Summary
Alternative splicing is a prevalent gene-regulatory mechanism, with over 95% of multi-exon human genes estimated to be alternatively spliced. Here, we describe a tissue-specific, developmentally regulated, highly conserved, and disease-associated alternative splicing event in exon 7 of the eyes absent homolog 3 (Eya3) gene. We discovered that EYA3 expression is vital to the proliferation and differentiation of myoblasts. Genome-wide transcriptomic analysis and mass spectrometry-based proteomic studies identified SIX homeobox 4 (SIX4) and zinc finger and BTB-domain containing 1 (ZBTB1), as major transcription factors that interact with EYA3 to dictate gene expression. EYA3 isoforms differentially regulate transcription, indicating that splicing aids in temporal control of gene expression during muscle cell differentiation. Finally, we identified RNA-binding fox-1 homolog 2 (RBFOX2) as the main regulator of EYA3 splicing. Together, our findings illustrate the interplay between alternative splicing and transcription during myogenesis.
Subject areas: Physiology, Molecular biology, Omics
Graphical abstract
Highlights
-
•
EYA3 expression is critical for myoblast proliferation and differentiation
-
•
EYA3 interacts with transcription factors SIX4 or ZBTB1 to dictate gene expression
-
•
EYA3 isoforms differentially regulate transcription during myogenesis
-
•
RBFOX2 is the main RNA-binding protein regulating EYA3 splicing
Physiology; Molecular biology; Omics
Introduction
Alternative splicing is a pre-messenger RNA (pre-mRNA) processing mechanism in which exons are selectively included or excluded to generate different mature transcripts, thereby introducing extensive transcript diversity from a limited number of genes. The resulting mRNAs may contain regulatory domains (e.g., binding regions for microRNAs which can regulate mRNA stability) or result in the production of different protein isoforms with various functions. The advent of high-throughput sequencing several decades ago substantiated alternative splicing as a ubiquitous step in RNA processing, with over 95% of intron-containing human genes estimated to be alternatively spliced.1,2 More recent studies have uncovered that splicing patterns change during organ development and help to define unique gene expression profiles found in different bodily tissues.3,4,5 Adult brain, testes, heart, and skeletal muscle display particularly high levels of tissue-specific alternative splice isoform expression.3,6,7,8,9,10,11
Although functional studies are surprisingly limited, examinations of individual splicing events have revealed profound roles for alternative splice isoforms in the induction of myogenic gene expression and the acquisition of characteristic features of adult skeletal muscle.12,13,14,15,16,17,18 For example, the myocyte enhancer factor 2D (Mef2d) gene encodes a transcription factor and undergoes a switch in the use of two mutually exclusive exons during muscle cell differentiation (myogenesis).12,13 The MEF2Dα2 isoform is expressed in later stages of differentiation and is less sensitive to inhibitory phosphorylation by the protein kinase A (PKA), allowing for the activation of muscle-specific genes.13
Dysregulation of alternative splicing is common in muscular diseases.17,19,20 In humans with congenital myotonic dystrophy (CDM), myotonic dystrophy type 1 (DM1), or myotonic dystrophy type 2 (DM2), and in inherited myopathy of Great Danes (IMGD), splicing of the muscle-specific exon of bridging integrator 1 (BIN1) reverts from adult to fetal patterns concomitant with disruptions in membrane organization.17,19 Re-expression of the muscle-specific BIN1 isoform in patient-derived cells rescues transverse tubule (T-tubule) formation.17 Similarly, correcting mis-splicing of the chloride channel 1 (CIC1) rescued defects in muscle relaxation (myotonia) in two mouse models of DM1.20 In Duchenne muscular dystrophy, the correction of mis-splicing in dystrophin mRNA has proven to be highly effective in the clinic to alleviate disease symptoms.21,22 These examples illustrate that alternative splice isoforms have the capacity to contribute to muscle architecture, gene expression programs, and are potentially powerful therapeutic targets for muscle diseases. Therefore, the identification of the roles of tissue-specific splice isoforms is of high interest. Here, we investigate a previously uncharacterized splice variant of the eyes absent homolog 3 (Eya3) gene in the context of myogenesis.
The eyes absent homolog (EYA) family of proteins are transcriptional cofactors as well as bifunctional threonine and tyrosine phosphatases.23,24 The Drosophila homolog, Eya, is required for fly eye development, and ectopic expression of Eya in other tissues is sufficient to produce eye-like structures in the legs and antennae of flies.25 In vertebrates, the EYA family regulates the development of multiple organs including the kidney, ear, heart, skeletal muscle, and craniofacial tissues.24,26,27,28 In early skeletal muscle development, EYA3 plays a key role in the activation of the myogenic transcription factor, myogenin (MYOG).26,29,30,31 Mechanistically, EYA family members are translocated to the nucleus by interacting with the since oculis homeobox (SIX) family of transcription factors where they bind to the promoters of target genes.29,30 Still, the transcriptional targets of EYA3 have not been identified on a genome-wide basis, and it is unknown whether other regulatory factors contribute to EYA3 mediated transcriptional control.
In this work, we investigated an evolutionarily conserved, developmentally regulated, tissue-specific, and disease associated splicing event in the Eya3 gene. We found that both EYA3 splice variants (i.e., the isoforms including exon 7 (EYA3+ex7) or lacking exon 7 (EYA3Δex7)) are vital for muscle cell proliferation and fusion. Mass spectrometry-based analysis of EYA3 isoforms coupled with RNA-sequencing (RNA-seq) studies of EYA3 depleted cells revealed an uncharacterized mechanism by which EYA3 partners with the zinc finger and BTB-domain containing 1 (ZBTB1) and SIX homeobox 4 (SIX4) to regulate gene expression. We discovered that inclusion of exon 7 attenuates EYA3 transcriptional capacity which may facilitate myogenic transcriptional programs. Finally, we demonstrated that an increase in the expression of the RNA-binding fox-1 homolog 2 (RBFOX2) during myogenesis mediates the inclusion of Eya3 exon 7. Together, our findings implicate two highly evolutionarily conserved EYA3 isoforms in the establishment of the myogenic transcriptional program.
Results
Alternative splicing of EYA3 exon 7 is evolutionarily conserved, tissue-specific, and developmental stage-specific
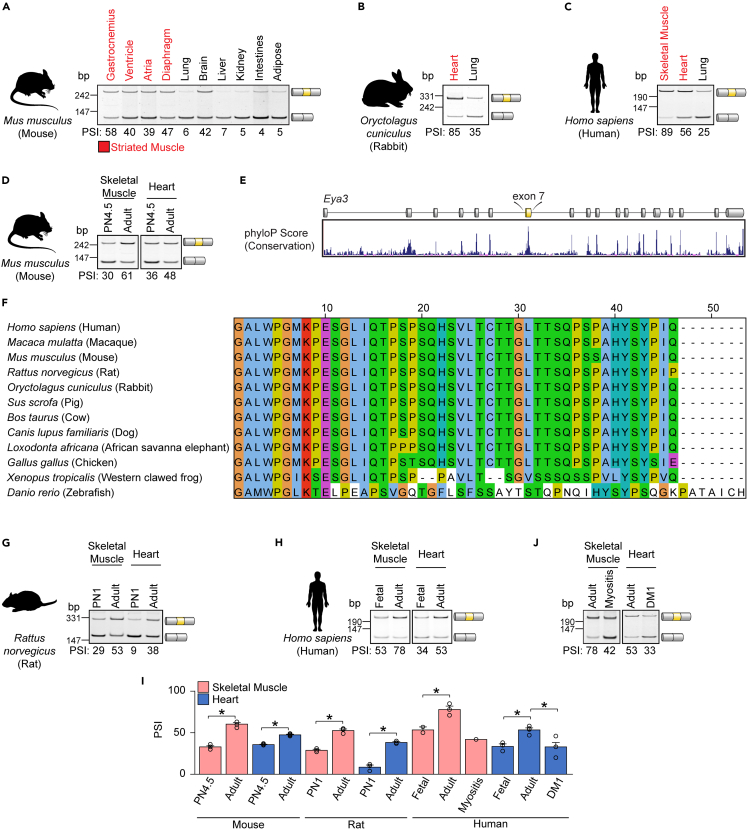
Inspection of previously generated RNA-seq data7 from cardiomyocytes freshly isolated from neonatal and adult mouse cardiac ventricular tissues revealed that exon 7 in the Eya3 gene is developmentally regulated by alternative splicing (Figure S1). Given that splice isoforms are often expressed in a tissue-specific manner, we first sought to comprehensively determine the splicing pattern of Eya3 in a broad set of tissues. We thus characterized Eya3 exon 7 splicing in a panel of adult mouse bodily tissues by reverse transcription PCR (RT-PCR) assays (Figure 1A). Primers were designed to hybridize to the constitutive exons (exon 6 and exon 8) flanking the alternative exon 7 of Eya3 pre-mRNA (Table S1), and samples were subjected to PCR amplification before being resolved by gel electrophoresis. The relative proportion of transcripts including the alternative region (Eya3 exon 7 in this case) was determined by densitometry and expressed as the percent spliced in (PSI).2 Expression of the Eya3 transcripts including exon 7 (Eya3+ex7) was prominent in gastrocnemius muscles, ventricle, atria, diaphragm, and brain, but was negligible in lung, liver, kidney, intestines, and adipose tissue (Figure 1A). Further examination of a panel of adult mouse striated muscle tissues indicated that Eya3+ex7 transcripts are expressed in the ventricles and atria, as well as in a variety of skeletal muscles, with the highest levels of exon 7 inclusion found in the tibialis anterior (TA, 73%), the gastrocnemius (72%), the extensor digitorum longus (EDL, 64%), and the soleus (54%) muscles and relatively lower levels in the ventricles (48%), atria (43%), and the flexor digitorum brevis (FDB, 42%) muscle (Figure S2). Similarly, in adult rabbit heart and human striated muscle, expression of Eya3+ex7 transcript was highly favored, compared to the isoform lacking exon 7 (Eya3Δex7) (Figures 1B and 1C). These data indicate that Eya3 pre-mRNA is alternatively spliced in a tissue-specific manner, with the highest levels of Eya3+ex7 expression in striated muscle and brain tissues.
Figure 1.
Alternative splicing of Eya3 exon 7 is evolutionarily conserved, tissue-specific, and developmental stage-specific
(A–C) Eya3 exon 7 splicing was profiled in a panel of adult mouse (A), rabbit (B), and human (C) tissues by reverse transcription PCR (RT-PCR). Striated muscle tissues are indicated in red.
(D) The inclusion of Eya3 exon 7 was determined by RT-PCR assays in striated muscle tissues from postnatal day 4.5 (PN4.5) and adult mice.
(E) Plot of basewise evolutionary conservation of the Eya3 gene as determined by phyloP score and visualized in the UCSC genome browser.
(F) Alignment of the Eya3 exon 7 amino acid sequence from 12 animal species. Colors based on the agreement with the consensus sequence and physiochemical properties of amino acids determined by Clustal X: hydrophobic residue (blue), positive charged residue (red), negative charged residue (magenta), polar residue (green), cysteine residue (pink), glycine residue (orange), proline residue (yellow), aromatic residue (cyan), non-conserved residue (white).
(G and H) The inclusion of Eya3 exon 7 was determined by RT-PCR assays at the indicated developmental timepoints in striated muscle tissues obtained from rats (G) and humans (H).
(I) Percent spliced in (PSI) of Eya3 exon 7 was determined by densitometry. Results are shown as mean ± SEM. ∗p < 0.05, Student’s T test. (n = 3–4), except in human fetal skeletal muscle (n = 2) and myositis (n = 1) samples, due to limited availability.
(J) The inclusion of Eya3 exon 7 was determined by RT-PCR assays in humans with myotonic dystrophy type 1 (DM1) and myositis. bp, base pairs. See also Figures S1–S5, Tables S1 and S2.
Because tissue-specific splicing patterns are often acquired during organ maturation, we next examined Eya3 exon 7 inclusion at a variety of developmental stages (Figure S3). In mouse neonatal and adult tissues, we observed a gradual increase in Eya3 exon 7 inclusion, up to 43% in adult brain (Figures S3A and S3G), 48% in adult heart, and 61% in adult skeletal muscle (Figure 1D). Notably, we did not observe developmental changes in Eya3 exon 7 inclusion in mouse lung, kidney, liver, spleen, or intestines (Figures S3B–S3G).
Phylogenetic sequence analysis (basewise conservation determined by phlyoP32) of the Eya3 gene revealed high conservation of the exonic sequences, with especially high scores estimated for sequences within and surrounding exon 7 (Figure 1E). Consistently, we observed a high degree of exon 7 amino acid sequence conservation amongst a diverse set of vertebrate species, including mammals, birds, amphibians, and fish (Figure 1F).
We next asked if the developmental regulation of Eya3 exon 7 splicing was also evolutionarily conserved in striated muscle tissues. Thus, we evaluated Eya3 exon 7 splicing patterns in rat, human, zebrafish, and chicken during development (Figures 1G, 1H, and S4). Strikingly, amongst all organisms examined, we observed a higher level of Eya3 exon 7 inclusion in adult striated muscle tissues compared to earlier stages of development (Figure 1I). These data demonstrate that the tissue-specific, developmentally regulated splicing of Eya3 exon 7 is highly conserved during evolution and, therefore, likely to have functional implications.
