Abstract
The ability to identify T cells that recognize specific peptide antigens bound to major histocompatibility complex (MHC) molecules has enabled enumeration and molecular characterization of the lymphocytes responsible for cell-mediated immunity. Fluorophore-labeled peptide:MHCI (p:MHCI) tetramers are well-established reagents for identifying antigen-specific CD8+ T cells by flow cytometry, but efforts to extend the approach to CD4+ T cells have been less successful perhaps owing to lower binding strength between CD4 and MHCII molecules. Here we show that p:MHCII tetramers engineered by directed evolution for enhanced CD4 binding outperform conventional tetramers for the detection of cognate T cells. Using the engineered tetramers, we identified about twice as many antigen-specific CD4+ T cells in mice immunized against multiple peptides than when using traditional tetramers. CD4 affinity enhanced p:MHCII tetramers therefore allow direct sampling of antigen-specific CD4+ T cells that cannot be accessed with conventional p:MHCII tetramer technology. These new reagents could provide a deeper understanding of the T cell repertoire.
ED SUMM:
The detection of antigen-specific CD4+ T cells is improved with engineered peptide:MHCII tetramers.
We hypothesized that the inherently weak affinity of CD4 for MHCII molecules 8,9 may contribute to the failure of p:MHCII tetramers to detect T cells that have low-affinity TCRs 7. To compensate for weak tetramer binding, we used directed evolution of the I-Ab MHCII molecule of C57BL/6 (B6) mice to produce mutant molecules with enhanced CD4 binding, reasoning that tetramers of these molecules would be better at detecting p:MHCII-specific T cells than conventional tetramers.
MHCII molecules are heterodimers of alpha and beta chains that interact to form a peptide-binding groove, which when occupied with a peptide is bound by TCRs 3, and a CD4 binding site on the MHCII beta chain, which consists in part of the amino acid residues E137, V142, I148, and L158 9,10. Based on this information, we generated a bacterial library of plasmids encoding an antigenic peptide called P5R 11 linked to I-Ab beta chains with random substitutions in the E137, V142, I148, and L158 codons (Fig. 1a) and flanked by flippase recognition target (FRT) sites 12. The plasmid library was introduced by transfection along with a plasmid encoding flippase into a CHO cell line containing the I-Ab alpha chain and a single FRT target site in the genome (Fig. 1b). Individual CHO cells then recombined a single I-Ab beta chain sequence into the target site. This strategy ensured that each CHO cell in the library displayed an I-Ab alpha/P5R:I-Ab beta heterodimer (referred to as P5R:I-Ab) with a unique combination of amino acids at the four targeted positions in the beta chain.
Fig.1 |. Identification of I-Ab molecules with enhanced binding to CD4.
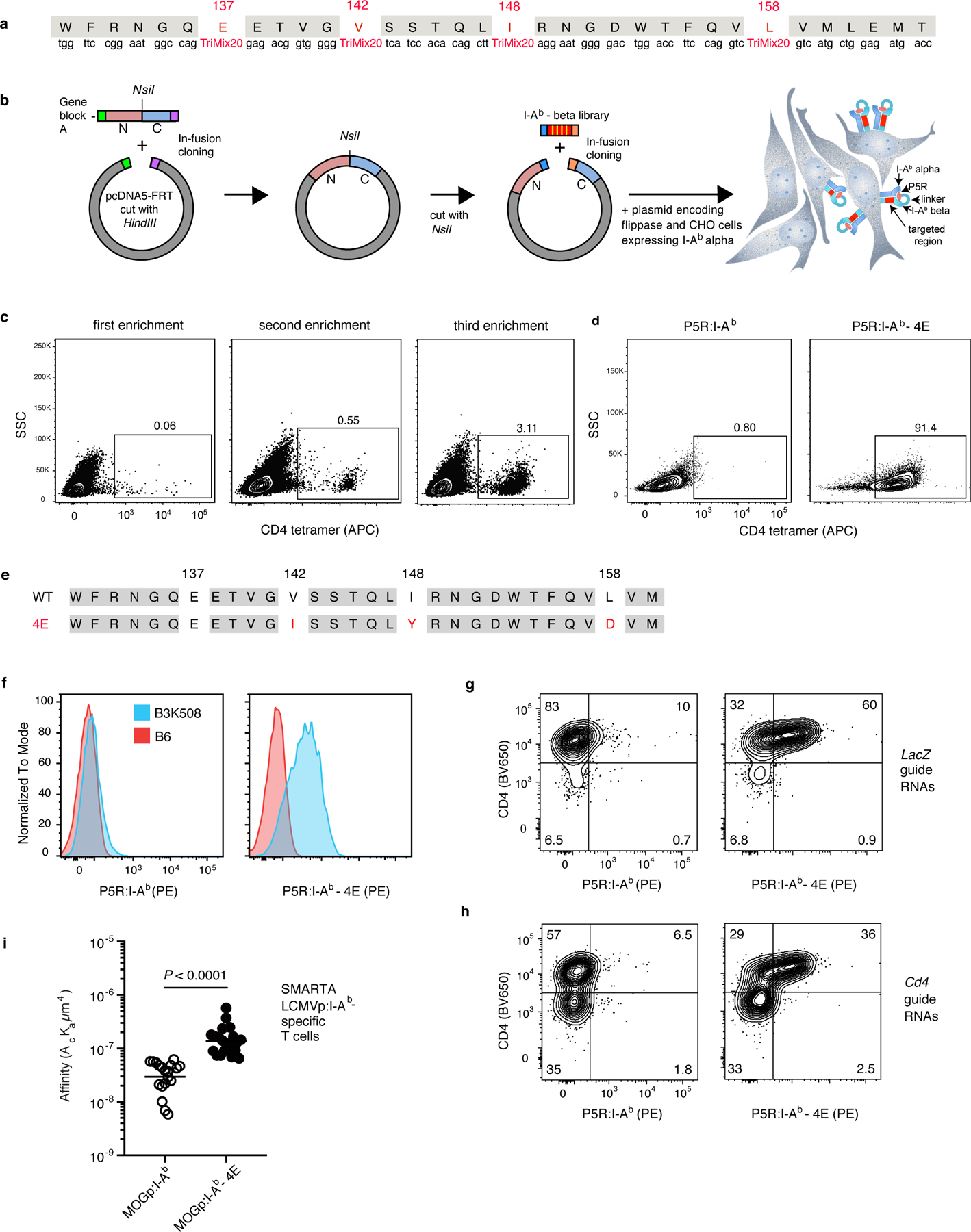
a, The region of the I-Ab beta chain targeted for generation of the mutant library. TriMix20 indicates a mixture of nucleotides encoding the following 20 codons (including CYS): AAA, AAC, ACT, ATC, ATG, CAG, CAT, CCG, CGT, CTG, GAA, GAC, GCT, GGT, GTT, TAC, TCT, TGC, TGG, TTC. b, Strategy for generation of a library of P5R:I-Ab beta chain vectors with substitutions at positions 137, 142, 148, and 158 and use of the vector mix to produce a library of I-Ab alpha chain-expressing CHO cells expressing a single P5R:I-Ab beta chain variant via flippase-mediated integration into a single FRT site. Yellow bars in the red segments indicate the four amino acid residues in the P5R:I-Ab beta chain that were targeted. N indicates the N-terminal portion of the I-Ab beta chain and C indicates the C-terminal portion of the I-Ab beta chain including the transmembrane domain and cytoplasmic tail. c, Flow cytometry plots of CHO populations from the transfected library stained with mouse CD4-streptavidin-APC tetramer, after the indicated rounds of enrichment with CD4-streptavidin-APC tetramer and APC antibody-conjugated magnetic beads. d, Flow cytometry plots of CD4-streptavidin-APC tetramer-stained CHO cells expressing wild-type P5R:I-Ab molecules, or a CHO clone expressing P5R:I-Ab-4E molecules. e, The amino acid sequences of the beta chains of P5R:I-Ab wild-type (top) or P5R:I-Ab-4E (bottom) molecules. Substitutions are shown in red. f, Flow cytometry histograms of CD4+ T cells from polyclonal B6 (red) or P5R:I-Ab-specific monoclonal B3K508 TCR transgenic mice (blue) stained with the indicated tetramers. g, h, B3K508 TCR transgenic T cells transduced with the retroviruses encoding either LacZ guide RNAs (g) or Cd4 guide RNAs (h) and stained with the indicated tetramers. Similar results were obtained in an independent experiment. i, Biomembrane probe force adhesion frequency assay measurements of the interactions between individual SMARTA TCR transgenic T cells and beads coated with the indicated p:I-Ab molecules. The two groups (n = 21 for each group from three independent experiments) were compared using an unpaired two-tailed Student’s t-test.
