Abstract
Escherichia coli O157:H7 is an important food-borne pathogen. Often E. coli O157:H7 is difficult to detect, because it is present sporadically at very low levels together with very high levels of competitor organisms which can be difficult to distinguish phenotypically. Cultural methods are time-consuming and give variable results in the detection of E. coli O157:H7. This study examined the performance of BAX for Screening/E. coli O157:H7, a new rapid method for the detection of E. coli O157:H7, against traditional and improved cultural methods and an immunodiffusion assay. All cultural methods demonstrated inadequacy in detecting the presence of E. coli O157:H7 in inoculated samples. The limitations of these cultural methods further complicate evaluation of screening methodologies. The BAX for Screening/E. coli O157:H7 assay outperformed the other methods, with a detection rate of 96.5%, compared to 39% for the best cultural method and 71.5% for the immunodiffusion method. The BAX for Screening/E. coli O157:H7 assay proved to be a rapid, highly sensitive test for the detection of low levels of E. coli O157:H7 in ground beef.
Escherichia coli O157:H7 is recognized as a significant food-borne pathogen. Hemorrhagic colitis caused by E. coli O157:H7 can lead to complications such as hemolytic uremic syndrome and thrombotic thrombocytopenic purpura, which may be fatal to young children or the aged. Further indication of the marked pathogenicity of this organism is epidemiological evidence that only a few cells are necessary to cause illness (9). Johnson et al. reported the presence of 0.9 to 4.3 CFU of E. coli O157:H7 per g in lots of ground beef that had been implicated in a 1993 outbreak of hemolytic uremic syndrome in humans (3, 11). U.S. Department of Agriculture Food Safety Inspection Service (FSIS) regulations declare that the presence of E. coli O157:H7 at or above 1 CFU/25 g constitutes adulterated and dangerous ground beef. Consequently, highly sensitive and reliable methods for the detection of E. coli O157:H7 are critical to ensure food safety.
Cultural methods often fail to detect the presence of low levels of E. coli O157:H7 in inoculated ground beef samples (13, 16). High levels of competing microflora in food samples compound the difficulty of isolating E. coli O157:H7. The common isolation strategy involves the use of one of several combinations of selective broths and agars, typically with indicators for enzymatic activities such as sorbitol fermentation. These isolation methods can take more than 48 h to complete and still require expert judgment and further testing to confirm the presence of E. coli O157:H7.
Immunoassay methods which yield a presumptive positive or negative screening result in 24 h or less have been developed. Typical sensitivities of immunoassays are approximately 106 CFU/ml, which may lead to false-negative results, particularly when initial levels of competing flora reach 105 to 106 CFU/g (7), a situation not uncommon in ground beef. In addition, positive results must be confirmed by isolation of pure colonies of the target bacteria. Screening of large numbers of ground beef samples has resulted in a high percentage of unconfirmed positive results, indicating either insufficient sensitivity of the isolation procedure or nonspecificity of the immunoassay (2, 5, 7). Firstenberg-Eden and Sullivan reported in their study of ground beef samples that 7.4% of EZ coli-positive samples did not yield E. coli O157:H7 even with the use of immunomagnetic separation and culturing to isolate the organism (7).
Previous evaluations of conventional methods for the detection of culture-based E. coli O157:H7 have operated in a range of 0.1 to >100 CFU/g, with the majority of these studies investigating levels higher than 1 CFU/g (2, 6, 10, 15, 16). Our study aimed for significantly lower inoculation levels (94% below 1 CFU/g) in order to further examine the effects of background flora on the detection rates of various assays and to evaluate the growth and detection rates near the regulatory limit of 1 CFU/25 g of ground beef.
