed in the murine model of sepsis following miR-21-oriented M2 polarization [118].
Another interesting interaction between miR-21 and the tumor microenvironment is the positive feedback between miR-21-induced M2 polarization and the epithelial-mesenchymal transition (EMT) process. The EMT transcription factor Snail activates miR-21 transcription, and miR-21-enriched exosomes secreted by HNSCC effectively inhibit the expression of M1 factors and promote M2 polarization, which can lead to angiogenesis and tumor progression [119]. In addition, Song et al. discovered another side of this feedback. They observed that exosomal miR-21-5p derived from EC109 and EC9706 tumor cells induce M2 macrophage transformation via the phosphatase and TENsin homolog deleted on chromosome 10 (PTEN)/Ak strain transforming (Akt)/STAT6 signaling pathway. Subsequently, TGF-β secretion by M2-polarized macrophages promotes esophageal cancer cell EMT through the TGF-β/Smad2 axis [59]. Along with these results, another study revealed that miR-21 integrates colony-stimulating factor 1 receptor (CSF-1R)/pTyr-721/PI3K axis signals, suppresses pro-inflammatory polarization, and induces the M2 phenotype. They observed that approximately 80 % of CSF-1R/miR-21 axis molecular targets are pro-inflammatory agents. Also, signal-regulatory protein beta1 (SIRPβ1) induces M2-associated responses as activators of the mitogen-activated protein kinase (MEK)/ERK1/2 pathway. Knockout of miR-21 reduced the expression of arginase 1 (Arg1), mannose receptor 1, IL-4Ra, and Fizz as alternative associated markers. Also, the administration of miR-21 inhibitors in mice increased the recruitment of inflammatory monocytes and the LPS responsiveness of peritoneal macrophages [60]. Furthermore, miR-21-mediated macrophage polarization was investigated in other inflammatory conditions, including chronic obstructive pulmonary disease (COPD), myocardial ischemia-reperfusion injury, and allogenic graft rejection [95,120,121]. Lu et al. proved that elevated miR-21 expression in cigarette smoke extract (CSE)-exposed RAW264.7 cells is correlated with M2 polarization and miR-21 inhibitors effectively reduce lung tissue degeneration in the murine model of COPD [95]. Also, Shen et al. proved that MSC-derived exosomal miR-21 polarized M1 macrophages to the M2 phenotype following IL-10 induction and IL-6 suppression and reduced inflammation in the murine model of myocardial ischemia-reperfusion injury [120]. In addition, Li et al. reported that adenosine deaminase acting on double-stranded RNA 1 (ADAR1) treatment in a murine model of allogeneic skin graft, efficiently suppresses mir-21 expression, downregulates Foxo1, induces IL-10 production that leads to M2 polarization, and impedes allogenic graft rejection [121].
However, a recent study demonstrated miR-21 involvement in pro-inflammatory M1 phenotype induction in macrophages. They reported that miR-21-transfected macrophages significantly upregulated macrophage inflammatory status and exosomes from these cells are rich in miR-21 and polarize naïve macrophages toward a pro-inflammatory phenotype partially in a PI3K- and NFκB-dependent manner [61].
The interaction between miR-21 and macrophage polarization seems to be an attractive therapeutic target for cancer and inflammatory disorders. Inhibiting miR-21 using antagomirs or pharmacologic agents could effectively induce antitumor immune responses and inflammatory M1 macrophages while administering strategies that upregulate miR-21 expression is crucial to suppress inflammation and induce M2 polarization to resolve inflammation-related injuries.
2.2. miR-34a
Both M1 polarization and M2 induction were observed to be mediated by miR-34a (Fig. 3). KLF4 was identified as the main target of miR-34a to promote M1 polarization. Overexpressed miR-34a in the adipose tissue aggravates obesity-induced systemic inflammation via suppressing KLF4, accumulating M1 pro-inflammatory macrophages [63]. In addition, adipocyte-derived exosomal miR-34a suppressed M2 polarization and enhanced metabolic dysregulation following uncontrolled obesity-associated inflammation [64]. The miR-34a suppression improved lung inflammation and histological symptoms in the murine acute respiratory distress syndrome (ARDS) model following KLF4 overexpression, which led to M2 phenotype polarization [62]. In addition, miR-34a overexpression in macrophages co-cultured with non-small cell lung cancer (NSCLC) cells leads to M2-to-M1 polarization following KLF4 suppression and decreases cell proliferation and clonogenic potential [122].
Fig. 3.
Schematic representations of potential targets and clinical outcomes of miR-34a involvement in macrophage polarization toward both pro-inflammatory and anti-inflammatory subsets.
The other side of miR-34a-macrophage polarization interactions promotes M2 macrophages. Recent studies showed that miR-34a blocks pro-inflammatory responses in LPS-stimulated macrophages by targeting Notch1, critical for producing LPS-mediated inflammatory cytokines, e.g., TNF-α and IL-6 [67]. In addition, adipocyte-derived exosomal miR-34a potentially targets NOD-like receptor P3 (NLRP3) and suppresses M1 polarization in the murine model of titanium-induced osteolysis [66]. Also, a recent study demonstrated that prolonged exposure to Benzo-a-pyrene (BaP) and Dibutyl phthalate (DBP) environmental pollutants in the rat model of hepatic injuries leads to aggravated hepatic inflammation as result of a deregulated M1/M2 balance. Further investigations revealed that downregulating miR-34a-5p, followed by upregulation in Notch signaling; induce M1 polarization and results in uncontrolled inflammatory responses [65].
Totally, it seems that miR-34a is an important regulator of macrophage polarization and controversial findings in promoting M1 or M2 phenotype may be due to variations in study models and conditions. Hence, further in-depth and mechanistic investigations are required to clarify the exact role of miR-34a in macrophage polarization and administer it in human clinical trials as a therapeutic strategy.
2.3. miR-124
The anti-inflammatory potential of miR-124 in polarizing macrophages toward the M2 phenotype was proved following miR-124 mimic transfection into RAW264.7 cells that suppressed LPS-induced IL-6 and TNF-α production and directly targeted STAT3 and TNF-α converting enzyme (TACE) [71]. In addition, miR-124 upregulation in both IL-4- and IL-13-treated macrophages was reported, while miR-124 knockdown suppressed M2 markers (Ym1 and CD206) and induced M1 hallmarks (TNF, iNOS, and CD86) [123]. Under normal physiological conditions, most tissue-resident macrophages appear to have an alternative-like phenotype. In the central nervous system (CNS), microglia, as CNS-resident macrophages, express alternative-associated genes, including Ym1, Fizz 1, IL-10, and IL-10 [124]. In brain microglia, elevated expression of miR-124 leads to direct targeting of C/EBPα and PU.1 in downstream that control M2 alternative polarization and is suggested to be a key regulator in the CNS microenvironment [70]. Further analysis in experimental autoimmune encephalomyelitis (EAE) as a murine model of multiple sclerosis (MS) revealed the therapeutic potential of miR-124 for reducing total numbers of macrophages, activating CD45high microglia, ameliorating inflammation and clinical symptoms, and also increasing CNS recovery [70]. In addition, administering the liposomal miR-124 in a murine model of ischemic stroke showed the therapeutic efficacy of miR-124 in a time-dependent manner. Application of a therapeutic regimen in the sub-acute phase and 48 h after stroke significantly reduced inflammatory microglia, while no significant effects were observed when miR-124 was administered ten days after stroke [68]. A similar therapeutic potential of miR-124-3p was observed in the spinal cord ischemia-reperfusion injury (SCIRI) murine model. In this case, BM-MSC-derived exosomal miR-124-3p targets endoplasmic reticulum to nucleus signaling 1 (Ern1) and induces M2 polarization, which results in amelioration of SCIRI-induced neuronal injuries [69].
In addition to the CNS, miR-124-polarized alternative macrophages are essential in regulating allergic immune responses. The miR-124 overexpression was observed in IL-4- and IL-13-exposed RAW264.7 and BMDM cells. Further investigations showed that the miR-124 inhibitor suppresses Ym1 and CD206 expression while upregulating CD86, NOS2, and TNF-α levels. In addition, increased numbers of CD14+ CD16+ intermediate monocytes with high levels of miR-124 were observed in allergic bronchial asthma patients [123]. The application of miR-124 mimics suppressed LPS-associated myeloperoxidase activity and relieved acute lung injuries in animal models. Functionally, the adhesion molecule-1 (ICAM-1) suppresses monocyte chemoattractant protein-1 (MCP-1) expression through miR-124 upregulation and induces M2 polarization [125].
Collectively, it seems miR-124-induced M2 polarization could potentially be administered for various inflammatory conditions by promoting alternative macrophages and suppressing the M1 inflammatory phenotype.
2.4. miR-125
The miR-125 family includes miR-125a and miR-125b, which are widely involved in regulating macrophage polarization. Recent studies showed that miR-125a-5p is upregulated in LPS-stimulated macrophages, and the mimic transfection increased the expression of classic phenotype-associated transcriptions via targeting A20 [48]. The ubiquitin-editing enzyme A20, also known as tumor necrosis factor alpha-induced protein 3 (TNFAIP3), is a negative regulator of the NFκB signaling pathway in different immune cells [126]. On the other hand, various studies have reported miR-125a-mediated M2 polarization. Banerjee et al. observed higher miR-125a expression in alternative macrophages compared to a classic phenotype that suppresses M1 activation and bactericidal activities while promoting alternative polarization and phagocytic potential for apoptotic cell ingestion through targeting KLF13 [72]. E26 transformation-specific variant six genes (ETV6) was another identified miR-125a-5p target for inducing M2 polarization and facilitating bone regeneration [74]. Also, administering BM-MSC-derived exosomal miR-125a in a murine model of spinal cord injury (SCI) showed neuroprotective effects by inducing M2 polarization and suppressing the M1 phenotype via targeting IRF5 [73]. In addition, Shi et al. administered Dioscin in the murine model of dextran sodium sulfate (DSS)-induced colitis. They reported suppressed M1 and facilitated M2 polarization followed by increased levels of miR-125a-5p that improved intestinal epithelial barrier function and ameliorated inflammatory bowel disease (IBD) symptoms [75].
Overexpressed miR-125b, which plays an essential role in improving antigen presentation, T cell activation, and antitumor responses, was observed in M1 macrophages [127]. The miR-125b activates pro-inflammatory macrophages by targeting IRF4 and TNFAIP3 [76,77]. In this case, recent studies showed that tumor cell-derived exosomal miR-125b reprogrammed macrophages to a classically activated phenotype with elevated levels of co-stimulatory factors and increased responses to the IFN-γ [78]. Also, the active form of vitamin D, 1, 25 (OH)2 D3, alleviates colitis through downregulating miR-125b, promoting M1-to-M2 macrophage polarization, and regulating macrophage subsets [128,129]. On the other hand, multiple studies have observed miR-125b suppression following LPS stimulation [130,131]. Also, it seems that miR-125b targets 3′UTR of TNF-α transcript, reduces stability, and negatively regulates inflammatory responses [[132], [133], [134]].