We also generated a mouse CD4/streptavidin-fluorophore tetramer and used it to select CD4 tetramer-binding cells from the CHO cell population that had been transfected with the library of P5R:mutant I-Ab molecules. CD4 tetramer-binding cells were enriched with several rounds of selection with magnetic beads conjugated with antibodies specific for the fluorophore component of the CD4/streptavidin-fluorochrome tetramer (Fig. 1c). Single CD4 tetramer-binding cells were sorted by flow cytometry after the third enrichment and expanded. Most of the enriched CHO clones stained uniformly with the CD4 tetramer, while CHO cells expressing conventional P5R:I-Ab molecules did not (Fig. 1d). Sequencing of genomic DNA revealed that multiple clones from the library contained I-Ab beta chains with the same V142I, I148Y, and L158D mutations (Fig. 1e). The I-Ab molecules containing these substitutions will be referred to as I-Ab-4E molecules.
P5R:I-Ab wild-type and P5R:I-Ab-4E tetramers linked to streptavidin-fluorophores were then generated and used to assess the capacity of these reagents to bind to CD4+ T cells from B3K508 TCR transgenic mice, which express a TCR that binds to P5R:I-Ab molecules with a low affinity (KD = 93 µM) 11. The conventional P5R:I-Ab tetramer bound to B3K508 T cells barely above background staining of the polyclonal B6 CD4+ T cell population (Fig. 1f). In contrast, the P5R:I-Ab-4E tetramer bound to B3K508 T cells in an amount that almost achieved baseline separation from the B6 population. Importantly, the P5R:I-Ab-4E tetramer did not bind to the total B6 T cell population appreciably better than the conventional tetramer (Fig. 1f, red histograms). Thus, although the P5R:I-Ab-4E tetramer bound cells expressing a P5R:I-Ab-specific TCR, it did not bind to the vast majority of polyclonal T cells in B6 mice. This finding rules out the possibility that the P5R:I-Ab-4E molecules bind all CD4+ T cells in a peptide-independent fashion.
We then determined whether the enhanced binding of P5R:I-Ab-4E molecules to B3K508 T cells was dependent on CD4 as expected based on how these molecules were selected. Retroviral vectors encoding Cas9, guide RNAs complementary to the LacZ or Cd4 genes, and a fluorescent reporter were transduced into B3K508 T cell blasts. The B3K508 population transduced with LacZ guide RNAs consisted of greater than 90% CD4+ cells most which bound to the P5R:I-Ab-4E not the wild-type P5R:I-Ab tetramer (Fig. 1g). The B3K508 population transduced with Cd4 guide RNAs contained a much larger subpopulation of CD4‒ cells as expected based on CRISPR/Cas9-mediated deletion of the Cd4 gene (Fig. 1h). These CD4-negative cells did not bind to P5R:I-Ab-4E tetramer, while many of the residual CD4+ cells did. These results demonstrate that the enhanced binding of P5R:I-Ab-4E tetramer to a low affinity P5R:I-Ab-specific TCR depends on CD4.
The affinity of CD4 molecules for I-Ab-4E or conventional I-Ab molecules was then assessed using the biomembrane force probe assay 7,13. SMARTA TCR transgenic T cells 14, which express CD4 and a TCR specific for a peptide (GP66) from the glycoprotein from Lymphocytic Choriomeningitis Virus 15 were used for this analysis. The I-Ab molecules used in this case, however, contained a peptide from mouse myelin oligodendrocyte glycoprotein (MOGp) that shares only one amino acid in the nonamer core 16 with the GP66 peptide and is thus unlikely to be an agonist for the SMARTA TCR. MOGp:I-Ab-4E or MOGp:I-Ab molecules were coated onto the surfaces of glass beads attached to biotinylated red blood cells (RBCs). Individual MHCII-coated bead/RBC complexes were aspirated onto one pipette and individual SMARTA TCR CD4+ transgenic T cells were aspirated onto an opposing pipette. The pipettes were moved together such that the SMARTA T cell contacted the MHCII-coated glass bead and then pulled apart. Deformation of the RBC:bead complex was detected by microscopy and the adhesion frequency and p:MHCII and TCR density were used to calculate the 2-dimensional affinity. As shown in Fig. 1i, bead-bound MOGp:I-Ab-4E molecules bound to SMARTA T cells with a 10-fold higher affinity than bead-bound MOGp:I-Ab molecules. As only MOGp:I-Ab-4E molecules were on the bead and the SMARTA TCR can bind weakly to MOGp:I-Ab-4E molecules at best, this affinity probably reflects CD4-I-Ab-4E interactions. These measurements indicate that I-Ab-4E molecules bind to CD4 with an affinity approaching that of MHCI molecules for CD8 17.
