Abstract
HnRNP K is a heterogeneous nuclear ribonucleoprotein and has been identified as an oncogene in most solid tumors via regulating gene expression or alternative splicing of genes by binding both DNA and pre-mRNA. However, how hnRNP K affects tumorigenesis and regulates the gene expression in cervical cancer (CESC) remains to be elucidated. In these data, higher expression of hnRNP K was observed in CESC and was negatively correlated with the patient survival time. We then overexpressed hnRNP K (hnRNP K-OE) and found that its overexpression promoted cell proliferation in HeLa cells (P = 0.0052). Next, global transcriptome sequencing (RNA-seq) experiments were conducted to explore gene expression and alternative splicing profiles regulated by hnRNP K. It is shown that upregulated genes by hnRNP K-OE were associated with inflammatory response and an apoptotic process of neuron cells, which involves in cancer. In addition, the alternative splicing of those genes regulated by hnRNP K-OE was associated with transcriptional regulation. Analysis of the binding features of dysregulated transcription factors (TFs) in the promoter region of the inflammatory response genes regulated by hnRNP K revealed that hnRNP K may modulate the expression level of genes related to inflammatory response by influencing the alternative splicing of TFs. Among these hnRNP K-TFs-inflammatory gene regulatory networks, quantitative reverse transcription polymerase chain reaction (RT-qPCR) experiments and gene silencing were conducted to verify the hnRNP K-IRF1-CCL5 axis. In conclusion, the hnRNP K-TFs-inflammatory gene regulatory axis provides a novel molecular mechanism for hnRNP K in promoting CESC and offers a new therapeutic target.
Keywords: hnRNP K, RNA-seq, inflammatory response, transcriptional regulation, alternative splicing, cervical cancer
Impact Statement
We for the first time used RNA-seq methodology to reveal how hnRNP K functions in the transcription and alternative splicing regulation in HeLa cells. The results showed that hnRNP K influenced the proliferation of HeLa cells. On the molecular level, the results suggested that hnRNP K positively modulates the splicing of genes involved in transcriptional process of and the RNA level of genes implicated in inflammation. More importantly, analysis of transcription factors (TFs) and binding motifs indicated that several TFs regulated by hnRNP K at splicing level bind to the promoter of differentially expressed genes (DEGs) associated with inflammatory response. Therefore, we speculate that hnRNP K regulates alternative splicing of TFs, which then bind to inflammatory genes and change its expression. The hnRNP K-TFs-inflammatory genes axis provides novel molecular mechanisms of hnRNP K involved in HeLa cells and offers new treatment targets in the future.
Introduction
Cancer remains a global major public health issue and effective treatments for cancer are still being investigated. Cervical cancer (CESC) ranks third in cancer incidence worldwide and is of the top four leading risk factors for mortality among women. 1 In spite of advances in disease detection and the application of vaccines against human papillomavirus (HPV) that can effectively prevent the disease,2 –5 there has been no significant improvement in the therapeutic effect for CESC during the past few decades.6,7 Currently, chemotherapy, 8 surgery, 9 and radiotherapy 10 are the most therapeutic regimen for CESC. However, resistance to chemotherapy of CESC cells results in poor treatment efforts. Hence, it is of great importance to provide a novel approach to cancer treatment through disclosing the mechanism of CESC development and singling out certain targets.