2.5. miR-146
The miR-146 family consists of two evolutionally conserved members. The miR-146a is highly expressed in M2 macrophages and is crucial in modulating macrophage polarization by targeting Notch1 and PPARγ and promoting the M1 subset [80]. Also, a recent study showed that particular matter 2.5 (PM2.5) upregulates miR-146a-5p to induce M1 macrophage polarization by targeting SIRT1 [81]. Primarily, miR-146a was introduced as a negative feedback regulator of macrophage activation by targeting TLR signaling downstream molecules such as IL-1 receptor-associated kinase 1 (IRAK1), TNF receptor-associated factor 6 (TRAF6), and NFκB [[135], [136], [137]]. Chen et al. reported that neonatal murine cardiomyocyte-derived exosomal miR-146a-5p significantly increased VEGFA expression and promoted M2-to-M1 macrophage polarization via targeting TRAF6. Interestingly, unlike the miRNA mimic, exosomal miR-146a-5p reduced iNOS and TNF-α. Hence, it seems that cardiomyocyte-derived exosomal miRNA promotes M1 polarization and induces inflammatory responses by targeting TRAF6 [79]. In contrast to above-mentioned results, different results were observed in other studies. Human umbilical cord blood (UCB)-derived MSC administration leads to significant upregulation in miR-146a-5p that shifts the M1-to-M2 macrophage phenotype by suppressing the STAT1/TRAF6 signaling pathway and reviving renal function in the murine model of the streptozotocin (STZ)-induced model of nephropathy diabetes [82].
The miR-146b is on the opposite side; it is highly expressed in M1 macrophages and regulates macrophage polarization by targeting IRF5 and suppressing the M1 phenotype [84]. The miR-146b is an IL-10-dependent miRNA in human and murine systems that targets multiple factors, including TLR4, MyD88, IRAK-1, and TRAF6, promoting anti-inflammatory status. Multiple lines of evidence emphasize that miR-146b overexpression in THP-1 cells dramatically reduces the LPS-dependent secretion of pro-inflammatory cytokines and chemokines, including TNF-α, IL-6, IL-8, CCL2, CCL3, CCL7, and CXCL10 [83]. In addition, Zhang et al. demonstrate that miR-146b ameliorates inflammation by suppressing IRF5, and miRNA variants are related to endometriosis and associated pains [85]. Similar results were observed in severe fever with thrombocytopenia syndrome virus (SFTSV) infection where increased expression of miR0146b targets STAT1 and promotes M2 macrophage polarization to facilitate virus shedding and expansion [138].
2.6. miR-155
The miR-155 is a multifunctional miRNA that plays multiple roles in inflammation and immune responses [139]. Various stimulants including IFN-β, TNF-α, IL-1, TLR2, TLR3, TLR4, and TLR9 signals, and specific antigens, upregulating miR-155 expression [132,140]. The crucial role of miR-155 in macrophage polarization was proved following the effects of miRNA knockdown on the M1-to-M2 transition [141]. Yi-Hong et al. showed that Toxoplasma gondii infection induces M1 macrophage polarization through miR-155 upregulation [86]. In addition, Zhang et al. reported that obese mice adipocyte-derived exosomal miR-155 promotes M1 macrophage and regulates insulin signaling and glucose uptake by adipocytes [90]. In this regard, pharmacologic regulation of miR-155 seems to be an interesting approach. For example, using resveratrol suppressed miR-155 expression and promoted M2 microglia polarization to ameliorate neuro-inflammation induced following cerebral ischemia [142]. In addition, the critical role of Cypermethrin (CYM), a type II pyrethroid, in facilitating tumor metastasis is exerted by suppressing miR-155 expression and M2 macrophage promotion [143].
Various studies investigated potential targets for miR-155 as a pro-inflammatory miRNA that promotes M1 macrophage polarization. They identified multiple vital genes such as SOCS1, Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1), and B-cell lymphoma-6 protein (Bcl6), which are negative regulators of inflammatory responses [[87], [88], [89],91,92]. Xu et al. revealed the potential role of the SOCS1/miR-155 axis in PI3K/Akt1-mediated classic macrophage polarization in mouse models of Staphylococcus aureus-induced respiratory infection [144]. In addition, another study reported significantly increased expression of miR-155 in GM-CSF/IFN-γ/LPS-polarized primary human BMDMs and microglia, while miR-155 downregulates SOCS1 and influences the expression of pro-inflammatory cytokines and co-stimulatory surface markers [145]. Shi et al. observed miR-155-5p overexpression following the co-culture of PBMC-derived macrophages with knee osteoarthritis synovial fluid (KOA SF) that induces M1 macrophage polarization through targeting SOCS1 and suppresses macrophage apoptosis via targeting caspase 3 [146]. Hu et al. implicated miR-155 inhibition in reducing M1-induced sympathetic neuronal remodeling and ventricular arrhythmia. Mechanistically, miR-155 suppression reduces M1 polarization and SOCS1/NFκB-dependent inflammatory responses that result in neuronal growth factor (NGF) upregulation [147]. These results highlighted the importance of miR-155-mediated macrophage polarization through targeting SOCS1 in immune responses against bacterial pathogens and CNS adaptive immunity. O'Connell and colleagues revealed that SHIP1, a negative regulator of TLR pathways, was targeted and suppressed by miR-155 in LPS-stimulated wild-type primary macrophages [148]. In addition, in pro-inflammatory macrophages, miR-155 directly targeted Bcl6 as an essential negative regulator of the NFκB pathway [88]. Recent studies have implied that the role of miR-155 in macrophage polarization is not limited to M1 promotion but influences M2 markers and suppresses alternative hallmarks [149]. The C/EBPβ, considered an alternative feature that regulates Arg-1 expression, is suppressed by miR-155 through the interacting 3′UTR region [150]. Arranz and colleagues observed that Akt protein kinases primarily influence the polarization of mouse peritoneal macrophages. In this case, Akt1 deletion induces the M1 phenotype, and Akt2 deletion promotes the alternative phenotype, and all these processes are mediated by the miR-155-C/EBPβ interaction [130,151]. Also, Zhang et al. showed that miR-155 inhibited C/EBPβ activation and subsequent M2 macrophage polarization, leading to choroidal neovascularization (CNV) suppression [93]. Another identified target of miR-155 was the TGF-β/Smad signaling pathway. Smad2 targets IL-13Rα1 and suppresses STAT6 activation in human primary macrophages [51,152].
Although miR-155 was introduced as a pro-inflammatory macrophage inducer, multiple lines of evidence implied that miR-155 negatively regulates inflammatory processes via targeting crucial elements involved in pro-inflammatory signal transduction. Tili et al. reported that by inducing TNF-α production, miR-155 directly targets transcripts of various anti-inflammatory and pro-apoptotic proteins involved in the LPS signaling pathway, such as Fas-associated death domain protein, IKKε, and receptor-interacting serine-threonine kinase 1. Hence, it is suggested that miR-155 acts as both a negative and positive regulator of LPS signaling [132]. In this line, Tange et al. identified MyD88, which is involved in the negative regulation of Helicobacter pylori-induced inflammation, as a target gene for miR-155 [153].
It seems that miR-155 exerts pleiotropic roles under different physiological and pathological conditions and promotes macrophage polarization toward both pro-inflammatory and anti-inflammatory phenotypes under different conditions (Fig. 4a). Hence, targeting and manipulating miR-155 could be a therapeutic approach to modulating immune hemostasis (Fig. 4b).
Fig. 4.
(A) Schematic representations of miR-155 potential targets to induce M1 or M2 macrophage polarization under different circumstances; (B) Targeting miR-155 by pharmacological agents to interfere macrophage polarization as therapeutic aproaches
2.7. miR-9
It was reported that miR-9 overexpression, followed by downregulation of the extracellular signal-regulated kinase 1/2 (ERK1/2) phosphatase Dusp6 by CCL2, could increase macrophage-mediated inflammatory responses [94]. Also, miR-9-5p targets NF-κB, adenosine monophosphate kinase (AMPK), and sirtuin 1 (SIRT1) to regulate macrophage polarization in both in vitro and in vivo models. Activated NF-κB and AMPK and suppressed SIRT1 expression led to miR-9-mediated M1 induction and M2 suppression that promoted osteoarthritis progression in the mouse model [154]. In this case, miR-9-associated strategies such as miR-9 antagomir could be considered a potential therapeutic method for osteoarthritis. The PPARδ, a well-known lipid and glucose hemostasis regulator, is another identified target for miR-9 to promote the M1 phenotype. The LPS-exposed human primary monocytes showed elevated levels of miR-9 and suppressed PPARδ expression. This situation was reversed following anti-miR-9 transfection [96]. In this case, Tong et al. reported that miR-9-enriched exosomes derived from human papillomavirus (HPV)-infected head and neck squamous cell carcinoma (HNSCC) downregulate PPARδ expression and induce M1 polarization, which finally increases tumor radiosensitivity [97].
2.8. miR-181
The potential role of miR-181b in promoting M2 macrophage polarization through various targets was proven in different studies. Although Notch1 was introduced as the direct target of miR-181b to induce the M2 phenotype and reduce atherosclerotic plaque vulnerability [99], another mechanism was identified for miR-181b to promote M2 and suppress M1 polarization in atherosclerosis. The miR-181b activates KLF4 and PIAS1 SUMOylation in macrophages, leading to M1-to-M2 polarization [155]. In addition, exosomal miRNA in different in vitro and in vivo models implied M2 promotion by miR-181b. It was observed that exosomal miR-181b significantly induces the M2 phenotype, inhibits inflammation, and increases osteointegration through suppressing protein kinase C delta type (PRKCD) and activating p-Akt. Also, they showed that miR-181b-mediated M2 polarization indirectly promotes osteogenic migration and differentiation via secreting VEGF and bone morphogenetic protein 2 (BMP-2) [100]. In another study, Ma et al. showed miR-181b in exosomes isolated from the serum of NSCLC patients and NSCLC cells. Co-culture of macrophages with exosomal miR-181b promotes M2 polarization through the JAK2/STAT3 signaling pathway [156].
2.9. miR-200c
The miR-200c belongs to the miR-200 family, and this miR's aberrant expression is associated with tumor progression, metastasis, and drug resistance in various types of tumors, including lung, ovarian, and colorectal cancer [103]. Following miR-200c restoration in triple-negative breast cancer cells (TNBC), the levels of IL-10 and plasminogen activator inhibitor 2 (PAI-2) were increased, which promoted tumor-associated macrophages (TAM) to an M2-like phenotype that led to the malignant progression of the tumor [101]. Also, Xiong et al. reported that miR-200c was overexpressed in exosomes isolated from the serum of ovarian cancer patients. In this case, miR-200c induces M2 macrophage polarization by targeting KLF6 and promotes tumor cell proliferation and invasion [102].