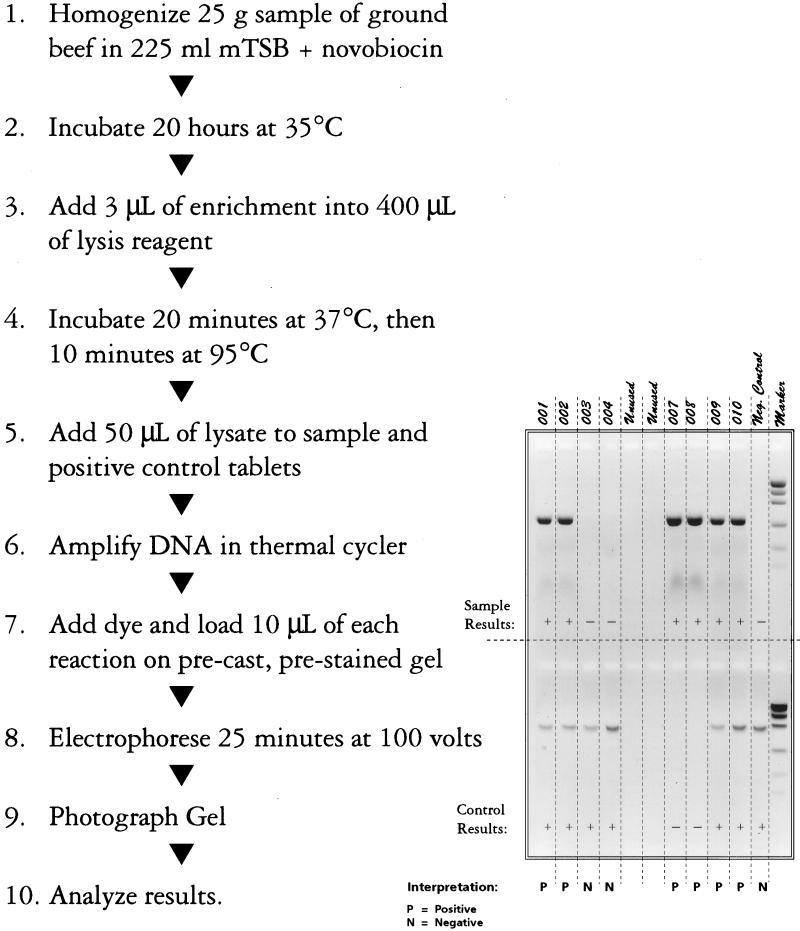
New selective media have been developed to increase the effectiveness of E. coli O157:H7 isolation (16, 17), as have various other methods (8). BAX for Screening/E. coli O157:H7 (Qualicon, Wilmington, Del.) (Fig. 1) is a commercial genetics-based assay (1) using the technology of PCR (12). The study reported here compares the performance of the BAX system assay with traditional culture, enhanced selective agars, and an immunodiffusion method. Because the natural occurrence of E. coli O157:H7 is both sporadic and usually at low levels, the study emphasized the detection of E. coli O157:H7 from ground beef samples inoculated with very few organisms (fewer than 3 CFU/g).
FIG. 1.
Bax for Screening/E. coli O157:H7 assay procedure.
MATERIALS AND METHODS
Ground beef samples (25 g each) were obtained from various local retail outlets in Madison, Wis. Prior to inoculation, each isolate was grown up in brain heart infusion broth (incubation at 35°C for 16 to 18 h) and transferred twice. Cultures prepared in this fashion were serially (1 to 10) diluted in 0.1% peptone diluent prior to addition to the meatballs. Aliquots (0.1 ml) of the appropriate dilution were added to individual meatballs, and the inoculum was allowed to soak in for 5 min. Inoculation levels were confirmed by spread plating 0.1-ml portions of the dilutions onto Trypticase soy agar plates and incubating at 35°C for 46 to 48 h.
