Abstract
Aims
The validated HCM Risk-Kids model provides accurate individualized estimates of sudden cardiac death risk in children with hypertrophic cardiomyopathy (HCM). A second validated model, PRIMaCY, also provides individualized estimates of risk, but its performance and clinical impact has not been independently investigated. The aim of this study was to investigate the clinical impact of using the PRIMaCY sudden cardiac death (SCD) risk model in childhood HCM.
Methods and results
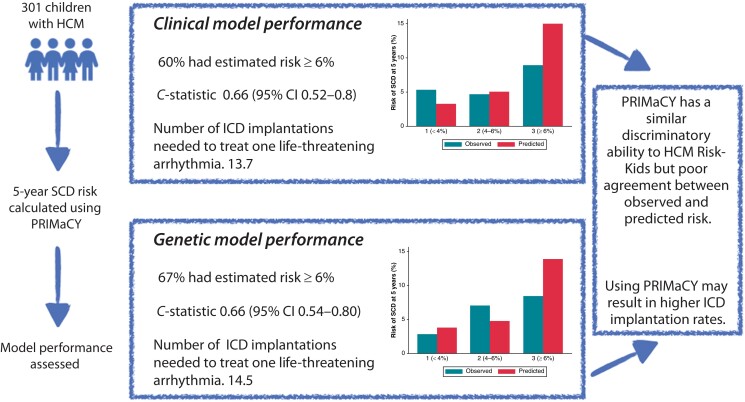
The estimated 5-year SCD risk was calculated for children meeting diagnostic criteria for HCM in a large single-centre cohort using PRIMaCY (clinical and genetic) and HCM Risk-Kids model, and model performance was assessed. Three hundred one patients [median age 10 (interquartile range 4–14)] were followed up for an average of 4.9 (±3.8) years, during which 30 (10.0%) reached the SCD or equivalent event endpoint. Harrell’s C-statistic for the clinical and genetic models was 0.66 [95% confidence interval (CI) 0.52–0.8] and 0.66 (95% CI 0.54–0.80) with a calibration slope of 0.19 (95% CI 0.04–0.54) and 0.26 (95% CI −0.03–0.62), respectively. The number needed to treat to potentially treat one life-threatening arrhythmia for the PRIMaCY clinical, PRIMaCY genetic, and HCM Risk-Kids models was 13.7, 14.5, and 9.4, respectively.
Conclusion
Although PRIMaCY has a similar discriminatory ability to that reported for HCM Risk-Kids, estimated risk estimates did not correlate well with observed risk. A higher proportion of patients met implantable cardioverter-defibrillator thresholds using PRIMaCY model compared with HCM Risk-Kids. This has important clinical implications as these patients will be exposed to a lifetime risk of complications and inappropriate therapies.
Keywords: Sudden death, Childhood, Risk, Prediction, Cardiomyopathy, Implantable cardioverter-defibrillator
Graphical Abstract
Graphical Abstract.
Graphical representation of the clinical impact of using different paediatric risk models in a cohort of patients aged 1–16 years.
What’s new?
We independently confirm that PRIMaCY has a similar ability to HCM Risk-Kids for distinguishing between patients at high and low risk for sudden death events.
However, risk estimates appear to be overestimated for some patients, with two-thirds meeting thresholds for implantable cardioverter-defibrillator (ICD) implantation.
Using the PRIMaCY model could lead to an increased number of patients undergoing ICD implantations, which are associated with a life-long risk of complications.
Introduction
Sudden cardiac death (SCD) is the most frequent mode of death in childhood hypertrophic cardiomyopathy (HCM), and identification of those at highest risk is a cornerstone of clinical management.1–3 Traditional guideline-endorsed approaches to risk stratification, which are based on the summation of different clinical risk factors, have been shown to have only modest discriminatory ability.2 In 2019, we published the first validated risk prediction model for childhood HCM (HCM Risk-Kids), developed and validated in a large multicentre consortium, that uses readily available clinical predictors to estimate 5-year SCD risk.4,5 This has subsequently been externally validated in a large cohort and in two smaller independent studies, confirming its superior performance compared with traditional risk stratification methods,6–8 and its use has been recommended in the 2023 European Society of Cardiology (ESC) guidelines for the management of cardioimyopathies9 and the 2022 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death.10,11 A second model (PRIMaCY) has more recently been described, with similar reported performance to HCM Risk-Kids in an external validation study,5 although this has not been confirmed in an independent external cohort. Whilst there are many similarities between the two risk models, important differences exist in the approach to risk factor selection, resulting in different risk predictor variables, and the age for which they are validated. Whilst HCM Risk-Kids was designed to be used up to the age of 16 years, PRIMaCY has been validated up to 18 years, resulting in an overlap with the widely used and ESC guideline-endorsed9 adult HCM risk-SCD calculator (for patients aged 16 and above).12 The aim of this study was, therefore, to perform the first independent external validation of the PRIMaCY model in a large single-centre paediatric HCM cohort, allowing comparison with HCM Risk-Kids.