EYA3 exon 7 is mis-spliced in human tissues and mouse models of striated muscle diseases
Because in disease contexts adults can express fetal splice variants,17,19,20,33 we evaluated Eya3 splicing in the skeletal muscle of adult humans diagnosed with myositis, an inflammatory muscle-wasting disease34 and in the hearts of adult individuals with DM1. Myositis and DM1 adult samples (Table S2) exhibited lower levels of exon 7 inclusion in comparison with controls (Figures 1I and 1J). Eya3 exon 7 inclusion levels in DM1 (33%) and myositis (42%) samples were similar to those observed in fetal hearts (34%) and fetal skeletal muscles (53%), respectively, indicating a complete reversal of Eya3 isoform ratios in these disease settings (Figures 1H and 1I). We further examined Eya3 splicing in ventricles obtained from mice subjected to transverse aortic constriction (TAC) surgeries, an established experimental model for pressure overload-induced heart failure (Figure S5). One response to TAC is cardiac remodeling which reduces the ejection fraction, a measurement that quantifies the percentage of blood in the ventricles that is pumped into circulation with each heartbeat. The ejection fraction serves as an indication of the degree of heart failure, with the most severely failing hearts displaying the lowest ejection fractions. Failing hearts displayed lower levels of Eya3 exon 7 inclusion compared to sham controls (Figure S5A) and we observed a high correlation between the ejection fraction and the inclusion levels of Eya3 exon 7 (Figure S5B). Together, these results indicate that the developmentally regulated splicing of Eya3 exon 7 is reverted to fetal patterns in striated muscle diseases.
EYA3 regulates the proliferation and differentiation of myoblasts
EYA3 contains an N-terminal transactivation domain (TAD) and a threonine phosphatase domain (pT-P) domain (Figure S6), although whether the pT-P activity is intrinsic or results only from close associations with other phosphatases is unclear.35,36,37 The C-terminal region is broadly termed the EYA domain (ED) and contains a tyrosine phosphatase domain (pY-P) characteristic of all EYA family members.23 The inclusion of Eya3 exon 7 produces a 46 amino acid in-frame insertion within the TAD and adjacent to the pT-P domain (Figure S6). Given that throughout all organisms examined, we observed the highest level of Eya3 exon 7 inclusion in skeletal muscle tissues, and that developmentally regulated splicing often has profound influences on tissue maturation and function, we next sought to identify the function of EYA3 isoforms in skeletal muscle cell differentiation.
To examine the cellular roles of EYA3 isoforms during early muscle development, we utilized the well-described C2C12 murine myoblast cell line.38,39 C2C12 cells can be differentiated through serum reduction, which promotes fusion of myoblasts to form multinucleated myotubes. Myotubes morphologically resemble, and share characteristics with, primitive muscle fibers and are thus commonly used as a model for myogenesis.
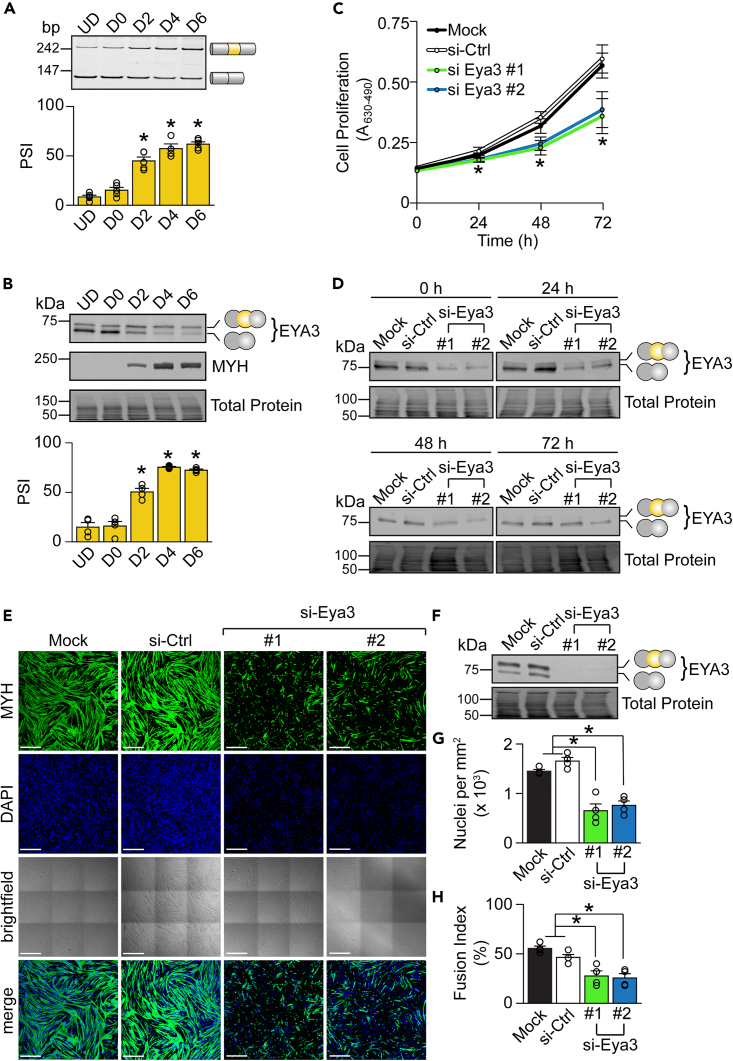
First, we measured the splicing of Eya3 exon 7 during C2C12 cell differentiation over the course of six days and observed a gradual increase in the expression of Eya3+ex7 isoform at the mRNA (62% at differentiation day (D) 6) and protein (72% at D6) levels (Figures 2A and 2B). The PSI values were similar between D4 and D6 myotubes, indicating that inclusion of exon 7 peaks and stabilizes at D4. Notably, exon inclusion at D4 (57% RNA, 76% protein) and D6, were similar to those observed in adult skeletal muscle tissues (ranging from 54% in soleus to 73% in TA) (Figure S2B), indicating that the C2C12 cell differentiation is an appropriate model to further examine the role of EYA3 splice isoforms.
Figure 2.
EYA3 isoforms contribute to myoblast proliferation and fusion
(A and B) The splicing pattern of Eya3 exon 7 during C2C12 cell differentiation at the RNA level was determined by reverse transcription PCR (RT-PCR) (n = 3–5) (A) and at the protein level by western blotting (n = 4) (B).
(C and D) Control (si-Ctrl) and EYA3 depleted (si-Eya3) myoblasts were subjected to colorimetric cellular proliferation assays (n = 4–7) (C) and Western blot studies (D) over the course of 72 h.
(E) Immunofluorescence detection of myosin heavy chain (MYH, green) and nuclei (DAPI, blue) was followed by confocal microscopy in control and EYA3 depleted myotubes. A tile scan was used to capture nine images per condition and were then stitched using Zen Black (Zeiss) image acquisition software. Scale bars = 500 μm.
(F) Protein lysates obtained from control and EYA3 depleted myotubes were analyzed by western blotting in parallel studies to those shown in panel E. See also Figures S6 and S7.
(G and H) Images were scored for nuclei per unit area (n = 4) (G) and fusion index (n = 4) (H). A, absorbance. bp, base pairs. D, days of differentiation. PSI, percent spliced in. UD, undifferentiated. Results are shown as mean ± SEM. ∗p < 0.05 (versus UD) (A, B), ∗p < 0.05 (both si-Eya3 #1 and si-Eya3 #2 versus si-Ctrl) (C), ∗p < 0.05 (versus si-Ctrl and mock) (G, H), Student’s T test.
To interrogate the function of EYA3 during myogenesis, we depleted both EYA3 isoforms in myoblasts using two independent small interfering RNAs (si-RNAs) (si-Eya3 #1, si-Eya3 #2) with knockdown efficiencies of 77% (si-Eya3 #1) and 75% (si-Eya3 #2) following differentiation (Figure S7). Consistent with previous reports in cancer cell lines and myoblasts,26,40 EYA3 depletion inhibited cellular proliferation over the course of 72 h (Figures 2C and 2D), prevented the formation of elongated myotubes (Figures 2E and 2F), reduced the number of nuclei within each field of view (Figure 2G), and inhibited myoblast fusion (Figure 2H).
Mitosis-related genes are regulated by EYA3 during myoblast differentiation
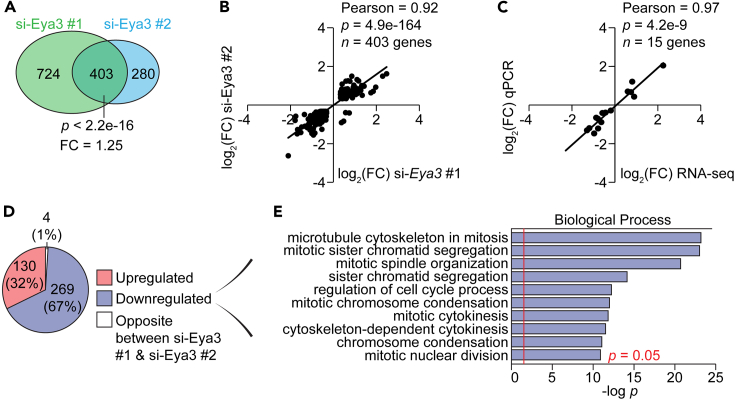
Because the depletion of EYA3 produced profound defects in myoblast growth and myotube morphology (Figures 2C–2H), and EYA3 is a known transcriptional cofactor,26,29,30,31 we next examined the transcriptional programs regulated by EYA3 in muscle cells by RNA-seq. Analysis of RNA-seq data generated from myotubes treated with si-Eya3 #1 or si-Eya3 #2 revealed widespread changes in gene expression with a significant overlap of the differentially expressed genes (defined as those with a fold change |FC| > 1.25; adjusted p value (p-adj) < 0.05 compared with controls) among both Eya3 si-RNAs (Figure 3A). The overlap between si-Eya3 #1 and si-Eya3 #2 contained 403 genes and represent high confidence candidates for EYA3 regulation (Figure 3A and Data S1). Moreover, the log2(FC) in gene expression for these candidates were highly correlated (Pearson = 0.92) (Figure 3B), indicating that the detected transcriptional changes are reproducible between two independent si-RNAs. A set of 15 (5 upregulated and 10 downregulated) EYA3 regulated genes were validated by real time quantitative PCR (qPCR) analysis (Table S3) in independently generated samples (Figure 3C). The high degree of correlation between gene expression values determined by RNA-seq studies and qPCR assays (Pearson = 0.97) (Figure 3C) indicated robust and accurate detection of EYA3 regulated genes. The majority (67%, 269 genes) of the differentially expressed genes were downregulated in EYA3 depleted myotubes, compared with those that were upregulated (32%, 130 genes) (Figure 3D). Genes encoding proteins that orchestrate mitotic division and the cell cycle were enriched among those being activated by EYA3 (Figure 3E).
Figure 3.
EYA3 promotes expression of genes involved in cell division
(A) Venn diagram displaying overlapping gene expression changes in response to EYA3 depletion among two independent small interfering RNAs (si-RNAs) (si-Eya3 #1 and si-Eya3 #2).
(B) Scatterplot of the relationship between log2 fold changes (FC) in gene expression for si-Eya3 #1 and si-Eya3 #2 based on the computational analysis of our RNA-sequencing (RNA-seq) data.
(C) Scatterplot displaying the correlation between log2(FC) in gene expression computed by bioinformatic analysis of the RNA-seq data and the experimental values determined by real time quantitative PCR (qPCR). See also Tables S3 and S4.
(D) The set of overlapping differentially expressed genes (403) were classified based on whether they were upregulated in response to si-Eya3 #1 and si-Eya3 #2 treatment (upregulated, pink), downregulated in response to si-Eya3 #1 and si-Eya3 #2 treatment (downregulated, purple), or regulated in opposite directions (opposite between si-Eya3 #1 & si-Eya3 #2, white) treatment.
(E) Gene ontology analysis (Biological Process) of the 269 downregulated genes in response to si-Eya3 #1 and si-Eya3 #2 treatment. p = 0.05 is indicated by the red line. A Fisher’s exact test was performed in panels A and E. A significance test for correlation was performed in panels B and C.
In summary, EYA3 can induce or inhibit gene expression; however, within the context of our study EYA3 may function more often as a transcriptional coactivator and to a lesser extent as a transcriptional corepressor. The fact that EYA3 acts as a coactivator of genes related to cellular division may explain why myoblast proliferation was inhibited in EYA3 depleted cells.