On the other hand, the antitumor potential of miR-200c via suppressing the M1-to-M2 shift was observed in different studies [157]. William et al. showed that miR-200c significantly suppressed the proliferation of mouse mammary epithelial tumor cell-1 (MET-1) in vivo. In this case, overexpressed miR-200c enhanced GM-CSF expression and induced antitumor M1 macrophage polarization, regulating EMT toward epithelial signatures and prognosis for better survival in breast cancer patients [104]. In addition, Raue et al. reported that miR-200c overexpression reduced the expression of migration-associated miRNAs and inhibited the tumor infiltration potential of macrophages [103].
The miR-223 expression is associated with the M2 phenotype, and miRNA downregulation promotes LPS-induced IL-1β and IL-6 release [105]. The miR-223 overexpression in RAW264.7 macrophages suppresses LPS-induced IL-6 and IL-1β secretion through targeting STAT3 [106]. PPARγ is a crucial regulator of macrophage activation that induces miR-223 expression following IL-4 and IL-13 stimulation of murine macrophages. Consistent with these results, miR-223 ablation in the murine model inactivated PPARγ-regulated anti-inflammatory responses in macrophages [158]. In addition to alternative phenotype promotion, miR-223 suppresses pro-inflammatory polarization by suppressing the NFκB/JNK axis by inhibiting PBX/Knotted 1 Homeobox 1 (Pknox1) [109]. The miR-223 suppresses M1 macrophage polarization by targeting Pknox1 and protects against inflammation and coxsackievirus B3 (CVB3)-induced injuries [110]. The miR-223 is widely expressed in bone marrow and adipose-isolated macrophages and is crucial in regulating adipose tissue inflammation. Dramatic miR-223 overexpression was observed in IL-4-treated BMDMs, while LPS stimulation poorly decreased miRNA expression. In addition, miR-223−/− macrophages showed an increased response to LPS, and PPARγ and Arg-1 expression was significantly reduced. MiR-223 deficiency promotes inflammatory responses and downregulates insulin signaling in the adipose tissue of fat-diet mice. It leads to deregulated adipokine expression [109]. The hypoxia-induced factor-1 alpha (HIF-1α) and C/EBPβ are other identified targets for miR-223. Dang et al. reported that miR-223 interferes with glycolysis pathways by targeting HIF-1α and induces anti-inflammatory responses due to suppressed M1 macrophage polarization [107]. Also, it was shown that miR-223-mediated C/EBPβ suppression inhibits inflammatory macrophage differentiation from human circulatory monocytes and THP-1 cells [108].
2.10. miR-511
In humans and mice, miR-511-3p expressed by the fifth intron of the mannose receptor C-type 1 (MRC1) gene encoding CD206 and transcriptionally co-regulated by MRC1 is involved in macrophage activation [159,160]. Recent studies have implied that miR-511-3p controls macrophage-mediated antimicrobial responses and increases intestinal inflammation [161]. Also, miR-511-3p overexpression using miRNA mimics inhibits M1 macrophage polarization and promotes the M2 phenotype through interacting prostaglandin D2 synthase (PTGDS) [113]. Although, CCL2, latent transforming growth factor β binding protein 1 (LTBP1), and Rho-associated coiled-coil containing protein kinase 2 (ROCK2) are direct targets, TLR4 and C/EBPα are indirect targets of miR-511-3p [112]. Squadrito et al. reported robust and bioactive expression of miR-511-3p in alternative macrophages. In addition, miRNA overexpression widely and precisely tunes down the expression of alternative-associated genes [159]. The miR-511-3p directly and selectively targets ROCK2 serine-threonine kinase [111] and phosphorylates IRF4 as an M2 macrophage inducer [162,163]. Unexpectedly, enforced miR-511-3p activity in TAMs ameliorates pro-tumor genetic programs in MRC1+ TAMs and inhibits blood vessel formation and tumor progression [159]. These results suggest that, although promoting alternative macrophages, miR-511-3p could reprogram the pro-tumor phenotype to the anti-tumor subset. Also, Karo-Atar and colleagues demonstrated that miR-511 expression was increased following IL-4 and IL-13 stimulation in both in vitro and in vivo mouse models of respiratory allergic disease. In addition, global transcriptome analysis following miR-511 overexpression suggested that this miRNA modulates various activities of alternative macrophages, such as cellular proliferation, metabolism, and immune responses [164].
2.11. Let-7 family
The Let-7 family was among the first identified tumor-suppressive miRNAs, including 12 highly-conserved genes that encode nine different miRNAs, including Let-7a-i and miR-98 [165]. Banerjee et al. observed higher levels of Let-7c in alternative BMDMs and alveolar macrophages than in classic phenotypes. In addition, Let-7c mimics transfection into BMDMs, significantly reduces classic hallmarks, and decreases bactericidal activities while promoting alternative polarization and potential apoptotic cell engulfment. Also, Let-7c siRNA induces M1 polarization in BMDMs. The mechanistic analysis identified C/EBPδ as the direct target of Let-7c [114]. Further investigations highlighted the critical role of Let-7c in regulating macrophage polarization. Zhang et al. showed that epigenetic loss of Let-7c upregulates EZH2-induced PAK1 expression and promotes pro-inflammatory polarization [166]. Another study showed that Let-7e upregulates in LPS-stimulated macrophages and overexpresses Let-7e in Akt1−/− macrophages, which influences sensitivity and tolerance to LPS through targeting TLR4 [130]. Kumar et al. observed that Let-7f downregulation suppressed inflammatory responses by directly targeting A20 in macrophages involved in Mycobacterium tuberculosis infection, while overexpression of Let-7f significantly increased TNF-α and IL-1β production by inflammatory polarized macrophages [115]. The miR-98 downregulation following LPS treatment intensifies TLR4-triggered IL-10 production, while miR-98 overexpression reverses these effects. Hence, it seems that miR 98 controls pro-inflammatory immune responses [116]. Li et al. reported that miR-98 induces M2-to-M1 polarization and inhibits tumor invasion and TAM-induced EMT in hepatocellular carcinoma [116]. Also, Peng et al. reported that miR-98-5p knockout increases tribbles homolog 1 (Trib1) expression and induces M2 polarization, which relieves symptoms in the murine model of IBD [167].
2.12. Other miRNAs
Multiple lines of evidence indicated the role of other miRNAs in regulating macrophage polarization through limited studies that are summarized in Table .2.
Table 2.
Summary of other miRNAs involved in macrophage polarization.
| Type of miRNA | Target | Effects on M1/M2 polarization | Consequences of macrophage polarization | Ref. |
|---|---|---|---|---|
| miR-10a-5p | pro-inflammatory genes | M2 ↑ |
|
[168] |
| miR-16 | IKKα/NFκB and PD-L1 | M1 ↑ |
|
[169,170] |
| miR-16-5p | TLR4 | M2 ↑ |
|
[171] |
| miR-19 | RORα | M1 ↑ |
|
[172] |
| miR-19a-3p | STAT1/IRF1 | M1 ↓ |
|
[165] |
| miR-19b-3p | PTPRD | M2 ↑ |
|
[173] |
| miR-22-3p | IRF5 | M2 ↑ |
|
[174] |
| miR-23 | IRF1 and Pknox1 | M1 ↓ |
|
[175] |
| miR-23a | A2 | M1 ↑ |
|
[176] |
| miR-24 | PLCB, NFκB, CHI3L1, and MAPK | M2 ↑ |
|
[177,178] |
| miR-26 | KLF4 CREB-C/EBPβ MKP1 |
M1 ↑ |
|
[179,180] |
| miR-27a | TLR2/4 and NFκB | M2 ↑ |
|
[181,182] [58] |
| PPARγ | M1 ↑ |
|
[58] | |
| miR-29a-3p | SOCS1/STAT6 | M2 ↑ |
|
[183] |
| miR-30b-5p | UBE2D2/KAT2B | M1 ↑ | _ | [184] [185] |
| mir-30d-5p | NF-κB, COSC1, SIRT1, and NLRP3 | M1 ↑ |
|
|
| miR-32 | PTEN | M2 ↑ |
|
[186] |
| miR-101 | ABCA1 MKP1/p38/JNK |
M1 ↑ | _ | [166,187] |
| miR-103 | STAT1/IRF1 | M2 ↑ |
|
[188] |
| miR-106b-5p | IRF1/IFN-β | M2 ↑ |
|
[189] |
| miR-127 | BCL-6 and Dusp1 phosphatase | M1 ↑ | _ | [190] |
| miR-130 | PPARγ | M1 ↑ |
|
[191,192] |
| miR-132 | STAT3 and NFκB | M1 ↓ |
|
[193,194] |
| Mycbp2 E3 ubiquitin ligase | M2 ↑- |
|||
| miR-135 | MAPK6 | M2 ↑ |
|
[98] |
| miR-138-5p | KDM68 | M2 ↑ |
|
[195] |
| miR-144-5p | TLR2 and OLR1 | M2 ↑ |
|
[196] |
| miR-145-5p | IL-16 | M2 ↑ |
|
[197,198] |
| miR-148-3p | PTEN/Akt SIRPα |
M1 ↑ |
|
[[199], [200], [201]] |
| miR-150 | STAT1 | M2 ↑ |
|
[202] |
| miR-182-5p | TLR4 FOXO1 |
M2 ↑ |
|
[203,204] |
| miR-188-3p | KLF2 ABCA1 RANTES OLR1 |
M1 ↓ |
|
[205] |
| miR-192-5p | EREG | M2 ↑ |
|
[206] |
| miR-195 | CX3CL1 and CX3CR1 | M2 ↑ |
|
[207] |
| miR-202-5p | MATN2 | M2 ↑ |
|
[208] |
| miR-205 | PI3K/Akt/mTOR | M2 ↑ |
|
[209] |
| miR-214-3p | GSK3B | M2 ↑ |
|
[210] |
| miR-216a | Smad3/NFκB | M1 ↑ |
|
[211,212] |
| TLR4/NFκB/PI3K/Akt | M2 ↑ |
|
||
| miR-217 | JAK3/STAT3 | M2 ↓ |
|
[213] |
| miR-221 | SOCS1, STAT3, and STAT1 | M1 ↑ |
|
[214] |
| miR-222-3p | STAT3/SOCS3 | M2 ↑ |
|
[215] |
| miR-301a-3p | PTEN/PI3K | M2 ↑ |
|
[[216], [217], [218]] |
| miR-302a | METTL3/SOCS2 | M1 ↑ |
|
[219] |
| miR-330-5p | Tim-3 | M2 ↑ |
|
[220] |
| miR-375 | KLF4 | M1 ↑ |
|
[221,222] |
| miR-382-5p | CDK8 and STAT1 | M2 ↑ |
|
[223] |
| miR-448 | TLR4 | M1 ↓ |
|
[224] |
| miR-471-3p | SIRT1 | M1 ↑ | _ | [225] |
| miR-494 | Nrdp1 | M1 ↑ |
|
[226,227] |
| miR-494-3p | TBL1X and LRP6 |
|
[228] | |
| miR-495 | FTO | M1 ↑ |
|
[229] |
| miR-498 | MDM2/ATF3 | M1 ↑ |
|
[230] |
| miR-505-5p | TMEM229B | M2 ↑ |
|
[231] |
| miR-506 | STAT3 | M1 ↑ |
|
[232] |
| miR-520a-3p | UVRAG | M1 ↑ |
|
[233] |
| miR-657 | FAM46C | M1 ↑ |
|
[234] |
| miR-720 | GATA3 | M1 ↑ | _ | [235] |
| miR-744-5p | TGF-β1 and MAPK | M2 ↓ |
|
[236] |
| miR-934 | PTEN/PI3K/Akt | M2 ↑ |
|
[237] |
| miR-6869-5p | PTPRO | M2 ↑ |
|
[238] |
Abbreviation AAA, abdominal aortic aneurysm; ABCA1, ATP-binding cassette transporter A1; Akt, Ak strain transforming; ALI, acute lung injury; AMD, age-related macular degeneration; ATF3, activating transcription factor 3; BPD, bronchopulmonary dysplasia; CBH, chronic brain hypo perfusion; CDK, cyclin-dependent kinase; CREB-C/EBPβ, cyclic-AMP-response element-binding protein-CCAAT/enhancer-binding protein-beta; CRLM, colorectal cancer liver metastasis; EMT, epithelial-mesenchymal transition; EREG, epiregulin; FTO, Fat mass and obesity associated; FOXO1, forkhead box transcription factor 1; GSK3B, glycogen synthase kinase 3 beta; HUVEC, human umbilical vein epithelial cell; I/R, ischemia/reperfusion; ICH, intracerebral hemorrhagic;IFN, interferon; IL, interleukin; IRF, interferon regulatory factor; JAK, Janus kinase; KLF, Kruppel-like factor; LPS, lipopolysaccharide; LRP6, low-density lipoprotein receptor-related protein 6;MATN2, matrilin-2; MAPK, mitogen-activated protein kinase; MDM2, murine double minute 2; METTL3, methyltransferase-like 3; MI, myocardial infarction; miR, microRNA; MKP1, mitogen-activated protein kinase phosphatase 1; NF-κB, nuclear factor-kappa B; NLRP3, NOD-like receptor P3; OLR1, ox-LDL receptor 1; PDAC, pancreatic ductal adenocarcinoma; PI3K, phosphoinositide 3-kinase; Pknox1, PBX/Knotted 1 Homebox 1; PLCB3, 1-phosphatidylinositol-4, 5-bisphosphate phosphodiesterase beta-3; PPAR, peroxisome proliferator-activated receptor; PTPRD, protein tyrosine phosphatase receptor type D; PTPRO, protein tyrosine phosphatase receptor type O; PTEN, phosphatase and TENsin homolog deleted on chromosome 10; RANTES, Regulated on Activation, Normal T Expressed and Secreted; RSA, recurrent spontaneous abortion; SCI, spinal cord injury; SIRPα, signal-regulatory protein alpha; SIRT1, sirtuin1; SOCS, suppressor of cytokine signaling; STAT, signal transducer and activator of transcription; T2D, type-2 diabetes; TAM, tumor-associated macrophage; TBL1X, transducing (beta)-like 1X-linked; TGF-β, transforming growth factor-beta; TLR, toll-like receptor; TMEM229B, protein transmembrane 229B; UVRAG, UV radiation resistance-associated gene protein; VSMC, vascular smooth muscle cell.
