Abstract
Rho GTPASE-activating protein 23 (ARHGAP23) is known to activate RHO-GTPase and has an important role in the infiltration and metastasis of tumors. Although previous studies suggested its involvement in certain human cancers, its role in pan-cancer remains unclear. In the present study, the expression, prognosis and potential functions of ARHGAP23 in pan-cancer were evaluated through various public databases such as Human Protein Atlas, Tumor IMmune Estimation Resource, Gene Set Co-Expression Analysis, Gene Expression Profiling Interactive Analysis, cBio Cancer Genomics Portal, Tumor-Immune System Interactions Database (TISIDB) and others. Through these data combined with a variety of biological information analysis methods, the potential role of ARHGAP23 as a carcinogenic gene was explored in the present study. The present analysis revealed that ARHGAP23 expressed abnormalities in >10 tumors, which was associated with differences in prognosis. Furthermore, the findings of the present study indicated that ARHGAP23 is associated with DNA methylation and multiple immune cell infiltrations in these tumors. ARHGAP23 expression was related to clinical prognosis, DNA methylation and immune infiltration. These findings support the potential of ARHGAP23 as a prognostic biomarker and a molecular target for cancer treatment.
Keywords: Rho GTPASE-activating protein 23, prognosis, pan-cancer, biomarker, pancreatic adenocarcinoma
Introduction
Cancer is a major global health issue and remains the most common cause of death worldwide (1). Although there has been significant development in diagnosis and treatment, which has decreased the mortality rate of most tumors, the overall incidence and mortality rate remains high (2). Exploring valuable oncogenes, which is made possible through the use of public databases such as The Cancer Genome Atlas (TCGA), is therefore crucial to understand the mechanisms of carcinogenesis and tumor progression.
The Rho GTPase-activating protein 23 (ARHGAP23) gene belongs to the ARH family of genes and encodes a GTPase in the Rho family. Transmembrane receptors transmit extracellular signals to activate Rho family GTPase, which functions by binding to downstream effectors. ARHGAP23 is also a novel Rho-GTPase regulator. ARHGAP23 mRNA was revealed to be expressed in several tissues including the placenta, prostate, hippocampus, brain medulla, brain tumor, salivary gland tumor, bladder tumor and head and neck neoplasms (3).
ARHGAP23 belongs to the Rho family of small GTPases. Rho-GTPases are involved in controlling various signal transduction pathways in all eukaryotic cells. Rho-GTPases cycle between GDP-bound inactive state and GTP-bound active state. It is an essential molecular switch that regulates cytoskeletal recombination, gene transcription and cell cycle progression (4). Considering that invasion and migration are crucial features of malignant tumors and that Rho-GTPases are molecular switches of cytoskeletal rearrangement and other processes, Rho-GTPases are closely related to malignant transformation, proliferation, invasion and metastasis of tumor cells (5,6). Abnormal activation and expression of Rho GTPases are associated with invasive metastasis of a large number of human cancers and Rho GTPases can be used as therapeutic targets for a variety of tumors (7). Similar to other proteins in the Rho-GTPases family, ARHGAP23 interacts with transmembrane receptors, negatively regulates the Rho/Rac/cell division cycle and is involved in transcriptional regulation, cell migration, survival, adhesion and proliferation (6). Therefore, it is necessary to study the relationship between ARHGAP23 and pan-cancer for a deep understanding of the relationship between Rho-GTPases and tumors. In the present study, several databases were explored, including TCGA, cBio Cancer Genomics Portal (cBioPortal) and Human Protein Atlas (HPA), to obtain information on the expression and prognosis of ARHGAP23 in different types of malignant tumors, as well as the immune infiltration and DNA methylation levels of ARHGAP23 in various tumors. Finally, the pathway of ARHGAP23 in tumorigenesis and development was investigated.
Materials and methods
Analysis of ARHGAP23 gene expression
Human Protein Atlas (HPA) (https://www.proteinatlas.org/) was used to examine the ARHGAP23 mRNA expression levels in normal tissues and cells.
Tumor IMmune Estimation Resource version 2 (TIMER2) (http://timer.cistrome.org/) ‘Diff Exp’ was used to investigate the differences in the ARHGAP23 expression levels between tumor and non-tumor tissues of various tumor types. ARHGAP23 expression levels were also evaluated between different molecular subgroups of breast cancer, between HPV-positive and HPV-negative head and neck squamous cell carcinomas (HNSCs), and between primary and metastatic skin cutaneous melanoma (SKCM).
Gene Expression Profiling Interactive Analysis 2 (GEPIA2) (http://gepia2.cancer-pku.cn/#index) was used to synthesize the tumor and the corresponding normal samples from TCGA and Genotype-Tissue Expression (GTEx) databases, respectively. The expression of 33 types of tumors in cancer and pre-cancer samples were analyzed. GEPIA2 and the Gene Set Cancer Analysis (GSCA) database (http://bioinfo.life.hust.edu/GSCA/#/) were used to evaluate the ARHGAP23 expression levels at the different stages of cancer.
Association between ARHGAP23 and prognosis
Survival data on the tumor samples from the TCGA database were obtained to investigate the association between the overall survival (OS), disease-specific survival (DSS), progression-free interval (PFI) and prognosis of the patients. The Kaplan-Meier analysis and log-rank test were used for each type of cancer. The R packages ‘survival’ and ‘survminer’ were used to plot survival curves.
Association between ARHGAP23 expression levels and methylation
cBioPortal (http://www.cbioportal.org/) within the TCGA database had tumor genetic data, including 32 types of tumors, a total of 10,953 samples and can provide researchers with multi-dimensional visualization data. In the present study, cBioPortal was selected to analyze the type and frequency of the ARHGAP23 gene mutations in all tumor types in ‘OncoPrint’ and ‘CancerTypesSummary’. ‘OncoPrint’ indicates the mutation, copy number and expression level of the target gene in all samples. The ‘CancerTypesSummary’ indicates the mutation rate of target genes in generalized cancer.
The University of Alabama at Birmingham cancer data analysis (UALCAN; http://ualcan.path.uab.edu/) portal is an interactive web portal for TCGA data analysis, survival analysis and superficial analysis. In the present study, UALCAN was used to study the promoter methylation levels of ARHGAP23 in cancer.
Expression of ARHGAP23 in tumor molecular subtypes and immune subtypes
The TISIDB (http://cis.hku.hk/TISIDB/) is an integrated repository portal for tumor immune system interactions. In the present study, the TISIDB was used to explore the association between ARHGAP23 expression levels and molecular or immune subtypes in pan-cancer.
Association between ARHGAP23 expression and immunity
CIBERSORT (https://cibersort.stanford.edu/) was used to calculate the 22 types of immune cells via a cancer relative score, which allows for the prediction of the immune cell phenotype.