EYA3Δex7 and EYA3+ex7 isoforms localize to the nucleus where they bind to transcription factors
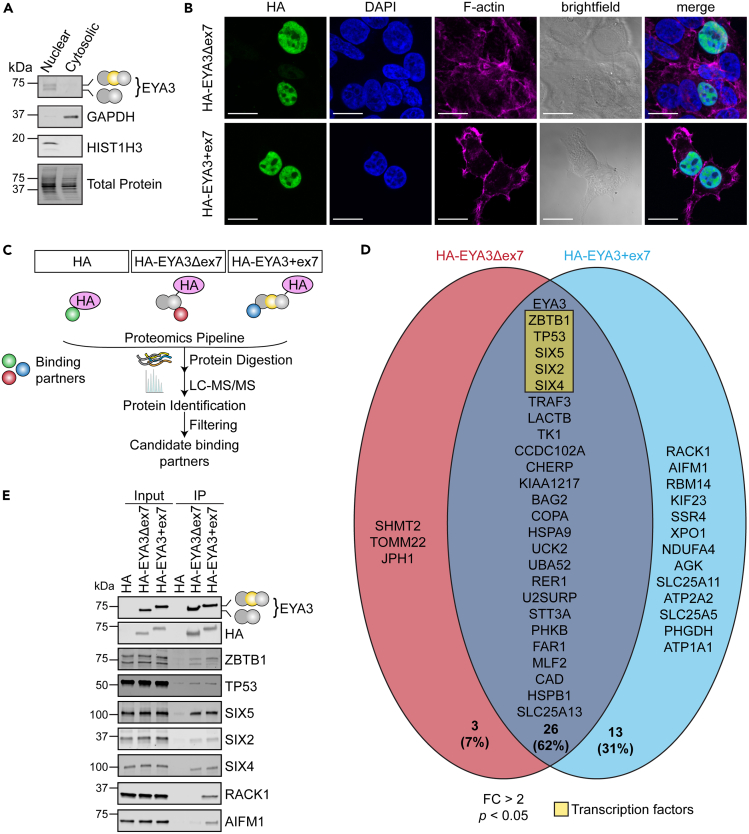
To further uncover the functions of EYA3 splice isoforms, we examined their intracellular distribution. Subcellular fractionation of C2C12 myotubes revealed that both EYA3 isoforms localize to the nucleus, and neither isoform was detected in the cytoplasmic fraction (Figure 4A). This observation was confirmed by heterologous expression of hemagglutinin (HA)-tagged EYA3 isoforms followed by immunofluorescence assays and confocal microscopy studies in HEK293T cells (Figure 4B). Therefore, EYA3Δex7 and EYA3+ex7 isoforms exhibit a similar intracellular distribution. These findings also support the hypothesis that the changes in transcriptional programs are the results of direct regulation by EYA3 isoforms in the nucleus.
Figure 4.
Transcription factors interact with EYA3 splice isoforms in the nucleus
(A) Nuclear and cytoplasmic extracts from C2C12 myotubes were analyzed by Western blot assays.
(B) HEK293T cells were transfected with plasmids encoding HA-tagged EYA3 isoforms and then evaluated by immunofluorescence and confocal microscopy. Cells were stained for HA (green), nuclei (DAPI, blue), and F-actin (Phalloidin, magenta). Scale bars = 20 μm.
(C) Schematic of the strategy used for the co-immunoprecipitation assays followed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) and data analysis to determine EYA3 isoform binding partners.
(D) Venn diagram displaying the overlapping and unique binding partners of EYA3 splice isoforms (FC > 2; p < 0.05). Identified binding partners with known functions as transcription factors are highlighted in yellow. Percent of interactors in each field of the Venn diagram are displayed.
(E) Validation of EYA3 isoform binding partners determined by immunoprecipitation (IP) followed by western blotting. AIFM1, apoptosis inducing factor mitochondria associated 1. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. HA, hemagglutinin epitope tag. HIST1H3, histone 3. PSI, percent spliced in. RACK1, receptor for activated C kinase 1. SIX2, SIX homeobox 2. SIX4, SIX homeobox 4. SIX5, SIX homeobox 5. TP53, tumor protein p53. ZBTB1, zinc finger and BTB-domain containing 1.
To elucidate the mechanism by which EYA3 regulates transcription, we identified the binding partners of each EYA3 isoform via co-immunoprecipitation in HEK293T cells followed by mass spectrometry-based proteomic analysis (Figures 4C and 4D and Data S2). Antibodies recognizing the individual EYA3 isoforms are not available. Thus, HEK293T cells expressing each (HA)-tagged EYA3 splice variant were used in this experiment. Interestingly, 26 binding partners (defined as those with FC > 2 and p < 0.05 compared to HA epitope tag control) were shared among the EYA3 isoforms, including five transcription factors: ZBTB1, tumor protein p53 (TP53), SIX5, SIX2, and SIX4 (Figure 4D). The binding of each EYA3 isoform with these five transcription factors was validated by western blotting (Figure 4E). From the 42 total binding partners that we identified, three (7%) and thirteen (31%) were unique interactors for EYA3Δex7 and EYA3+ex7, respectively. The exclusive binding of EYA3+ex7 with receptor for activated C kinase 1 (RACK1) and apoptosis inducing factor mitochondria associated 1 (AIFM1) was validated by western blotting (Figure 4E). Therefore, inclusion of exon 7 facilitates interactions with different proteins and may contribute to distinct functions of EYA3 splice isoforms.
EYA3 isoforms partner with ZBTB1 and SIX4 to regulate gene expression
The members of the SIX family of transcription factors (SIX1-SIX5) have been reported to partner with EYA family members during organogenesis of the kidney, ear, heart, skeletal muscle, and craniofacial tissues.24,26,27,28 In C2C12 cells, the most prominently expressed SIX family member (as determined by RNA-seq of control myotubes) that was also detected as a binding partner of EYA3 was SIX4 (Table S4). ZBTB1 was the most highly enriched protein detected for both EYA3Δex7 and EYA3+ex7 (Data S2) and has not been previously described as a binding partner of EYA3. The large family of ZBTB transcription factors are critical coordinators of blood lineage differentiation.41 ZBTB1 represses the expression of genes involved in the specification of myeloid fates but has also been demonstrated to promote the transcription of the asparagine synthetase locus in T lymphocytes.42,43 Considering these observations, we hypothesized that EYA3 regulates transcription by partnering with ZBTB1 and by partnering with SIX4.
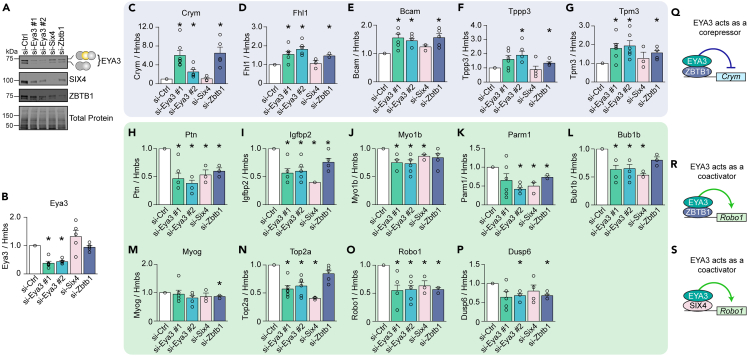
To examine whether ZBTB1 or SIX4 are required for the expression of EYA3 target genes, we depleted SIX4 or ZBTB1 in C2C12 myoblasts and differentiated them into myotubes for four days (Figures 5A and 5B). Next, we quantified the expression of a set of EYA3 regulated genes (Figures 5C–5P). Genes upregulated in response to EYA3 depletion were also upregulated in ZBTB1 but not SIX4 depleted myotubes (Figures 5C–5G). Interestingly, genes that were downregulated upon EYA3 depletion were also downregulated in ZBTB1 and in SIX4 depleted myotubes (Figures 5H–5P). To confirm that most of our results were not attributed to differentiation defects upon EYA3 depletion, we selected a subset of our proposed target genes and analyzed them in undifferentiated myoblasts (48 h after si-RNA delivery) (Figure S8). For the majority of tested genes, we observed the same trend in myoblasts as in differentiated myotubes, further substantiating these as direct targets of EYA3, and not simply the result of indirect effects from alterations in muscle cell differentiation. Together, these findings led us to propose two models for gene expression regulation by EYA3: (i) ZBTB1 and EYA3 partner to repress the transcription of a set of genes independently of SIX4 (EYA3 thus acts as a corepressor) (Figure 5Q), (ii) EYA3 interacts with ZBTB1 to promote the transcription of a set of genes in muscle cells (acting as a coactivator) (Figure 5R), or EYA3 interacts with SIX4 to promote the transcription of a set of genes in muscle cells (acting as a coactivator) (Figure 5S).
Figure 5.
ZBTB1 and SIX4 partner with EYA3 to regulate gene expression
(A) EYA3, SIX4, and ZBTB1 expression levels were determined by western blotting following the delivery of small interfering RNAs (si-RNA) in C2C12 myoblasts that were then differentiated for four days.
(B–P) Real time quantitative PCR (qPCR) analysis of differentially expressed genes upon EYA3, SIX4, or ZBTB1 depletion (n = 3–6): Eya3 (B), Crym, crystallin mu (C), Fhl1, four and a half LIM domains 1 (D), Bcam, basal cell adhesion molecule (E), Tppp3, tubulin polymerization promoting protein family member 3 (F), Tpm3, tropomyosin 3 (G), Ptn, pleiotrophin (H), Igfbp2, insulin-like growth factor binding protein 2 (I), Myo1b, myosin IB (J), Parm1, prostate androgen-regulated mucin-like protein 1 (K), Bub1b, BUB1 mitotic checkpoint serine/threonine kinase B (L), Myog, myogenin (M), Top2a, DNA topoisomerase II alpha (N), Robo1, roundabout guidance receptor 1 (O), and Dusp6, dual specificity phosphatase 6 (P). See also Figure S8 and Table S3.
(Q and R) Proposed models for the regulation of gene expression by EYA3 together with ZBTB1 as a transcriptional corepressor (Q), with ZBTB1 as a transcriptional coactivator (R), or with SIX4 as a transcriptional coactivator (S). Hmbs, hydroxymethylbilane synthase. Results are shown as mean ± SEM. ∗p < 0.05 (versus si-Ctrl), Student’s T test.
Inclusion of exon 7 attenuates EYA3 transcriptional capacity and facilitates proper gene expression during myogenesis
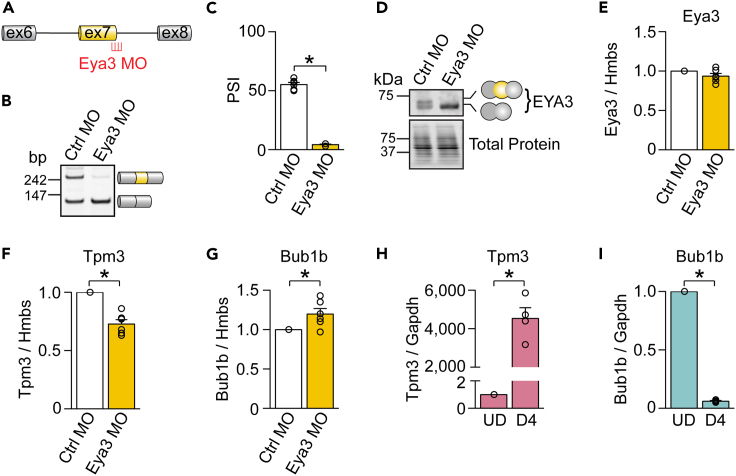
Next, we sought to investigate whether EYA3 isoforms differ in their capacity to regulate gene expression (either as a coactivator or as a corepressor). To test this possibility, we modulated endogenous Eya3 exon 7 splicing using a morpholino antisense oligonucleotide (MO) designed to hybridize to the downstream splice site of Eya3 exon 7 (Figure 6A). Myoblasts were treated with the Eya3 exon 7 MO and then differentiated into myotubes for four to five days. Eya3 MO strongly repressed inclusion of exon 7 at the RNA (Figures 6B and 6C) and protein levels (Figure 6D) but did not alter the overall Eya3 mRNA expression levels (Figure 6E). However, blocking the expression of EYA3+ex7 isoform altered the expression of two genes (Tpm3 and Bub1b) that are regulated during myogenesis (upregulated and downregulated, respectively) and are responsive (repressed and activated, respectively) to EYA3 depletion. Specifically, we observed lower expression of tropomyosin 3 (Tpm3) (Figure 6F) and increased expression of BUB1 mitotic checkpoint serine/threonine kinase B (Bub1b) (Figure 6G) mRNAs.
Figure 6.
Inclusion of EYA3 exon 7 attenuates its transcriptional capacity as a coactivator and corepressor
(A) Schematic of the hybridization of a morpholino (MO) targeting the downstream splice site of Eya3 exon 7 (Eya3 MO).
(B–D) Inclusion of Eya3 exon 7 was determined via reverse transcription PCR (RT-PCR) at the RNA level (n = 6) (B and C) and via Western blot at the protein level (D) following treatment with a control morpholino (Ctrl MO) or Eya3 MO in myoblasts that were then differentiated into myotubes.
(E) Real time quantitative PCR (qPCR) analysis of Eya3 mRNA expression in response to Eya3 MO (n = 6) treatment.
(F and G) qPCR analysis of tropomyosin 3 (Tpm3) (F) and BUB1 mitotic checkpoint serine/threonine kinase B (Bub1b) (G) mRNA expression in response to Eya3 MO (n = 6) treatment.
(H and I) qPCR analysis of Tpm3 (H) and Bub1b (I) mRNA expression in undifferentiated C2C12 myoblasts (UD) and myotubes at differentiation day 4 (D4) (n = 4). bp, base pairs. Gapdh, glyceraldehyde-3-phosphate dehydrogenase. Hmbs, hydroxymethylbilane synthase. Results are shown as mean ± SEM. ∗p < 0.05, Student’s T test.
Tpm3 mRNA is upregulated during myogenesis (Figure 6H) and TPM3 protein is primarily expressed in slow-twitch muscle fibers where it plays a central role in muscle contraction by controlling actin filament sliding across the sarcomere in a calcium dependent manner.44 Bub1b mRNA is downregulated during myogenesis (Figure 6I). BUB1B protein is vital for the establishment of kinetochore-microtubule attachments required for mitotic checkpoint signaling and faithful chromosomal segregation during mitosis.45 Therefore, our discoveries raise the possibility that an important function of EYA3 alternative splicing in the context of skeletal muscle is to modulate the transcription of genes during myogenesis.