Dysregulation of many RNA-binding proteins (RBPs) is discovered during the progression of various tumors, 11 since RBPs play important role in mRNA processing, including mRNA stability, 12 transport, 13 alternative splicing, 14 and translation, 15 they are commonly regarded as therapeutic targets and diagnostic indicators. 16 Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is an RBP and belongs to the subfamily of hnRNPs with a size of 65 kDa. Most of hnRNPs are present in nucleus, while hnRNP K seems to shuttle between cytosol and nucleus; 17 therefore, hnRNP K participates in multiple biological and cellular processes, especially in transcriptional and post-transcriptional regulation. For example, hnRNP K was known as a transcription factor with an ability to bind to a particular DNA sequence and to interact with other transcription regulators. 18 Meanwhile, hnRNP K can regulate pathways in carcinogenesis and tumor suppression. 19 Most interestingly, sumoylation of hnRNP K induces p53 transcriptional activation. 20 In addition to transcription regulation, hnRNP K can also function as gene splicing mediator. HnRNP K includes the third exon of MRPL33 in colorectal cancer tissue and is involved in tumor progression. 21 Peng et al. 22 suggested that upregulation of hnRNP K in gastric cancer (GC) activates the expression of SRSF1, which is a famous splicing factor. Then, SRSF1 induces the expression of the isoform of oncogene CD44, and ultimately promotes the production, migration, and invasion of GC cell. As an oncogene, hnRNP K has been widely investigated in multiple solid cancers, with cancers of lung, 19 prostate,23 –25 colon, 26 renal cell, 27 and the nasopharynx included, 28 which indicates the close connection between hnRNP K and tumor formation and development. However, the function of hnRNP K in CESC was rarely reported. Zhang et al. 29 found that nujiangexathone, a compound from Garcinia nujiangensis, can inhibit the progression of CESC by downregulating hnRNP K, eventually causing cell-cycle arrest. One recent study demonstrated hnRNP K promotes malignant phenotypes of HeLa cells by regulating lncRNA-LINC00263, 30 while the precise mechanism of hnRNP K implicated in CESC remained elusive. As an RBP, whether hnRNP K also has considerable roles in the CESC development by modulating transcriptome profile associated with cancer needs to be further investigated.
To further explore the mechanisms and functions of hnRNP K in CESC, it was overexpressed in HeLa cells, which could significantly promote HeLa cell proliferation. RNA sequencing was performed to discover hnRNP K–modulated splicing and gene expression pattern in HeLa cells. Functional enrichment analysis revealed that differentially expressed genes (DEGs) were significantly enriched in inflammation pathways. On the contrary, alternative splicing genes regulated by hnRNP K (RASGs) were significantly enriched in the pathways of transcriptional regulation. Combined with analyzing the target genes of transcription factors (TFs) regulated by hnRNP K, the motifs of those TFs were observed in the promoter regions of inflammatory response DEGs. The results suggest that hnRNP K mediates the transcription of inflammatory-response-related genes and this regulation may be mediated by the splicing of TFs. The hnRNP K-TFs-inflammatory genes axis potentially sheds light on the mechanisms of hnRNP K implicated in CESC and offers new therapy target in the future.
Materials and methods
Construction of plasmid and small interfering RNA
Pairs of primers were prepared by CE Design V1.04. Every primer included a set of sequences specific to each gene and the vector pIRES-hrGFP-1a. GFP, which is a fluorescence label contained in plasmid, is widely used as genetically encoded fluorescent fusion tags. The primers are listed below.
F-primer: agcccgggcggatccgaattcATGGAAACTGAACAGCCAG
R-primer: gtcatccttgtagtcctcgagGAAAAACTTTCCAGAATACTGC
The pIRES-hrGFP-1a vector was digested in EcoRI (NEB, 3101S, England) and XhoI (NEB, R0146V, England) at 37°C for 2–3 h. A 1.0% agarose gel was used to run the enzyme-digested vector. And Qiagen column kit was used to purify the vector. With TRIzol reagent, total RNA was separated from HeLa cells. After purification, cDNA was generated by the reverse transcription of RNA. The fragment inserted was synthesized via amplification of polymerase chain reaction (PCR). By the ClonExpress® II One Step Cloning Kit (Vazyme, C112, China), both the PCR-mediated insertion and a linearized vector digested via EcoRI and XhoI (NEB) were ligated in a PCR microtube. Chemical transformation was used to introduce plasmids into the Escherichia coli strain. Placed on the LB agar plates with 1 µL/mL ampicillin, the cells were incubated for one night. To screen out the colonies, colony PCR was performed with universal primers (on the backbone vector) by 28 cycles. For IRF1 silencing, small interfering RNA (siIRF1) GCUACACAGUUCCAGGCUATT (sense) and non-targeting control siRNA (siNegative): 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) were obtained from Gemma (Suzhou, China).