Analysis of ARHGAP23-protein interaction
The Gene Multiple Association Network Integration Algorithm (GeneMANIA) database (http://www.genemania.org) allows researchers to find similar genes of target genes based on a large amount of genomics and proteomics data. The Biological General Repository for Interaction Datasets (BioGRID; https://thebiogrid.org/) can be used to analyze protein-protein interaction (PPI) networks. GeneMANIA and BioGRID were used to obtain genes similar to ARHGAP23 and construct PPI networks.
Enrichment analysis of ARHGAP23 associated genes
The ‘Most Similar Genes’ module of GEPIA2 was used to extract 100 genes that were most similar to ARHGAP23. The Gene Ontology (GO) database (http://geneontology.org) and the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/) enrichment analysis were used to investigate the biological functions and signaling pathways associated with ARHGAP23 in TCGA tumors. P<0.05 was considered to indicate a statistically significant difference.
Specimen collection
Samples from patients that underwent a pancreaticoduodenectomy in the First Hospital of Lanzhou University (Lanzhou, China) from January 2018 to January 2022 were collected. The specimens were placed in 10% formalin for 4-6 h at room temperature and then embedded in paraffin. Patients were excluded from the present study if diagnosed with: i) Tumors in other organs; ii) a primary unknown cancer; and iii) had a previous history of cancer treatment. All patients provided written informed consent and had complete clinical data. The clinicopathological data of the patients are presented in Table I. The present study was approved by the Ethics Committee of the First Hospital of Lanzhou University (approval no. LDYYLL2022-196; Lanzhou, China). It was conducted in accordance with the ethical guidelines of the Declaration of Helsinki (2013).
Table I.
The association between ARHGAP23 and clinicopathological characteristics in pancreatic adenocarcinoma.
| Patient characteristics | Frequency (n) | Percentage (%) |
|---|---|---|
| Age, years | ||
| ≤60 | 23 | 46 |
| >61.00 | 27 | 54 |
| Sex | ||
| Male | 30 | 60 |
| Female | 20 | 40 |
| Differentiation | ||
| Well | 4 | 8 |
| Moderate | 11 | 22 |
| Moderate-Poor | 35 | 70 |
| Site | ||
| Body and tail of pancreas | 7 | 14 |
| Head of pancreas | 43 | 86 |
| T | ||
| 1 | 11 | 22 |
| 2 | 31 | 62 |
| 3 | 8 | 16 |
| N | ||
| 0 | 20 | 40 |
| 1 | 20 | 40 |
| 2 | 9 | 18 |
| ARHGAP23 score | ||
| ≤150 | 33 | 66 |
| >151.00 | 17 | 34 |
ARHGAP23, Rho GTPASE-activating protein 23.
Immunohistochemistry
Immunohistochemistry was conducted according to the Elivision Plus test kit protocol (RAMCON). Continuous sections of paraffin-embedded tissue were 4-µm-thick. All paraffin sections were dewaxed in xylene and rehydrated in alcohol of descending series. Samples were then incubated with 3% hydrogen peroxide at room temperature for 10 min, with citrate buffer for antigen repair. The samples were washed three times with 1X PBS for 10 min and all sections were blocked with 10% goat serum (cat. no. SL038; Beijing Solarbio Science & Technology Co., Ltd.) for 20 min at room temperature to prevent non-specific binding. A polyclonal antibody for rabbit anti-ARHGAP23 (cat. no. YT0319; ImmunoWay Biotechnology Company) diluted at 1:200 was used overnight at 4˚C. Subsequently, samples were washed three times with 1X PBS for 10 min. The HRP-conjugated goat anti-rabbit IgG secondary antibody (1:500; PR30011; Proteintech Group, Inc.) was added and incubated at room temperature for 1 h, before being washed three times with 1X PBS for 10 min. Finally, DAB chromogenic agent was used for color development and the samples were rinsed with tap water, rescaled and dehydrated. The slides were observed under a light microscope.