RBFOX2 regulates EYA3 exon 7 alternative splicing
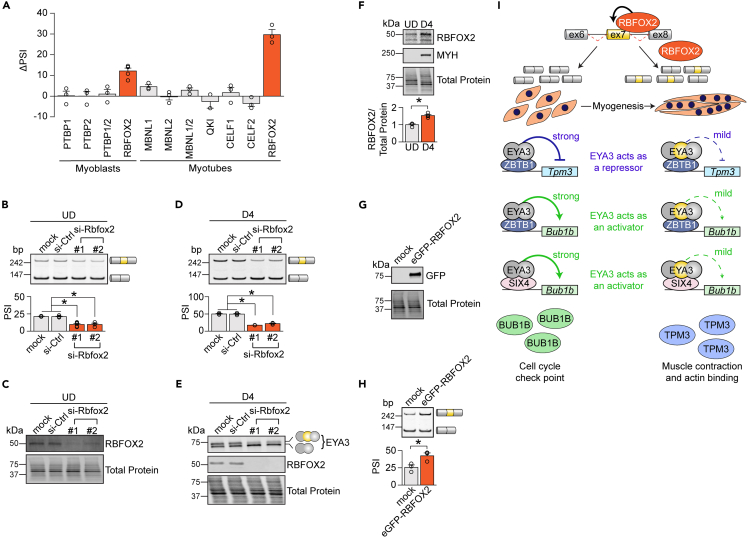
To determine the trans-regulatory factors that might control alternative splicing of Eya3 pre-mRNA, we depleted eight RNA-binding proteins (RBPs) using si-RNAs (Figures 7A–7E and S9). Candidate RBPs were selected based on the following criteria: (i) they were previously demonstrated to regulate alternative splicing in striated muscles and (ii) they are expressed in C2C12 cells.7,12,14,46,47,48 We observed no significant differences in Eya3 exon 7 splicing in response to the depletion of the polypyrimidine tract binding protein 1 (PTBP1), the polypyrimidine tract binding protein 2 (PTBP2), the muscleblind like splicing regulator 1 (MBNL1), the muscleblind like splicing regulator 2 (MBNL2), the KH domain containing RNA binding (QKI), the CUGBP Elav-like family member 1 (CELF1), or the CUGBP Elav-like family member 2 (CELF2) (Figures 7A and S9); however, the depletion of RBFOX2 in both myoblasts and myotubes drastically decreased the inclusion of Eya3 exon 7 at the mRNA level (Figures 7A–7D). The decrease in EYA3+exon7 expression following RBFOX2 depletion was also evident at the protein level (Figure 7E). In accordance with previous studies,12,14 we observed that RBFOX2 levels are 53% higher in myotubes compared to myoblasts (Figure 7F). To investigate the effect of ectopic overexpression of RBFOX2, we transfected myoblasts with a plasmid containing eGFP-tagged RBFOX2. Eya3 exon 7 inclusion levels increased from 26% in control conditions to 42% when RBFOX2 was overexpressed (Figures 7G and 7H). These findings suggest that the upregulation of RBFOX2 expression during myogenesis mediates the increase in inclusion of Eya3 exon 7.
Figure 7.
Eya3 alternative splicing is regulated by RBFOX2
(A) Differences in Eya3 exon 7 percent spliced in (ΔPSI) were defined as the differences between the PSI in si-Ctrl and RNA-binding protein (RBP) knockdown cells (i.e., si-Ctrl – si-RBP) in myoblasts and myotubes as determined by reverse transcription PCR (RT-PCR).
(B–E) Inclusion of Eya3 exon 7 in myoblasts (UD) (B) and myotubes (D4) (D) was determined by RT-PCR following RBFOX2 depletion (n = 3–5). RBFOX2 and EYA3 expression levels in myoblasts (UD) and myotubes (D4) were assayed via western blotting (C and E, respectively).
(F) RBFOX2 expression levels in myoblasts (UD) and myotubes (D4) were analyzed via Western blot assays (n = 4).
(G and H) Myoblasts were transfected with a plasmid encoding eGFP-tagged RBFOX2. Expression of eGFP-RBFOX2 was determined by western blotting (G) and splicing of Eya3 exon 7 was measured by RT-PCR assays (H) (n = 3).
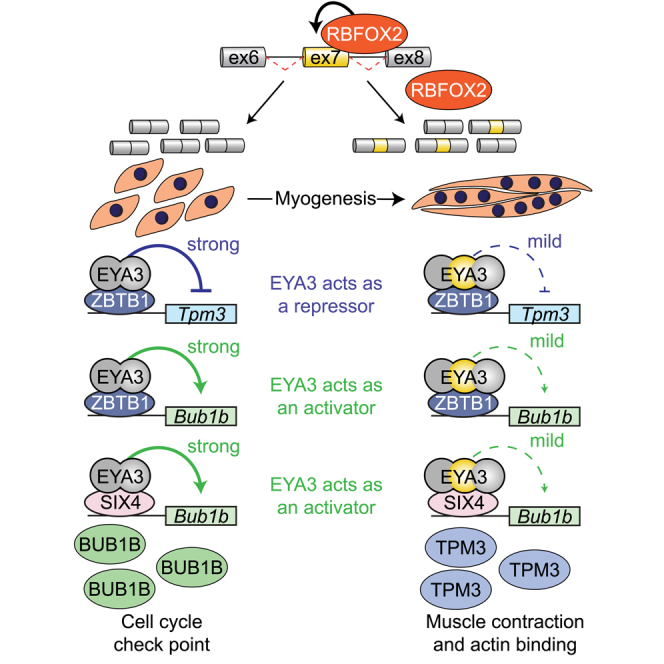
(I) Proposed model of how Eya3 exon 7 splicing is regulated during myogenesis and its functional implications. bp, base pairs. Bub1b, BUB1 mitotic checkpoint serine/threonine kinase B. CELF1, CUGBP Elav-like family member 1. CELF2, CUGBP Elav-like family member 2. ex, exon. MBNL1, muscleblind like splicing regulator 1. MBNL2, muscleblind like splicing regulator 2. PTBP1, polypyrimidine tract binding protein 1. PTBP2, polypyrimidine tract binding protein 2. QKI, KH domain containing RNA binding. RBFOX2, RNA binding fox-1 homolog 2. SIX4, SIX homeobox 4. Tpm3, tropomyosin 3. ZBTB1, zinc finger and BTB-domain containing 1. Results are shown as mean ± SEM. ∗p < 0.05 (versus si-Ctrl and mock) (A, B, D), ∗p < 0.05 (versus UD) (F), ∗p < 0.05 (versus mock) (H), Student’s T test.
Discussion
The specification of the myogenic lineage is regulated at multiple levels by a hierarchy of transcription factors, extracellular signals, dynamic epigenetic modifications, and microRNAs.49,50,51,52,53 EYA and SIX family members are known to form an important transcriptional complex during the progression of myogenesis. Still, many of the transcriptional targets and additional participants in EYA-controlled gene expression were previously unknown. Our studies reveal an unknown role for ZBTB1 as a myogenic transcription factor, define the EYA3 target genes on a genome-wide basis, and expose an additional layer of regulation by alternative splicing in the temporal control of gene expression.
The activation of a major myogenic transcription factor, MYOG, is controlled by the binding of the SIX-EYA complex to its promoter.29,30 MYOG then coordinates the expression of genes that encode proteins involved in terminal myogenic differentiation. Our study identifies a previously unknown role for ZBTB1 in the regulation of the myogenic transcriptional program by modulating the expression of genes driving muscle function, including Myog, crystallin mu (Crym), and Tpm3. Several ZBTB family members dictate commitment to various cellular fates during blood lineage specification.54,55 Our findings imply that ZBTB1 plays an analogous role in the skeletal muscle context, by partnering with EYA3 to enforce proper gene expression during myogenesis.
We found that EYA3 can act as a corepressor or a coactivator depending on the transcriptional target (Figure 5). For example, EYA3 is a coactivator of the expression of Bub1b (Figure 5L) and a corepressor of Tpm3 transcription (Figure 5G). During the G2/M transition, BUB1B levels peak to ensure the faithful segregation of chromosomes before commitment to mitosis.56 BUB1B expression is strongly repressed during myogenesis (Figure 6I), consistent with the fact that mature muscle fibers are post-mitotic. In contrast, Tpm3 gene encodes the slow muscle alpha (α)-tropomyosin and is upregulated later in myogenesis (Figure 6H), when fiber-type specialization occurs. By blocking the inclusion of Eya3 exon 7 during myoblast differentiation, we demonstrated that EYA3+ex7 displays a reduced capacity to activate or repress the Bub1b and Tpm3 loci, respectively (Figures 6F and 6G). These findings support a model in which expression of EYA3Δex7 isoform in myoblasts serves to strongly promote Bub1b and repress Tpm3 transcription (Figure 7I, left). During skeletal muscle cell differentiation, the induction of EYA3+ex7 isoform expression by RBFOX2 attenuates the transcriptional activation of Bub1b (thus reducing its expression) and the transcriptional repression of Tpm3 (thus increasing its expression) (Figure 7I, right). In sum, inclusion of exon 7 mitigates the coregulatory capacities of EYA3, thereby supporting gene expression at appropriate temporal windows. Notably, slow twitch fibers are disproportionately affected in muscles of humans with DM1 and muscle biopsies from patients with chronic myositis display fewer slow twitch fibers than healthy individuals.57,58 In this context, our discoveries raise the possibility that splicing reversion of Eya3 exon 7 (Figure 1J) contributes to the imbalance in fiber type composition observed in muscle diseases.
Our work expands upon a growing number of alternative splice variants with unique transcriptional capacities.59,60,61,62,63 Like exon 7 of Eya3, alternative regions often fall within the transactivation domain of transcriptional regulators.59,60,61,64 In some cases, differences in transcriptional activation can result from a weaker association with DNA or other regulatory factors.61 Here we identified 16 proteins that bind to either EYA3+ex7 or EYA3Δex7 splice isoforms exclusively (Figure 4D and Data S2). For example, RACK1 interacts with EYA3+ex7 form but not EYA3Δex7 isoform. RACK1 has been demonstrated to associate with and repress transcriptional activation by the basic-helix-loop-helix ARNT like 1 (BMAL1) transcription factor.65 Therefore, the association between EYA3+ex7 isoform and RACK1 may serve to inhibit the transcription of EYA3 target genes such as Bub1b.
Previous work has also revealed that the dephosphorylation of a repressor complex by the EYA3 pT-P domain is an essential step in the activation of SIX target genes.26 Since the alternative region of EYA3 is directly proximal to its pT-P domain, another possibility is that differences in gene activation or repression result from differences in phosphatase activity between EYA3 variants.
Limitations of the study
Correction of splicing abnormalities has proved effective in treating muscular diseases (Duchenne muscular dystrophy)21,22 and neuromuscular disorders (spinal muscular atrophy).66,67 Mounting evidence indicates a central role for widespread splicing regulation during muscle development and dysregulation in muscular diseases.8,17,20,68 Our work provided significant insights into how splicing regulation of a transcription factor influences its interaction with other proteins and functions during myogenesis. However, a greater understanding of the molecular regulatory mechanisms and functions of splice isoforms is urgently required for the identification of relevant therapeutic targets.