3. Concluding remarks and future prospective
The miRNAs epigenetically regulate hundreds of genes involved in various physiological and pathological processes. The role of miRNAs in macrophage polarization has attracted much attention. Recent studies proved that over 60 miRNAs influence multiple adaptor proteins and transcription factors to regulate macrophage polarization. Hence, alterations in miRNA expression could affect M1 and M2 phenotype switching. Applying specific miRNA mimics or antagomirs could control the immune response and inflammation by regulating macrophage polarization under different physiological and pathological conditions. For example, antagomirs inhibiting pro-inflammatory miRNA promote M2 macrophage polarization, limiting inflammation and related injuries. On the other hand, M1-associated miRNA overexpression could effectively enhance antitumor immune responses and limit tumor proliferation, invasion, and metastasis. In this case, various studies implicated miRNAs in human clinical trials. A liposomal miR-34a mimic called MRX34 was administered on advanced solid tumor patients in phase I clinical trial [239]; an anti-miR-17 oligonucleotide called RGLS4326 was applied for polycystic kidney disease [240]; an anti-miR-12 called Miravirsen was used for hepatitis C virus (HCV) infection [241]; and for cardiovascular disease and wound healing, MRG-110, an anti-miR92a, was administered [242]. Unlike single-gene therapy, miRNAs are considered more effective and suitable therapeutic approaches. In this case, each miRNA could target multiple genes involved in macrophage polarization. Also, each gene could be targeted by multiple miRNAs. Hence, implicating miRNA mimics or targeting miRNAs using antagomirs holds promise for therapeutic approaches through regulating macrophage polarization. The other importance of miRNA-macrophage interactions is their biomarker potential. For example, monitoring the macrophage polarization status in in vivo disease conditions or even in vitro models could be served by miRNA potential. In addition, the development of technologies associated with miRNA modification and delivery systems is potential approach that could provide insights into the dynamic changes in macrophage polarization through miRNA regulation in deregulated conditions and also helps translate the primary findings into clinical applications.
Finally, identifying the exact mechanism of miRNA-macrophage polarization interaction seems necessary. Much more research should be conducted to overcome limitations in miRNA-based therapeutic approaches.
Funding
‘Not applicable.’
Data availability statement
No data was used for the research described in the article.
CRediT authorship contribution statement
Shaho Khayati: Conceptualization, Writing – original draft. Sajad Dehnavi: Conceptualization, Writing – original draft, Writing – review & editing. Mahvash Sadeghi: Writing – original draft, Writing – review & editing. Jalil Tavakol Afshari: Writing – review & editing. Seyed-Alireza Esmaeili: Writing – original draft. Mojgan Mohammadi: Conceptualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Yang J., et al. Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark. Res. 2014;2(1):1–9. doi: 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sica A., et al. Macrophage polarization in pathology. Cell. Mol. Life Sci. 2015;72(21):4111–4126. doi: 10.1007/s00018-015-1995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapouri‐Moghaddam A., et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018;233(9):6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 4.Epelman S., Lavine K.J., Randolph G.J. Origin and functions of tissue macrophages. Immunity. 2014;41(1):21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Essandoh K., et al. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46(2):122. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H., et al. Intravenous tolerance modulates macrophage classical activation and antigen presentation in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2009;208(1–2):54–60. doi: 10.1016/j.jneuroim.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X.q., et al. Emerging role of micro RNA s in regulating macrophage activation and polarization in immune response and inflammation. Immunology. 2016;148(3):237–248. doi: 10.1111/imm.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray P.J. Macrophage polarization. Annu. Rev. Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 9.Moradi‐Chaleshtori M., et al. Tumor‐derived exosomal microRNAs and proteins as modulators of macrophage function. J. Cell. Physiol. 2019;234(6):7970–7982. doi: 10.1002/jcp.27552. [DOI] [PubMed] [Google Scholar]
- 10.Li C., et al. Macrophage polarization and meta-inflammation. Transl. Res. 2018;191:29–44. doi: 10.1016/j.trsl.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sierra-Filardi E., et al. Heme Oxygenase-1 expression in M-CSF-polarized M2 macrophages contributes to LPS-induced IL-10 release. Immunobiology. 2010;215(9–10):788–795. doi: 10.1016/j.imbio.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Van Tits L., et al. Oxidized LDL enhances pro-inflammatory responses of alternatively activated M2 macrophages: a crucial role for Krüppel-like factor 2. Atherosclerosis. 2011;214(2):345–349. doi: 10.1016/j.atherosclerosis.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 13.Tian S., et al. HMGB1 exacerbates renal tubulointerstitial fibrosis through facilitating M1 macrophage phenotype at the early stage of obstructive injury. Am. J. Physiol. Ren. Physiol. 2015;308(1):F69–F75. doi: 10.1152/ajprenal.00484.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivshankar P., et al. Caveolin-1 deletion exacerbates cardiac interstitial fibrosis by promoting M2 macrophage activation in mice after myocardial infarction. J. Mol. Cell. Cardiol. 2014;76:84–93. doi: 10.1016/j.yjmcc.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon S., Martinez F.O. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Murray P.J., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills C.D., Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. J. Innate Immun. 2014;6(6):716–726. doi: 10.1159/000364945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toshchakov V., et al. TLR4, but not TLR2, mediates IFN-β–induced STAT1α/β-dependent gene expression in macrophages. Nat. Immunol. 2002;3(4):392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 19.Krausgruber T., et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 2011;12(3):231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 20.Ní Gabhann J., et al. Btk regulates macrophage polarization in response to lipopolysaccharide. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eun S.Y., et al. LPS potentiates nucleotide-induced inflammatory gene expression in macrophages via the upregulation of P2Y2 receptor. Int. Immunopharm. 2014;18(2):270–276. doi: 10.1016/j.intimp.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Arnold C.E., et al. A critical role for suppressor of cytokine signalling 3 in promoting M 1 macrophage activation and function in vitro and in vivo. Immunology. 2014;141(1):96–110. doi: 10.1111/imm.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sierra-Filardi E., et al. Activin A skews macrophage polarization by promoting a proinflammatory phenotype and inhibiting the acquisition of anti-inflammatory macrophage markers. Blood, The Journal of the American Society of Hematology. 2011;117(19):5092–5101. doi: 10.1182/blood-2010-09-306993. [DOI] [PubMed] [Google Scholar]
- 24.Mantovani A., et al. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013;229(2):176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani A., et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Zhong B., et al. PDCD4 modulates markers of macrophage alternative activation and airway remodeling in antigen‐induced pulmonary inflammation. J. Leukoc. Biol. 2014;96(6):1065–1075. doi: 10.1189/jlb.3A0313-136RRR. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia S., et al. Rapid host defense against Aspergillus fumigatus involves alveolar macrophages with a predominance of alternatively activated phenotype. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0015943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantovani A., et al. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 29.Martinez F.O., Helming L., Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27(1):451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 30.Brombacher F., et al. Macrophages and Dendritic Cells. Springer; 2009. Analyzing classical and alternative macrophage activation in macrophage/neutrophil-specific IL-4 receptor-alpha-deficient mice; pp. 225–252. [DOI] [PubMed] [Google Scholar]
- 31.Chawla A. Control of macrophage activation and function by PPARs. Circ. Res. 2010;106(10):1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luzina I.G., et al. Regulation of inflammation by interleukin‐4: a review of “alternatives”. J. Leukoc. Biol. 2012;92(4):753–764. doi: 10.1189/jlb.0412214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouhlel M.A., et al. PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metabol. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Roy S. miRNA in macrophage development and function. Antioxidants Redox Signal. 2016;25(15):795–804. doi: 10.1089/ars.2016.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghafouri-Fard S., et al. The impact of non-coding RNAs on macrophage polarization. Biomed. Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.112112. [DOI] [PubMed] [Google Scholar]
- 36.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 37.Moghiman T., et al. Therapeutic angiogenesis with exosomal microRNAs: an effectual approach for the treatment of myocardial ischemia. Heart Fail. Rev. 2021;26:205–213. doi: 10.1007/s10741-020-10001-9. [DOI] [PubMed] [Google Scholar]