Dyeing evaluation
All immunohistochemically stained sections were performed blind by two experienced pathologists. The expression of ARHGAP23 was evaluated according to the proportion of positive stained cells. In addition, expression levels were scored according to staining intensity (0, negative; 1, weak; 2, moderate; and 3, strong). Score=the proportion of positive stained cells x staining intensity x100. A final score of >150 was considered high expression, while a score of ≤150 was considered low expression.
Statistical analysis
SPSS (version 25.0; IBM Corp.) was used for statistical analysis. R v4.2.2 (RStudio, Inc.) was used to conduct statistical analysis and graph visualization. The data were analyzed using unpaired Student's t-test or one-way ANOVA followed by Tukey's post hoc test (for multiple-group comparison) single factor analysis. All results are presented as mean ± standard deviation. All experiments were repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of ARHGAP23 in pan-cancer
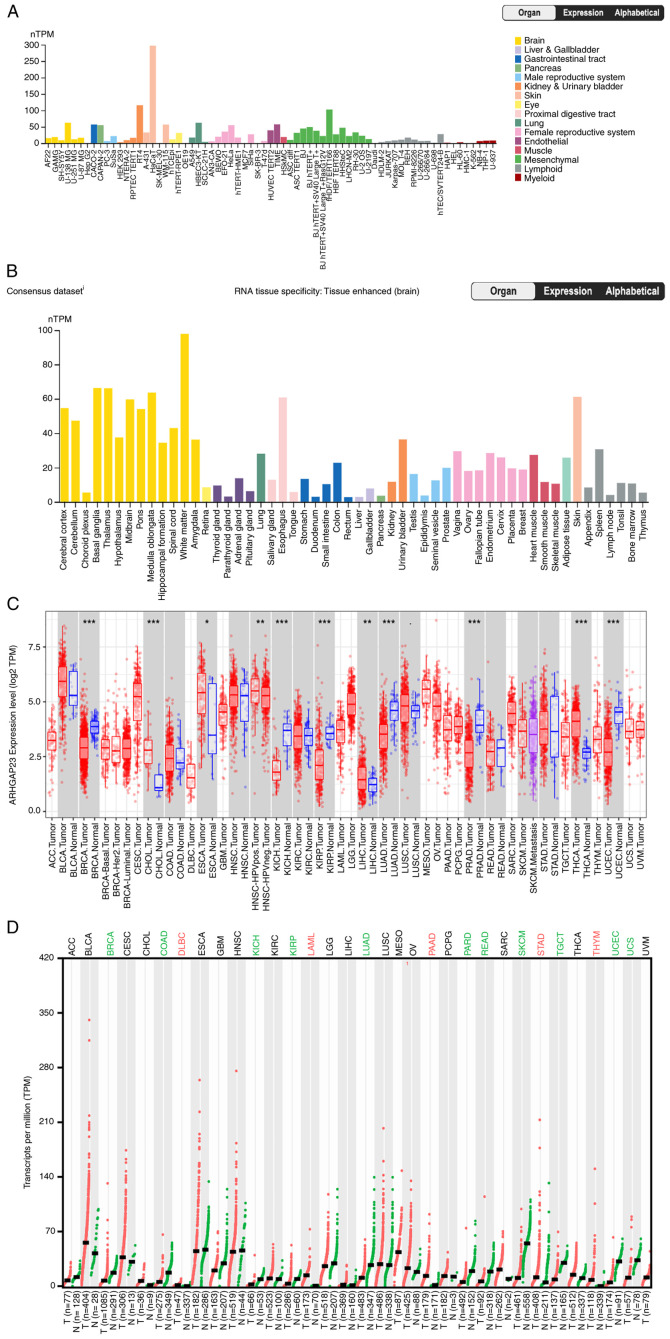
After analyzing data from the HPA database in all normal cells, it was observed that the expression of ARHGAP23 was relatively high in kidney, urinary bladder and skin cell lines, whereas it was low in lymphoid and myeloid cell lines. The HaCaT and RT4 cell lines exhibited the highest ARHGAP23 expression (Fig. 1A). Further analysis revealed that ARHGAP23 was expressed in all tissues and highly expressed in the white matter, basal ganglia and thalamus, indicating that ARHGAP23 was mostly expressed in brain-related tissues (Fig. 1B). The expression level of ARHGAP23 in pan-cancer was examined using TIMER2. The expression level of ARHGAP23 in various tumors was lower than in normal tissues, including breast invasive carcinoma (BRCA), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), lung adenocarcinoma (LUAD), pheochromocytoma and paraganglioma (PCPG) and uterine corpus endometrial carcinoma (UCEC), while cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC) and thyroid carcinoma (THCA) were significantly upregulated (Fig. 1C). Due to the lack of matched normal tissues in some tumors in TCGA database, the present study further examined the expression of ARHGAP23 by using GEPIA2, which contains TCGA and Genotype-Tissue Expression datasets. The results revealed that lymphoid neoplasm diffuse large b-cell lymphoma, acute myeloid leukemia, pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD) and thymoma (THYM) exhibited significantly high expression. The expression levels in BRCA, colon adenocarcinoma (COAD), KICH, KIRP, LUAD, prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), SKCM, testicular germ cell tumors (TGCT), UCEC and uterine carcinosarcoma (UCS) were lower than those in normal tissues (Fig. 1D). In addition, mRNA expression levels of ARHGAP23 were higher in COAD, ESCA, kidney renal clear cell carcinoma (KIRC) and UCEC (Fig. 2A-E).
Figure 1.
