Abstract
The phyA gene encoding an extracellular phytase from the thermophilic fungus Thermomyces lanuginosus was cloned and heterologously expressed, and the recombinant gene product was biochemically characterized. The phyA gene encodes a primary translation product (PhyA) of 475 amino acids (aa) which includes a putative signal peptide (23 aa) and propeptide (10 aa). The deduced amino acid sequence of PhyA has limited sequence identity (ca. 47%) with Aspergillus niger phytase. The phyA gene was inserted into an expression vector under transcriptional control of the Fusarium oxysporum trypsin gene promoter and used to transform a Fusarium venenatum recipient strain. The secreted recombinant phytase protein was enzymatically active between pHs 3 and 7.5, with a specific activity of 110 μmol of inorganic phosphate released per min per mg of protein at pH 6 and 37°C. The Thermomyces phytase retained activity at assay temperatures up to 75°C and demonstrated superior catalytic efficiency to any known fungal phytase at 65°C (the temperature optimum). Comparison of this new Thermomyces catalyst with the well-known Aspergillus niger phytase reveals other favorable properties for the enzyme derived from the thermophilic gene donor, including catalytic activity over an expanded pH range.
Phytases (myo-inositol hexakisphosphate phosphohydrolases; EC 3.1.3.8) catalyze the hydrolysis of phytic acid (myo-inositol hexakisphosphate) to the mono-, di-, tri-, tetra-, and pentaphosphates of myo-inositol and inorganic phosphate. A broad range of microorganisms, including bacteria (20), yeasts (2), and filamentous fungi (10, 19, 27), produce phytases.
Phytic acid is the primary storage form of phosphate in cereal grains, legumes, and oilseeds, such as soy, which are the principal components of animal feeds. However, monogastric animals are unable to metabolize phytic acid and largely excrete it in their manure. Therefore, the presence of phytic acid in animal feeds for chickens and pigs is undesirable, because the phosphate moieties of phytic acid chelate essential minerals and possibly proteins, rendering the nutrients unavailable. Since phosphorus is an essential element for the growth of all organisms, livestock feed must be supplemented with inorganic phosphate. There are a number of published reports (12, 16, 18, 26) describing the use of phytases in the feeds of monogastric animals and in human food.
When phytic acid is not metabolized by monogastric animals the phosphate level in the manure can also create disposal problems. The amount of manure produced worldwide has increased significantly as a result of increased livestock production. Environmental pollution with high-phosphate manure has caused problems in various locations around the world due to the accumulation of phosphate, particularly in bodies of water. Consequently, animal feed distributors in Europe have begun to formulate feed products with supplemental phytase in order to improve feedlot productivity and decrease phosphate waste. Thus, phytases are also useful for reducing the amount of phytate in manure (13, 18). The current commercial feed supplement is a recombinant Aspergillus niger (previously Aspergillus ficuum) phytase produced in Aspergillus niger (27) or Aspergillus oryzae (i.e., Phytase Novo [13]).
There is a definite commercial need for second-generation phytases with improved properties (e.g., higher thermostability and catalytic efficiency) that can be produced in commercially significant quantities. Our objectives were to identify, clone, and characterize a phytase from a thermophilic fungus in anticipation that this enzyme would offer superior biochemical properties.
MATERIALS AND METHODS
DNA extraction and hybridization analysis.
Total cellular DNA was extracted from Thermomyces lanuginosus CBS 586.94 by the procedure described by Timberlake and Bernard (21). Genomic DNA samples were analyzed by Southern hybridization (6) under conditions of low stringency (i.e., 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 25% formamide, 0.3% sodium dodecyl sulfate [SDS]). A phytase-specific probe fragment comprising the Aspergillus niger phyA coding region (approximately 1.6 kb) was radiolabeled by nick translation (11) with [α-32P]dCTP (Amersham, Arlington Heights, Ill.) and added to the hybridization buffer at an activity of approximately 106 cpm per ml. The hybridization and washing conditions have been described previously (4).
DNA libraries and identification of phytase clones.