Methods
The study cohort comprised consecutively evaluated patients aged 1–18 years meeting diagnostic criteria for HCM from a single quaternary referral centre (Great Ormond Street Hospital Center for Inherited Cardiovascular Diseases). Included patients were evaluated between 1995 and 2020. Patients were excluded if they met criteria for secondary prevention ICD implantation [history of ventricular fibrillation (VF) or sustained ventricular tachycardia (VT)], had known inborn errors of metabolism or syndromic disease, or <1 month follow-up. This cohort includes patients used to develop the HCM Risk-Kids model. None of the patients were included in the development of the PRIMaCY model.5
Patient assessment and data collection
Anonymized clinical data were collected retrospectively from baseline evaluation, including demographics, symptoms, pedigree analysis, ambulatory and 12-lead electrocardiography (ECG), and 2-dimensional (2D) Doppler and colour transthoracic echocardiogram. Predictor variables for the PRIMaCY5 risk model were recorded at the time of, or prior to, baseline evaluation. Specifically, age; unexplained syncope; family history of SCD; non-sustained ventricular tachycardia (NSVT) on ambulatory ECG recordings; measures of LV hypertrophy [interventricular septal thickness (IVST) and posterior wall thickness (PWT)] as absolute 2D measurements (mm) and body surface area–corrected Z scores; left atrial (LA) diameter Z score; peak resting left ventricular outflow tract (LVOT) gradient; and the results of genetic testing [pathogenic (P) or likely pathogenic (LP) variant, variant of unknown significance (VUS) and no pathogenic variants] were recorded. Patients harbouring P/LP variants were considered genotype positive. All reported genetic variants were re-classified according to the American College of Medical Genetics (ACMG) guidelines.13 The definition of all predictor variables has been previously described and is detailed in the Supplementary methods.5
Clinical outcomes
The primary study endpoint was SCD or an equivalent event (aborted cardiac arrest, appropriate ICD therapy for a ventricular tachyarrhythmia, or sustained VT with haemodynamic compromise), as previously described.5
Statistical analysis
Statistical analysis was performed using Stata statistical software (version 17) and Python (version 3.8). Variables are described as mean (±standard deviation, SD), median (interquartile range, IQR), counts, or percentages, as appropriate. Follow-up time was calculated from the time of baseline evaluation to the date of reaching study endpoint, death from another cause, or most recent evaluation. The Kaplan–Meier method was used to estimate the incidence of reaching the study endpoint. This was a complete case analysis. Patients with missing data for any risk predictor variables were excluded from the study to allow calculation of estimates for 5-year SCD risk using the PRIMaCY model.
Estimating 5-year sudden cardiac death risk
Follow-up was censored at 5 years, and the estimated 5-year risk of SCD was calculated for each individual patient using the PRIMaCY (aged 1–18 years) and HCM Risk-Kids (aged 1–16 years) model. For patients who had undergone genetic testing, estimates were calculated using both the PRIMaCY genetic and clinical models. Risk estimates were classified as low (<4%), intermediate (4–6%), or high (≥6%) risk, reflecting current ESC guidelines.9,10 Secondary analysis using a threshold of >8.3% to define high risk, as reported in the PRIMaCY development manuscript, is reported in the Supplementary results.
Model validation
Harrell’s C-index14 and Uno’s C-index15 were used to measure how well the PRIMaCY model discriminated between low- and high-risk patients (a value of 1 indicates perfect discrimination, whilst a value of 0.5 indicates no discrimination). Model calibration was described graphically for patients in low-, intermediate-, and high-risk groups and using calibration plots. Ninety-five per cent confidence intervals (CIs) were obtained using a bootstrap procedure with 10 000 iterations of random sampling with replacement. Model validation was performed separately for the clinical and genetic PRIMaCY models. Model performance measures are not reported for HCM Risk-Kids in this population as this cohort of patients was previously used to develop or validate the model, meaning any estimates could be affected by over-fitting of the data.
Comparison of the clinical impact of different paediatric risk models
The proportion of patients classified as high (≥6%), intermediate (4–6%), or low (<4%) risk by each risk model is described graphically. The number needed to treat for one appropriate ICD therapy was calculated for each risk model using a threshold for ICD implantation of ≥6%.
Ethics
Local ethical approval was given from the research office, Great Ormond Street Hospital, with waiver of informed consent for retrospective, anonymized data (R&D number 19HL04 and REC21/NI/0122).
Results
Three hundred one patients (male n = 195, 64.8%) met diagnostic criteria for HCM with a median age of 10 years (IQR 4, 14) at baseline assessment. One hundred and forty-five (48.2%) were probands, and 172 (57.2%) had a family history of HCM. Eighty patients (26.6%) were on medical therapy [β-blockers, n = 68 (22.5%); disopyramide, n = 16 (5.3%); calcium channel blockers, n = 11(3.7%); amiodarone, n = 2 (0.7%); angiotensin-converting enzyme (ACE) inhibitor, n = 2 (0.7%); diuretics, n = 1 (0.3%)]. Two hundred and seven (68.8%) had undergone genetic testing, of whom 143 (69.1%) had a P/LP variant. Table 1 and Supplementary material online, TableS1 describe the clinical phenotype and clinical risk factors for SCD.