Differential expression of ARHGAP23. (A) Expression of ARHGAP23 in various normal cell lines. (B) Expression of ARHGAP23 in various tissues. (C) Expression of ARHGAP23 in pan-cancer based on Tumor IMmune Estimation Resource version 2 exploration. (D) Analysis of ARHGAP23 in tumor pairs and normal samples in Gene Expression Profiling Interactive Analysis 2 expression in pan-cancer. ARHGAP23, Rho GTPASE-activating protein 23; nTPM, number of transcripts per million. *P<0.05, **P<0.01 and ***P<0.001.
Figure 2.
Expression levels of ARHGAP23 at different stages of different tumors. (A) The expression levels of ARHGAP23 in different stages of pan-cancer were investigated by Gene Set Cancer Analysis database. (B-E) Gene Expression Profiling Interactive Analysis 2 database was used to explore the differential expression of ARHGAP23 in different stages of tumor in COAD, ESCA, KIRC and UCEC. ARHGAP23, Rho GTPASE-activating protein 23; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; KIRC, kidney renal clear cell carcinoma; UCEC, uterine corpus endometrial carcinoma.
Prognostic value of ARHGAP23 in tumors
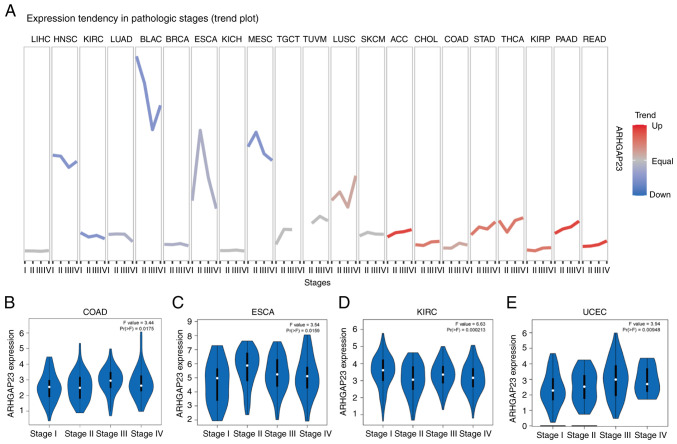
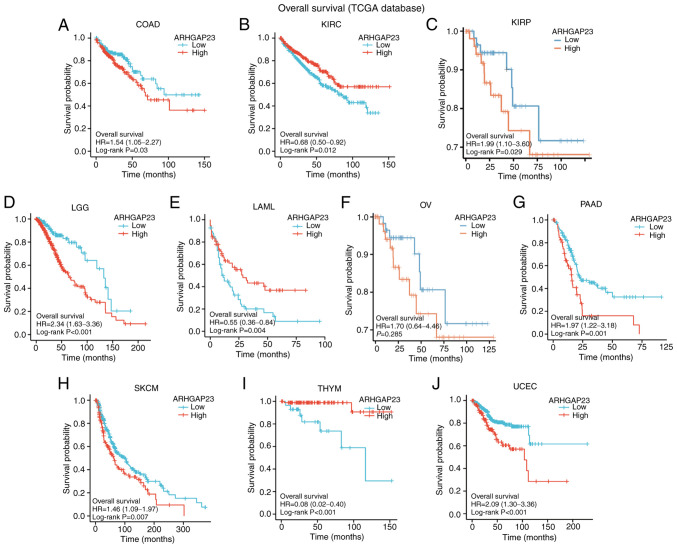
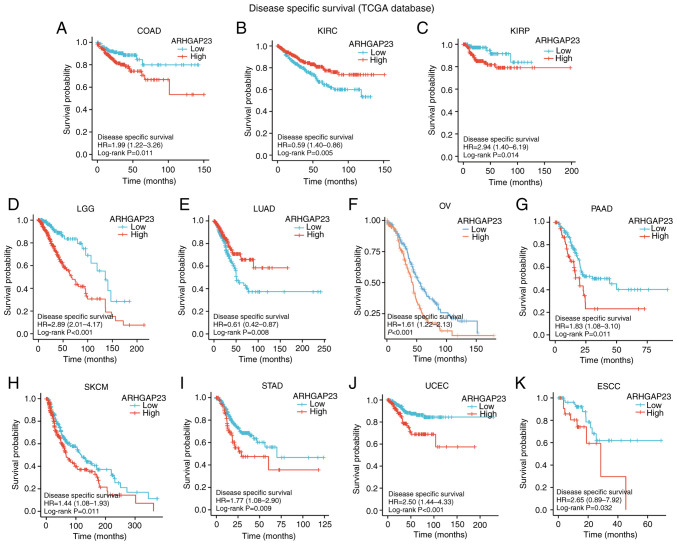
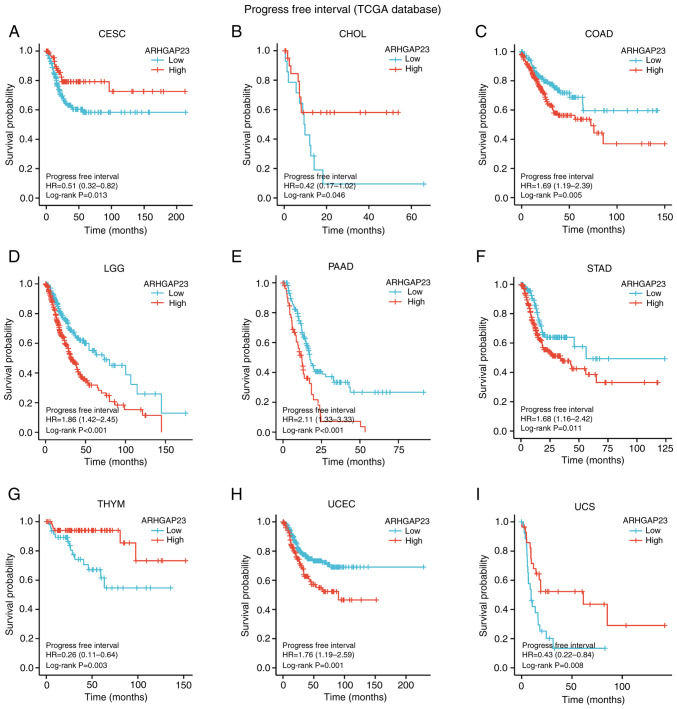
Survival indicators such as OS, DSS and PFI were utilized to investigate the correlation between ARHGAP23 expression and prognosis. Kaplan-Meier survival analysis demonstrated that in patients with COAD (P=0.03), KIRP (P=0.029), brain lower grade glioma (LGG; P<0.001), ovarian serous cystadenocarcinoma (OV; P=0.002), PAAD (P=0.001), SKCM (P=0.007) and UCEC (P<0.001), low expression of ARHGAP23 was associated with improved OS, while high expression of ARHGAP23 had improved OS in KIRC (P=0.012) and THYM (P<0.001) (Fig. 3). Patients with KIRC (P=0.005) or LUAD (P=0.008) and high expression of ARHGAP23 had improved DSS. Patients with COAD (P=0.011), KIRP (P=0.014), LGG (P<0.001), OV (P<0.001), PAAD (P=0.011), SKCM (P=0.011), STAD (P=0.009), UCEC (P<0.001), or esophageal squamous cell carcinoma ESCC (P=0.032) and low expression of ARHGAP23 had improved DSS (Fig. 4). ARHGAP23 high expression is positively correlated with improved PFI in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), CHOL, THYM and UCS (P<0.05). The low expression of ARHGAP23 was correlated with PFI of COAD (P=0.005), LGG (P<0.001), PAAD (P<0.001), STAD (P=0.011) and UCEC (P=0.001) (Fig. 5).
Figure 3.
Relationship between ARHGAP23 expression and OS. (A-J) Kaplan-Meier analysis of the association between ARHGAP23 expression and OS. ARHGAP23, Rho GTPASE-activating protein 23; OS, overall survival; HR, hazard ratio; COAD, colon adenocarcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, lower grade glioma; LAML, acute myeloid leukemia; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma.
Figure 4.
Relationship between ARHGAP23 expression and DSS. (A-K) Kaplan-Meier analysis of the association between ARHGAP23 expression and DSS. ARHGAP23, Rho GTPASE-activating protein 23; DSS, disease-specific survival; HR, hazard ratio; COAD, colon adenocarcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, lower grade glioma; LUAD, lung adenocarcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; ESCC, esophageal squamous cell carcinoma.
Figure 5.
Relationship between expression of ARHGAP23 and PFI. (A-I) Kaplan-Meier analysis of the association between ARHGAP23 expression and PFI. ARHGAP23, Rho GTPASE-activating protein 23; PFI, progression-free interval; HR, hazard ratio; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; LGG, lower grade glioma; PAAD, pancreatic adenocarcinoma; STAD, stomach adenocarcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma.