Although we addressed some of our questions in tissue samples, most of our functional and molecular assays were performed using cell lines. C2C12 cells are a great model to learn about muscle biology, but, obviously, more physiological studies using animal models will be the next step in this line of research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-EYA3 | Bethyl Laboratories | Cat# A302-689A; RRID:AB_10752432 |
| Mouse monoclonal anti-HA Tag (clone 2-2.2.14) | Thermo Fisher Scientific | Cat# 26183; RRID:AB_10978021 |
| Mouse monoclonal anti-MYH (clone B-5) | Santa Cruz Biotechnology | Cat# sc-376157; RRID:AB_10989398 |
| Mouse monoclonal anti-RBM9 (RBFOX2) (clone F-8) | Santa Cruz Biotechnology | Cat# sc-271407; RRID:AB_10612209 |
| Rabbit monoclonal anti-PTBP1 (clone EPR9048(B)) | Abcam | Cat# ab133734; RRID:AB_2814646 |
| Rabbit monoclonal anti-PTBP2 (clone EPR9890) | Abcam | Cat# ab154787; RRID:AB_2923480 |
| Mouse monoclonal anti-MBNL1 (clone D-4) | Santa Cruz Biotechnology | Cat# sc-515374 |
| Mouse monoclonal anti-MBNL2 (clone 3B4) | Santa Cruz Biotechnology | Cat# sc-136167; RRID:AB_2140469 |
| Mouse monoclonal anti-CUG-BP1 (CELF1) (clone 3B1) | Santa Cruz Biotechnology | Cat# sc-20003; RRID:AB_627319 |
| Mouse monoclonal anti-CUG-BP2 (CELF2) (clone 1H2) | Santa Cruz Biotechnology | Cat# sc-47731; RRID:AB_2086271 |
| Rabbit monoclonal anti-QK1 (clone EPR7306) | Abcam | Cat# ab126742; RRID:AB_11129508 |
| Mouse monoclonal anti-GAPDH (clone G-9) | Santa Cruz Biotechnology | Cat# sc-365062; RRID:AB_10847862 |
| Rabbit polyclonal anti-Histone H3 | Abcam | Cat# ab1791; RRID:AB_302613 |
| Rabbit polyclonal anti-ZBTB1 | Proteintech | Cat# 26287-1-AP; RRID:AB_2880463 |
| Rabbit polyclonal anti-SIX4 | Proteintech | Cat# 21305-1-AP; RRID:AB_10860258 |
| Rabbit polyclonal anti-p53 | Cell Signaling Technology | Cat# 9282; RRID:AB_331476 |
| Rabbit polyclonal anti-SIX2 | Proteintech | Cat# 11562-1-AP; RRID:AB_2189084 |
| Rabbit polyclonal anti-SIX5 | Proteintech | Cat# 22938-1-AP; RRID:AB_2879186 |
| Mouse monoclonal anti-RACK1 (clone B-3) | Santa Cruz Biotechnology | Cat# sc-17754; RRID:AB_2247471 |
| Mouse monoclonal anti-AIFM1 (clone E-1) | Santa Cruz Biotechnology | Cat# sc-13116; RRID:AB_626654 |
| Goat polyclonal anti-Rabbit IgG | Thermo Fisher Scientific | Cat# SA5-35571; RRID:AB_2556775 |
| Goat polyclonal anti-Mouse IgG | Thermo Fisher Scientific | Cat# SA5-10176; RRID:AB_2556756 |
| Goat polyclonal anti-Mouse Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11001; RRID:AB_2534069 |
| Critical commercial assays | ||
| CellTiter 96 AQueous One Solution Cell Proliferation Assay | Promega | Cat# G3580 |
| High-capacity cDNA reverse transcription kit | Applied Biosystems | Cat# 4368813 |
| Taqman Fast Advanced Master Mix | Applied Biosystems | Cat# 4444557 |
| Deposited data | ||
| Raw RNA-seq data files | This paper | GSE212436 |
| Proteomic datasets | This paper | PXD037572 |
| Experimental models: Cell lines | ||
| Mouse: C2C12 | ATCC | Cat# CRL-1772; RRID:CVCL_0188 |
| Human: HEK293T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Experimental models: Organisms/strains | ||
| Mouse: FVB/NJ | The Jackson Laboratory | Cat# 001800; RRID:IMSR_JAX:001800 |
| Mouse: C57BL/6J | The Jackson Laboratory | Cat# 000664; RRID:IMSR_JAX:000664 |
| Rat: Crl:SD | Charles River | RRID:RGD_737891 |
| Chicken: fertilized eggs | Pilgrim’s Farms | N/A |
| Zebrafish: AB | ZIRC | RRID:ZIRC_ZL1 |
| Oligonucleotides | ||
| Morpholino: negative control MO CCTCT TACCTCAGTTACAATTTATA |
Gene Tools | N/A |
| Morpholino: Eya3 exon 7 MO ACACAACCATGCCTATTTACCTTGA |
Gene Tools | N/A |
| siRNA targeting sequence: Ptbp1 GGUGUGGUCAAAGGCUUCAAGUUCU |
ThermoFisher Scientific | siRNA ID: MSS276537 |
| siRNA targeting sequence: Ptbp1 CCUCUGGAGACAGCCAGCCUUCACU |
ThermoFisher Scientific | siRNA ID: MSS276539 |
| siRNA targeting sequence: Ptbp2 GGUGGCAAUACAGUCCUGUUGGUUA |
ThermoFisher Scientific | siRNA ID: MSS225938 |
| siRNA targeting sequence: Ptbp2 GGGCACUGUGAAAGCAUUUAAGUUU |
ThermoFisher Scientific | siRNA ID: MSS225940 |
| siRNA targeting sequence: Qki GAGCGGUUGAAGAAGUGAAGAAGUU |
ThermoFisher Scientific | siRNA ID: MSS208338 |
| siRNA targeting sequence: Qki GAGCGGCUGCUGGACGAAGAAAUUA |
ThermoFisher Scientific | siRNA ID: MSS276677 |
| siRNA targeting sequence: Mbnl1 CCACAGCCAACCAGAUACCCAUAAU |
ThermoFisher Scientific | siRNA ID: MSS226392 |
| siRNA targeting sequence: Mbnl1 GCAUUUCUCCCACCAGGCUCAAUAU |
ThermoFisher Scientific | siRNA ID: MSS226393 |
| siRNA targeting sequence: Mbnl2 GAGAUUAAUGGGAGGAACAAUUUGA |
ThermoFisher Scientific | siRNA ID: MSS272396 |
| siRNA targeting sequence: Mbnl2 GCGUUGCAUGAGGGAGAAAUGCAAA |
ThermoFisher Scientific | siRNA ID: MSS200587 |
| siRNA targeting sequence: Celf1 GGACAGAUUGAAGAGUGCCGGAUAU |
ThermoFisher Scientific | siRNA ID: MSS203372 |
| siRNA targeting sequence: Celf1 CCAUGAACGGCUUUCAAAUUGGAAU |
ThermoFisher Scientific | siRNA ID: MSS203374 |
| siRNA targeting sequence: Celf2 CAGAGUAAAGGUUGUUGUUUCGUAA |
ThermoFisher Scientific | siRNA ID: MSS274200 |
| siRNA targeting sequence: Celf2 GCUGGAGCCACUGUCGGAUUGAAUA |
ThermoFisher Scientific | siRNA ID: MSS204012 |
| siRNA targeting sequence: Eya3 GGACCCAACAGUAGUAAUU |
Dharmacon | Cat# D-065562-01-0002 |
| siRNA targeting sequence: Eya3 UGAGGGAAAUCUACGACAA |
Dharmacon | Cat# D-065562-02-0002 |
| siRNA targeting sequence: Six4 GCAAGCAAAUGUACUGUGU |
Dharmacon | Cat# D-057996-01-0002 |
| siRNA targeting sequence: Zbtb1 UAUCAGCGCUGUCAUUUGA | Dharmacon | Cat# D-057238-02-0002 |
| siRNA targeting sequence: Rbfox2 GCAAAUGGCUGGAAGUUAA |
Dharmacon | Cat# D-051552-01-0002 |
| siRNA targeting sequence: Rbfox2 CGAGAAUAGUGCUGAUGCA |
Dharmacon | Cat# D-051552-04-0002 |
| siRNA targeting sequence: Luciferase Reporter Control GCACUCUGAUUGACAAAUACGAUUU |
Invitrogen | Cat# 12935146 |
| siRNA targeting sequence: siGENOME Non-Targeting Control UAGCGACUAAACACAUCAA |
Dharmacon | Cat# D-001210-01-20 |
|
M. musculus Eya3 alternative exon primer forward: TCCTGGGCAGACTCAGTACC |
Sigma Aldrich | N/A |
|
M. musculus Eya3 alternative exon primer reverse: GGTATCAGGCTGGCATTTGT |
Sigma Aldrich | N/A |
|
H. sapiens EYA3 alternative exon primer forward: GCAGACTCAACCCTATGCTG |
Sigma Aldrich | N/A |
|
H. sapiens EYA3 alternative exon primer reverse: TATCAGGCTGGCATTTGTGC |
Sigma Aldrich | N/A |
|
O. cuniculus EYA3 alternative exon primer forward: GGCAGACTCAGTACCAGACA |
ThermoFisher Scientific | N/A |
|
O. cuniculus EYA3 alternative exon primer reverse: TGGTTTGAGATGCTGGCAAC |
ThermoFisher Scientific | N/A |
|
R. norvegicus Eya3 alternative exon primer forward: CACACATCCTCTCGGTTCCT |
ThermoFisher Scientific | N/A |
|
R. norvegicus Eya3 alternative exon primer reverse: GTGGATATCAGGCTGGCATT |
ThermoFisher Scientific | N/A |
|
D. rerio eya3 alternative exon primer forward: ATGCCAGTCAAGTGGCTTTC |
Sigma Aldrich | N/A |
|
D. rerio eya3 alternative exon primer reverse: CTGTGGTCACTGCGGTTG |
Sigma Aldrich | N/A |
|
G. gallus EYA3 alternative exon primer forward: AATGAGCCCTTACCCTGGTC |
Sigma Aldrich | N/A |
|
G. gallus EYA3 alternative exon primer reverse: ACTGGACTGGCATTGGTAGT |
Sigma Aldrich | N/A |
| Recombinant DNA | ||
| Plasmid: pEGFP rbFOX2 | Addgene | Addgene plasmid # 63086; RRID:Addgene_63086 |
| Plasmid: HA-EYA3-long (+exon 7) | This paper | pHW14-pLenti-Eya3-Long |
| Plasmid: HA-EYA3-short (-exon 7) | This paper | pHW13-pLenti-Eya3-Short |
| Plasmid: HA-empty | This paper | pHW19-pLenti-HA |
| Software and algorithms | ||
| ImageJ version 1.51w for Windows | Schneider et al.69 |
https://imagej.net/ RRID:SCR_003070 |
| GraphPad Prism version 10.0.3 for Windows | GraphPad Software |
www.graphpad.com RRID:SCR_002798 |
| STAR version 2.7.6a | Dobin et al.70 | https://github.com/alexdobin/STAR |
| Salmon version 1.4.0 | Patro et al.71 | https://github.com/COMBINE-lab/salmon |
| DESeq2 package | Love et al.72 | https://github.com/thelovelab/DESeq2 |
| Image Lab version 6.0.1 for Windows | Bio-rad Software |
www.bio-rad.com RRID:SCR_014210 |
| Zen Black version 14.0.0.201 imaging software for Windows | Zeiss Software |
https://www.zeiss.com RRID:SCR_018163 |
| Other | ||
| QuantStudio™ 7 Flex Real-Time PCR System | Applied Biosystems | Cat# 4485696 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jimena Giudice (jimena_giudice@med.unc.edu).
Materials availability
This study generated three unique plasmids for Eya3 splice isoforms fused to HA tag and a control plasmid. See key resources table.
Experimental model and study participant details
Cell lines and culture maintenance
C2C12 myoblasts and HEK293T cells (American Type Culture Collection, ATCC) were maintained in growth media containing Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/mL), streptomycin (100 μg/mL), and glutamine (2 mM) in a humidified incubator at 37°C under 5% CO2. C2C12 myoblasts were regularly passaged to maintain 30-40% confluency, to prevent spontaneous differentiation. To differentiate myoblasts, cells were grown to 80-100% confluency, washed with phosphate buffered saline (PBS), and the growth media was replaced with differentiation media containing DMEM supplemented with 2% horse serum, penicillin (100 units/mL), streptomycin (100 μg/mL), and glutamine (2 mM). Differentiation media was replaced every 48 h.
Animal models for experiments
Animal protocols and experiments were approved by The University of North Carolina at Chapel Hill (UNC-Chapel Hill) or Baylor College of Medicine Institutional Animal Care and Use Committee. Animal accommodation was provided for in AAALAC accredited facilities compliant with approved regulatory guidelines for animal welfare.
Rat
Neonatal and adult rat (Rattus norvegicus) skeletal muscle was donated for this work from a UNC-Chapel Hill colony of Sprague Dawley rats. Neonatal (PN1) and adult (3-month-old) rats were euthanized by carbon dioxide inhalation followed by decapitation (for PN1) or cervical dislocation (for adults). Tissues were harvested and snap frozen in liquid nitrogen.
Rabbit
Adult rabbit (Oryctolagus cuniculus) heart and lung total RNAs were purchased commercially (Zyagen, TD-801, TD-601).
Chicken
Chicken (Gallus gallus) embryonic skeletal muscle was donated for this work from fertilized eggs (Pilgrim’s Farms, Sanford, NC, USA) maintained in a humidified incubator at 38°C until embryonic day 12 (E12) based upon previously established developmental staging guidelines.73 The eggs were windowed74 to expose the embyo and skeletal muscle tissue was harvested from both legs, placed in TRIzol reagent, and snap frozen in liquid nitrogen. Total RNA from adult chicken skeletal muscle was purchased commercially (Zyagen, CR-102).
Zebrafish
Zebrafish (Danio rerio) embryos and adult tissue were donated for this work from a UNC-Chapel Hill colony of wild-type (AB strain) fish. Embryos staged at 24 h post-fertilization (hpf),75 were placed in TRIzol reagent, and snap frozen in liquid nitrogen. Adult fish were euthanized by immersion in sodium bicarbonate buffered tricaine methanesulfonate (Sigma) for 10 min following the respiratory cessation of opercular movement.76 Fish were placed on ice, the tail was removed, skin was removed, and skeletal muscle was harvested and placed in TRIzol reagent and snap frozen in liquid nitrogen.
Human RNA samples
Total RNA from fetal human heart and skeletal muscle (Clontech, Invitrogen, Statgene, Cell Applications, Takara, Amsbio), adult human heart and skeletal muscle (Ambion, Amsbio, Takra, Cell Applications), and an adult myositis skeletal muscle sample (Origene, CR559548) were sourced commercially. Access to human fetal RNA samples and human myositis RNA samples was limited due to reduced commercial and institution availability. Human DM1 RNA samples were derived from autopsies from four individuals whose major clinical findings are summarized in Table S2.
Mouse
Neonatal and adult mice (Mus musculus) were bred from a UNC-Chapel Hill colony of FVB/NJ mice originally purchased from The Jackson Laboratory. Neonatal (PN4.5) and adult (4-month-old) mice were euthanized by isoflurane inhalant followed by secondary physical euthanasia.68 Harvested tissues were immediately snap frozen. Three- to four-month-old wild-type C57BL/6J male mice were subjected to transverse aortic constriction or sham surgeries followed by serial echocardiography prior to euthanasia (see Transthoracic echocardiography).