Table 1.
Baseline demographics and clinical risk factors for SCD
| Clinical characteristic | |
|---|---|
| Age (years) at baseline evaluation (median, IQR) | 10 (4, 14) |
| Male sex | 195 (64.8%) |
| Family history of HCM | 171 (56.8%) |
| NYHA > 1 | 41 (27.2%) |
| Family history of SCD (n, %) | 15 (5%) |
| Genetic testing performed (n, %) | 207 (68.8%) |
| Pathogenic/likely pathogenic variant (n, %) | 143 (69.1%) |
| VUS (n, %) | 21 (10.1%) |
| No variants identified (n, %) | 43 (20.8%) |
| Unexplained syncope within 6 months of baseline assessment (n, %) | 11 (3.7%) |
| NSVT on ambulatory ECG within 6 months of baseline assessment (n, %) | 11 (3.7%) |
| LVMWT Z score (median, IQR) | 9.3 (5.4, 15.5) |
| IVST Z score (median, IQR) | 7.6 (3.1, 14.2) |
| LVPWT Z score (median, IQR) | 2.5 (−0.1 to 6.1) |
| LA diameter Z score (mean, ±SD) | 1.6 (±2.3) |
| Maximal LVOT gradient, mmHg (median, IQR) | 8 (5, 16) |
ECG, electrocardiography; IVST, interventricular septal thickness; IQR, interquartile range; LA, left atrial; LVMWT, left ventricular maximal wall thickness; LVOT, left ventricular outflow tract; LVPWT, left ventricular posterior wall thickness; NSVT, non-sustained ventricular tachycardia; NYHA, New York Heart Association; SCD, sudden cardiac death; VUS, variant of unknown significance.
Clinical follow-up
Patients were followed up for a mean of 4.9 (±3.8) years, during which 22 (7.3%) died [SCD, n = 9 (40.9%); CCF, n = 2 (9.1%); other cardiovascular (CV), n = 4 (18.2%); non-CV, n = 4 (18.2%); unknown, n = 3 (13.6%)], with a corresponding 1- and 5-year survival of 98.6% (95% CI 