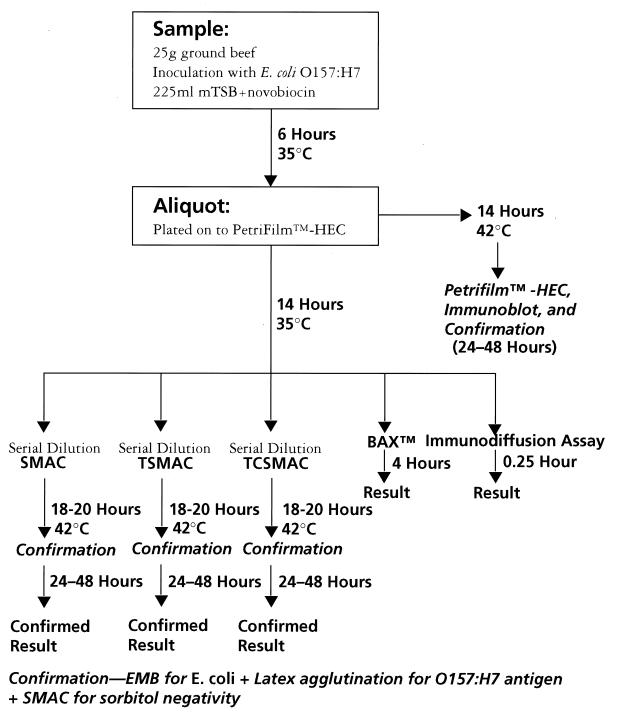
These samples were inoculated at high (7 to 64 CFU/25 g [0.28 to 2.56 CFU/g]) and low (0.7 to 6.4 CFU/25 g [0.03 to 0.26 CFU/g]) levels with 1 of 10 E. coli strains (9 O157:H7 strains and 1 O157:HNM strain [Table 1]). These ten strains were previously isolated from food or during outbreak investigations (4). All strains were received from A. M. Sharar, Microbiology Division, FSIS, U.S. Department of Agriculture. A 225-ml portion of modified Trypticase soy broth (mTSB) plus novobiocin was added to each sample (Fig. 2). Sample enrichments were incubated for 6 h, and an aliquot was removed and reincubated for 14 h at 35°C. The 6-hour aliquot was plated onto Petrifilm-HEC (3M Co., Minneapolis, Minn.) and incubated for 14 h at 42°C. After 18 to 20 h, aliquots of enrichment cultures were removed, serially diluted (1:10), and plated onto sorbitol MacConkey agar (SMAC), tellurite SMAC (TSMAC), and tellurite cefixime SMAC (TCSMAC) (16, 17). Agar plates were incubated at 42°C overnight (18 to 20 h). Aliquots of enrichment cultures were also removed and processed in parallel through a commercial immunodiffusion system assay (VIP BioControl Systems, Inc., Bellevue, Wash.) and through the BAX system assay according to the manufacturer’s directions. Petrifilm-HEC plates were immunoblotted, and positive colonies were picked for confirmation (11, 15). Up to six characteristic colonies were picked on as many as three dilution plates from each of the three agars and Petrifilm for confirmation.
TABLE 1.
Samples used in the study
| Straina | Meat sampleb | Inoculation level (CFU/25 g) | Strain | Meat sample | Inoculation level (CFU/25 g) | Strain | Meat sample | Inoculation level (CFU/25 g) | Strain | Meat sample | Inoculation level (CFU/25 g) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C | 1.5 | 4 | A | 2.3 | 7 | C | 1.5 | 10 | C | 1 | |||
| 1 | C | 15 | 4 | A | 23 | 7 | C | 15 | 10 | C | 10 | |||
| 1 | D | 1.5 | 4 | B | 2.3 | 7 | D | 1.5 | 10 | D | 1 | |||
| 1 | D | 15 | 4 | B | 23 | 7 | D | 15 | 10 | D | 10 | |||
| 1 | E | 0.8 | 4 | E | 1.7 | 7 | E | 1.5 | 10 | G | 2.3 | |||
| 1 | E | 8 | 4 | E | 17 | 7 | E | 15 | 10 | G | 23 | |||
| 1 | F | 0.8 | 4 | F | 1.7 | 7 | F | 15 | 10 | H | 23 | |||
| 1 | F | 8 | 4 | F | 17 | 7 | F | 1.5c | 10 | H | 2.3c | |||
| 1 | I | 16 | 4 | I | 2.2 | 7 | I | 1.5 | 10 | K | 1.3 | |||
| 1 | I | 1.6c | 4 | I | 22 | 7 | I | 15 | 10 | K | 13 | |||
| 1 | J | 16 | 4 | J | 2.2 | 7 | J | 15 | 10 | L | 1.3 | |||
| 1 | J | 1.6c | 4 | J | 22 | 7 | J | 1.5c | 10 | L | 13 | |||
| 1 | M | 1.9c | 4 | M | 18 | 7 | M | 13 | 10 | O | 1.3 | |||
| 1 | M | 19c | 4 | M | 1.8c | 7 | M | 1.3c | 10 | O | 13 | |||