Correlation between ARHGAP23 and molecular or immune subtypes of tumors
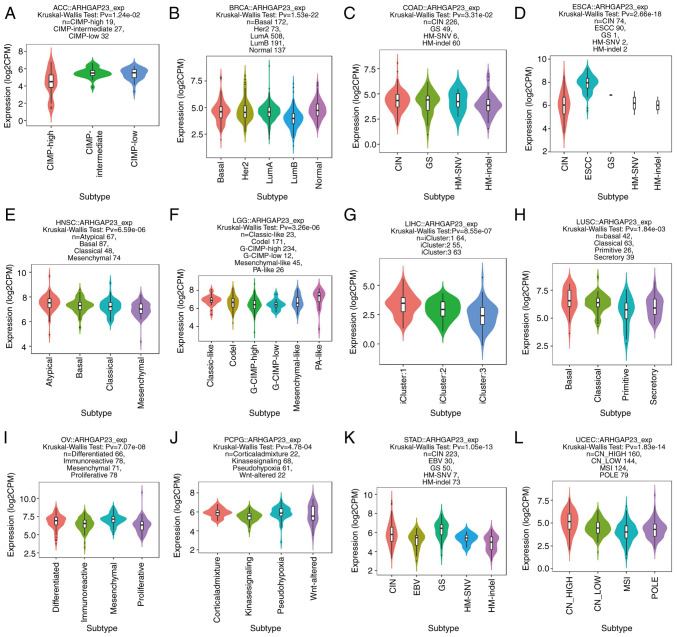
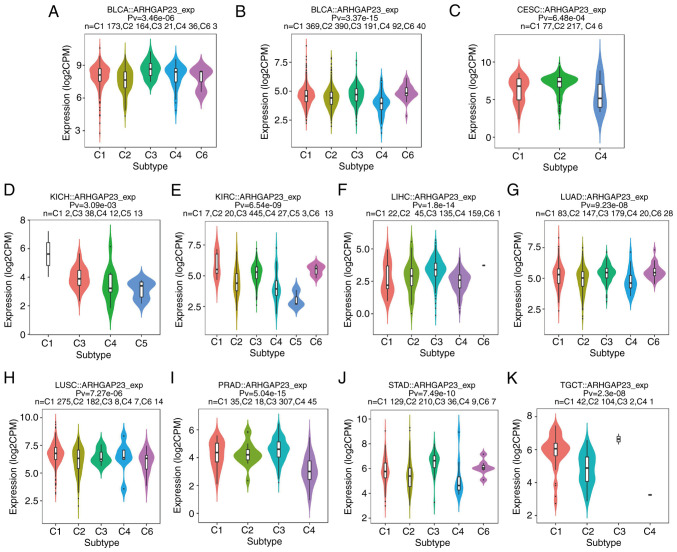
The relationship between the expression of ARHGAP23 and its molecular subtypes in pan-carcinoma was investigated by using the TISIDB database. In adrenocortical carcinoma, the expression of CpG island methylator phenotype-intermediate was the highest (Fig. 6A); in BRCA, the immune subtype of LumA demonstrated the highest expression (Fig. 6B); in COAD, the expression of the genomically stable (GS) subtype was higher than that of other subtypes (Fig. 6C); while in ESCA, the expression of ESCC subtype was the highest (Fig. 6D). The number of ESCC were also the highest (Fig. 6D) and the atypical subtypes were the highest in HNSC (Fig. 6E). The pilocytic astrocytoma-like subtypes were the highest in LGG (Fig. 6F), while LIHC only had three subtypes (iCluster: 1, 2 and 3) (Fig. 6G). The expression of basal was the highest in lung squamous cell carcinoma (LUSC) (Fig. 6H) and mesenchymal subtype was the highest in OV (Fig. 6I). It was observed that the expression of the corticalad mixture was the highest in PCPG (Fig. 6J), while the expression of the GS subtype in STAD was the highest (Fig. 6K). Finally, in UCEC, the expression and number of CN_HIGH was the highest (Fig. 6L). Subsequently, the expression of ARHGAP23 and its relationship to immune subtypes (C1, wound healing; C2, IFN-γ dominant; C3, inflammatory; C4, lymphocyte depleted; C5, immunologically quiet; and C6, TGF-β dominant) was investigated and ARHGAP23 was expressed differently in bladder urothelial carcinoma (BLCA), BRCA, CESC, KICH, KIRC, LIHC, LUAD, LUSC, PRAD, STAD, TGCT and 11 other types of cancer. Among BLCA and LUSC, C1 was the most expressed subtype (Fig. 7A and H). Among KICH, KIRC, LUAD and PRAD, the expression of the C3 subtype was the highest (Fig. 7D, E, G and I). Among BRCA, CESC, STAD and TGCT, C2 subtype was the highest expressed subtype (Fig. 7B, C, J and K). In LIHC, the expression of C4 subtype was the highest and also the highest (Fig. 7C).
Figure 6.
Association between ARHGAP23 expression and molecular subtypes in The Cancer Genome Atlas tumors. (A) ACC; (B) BRCA; (C) COAD; (D) ESCA; (E) HNSC; (F) LGG; (G) LIHC; (H) LUSC; (I) OV; (J) PCPG; (K) STAD and (L) UCEC. ARHGAP23, Rho GTPASE-activating protein 23; ACC, adrenocortical carcinoma; CIMP, CpG island methylator phenotype; BRCA, breast invasive carcinoma; LumA, luminal A; LumB, luminal B; COAD, colon adenocarcinoma; CIN, chromosomal instability; GS, genomically stable; HM-SNV, hypermutated-single-nucleotide variant predominant; HM-indel, hypermutated-insertion deletion mutation; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LGG, lower grade glioma; G-CIMP, glioma-CIMP; PA, pilocytic astrocytoma; LIHC, liver hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PCPG, pheochromocytoma and paraganglioma; STAD, stomach adenocarcinoma; EBV, Epstein-Barr virus; UCEC, uterine corpus endometrial carcinoma; CN, copy number; MSI, microsatellite instability.
Figure 7.
Association between ARHGAP23 expression and immune subtypes in The Cancer Genome Atlas tumors. (A) BLCA; (B) BRCA; (C) CESC; (D) KICH; (E) KIRC; (F) LIHC; (G) LUAD; (H) LUSC; (I) PRAD; (J) STAD and (K) TGCT. ARHGAP23, Rho GTPASE-activating protein 23; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors.
Relationship between ARHGAP23 expression mutation and DNA methylation in tumors
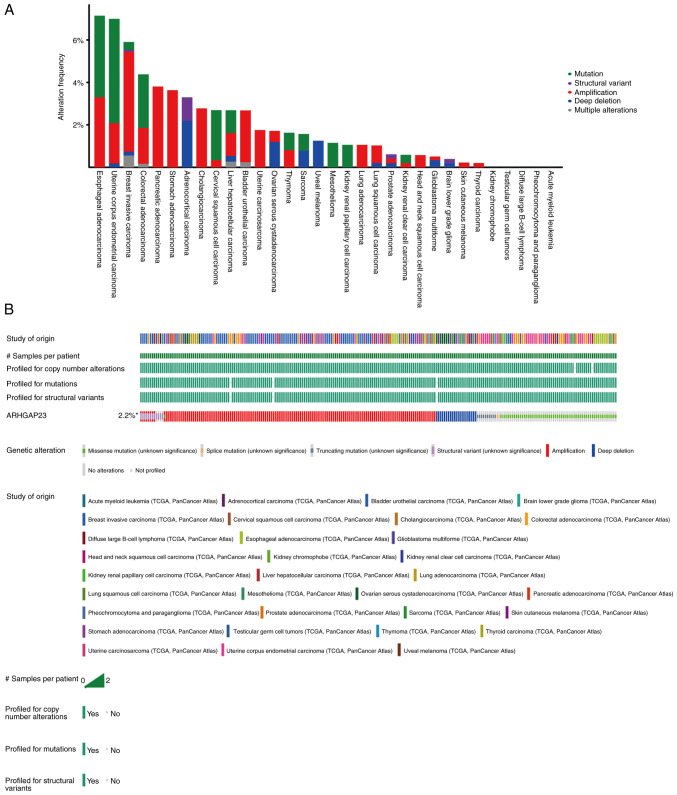
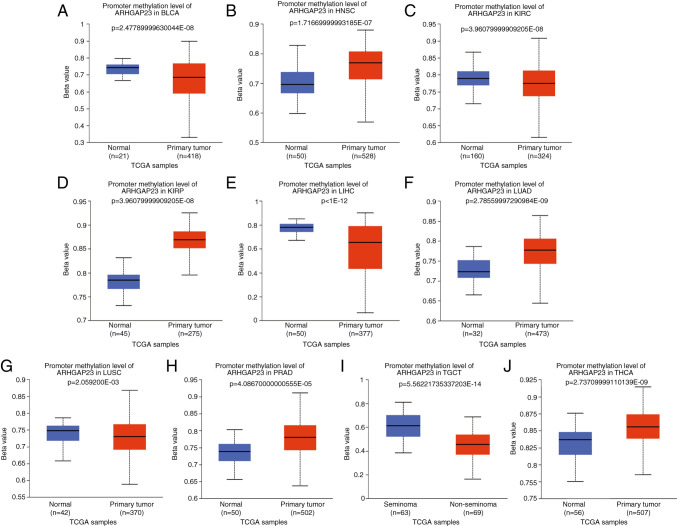
To explore the gene mutation of ARHGAP23 in various cancers, the cBioPortal platform based on TCGA data was used to analyze the mutation status of each tumor. ARHGAP23 had the highest amplification of all mutation types, with ESCA and UCEC having the highest mutation rates (>6%). The mutation type in both PAAD and STAD was solely amplification (Fig. 8). Abnormal DNA methylation is a common epigenetic disorder in tumors, which is closely related to their occurrence. The present study compared the methylation values of ARHGAP23 between normal and tumor tissues using the UALCAN online tool. The results revealed that the promoter methylation level of ARHGAP23 was significantly decreased in several tumor tissues, including BLCA (Fig. 9A), KIRC (Fig. 9C), LIHC (Fig. 9E), LUSC (Fig. 9G) and TGCT (Fig. 9I), where it was significantly increased in HNSC (Fig. 9B), KIRP (Fig. 9D), LUAD (Fig. 9F), PRAD (Fig. 9H) and THCA (Fig. 9J).