96.4–99.5%) and 95.2% (95% CI 91.3–97.4%), respectively. Five patients (1.7%) underwent cardiac transplantation, 14 (4.7%) had a LV septal myectomy, and 80 (26.6%) had an ICD implanted for primary (n = 70, 87.5%) or secondary (n = 10, 12.5%) prevention. Thirty patients experienced a life-threatening arrhythmic event [SCD, n = 9 (30%); appropriate ICD therapy for ventricular tachyarrhythmia, n = 2 (6.7%); resuscitated cardiac arrest, n = 8 (26.7%); or sustained VT, n = 11 (36.7%)], with an overall incidence of 1.90 (95% CI 1.315–2.759) per 100 patient-years.
Validation of the PRIMaCY model
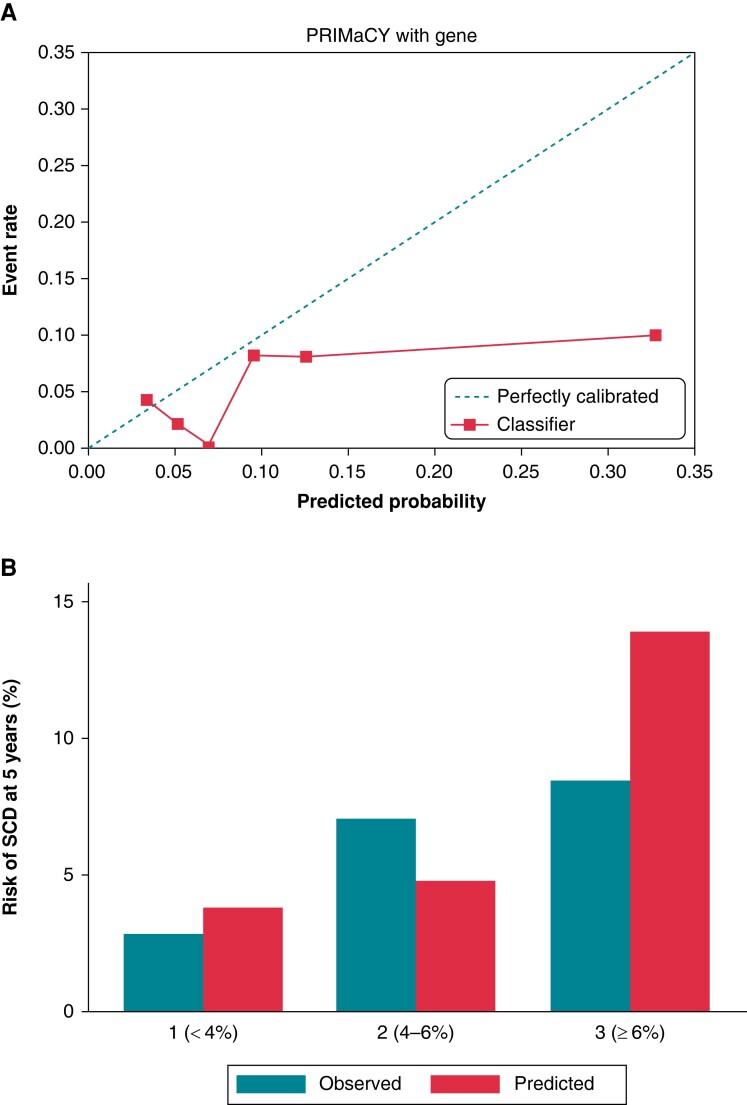
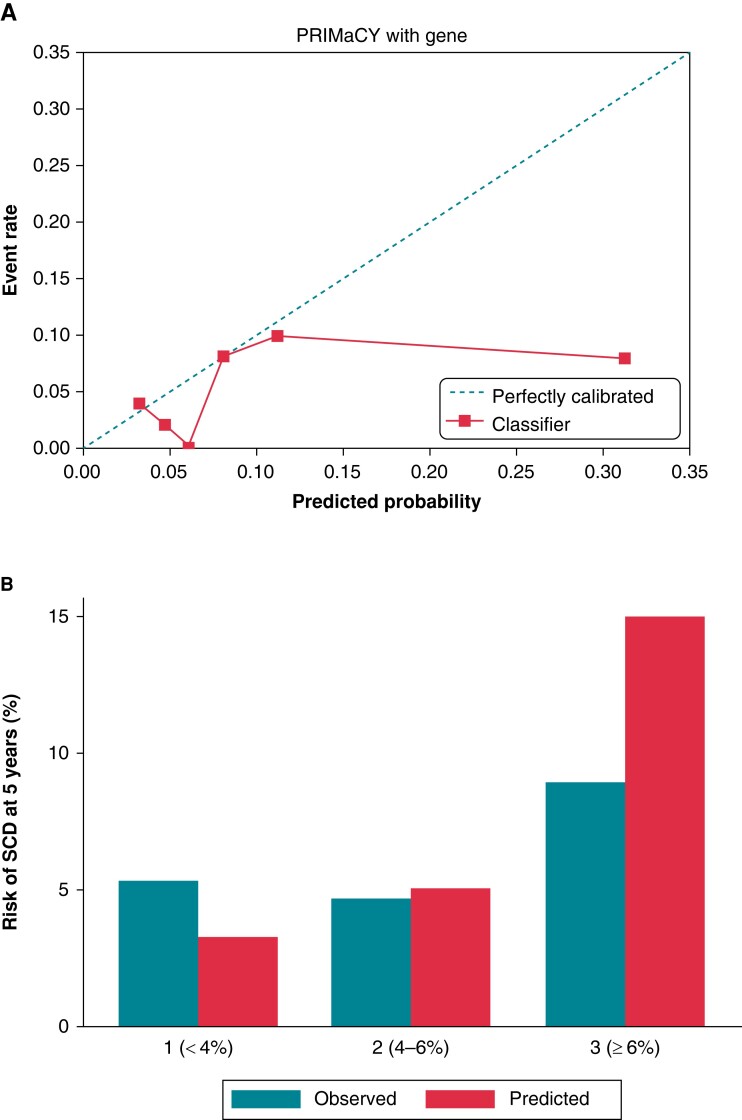
Seventeen arrhythmic events occurred within 5 years’ follow-up. The Harrell’s and Uno C-statistics for the PRIMaCY genetic model were 0.66 (95% CI 0.54–0.80) and 0.60 (95% CI 0.53–0.79), respectively, with a calibration slope of 0.26 (95% CI −0.03–0.62) (Figure 1A). The Harrell’s and Uno C-statistics for the PRIMaCY clinical model were 0.66 (95% CI 0.52–0.8) and 0.61 (95% 0.51–0.80), respectively, with a calibration slope 0.19 (95% CI 0.04–0.54) (Figure 2A). Figures 1B and 2B graphically compare the observed and predicted risk for patients in different risk groups.
Figure 1.
Model validation for PRIMaCY genetic model (A) calibration slope and (B) bar chart comparing observed vs. predicted 5-year SCD risk by risk group. SCD, sudden cardiac death.
Figure 2.
Model validation for PRIMaCY clinical model (A) calibration slope and (B) bar chart comparing observed vs. predicted 5-year SCD risk by risk group. SCD, sudden cardiac death.