| 1 | N | 19 | 4 | N | 1.8 | 7 | N | 1.3 | 10 | P | 1.3 | |||
| 1 | N | 1.9c | 4 | N | 18 | 7 | N | 13 | 10 | P | 13 | |||
| 1 | Q | 5.9 | 4 | Q | 1.8 | 7 | Q | 5.1 | 10 | S | 1.6c | |||
| 1 | Q | 59 | 4 | Q | 18 | 7 | Q | 51 | 10 | S | 16c | |||
| 1 | R | 5.9 | 4 | R | 18 | 7 | R | 5.1 | 10 | T | 16 | |||
| 1 | R | 59 | 4 | R | 1.8c | 7 | R | 51 | 10 | T | 1.6c | |||
| 2 | A | 1.4 | 5 | A | 1.1 | 8 | C | 1.3 | ||||||
| 2 | A | 14c | 5 | A | 11 | 8 | C | 13 | ||||||
| 2 | B | 1.4c | 5 | B | 1.1 | 8 | D | 13 | ||||||
| 2 | B | 14c | 5 | B | 11 | 8 | D | 1.3c | ||||||
| 2 | E | 3.1c | 5 | G | 1.6 | 8 | G | 1.3 | ||||||
| 2 | E | 31c | 5 | G | 16 | 8 | G | 13 | ||||||
| 2 | F | 3.1c | 5 | H | 1.6 | 8 | H | 13 | ||||||
| 2 | F | 31c | 5 | H | 16 | 8 | H | 1.3c | ||||||
| 2 | I | 1.4c | 5 | K | 1.9 | 8 | K | 0.86 | ||||||
| 2 | I | 14c | 5 | K | 19 | 8 | K | 8.6 | ||||||
| 2 | J | 1.4c | 5 | L | 1.9 | 8 | L | 0.86 | ||||||
| 2 | J | 14c | 5 | L | 19 | 8 | L | 8.6 | ||||||
| 2 | M | 2c | 5 | O | 1.9 | 8 | O | 2 | ||||||
| 2 | M | 20c | 5 | O | 19 | 8 | O | 20 | ||||||
| 2 | N | 2c | 5 | P | 1.9 | 8 | P | 2 | ||||||
| 2 | N | 20c | 5 | P | 19 | 8 | P | 20 | ||||||
| 2 | Q | 6.4c | 5 | S | 2.5c | 8 | S | 2.3 | ||||||
| 2 | Q | 64c | 5 | S | 25 | 8 | S | 23 | ||||||
| 2 | R | 6.4c | 5 | T | 2.5 | 8 | T | 2.3 | ||||||
| 2 | R | 64c | 5 | T | 25 | 8 | T | 23 | ||||||
| 3 | C | 1 | 6 | A | 1.7 | 9 | C | 1.1 | ||||||
| 3 | C | 10 | 6 | A | 17 | 9 | C | 11 | ||||||
| 3 | D | 1 | 6 | B | 1.7 | 9 | D | 1.1 | ||||||
| 3 | D | 10 | 6 | B | 17 | 9 | D | 11 | ||||||
| 3 | E | 1.9 | 6 | G | 1.5 | 9 | G | 0.7 | ||||||
| 3 | E | 19 | 6 | G | 15 | 9 | G | 7.3 | ||||||
| 3 | F | 19 | 6 | H | 1.5 | 9 | H | 0.7 | ||||||
| 3 | F | 1.9c | 6 | H | 15 | 9 | H | 7.3 | ||||||
| 3 | I | 0.9 | 6 | K | 0.98 | 9 | K | 1.6 | ||||||
| 3 | I | 9 | 6 | K | 9.8 | 9 | K | 16 | ||||||
| 3 | J | 9 | 6 | L | 0.98 | 9 | L | 1.6 | ||||||
| 3 | J | 0.9c | 6 | L | 9.8 | 9 | L | 16 | ||||||
| 3 | M | 12 | 6 | O | 1.2 | 9 | O | 1.8 | ||||||
| 3 | M | 1.2c | 6 | O | 12 | 9 | O | 18 | ||||||
| 3 | N | 1.2 | 6 | P | 1.2 | 9 | P | 1.8 | ||||||
| 3 | N | 12 | 6 | P | 12 | 9 | P | 18 | ||||||
| 3 | Q | 4.5 | 6 | S | 33 | 9 | S | 1.3 | ||||||
| 3 | Q | 45 | 6 | S | 3.3c | 9 | S | 13 | ||||||
| 3 | R | 45 | 6 | T | 3.3 | 9 | T | 13 | ||||||
| 3 | R | 4.5c | 6 | T | 33 | 9 | T | 1.3c |
a Strains: 1, FSIS 014-90 (E. coli O157:HNM); 2, FSIS 062-93 (E. coli O157:H7); 3, FSIS 063-93 (E. coli O157:H7); 4, FSIS 064-93 (E. coli O157:H7); 5, FSIS 380-94 (E. coli O157:H7 [4]); 6, FSIS 300-94 (E. coli O157:H7); 7, FSIS 065-93 (E. coli O157:H7); 8, FSIS 264-94 (E. coli O157:H7); 9, FSIS 311-94 (E. coli O157:H7); 10, FSIS 413-95 (E. coli O157:H7).