Cell culture and transfection
Both HeLa and A549 cells were obtained from the Institute of Biochemistry and Cell Biology (Shanghai, China). Cell culture conditions and transfection methods were followed from a previously published paper. 31
The efficiency of overexpressed hnRNP K (hnRNP K-OE) and IRF1 knockdown was assessed using quantitative reverse transcription polymerase chain reaction (RT-qPCR) method with an internal control gene GAPDH. First, RT-qPCR was performed to determine hnRNP K expression and GAPDH. The density of hnRNP K and IRF1 was standardized to the GAPDH level according to the 2–ΔΔCTcalculation method, which was reported by Livak and Schmittgen in 2001. 32 The primers were presented in Supplemental Table S1. What’s more, Western blot (WB) was also performed, the purpose of which was to evaluate the efficiency of hnRNP K overexpression; anti-hnRNP K (11426-1-AP, 1:1000; Proteintech, China) was used against hnRNP K and GADPH was used as internal control.
Cell proliferation and apoptosis experiments
To assess how hnRNP K overexpression or IRF1 silencing impacts cell proliferation, an MTT assay experiment was conducted as described previously, 33 in which MTT-produced crystals were melted in dimethyl sulfoxide (DMSO). Then, the optical density (OD) values were determined at an optical wavelength of 570 nm. For cell apoptosis, 105 cells were seeded into 24-well culture plates and incubated at 37°C and 5% CO2 for 24 h. Then, the apoptosis level was assessed by flow cytometry (CytoFLEX; Beckman, USA) following one published paper. 34
RNA-seq experiment
Total RNA from HeLa cell was extracted using TRIzol (15596-018, Ambion, USA). The total RNA purification, genomic DNA removal, quantity and quality determination of purified RNA, and RNA integrity were conducted or assessed following the published methods. 31
Before sequencing, 1 μg total RNA was utilized to construct RNA-seq library of each sample. First, conjugated magnetic beads with oligo (dT) (61005, Invitrogen; Thermo Fisher Scientific, Inc., USA) were used to purify and concentrate the polyA mRNAs. The fragmented RNAs were end-repaired, followed by ligating at the 5′ end at 95°C. Then, purified RNAs were reverse-transcribed (RT) to cDNAs and stored at −80°C after amplifying. The libraries were constructed to perform paired-end high-throughput sequencing with 150 nt on the HiSeq X Ten system (Illumina, USA).
RNA-seq data and DEGs analysis
First, raw reads with unknown bases were trimmed off/eliminated. Then, sequencing adaptors and short reads not more than 16 nt were also discarded with FASTX-Toolkit (version 0.0.13). Clean reads were aligned onto the human genome (GRCh38) using TopHat2 35 software. We counted uniquely mapped reads to calculate FPKM (Fragments Per Kilobase of exon per Million reads) values for detected genes as their expression level. 36 The DEGs were screened out using the R Bioconductor package edgeR 37 with false discovery rate (FDR) < 0.05 and fold change (FC) ⩾ 2 or ⩽ 0.5 as criteria.
Alternative splicing analysis
To identify differential alternative splicing events (ASEs), regulated ASEs (RASEs) were determined with ABLas program. 38 According to the uniquely mapped reads and splice junction reads, ASEs were commonly divided into eight types, which contains exon skipping (ES), mutually exclusive exons (MXES), the MXE combined with alternative 5′promoter (5pMXE), the MXE combined with alterative 3′promoter (3pMXE), intron retention (IR), mutually exclusive 3′ untranslated regions (UTRs), alternative 5′ splice site (A5SS) and alternative 3′ splice site (A3SS). To identify hnRNP K-RASEs, Fisher’s exact test was utilized to count the statistical P value. The RASE difference was defined as ratio between alternatively spliced reads and constitutively spliced reads. Ratio > 0.2 and P value < 0.05 were the thresholds to identify RASEs.
DEGs and RASEs verification
To verify identified DEGs and RASEs from the RNA-seq data, RT-qPCR experiment was conducted. We designed specific primer sequences for RASEs, following one published study. 39 The primer sequences utilized in qPCR are shown in Supplemental Table S1. The detailed experimental procedures had been described in the above part.