Figure 8.
Mutation of ARHGAP23. (A) Frequency distribution of ARHGAP23 alteration. (B) OncoPrint visual summary of alterations in a query of ARHGAP23 from cBio Cancer Genomics Portal. ARHGAP23, Rho GTPASE-activating protein 23.
Figure 9.
Methylation levels of ARHGAP23. (A) BLCA; (B) HNSC; (C) KIRC; (D) KIRP; (E) LIHC; (F) LUAD; (G) LUSC; (H) PRAD; (I) TGCT and (J) THCA. ARHGAP23, Rho GTPASE-activating protein 23; TCGA, The Cancer Genome Atlas; BLCA, bladder urothelial carcinoma; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma.
Effects of ARHGAP23 on the regulation of immune cell infiltration
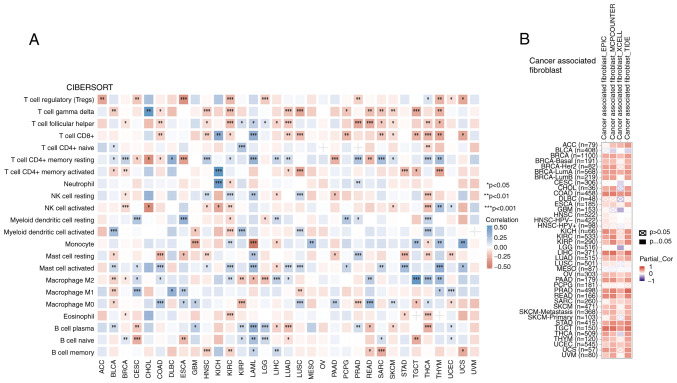
CIBERSORT results revealed that for all tumor types, except OV and uveal melanoma, there was an association between the infiltration level of immune cells and the expression of ARHGAP23. THYM was closely associated with T cells other than T cell CD4+ naive (Fig. 10A). A total of four algorithms were used to calculate the relationship between ARHGAP23 expression and cancer-associated fibroblasts (CAFs). It was observed that in CAFs, ARHGAP23 was positively associated with immune cells in BRCA, COAD, KIRC, KIRP, LIHC, LUAD, OV, PAAD, PRAD, READ, SKCM-metastasis, STAD, TGCT and UCEC (Fig. 10B).
Figure 10.
Relationship between ARHGAP23 expression and immune cells in pan-cancer. (A) CIBERSORT predicts the relationship between ARHGAP23 expression and immune cell infiltration in different cancers. (B) The relationship between ARHGAP23 expression and immune infiltration of cancer-associated fibroblasts. *P<0.05, **P<0.01 and ***P<0.001. ARHGAP23, Rho GTPASE-activating protein 23.
ARHGAP23-related gene enrichment analysis
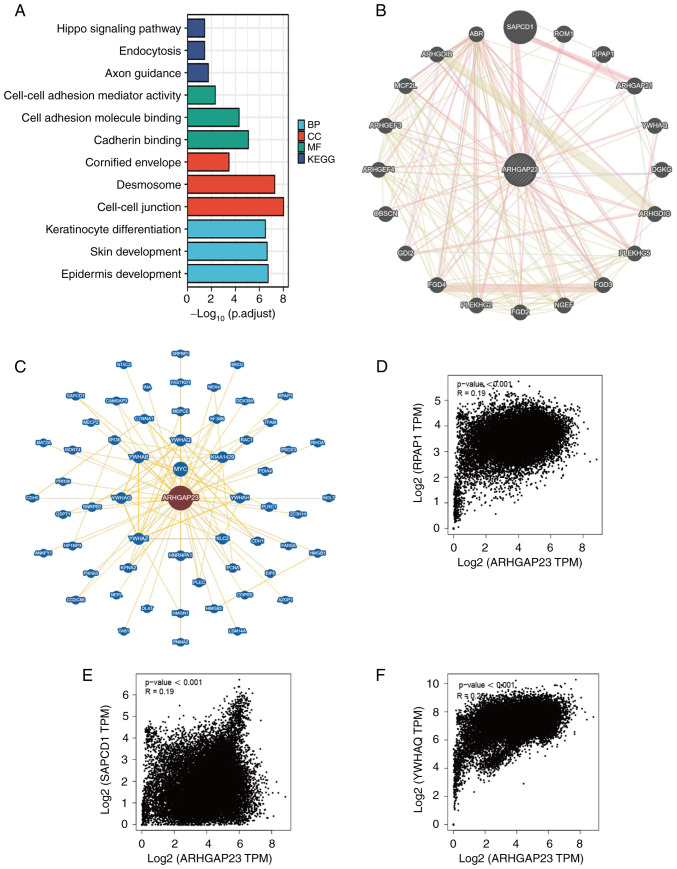
To investigate the role of ARHGAP23 in tumor pathogenesis, the top 100 genes similar to ARHGAP23 were extracted from the GEPIA2 based on TCGA database. GO and KEGG analyses were then performed on these 100 genes and ARHGAP23. The results of the GO analysis demonstrated that the biological processes subontology mainly included keratinocyte differentiation, skin development and epidermis development. Cellular component mainly included cell-cell junction, desmosome and cornified envelope. Molecular function mainly involved cadherin binding, cell adhesion molecule binding and cell-cell adhesion mediator activity. KEGG pathways are mainly enriched in axon guidance, endocytosis and Hippo signaling pathway (Fig. 11A). The protein-protein interaction network diagram was then constructed using GeneMANIA and BioGRID. Functional analysis in GeneMANIA exhibited that ARHGAP23 and its similar genes were mainly associated with Rho GTPase binding and Ras guanyl-nucleotide exchange factor activity pathways (Fig. 11B). The main functions of ArhGap23-related genes in BioGRID were concentrated on Rho GTPase Activator Activity (Fig. 11C). The same genes SAPCD1, YWHAQ and RPAP1 were detected in GeneMANIA and BioGRID, and it was further verified that the expression of ARHGAP23 was highly positively associated with SAPCD1, YWHAQ and RPAP1 (Fig. 11D-F).