The capacity of p:I-Ab-4E tetramers to detect polyclonal CD4+ T cells was then tested in B6 mice immunized with the peptides of interest. Five peptides that bind I-Ab were chosen for study – P5R, which was in the set of mutant I-Ab molecules from which I-Ab-4E molecules were selected (Fig. 1), another non-mouse peptide called 2W known for its immunogenicity 18,19, GP66, OVA3, a peptide from the well-studied antigen chicken ovalbumin 20, and mouse MOGp. B6 mice were immunized with these peptides emulsified in complete Freund’s adjuvant (CFA). Seven days later, lymph node and spleen cell samples from individual mice were split and half of each sample was stained with p:I-Ab-allophycocyanin (APC) and p:I-Ab-phycoerythrin (PE) tetramers and half with p:I-Ab-4E-APC and p:I-Ab-4E-PE tetramers. Split samples from the same animals were used to reduce mouse to mouse variability and staining with the same tetramers conjugated with different fluorochromes was done to enhance the TCR specificity of the assay 16. The tetramer-bound cells were then enriched with magnetic particles coated with fluorophore antibodies to capture all of the tetramer-binding cells in the samples 19. The enriched samples were then analyzed by flow cytometry for tetramer-binding cells by gating sequentially on cells with the light scatter properties of single live lymphocytes that expressed the T cell marker CD90 but not a set of non-T cell lineage markers and expressed CD4 but not CD8 (Supplementary Fig. 1).
The CD4+ populations enriched with wild-type I-Ab tetramers from the spleen and lymph nodes of mice immunized with P5R, 2W, OVA3, GP66, or MOGp (Fig. 2a) consisted on average (rounded to two significant figures) of 5,600, 85,000, 2,200, 33,000, or 1,200 CD4+ tetramer-binding effector T cells per mouse. The variability in the number of effector T cells detected with these tetramers in peptide-primed mice is related to differences in the number of naïve cells specific for each of these epitopes 16. The relatively small response of the MOGp:I-Ab tetramer-binding population is likely related to immune tolerance mechanisms that operate for this self-epitope 21. The CD4+ populations enriched with I-Ab-4E tetramers from the spleen and lymph nodes of mice immunized with P5R, 2W, OVA3, GP66, or MOGp consisted on average of 9,700, 110,000, 4,600, 63,000, or 5,200 tetramer-binding effector cells (Fig. 2b), which represented 1.7-, 1.3-, 2.1-, 1.9, or 4.2-fold increases, respectively, over the numbers detected with the comparable conventional I-Ab tetramers (Fig. 2c). The majority of CD4+ T cells detected with the I-Ab or I-Ab-4E tetramers had the CD44hi phenotype of effector T cells (Fig. 2d). Thus, p:I-Ab-4E tetramers were superior to I-Ab tetramers at detecting epitope-specific CD4+ effector T cell populations expanded by immunization.
Fig. 2 |. Detection of polyclonal CD4+ T cells from immunized mice using p:I-Ab-4E tetramers.
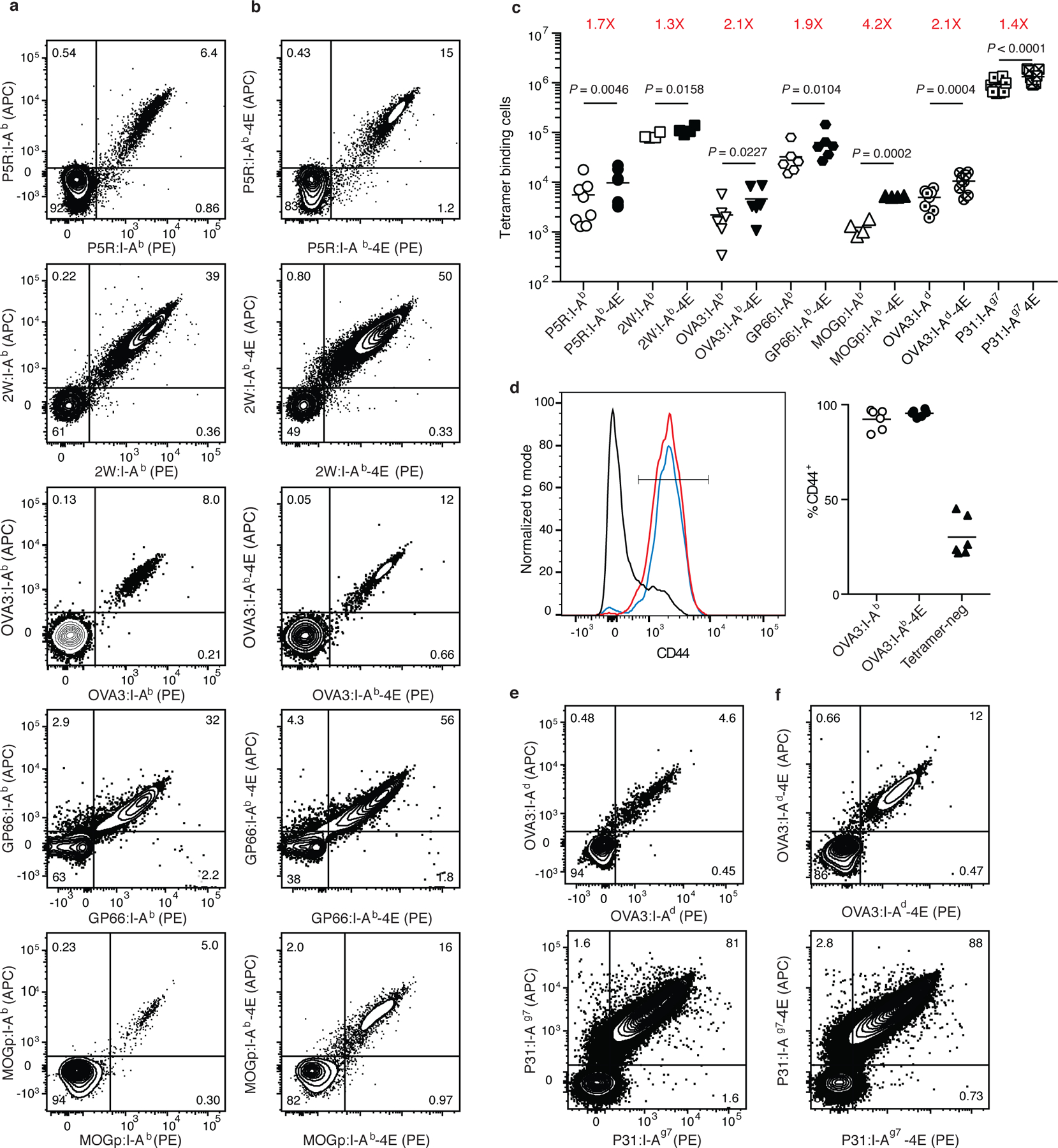