Clinical impact of using PRIMaCY genetic and clinical models
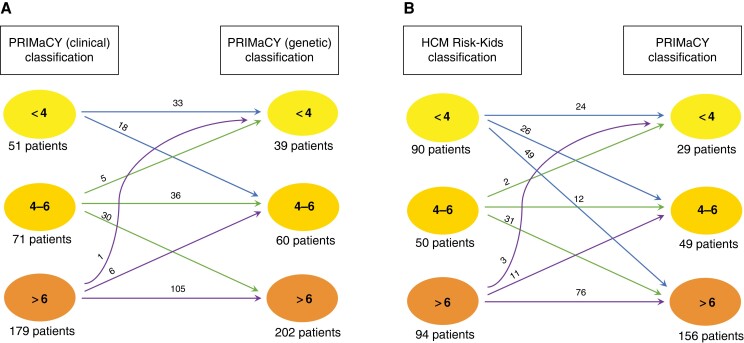
A comparison of predicted risk using the PRIMaCY genetic and clinical models is shown in Figure 3A. For patients who had undergone genetic testing (n = 207), 48 patients (23.2%) were assigned to a higher risk group by the PRIMaCY model following inclusion of genetic testing results. Table 2 and Supplementary material online, TableS2 summarizes the clinical impact of using the risk models on ICD implantation decisions and the prediction of arrhythmic events. The number needed to treat one lethal ventricular arrhythmia was 14.5 and 13.7 for the genetic and clinical models, respectively.
Figure 3.
Comparison of 5-year estimated risk for patients using (A) PRIMaCY genetic and PRIMaCY clinical model and (B) PRIMaCY clinical and HCM Risk-Kids.
Table 2.
Clinical impact of using PRIMaCY risk models for guiding ICD implantation in childhood HCM
| Cohort | Proportion above threshold ≥6 for ICD implantation | Proportion of arrhythmic events predicted | Number needed to treat | |
|---|---|---|---|---|
| 1–18 years at baseline | PRIMaCY (genetics) | 202 (67.1%) | 14/17 (82.4%) | 14.5 |
| PRIMaCY (clinical) | 179 (59.5%) | 13/17 (76.5%) | 13.7 | |
| 1–16 years at baseline | PRIMaCY (genetics) | 158 (67.5%) | 13/16 (81.3%) | 12.2 |
| PRIMaCY (clinical) | 135 (57.7%) | 12/16 (75%) | 11.3 | |
| HCM Risk-Kids | 94 (40.2%) | 10/16 (62.5%) | 9.4 |
Comparison of the clinical impact of using PRIMaCY vs. HCM Risk-Kids model
A comparison of predicted risk using the PRIMaCY (clinical) and HCM Risk-Kids model for patients aged 1–16 years (n = 234) at baseline is shown in Figure 3B. There was between-model agreement in the calculated risk group between PRIMaCY (clinical) and HCM Risk-Kids for 118 patients (50.4%). Of the remaining 116 patients, 101 (87.1%) were assigned to a higher estimated risk group using the PRIMaCY model. Table 2 summarizes the clinical impact of using PRIMaCY or HCM Risk-Kids on ICD implantation and the prediction of arrhythmic events.
Discussion
In this study, we have shown that, although PRIMaCY has a similar ability to discriminate between high- and low-risk patients as HCM Risk-Kids, model calibration was poor, with the model appearing to over-estimate risk for some patients. Two-thirds of patients had an estimated risk ≥6%, which could result in higher overall rates of ICD implantation.
Clinical impact of using different paediatric risk models for sudden cardiac death
The development of paediatric-specific risk models that allow clinicians to calculate individualized estimates of 5-year risk represents a major advance in the management of HCM in childhood and offers an opportunity for personalization of ICD implantation decision-making, in line with accepted practice for adult patients.4,5,12,16 Indeed, the new 2023 cardiomyopathy guidelines specifically recommend their use for the risk stratification of sudden death events in childhood disease. This study is the first independent external validation of the PRIMaCY risk model and confirms it to have superior discriminatory ability compared with traditional methods of risk stratification (C-statistic 0.66 vs. 0.62)2 and not dissimilar to that previously reported for HCM Risk-Kids (Uno’s C-index for HCM Risk-kids 0.71 vs. 0.61 for PRIMaCY).8 However, despite showing a similar ability to discriminate between high- and low-risk patients, we report a significant difference in estimated and observed risk (calibration) using the PRIMaCY model. As highlighted in the 2023 ESC Guidelines for the Management of Cardiomyopathies, defining universal thresholds for acceptable risk is challenging, particularly in children. The thresholds chosen to represent low, medium, and high risk in this study reflect those endorsed by the new 2023 ESC cardiomyopathy guidelines9 and 2022 ESC ventricular arrhythmia guidelines.10 The original PRIMaCY study did not recommend thresholds for ICD implantation but split the cohort into tertiles of risk for demonstration of model performance. Importantly, the chosen threshold does not affect the estimates of model performance reported in this manuscript (i.e. C-statistic and model calibration). It is beyond the scope of this manuscript to determine the appropriate threshold for ICD implantation in childhood. Indeed, the strength of personalized risk prediction models lies in their use as part of an overt shared decision-making process that is based on real-world data as well as individual preferences, beliefs, circumstances, and values and includes acknowledgment of gaps in evidence.9 However, this study suggests that the PRIMaCY model may provide higher SCD risk estimates compared with HCM Risk-Kids in a substantial proportion of individuals leading to a higher rate of ICD implantation. Two-thirds met the threshold of ≥6% for ICD implantation using the PRIMaCY model in this population compared with 40% using HCM Risk-Kids. The number needed to potentially treat one life-threatening arrhythmia for the PRIMaCY clinical, PRIMaCY genetic, and HCM Risk-Kids models was 13.7, 14.5, and 9.4, respectively. This is the case even if using a higher threshold for ICD implantation of 8.3%. Children are recognized to be at higher risk of ICD-related complications, with up to a third experiencing a complication or inappropriate therapy within 5 years of follow-up.17 As these patients are undergoing ICD implantation at a young age, this risk will continue into early adulthood and beyond. Although no model will provide perfect risk estimates, an awareness of differences between different published risk models is important for clinicians and families to facilitate this process of informed shared decision-making. Incorporation of estimates of ICD complication rates in future iterations of risk models would be useful, although risk factors for ICD complications are currently poorly understood.17