Transverse aortic constriction
Surgeries were performed in accordance with established protocols.77 Briefly, 2% isoflurane in 100% O2 (0.8 L/min) was used to anesthetize the mice. Endotracheal intubation and ventilation were used to maintain 1.5-2.0% isoflurane levels throughout the entire procedure. Anterior thoracotomies were performed to visualize the aortic arch. Sutures against a 28-gauge needle were placed between the first and second trunk of the aortic arch. To ensure consistent constriction levels, quantifications of alterations in Doppler velocities of the right and left carotid arteries were determined seven days after surgery.78 Sham surgeries were performed in an identical manner, but without constriction of the transverse aorta. Both left and right ventricles were harvested and flash-frozen in liquid nitrogen.
Transthoracic echocardiography
While mice were anesthetized with isoflurane and had a heart rate >400 beats per minute (bpm), the VisualSonics VeVo 770 Imaging System (VisualSonics, Toronto, Canada) was used to perform echocardiography to record systolic and diastolic function of the heart.79 To avoid confounding effects of hypothermia, body temperature was maintained in a range (37.0 ± 1.0)°C.
Method details
Morpholino (MO) delivery
Following trypsinization, C2C12 myoblasts were washed with PBS and resuspended in R buffer (Thermo Fisher Scientific) at a concentration of 107 cells/mL. Myoblasts were transfected with the following MOs (Gene Tools): (i) a standard control MO, which targets a mutation in an intron of the human beta-globin gene and is the most widely used negative control MO (CCT-CTT-ACC-TCA-GTT-ACA-ATT-TAT-A, Ctrl MO), or (ii) a MO designed to hybridize to the 5’ splice site of Eya3 exon 7 (ACA-CAA-CCA-TGC-CTA-TTT-ACC-TTG-A, Eya3 MO). Myoblasts were combined with 20 μM MO being subjected to electroporation using a Neon Transfection System (Thermo Fisher Scientific) with the following parameters: 1650 V, 10 ms, 3 pulses. Following electroporation, MO transfected C2C12 myoblasts were placed in six-well plates, maintained in growth media until they reached 80-100% confluency, and then differentiated (as described in the section “Cell lines and culture maintenance”) for four to five days before protein and RNA extraction.
Plasmid construction and delivery
The expression plasmid for N-terminal eGFP-tagged RBFOX2 was a gift from Dr. Nicolas Charlet-Berguerand (Addgene plasmid #63086; http://n2t.net/addgene:63086; RRID:Addgene_63086). Inserts containing the HA-tagged EYA3 isoforms or HA tag alone were cloned into the pLenti CMV plasmid. Constructs were verified by sanger sequencing. Cells were cultured in six well plates (100,000 C2C12 myoblasts per well or 600,000 HEK293T cells per well) or 15 cm plates (4,000,000 HEK293T cells per plate) in DMEM supplemented with 10% FBS and glutamine (2 mM). Lipofectamine3000 (Invitrogen, L3000015) was used to deliver plasmids according to the manufacturer’s protocol.
si-RNA delivery
Cells were cultured in six well plates (100,000 C2C12 myoblasts per well) in DMEM supplemented with 10% FBS and 2 mM glutamine. The next day, si-RNAs (Table S5) targeting Ptbp1 (Invitrogen, MSS276537, MSS276539), Ptbp2 (Invitrogen, MSS225938, MSS225940), Qki (Invitrogen, MSS208338, MSS276677), Mbnl1 (Invitrogen, MSS226392, MSS226393), Mbnl2 (Invitrogen, MSS272396, MSS200587), Celf1 (Invitrogen, MSS203372, MSS203374), Celf2 (Invitrogen, MSS274200, MSS204012), Eya3 (Dharmacon, D-065562-01, D-065562-02), Six4 (Dharmacon, D-057996-01-0002), or Zbtb1 (Dharmacon, D-057238-02-0002) were delivered using the Lipofectamine RNAiMax transfection reagent (Invitrogen, #13778075) according to the manufacturer’s protocol. To avoid previously reported differentiation defects in C2C12 cells following RBFOX2 depletion,12 myoblasts were differentiated for two days prior to transfection with si-RBFOX2 (Dharmacon, D-051552-01-0002, D-051552-04-0002), and then differentiated for two additional days prior to sample collection. Protein lysates and RNA were prepared from myoblasts 24 h after PTBP1 and PTBP2 depletion. After EYA3, SIX4, ZBTB1, QKI, MBNL1, MBNL2, CELF1, and CELF2 si-RNA depletion, C2C12 myoblasts were maintained in six-well plates in growth media until they reached 80-100% confluency and then differentiated for four to six days prior to collection of RNA and protein lysates. Negative control si-RNAs (si-Ctrl) were obtained from the same manufacturer as their respective targeting si-RNAs. For experiments using si-RNAs from Invitrogen, an si-RNA targeting the luciferase gene (Invitrogen, #12935146) was used as a negative control. For experiments using si-RNAs from Dharmacon, a non-targeting si-RNA #1 (Dharmacon, D-001210-01-20) was used as a negative control.
Tissue homogenization, RNA extraction, and cDNA synthesis
Tissues were flash frozen in liquid nitrogen and transferred into tubes containing 1.4 mm ceramic beads (Lysing Matrix D, MP Biomedical) with TRIzol (Invitrogen). Tissues were pulverized using a Precellys-24 homogenizer (Bertin Instruments) with the following settings: 6,500 r.p.m. for 20 s intervals. For RT-PCR and qPCR assays, RNA was extracted using TRIzol and purified via phenol-chloroform extraction, according to the manufacturer’s protocol. For RNA-seq studies, RNA was extracted using the RNeasy mini kit (QIAGEN, #74104) according to the manufacturer’s protocol. To determine RNA concentration and quality, a NanoDrop Lite Spectrophotometer (ND-LITE, Thermo Fisher Scientific) was used. cDNA was prepared by reverse-transcribing RNA using the High-capacity cDNA reverse transcription kit (Applied Biosystems, #4368813) with RNase inhibitors (Applied Biosystems, N8080119). The following thermocycler parameters were used: (i) 25°C for 10 min, (ii) 37°C for 120 min, (iii) 85°C for 5 min, (iv) 4°C hold.
RNA-seq, RNA-seq data analysis, and gene ontology analysis
Three replicates for each treatment condition were generated for the RNA-seq studies. An HTX Synergy plate reader (BioTek) equipped with the Take3 micro-volume plate accessory (BioTek) and electrophoresis on a TapeStation instrument (Agilent) were used to assess RNA quality. All samples passed the following quality thresholds: (a) A260nm/A280nm ≥ 2.0, (b) A260nm/A230nm ≥ 1.5, (c) DV200 ≥ 95, and (d) RNA integrated number (RIN) ≥ 9.3. KAPA mRNA capture beads (Roche, #07962207001) were used to isolate polyadenylated RNAs. cDNA libraries were generated using the KAPA stranded mRNA-Seq kit. Samples were pooled and analyzed using an Illumina HiSeq4000 instrument with 75 bp paired end reads. Paired-end reads were aligned to the mouse genome (mm10) using STAR v2.7.6a.70 Salmon v1.4.071 was used to determine gene-level read counts. The DESeq2 package72 in R Studio was used to quantify differential gene expression. Differentially expressed genes were defined as those with |FC| > 1.25 (meaning FC > 1.25 for upregulation or FC < 1/1.25 for downregulation) and p-adj < 0.05. Enrichr80,81,82 was used to analyze differentially expressed genes. Reported p-values were based on Fisher’s exact test computed by Enrichr software.80,81,82
Alternative splicing evaluation by RT-PCR
Primers were designed to bind to constitutive exons flanking the alternative exon of Eya3 gene using the UCSC genome browser83 and Primer 3 (Table S1). Primers were diluted to a final concentration of 0.5 μM in GoTaq reagent (Promega, M7123) and nuclease-free water. The following thermocycling parameters were used for PCR amplification: (i) 95°C for 1 min 15 s, (ii) 28 cycles of 95°C for 45 s, 57°C for 45 s, 72°C for 1 min, (iii) 72°C for 10 min, (iv) 4°C hold. The PCR products were resolved via 6% polyacrylamide gel electrophoresis in TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) at 140 V for 1-2 h. Gels were incubated for 10 min in an ethidium bromide solution (0.4 μg/mL) at room temperature prior to imaging on the ChemiDocTM XRS+ (Biorad). Densitometry quantifications were performed using the Biorad Image Lab software. The product-length dependent staining bias of ethidium bromide was corrected using a standard curve based on the pUC19 ladder (Thermo Fisher Scientific, SM0221). The PSI was determined via the following equation:
qPCR
TaqMan® probes (Table S3) (Applied Biosystems) and cDNA were combined in Taqman Fast Advanced Master Mix (Applied Biosystems), according to the manufacturer’s instructions. Samples were analyzed in 96-well plates using an Applied Biosystems QuantStudio 7 Flex Real-Time PCR system. The following thermocycling parameters were used: (i) 95°C for 20 s and (ii) 40 cycles of 95°C for 1 s, 60°C for 20 s. To control for variation in input cDNA concentration, cycle threshold (CT) values of the gene targets were normalized to the CT of the hydroxymethylbilane synthase (Hmbs) control gene or glyceraldehyde-3-phosphate dehydrogenase (Gapdh) control gene from the same sample. All samples were evaluated in quadruplicate.
Cellular proliferation assays
Transfected C2C12 cells were plated at a density of 2,000 cells per well in 96-well plates. Cell viability was measured over the course of 72 h using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, G3580), according to manufacturer’s instructions.
Subcellular fractionation
Fractionation was performed by sucrose density gradient centrifugation.84 Briefly, C2C12 cells were homogenized in STM buffer (250 mM sucrose, 50 mM Tris-HCl, 5 mM MgCl2) containing protease inhibitors before being centrifuged at 800 xg for 15 min. The supernatant, containing the cytoplasmic fraction, was further purified by centrifugation, and precipitated in equal volume of acetone at -20°C for 1 h. The pellet, containing the nuclear fraction, was resuspended in STM buffer, centrifuged at 500 xg for 15 min, and the resulting pellet was resuspended in STM buffer.
Protein lysate preparation
Cells were first washed with ice cold PBS before being lysed in ice cold RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), pH 7.5) containing protease and phosphatase inhibitors (Thermo Fisher Scientific). A cell scraper was used to facilitate cell detachment and harvesting. Whole cell lysates were incubated on ice for 15 min, sonicated in an ice bath at 75 V for 3 min (30 s on, 30 s off), incubated on ice for an additional 15 min, and centrifuged for 10 min at 14,000 r.p.m at 4°C. Protein fractions were isolated and transferred into fresh tubes. The BCA protein assay kit (Prometheus Protein Biology Products, #18-441) was used to estimate protein concentration.
Co-immunoprecipitation assays
Protein lysates were collected with IP lysis buffer (Pierce, #87787) in low protein binding collection tubes (Thermo Fisher Scientific, #90410). Samples were precleared by incubating 25 mg of total protein with 50 μL of washed A/G magnetic beads (Pierce, #88802) for 20 min at room temperature on a rotating platform. A magnetic stand was used to isolate beads and the supernatant was transferred into a tube containing 5 μg antibody against the HA epitope (anti-HA from Thermo Fisher Scientific, #26183). Samples were incubated overnight at 4°C on a rotating platform. The next day, samples were transferred into tubes containing 25 μL washed A/G magnetic beads and were incubated at room temperature for 1 h on a rotating platform. The supernatant was collected as flow-through and beads were subjected to three washes with ice cold IP lysis buffer followed by four washes with ice cold TBST (pH 8.0). Samples were submitted for mass spectrometry analysis or were eluted by incubating beads with 150 μL HA synthetic peptide (Thermo Fisher Scientific, #26184) at 37°C for 10 min.
Sample preparation for mass spectrometry
Immunoprecipitated protein samples were subjected to on-bead trypsin digestion.85 Briefly, after the last wash buffer step during affinity purification, beads were resuspended in 50 μL of 50 mM ammonium bicarbonate (pH 8). On-bead digestion was performed by adding 1 μg trypsin and incubating, while shaking, overnight at 37°C. The next day, 0.5 μg trypsin was added and incubated at 37°C for an additional 3 h. Beads were pelleted, and supernatants transferred into fresh tubes. The beads were washed twice with 100 μL LC-MS grade water, and washes added to the original supernatants. Samples were acidified by adding formic acid to final concentration of 2%. Peptides were desalted using peptide desalting spin columns (Thermo Fisher Scientific), lyophilized, and stored at -80°C until further analysis.
Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis
The peptide samples were analyzed by LC-MS/MS using an Easy nLC 1200 coupled to a QExactive HF mass spectrometer (Thermo Fisher Scientific). Samples were injected onto an Easy Spray PepMap C18 column (75 μm id × 25 cm, 2 μm particle size) (Thermo Fisher Scientific) and separated over a 2 h method. The gradient for separation consisted of 5–45% mobile phase B at a 250 nL/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in acetonitrile (ACN). The QExactive HF was operated in data-dependent mode where the 15 most intense precursors were selected for subsequent fragmentation. Resolution for the precursor scan (m/z 300–1600) was set to 120,000, while MS/MS scans resolution was set to 15,000. The normalized collision energy was set to 27% for higher-energy collisional dissociation (HCD). Peptide match was set to preferred, and precursors with unknown charge or a charge state of 1 and ≥ 7 were excluded.