a-c, Representative flow cytometry contour plots of CD4+ T cells in split samples from the spleen and lymph nodes of individual B6 mice immunized with P5R, 2W, OVA3, GP66, or MOGp peptides in CFA seven days earlier, after enrichment with either p:I-Ab (a) or p:I-Ab-4E (b) tetramers. c, Scatter plots showing the number of CD4+ T cells detected with the indicated tetramers in split samples from individual mice (n = 4–9, from 2 independent experiments) with horizontal bars at the mean values. Mean values were compared with a paired Students t-test. P values are shown for each pair. d, Representative flow cytometry histograms of CD44 expression by OVA3:I-Ab (red), OVA3:I-Ab-4E (blue) or tetramer-negative (black) CD4+ T cells from B6 mice immunized with OVA peptide in CFA, and a scatterplot of the percentage of the T cells in the indicated populations that had a CD44high activated phenotype. e, f, Representative flow cytometry contour plots of CD4+ T cells in split samples from the spleen and lymph nodes of individual BALB/c (top row) or NOD (bottom row) mice immunized with OVA3 or P31 peptides in CFA seven days earlier, after enrichment with either p:I-Ab or p:I-Ag7 (e) or p:I-Ab-4E or p:I-Ag7-4E (f) tetramers.
Strains of mice other than the B6 are useful for immunological studies. We therefore determined whether the 4E mutations improve the utility of tetramers of allelic forms of MHCII molecules from other common mouse strains. The V142I, I148Y, and L158D mutations were thus introduced into the I-Ad beta chain of BALB/c and the I-Ag7 beta chain of NOD mice and incorporated into tetramers containing OVA3 20 or P31, a variant of an insulin B chain peptide 22, respectively. The OVA3:I-Ad-4E tetramer detected 2.1-fold more cells in OVA peptide/CFA-primed BALB/c mice (12,000 versus 6,000 cells), while the P31:I-Ag7-4E tetramer detected 1.4-fold more cells in P31/CFA-primed NOD mice (1,400,000 versus 1,000,000 cells) than the comparable wild-type tetramers (Fig. 2c, e, f). The large response to the P31 peptide is related to the fact that the P31:I-Ag7-specific naive T cell population is extraordinarily large 23. Thus, p:I-Ad-4E and p:I-Ag7-4E tetramers were superior to the wild-type versions at detecting epitope-specific CD4+ effector T cell populations.
We performed additional experiments to determine whether the cells detected with p:I-A-4E tetramers were truly specific for the peptide of interest. We took advantage of the fact that TCR signaling after immunization with peptide causes peptide-specific naïve T cells to proliferate and increase expression of CD44. B6 mice were immunized with P5R or MOGp with the expectation that P5R-immunized mice would contain an expanded population of CD44hi P5R:I-Ab-specific effector T cells and a small population of CD44lo MOGp:I-Ab-specific naive T cells, while mice immunized with MOGp would have the reciprocal pattern. The results of p:I-Ab-4E tetramer staining fit these expectations. B6 mice immunized with P5R contained an expanded population of CD44hi P5R:I-Ab-4E tetramer-binding cells but a small population of CD44lo MOGp:I-Ab-4E tetramer-binding cells, while mice immunized with MOGp contained an expanded population of CD44hi MOGp:I-Ab-4E tetramer-binding cells but a small population of CD44lo P5R:I-Ab-4E tetramer-binding cells (Fig. 3a–c). In either type of immunized mice, P5R:I-Ab-4E tetramer-binding cells did not bind MOGp:I-Ab-4E tetramer and MOGp:I-Ab-4E tetramer-binding cells did not bind P5R:I-Ab-4E tetramer (Fig. 3a, b). The facts that a p:I-Ab-4E tetramer only detected activation of T cells in mice injected with the relevant peptide and did not bind to T cells specific for a different p:I-Ab ligand are evidence that p:I-Ab-4E tetramer binding is peptide-specific.
Fig. 3 |. Detection of polyclonal CD4+ effector T cells p:I-Ab-4E tetramers is peptide-specific.