- 38.Simpson L.J., et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol. 2014;15(12):1162–1170. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Necsulea A., Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 2014;15(11):734–748. doi: 10.1038/nrg3802. [DOI] [PubMed] [Google Scholar]
- 40.Moghaddam A.S., et al. Cardioprotective microRNAs: lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis. 2019;285:1–9. doi: 10.1016/j.atherosclerosis.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Das A., et al. Monocyte and macrophage plasticity in tissue repair and regeneration. Am. J. Pathol. 2015;185(10):2596–2606. doi: 10.1016/j.ajpath.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baltimore D., et al. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 43.Gentner B., et al. Identification of hematopoietic stem cell–specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med. 2010;2(58):58ra84. doi: 10.1126/scitranslmed.3001522. 58ra84. [DOI] [PubMed] [Google Scholar]
- 44.Friedman A. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 45.Ghani S., et al. Macrophage development from HSCs requires PU. 1-coordinated microRNA expression. Blood. The Journal of the American Society of Hematology. 2011;118(8):2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radmanesh F., et al. The immunomodulatory effects of mesenchymal stromal cell‐based therapy in human and animal models of systemic lupus erythematosus. IUBMB Life. 2020;72(11):2366–2381. doi: 10.1002/iub.2387. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., et al. Expression profiles of miRNAs in polarized macrophages. Int. J. Mol. Med. 2013;31(4):797–802. doi: 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- 48.Graff J.W., et al. Identifying functional microRNAs in macrophages with polarized phenotypes. J. Biol. Chem. 2012;287(26):21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cobos Jiménez V., et al. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol. Genom. 2014;46(3):91–103. doi: 10.1152/physiolgenomics.00140.2013. [DOI] [PubMed] [Google Scholar]
- 50.Melton D.W., et al. Dynamic macrophage polarization-specific miRNA patterns reveal increased soluble VEGF receptor 1 by miR-125a-5p inhibition. Physiol. Genom. 2016;48(5):345–360. doi: 10.1152/physiolgenomics.00098.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Nunez R.T., Louafi F., Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1) J. Biol. Chem. 2011;286(3):1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An Y., Yang Q. MiR-21 modulates the polarization of macrophages and increases the effects of M2 macrophages on promoting the chemoresistance of ovarian cancer. Life Sci. 2020;242 doi: 10.1016/j.lfs.2019.117162. [DOI] [PubMed] [Google Scholar]
- 53.Xiao L., et al. Endometrial cancer cells promote M2-like macrophage polarization by delivering exosomal miRNA-21 under hypoxia condition. Journal of Immunology Research. 2020:2020. doi: 10.1155/2020/9731049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren W., et al. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J. Exp. Clin. Cancer Res. 2019;38(1):1–14. doi: 10.1186/s13046-019-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asangani I.A., et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 56.Hilliard A., et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol. 2006;177(11):8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 57.Sheedy F.J., et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11(2):141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 58.Yao F., et al. Adipogenic miR-27a in adipose tissue upregulates macrophage activation via inhibiting PPARγ of insulin resistance induced by high-fat diet-associated obesity. Exp. Cell Res. 2017;355(2):105–112. doi: 10.1016/j.yexcr.2017.03.060. [DOI] [PubMed] [Google Scholar]
- 59.Song J., et al. Esophageal cancer-derived extracellular vesicle miR-21-5p contributes to EMT of ESCC cells by disorganizing macrophage polarization. Cancers. 2021;13(16):4122. doi: 10.3390/cancers13164122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caescu C.I., et al. Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood. The Journal of the American Society of Hematology. 2015;125(8):e1–e13. doi: 10.1182/blood-2014-10-608000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madhyastha R., et al. MicroRNA 21 elicits a pro-inflammatory response in macrophages, with exosomes functioning as delivery vehicles. Inflammation. 2021;44:1274–1287. doi: 10.1007/s10753-021-01415-0. [DOI] [PubMed] [Google Scholar]
- 62.Khan M.J., et al. Inhibition of miRNA-34a promotes M2 macrophage polarization and improves LPS-induced lung injury by targeting Klf4. Genes. 2020;11(9):966. doi: 10.3390/genes11090966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan Y., et al. miR-34a aggravates obesity-induced adipose inflammation and metabolic dysfunction via blocking polarization of anti-inflammatory M2 macrophage. Diabetes. 2018;67(Supplement_1) [Google Scholar]
- 64.Pan Y., et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Invest. 2019;129(2):834–849. doi: 10.1172/JCI123069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen W., et al. Long-term co-exposure DBP and BaP causes imbalance in liver macrophages polarization via activation of Notch signaling regulated by miR-34a-5p in rats. Chem. Biol. Interact. 2022;359 doi: 10.1016/j.cbi.2022.109919. [DOI] [PubMed] [Google Scholar]
- 66.Gao X.R., et al. miR‐34a carried by adipocyte exosomes inhibits the polarization of M1 macrophages in mouse osteolysis model. J. Biomed. Mater. Res. 2021;109(6):994–1003. doi: 10.1002/jbm.a.37088. [DOI] [PubMed] [Google Scholar]
- 67.Jiang P., et al. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp. Cell Res. 2012;318(10):1175–1184. doi: 10.1016/j.yexcr.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 68.Hamzei Taj S., et al. Dynamic modulation of microglia/macrophage polarization by miR-124 after focal cerebral ischemia. J. Neuroimmune Pharmacol. 2016;11(4):733–748. doi: 10.1007/s11481-016-9700-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li R., et al. Bone marrow mesenchymal stem cell-derived exosomal microRNA-124-3p attenuates neurological damage in spinal cord ischemia-reperfusion injury by downregulating Ern1 and promoting M2 macrophage polarization. Arthritis Res. Ther. 2020;22(1):1–14. doi: 10.1186/s13075-020-2146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ponomarev E.D., et al. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α–PU. 1 pathway. Nat. Med. 2011;17(1):64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Y., et al. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res. 2013;23(11):1270–1283. doi: 10.1038/cr.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banerjee S., et al. miR-125a-5p regulates differential activation of macrophages and inflammation. J. Biol. Chem. 2013;288(49):35428–35436. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang Q., et al. Bone marrow mesenchymal stem cell-derived exosomal microRNA-125a promotes M2 macrophage polarization in spinal cord injury by downregulating IRF5. Brain Res. Bull. 2021;170:199–210. doi: 10.1016/j.brainresbull.2021.02.015. [DOI] [PubMed] [Google Scholar]
- 74.He W., Zhang N., Lin Z. MicroRNA-125a-5p modulates macrophage polarization by targeting E26 transformation-specific variant 6 gene during orthodontic tooth movement. Arch. Oral Biol. 2021;124 doi: 10.1016/j.archoralbio.2021.105060. [DOI] [PubMed] [Google Scholar]
- 75.Shi L., et al. Dioscin ameliorates inflammatory bowel disease by up‐regulating miR‐125a‐5p to regulate macrophage polarization. J. Clin. Lab. Anal. 2022 doi: 10.1002/jcla.24455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhuri A.A., et al. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011;187(10):5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim S.-W., et al. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc. Natl. Acad. Sci. USA. 2012;109(20):7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su M.-J., Aldawsari H., Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci. Rep. 2016;6(1):1–15. doi: 10.1038/srep30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C., et al. Disease markers; 2022. Role of Cardiomyocyte-Derived Exosomal MicroRNA-146a-5p in Macrophage Polarization and Activation; p. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang C., et al. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264. 7 macrophages. Int. Immunopharm. 2016;32:46–54. doi: 10.1016/j.intimp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 81.Zhong Y., et al. PM2. 5 upregulates MicroRNA-146a-3p and induces M1 polarization in RAW264. 7 cells by targeting Sirtuin1. Int. J. Med. Sci. 2019;16(3):384. doi: 10.7150/ijms.30084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., et al. MicroRNA-146a-5p-modified human umbilical cord mesenchymal stem cells enhance protection against diabetic nephropathy in rats through facilitating M2 macrophage polarization. Stem Cell Res. Ther. 2022;13(1):1–16. doi: 10.1186/s13287-022-02855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Curtale G., et al. Negative regulation of Toll-like receptor 4 signaling by IL-10–dependent microRNA-146b. Proc. Natl. Acad. Sci. USA. 2013;110(28):11499–11504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng L., et al. Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine. 2016;14:83–96. doi: 10.1016/j.ebiom.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Z., et al. miR-146b level and variants is associated with endometriosis related macrophages phenotype and plays a pivotal role in the endometriotic pain symptom. Taiwan. J. Obstet. Gynecol. 2019;58(3):401–408. doi: 10.1016/j.tjog.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 86.Yi-Hong C., Jing L., Lu H. MicroRNA-155 induces macrophage polarization to M1 in Toxoplasma gon-dii infection. Zhongguo xue xi Chong Bing Fang zhi za zhi= Chinese Journal of Schistosomiasis Control. 2019;30(6):652–655. doi: 10.16250/j.32.1374.2018125. [DOI] [PubMed] [Google Scholar]
- 87.Lu Z.-J., et al. MicroRNA-155 promotes the pathogenesis of experimental colitis by repressing SHIP-1 expression. World J. Gastroenterol. 2017;23(6):976. doi: 10.3748/wjg.v23.i6.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nazari-Jahantigh M., et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Invest. 2012;122(11):4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ye J., et al. Mediators of inflammation; 2016. miR-155 Regulated Inflammation Response by the SOCS1-STAT3-PDCD4 axis in Atherogenesis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y., et al. Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J. Mol. Cell Biol. 2016;8(6):505–517. doi: 10.1093/jmcb/mjw040. [DOI] [PubMed] [Google Scholar]