Genomic DNA libraries were constructed with the bacteriophage cloning vector λZipLox (Life Technologies, Gaithersburg, Md.) with Escherichia coli Y1090ZL cells (Life Technologies) as a host for plating and purification of recombinant bacteriophages and E. coli DH10Bzip (Life Technologies) for excision of individual pZL1-phytase clones. Total cellular DNA was partially digested with Tsp509I and size fractionated on 1% agarose gels. DNA fragments migrating in the range of 3 to 7 kb were excised and eluted from the gel with Prep-a-Gene reagents (Bio-Rad Laboratories, Hercules, Calif.). The eluted DNA fragments were ligated with EcoRI-cleaved and dephosphorylated λZipLox vector arms (Life Technologies), and the ligation mixtures were packaged with commercial packaging extracts (Stratagene, La Jolla, Calif.). The packaged DNA libraries were plated and amplified in E. coli Y1090ZL cells (Life Technologies). Approximately 30,000 plaques from the library were screened by plaque hybridization with the radiolabeled phytase probe. One positive clone which hybridizes strongly to the probe was picked and purified twice in E. coli Y1090ZL cells. The phytase clone was subsequently excised from the λZipLox vector as a pZL1-phytase clone (5) and designated pMWR46.
Molecular analysis of the T. lanuginosus phytase gene.
Restriction mapping of pMWR46 was performed by standard methods (11). DNA sequencing of the phytase clones was performed with model 373A automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.) by the primer-walking technique with dye-terminator chemistry (7). In addition to the lac forward and lac reverse primers, specific oligonucleotide sequencing primers were synthesized on an Applied Biosystems model 394 DNA-RNA synthesizer according to the manufacturer’s instructions.
Construction of the phytase expression vector pMWR48.
The coding region of the T. lanuginosus phyA gene was amplified by PCR with the forward primer 5′-ATTTAAATGGCGGGGATAGGTTTGG-3′ and the reverse primer 5′-CTTAATTAATCAAAAGCAGCGATCCC-3′. The sense primer incorporated the first in-frame ATG and extends 16 bp downstream. The antisense primer incorporated a region 14 bp upstream of the translational stop codon and extends through the stop codon. To facilitate the cloning of the amplified fragment, the sense and antisense primers contain a SwaI and a PacI restriction site, respectively. The amplified product was digested with SwaI and PacI and ligated with pDM181 (also digested with SwaI and PacI), a plasmid which provides the Fusarium oxysporum trypsin gene promoter and terminator and the bar resistance cassette (3). The resulting expression vector was designated pMWR48.
Transformation of Fusarium venenatum and analysis of transformants.
Transformation protocols and methods for purification of F. venenatum (28) transformants are described by Royer et al. (15). Mycelia from primary transformants were used to inoculate shake flasks containing 25 ml of M400 Da medium (50 g of maltodextrin, 2 g of MgSO4 · 7H2O, 2 g of KH2PO4, 4 g of citric acid, 8 g of yeast extract, 2 g of urea, and 0.5 ml of trace metal solution per liter [15]) and incubated with shaking at 30°C. One milliliter of culture supernatant was harvested at 4, 5, and 7 days and stored at 4°C. Phytase activity was assayed as described below. Spores from the primary transformants producing the highest phytase activity were generated by inoculating 20 ml of R medium (12.1 g of NaNO3/liter, 50 g of succinic acid/liter, 20 ml of 50× Vogel’s salts, 25 mM NaNO3 [pH 6.0] [15]) with mycelia and incubating it at 30°C with shaking for 2 to 3 days. Single spores were isolated by spreading 150 ml of spore culture onto manipulator plates (1X Vogel’s salts, 25 ml of NaNO3, 2.5% sucrose, 2% Noble agar) containing 5 mg of Basta [phosphinothricin or 2-amino-4-(hydroxymethyl-phosphinyl)butanoic acid; Hoechst-Schering, Rodovre, Denmark] per ml and using a micromanipulator to transfer single spores to a clear region of the plate. After 3 days of growth at room temperature, the germinated spores were transferred to individual Vogel plates containing 5 mg of Basta/ml. Shake flasks containing 25 ml of M400Da medium plus 5 mg of Basta/ml were inoculated in duplicate with mycelial plugs from each single-spore isolate and incubated at 30°C. The best single-spore isolate was selected based on assay of the secreted enzymatic activity, where the transformants produced >150-fold more phytase activity than an untransformed control.
Protein purification.
The best F. venenatum transformant was run in two 2-liter fermentors with a standard protocol (3). The frozen cell-free broth (1,700 ml) was thawed, clarified by centrifugation, and concentrated on a hollow-fiber Amicon filtration unit with an S1Y10 filter to a volume of 350 ml. The sample was adjusted to pH 7, diluted to a conductivity of 2 mS, and chromatographed at room temperature on a 75-ml-bed-volume Q-Sepharose Big Beads column (Pharmacia), which had been equilibrated in 20 mM Tris-Cl, pH 7. The column was developed at 5 ml/min with the equilibration buffer until the effluent A280 had decreased to near baseline. The column was then developed at 5 ml/min with a 600-ml gradient of 0 to 0.6 M NaCl in the same buffer. The bound enzyme activity was found to elute in fractions corresponding to ca. 0.2 M NaCl.