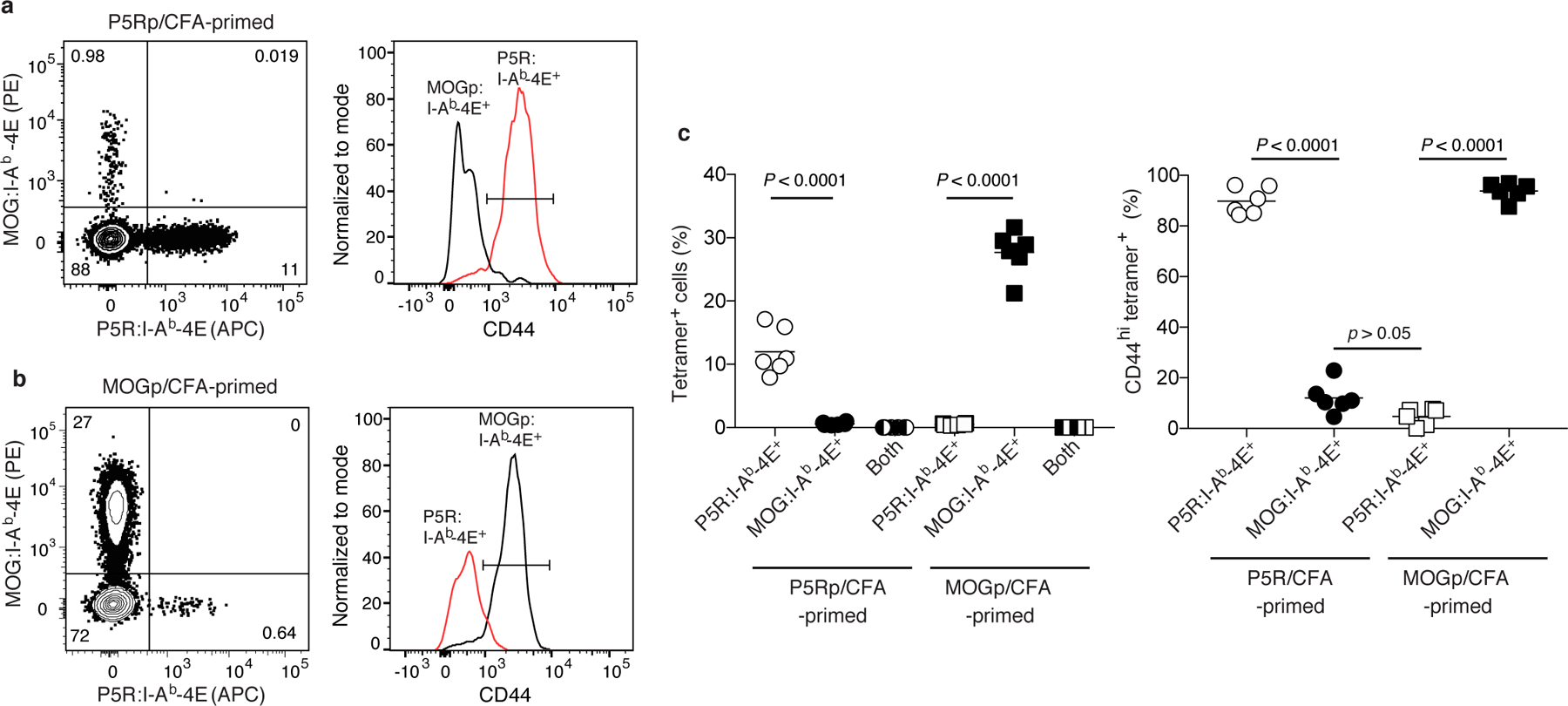
a, b, Representative flow cytometry contour plots of tetramer staining (left) and histograms of CD44 expression (right) by CD4+ T cells in split samples from P5R:I-Ab-4E and MOGp:I-Ab-4E tetramer-enriched samples of spleen and lymph nodes from B6 mice immunized with P5R (a) or MOGp (b) in CFA seven days earlier. c, Scatter plots showing the percentage of CD4+ T cells (left) from individual mice (n = 6, from 2 independent experiments) detected with the indicated tetramers in tetramer-enriched samples or the percentage of CD44hi cells among the indicated tetramer-binding populations (right) with horizontal bars at the mean values. Mean values were compared with by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. P values are shown.
Although tetramer technology has transformed T cell immunology, it has been limited to some degree by the failure these reagents to detect CD4+ T cells with TCRs at the low but biologically relevant end of the affinity spectrum 7. Our results show that this problem can be reduced with p:MHCII tetramers that bind to CD4 better than natural p:MHCII tetramers, offering an alternative to approaches based on increased MHCII valency 24. It remains to be seen, however, whether even CD4 affinity enhanced p:MHCII tetramers detect all CD4+ T cells capable of responding to a given peptide.
Our results and those of Williams et al. 25 showing that T cells expressing an evolved CD4 molecule with an increased capacity to bind wild-type MHCII could respond to lower doses of peptide than T cells expressing wild-type CD4 molecules provide evidence for a window in which CD4-MHCII interactions can be improved without a loss of TCR specificity for the peptide component of the p:MHCII complex. These conclusions were reached, however, from a limited set of experimental conditions, and thus more research is needed to confirm that CD4 affinity enhanced p:MHCII tetramers lack non-specific binding in all cases.
The CD4 affinity-enhanced tetramer-based approach should enable the study of epitope-specific CD4+ T cells that were invisible in previous studies. This could be a significant development since such T cells may have different functions than the ones previously detected with conventional p:MHCII tetramers. In addition, the peptide-bound CD4 affinity-enhanced MHCII molecules described here could eventually be used as therapeutics to either activate or inhibit epitope-specific T cells more potently than natural MHCII molecules.
METHODS
Mice
Female B6, BALB/c, NOD, and B6.Cg-Ptprca Pepcb Tg(TcrLCMV)1Aox/PpmJ (SMARTA) TCR transgenic mice 14 most 6–8 weeks old were purchased from the Jackson Laboratory or the National Cancer Institute Mouse Repository. Eric Huseby (University of Massachusetts) provided B3K508 TCR transgenic 11 Rag−/− mice. Mice were housed in specific pathogen-free conditions at the University of Minnesota or University of Utah. Experiments were conducted in accordance with University of Minnesota IACUC protocol# 1612-34360A, approved 9/23/2019.
Immunizations
Mice were immunized subcutaneously by tail base injection of 10 µg of P5R (EAQKARANKAVDKA) 11, 2W (EAWGALANWAVDSA) 18, MOG peptide 35–55 (MEVGWYRPPFSRVVHLYRNGK) 26, OVA peptide 323–339 (ISQAVHAAHAEINEAGR) 20, P31 (YVRPLWVRME), or GP66 (DIYKGVYQFKSV) 15 peptides emulsified in 50 µl of CFA.
Production of CHO cells stably expressing the I-Ab alpha chain
The Flp-In-CHO cell line (Thermo Fisher) was grown in Ham’s F12 media (Thermo Fisher) supplemented with 10% fetal bovine serum (FBS, 2 mM L-glutamine, 1% Pen-Strep and 100 µg/mL Zeocin (Thermo Fisher). A sequence encoding the full-length I-Ab alpha chain (NCBI Ref seq: NM_010378.2) was cloned into the pcDNA3.1-Neo vector (Invitrogen) and transfected into Flp-In CHO cells using Lipofectamine (ThermoFisher). Two days after transfection, cells were washed once with PBS and complete Ham’s media containing 1 mg/ml G418 Geneticin (Thermofisher) was added. Resistant cells expressing the I-Ab alpha chain were selected and expanded for further co-transfection with the I-Ab beta chain library as described below.