- 91.Cai X., et al. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J. Mol. Cell Biol. 2012;4(5):341–343. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 92.Wang P., et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010;185(10):6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 93.Zhang P., et al. MicroRNA-155 inhibits polarization of macrophages to M2-type and suppresses choroidal neovascularization. Inflammation. 2018;41(1):143–153. doi: 10.1007/s10753-017-0672-8. [DOI] [PubMed] [Google Scholar]
- 94.Lorenzo-Pouso A.I., et al. Autophagy in periodontal disease: evidence from a literature review. Arch. Oral Biol. 2019;102:55–64. doi: 10.1016/j.archoralbio.2019.03.029. [DOI] [PubMed] [Google Scholar]
- 95.Lu J., Xie L., Sun S. The inhibitor miR-21 regulates macrophage polarization in an experimental model of chronic obstructive pulmonary disease. Tob. Induc. Dis. 2021;19 doi: 10.18332/tid/140095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thulin P., et al. MicroRNA-9 regulates the expression of peroxisome proliferator-activated receptor δ in human monocytes during the inflammatory response. Int. J. Mol. Med. 2013;31(5):1003–1010. doi: 10.3892/ijmm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tong F., et al. HPV+ HNSCC-derived exosomal miR-9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett. 2020;478:34–44. doi: 10.1016/j.canlet.2020.02.037. [DOI] [PubMed] [Google Scholar]
- 98.Wang R., Xu B. TGF-β1-modified MSC-derived exosomal miR-135b attenuates cartilage injury via promoting M2 synovial macrophage polarization by targeting MAPK6. Cell Tissue Res. 2021;384(1):113–127. doi: 10.1007/s00441-020-03319-1. [DOI] [PubMed] [Google Scholar]
- 99.An T.-H., et al. MiR-181b antagonizes atherosclerotic plaque vulnerability through modulating macrophage polarization by directly targeting Notch1. Mol. Neurobiol. 2017;54(8):6329–6341. doi: 10.1007/s12035-016-0163-1. [DOI] [PubMed] [Google Scholar]
- 100.Liu W., et al. A novel delivery nanobiotechnology: engineered miR-181b exosomes improved osteointegration by regulating macrophage polarization. J. Nanobiotechnol. 2021;19(1):1–18. doi: 10.1186/s12951-021-01015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meng Z., et al. miR-200c/PAI-2 promotes the progression of triple negative breast cancer via M1/M2 polarization induction of macrophage. Int. Immunopharm. 2020;81 doi: 10.1016/j.intimp.2019.106028. [DOI] [PubMed] [Google Scholar]
- 102.Xiong J., et al. MiR-200b is upregulated in plasma-derived exosomes and functions as an oncogene by promoting macrophage M2 polarization in ovarian cancer. J. Ovarian Res. 2021;14(1):1–10. doi: 10.1186/s13048-021-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raue R., et al. MicroRNA-200c attenuates the tumor-infiltrating capacity of macrophages. Biology. 2022;11(3):349. doi: 10.3390/biology11030349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams M.M., et al. MicroRNA-200c restoration reveals a cytokine profile to enhance M1 macrophage polarization in breast cancer. NPJ Breast Cancer. 2021;7(1):1–13. doi: 10.1038/s41523-021-00273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiao P., et al. miR-223: an effective regulator of immune cell differentiation and inflammation. Int. J. Biol. Sci. 2021;17(9):2308. doi: 10.7150/ijbs.59876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Q., et al. 2012. Inducible microRNA-223 Down-Regulation Promotes TLR-Triggered IL-6 and IL-1β Production in Macrophages by Targeting STAT3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dang C.P., Leelahavanichkul A. Over-expression of miR-223 induces M2 macrophage through glycolysis alteration and attenuates LPS-induced sepsis mouse model, the cell-based therapy in sepsis. PLoS One. 2020;15(7) doi: 10.1371/journal.pone.0236038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ismail N., et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. The Journal of the American Society of Hematology. 2013;121(6):984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhuang G., et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125(23):2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 110.Gou W., et al. MiR-223/Pknox1 axis protects mice from CVB3-induced viral myocarditis by modulating macrophage polarization. Exp. Cell Res. 2018;366(1):41–48. doi: 10.1016/j.yexcr.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 111.Knipe R.S., Tager A.M., Liao J.K. The Rho kinases: critical mediators of multiple profibrotic processes and rational targets for new therapies for pulmonary fibrosis. Pharmacol. Rev. 2015;67(1):103–117. doi: 10.1124/pr.114.009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saradna A., et al. Macrophage polarization and allergic asthma. Transl. Res. 2018;191:1–14. doi: 10.1016/j.trsl.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou Y., et al. Mannose receptor modulates macrophage polarization and allergic inflammation through miR-511-3p. J. Allergy Clin. Immunol. 2018;141(1):350–364. doi: 10.1016/j.jaci.2017.04.049. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Banerjee S., et al. MicroRNA let-7c regulates macrophage polarization. J. Immunol. 2013;190(12):6542–6549. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kumar M., et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe. 2015;17(3):345–356. doi: 10.1016/j.chom.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 116.Liu Y., et al. MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Lett. 2011;585(12):1963–1968. doi: 10.1016/j.febslet.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 117.Sheedy F.J. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yao M., et al. Exosomal miR-21 secreted by IL-1β-primed-mesenchymal stem cells induces macrophage M2 polarization and ameliorates sepsis. Life Sci. 2021;264 doi: 10.1016/j.lfs.2020.118658. [DOI] [PubMed] [Google Scholar]
- 119.Hsieh C.-H., Tai S.-K., Yang M.-H. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering MiR-21-abundant exosomes. Neoplasia. 2018;20(8):775–788. doi: 10.1016/j.neo.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shen D., He Z. Mesenchymal stem cell-derived exosomes regulate the polarization and inflammatory response of macrophages via miR-21-5p to promote repair after myocardial reperfusion injury. Ann. Transl. Med. 2021;9(16) doi: 10.21037/atm-21-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li J., et al. ADAR1 attenuates allogeneic graft rejection by suppressing miR‐21 biogenesis in macrophages and promoting M2 polarization. Faseb. J. 2018;32(9):5162–5173. doi: 10.1096/fj.201701449R. [DOI] [PubMed] [Google Scholar]
- 122.Arora S., Ali S., Syed M.A. Eur Respiratory Soc; 2020. MiR-34a Favours Macrophage Polarization Switch from M2 to M1 Phenotype in Non Small Cell Lung Cancer (NSCLC) [Google Scholar]
- 123.Veremeyko T., et al. IL-4/IL-13-dependent and independent expression of miR-124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ponomarev E.D., et al. CNS-derived interleukin-4 is essential for the regulation of autoimmune inflammation and induces a state of alternative activation in microglial cells. J. Neurosci. 2007;27(40):10714–10721. doi: 10.1523/JNEUROSCI.1922-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gu W., et al. ICAM-1 regulates macrophage polarization by suppressing MCP-1 expression via miR-124 upregulation. Oncotarget. 2017;8(67) doi: 10.18632/oncotarget.22948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wertz I.E., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 127.Ganesh S., et al. Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials. 2013;34(13):3489–3502. doi: 10.1016/j.biomaterials.2013.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu X., et al. 1, 25-Dihydroxyvitamin D regulates macrophage polarization and ameliorates experimental inflammatory bowel disease by suppressing miR-125b. Int. Immunopharm. 2019;67:106–118. doi: 10.1016/j.intimp.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 129.Yazdanpanah E., et al. Vitamin D3 alters the expression of toll‐like receptors in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. J. Cell. Biochem. 2017;118(12):4831–4835. doi: 10.1002/jcb.26155. [DOI] [PubMed] [Google Scholar]
- 130.Androulidaki A., et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31(2):220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Murphy A.J., Guyre P.M., Pioli P.A. Estradiol suppresses NF-κB activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J. Immunol. 2010;184(9):5029–5037. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tili E., et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 133.Huang H.C., et al. miRNA‐125b regulates TNF‐α production in CD14+ neonatal monocytes via post‐transcriptional regulation. J. Leukoc. Biol. 2012;92(1):171–182. doi: 10.1189/jlb.1211593. [DOI] [PubMed] [Google Scholar]
- 134.Rajaram M.V., et al. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. USA. 2011;108(42):17408–17413. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Taganov K.D., et al. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vergadi E., et al. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J. Immunol. 2014;192(1):394–406. doi: 10.4049/jimmunol.1300959. [DOI] [PubMed] [Google Scholar]
- 137.Zeng Z., et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp. Lung Res. 2013;39(7):275–282. doi: 10.3109/01902148.2013.808285. [DOI] [PubMed] [Google Scholar]
- 138.Zhang L., et al. Severe fever with thrombocytopenia syndrome virus-induced macrophage differentiation is regulated by miR-146. Front. Immunol. 2019;10:1095. doi: 10.3389/fimmu.2019.01095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Faraoni I., et al. miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta, Mol. Basis Dis. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 140.O'Connell R.M., et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zonari E., et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. The Journal of the American Society of Hematology. 2013;122(2):243–252. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 142.Ma S., et al. Resveratrol promoted the M2 polarization of microglia and reduced neuroinflammation after cerebral ischemia by inhibiting miR-155. Int. J. Neurosci. 2020;130(8):817–825. doi: 10.1080/00207454.2019.1707817. [DOI] [PubMed] [Google Scholar]
- 143.Huang F., et al. Cypermethrin promotes lung cancer metastasis via modulation of macrophage polarization by targeting MicroRNA-155/Bcl6. Toxicol. Sci. 2018;163(2):454–465. doi: 10.1093/toxsci/kfy039. [DOI] [PubMed] [Google Scholar]
- 144.Xu F., et al. Akt1-mediated regulation of macrophage polarization in a murine model of Staphylococcus aureus pulmonary infection. J. Infect. Dis. 2013;208(3):528–538. doi: 10.1093/infdis/jit177. [DOI] [PubMed] [Google Scholar]