The collected activity peak was concentrated by ultrafiltration with a PM-10 membrane to a volume of 25 ml, diluted to a conductivity of 0.9 mS, and chromatographed at 4 ml/min on a MonoQ HR 10/16 column which had been equilibrated in 20 mM MOPS (morpholinepropanesulfonic acid), pH 7. The column was developed with 80 ml of starting buffer and then with a 400-ml gradient of 0 to 0.5 M NaCl in the same buffer. Enzyme activity was detected in fractions by using the p-nitrophenyl phosphate measurement described below. The active fractions were also analyzed with a Novex 10 to 27% gradient SDS-polyacrylamide gel, and the fractions were combined if judged by electrophoresis to be substantially purified.
The peak fractions were combined, concentrated with an Amicon PM-10 membrane by ultrafiltration, and exchanged into 20 mM MES (morpholine ethanesulfonic acid), pH 5.5. The sample conductivity was 1.1 mS. One-third of this sample was chromatographed at 1 ml/min on a Mono S HR 5/5 column (Pharmacia) which had been equilibrated in the same buffer. The column was developed with 5 ml of starting buffer and then with a 25-ml linear gradient of 0 to 0.6 M sodium chloride in the same buffer. The active fractions were combined after electrophoretic analysis to eliminate those which contained trace contaminants.
Physicochemical characterization.
Isoelectric focusing (IEF) was performed with a Novex pH 3 to 7 IEF gel according to the instructions of the manufacturer. IEF standards from both Pharmacia and Bio-Rad were used to calibrate the gel.
The protein extinction coefficient was determined experimentally by quantitative amino acid analysis with a Hewlett-Packard AminoQuant system. The analysis assumed 49,700 for the protein molecular weight, based on the translated gene sequence for the mature protein.
Amino-terminal sequence analysis was performed on an Applied Biosystems 476A sequencer.
Enzyme assays.
Phytase activity was measured by two different methods. During purification, fractions were rapidly evaluated by measuring the rate of p-nitrophenyl phosphate hydrolysis at 405 nm with 10 mM substrate in 0.2 M sodium citrate, pH 5.5, at 30°C with a plate reader (Thermomax; Molecular Devices).
Enzyme kinetics studies performed on purified enzyme samples were accomplished by the assay of inorganic phosphate liberated from corn phytic acid (Sigma catalog no. P 8810). Exhaustive phytate hydrolysis was accomplished by incubating 0.5 or 0.1% phytic acid with enzyme (1 U/ml) in 0.2 M sodium citrate, pH 5.5, at 37°C. Aliquots were removed over a period of 10 h and analyzed (see below) for kinetics of phosphorus release. Ten hours was found to be sufficient for the completion of product formation. Standard enzyme kinetics reactions were carried out for 30 min at 37°C in 0.5% (wt/wt) phytic acid. The reaction was quenched by the addition of an equal volume of 15% (wt/wt) trichloroacetic acid. After cooling, 100 μl of the resulting mixture was diluted in 1 ml of water. The sample was incubated at 50°C for 5 min. Color reagent (1 ml) was added, and the 50°C incubation was continued for 15 min. The absorbance of a 200-μl aliquot was measured at 690 nm with a microplate reader. The color reagent was composed of 6 N sulfuric acid–water–2.5% (wt/vol) hepta-ammonium molybdate–10% ascorbate (aqueous) in a ratio of 1:2:1:1 and was prepared fresh daily. Quantitation was based on a standard curve generated with a 10 mM sodium monobasic phosphate standard. One unit is defined as 1 μmol of inorganic phosphate released per min with 0.5% phytic acid in 0.2 M sodium citrate, pH 5.5, at 37°C.
Steady-state kinetics measurements were made by substrate titration. Phytate concentrations were 2.16, 1.08, 0.541, 0.216, 0.108, and 0.0758 mM for Km determination. Phytate concentrations of 1.08, 0.541, 0.216, and 0.108 mM in the presence or absence of 1 mM sodium monobasic phosphate were used to evaluate product inhibition.
Thermostability measurement.
Phytase samples were dissolved at 100 U per ml in 0.2 M sodium citrate, pH 5.5. One hundred-microliter aliquots of each enzyme solution were incubated for 20 min in a water bath at 37, 45, 50, 55, 60, 65, 70, and 75°C. After the heat treatment, the samples were stored at 0°C until activity assays were performed. Each sample was diluted 1:80 in 0.2 M sodium citrate, pH 5.5, containing 0.01% (wt/wt) Tween 20, and the standard activity assay was performed.
pH-activity measurement.