Mass spectrometry data analysis
Raw data files were processed using MaxQuant (version 1.6.12.0) and searched against the reviewed human database (containing 20,203 entries), appended with a contaminants database, using Andromeda within MaxQuant. Enzyme specificity was set to trypsin, up to two missed cleavage sites were allowed, and methionine oxidation and N-terminus acetylation were set as variable modifications. A 1% false discovery rate (FDR) was used to filter all data. Match between runs was enabled (5 min match time window, 20 min alignment window), and a minimum of two unique peptides was required for label-free quantitation using the label-free quantification (LFQ) intensities. Perseus (version 1.6.14.0) was used for further processing.86 Only proteins with >1 unique+razor peptide were used for LFQ analysis. Proteins with 50% missing values were removed and missing values were imputed. Log2(FC) ratios were calculated using the averaged log2(LFQ) intensities and students t-test performed for each pair-wise comparison. FC was computed from log2(FC). To control for the immunoprecipitation peptides that interact with beads or the HA tag, we performed immunoprecipitations from lysates derived from cells transfected with an identical plasmid backbone but containing the HA tag alone (control samples). Proteins with p-value < 0.05 and FC > 2 compared with those detected in the control samples, and with < 2 hits in the contaminant repository for affinity purification database were considered interactors.
Western blotting
Protein samples and loading buffer (50 mM Tris-HCl pH 6.8, 12.5 mM EDTA, 10% glycerol, 2% SDS, 0.02% bromophenol blue, 360 mM beta-mercaptoethanol) were combined before being boiled for 5 min. Denatured samples were resolved by SDS polyacrylamide gel electrophoresis (SDS-PAGE) using the Mini-PROTEAN Tetra vertical electrophoresis system (BioRad) and TGX Stain-free gels (BioRad). The following parameters were used for electrophoresis: (i) 90 V for 30 min, (ii) 130 V for 45-75 min. Trihalo fluorophore compounds in TGX gels were activated 2.5 min under UV light to visualize fluorescent compounds bound to tryptophan residues in protein samples using a ChemiDocTM XRS+ imaging system (BioRad). The Mini Trans-Blot Cell system (BioRad) was used to transfer proteins into Amersham Hybond Low Fluorescence 0.2 μm PVDF membranes (GE Healthcare Life Sciences, #10600022). Following protein transfer, total transferred protein was imaged using the ChemiDocTM XRS+ imaging system (BioRad) before being blocked with a 5% nonfat dry milk solution in tris-buffered saline with tween (TBST) (20 mM Tris-base, 137 mM NaCl, 0.1% tween 20, pH 7.6) for 1 h at room temperature. Blots were then rinsed with TBST, and incubated overnight at 4°C with primary antibodies diluted in 1% bovine serum albumin (BSA) in TBST: anti-EYA3 (Bethyl Laboratories, A302-689A-T, 1:500), anti-HA (Thermo Fisher Scientific, #26183, 1:500 or 1:1,000), anti-myosin heavy chain (MYH) (Santa Cruz Biotechnology, sc-376157, 1:500), anti-RBFOX2 (Santa Cruz Biotechnology, sc-271407, 1:500), anti-PTBP1 (Abcam, ab133734, 1:500), anti-PTBP2 (Abcam, ab154787, 1:500), anti-MBNL1 (Santa Cruz Biotechnology, sc-515374, 1:500), anti-MBNL2 (Santa Cruz Biotechnology, sc-136167, 1:500), anti-CELF1 (Santa Cruz Biotechnology, sc-20003, 1:200), anti-CELF2 (Santa Cruz Biotechnology, sc-47731, 1:200), anti-QKI (Abcam, ab126742, 1:500), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, sc-365062, 1:500), anti-histone H3 (Abcam, ab1791, 1:500 or 1:1000), anti-ZBTB1 (Proteintech, #26287-1-AP, 1:250), anti-SIX4 (Proteintech, #21305-1-AP, 1:250), anti-TP53 (Cell Signaling Technology, #9282, 1:250), anti-SIX2 (Proteintech, #11562-1-AP, 1:250), anti-SIX5 (Proteintech, #22938-1-AP, 1:250), anti-RACK1 (Santa Cruz Biotechnology, sc-17754, 1:250), anti-AIFM1 (Santa Cruz Biotechnology, sc-13116, 1:200). Primary antibody solutions were removed, and blots were subjected to three, 10 min washes with TBST before being incubated with an anti-rabbit Dylight 800 secondary antibody (Thermo Fisher Scientific, SA5-35571) or anti-mouse Dylight 800 secondary antibody (Thermo Fisher Scientific, SA5-10176) solution diluted (1:10,000) in 1% BSA in TBST for 1 h in light blocking boxes. Blots were subjected to three, 10 min washes with TBST prior to imaging on a Li-Cor Odyssey CLx Imager. Target signals were quantified via densitometry using ImageJ or Biorad Image Lab software and total transferred protein was quantified using the Biorad Image Lab software. For quantifications, target signals were normalized to total transferred protein (full lanes) from the same sample.
Immunofluorescence confocal microscopy and image processing
For fixation, cells were incubated with 4% paraformaldehyde diluted in PBS for 20 min with agitation before being subjected to three, 10 min washes with PBS. Cells were incubated in blocking solution, containing 1% BSA, and 0.3% Triton in PBS for 1 h. Samples were incubated overnight at 4°C with primary antibodies diluted in blocking solution: anti-MYH, (Santa Cruz Biotechnology, sc-376157, 1:150). Cells were subjected to three, 10 min washes with PBS before being incubated with goat anti-mouse IgG (H+L) Alexa Fluor 488 from Thermo Fisher Scientific (A-11001) diluted (1:150) in blocking solution for 1 h at room temperature. Cells were subjected to three washed three, 10 min washes with PBS before being incubated with DAPI (2 μM) diluted in PBS for 5 min, followed by three, 10 min washes, all at room temperature. A Zeiss 880 confocal microscope equipped with a Plan-Apo 10X objective (NA = 0.45, working distance = 2.0 mm), Plan-Apo 63x objective (NA = 1.4, oil), and Zen Black (Zeiss) image acquisition software were used to capture images. For C2C12 cell studies, a 3x3 tile scan was performed to capture images from nine neighboring fields and stitched using Zen Black software. The following lasers and fluorophore pairs were used: argon multiline laser at 488 nm (Alexa Fluor 488), a 561 nm laser diode (Alexa Fluor 555), a helium-neon laser at 633 (Alexa Fluor 633 Phalloidin), and a 405 nm laser diode (DAPI). Images of C2C12 cells were captured with the following emission filters: band-pass 490-615 nm (Alexa Fluor 488), and band pass 410-514 nm (DAPI). Images of HEK293T cells were captured with the following emission filters: band-pass 490-540 nm (Alexa Fluor 488), band-pass 570-615 nm (Alexa Fluor 555), band-pass 638-747 nm (Alexa Fluor 633 Phalloidin), and band pass 410-514 nm (DAPI). Images were processed using ImageJ software. Fusion index was calculated as the percent of nuclei within MYH expressing myotubes with three or more nuclei. Cell counts were determined using the Cell Counter plugin in ImageJ.
Quantification and statistical analysis
In most cases, p-values were determined via unpaired Student’s T test (two-tailed) with those below 0.05 considered statistically significant. The exceptions to this approach were: (i) the analyses of overlap between differentially expressed genes following si-Eya3 #1 and si-Eya3 #2 treatment and GO enrichment analysis which were determined by a Fisher’s exact test, and (ii) the analyses of correlation, which were based on a T distribution under the null hypothesis of Pearson (ρ) = 0. Data are reported as mean ± standard error of the mean (SEM).
Acknowledgments
We thank the following laboratories at The University of North Carolina at Chapel Hill for their donation of skeletal muscle tissues from different species: (i) Dr. Kathleen M. Caron (zebrafish embryos and adult), Dr. Michael Bressan (chicken embryos), Dr. Natasha Snider (adult rat), and Dr. Sarah Cohen (neonatal rat). We thank the laboratory of Dr. Thomas A. Cooper at Baylor College of Medicine for their donation of DM1 cDNA samples. We would like to thank Dr. Joan Taylor (The University of North Carolina at Chapel Hill) for providing feedback on this project. We thank Matt Soloway of the UNC Center for Bioinformatics for their technical support in the deposition of RNA-seq datasets in GEO. The work in Dr. Giudice’s laboratory is supported by The University of North Carolina at Chapel Hill (start-up funds and a Jefferson Pilot Award, J.G.), the National Institutes of Health (NIH-NIGMS R01GM130866, J.G.), the American Heart Association (19CDA34660248, J.G.), and the National Science Foundation (NSF, CAREER Award, number 2239056, J.G.). H.J.W. was supported by a merit-doctoral fellowship from the Graduate School at The University of North Carolina at Chapel Hill, a NIH-NIGMS training award (T32GM119999), and by the National Science Foundation (NSF, DGE-1650116). We acknowledge the support of the Genetics and Molecular Biology Curriculum (GMB), the Program in Translational Medicine, and the Mechanistic, and the Interdisciplinary Biology (MiBio) Graduate Training Program, all at The University of North Carolina at Chapel Hill.
We gratefully acknowledge the technical support from the High Throughput Sequencing Facility at The University of North Carolina at Chapel Hill. This facility is supported by the University Cancer Research Fund, Comprehensive Cancer Center Core Support grant (P30-CA016086), and UNC Center for Mental Health and Susceptibility grant (P30-ES010126). Microscopy was performed at the Hooker Imaging Core Facility at The University of North Carolina at Chapel Hill which is supported in part by the P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. Mass spectrometry studies were conducted at the UNC Proteomics Core Facility, which is supported in part by the NCI Center Core Support Grant (P30-CA016086) to the UNC Lineberger Comprehensive Cancer Center. Organism silhouettes used in some figures were generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license and PhyloPic under the Public Domain Dedication 1.0 license.
Author contributions
Conceptualization, H.J.W. and J.G.; investigation, H.J.W. and R.E.B.; formal analysis, H.J.W., R.E.B., M.S., C.A.M., and L.E.H.; writing – original draft, H.J.W and J.G.; writing – review & editing, H.J.W., R.E.B., M.S., C.A.M., X.H.T.W., L.E.H., and J.G.; funding acquisition, J.G.; resources, X.H.T.W., L.H., and J.G.; supervision, J.G.
Declaration of interests
X.H.T.W. is a founding partner of Elex Biotech, a start-up company that develops drug molecules that target ryanodine receptors to treat cardiac arrhythmias. X.H.T.W. is also a consultant to Rocket Pharmaceuticals. The remaining authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108258.
Supplemental information
Differentially expressed genes were defined as those with FC > 1.25, p-adj <0.05 in both si-Eya3 #1 and si-Eya3 #2.
Interactors were defined as those with FC > 2 and p < 0.05, compared to HA tag control.
Data and code availability
-
•
Raw RNA-seq data files have been deposited at NCBI Gene Expression Omnibus (GEO) and the proteomic datasets generated and analyzed in this study have been deposited at the Proteomics Identification Database (PRIDE) repository. These deposited data are publicly available as of the date of this publication and accession codes are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required concerning the data reported in this paper is available from the lead contact upon request.