b Meat samples (type of meat, percent lean, aerobic plate count [CFU per gram]): A, ground round, >85, 4.6 × 105; B, ground chuck, 80 to 85, 4.9 × 105; C, ground chuck, 80 to 85, 8.3 × 105; D, ground beef, 75, 1.5 × 106; E, ground chuck, 80 to 85, 2.4 × 106; F, ground beef, 75, 2.0 × 106; G, ground round, >85, 1.7 × 106; H, ground beef, 75, 3.9 × 106; I, ground chuck, 80 to 85, 6.9 × 105; J, ground beef, 75, 8.0 × 106; K, ground beef, 75, 2.2 × 105; L, ground chuck, 80 to 85, 7.0 × 103; M, ground chuck, 80 to 85, 4.2 × 106; N, ground round, >85, 1.6 × 103; O, ground beef, 75, 6.3 × 103; P, ground chuck, 80 to 85, 4.5 × 104; Q, ground chuck, 80 to 85, 7.0 × 105; R, ground beef, 75, 8.0 × 106; S, ground beef, 75, 2.6 × 106; T, ground chuck, 80 to 85, 2.3 × 105.
c The sample failed to yield a positive result by any method.
FIG. 2.
Experiment design. EMB, eosin-methylene blue.
Confirmation included isolation on SMAC, streaking onto eosin-methylene blue agar for E. coli confirmation, and latex agglutination testing for O157 and H7 antigens (RIM O157:H7; Remel, Lenexa, Kans.).
Note that the enrichment method used here is not that recommended by 3M for the Petrifilm-HEC immunoblot. Differences include the use of mTSB plus novobiocin (specified for the immunodiffusion assay) rather than modified E. coli plus novobiocin and the elimination of shaking during the 6-h initial incubation. For these reasons, Petrifilm-HEC is regarded as a supplemental tool for the recovery of E. coli O157:H7 and not as a screening (detection) method.
The BAX system assay is a simple, PCR-based method employing tableted PCR reagents. Primer sequences specific for O157:H7 were identified following screening against a panel consisting of 113 target and 176 non-O157:H7 E. coli strains (1). The primer set chosen amplified a specific 530-bp product in 99% of the target strains and 0% of the nontarget strains. These primers were tableted along with Taq polymerase, excipients, and deoxynucleoside triphosphates. The assay method consists of an overnight enrichment of homogenized ground beef in mTSB plus novobiocin, a simple sample lysate preparation, direct addition of 50 μl of the lysate to a tablet, and thermal cycling followed by agarose gel detection. Lysate preparation consists of 3 μl of enrichment solution added to 40 μl of buffer containing protease. The solution is treated at 37°C for 20 min and at 95°C for 10 min and then chilled for 5 min.
Fifty microliters of lysate is added to the PCR sample and positive control (containing positive control DNA construct) tablets in 0.2-ml tubes. The tubes are cycled in a GeneAmp 9600 thermal cycler (Perkin-Elmer, Branchburg, N.J.) for 35 cycles of a 94°C, 15-s denaturation followed by annealing temperature-time combinations of 70°C-2 min or 72°C-3 min. Upon completion of cycling, 10 μl of loading dye solution was added to each tube. Next, 10 μl of each sample was loaded onto a precast, prestained (with ethidium bromide), 2% agarose gel (FMC) and subjected to electrophoresis for 25 min at 100 V in 0.5× Tris-borate-EDTA buffer. A DNA molecular weight standard (Gibco BRL, Gaithersburg, Md.) was included on each gel. This standard (mass ladder) has fragment sizes of 100, 200, 400, 800, 1,200, and 2,000 bp.