To attain a buffering range between pHs 2 and 7, a three-component 125 mM glycine-acetate-citrate buffer was employed. The buffer components were combined at final concentrations of 42 mM per component, and phytic acid was added as a solid to 1% (wt/wt). This mixture was adjusted to pH 7 with concentrated HCl, and a 10-ml aliquot was taken. This process was repeated for every 0.5 pH units through pH 2.
Enzyme stock solutions of 20 U per ml were prepared in 20 mM MES buffer, pH 5.5. Substrate (1% [wt/wt]; 850 μl) in buffer at a given pH was combined with 100 μl of water and 50 μl of enzyme stock solution and incubated for 30 min at 37°C. Subsequently, the enzyme reaction was quenched with 1 ml of 15% trichloroacetic acid and quantitated by the standard method.
Temperature-activity measurement.
Enzyme stock solutions of 12.5 U per ml were prepared in 0.2 M sodium citrate buffer, pH 5.5. Two hundred fifty microliters of 1% phytic acid substrate was added to a 1.7-ml Eppendorf tube followed by 240 μl of 0.2 M sodium citrate buffer, pH 5.5. This solution was vortexed and placed in a water bath at the designated temperature. After 20 min of equilibration in the water bath, the mixture was vortexed and 10 μl of phytase solution was added. The sample was vortexed and incubated in the water bath for an additional 30 min, and then the reaction was quenched with 1 ml of 15% trichloroacetic acid and quantitated by the standard method.
Nucleotide sequence accession number.
The complete phyA gene sequence has been deposited in GENESEQN as accession no. T90070.
RESULTS
Cloning of phytase gene sequences from T. lanuginosus.
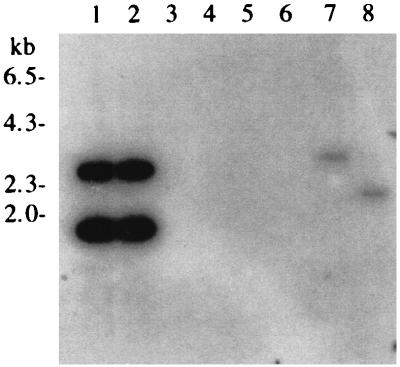
Southern blotting experiments indicated that an Aspergillus phytase gene fragment could be used as a probe to identify phytase gene-specific fragments in T. lanuginosus genomic DNA (Fig. 1). We screened 30,000 plaques from a genomic library of T. lanuginosus DNA constructed in λZipLox for hybridization with the Aspergillus phytase gene probe. Several positive clones were picked and excised by an in vivo-excision protocol (5).
FIG. 1.
Autoradiogram from Southern hybridization analysis of T. lanuginosus genomic DNA with an Aspergillus phytase gene probe. Lanes 1 and 2, A. niger genomic DNA digested with BamHI and BamHI plus PstI, respectively; lanes 3 and 4, Myceliophthora thermophila genomic DNA digested with BamHI and BamHI plus PstI, respectively; lanes 5 and 6, Thielavia terrestris genomic DNA cleaved with BamHI and BamHI plus PstI, respectively; lanes 7 and 8, T. lanuginosus genomic DNA cut with BamHI and BamHI plus PstI, respectively.
Analysis of the T. lanuginosus phyA gene.
DNA sequencing of one T. lanuginosus phytase clone (pMWR46) showed an open reading frame similar to the A. niger phytase gene. The positions of introns and exons within the phyA gene were assigned based on comparison of the deduced amino acid sequence with the deduced amino acid sequence of the corresponding A. niger phytase gene product. On the basis of this analysis, the T. lanuginosus phytase gene is comprised of two exons (47 and 1,377 bp), which are separated by a small intron (56 bp). The size and composition of the intron is consistent with those of other fungal genes (9) in that all contain consensus splice donor and acceptor sequences as well as a near approximation of the consensus lariat sequence (RCTRAC) near the 3′ end of each intervening sequence.