Molecular evolution of the I-Ab beta chain
A library of I-Ab beta chains with random substitutions in the codons for amino acids 137, 142, 148, and 158 (Fig. 1a) was generated in bacteria using standard molecular biology techniques (Fig. 1b). Briefly, the pcDNA5/FRT vector (Thermo Fisher) was linearized with HindIII (NEB) and gel purified. An 850 bp gene block (Gene block A shown below) containing coding sequences for the N-terminal and C-terminal flanking regions with an NsiI restriction site in the middle (underlined) and 15 bp overlapping segments at the ends was obtained from Integrated DNA Technologies.
Gene Block A:
GTTTAAACTTAAGCTTATGGCTCTGCAGATCCCCAGCCTCCTCCTCTCAGCTGCTGTG
GTGGTGCTGATGGTGCTGAGCAGCCCCGGGACTGAGGGCGGAGATTCCGAGGCCCA
GAAGGCCCGCGCCAACAAGGCCGTGGACAAGGCCGGCGGCGGAGGTACTAGTGGC
GGTGGAAGTGGAGGGTCTGAAAGGCATTTCGTGTACCAGTTCATGGGCGAGTGCTA
CTTCACCAACGGGACGCAGCGCATACGATATGTGACCAGATACATCTACAACCGGG
AGGAGTACGTGCGCTACGACAGCGACGTGGGCGAGCACCGCGCGGTGACCGAGCTG
GGGCGGCCAGACGCCGAGTACTGGAACAGCCAGCCGGAGATCCTGGAGCGAACGC
GGGCCGAGCTGGACACGGTGTGCAGACACAACTACGAGGGGCCGGAGACCCACAC
CTCCCTGCGGCGGCTTGAACAGCCCAATGTCGTCATCTCCCTGTCCAGGACAGAGGC
CCTCAACCACCACAACACTCTGGTCTGCTCAGTGACAGATTTCTACCCAGCCAAGAT
CAAAGTGCGCTGGTTCCGGAATGGCCAGATGCATGTCATGCTGGAGATGACCCCTCG
GCGGGGAGAGGTCTACACCTGTCACGTGGAGCATCCCAGCCTGAAGAGCCCCATCA
CTGTGGAGTGGAGGGCACAGTCTGAGTCTGCCTGGAGCAAGATGTTGAGCGGCATC
GGGGGCTGCGTGCTTGGGGTGATCTTCCTCGGGCTTGGCCTTTTCATCCGTCACAGG
AGTCAGAAAGGACCTCGAGGCCCTCCTCCAGCAGGGCTCCTGCAGTGAAGCTTGGT
ACCGAGC
Gene Block A was cloned into the HindIII-digested pcDNA5/FRT vector using In-Fusion cloning (Takara Bio USA). The resulting construct was sequence verified and linearized with NsiI, gel purified and PCR amplified with forward primer: 5’-GTCATGCTGGAGATGACCCCTCGG-3’ and reverse primer 5’- CTGGCCATTCCGGAACCAGCG-3’ to generate 17 bp overlaps for In-Fusion cloning of inserts containing the library. Single stranded oligonucleotides randomized at the indicated positions in Fig. 1a were obtained from Integrated DNA Technologies and converted to double stranded DNA with a reverse primer 5’-GTCATCTCCAGCATGAC-3’. Seventeen bp overlapping regions at the ends were included in this 100 bp fragment for In-Fusion cloning into the vector generated as described above. The fused products were transformed into Stellar competent Escherichia coli cells (Takara Bio USA) that were scaled up and selected in media containing carbenicillin. Some transformants were also selected on carbenicillin-containing plates, and plasmids from 29 E. coli clones were sequenced to confirm mutagenesis of the targeted sites. Plasmids were then isolated from the collection of E. coli cells grown in liquid culture and co-transfected with the pOG44 plasmid encoding flippase into a CHO cell line containing a single FRT target site and stably expressing the I-Ab alpha chain. Transfectants were selected by growth in hygromycin (500 µg/ml). During selection, flippase recombined the FRT site flanked DNA molecules encoding P5R:I-Ab beta chains into the FRT target site, one recombination per cell. The transfected population was then enriched for cells capable of binding to a mouse CD4-streptavidin-APC tetramer using APC-conjugated magnetic beads. Tetramer-binding single cells were then sorted by flow cytometry and their P5R:I-Ab beta chain DNA sequences were determined. The dominant clone in the population, which was named P5R:I-Ab-4E, contained V142I, I148Y, and L158D substitutions and was chosen for further study.
CD4 tetramer generation
Regions encoding the signal peptide and the extracellular domain from Cd4 cDNA ORF Clone (Sino Biological, Cat #: MG50134-CH, NCBI Ref Seq: NM_013488.2) was amplified using forward primer TD/mu.CD4-EcoR1-for: 5’ – CCCCCGAATTCATGTGCCGAGCCATCTCTCTTA – 3’ and reverse primer
TD/mu.CD4-Sal1-rev: 5’ – CCCCCGTCGACTTAATGATGATGATGATGATGTTCATGC CATTCAATTTTCTGCGCTTCAAAAATATCGTTCAGGCCGCTGCCGCCGCTGCCGCC TGTCTGGTTCACCCCTCTGGAT – 3’ using Platinum Pfx DNA Polymerase (Thermo Fisher). The reverse primer was designed to code for an AVI tag followed by a 6x His tag. The 1,288 bp amplicon was purified using MinElute PCR purification kit (Qiagen) and double digested with EcoRI (NEB) and Sal1(NEB) in CutSmart buffer. Purified restriction digested insert was ligated to similarly digested and purified pRMHa3 vector 27 using T4 DNA ligase (Thermo Fisher). The sequence verified construct of mu.CD4-pRMHa3 was co-transfected into Drosophila S2 cells with p18-BirA ligase and pCO-Blast plasmids. Transfection, selection, culture scale up, induction, purification and tetramerization were done as described below for pMHCII tetramer generation.