- 145.Moore C.S., et al. miR‐155 as a multiple sclerosis–relevant regulator of myeloid cell polarization. Ann. Neurol. 2013;74(5):709–720. doi: 10.1002/ana.23967. [DOI] [PubMed] [Google Scholar]
- 146.Li G.-S., Cui L., Wang G.-D. miR-155-5p regulates macrophage M1 polarization and apoptosis in the synovial fluid of patients with knee osteoarthritis. Exp. Ther. Med. 2021;21(1):1. doi: 10.3892/etm.2020.9500. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu J., et al. Inhibition of microRNA-155 attenuates sympathetic neural remodeling following myocardial infarction via reducing M1 macrophage polarization and inflammatory responses in mice. Eur. J. Pharmacol. 2019;851:122–132. doi: 10.1016/j.ejphar.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 148.O'Connell R.M., et al. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ruggiero T., et al. LPS induces KH‐type splicing regulatory protein‐dependent processing of microRNA‐155 precursors in macrophages. Faseb. J. 2009;23(9):2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 150.He M., et al. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPβ. Cell. Mol. Immunol. 2009;6(5):343–352. doi: 10.1038/cmi.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arranz A., et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc. Natl. Acad. Sci. USA. 2012;109(24):9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Louafi F., Martinez-Nunez R.T., Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J. Biol. Chem. 2010;285(53):41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tang B., et al. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584(8):1481–1486. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 154.Wang J., et al. MiR-9-5p promotes M1 cell polarization in osteoarthritis progression by regulating NF-κB and AMPK signaling pathways by targeting SIRT1. Int. Immunopharm. 2021;101 doi: 10.1016/j.intimp.2021.108207. [DOI] [PubMed] [Google Scholar]
- 155.Wang Z., et al. Hypermethylation of miR-181b in monocytes is associated with coronary artery disease and promotes M1 polarized phenotype via PIAS1-KLF4 axis. Cardiovasc. Diagn. Ther. 2020;10(4):738. doi: 10.21037/cdt-20-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ma J., et al. Genes & Genomics; 2022. The Role of Exosomal miR-181b in the Crosstalk between NSCLC Cells and Tumor-Associated Macrophages; pp. 1–16. [DOI] [PubMed] [Google Scholar]
- 157.Li N., et al. miR‐21a negatively modulates tumor suppressor genes PTEN and miR‐200c and further promotes the transformation of M2 macrophages. Immunol. Cell Biol. 2018;96(1):68–80. doi: 10.1111/imcb.1016. [DOI] [PubMed] [Google Scholar]
- 158.Ying W., et al. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Invest. 2015;125(11):4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Squadrito M.L., et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012;1(2):141–154. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 160.Squadrito M.L., et al. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34(7):350–359. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Heinsbroek S.E., et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. 2016;9(4):960–973. doi: 10.1038/mi.2015.113. [DOI] [PubMed] [Google Scholar]
- 162.Ostuni R., et al. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36(4):229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 163.Raggi C., et al. Cancer stem cells and tumor-associated macrophages: a roadmap for multitargeting strategies. Oncogene. 2016;35(6):671–682. doi: 10.1038/onc.2015.132. [DOI] [PubMed] [Google Scholar]
- 164.Karo-Atar D., et al. MicroRNA profiling reveals opposing expression patterns for miR-511 in alternatively and classically activated macrophages. J. Asthma. 2015;52(6):545–553. doi: 10.3109/02770903.2014.988222. [DOI] [PubMed] [Google Scholar]
- 165.Zhu X., et al. MiR-19a-3p suppresses M1 macrophage polarization by inhibiting STAT1/IRF1 pathway. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.614044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang W., et al. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-κB pathway. Cell Death Differ. 2015;22(2):287–297. doi: 10.1038/cdd.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Peng Y., et al. MiR-98-5p expression inhibits polarization of macrophages to an M2 phenotype by targeting Trib1 in inflammatory bowel disease. Acta Biochim. Pol. 2020;67(2):157–163. doi: 10.18388/abp.2020_5152. [DOI] [PubMed] [Google Scholar]
- 168.Cho Y.K., et al. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol. Metabol. 2019;29:86–98. doi: 10.1016/j.molmet.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jang J.-Y., et al. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. 2013;13(1):1–12. doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jia X., et al. MiR‐16 regulates mouse peritoneal macrophage polarization and affects T‐cell activation. J. Cell Mol. Med. 2016;20(10):1898–1907. doi: 10.1111/jcmm.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tian J., et al. Exosomal microRNA-16-5p from adipose mesenchymal stem cells promotes TLR4-mediated M2 macrophage polarization in septic lung injury. Int. Immunopharm. 2021;98 doi: 10.1016/j.intimp.2021.107835. [DOI] [PubMed] [Google Scholar]
- 172.Shin C.-H., et al. Exosomal miRNA-19a and miRNA-614 induced by air pollutants promote proinflammatory M1 macrophage polarization via regulation of RORα expression in human respiratory mucosal microenvironment. J. Immunol. 2020;205(11):3179–3190. doi: 10.4049/jimmunol.2000456. [DOI] [PubMed] [Google Scholar]
- 173.Chen J., et al. Tumor‐derived exosomal miR‐19b‐3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin. Transl. Med. 2021;11(9):e478. doi: 10.1002/ctm2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fang H., et al. MicroRNA‐22‐3p alleviates spinal cord ischemia/reperfusion injury by modulating M2 macrophage polarization via IRF5. J. Neurochem. 2021;156(1):106–120. doi: 10.1111/jnc.15042. [DOI] [PubMed] [Google Scholar]
- 175.Chen Z., et al. Pioglitazone decreased renal calcium oxalate crystal formation by suppressing M1 macrophage polarization via the PPAR-γ-miR-23 axis. Am. J. Physiol. Ren. Physiol. 2019;317(1):F137–F151. doi: 10.1152/ajprenal.00047.2019. [DOI] [PubMed] [Google Scholar]
- 176.Zhang Y., et al. Extracellular vesicle-encapsulated microRNA-23a from dorsal root ganglia neurons binds to A20 and promotes inflammatory macrophage polarization following peripheral nerve injury. Aging (Albany NY) 2021;13(5):6752. doi: 10.18632/aging.202532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhu F., et al. 2021. Human Umbilical Cord Mesenchymal Stem Cells Derived Exosomes Attenuate Injury of Myocardial Infarction by miR-24-3p-Promoted M2 Macrophage Polarization. [DOI] [PubMed] [Google Scholar]
- 178.Jingjing Z., et al. MicroRNA-24 modulates Staphylococcus aureus-induced macrophage polarization by suppressing CHI3L1. Inflammation. 2017;40(3):995–1005. doi: 10.1007/s10753-017-0543-3. [DOI] [PubMed] [Google Scholar]
- 179.Sahu S.K., et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 2017;13(5) doi: 10.1371/journal.ppat.1006410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu X., et al. Inhibition of PTP1B promotes M2 polarization via MicroRNA-26a/MKP1 signaling pathway in murine macrophages. Front. Immunol. 2019;10:1930. doi: 10.3389/fimmu.2019.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Saha B., et al. Alcohol-induced miR-27a regulates differentiation and M2 macrophage polarization of normal human monocytes. J. Immunol. 2015;194(7):3079–3087. doi: 10.4049/jimmunol.1402190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang J., et al. Mesenchymal stem cell–derived extracellular vesicles alleviate acute lung injury via transfer of miR-27a-3p. Crit. Care Med. 2020;48(7):e599–e610. doi: 10.1097/CCM.0000000000004315. [DOI] [PubMed] [Google Scholar]
- 183.Cai J., et al. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am. J. Physiol. Cell Physiol. 2019;316(5):C731–C740. doi: 10.1152/ajpcell.00366.2018. [DOI] [PubMed] [Google Scholar]
- 184.Qi X., et al. miR-30b-5p releases HMGB1 via UBE2D2/KAT2B/HMGB1 pathway to promote pro-inflammatory polarization and recruitment of macrophages. Atherosclerosis. 2021;324:38–45. doi: 10.1016/j.atherosclerosis.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 185.Jiao Y., et al. Exosomal miR-30d-5p of neutrophils induces M1 macrophage polarization and primes macrophage pyroptosis in sepsis-related acute lung injury. Crit. Care. 2021;25(1):1–15. doi: 10.1186/s13054-021-03775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bao L., Li X. MicroRNA-32 targeting PTEN enhances M2 macrophage polarization in the glioma microenvironment and further promotes the progression of glioma. Mol. Cell. Biochem. 2019;460(1):67–79. doi: 10.1007/s11010-019-03571-2. [DOI] [PubMed] [Google Scholar]
- 187.Zhu Q.-Y., et al. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J. Immunol. 2010;185(12):7435–7442. doi: 10.4049/jimmunol.1000798. [DOI] [PubMed] [Google Scholar]
- 188.Zhu X., et al. MiR-103 protects from recurrent spontaneous abortion via inhibiting STAT1 mediated M1 macrophage polarization. Int. J. Biol. Sci. 2020;16(12):2248. doi: 10.7150/ijbs.46144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shi Y., et al. miR-106b-5p inhibits IRF1/IFN-β signaling to promote M2 macrophage polarization of glioblastoma. OncoTargets Ther. 2020;13:7479. doi: 10.2147/OTT.S238975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ying H., et al. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J. Immunol. 2015;194(3):1239–1251. doi: 10.4049/jimmunol.1402088. [DOI] [PubMed] [Google Scholar]
- 191.Moradi-Chaleshtori M., et al. Transfer of miRNA in tumor-derived exosomes suppresses breast tumor cell invasion and migration by inducing M1 polarization in macrophages. Life Sci. 2021;282 doi: 10.1016/j.lfs.2021.119800. [DOI] [PubMed] [Google Scholar]
- 192.Guo Q., et al. miR‐130b‐3p regulates M1 macrophage polarization via targeting IRF1. J. Cell. Physiol. 2021;236(3):2008–2022. doi: 10.1002/jcp.29987. [DOI] [PubMed] [Google Scholar]
- 193.Liu F., et al. miR-132 inhibits lipopolysaccharide-induced inflammation in alveolar macrophages by the cholinergic anti-inflammatory pathway. Exp. Lung Res. 2015;41(5):261–269. doi: 10.3109/01902148.2015.1004206. [DOI] [PubMed] [Google Scholar]