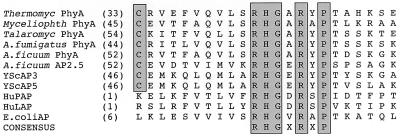
The deduced amino acid sequence of the T. lanuginosus gene product shows the characteristics of an extracellular fungal enzyme with a cleavable signal sequence. Based on the rules of von Heijne (25), the first 22 amino acids of PhyA likely comprise a secretory signal peptide which directs the nascent polypeptide into the endoplasmic reticulum. Amino-terminal amino acid sequencing suggests that the next 10 amino acids constitute a propeptide which terminates with a dibasic cleavage site (LysLys). The mature PhyA is an acidic protein (predicted isoelectric point, 5.4) composed of 452 amino acids (molecular mass, 51 kDa). The amino acid sequence also contains the active-site motif RHGXRXP, which is shared by other known phytases and acid phosphatases (Fig. 2) (23, 27). Lastly, the deduced amino acid sequence of the mature PhyA has approximately 47.5% identity with the phytase from A. niger (GenBank accession no. M94550).
FIG. 2.
Alignment of putative active-site regions of acid phosphatases (AP) and phytases from various species. The M. thermophila (Myceliophth; TREMBL O00107), Talaromyces thermophilus (Talaromyc; TREMBL O00096), A. fumigatus (TREMBL O00092), A. ficuum (A. niger) (SwissProt P34752 and P34754), Saccharomyces cerevisiae (YScAP3 and -5; SwissProt P24031 and P00635), human (HuPAP and HuLAP; SwissProt P15309 and P11117), and E. coli (SwissProt P07102) sequences were obtained from the databases indicated. The numbers in parentheses are the starting amino acid positions from the mature proteins for the sequences compared. Identical amino acids are boxed. Thermocyl, T. lanuginosus.
Analysis of F. venenatum transformants expressing T. lanuginosus phytase.
F. venenatum has recently been developed as an efficient fungal host for the production of heterologous proteins (15). Culture supernatants from 14 of the 17 primary transformants of pMWR48 were positive when assayed for phytase activity. Two primary transformants with the highest phytase activity were selected for single-spore isolation, and nine single-spore isolates were obtained.
Physicochemical characterization of the recombinant phytase.
The purified T. lanuginosus phytase was apparently homogeneous in SDS-polyacrylamide gel electrophoresis, with a single component corresponding to a molecular weight of 60,000. The protein sample contained numerous components in IEF analysis ranging from pH 4.7 to 5.2. In contrast to the T. lanuginosus phytase, recombinant A. niger phytase is composed of a single major component with a pI near 4.9 and two minor bands around pI 4.7.
Amino-terminal sequence analysis of the purified T. lanuginosus enzyme identified three components: the major component (ca. 60%) is H2N-His-Pro-Asn-Val-Asp-Ile-Ala-Arg-His-Trp-Gly-Gln…, which corresponds to a Kex2 cleavage site at position 34 in the primary translation product. Two minor sequences, H2N-Gly-Glu-Asp-Glu-Pro-Phe-Val-Arg-Val-Leu-Val-Asn…(ca. 30%) and H2N-Ser-Glu-Glu-Glu-Glu-Glu-Gly-Glu-Asp-Glu-Pro-Phe…(ca. 10%), correspond to internal cleavage sites near the COOH terminal of the protein at positions 428 and 435 in the primary translation product. The observation that our protein sequence data exactly match the predicted translation product of the T. lanuginosus gene and the finding that untransformed Fusarium host strains produce 2 orders of magnitude less enzyme activity both argue strongly that we have isolated a heterologous gene product.
The specific activities for the two recombinant phytases (i.e., those of T. lanuginosus and A. niger) were 91 and 180 U/mg, respectively, under standard assay conditions at pH 5.5. At its pH 6 optimum T. lanuginosus phytase had a specific activity of 110 μmol of inorganic phosphate released per min per mg of protein at 37°C. Exhaustive enzymatic hydrolysis of phytic acid revealed that A. niger and T. lanuginosus phytases released identical amounts (70%) of the total theoretically available phosphorus. Steady-state kinetic measurements disclosed that the apparent Km of T. lanuginosus phytase is approximately 110 μM with respect to phytate while A. niger has an apparent Km of 200 μM. There was a faint indication of excess substrate inhibition at the 2.16 mM substrate concentration, perhaps congruent with the report of inhibition above 2 mM for A. niger phytase (22). Steady-state kinetics measurements with 1 mM phosphate present failed to reveal any type of inhibition with this product. We estimate that the Ki for phosphate must exceed 3 mM to be undetectable in our experiments. In contrast Ullah (22) has reported that phosphate is a competitive inhibitor, with a Ki of 1.9 mM.