Impact of including genetics in risk stratification for childhood hypertrophic cardiomyopathy
Patients with a confirmed disease-causing sarcomeric variant have previously been reported to have earlier disease onset and worse long-term outcomes including an increased risk of arrhythmias.3,18–20 PRIMaCY5 is the first model to have attempted to include genotype status as a risk predictor for SCD risk, although its inclusion in the risk model did not significantly improve model performance in the original report.5 When comparing the results of the genetic and clinical models in this cohort, incorporation of the genetic testing result increased the absolute estimated risk in almost 75% of patients, leading to a 20% increase in the proportion of patients meeting the threshold for ICD implantation. This is despite one-fifth of patients having no disease-causing variants identified and others having variants historically considered more ‘benign’. Including genetic information did not significantly improve the discrimination or calibration of the model. Of note, patients seen in more recent eras were more likely to have undergone genetic testing and gene panels are likely to have changed over time. However, the proportion of patients who had undergone genetic testing was higher than reported in the initial PRIMaCY development or validation cohort. It is possible that the lack of difference in model performance could be explained by the fact that the significant heterogeneity at both the gene level and between individual variants in sarcomeric disease is not taken into account with this approach.21,22 It is likely that variants in specific regions of individual genes (e.g. the converter region in MYH723), or even specific individual variants, are associated with an increased SCD risk in childhood, but such genotype–phenotype correlations may well be limited by sample size. Future studies exploring the role of genetics in risk stratification are required; nonetheless, an awareness of the effect of including genetic testing results on PRIMaCY risk estimates is important for clinicians when using the model in clinical practice.
Limitations
Patients with incomplete data for risk predictor variables were excluded from this study to allow for PRIMaCY risk estimates to be calculated. Whilst this could be considered a strength of the study, as no missing data were imputed, the results may not be representative of the wider childhood HCM population. However, reassuringly, the clinical population characteristics and event rate are comparable with recently published large multicentre population studies and the PRIMaCY development cohort.3–5 This includes the proportion of patients with a family history (57.2% vs. 48% in PRIMaCY development cohort), unexplained syncope (3.7% vs. 3.0%), and NSVT on ambulatory ECG monitoring (3.1% vs. 3.7%). A higher proportion of patients were receiving β-blocker therapy in the PRIMaCY development cohort (22.5% vs. 59.1%). However, there is no strong evidence to support the role of β-blocker therapy in SCD risk for adult or paediatric populations.9 The number of patients included in this study is greater than that used for external validation in the original PRIMaCY study, yet the small sample size and low event rate resulted in wide CIs for measures of model performance, reflecting uncertainty in the estimates. Confidence intervals were not reported in the original PRIMaCY study, so it is not possible to compare this with our results. Inherent to the retrospective, longitudinal design of this study, patients were recruited over a long-time period. Patients presenting in the earliest era (pre-2000) had higher maximal wall thickness Z scores and larger LA diameters but did not otherwise differ in terms of baseline clinical characteristics. This could be a result of more recent patients being diagnosed at an earlier time point through family screening. However, no era effect was seen for arrhythmic events. Data on ethnicity were not collected in this study, but future studies investigating the impact of ethnicity on outcomes, including sudden death risk, in childhood disease would be valuable. Only two-fifths of the reported cohort had a follow-up of 5 years or longer. Both available risk models predict the risk of a SCD event occurring within 5 years, meaning patients with shorter follow-up times could still reach the endpoint within 5 years. This could affect the estimates of predictive accuracy including the number needed to treat but is a limitation shared with the previously published external validation studies of both HCM Risk-Kids8 and PRIMaCY,5 which had median follow-up of median 5.3 (IQR 2.6–8.3) and 3.9 (1.5–6.7) years, respectively. Fourteen patients (4.7%) underwent LV septal myectomy during follow-up. The impact of septal myectomy on risk is poorly understood in both adult and paediatric populations, and neither the paediatric risk models (HCM Risk-Kids or PRIMaCY) nor adult risk models (HCM Risk-SCD) have been validated before and after myectomy. Risk models are currently used in this patient group in clinical practice, although they should be used with caution. This study reports the clinical impact of calculated estimated risks on decision-making, falsely assuming that the estimated risk is the only tool used by clinicians when determining if a patient should undergo primary ICD implantation. In reality, ICD implantation decisions are affected by clinical-, societal-, and healthcare-related factors not accounted for in this study and future prospective, multicentre studies following model implementation in clinical practice will be required to evaluate the true effect of the models on clinical decision-making.