Figure 11.
Enrichment analysis of ARHGAP23 related genes. (A) Gene Ontology and KEGG analysis of the top 100 genes similar to ARHGAP23 obtained based on GEPIA2. (B) The gene-gene interaction network of ARHGAP23 from Gene Multiple Association Network Integration Algorithm. (C) The gene-gene interaction network of ARHGAP23 from BioGRID. (D-F) GEPIA2 analyzed the association between ARHGAP23 and suppressor APC domain containing 1, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta and RPAP1 in all the samples from The Cancer Genome Atlas and Genotype-Tissue Expression. ARHGAP23, Rho GTPASE-activating protein 23; GEPIA2, Gene Expression Profiling Interactive Analysis 2; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological processes; MF, molecular function; CC, cellular component; TPM, transcripts per million.
ARHGAP23 is highly expressed in PAAD and is associated with poor prognosis
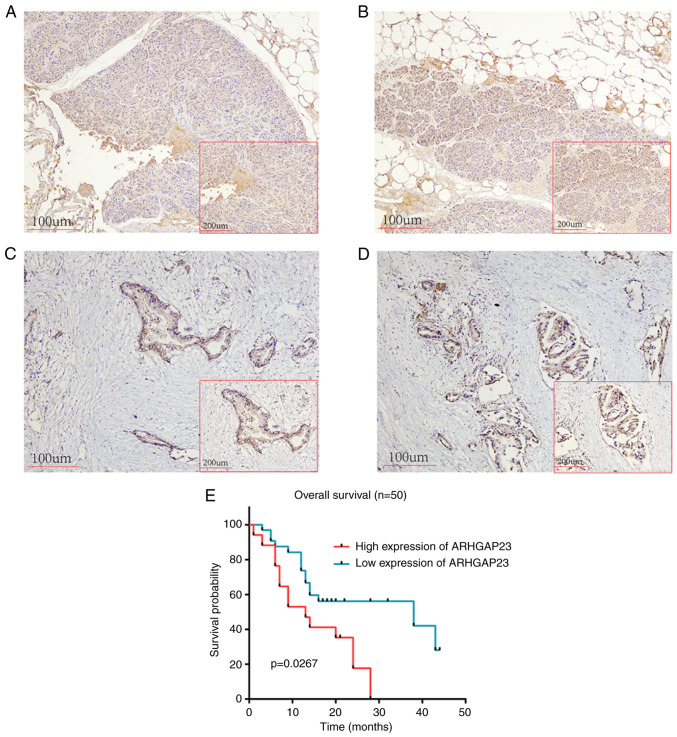
Immunohistochemical results of 50 pairs of tumor and normal tissues from patients with PAAD revealed that the expression level of ARHGAP23 protein in PAAD was upregulated (Fig. 12A-D). According to the expression of ARHGAP23, patients were evenly divided into high- and low-expression groups, in which patients with low expression had improved prognoses (Fig. 12E). In multivariate analysis, the expression of ARHGAP23 was associated with OS (Table II).
Figure 12.
Expression and prognostic value of ARHGAP23 in PAAD. (A and B) Negative staining of ARHGAP23 in adjacent tissues (magnification, x100). (C and D) Positive staining of ARHGAP23 in PAAD tissues (magnification, x100). (E) Survival analysis of low and high ARHGAP23 expression groups. ARHGAP23, Rho GTPASE-activating protein 23, PAAD, pancreatic adenocarcinoma.
Table II.
Multivariate analyses of factors associated with overall survival.
| Clinicopathological factor | HR | 95% CI of HR | P-value | |
|---|---|---|---|---|
| ARHGAP23 (>150) | 6.29 | 2.58 | 15.31 | <0.001 |
| Age (>60) | 0.87 | 0.41 | 1.84 | 0.71 |
| Sex (female vs. male) | 1.04 | 0.46 | 2.33 | 0.93 |
| Differentiation | ||||
| Well | 0.51 | |||
| Moderate | 0.65 | 0.14 | 2.97 | 0.58 |
| Moderate-Poor | 1.22 | 0.36 | 4.22 | 0.75 |
| Site (Head of pancreas vs. body and tail of pancreas) | 1.43 | 0.50 | 4.11 | 0.50 |
| T stage | ||||
| 1 | 0.24 | |||
| 2 | 2.37 | 0.80 | 7.06 | 0.12 |
| 3 | 2.75 | 0.76 | 9.90 | 0.12 |
| N stage | ||||
| 0 | 0.64 | |||
| 1 | 1.48 | 0.65 | 3.40 | 0.35 |
| 2 | 1.20 | 0.37 | 3.87 | 0.76 |
HR, hazard ratio; CI, confidence interval; ARHGAP23, Rho GTPASE-activating protein 23.
Discussion
Because of the rapid progress of bioinformatics algorithms and the continuous development of different databases, the study of the biomarkers of pan-cancer and their related functions has become easier (8). The present study aimed to comprehensively analyze the characteristics of ARHGAP23 in 33 types of cancer, aiming to explore the value of ARHGAP23 in cancer prognosis, progression and treatment.