During development of the BAX system assay, sensitivity studies that demonstrated the ability of the assay to produce a positive band for 105 CFU of E. coli O157:H7 per ml in the presence of 109 CFU of background flora per ml in the enriched sample were carried out (data not shown).
Twenty ground beef samples with various fat contents were processed. Initial aerobic plate counts, inoculation levels, inoculation strains, and ground beef types for the samples are given in Table 1. Each of the 10 strains was inoculated five times at two levels in various portions of ground beef to yield a total of 200 samples. Half of these samples were inoculated at levels higher than 7 CFU/25 g (0.28 CFU/g), and the other half were inoculated at levels below 6.4 CFU/25 g (0.26 CFU/g).
RESULTS
Overview.
As previously stated, 200 ground beef samples were prepared and inoculated at very low levels. We subjected inoculated samples to a battery of detection and recovery procedures; 42 samples were negative by all detection procedures. Successfully inoculating samples at these extremely low but significant levels was difficult. It is not unusual to fail to detect or to recover pathogens inoculated into samples at levels of <5 CFU/25 g. This is largely due to statistical considerations. Of the 200 samples in this study, 98 were inoculated at <5.0 CFU/25 g. By using a Poisson distribution to calculate the probability of X = 0 cells in an inoculum of λ concentration, 20 samples are predicted to be uninoculated and thus to yield negative results by all methods studied. In addition, one strain (FSIS-062-93 [strain 2 in Table 1]) failed to produce a positive result by any method for 19 of 20 samples inoculated with 1.4 to 64 CFU/25 g (0.056 to 2.56 CFU/g). We suspect that this strain had difficulty competing with the other microflora present in the ground beef samples. These 19 failures combined with the 20 statistically predicted failures account for 39 of the 42 inoculated samples that failed to yield a positive result despite the use of extensive confirmation procedures. We are forced to conclude that inoculated samples yielding negative results by every procedure tested did not receive any viable E. coli O157.
An additional recovery method using 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide (BCIG) in SMAC (14) was also examined. This method, as recommended by FSIS, uses 150- by 15-mm petri plates. Because of the high cost of BCIG, our experiment used standard laboratory plates (100 by 15 mm). Use of standard plates for SMAC-BCIG resulted in recovery of only 13 of 40 inoculated samples (data not shown). The high level of background organisms present in the enrichment broth may account for this unacceptable performance. As this method failed to yield any significant improvement in recovery rate compared with other cultural techniques employed in this study, its use was not continued.
Tabulated positive results are given in Table 2. Positive results for BAX system assays are indicated by the presence of a band in the appropriate location, regardless of intensity (Fig. 1). Immunodiffusion results are considered positive by the presence of a band in the sample location as indicated in the manufacturer’s instructions. Occasionally, immunodiffusion results were rerun due to a very weak reaction. In some instances, reruns were positive, and in other cases, they were negative; in each case, the rerun results were recorded for tabulation. Results from the SMAC, TSMAC, TCSMAC, and Petrifilm-HEC methods are considered positive if a confirmed E. coli O157:H7 colony was identified. As previously explained, 42 samples were negative by all methods. Therefore, all calculations are based on the 158 inoculated samples which were positive by at least one method, rather than on 200 samples.
TABLE 2.
Screening results for inoculated samples found positive by any detection or recovery method
| Data seta | No. (%) positive
|
|||||
|---|---|---|---|---|---|---|
| Expected | BAX system | Immunodiffusion | TCSMAC | TSMAC | SMAC | |
| Total | 158 (100) | 151 (96.5) | 113 (71.5) | 61 (39) | 61 (39) | 28 (17.7) |
| High only | 88 (100) | 88 (100) | 72 (81.8) | 39 (44.3) | 37 (42) | 22 (25) |
| Low only | 70 (100) | 63 (90) | 41 (58.5) | 22 (31.4) | 24 (34) | 6 (8.5) |
a High, inoculation level of above 0.28 CFU/g; low, inoculation level of 0.03 to 0.26 CFU/g.