WB experiment
Except for RT-qPCR, WB experiment was conducted to assess the protein level of CCL5 in A549 cells and HeLa cells; total protein was obtained from hnRNP K-OE and control cells with radioimmunoprecipitation assay (RIPA) buffer. The western assay was performed according to the description of previous study. 40 The information of primary antibodies used in experiment was as follows: CCL5 (1:500; Sigma, USA) and GAPDH (A19056, 1:1000; ABclonal, China). Bound secondary antibody (HRP Goat Anti-Rabbit IgG [H + L], AS014 1:10000; ABclonal, China) was identified with enhanced chemiluminescence (ECL) reagent (32106; Thermo Scientific, USA). We used GAPDH or β-actin as internal controls.
Other statistical analysis
Functional enrichment statistics of selected gene sets were predicted using GO and KEGG pathway annotation with the KOBAS 2.0 server. 41 P-value was calculated by hypergeometric test and was then corrected by Benjamini–Hochberg FDR controlling procedure to identify the enrichment. Unpaired Student’s t-test was conducted to make comparisons between two groups. P-value < 0.05 was regarded as significant difference. The statistical values were presented as the mean ± standard deviation with at least three biological replicates, except for the RNA-seq.
Results
Expression of hnRNP K and prognostic value in CESC
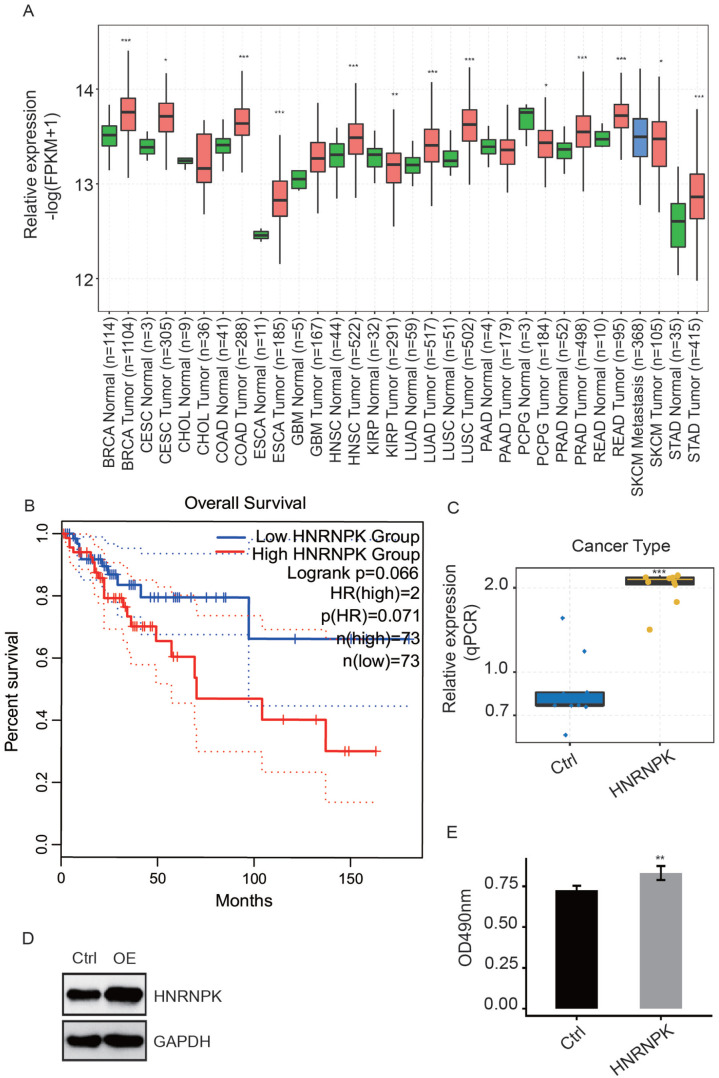
HnRNP K is an extensively expressed gene in multiple cancers; its expression between tumor and normal tissue in different cancer types from TCGA is shown in Figure 1(A) using GEPLA2, a web-based tool. HnRNP K had a higher expression level in tumor than in normal tissues in 12 of 16 cancer types, indicating that hnRNP K was inclined to act as an oncogene. Furthermore, we assessed the prognostic value of hnRNP K in CESC based on TCGA samples and found that elevated hnRNP K expression was correlated with poor prognosis of patients with CESC (Figure 1(B)).