References
- 1.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merkin J., Russell C., Chen P., Burge C.B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:1–15. doi: 10.1186/GB-2004-5-10-R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan Z., Garrett-Engele P.W., Johnson J.M., Castle J.C. Evolutionarily conserved and diverged alternative splicing events show different expression and functional profiles. Nucleic Acids Res. 2005;33:5659–5666. doi: 10.1093/NAR/GKI834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthès P., Kokkinaki M., Nef S., Gnirke A., et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/J.CELREP.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Giudice J., Xia Z., Wang E.T., Scavuzzo M.A., Ward A.J., Kalsotra A., Wang W., Wehrens X.H.T., Burge C.B., Li W., Cooper T.A. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat. Commun. 2014;5:3603. doi: 10.1038/ncomms4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinegar A.E., Xia Z., Loehr J.A., Li W., Rodney G.G., Cooper T.A. Extensive alternative splicing transitions during postnatal skeletal muscle development are required for calcium handling functions. Elife. 2017;6:e27192. doi: 10.7554/eLife.27192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazin P., Xiong J., Liu X., Yan Z., Zhang X., Li M., He L., Somel M., Yuan Y., Phoebe Chen Y.P., et al. Widespread splicing changes in human brain development and aging. Mol. Syst. Biol. 2013;9:633. doi: 10.1038/MSB.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X., Chen M.H., Wu X., Kodani A., Fan J., Doan R., Ozawa M., Ma J., Yoshida N., Reiter J.F., et al. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell. 2016;166:1147–1162.e15. doi: 10.1016/J.CELL.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng S., Gray E.E., Chawla G., Porse B.T., O’Dell T.J., Black D.L. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15:381–388. doi: 10.1038/NN.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R.K., Xia Z., Bland C.S., Kalsotra A., Scavuzzo M.A., Curk T., Ule J., Li W., Cooper T.A. Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol. Cell. 2014;55:592–603. doi: 10.1016/j.molcel.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S., Faralli H., Yao Z., Rakopoulos P., Palii C., Cao Y., Singh K., Liu Q.C., Chu A., Aziz A., et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 2013;27:1247–1259. doi: 10.1101/GAD.215400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinkle E.R., Wiedner H.J., Torres E.V., Jackson M., Black A.J., Blue R.E., Harris S.E., Guzman B.B., Gentile G.M., Lee E.Y., et al. Alternative splicing regulation of membrane trafficking genes during myogenesis. RNA. 2022;28:523–540. doi: 10.1261/rna.078993.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giudice J., Loehr J.A., Rodney G.G., Cooper T.A. Alternative splicing of four trafficking genes regulates myofiber structure and skeletal muscle physiology. Cell Rep. 2016;17:1923–1933. doi: 10.1016/j.celrep.2016.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulay G., Lainé J., Lemaître M., Nakamori M., Nishino I., Caillol G., Mamchaoui K., Julien L., Dingli F., Loew D., et al. Alternative splicing of clathrin heavy chain contributes to the switch from coated pits to plaques. J. Cell Biol. 2020;219:e201912061. doi: 10.1083/jcb.201912061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fugier C., Klein A.F., Hammer C., Vassilopoulos S., Ivarsson Y., Toussaint A., Tosch V., Vignaud A., Ferry A., Messaddeq N., et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 18.Blue R.E., Koushik A., Engels N.M., Wiedner H.J., Cooper T.A., Giudice J. Modulation of alternative splicing of trafficking genes by genome editing reveals functional consequences in muscle biology. Int. J. Biochem. Cell Biol. 2018;105:134–143. doi: 10.1016/j.biocel.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böhm J., Vasli N., Maurer M., Cowling B.S., Shelton G.D., Kress W., Toussaint A., Prokic I., Schara U., Anderson T.J., et al. Altered splicing of the BIN1 muscle-specific exon in humans and dogs with highly progressive centronuclear myopathy. PLoS Genet. 2013;9:e1003430. doi: 10.1371/journal.pgen.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P., et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 23.Roychoudhury K., Hegde R.S. The eyes absent proteins: unusual HAD family tyrosine phosphatases. Int. J. Mol. Sci. 2021;22:3925. doi: 10.3390/IJMS22083925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tadjuidje E., Hegde R.S. The Eyes absent proteins in development and disease. Cell. Mol. Life Sci. 2013;70:1897–1913. doi: 10.1007/S00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonini N.M., Leiserson W.M., Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 26.Li X., Oghi K.A., Zhang J., Krones A., Bush K.T., Glass C.K., Nigam S.K., Aggarwal A.K., Maas R., Rose D.W., Rosenfeld M.G. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/NATURE02083. [DOI] [PubMed] [Google Scholar]
- 27.Xu P.X., Adams J., Peters H., Brown M.C., Heaney S., Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 28.Zou D., Silvius D., Rodrigo-Blomqvist S., Enerbäck S., Xu P.X. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev. Biol. 2006;298:430–441. doi: 10.1016/J.YDBIO.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohto H., Kamada S., Tago K., Tominaga S.-I., Ozaki H., Sato S., Kawakami K. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H., Stavnezer E. Ski regulates muscle terminal differentiation by transcriptional activation of Myog in a complex with Six1 and Eya3. J. Biol. Chem. 2009;284:2867–2879. doi: 10.1074/JBC.M807526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda K., Watanabe Y., Ohto H., Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/GR.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freyermuth F., Rau F., Kokunai Y., Linke T., Sellier C., Nakamori M., Kino Y., Arandel L., Jollet A., Thibault C., et al. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat. Commun. 2016;7:11067. doi: 10.1038/ncomms11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundberg I.E., De Visser M., Werth V.P. Classification of myositis. Nat. Rev. Rheumatol. 2018;14:269–278. doi: 10.1038/NRRHEUM.2018.41. [DOI] [PubMed] [Google Scholar]
- 35.Vartuli R.L., Zhou H., Zhang L., Powers R.K., Klarquist J., Rudra P., Vincent M.Y., Ghosh D., Costello J.C., Kedl R.M., et al. Eya3 promotes breast tumor-associated immune suppression via threonine phosphatase-mediated PD-L1 upregulation. J. Clin. Invest. 2018;128:2535–2550. doi: 10.1172/JCI96784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Zhou H., Li X., Vartuli R.L., Rowse M., Xing Y., Rudra P., Ghosh D., Zhao R., Ford H.L. Eya3 partners with PP2A to induce c-Myc stabilization and tumor progression. Nat. Commun. 2018;9:1047. doi: 10.1038/s41467-018-03327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sano T., Nagata S. Characterization of the threonine-phosphatase of mouse eyes absent 3. FEBS Lett. 2011;585:2714–2719. doi: 10.1016/j.febslet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Yaffe D., Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 39.Blau H.M., Pavlath G.K., Hardeman E.C., Chiu C.P., Silberstein L., Webster S.G., Miller S.C., Webster C. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 40.Pandey R.N., Rani R., Yeo E.J., Spencer M., Hu S., Lang R.A., Hegde R.S. The eyes absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Z.Y., He T.T., Gao X.M., Zhao Y., Wang J. ZBTB transcription factors: key regulators of the development, differentiation and effector function of T cells. Front. Immunol. 2021;12:713294. doi: 10.3389/FIMMU.2021.713294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Lu Y., Cao X., Zhen T., Kovalovsky D. Zbtb1 prevents default myeloid differentiation of lymphoid-primed multipotent progenitors. Oncotarget. 2016;7:58768–58778. doi: 10.18632/ONCOTARGET.11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams R.T., Guarecuco R., Gates L.A., Barrows D., Passarelli M.C., Carey B., Baudrier L., Jeewajee S., La K., Prizer B., et al. ZBTB1 regulates asparagine synthesis and leukemia cell response to L-asparaginase. Cell Metabol. 2020;31:852–861.e6. doi: 10.1016/j.cmet.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marston S., Memo M., Messer A., Papadaki M., Nowak K., Mcnamara E., Ong R., El-Mezgueldi M., Li X., Lehman W. Mutations in repeating structural motifs of tropomyosin cause gain of function in skeletal muscle myopathy patients. Hum. Mol. Genet. 2013;22:4978–4987. doi: 10.1093/HMG/DDT345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lampson M.A., Kapoor T.M. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 2005;7:93–98. doi: 10.1038/NCB1208. [DOI] [PubMed] [Google Scholar]
- 46.Hall M.P., Nagel R.J., Fagg W.S., Shiue L., Cline M.S., Perriman R.J., Donohue J.P., Ares M. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 2013;19:627–638. doi: 10.1261/rna.038422.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A., Swanson M.S. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang E.T., Ward A.J., Cherone J.M., Giudice J., Wang T.T., Treacy D.J., Lambert N.J., Freese P., Saxena T., Cooper T.A., Burge C.B. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25:858–871. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bentzinger C.F., Wang Y.X., Rudnicki M.A. Building muscle: molecular regulation of myogenesis. Cold Spring Harbor Perspect. Biol. 2012;4:a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sincennes M.-C., Brun C.E., Rudnicki M.A. Concise review: Epigenetic regulation of myogenesis in health and disease. Stem Cells Transl. Med. 2016;5:282–290. doi: 10.5966/sctm.2015-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirai H., Verma M., Watanabe S., Tastad C., Asakura Y., Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J. Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J.F., Tao Y., Li J., Deng Z., Yan Z., Xiao X., Wang D.Z. MicroRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedner H.J., Torres E.V., Blue R.E., Tsai Y.H., Parker J., Giudice J. SET domain containing 2 (SETD2) influences metabolism and alternative splicing during myogenesis. FEBS J. 2022;289:6799–6816. doi: 10.1111/febs.16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enders A., Stankovic S., Teh C., Uldrich A.P., Yabas M., Juelich T., Altin J.A., Frankenreiter S., Bergmann H., Roots C.M., et al. ZBTB7B (Th-POK) regulates the development of IL-17-producing CD1d-restricted mouse NKT cells. J. Immunol. 2012;189:5240–5249. doi: 10.4049/JIMMUNOL.1201486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao A.P., Ishizuka I.E., Kasal D.N., Mandal M., Bendelac A. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 2017;8:863. doi: 10.1038/S41467-017-00882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davenport J.W., Fernandes E.R., Harris L.D., Neale G.A., Goorha R. The mouse mitotic checkpoint gene Bub1b, a novel Bub1 family member, is expressed in a cell cycle-dependent manner. Genomics. 1999;55:113–117. doi: 10.1006/GENO.1998.5629. [DOI] [PubMed] [Google Scholar]
- 57.Loell I., Helmers S.B., Dastmalchi M., Alexanderson H., Munters L.A., Nennesmo I., Lindroos E., Borg K., Lundberg I.E., Esbjörnsson M. Higher proportion of fast-twitch (type II) muscle fibres in idiopathic inflammatory myopathies - evident in chronic but not in untreated newly diagnosed patients. Clin. Physiol. Funct. Imag. 2011;31:18–25. doi: 10.1111/J.1475-097X.2010.00973.X. [DOI] [PubMed] [Google Scholar]
- 58.Talbot J., Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley interdisciplinary reviews. Dev. Biol. 2016;5:518–534. doi: 10.1002/WDEV.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baudry D., Faussillon M., Cabanis M.O., Rigolet M., Zucker J.M., Patte C., Sarnacki S., Boccon-Gibod L., Junien C., Jeanpierre C. Changes in WT1 splicing are associated with a specific gene expression profile in Wilms’ tumour. Oncogene. 2002;21:5566–5573. doi: 10.1038/SJ.ONC.1205752. [DOI] [PubMed] [Google Scholar]
- 60.Ng I.H.W., Ng D.C.H., Jans D.A., Bogoyevitch M.A. Selective STAT3-α or -β expression reveals spliceform-specific phosphorylation kinetics, nuclear retention and distinct gene expression outcomes. Biochem. J. 2012;447:125–136. doi: 10.1042/BJ20120941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi S., Lee H.S., Cho N., Kim I., Cheon S., Park C., Kim E.-M., Kim W., Kim K.K. RBFOX2-regulated TEAD1 alternative splicing plays a pivotal role in Hippo-YAP signaling. Nucleic Acids Res. 2022;50:8658–8673. doi: 10.1093/nar/gkac509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee K.S., Cao Y., Witwicka H.E., Tom S., Tapscott S.J., Wang E.H. RNA-binding protein muscleblind-like 3 (MBNL3) disrupts myocyte enhancer factor 2 (Mef2) β-exon splicing. J. Biol. Chem. 2010;285:33779–33787. doi: 10.1074/JBC.M110.124255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu B., Ramachandran B., Gulick T. Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J. Biol. Chem. 2005;280:28749–28760. doi: 10.1074/JBC.M502491200. [DOI] [PubMed] [Google Scholar]
- 64.Dolfini D., Andrioletti V., Mantovani R. Overexpression and alternative splicing of NF-YA in breast cancer. Sci. Rep. 2019;9:12955. doi: 10.1038/S41598-019-49297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robles M.S., Boyault C., Knutti D., Padmanabhan K., Weitz C.J. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science (New York, N.Y.) 2010;327:463–466. doi: 10.1126/SCIENCE.1180067. [DOI] [PubMed] [Google Scholar]
- 66.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/HMG/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 67.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/PNAS.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith J.A., Curry E.G., Blue R.E., Roden C., Dundon S.E.R., Rodríguez-Vargas A., Jordan D.C., Chen X., Lyons S.M., Crutchley J., et al. FXR1 splicing is important for muscle development and biomolecular condensates in muscle cells. J. Cell Biol. 2020;219:e201911129. doi: 10.1083/jcb.201911129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamburger V., Hamilton H.L. A series of normal stages in the development of the chick embryo. 1951. Dev. Dynam. 1992;195:231–272. doi: 10.1002/AJA.1001950404. [DOI] [PubMed] [Google Scholar]
- 74.Goudy J., Henley T., Méndez H.G., Bressan M. Simplified platform for mosaic in vivo analysis of cellular maturation in the developing heart. Sci. Rep. 2019;9:10716. doi: 10.1038/S41598-019-47009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dynam. 1995;203:253–310. doi: 10.1002/AJA.1002030302. [DOI] [PubMed] [Google Scholar]
- 76.Matthews M., Varga Z.M. Anesthesia and euthanasia in zebrafish. ILAR J. 2012;53:192–204. doi: 10.1093/ILAR.53.2.192. [DOI] [PubMed] [Google Scholar]
- 77.deAlmeida A.C., van Oort R.J., Wehrens X.H. Transverse aortic constriction in mice. J. Vis. Exp. 2010;21:1729. doi: 10.3791/1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reynolds J.O., Quick A.P., Wang Q., Beavers D.L., Philippen L.E., Showell J., Barreto-Torres G., Thuerauf D.J., Doroudgar S., Glembotski C.C., Wehrens X.H.T. Junctophilin-2 gene therapy rescues heart failure by normalizing RyR2-mediated Ca 2+ release. Int. J. Cardiol. 2016;225:371–380. doi: 10.1016/J.IJCARD.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Respress J.L., Wehrens X.H. Transthoracic echocardiography in mice. J. Vis. Exp. 2010;28:1738. doi: 10.3791/1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/GR.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dimauro I., Pearson T., Caporossi D., Jackson M.J. A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res. Notes. 2012;5:513. doi: 10.1186/1756-0500-5-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rank L., Herring L.E., Braunstein M. Evidence for the mycobacterial Mce4 transporter being a multiprotein complex. J. Bacteriol. 2021;203:e00685-20. doi: 10.1128/JB.00685-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/NMETH.3901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes were defined as those with FC > 1.25, p-adj <0.05 in both si-Eya3 #1 and si-Eya3 #2.
Interactors were defined as those with FC > 2 and p < 0.05, compared to HA tag control.
Data Availability Statement
-
•
Raw RNA-seq data files have been deposited at NCBI Gene Expression Omnibus (GEO) and the proteomic datasets generated and analyzed in this study have been deposited at the Proteomics Identification Database (PRIDE) repository. These deposited data are publicly available as of the date of this publication and accession codes are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required concerning the data reported in this paper is available from the lead contact upon request.