Conclusions
This study shows that PRIMaCY has a similar discriminatory ability to HCM Risk-Kids but there was poor agreement between observed and predicted risk. A higher proportion of patients had a risk of ≥6% using the PRIMaCY model compared with HCM Risk-Kids, of whom <1 in 10 experienced a life-threatening arrhythmic event. The higher proportion of patients meeting ICD thresholds may have important clinical implications, as patients will be exposed to a lifetime risk of complications and inappropriate therapies. This emphasizes the need for risk prediction models to be used as part of a systematic risk assessment and should include discussion of the limitations of calculated estimates with patients and parents, allowing an informed and shared decision to be made.
Supplementary Material
Contributor Information
Gabrielle Norrish, Centre for Inherited Cardiovascular Diseases, Zayed Centre for Research, Great Ormond Street Hospital, Great Ormond Street, London, WC1N 4JH, UK; Institute of Cardiovascular Sciences, University College London, 62 Huntley St, London, WC1E 6DD, UK.
Alexandros Protonotarios, Institute of Cardiovascular Sciences, University College London, 62 Huntley St, London, WC1E 6DD, UK; St Bartholomew’s Centre for Inherited Cardiovascular Diseases, St Bartholomew’s Hospital, London, UK.
Maria Stec, Centre for Inherited Cardiovascular Diseases, Zayed Centre for Research, Great Ormond Street Hospital, Great Ormond Street, London, WC1N 4JH, UK; 1st Department of Cardiology, Faculty of Medical Sciences in Katowice, Medical University of Silesia, Katowice, Poland.
Olga Boleti, Centre for Inherited Cardiovascular Diseases, Zayed Centre for Research, Great Ormond Street Hospital, Great Ormond Street, London, WC1N 4JH, UK; Institute of Cardiovascular Sciences, University College London, 62 Huntley St, London, WC1E 6DD, UK.
Ella Field, Centre for Inherited Cardiovascular Diseases, Zayed Centre for Research, Great Ormond Street Hospital, Great Ormond Street, London, WC1N 4JH, UK; Institute of Cardiovascular Sciences, University College London, 62 Huntley St, London, WC1E 6DD, UK.
Elena Cervi, Centre for Inherited Cardiovascular Diseases, Zayed Centre for Research, Great Ormond Street Hospital, Great Ormond Street, London, WC1N 4JH, UK.
Perry M Elliott, Institute of Cardiovascular Sciences, University College London, 62 Huntley St, London, WC1E 6DD, UK; St Bartholomew’s Centre for Inherited Cardiovascular Diseases, St Bartholomew’s Hospital, London, UK.
Juan P Kaski, Centre for Inherited Cardiovascular Diseases, Zayed Centre for Research, Great Ormond Street Hospital, Great Ormond Street, London, WC1N 4JH, UK; Institute of Cardiovascular Sciences, University College London, 62 Huntley St, London, WC1E 6DD, UK.
Supplementary material
Supplementary material is available at Europace online.
Funding
G.N., E.F. and J.P.K. are supported by Great Ormond Street Hospital Children’s Charity. E.F. and J.P.K. are supported by Max’s Foundation. J.P.K. is supported by a Medical Research Council Clinical (MRC)—National Institute for Health Research (NIHR) Clinical Academic Research Partnership (CARP) award (MR/T024062/1). This work is (partly) funded by the NIHR GOSH BRC.
Data availability
The data underlying this article cannot be shared publically as consent of dissemination of patient data was not obtained. G.N., M.S., and J.P.K. had access to all data and final responsibility for the submission of the manuscript.