To the best of the authors' knowledge, there are only a few studies on the role of ARHGAP23 in cancer development and progression. According to the results of the present study, mRNA expression of ARHGAP23 is low in several types of cancer compared with normal tissues. It was found that ARHGAP23 is highly expressed in PAAD and patients with PAAD and low expression of ARHGAP23 has improved OS, DSS, PFI and RFS. In some tumors, expression of ARHGAP23 is higher than that of paracancerous tissues, but low expression of ARHGAP23 is associated with an improved prognosis, and in others the opposite is true, possibly because the protein can be regulated at multiple levels. Second, the expression of this gene may change early in the development of cancer, and it is less certain during the development of cancer. Finally, it may be that high expression of ARHGAP23 is associated with cancer development, while low expression is also associated with cancer development, possibly because regulation of ARHGAP23 is passive (9). In PAAD, the expression of ARHGAP23 is significantly increased, and patients with high expression of ARHGAP23 have worse prognosis. Unfortunately, ARHGAP23 is not associated with the clinical characteristics of PAAD, which may be related to the small sample size of the present study. However, PAAD samples are difficult to collect, and all PAAD samples have been collected during the past five years. The sample size will be increased in the future.
The expression level of ARHGAP23 was correlated with the molecular subtypes of 12 tumors and the immune subtypes of 11 tumor types, among which ARHGAP23 was associated with the molecular subtypes and immune subtypes of BRCA, LIHC, LUSC and STAD. Therefore, ARHGAP23 is a promising prognostic marker and therapeutic target for tumor subtypes. Future studies focusing on unique molecular subtypes or immune subtypes of cancer may provide appropriate directions for exploring the clinical effects of ARHGAP23.
Numerous studies confirmed that mutations in the genome are associated with tumor progression and resistance to antitumor drugs (10,11). In the present study, ARHGAP23 mutations were found in 27 types of tumors, indicating that these mutations are involved in tumor development. Abnormal DNA methylation is associated with multiple tumor types (12,13). DNA methylation can lead to structural changes in chromosomes, potentially leading to tumorigenesis by shutting down tumor suppressor genes. In recent years, a large number of studies have demonstrated that abnormal DNA methylation is closely related to the occurrence, development and cell cancerization of tumors. DNA methylation level and specific gene methylation degree changes can be used as tumor diagnostic indicators. In the present study, evidence was provided that the expression of ARHGAP23 is associated with reduced DNA methylation in promoter regions in different tumor types, especially in TGCT.
The extensive interweaving of immune cells and tumor cells plays an important role in the invasion and metastasis of various malignant tumors (14). The present study revealed that the expression of ARHGAP23 is positively associated with a variety of immune cells, especially in KIRC and THCA. CAFs are permanently activated fibroblasts with a powerful tumor regulatory role (15). CAFs involve a series of biological processes that promote tumorigenesis, including not only tumor cell invasion and cancer stem cell renewal, but also chemotherapy resistance and immune cell evasion (16,17). In the present study, it was observed that the expression of ARHGAP23 was positively associated with CAF infiltration in most tumors. A previous study revealed that CAF plays an important role in tumor progression, including cancer cell cancerization, invasion, metastasis and chemotherapy resistance (16). Immune cells, including macrophages, natural killer cells, dendritic cells and T cells, play a dual role in the tumor microenvironment (18). The present study exhibited that ARHGAP23 was positively associated with CAF infiltration in a variety of tumors. These results elucidated the potential role and value of ARHGAP23 in tumor immunity.
Although the authors of the present study made great efforts to improve the study, there were inevitably certain limitations. First, only online databases were used to explore the expression and prognosis of ARHGAP23, without actual clinical data. Furthermore, ARHGAP23 was tentatively linked to cancer progression in various tumors and more experimental work is needed to determine the exact molecular function.
In conclusion, comprehensive bioinformatics analysis techniques were used in the present study to investigate the expression level, clinical prognosis, methylation value, genetic changes, and immunomodulatory effects of ARHGAP23 in pan-cancer. The findings of the present study indicated that ARHGAP23 may be a novel potential prognostic and immune-related biomarker for patients with cancer.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the 2022 Natural Science Foundation of Gansu Province (grant nos. 2022-0405-JCC-0319, 21JR7RA412 and 20JR5RA145), Key Project of Science and Technology Innovation Platform Fund of Gansu Provincial People's Hospital (grant no. 21gssya-4), Postdoctoral Fund of Gansu Provincial People's Hospital (grant nos. ZX-62000001-2022-010 and ZX-62000001-2022-055) and Key Talent Project of Gansu Province (grant no. 2022RCXM025).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL and PT designed the present study. XLiu and DW analyzed the data and wrote the manuscript. XLi and KY collected the data. LW and PT analyzed the data. YL and DW revised the manuscript. All authors read and approved the final version of the manuscript. YL and PT confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The present study was approved by The Ethics Committee of The First Hospital of Lanzhou University (approval no. LDYYLL2022-196; Lanzhou, China). All of the patients were fully informed about the study procedures and provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783–791. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Katoh M, Katoh M. Identification and characterization of human ARHGAP23 gene in silico. Int J Oncol. 2004;25:535–540. [PubMed] [Google Scholar]
- 4.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 5.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 6.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 7.Cardama GA, Gonzalez N, Maggio J, Menna PL, Gomez DE. Rho GTPases as therapeutic targets in cancer (Review) Int J Oncol. 2017;51:1025–1034. doi: 10.3892/ijo.2017.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder MP, Gonzalez-Perez A, Lopez-Bigas N. Visualizing multidimensional cancer genomics data. Genome Med. 2013;5(9) doi: 10.1186/gm413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kustatscher G, Grabowski P, Schrader TA, Passmore JB, Schrader M, Rappsilber J. Co-regulation map of the human proteome enables identification of protein functions. Nat Biotechnol. 2019;37:1361–1371. doi: 10.1038/s41587-019-0298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Yasukawa M, Chen K, Hu L, Broaddus RR, Ding L, Mardis ER, Spellman P, Levine DA, Mills GB, et al. Association of Somatic Mutations of ADAMTS genes with chemotherapy sensitivity and survival in high-grade serous ovarian carcinoma. JAMA Oncol. 2015;1:486–494. doi: 10.1001/jamaoncol.2015.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rong G, Yi Z, Ma F, Guan Y, Xu Y, Li L, Xu B. DNA damage response as a prognostic indicator in metastatic breast cancer via mutational analysis. Ann Transl Med. 2021;9(220) doi: 10.21037/atm-20-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C, Stirnimann CU, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176:98–112.e14. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 14.Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front Immunol. 2020;11(369) doi: 10.3389/fimmu.2020.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: Cancer-associated fibroblasts and their markers. Int J Cancer. 2020;146:895–905. doi: 10.1002/ijc.32193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galbo PM Jr, Zang X, Zheng D. Molecular features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clin Cancer Res. 2021;27:2636–2647. doi: 10.1158/1078-0432.CCR-20-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, Sirven P, Magagna I, Fuhrmann L, Bernard C, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 18.An Y, Liu F, Chen Y, Yang Q. Crosstalk between cancer-associated fibroblasts and immune cells in cancer. J Cell Mol Med. 2020;24:13–24. doi: 10.1111/jcmm.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.