Table 2 also contains data for two subsets of samples broken out by inoculation level. High refers to samples inoculated with E. coli O157:H7 levels of 7 to 64 CFU/25 g (0.28 to 2.56 CFU/g) per sample. Low refers to samples inoculated with between 0.7 and 6.4 CFU/25 g (between 0.03 and 0.26 CFU/g).
Conventional agar results.
With individual conventional agars, recovery rates ranged from 17.7 to 39% for all samples (Table 2), from 25 to 44.3% for the high inoculation levels, and from 8.5 to 31.4% for the low inoculation levels.
For the purpose of analysis, four methods—Petrifilm-HEC, TCSMAC, TSMAC, and SMAC—are considered recovery methods capable of yielding confirmed colonies. A total of 107 of 158 positive samples (Table 3) yielded a confirmed E. coli O157:H7 isolate by at least one of the four recovery procedures.
TABLE 3.
Comparison of confirmation data for BAX system assay versus immunodiffusion assay
| Data seta | No.
|
||
|---|---|---|---|
| BAX system positive | Immunodiffusion positive | Confirmed by isolation | |
| Total | 151 | 113 | 107 |
| High | 88 | 72 | 63 |
| Low | 63 | 41 | 44 |
a High, inoculation level of above 0.28 CFU/g; low, inoculation level of 0.03 to 0.26 CFU/g.
Immunodiffusion method results.
The immunodiffusion method recorded a combined 71.5% of the inoculated samples as positive (Table 2). This method detected 81.8% of the high-inoculation samples and 58.5% of the low-inoculation samples as positive.
Broths from 18 of the 107 culture-confirmed samples were negative with the immunodiffusion method, for a demonstrated false-negative rate of 16.8%. False negatives spanned both high- and low-inoculum subsets.
BAX system results.
At high inoculation levels (above 0.28 CFU/g), the BAX system assay detected 100% of the samples, with no false negatives, while at the low inoculation levels (0.03 to 0.26 CFU/g), the BAX system assay detected 90% of the samples. Overall, the BAX system assay reported a combined 96.5% of inoculated samples as positive (Table 2).
Of the 107 samples which yielded a confirmed positive result by cultural methods (Table 3), the BAX system assay reported five of those broths as negative, for a demonstrated false-negative rate of 4.7%. All false-negative samples were from the low-inoculum subset. The false-negative rates of the BAX system assay (4.7%) and the immunodiffusion method (16.8%) are significantly different (P = 0.05).
Even use of a combination of four recovery methods and definition of a positive as any positive result by any of the four methods yields an unacceptably low recovery rate. Because all of these positive results were obtained from inoculated samples, it is our contention that all of these BAX system-positive results are true positives. Our data demonstrate the extreme difficulty of recovering low but significant levels of E. coli O157:H7 by cultural methods. For samples detected as positive by the BAX system assay, additional culture-based methods, perhaps in conjunction with immunomagnetic capture techniques (7), may be required. Such additional confirmation may be examined in a future study.
DISCUSSION
The results of the detection of low inoculum levels of E. coli O157:H7 in ground beef on individual conventional agars were not encouraging. The data shown here also indicate that as contamination levels decrease, the ability to recover E. coli O157:H7 is also reduced. No single conventional agar had a high detection rate (the best individual agar had a recovery rate of only 39%). These data suggest that routine cultural isolation and recovery of E. coli O157:H7 in ground beef are best achieved with a combination of agars, not by relying on any single type. However, the effort needed to perform four isolation procedures in parallel is extreme and not feasible in routine laboratory operations. Additionally, even this extreme effort would have recovered only 67% (107 of 158) of the positive samples.
Given that it would be highly unusual for a laboratory to perform four conventional methods simultaneously, the need for a single, rapid, reliable detection method is increased. The BAX system assay gave positive results for 96.5% of samples that proved positive by any available test. This rate of detection is significantly better statistically than those of the other methods, especially at very low inoculation levels. The BAX system is easy to use and requires less labor than conventional methods. The timeliness and superior sensitivity illustrated here make the BAX for Screening/E. coli O157:H7 assay the ideal method for the detection of this potentially lethal pathogen in difficult food matrices such as ground beef.
ACKNOWLEDGMENTS
We thank D. Duescher, J. Siehr, and P. M. Mrozinski for technical assistance and A. M. Sharar, Microbiology Division, FSIS, USDA, for supplying the isolates used in this study.