References
- 1. Norrish G, Field E, McLeod K, Ilina M, Stuart G, Bhole Vet al. Clinical presentation and survival of childhood hypertrophic cardiomyopathy: a retrospective study in United Kingdom. Eur Heart J 2019;40:986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Norrish G, Ding T, Field E, McLeod K, Ilina M, Stuart Get al. A validation study of the European Society of Cardiology guidelines for risk stratification of sudden cardiac death in childhood hypertrophic cardiomyopathy. Europace 2019;21:1559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marston NA, Han L, Olivotto I, Day SM, Ashley EA, Michels Met al. Clinical characteristics and outcomes in childhood-onset hypertrophic cardiomyopathy. Eur Heart J 2021;42:1988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Norrish G, Ding T, Field E, Ziólkowska L, Olivotto I, Limongelli Get al. Development of a novel risk prediction model for sudden cardiac death in childhood hypertrophic cardiomyopathy (HCM Risk-Kids). JAMA Cardiol 2019;4:918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miron A, Lafreniere-Roula M, Steve Fan CP, Armstrong KR, Dragulescu A, Papaz Tet al. A validated model for sudden cardiac death risk prediction in pediatric hypertrophic cardiomyopathy. Circulation 2020;142:217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petryka-Mazurkiewicz J, Ziolkowska L, Kowalczyk-Domagala M, Mazurkiewicz L, Boruc A, Spiewak Met al. LGE for risk stratification in primary prevention in children with HCM. JACC Cardiovasc Imaging 2020;13:2684–6. [DOI] [PubMed] [Google Scholar]
- 7. Östman-Smith I, Sjöberg G, Alenius Dahlqvist J, Larsson P, Fernlund E. Sudden cardiac death in childhood hypertrophic cardiomyopathy is best predicted by a combination of electrocardiogram risk-score and HCMRisk-Kids score. Acta Paediatr 2021;110:3105–15. [DOI] [PubMed] [Google Scholar]
- 8. Norrish G, Qu C, Field E, Cervi E, Khraiche D, Klaassen Set al. External validation of the HCM Risk-Kids model for predicting sudden cardiac death in childhood hypertrophic cardiomyopathy. Eur J Prev Cardiol 2022;29:678–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arbelo E, Protonotarios A, Gimeno JR, Arbustini E, Barriales-Villa R, Basso Cet al. 2023 ESC guidelines for the management of cardiomyopathies. Eur Heart J 2023;44:3503–626. [DOI] [PubMed] [Google Scholar]
- 10. Zeppenfeld K, Tfelt-Hansen J, de Riva M, Winkel BG, Behr ER, Blom NAet al. 2022 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur Heart J 2022;43:3997–4126. [DOI] [PubMed] [Google Scholar]
- 11. Könemann H, Dagres N, Merino JL, Sticherling C, Zeppenfeld K, Tfelt-Hansen Jet al. Spotlight on the 2022 ESC guideline management of ventricular arrhythmias and prevention of sudden cardiac death: 10 novel key aspects. Europace 2023;25:euad091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Mahony C, Jichi F, Pavlou M, Monserrat L, Anastasakis A, Rapezzi Cet al. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM risk-SCD). Eur Heart J 2014;35:2010–20. [DOI] [PubMed] [Google Scholar]
- 13. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster Jet al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrell F. Regression modeling strategies. Switzerland: Springer; 2001. [Google Scholar]
- 15. Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 2011;30:1105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Mahony C, Jichi F, Ommen SR, Christiaans I, Arbustini E, Garcia-Pavia Pet al. International external validation study of the 2014 European Society of Cardiology guidelines on sudden cardiac death prevention in hypertrophic cardiomyopathy (EVIDENCE-HCM). Circulation 2018;137:1015–23. [DOI] [PubMed] [Google Scholar]
- 17. Norrish G, Chubb H, Field E, McLeod K, Ilina M, Spentzou Get al. Clinical outcomes and programming strategies of implantable cardioverter-defibrillator devices in paediatric hypertrophic cardiomyopathy: a UK national cohort study. Europace 2021;23:400–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby Det al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the sarcomeric human cardiomyopathy registry (SHaRe). Circulation 2018;138:1387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopes LR, Rahman MS, Elliott PM. A systematic review and meta-analysis of genotype-phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart 2013;99:1800–11. [DOI] [PubMed] [Google Scholar]
- 20. Sedaghat-Hamedani F, Kayvanpour E, Tugrul OF, Lai A, Amr A, Haas Jet al. Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: a meta-analysis on 7675 individuals. Clin Res Cardiol 2018;107:30–41. [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Giustiniani D, Arad M, Ortiz-Genga M, Barriales-Villa R, Fernández X, Rodríguez-García Iet al. Phenotype and prognostic correlations of the converter region mutations affecting the beta myosin heavy chain. Heart 2015;101:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pasquale F, Syrris P, Kaski JP, Mogensen J, McKenna WJ, Elliott P. Long-term outcomes in hypertrophic cardiomyopathy caused by mutations in the cardiac troponin T gene. Circ Cardiovasc Genet 2012;5:10–7. [DOI] [PubMed] [Google Scholar]
- 23. García-Giustiniani D, Arad M, Ortíz-Genga M, Barriales-Villa R, Fernández X, Rodríguez-García Iet al. Phenotype and prognostic correlations of the converter region mutations affecting the β myosin heavy chain. Heart 2015;101:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publically as consent of dissemination of patient data was not obtained. G.N., M.S., and J.P.K. had access to all data and final responsibility for the submission of the manuscript.