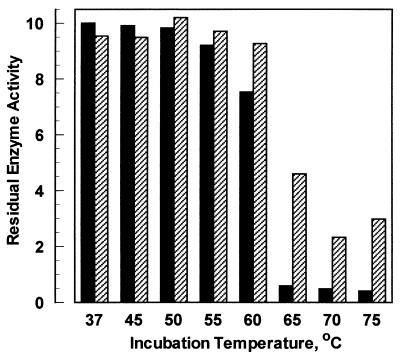
A comparison of enzyme thermostability profiles (Fig. 3) suggests that differences between the stabilities of the two enzymes are small. Neither enzyme is fully inactivated by a high-temperature incubation, and the residual activity profiles are consistent with partially reversible thermal denaturation (24). Differential scanning calorimetry (DSC) experiments reveal that the A. niger enzyme has a transition at 60°C while T. lanuginosus phytase unfolds at 69°C. Others have reported an Aspergillus fumigatus phytase which has an apparently greater propensity for reversible thermal denaturation (14), as measured by residual enzyme activity. However, there are no published data on thermal denaturation points for the A. fumigatus phytase or other phytase species.
FIG. 3.
Phytase thermal stability. Comparison of residual enzyme activity after a 20-min incubation at various temperatures. Full activity corresponds to 10 U. Solid bar, A. niger phytase; cross-hatched bar, T. lanuginosus phytase.
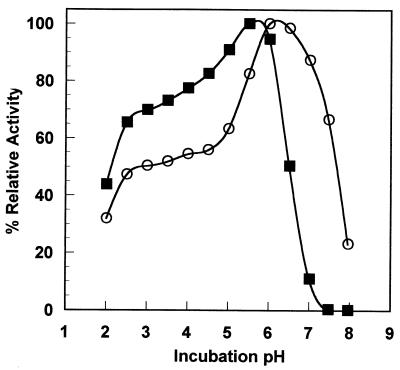
The pH-activity profile comparison of T. lanuginosus and A. niger phytases indicates substantial similarity between the pH profiles of the two enzymes (Fig. 4). However, the T. lanuginosus enzyme is active at neutral pH while the A. niger enzyme is not. We could not reproduce the earlier reports (e.g., reference 17) that A. niger phytase possesses two pH optima; employing a composite buffer, we measured a broad shoulder near pH 3. We note that there are very few cases of a single enzyme species possessing two pH optima. The earlier reports may originate from impure material which contains traces of the A. niger acid phosphatase (29), or they could be artifacts of employing more than one buffer to span the pH range.
FIG. 4.
Phytase pH-activity profile; comparison of relative enzyme activity at various incubation pHs. A relative activity of 100% corresponds to 1 and 1.21 μmol of inorganic phosphate released per min for A. niger and T. lanuginosus phytases, respectively. Solid square, A. niger phytase; open circles, T. lanuginosus phytase.
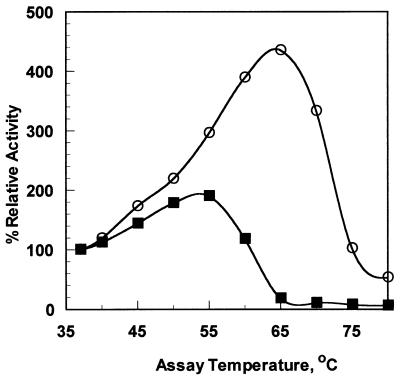
Measurement of enzyme activity as a function of temperature revealed a significant difference between the two enzymes (Fig. 5). T. lanuginosus phytase has maximum enzyme activity near 65°C and has partial activity even at 75°C. In contrast, A. niger phytase is essentially inactive at 65°C. These results are congruent with the DSC data for the two enzymes, which also indicate a 9°C stability improvement for the Thermomyces phytase.
FIG. 5.
Phytase temperature-activity measurement; observed enzyme activity as a function of incubation temperature. A relative activity of 100% corresponds to 0.125 μmol of inorganic phosphate released per min. Solid squares, A. niger phytase; open circles, T. lanuginosus phytase.
DISCUSSION
Enzyme activity at elevated temperatures may be relevant in applications such as saccharification (a high-temperature industrial process to generate high-fructose corn syrup), where others have reported that the addition of phytase improves carbohydrate yields (1). Figure 5 demonstrates that at 55°C, the optimal temperature for A. niger phytase, the Thermomyces phytase performs at 79% of the A. niger phytase turnover number (despite lower specific activity for Thermomyces phytase at 37°C) and at 60°C the Thermomyces phytase is operating at 67%-greater catalytic efficiency than the A. niger enzyme. The A. niger phytase is inactivated at 65°C, where Thermomyces phytase activity is maximal.