p:MHCII tetramers
Antigenic peptide sequences used for tetramer production were: P5R (EAQKARANKAVDKA), 2W (EAWGALANWAVDSA), GP66 (DIYKGVYQFKSV), MOGp (GWYRSPFSRVV), OVA3 (GHAAHAEINEA), and P31 (YVRPLWVRME). In most cases, single cistrons with a signal peptide, the antigenic peptide of interest, a flexible linker (GGGGTSGGGSGGS), wild-type I-Ab or I-Ab-4E beta chain sequences lacking the transmembrane domain and cytoplasmic tail, a short linker (GGGGS ), a basic leucine zipper domain (TTAPSAQLKKKLQALKKKNAQLKWKLQALKKKLAQ), and a 6x His epitope tag in N-terminal to C-terminal order were inserted into the pRMHa-3 vector with an upstream metallothionein promoter along the lines described by Moon et al. 19. p:I-A and p:I-Ag7 constructs were made in the same way except that wild-type p:I-Ad or p:I-Ag7 beta chain sequences or variants encoding V142I, I148Y, and L158D substitutions were used. The beta chain plasmids were co-transfected into Drosophila S2 cells via calcium phosphate with a pRMHa-3-based plasmid encoding a single cistron with a signal peptide, the I-Ab, I-Ad, or I-Ag7 alpha chain lacking the transmembrane domain and cytoplasmic tail, a short linker (GGGGS), an acidic leucine zipper domain (TTAPSAQLEKELQALEKENAQLEWELQALEKELAQ), a short linker (GGSGGS), BirA biotinylation signal sequence (GLNDIFEAQKIEWHE), and a 6x His epitope tag in N- to C-terminal order, along with another plasmid encoding a blasticidin resistance gene at a molar ratio of 9:9:1. A similar strategy was used for the MOGp:I-Ab, MOGp:I-Ab-4E, OVA3:I-Ab, OVA3:I-Ab-4E, OVA3:I-Ad, and OVA3:I-Ad-4E tetramers, except that linker sequence connecting the peptides to the beta chain was changed to include a cysteine residue (GCGGTSGGGSGGS). These beta chains were co-transfected with an I-A alpha chain plasmid with a substitution encoding a cysteine residue at position 72. Following translation, a disulfide bond is formed between the introduced cysteines, linking the beta and alpha chains and locking the OVA peptide into a specific binding register 28. Transfected cells were selected in blasticidin-containing media and induced with copper sulfate as described 19. Biotin-labeled p:I-A beta/I-A alpha and p:I-A-4E beta/I-A alpha heterodimeric monomers were purified from culture supernatants using nickel NTA and monovalent avidin columns as described 16. These biotin-labeled monomers were formed into tetramers by mixing in a 4:1 ratio with PE- or APC-conjugated streptavidin (Prozyme). The tetramers described here can be obtained upon request from the corresponding author or potentially in the future from a commercial source.
CRISPR/Cas9-based editing of the Cd4 gene
Two MSCV-based γ retroviral vectors were used to target the Cd4 gene as described by Kotov et al. 29. One vector encoded Cas9 and the fluorescent protein mNeongreen and the other either Cd4 or control LacZ guide RNAs (gRNAs) and the fluorescent protein mAmetrine 30. Both vectors were produced by modifying the LMP-Amt vector from S. Crotty (La Jolla Institute) as described by Kotov et al. 29. Platinum-E cells (Cell Biolabs) were grown in complete DMEM (Life Technologies) and transfected with the aforementioned plasmids. Virus-containing supernatants were harvested several days later. B3K508 T cells were cultured with complete IMDM (MilliporeSigma) containing IL-7 (Tonbo Biosciences) and then activated in plates coated with CD3 (2C11; Bio X Cell) and CD28 (37.51; Bio X Cell) antibodies. The cells were transduced with retroviral supernatant and polybrene (MilliporeSigma) 24 and 40 hours after activation and transferred to uncoated plates without CD3 or CD28 Abs for five days, the last three in complete IMDM-containing IL-7 (Tonbo Biosciences). The cells were then stained with P5R:I-Ab or P5R:I-Ab-4E tetramers and then BV786-labeled CD4 (RM4–5; BD Biosciences) antibody and a fixable viability dye (Ghost Dye Red 780; Tonbo Biosciences). The stained cells were analyzed by flow cytometry as described below.
Biomembrane force probe assay
The biomembrane force probe adhesion frequency assay was performed as described 13 using streptavidin beads coated with biotinylated MOGp:I-Ab or MOG:I-Ab-4E molecules attached to biotinylated red blood cells (RBCs) and naïve splenic CD4+ SMARTA TCR transgenic T cells purified using the MACS CD4+ T cell magnetic separation kit (Miltenyi Biotec).
Cell enrichment and flow cytometry
Single cell suspensions of spleen and lymph nodes from B3K508 mice were incubated in complete medium with 10 nM p:I-Ab or p:I-Ab-4E tetramers linked to streptavidin-PE at 37°C for 45 minutes. The tetramer concentration was chosen from a titration experiment showing that the number of P5R:I-Ab-4E tetramer-binding cells detected in P5R-immunized mice peaked around 10 nM and that P5R:I-Ab-4E tetramer detected 40 percent more T cells than conventional P5R:I-Ab tetramer at doses between two and 40 nM. Tetramer-stained samples were washed with cold PBS plus 4% FBS and stained with antibodies against surface markers and GhostDye Red 780 viability dye (Tonbo Biosciences). Single cell suspensions of spleen and lymph nodes from B6 mice were stained for 45 minutes at 37°C with 10 nM p:I-Ab or p:I-Ab-4E tetramers linked to streptavidin-PE or streptavidin-APC. The samples were then mixed with magnetic beads conjugated with PE or APC antibodies and enriched for bead-bound cells on magnetized columns. A portion was removed for bead-based counting as described 19. The samples then underwent surface staining on ice with a mixture of antibodies specific for B220 (APC-eF780-labeled clone RA3-6B2, eBioscience, Cat# 47-0452-82, Lot# 2025231), CD11b (APC-eF780-labeled clone MI-70, eBioscience, Cat# 47-0112-82, Lot# 2011193), CD11c (APC-eF780-labeled clone N418, eBioscience, Cat# 47-0114-82, Lot# 2026255), and F4/80 (APC-eF780-labeled clone BM8, eBioscience, Cat# 47-4801-82, Lot# 2025231); or B220 (eFluor 450-labeled clone RA3-6B2, eBioscience, Cat# 48-0452-82, Lot# 4339477), CD11c (eFluor 450-labeled clone N418, eBioscience, Cat# 48-0114-82, Lot# 4339477), CD11b (eFluor 450-labeled clone M1/70, eBioscience, Cat# 48-0112-82), and F4/80 (eFluor 450-labeled clone BM8, eBioscience, Cat# 48-4801-82); and CD90.2 (Brilliant Violet 605-labeled clone 53-2.1, Biolegend, Cat# 140317, Lot# B264060) or CD90.2 (Horizon-V500-labeled clone 53-2.1, BD Bioscience, Cat# 561616, Lot# 7180523); and CD4 (Brilliant Ultra Violet 395-labeled clone GK1.5, BD Bioscience, Cat# 563790, Lot# 8213920) or CD4 (Brilliant Violet 650-labeled clone RM4-5, Biolegend, Cat# 100555); and CD8a (Alexa Fluor 488-labeled clone 53-6.7, Biolegend, Cat# 100723, Lot# B254526); and CD44 (Alexa Flour 700-labeled clone IM7, eBioscience, Cat# 56-0441-82, Lot# 4329936) antibodies. In all cases, stained cells were analyzed on an LSR II or Fortessa (Becton Dickinson) flow cytometer. Data were analyzed with FlowJo software (TreeStar).