- 194.Wang Y., et al. Mesenchymal stem cell–secreted extracellular vesicles carrying TGF‐β1 up‐regulate miR‐132 and promote mouse M2 macrophage polarization. J. Cell Mol. Med. 2020;24(21):12750–12764. doi: 10.1111/jcmm.15860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xun J., et al. Cancer-derived exosomal miR-138-5p modulates polarization of tumor-associated macrophages through inhibition of KDM6B. Theranostics. 2021;11(14):6847. doi: 10.7150/thno.51864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shi X., et al. MiR-144-5p limits experimental abdominal aortic aneurysm formation by mitigating M1 macrophage-associated inflammation: suppression of TLR2 and OLR1. J. Mol. Cell. Cardiol. 2020;143:1–14. doi: 10.1016/j.yjmcc.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 197.Huang Y., et al. IL-16 regulates macrophage polarization as a target gene of mir-145-3p. Mol. Immunol. 2019;107:1–9. doi: 10.1016/j.molimm.2018.12.027. [DOI] [PubMed] [Google Scholar]
- 198.Su D. Up-regulation of MiR-145-5p promotes the growth and migration in LPS-treated HUVECs through inducing macrophage polarization to M2. J. Recept. Signal Transduction. 2021;41(5):434–441. doi: 10.1080/10799893.2020.1818095. [DOI] [PubMed] [Google Scholar]
- 199.Carson IV W.F., et al. Enhancement of macrophage inflammatory responses by CCL2 is correlated with increased miR-9 expression and downregulation of the ERK1/2 phosphatase Dusp6. Cell. Immunol. 2017;314:63–72. doi: 10.1016/j.cellimm.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Huang F., et al. miR-148a-3p mediates Notch signaling to promote the differentiation and M1 activation of macrophages. Front. Immunol. 2017;8:1327. doi: 10.3389/fimmu.2017.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ma D., et al. miR-148a affects polarization of THP-1-derived macrophages and reduces recruitment of tumor-associated macrophages via targeting SIRPα. Cancer Manag. Res. 2020;12:8067. doi: 10.2147/CMAR.S238317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Qiu M., et al. MicroRNA-150 deficiency accelerates intimal hyperplasia by acting as a novel regulator of macrophage polarization. Life Sci. 2020;240 doi: 10.1016/j.lfs.2019.116985. [DOI] [PubMed] [Google Scholar]
- 203.Xu J., et al. Hypoxic bone marrow mesenchymal stromal cells‐derived exosomal miR‐182‐5p promotes liver regeneration via FOXO1‐mediated macrophage polarization. Faseb. J. 2022;36(10) doi: 10.1096/fj.202101868RRR. [DOI] [PubMed] [Google Scholar]
- 204.Zhao J., et al. Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc. Res. 2019;115(7):1205–1216. doi: 10.1093/cvr/cvz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang X.-F., et al. MiR-188-3p upregulation results in the inhibition of macrophage proinflammatory activities and atherosclerosis in ApoE-deficient mice. Thromb. Res. 2018;171:55–61. doi: 10.1016/j.thromres.2018.09.043. [DOI] [PubMed] [Google Scholar]
- 206.An L., Yin F. MiR-192-5p suppresses M1 macrophage polarization via epiregulin (EREG) downregulation in gouty arthritis. Tissue Cell. 2021;73 doi: 10.1016/j.tice.2021.101669. [DOI] [PubMed] [Google Scholar]
- 207.Mao M., et al. MicroRNA-195 prevents hippocampal microglial/macrophage polarization towards the M1 phenotype induced by chronic brain hypoperfusion through regulating CX3CL1/CX3CR1 signaling. J. Neuroinflammation. 2020;17(1):1–20. doi: 10.1186/s12974-020-01919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang L., et al. MiR-202-5p promotes M2 polarization in allergic rhinitis by targeting MATN2. Int. Arch. Allergy Immunol. 2019;178(2):119–127. doi: 10.1159/000493803. [DOI] [PubMed] [Google Scholar]
- 209.He L., et al. 2021. Ovarian Cancer Cell-Derived Exosomal miR-205 Promotes M2 Macrophage Polarization and Ovarian Cancer Cell Metastasis by Activating the AKT/mTOR Signalling Pathway. [Google Scholar]
- 210.Peng L.Y., et al. MicroRNA‐214‐3p facilitates M2 macrophage polarization by targeting GSK3B. Kaohsiung J. Med. Sci. 2022;38(4):347–356. doi: 10.1002/kjm2.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yang S., et al. MicroRNA-216a promotes M1 macrophages polarization and atherosclerosis progression by activating telomerase via the Smad3/NF-κB pathway. Biochim. Biophys. Acta, Mol. Basis Dis. 2019;1865(7):1772–1781. doi: 10.1016/j.bbadis.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 212.Liu W., et al. Exosome-shuttled miR-216a-5p from hypoxic preconditioned mesenchymal stem cells repair traumatic spinal cord injury by shifting microglial M1/M2 polarization. J. Neuroinflammation. 2020;17(1):1–22. doi: 10.1186/s12974-020-1726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jiang B., et al. MiR-217 inhibits M2-like macrophage polarization by suppressing secretion of interleukin-6 in ovarian cancer. Inflammation. 2019;42(5):1517–1529. doi: 10.1007/s10753-019-01004-2. [DOI] [PubMed] [Google Scholar]
- 214.Cai M., et al. Mammary epithelial cell derived exosomal MiR-221 mediates M1 macrophage polarization via SOCS1/STATs to promote inflammatory response. Int. Immunopharm. 2020;83 doi: 10.1016/j.intimp.2020.106493. [DOI] [PubMed] [Google Scholar]
- 215.Ying X., et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7(28) doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Huang Y., et al. Endometriosis derived exosomal miR-301a-3p mediates macrophage polarization via regulating PTEN-PI3K axis. Biomed. Pharmacother. 2022;147 doi: 10.1016/j.biopha.2022.112680. [DOI] [PubMed] [Google Scholar]
- 217.Hsu L.-W., et al. MicroRNA-301a inhibition enhances the immunomodulatory functions of adipose-derived mesenchymal stem cells by induction of macrophage M2 polarization. Int. J. Immunopathol. Pharmacol. 2020;34 doi: 10.1177/2058738420966092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang X., et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer MetastasisTumor-promoting effects of hypoxic exosomal miR-301a. Cancer Res. 2018;78(16):4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 219.Zhong C., et al. Histone demethylase JMJD1C promotes the polarization of M1 macrophages to prevent glioma by upregulating miR‐302a. Clin. Transl. Med. 2021;11(9):e424. doi: 10.1002/ctm2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sun J., et al. miR-330-5p/Tim-3 axis regulates macrophage M2 polarization and insulin resistance in diabetes mice. Mol. Immunol. 2018;95:107–113. doi: 10.1016/j.molimm.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 221.Qiu Y., et al. MiR-375 silencing attenuates pro-inflammatory macrophage response and foam cell formation by targeting KLF4. Exp. Cell Res. 2021;400(1) doi: 10.1016/j.yexcr.2021.112507. [DOI] [PubMed] [Google Scholar]
- 222.Chen W., et al. Tanshinone IIA harmonizes the crosstalk of autophagy and polarization in macrophages via miR-375/KLF4 pathway to attenuate atherosclerosis. Int. Immunopharm. 2019;70:486–497. doi: 10.1016/j.intimp.2019.02.054. [DOI] [PubMed] [Google Scholar]
- 223.Lv Y., et al. MiR‐382‐5p suppresses M1 macrophage polarization and inflammatory response in response to bronchopulmonary dysplasia through targeting CDK8: involving inhibition of STAT1 pathway. Gene Cell. 2021;26(10):772–781. doi: 10.1111/gtc.12883. [DOI] [PubMed] [Google Scholar]
- 224.Zhao Q., et al. Suppression of TLR4 by miR-448 is involved in diabetic development via regulating macrophage polarization. J. Pharm. Pharmacol. 2019;71(5):806–815. doi: 10.1111/jphp.13048. [DOI] [PubMed] [Google Scholar]
- 225.Liu G., et al. The effect of miR-471-3p on macrophage polarization in the development of diabetic cardiomyopathy. Life Sci. 2021;268 doi: 10.1016/j.lfs.2020.118989. [DOI] [PubMed] [Google Scholar]
- 226.Zhao G., et al. TGF‐β3‐induced miR‐494 inhibits macrophage polarization via suppressing PGE 2 secretion in mesenchymal stem cells. FEBS Lett. 2016;590(11):1602–1613. doi: 10.1002/1873-3468.12200. [DOI] [PubMed] [Google Scholar]
- 227.Shao G., et al. MiRNA-494 enhances M1 macrophage polarization via Nrdp1 in ICH mice model. J. Inflamm. 2020;17(1):1–13. doi: 10.1186/s12950-020-00247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.van Ingen E., et al. Inhibition of microRNA-494-3p activates Wnt signaling and reduces proinflammatory macrophage polarization in atherosclerosis. Mol. Ther. Nucleic Acids. 2021;26:1228–1239. doi: 10.1016/j.omtn.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hu F., et al. MiR-495 regulates macrophage M1/M2 polarization and insulin resistance in high-fat diet-fed mice via targeting FTO. Pflueg. Arch. Eur. J. Physiol. 2019;471(11):1529–1537. doi: 10.1007/s00424-019-02316-w. [DOI] [PubMed] [Google Scholar]
- 230.Li D., et al. miR-498 inhibits autophagy and M2-like polarization of tumor-associated macrophages in esophageal cancer via MDM2/ATF3. Epigenomics. 2021;13(13):1013–1030. doi: 10.2217/epi-2020-0341. [DOI] [PubMed] [Google Scholar]
- 231.Zhao S., et al. MiR‐505 promotes M2 polarization in choroidal neovascularization model mice by targeting transmembrane protein 229B. Scand. J. Immunol. 2019;90(6) doi: 10.1111/sji.12832. [DOI] [PubMed] [Google Scholar]
- 232.Sun L., et al. MiR-506 suppresses tumor progression by reprogramming macrophage polarization in pancreatic ductal adenocarcinoma. Cancer Res. 2019;79(13_Supplement):3556. 3556. [Google Scholar]
- 233.Qi J.R., et al. MiR-520a-3p inhibited macrophage polarization and promoted the development of atherosclerosis via targeting UVRAG in apolipoprotein E knockout mice. Front. Mol. Biosci. 2021;7 doi: 10.3389/fmolb.2020.621324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang P., et al. Mediators of inflammation; 2019. miR-657 Promotes Macrophage Polarization toward M1 by Targeting FAM46C in Gestational Diabetes Mellitus; p. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhong Y., Yi C. MicroRNA-720 suppresses M2 macrophage polarization by targeting GATA3. Biosci. Rep. 2016;36(4) doi: 10.1042/BSR20160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Liu L., et al. Mesenchymal stem cell-derived extracellular vesicles prevent glioma by blocking M2 polarization of macrophages through a miR-744-5p/TGFB1-dependent mechanism. Cell Biol. Toxicol. 2022:1–17. doi: 10.1007/s10565-021-09652-7. [DOI] [PubMed] [Google Scholar]
- 237.Zhao S., et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020;13(1):1–19. doi: 10.1186/s13045-020-00991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang P., et al. MiR-6869-5p induces M2 polarization by regulating PTPRO in gestational diabetes mellitus. Mediat. Inflamm. 2021:2021. doi: 10.1155/2021/6696636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Beg M.S., et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. N. Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lee E.C., et al. Discovery and preclinical evaluation of anti-miR-17 oligonucleotide RGLS4326 for the treatment of polycystic kidney disease. Nat. Commun. 2019;10(1):4148. doi: 10.1038/s41467-019-11918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Janssen H.L., et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 242.Abplanalp W.T., et al. Efficiency and target derepression of anti-miR-92a: results of a first in human study. Nucleic Acid Therapeut. 2020;30(6):335–345. doi: 10.1089/nat.2020.0871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.