Enzyme thermal stability is also relevant in animal feed applications, where the enzyme is normally incorporated into the grains prior to pelletization and the feed briefly reaches processing temperatures of 85 to 90°C. In this circumstance a commercial phytase product must be able to withstand brief heating prior to encountering an animal’s digestive tract at 37°C. Our physicochemical data demonstrate an improvement of approximately 9°C in denaturation temperature for Thermomyces phytase versus the present A. niger product.
Animal-feeding trials with formulated phytase supplementation would involve testing a total of 300 broilers or piglets at two enzyme dosages plus a control without enzyme addition. Typically the apparent total-tract digestibility of dissolved matter, organic matter, nitrogen, calcium, and total phosphorus would be monitored at one or two points during an animal’s growth to determine the effect of enzyme dosage on feed intake and conversion. Such animal-feeding trials and the level of analysis required to present and evaluate the data are beyond the scope of this paper.
It is tempting to speculate about the structural origins of thermal stability in phytases. However, there is no obvious pattern to the sequence differences between phytases from thermophiles (represented by Myceliophthora, Talaromyces, and Thermomyces) and mesophiles (represented by A. niger and A. fumigatus). For example, there are no gross differences in protein structure, such as addition or deletion of secondary structure elements. Nor is there a systematic pattern to the sequence differences between the two representative enzymes; i.e., hydrophobic replacements, addition of salt bridges, addition of potential disulfide bonding sites, and deletion of asparagine or aspartate residues are not readily apparent. The most striking difference is the additional consensus N-linked glycosylation site present in the two Aspergillus enzymes (sequence position 231 in reference 27) but missing in the three thermophile examples. We believe that the most likely explanation which can be deduced for the sequence differences is derived from evolutionary rather than functional factors.
Recently the discovery of new industrial enzymes has focused on novel microbial sources representing extreme conditions (extremophiles). In many cases the genes encoding these interesting enzymes can be cloned without prior isolation of the catalyst or culturing of the donor microbe. However, heterologous production of the novel enzyme often results in extremely low yields of secreted product or accumulation of inactive material as inclusion bodies. Either of these outcomes is incompatible with the production economics required for commercialization. We have searched for new industrial catalysts from a constellation of thermophilic fungi that are more closely related than the extremophiles to the industrial fungal production strains which are available. We have successfully isolated enzymes with both improved thermal stability characteristics and the potential for high-level commercial production (4).
T. lanuginosus phytase is an alternative enzyme with performance advantages over the conventional A. niger enzyme in the form of stable enzyme activity at elevated temperatures and superior substrate saturation kinetics at physiological pH. A second-generation commercial enzyme may also benefit from protein engineering when a three-dimensional protein structure is available, as is the case for the A. fumigatus enzyme (8).
ACKNOWLEDGMENTS
We thank Carin Morris (Novo Nordisk Biotech) for performing phytase enzyme activity assays, Sam Johnstone (Novo Nordisk Biotech) for performing Fusarium fermentations, and Claus Crone Fuglsang (Novo Nordisk A/S) for measuring phytase stability by DSC.
REFERENCES
- 1.Antrim R L, Mitchinson C, Solheim L P. Method for liquefying starch. 1996. PCT patent application WO 96/28567. [Google Scholar]
- 2.Barbaric S, Kozulic B, Reis B, Mildner P. Physicochemical and kinetic properties of acid phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1984;259:878–883. [PubMed] [Google Scholar]
- 3.Berka R M, Rey M W, Klotz A V. Polypeptides having phytase activity and nucleic acids encoding same. 1997. PCT patent application WO 97/35017. [Google Scholar]
- 4.Berka R M, Schneider P, Golightly E J, Brown S H, Madden M, Brown K M, Halkier T, Mondorf K, Xu F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme produced in Aspergillus oryzae. Appl Environ Microbiol. 1997;63:3151–3157. doi: 10.1128/aem.63.8.3151-3157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Alessio J M, Bebee R, Hartley J L, Noon M C, Polayes D. Lambda ZipLoxTM: automatic subcloning of cDNA. Focus (Life Technologies, Gaithersburg, Md) 1992;14:76–79. [Google Scholar]