REFERENCES
- 1.Barbour W M, Ecret L D, Jensen M A, Hazel J W, Jackson R E, Stoltzfus A M, Tice G. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiol; 1996. A PCR-based method for the detection of E. coli O157:H7 from ground beef, abstr. P-5; p. 396. [Google Scholar]
- 2.Bennett A R, MacPhee S, Betts R P. Evaluation of methods for the isolation and detection of Escherichia coli O157:H7 in minced beef. Lett Appl Microbiol. 1995;20:375–379. doi: 10.1111/j.1472-765x.1995.tb01325.x. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Update: multistate outbreak of Escherichia coli O157:H7 infections from Western United States, 1992–1993. Morbid Mortal Weekly Rep. 1993;42:258–263. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Escherichia coli O157:H7 outbreak linked to commercially distributed dry cured salami—Washington and California, 1994. Morbid Mortal Weekly Rep. 1995;44:157–160. [PubMed] [Google Scholar]
- 5.Chapman P A, Siddons C A. Evaluation of a commercial enzyme immunoassay (EHEC-Tek) for detecting Escherichia coli O157:H7 in beef and beef products. Food Microbiol. 1996;13:175–182. [Google Scholar]
- 6.Deng M V, Fratamico P M. A multiplex PCR for rapid identification of Shiga-like toxin-producing Escherichia coli O157:H7 isolated from food. J Food Prot. 1996;59:570–576. doi: 10.4315/0362-028X-59.6.570. [DOI] [PubMed] [Google Scholar]
- 7.Firstenberg-Eden R, Sullivan N M. EZ coli rapid detection system: a rapid method for the detection of Escherichia coli O157:H7 in meat and other foods. J Food Prot. 1997;60:219–225. doi: 10.4315/0362-028X-60.3.219. [DOI] [PubMed] [Google Scholar]
- 8.Giese J. Rapid microbiological testing kits and instruments. Food Technol. 1995;49(7):64–70. [Google Scholar]
- 9.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 10.Jinneman K C, Trost P A, Hill W E, Weagant S D, Bryant J L, Kaysner C A, Wekell M M. Comparison of template preparation methods from foods for amplification of Escherichia coli O157 shiga-like toxins Type I and II DNA by multiplex polymerase chain reaction. J Food Prot. 1995;58:722–726. doi: 10.4315/0362-028X-58.7.722. [DOI] [PubMed] [Google Scholar]
- 11.Johnson J L, Rose B E, Sharar A K, Ransom G M, Lattuada C P, McNamara A M. Methods used for detection and recovery of Escherichia coli O157:H7 associated with a food-borne disease outbreak. J Food Prot. 1995;58:597–603. doi: 10.4315/0362-028X-58.6.597. [DOI] [PubMed] [Google Scholar]
- 12.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 13.Niroomand F, Lord C. Comparison of rapid techniques for the detection of Escherichia coli O157:H7. J Rapid Methods Automation Microbiol. 1994;3:85–96. [Google Scholar]
- 14.Okrend A J, Rose B E, Lattuada C P. Use of 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide in MacConkey sorbitol agar to aid in the isolation of Escherichia coli O157:H7 from ground beef. J Food Prot. 1990;53:941–943. doi: 10.4315/0362-028X-53.11.941. [DOI] [PubMed] [Google Scholar]
- 15.Okrend A J, Rose B E, Matner R. An improved screening method for the detection and isolation of Escherichia coli O157:H7 from meat, incorporating the 3M Petrifilm™ Test Kit-HEC for hemorrhagic Escherichia coli O157:H7. J Food Prot. 1990;53:936–940. doi: 10.4315/0362-028X-53.11.936. [DOI] [PubMed] [Google Scholar]
- 16.Weagant S D, Bryant J L, Jinneman K C. An improved rapid technique for isolation of Escherichia coli O157:H7 from foods. J Food Prot. 1995;58:7–12. doi: 10.4315/0362-028X-58.1.7. [DOI] [PubMed] [Google Scholar]
- 17.Zadik P M, Chapman P A, Siddons C A. Use of tellurite for the selection of verocytoxigenic Escherichia coli O157. J Med Microbiol. 1993;39:155–158. doi: 10.1099/00222615-39-2-155. [DOI] [PubMed] [Google Scholar]