Supplementary Material
The figure shows a representative gating sequence for a spleen and lymph node sample from an unimmunized B6 mouse after tetramer staining and magnetic enrichment. The sequence was designed to include cells with light scatter properties of lymphocytes (a), were singlets (b), were non-T lineage-negative CD90+ (c), viable (d), and CD4+ or CD8+ (e). Cells in the CD4+ gate are displayed in Figures 2 and 3.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grants R01 AI143826 and R01 AI039614 to M.K.J., F32 AI114050 to D.M, T32 AI083196 and T32 AI007313 to D.I.K., and R01 AI096879 to B.D.E.
Footnotes
ETHICS DECLARATION
Experiments were approved by the University of Minnesota IACUC and conducted in accordance with their policies.
DATA AVAILABILITY
The main data of this study are available within the article and its Supplementary Figures. Source data are provided with this paper. All other data are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Gray JI, Westerhof LM & MacLeod MKL The roles of resident, central and effector memory CD4 T-cells in protective immunity following infection or vaccination. Immunology (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis MM T cell receptor gene diversity and selection. Annu. Rev. Biochem 59, 475–496 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Rudolph MG, Stanfield RL & Wilson IA How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol 24, 419–466 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Altman JD et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96. (1996). [PubMed] [Google Scholar]
- 5.Doherty PC The tetramer transformation. J. Immunol 187, 5–6 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Crawford F, Kozono H, White J, Marrack P & Kappler J Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 8, 675–682 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Martinez RJ, Andargachew R, Martinez HA & Evavold BD Low-affinity CD4+ T cells are major responders in the primary immune response. Nat. Commun 7, 13848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Y, Kern P, Chang H & Reinherz E T cell receptor binding to a pMHCII ligand is dinetically distinct from and independent of CD4. J. Biol. Chem 276, 5659–5667 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Jonsson P et al. Remarkably low affinity of CD4/peptide-major histocompatibility complex class II protein interactions. Proc. Natl. Acad. USA 113, 5682–5687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XX et al. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex. Proc. Natl. Acad. USA 108, 15960–15965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govern CC, Paczosa MK, Chakraborty AK & Huseby ES Fast on-rates allow short dwell time ligands to activate T cells. Proc. Natl. Acad. USA 107, 8724–8729 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortensen R Overview of gene targeting by homologous recombination. Curr. Protoc. Mol. Bio.l Chapter 23, Unit 23 21 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Kolawole EM, Andargachew R, Liu B, Jacobs JR & Evavold BD 2D Kinetic analysis of TCR and CD8 coreceptor for LCMV GP33 epitopes. Front. Immunol 9, 2348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oxenius A, Bachmann MF, Zinkernagel RM & Hengartner H Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol 28, 390–400 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Oxenius A et al. Presentation of endogenous viral proteins in association with major histocompatibility complex class II: on the role of intracellular compartmentalization, invariant chain and the TAP transporter system. Eur. J. Immunol 25, 3402–3411 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Nelson RW et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 42, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyer JR et al. T cell receptor and coreceptor CD8 alphaalpha bind peptide-MHC independently and with distinct kinetics. Immunity 10, 219–225 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Rees W et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc. Natl. Acad. USA 96, 9781–9786. (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon JJ et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson JM, Jensen PE & Evavold BD DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323–339 epitope. J. Immunol 164, 4706–4712 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Malhotra D et al. Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol 17, 187–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masteller EL et al. Peptide-MHC class II dimers as therapeutics to modulate antigen-specific T cell responses in autoimmune diabetes. J. Immunol 171, 5587–5595 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Stratmann T et al. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. J. Clin. Invest 112, 902–914 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J et al. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc. Natl. Acad. USA 113, E1890–1897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams T et al. Development of T cell lines sensitive to antigen stimulation. J. Immunol. Methods 462, 65–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only REFERENCES
- 26.Slavin A et al. Induction of a multiple sclerosis-like disease in mice with an immunodominant epitope of myelin oligodendrocyte glycoprotein. Autoimmunity 28, 109–120 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Bunch TA, Grinblat Y & Goldstein LS Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res 16, 1043–1061 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon JJ et al. Quantitative impact of thymic selection on Foxp3+ and Foxp3- subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. USA 108, 14602–14607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotov DI et al. TCR affinity biases Th cell differentiation by regulating CD25, Eef1e1, and Gbp2. J. Immunol 202, 2535–2545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi YS et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol 190, 4014–4026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The figure shows a representative gating sequence for a spleen and lymph node sample from an unimmunized B6 mouse after tetramer staining and magnetic enrichment. The sequence was designed to include cells with light scatter properties of lymphocytes (a), were singlets (b), were non-T lineage-negative CD90+ (c), viable (d), and CD4+ or CD8+ (e). Cells in the CD4+ gate are displayed in Figures 2 and 3.
Data Availability Statement
The main data of this study are available within the article and its Supplementary Figures. Source data are provided with this paper. All other data are available from the corresponding author upon reasonable request.