- 6.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics, a manual for genetic engineering. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 7.Giesecke H, Obermaier B, Domedy H, Neubert W J. Rapid sequencing of the Sendai virus 6.8-kb large L gene through primer walking with an automated DNA sequencer. J Virol Methods. 1992;38:47–60. doi: 10.1016/0166-0934(92)90168-d. [DOI] [PubMed] [Google Scholar]
- 8.Grueninger-Leitch F, D’Arcy A, D’Arcy B, Chene C. Deglycosylation of proteins for crystallization using recombinant fusion protein glycosidases. Protein Sci. 1996;5:2617–2622. doi: 10.1002/pro.5560051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes in filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, England: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 10.Howson S J, Davis R P. Production of phytate-hydrolyzing enzyme by some fungi. Enzyme Microb Technol. 1983;5:377–382. [Google Scholar]
- 11.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 12.Nelson T S, Shieh T R, Wodzinski R J, Ware J H. Effect of supplemental phytase on the utilization of phytate phosphorus by chicks. J Nutr. 1971;101:1289–1294. doi: 10.1093/jn/101.10.1289. [DOI] [PubMed] [Google Scholar]
- 13.Novo Nordisk A/S. Phytase Novo cuts phosphorus in manure by 30% BioTimes. 1995;10:8–9. [Google Scholar]
- 14.Pasamontes L, Haiker M, Wyss M, Tessier M, van Loon A P G H. Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl Environ Microbiol. 1997;63:1696–1700. doi: 10.1128/aem.63.5.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Royer J C, Moyer D M, Reiwitch S G, Madden M S, Jensen E B, Brown S H, Yonker C C, Johnstone J A, Golightly E J, Yoder W T, Shuster J R. Fusarium graminearum A 3/5 as a novel host for heterologous protein production. Bio/Technology. 1995;13:1479–1483. doi: 10.1038/nbt1295-1479. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg A-S, Andersson H. Effect of dietary phytase on the digestion of phytate in stomach and small intestine of humans. J Nutr. 1988;118:469–473. doi: 10.1093/jn/118.4.469. [DOI] [PubMed] [Google Scholar]
- 17.Sandberg A-S, Hulthen L R, Türk M. Dietary Aspergillus niger phytase increases iron absorption in humans. J Nutr. 1996;126:476–480. doi: 10.1093/jn/126.2.476. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz G, Hoppe P P. Phytase enzyme to curb pollution from pigs and poultry. Feed Magazine. 1992;1992:22–26. [Google Scholar]
- 19.Shieh T R, Ware J H. Survey of microorganisms for the production of extracellular phytase. Appl Microbiol. 1968;6:1348–1351. doi: 10.1128/am.16.9.1348-1351.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu M. Purification and characterization of phytase from Bacillus subtilis (natto) N-77. Biosci Biotechnol Biochem. 1992;56:1266–1269. [Google Scholar]
- 21.Timberlake W E, Bernard E C. Organization of a gene cluster expressed specifically in the asexual spores of Aspergillus nidulans. Cell. 1981;26:29–37. doi: 10.1016/0092-8674(81)90030-1. [DOI] [PubMed] [Google Scholar]
- 22.Ullah A H J. Production, rapid purification and catalytic characterization of extracellular phytase from Aspergillus ficuum. Prep Biochem. 1988;18:443–458. doi: 10.1080/00327488808062543. [DOI] [PubMed] [Google Scholar]
- 23.Ullah A H J, Dischinger H C., Jr Identification of active-site residues in Aspergillus ficuum extracellular pH 2.5 optimum acid phosphatase. Biochem Biophys Res Commun. 1993;192:754–759. doi: 10.1006/bbrc.1993.1478. [DOI] [PubMed] [Google Scholar]
- 24.Ullah A H J, Mullaney E J. Disulfide bonds are necessary for structure and activity in Aspergillus ficuum phytase. Biochem Biophys Res Commun. 1996;227:311–317. doi: 10.1006/bbrc.1996.1506. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- 26.Wang H L, Swain E W, Hasseltine C W. Phytase of moulds used in oriental food fermentation. J Food Sci. 1980;45:1262–1266. [Google Scholar]
- 27.Wodzinski R J, Ullah A H J. Phytase. In: Neidleman S L, Laskin A I, editors. Advances in applied microbiology. Vol. 42. New York, N.Y: Academic Press, Inc.; 1996. pp. 263–302. [DOI] [PubMed] [Google Scholar]
- 28.Yoder W T, Christianson L M. Species-specific primers resolve members of Fusarium section Fusarium: taxonomic status of the edible “Quorn” fungus reevaluated. Fungal Genet Biol. 1998;23:68–80. doi: 10.1006/fgbi.1997.1027. [DOI] [PubMed] [Google Scholar]
- 29.Zyla K. The role of acid phosphatase activity during enzymic dephosphorylation of phytates by Aspergillus niger phytase. World J Microbiol Biotechnol. 1993;9:117–119. doi: 10.1007/BF00656531. [DOI] [PubMed] [Google Scholar]