Figure 1.
HnRNP K overexpression promotes the proliferation of HeLa cells. (A) Box plot showing the relative expression of hnRNP K in tumor and normal samples from TCGA. (B) Line plot showing the Kaplan–Meier analysis of the connection between hnRNP K expression and clinical prognosis based on TCGA data. (C-D) HnRNP K expression was validated by RT-qPCR (C) and WB (D) after treatment with hnRNP K-OE or control vector. (E) MTT assay demonstrated that HeLa cells being treated with hnRNP K-overexpression vector increased significantly. A comparison between control and hnRNP K-OE HeLa cells was made using Student’s t-test. **P < 0.01; ***P < 0.001. (A color version of this figure is available in the online journal.)
HnRNP K overexpression promotes HeLa cell proliferation
To uncover the mechanisms of hnRNP K functioning in CESC, an hnRNP K–overexpression HeLa cell model was constructed. The hnRNP K expression level was examined by conducting RT-qPCR and WB experiments (Figure 1(C) and (D)), indicating that hnRNP K was successfully overexpressed in HeLa cells. In conformity with the results of MTT assay (Figure 1(E)), the proliferation level was increased in hnRNP K–overexpressed HeLa cells in comparison with control (P = 0.0052), demonstrating that highly expressed hnRNP K promotes proliferation in HeLa cells.
HnRNP K–regulated HeLa transcriptomes
In order to comprehensively study hnRNP K–mediated regulation in HeLa cell, cDNA libraries were constructed with hnRNP K-OE cells and control cells (2 biological replicates). We then obtained four RNA-seq samples by high-throughput sequencing method.
The mean and standard deviation of raw data was 94.2 ± 4.4 million per sample. A total of 90.1 ± 4.9 million reads per sample, which were prepared for downstream bioinformatics analysis, were retained after discarding low-quality sequences and adaptor sequences. Then, we used TopHat2 to map the filtered reads onto the human GRCh38 genome, which revealed that 74.80–82.19% were mapped and 94.94–95.70% were uniquely mapped.
To show the expression values of the genes detected from the uniquely mapped reads, we calculated the FPKM values. In total, 26,247 genes were attained with FPKM > 0, among which 11,810 genes had FPKM > 1 in at least one sample. The principal component analysis (PCA) result revealed that the top two components account for 79% of the variation, and hnRNP K-OE and control samples were clearly separated from each other, indicating that hnRNP K-OE can extensively affect expression in HeLa cells (Supplemental Figure S1(A)).
HnRNP K selectively regulates DEGs associated with inflammatory response
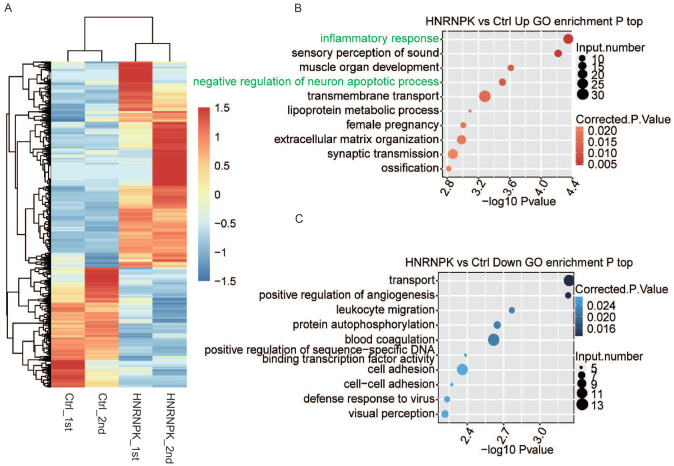
Based on gene expression values, we further studied genes that were significantly regulated by hnRNP K. To identify the DEGs, we used edgeR program 37 with criteria of absolute FC ⩾ 2 or ⩽ 0.5 and FDR < 0.05. A total of 2405 DEGs with 1519 upregulated and 886 downregulated genes were identified between hnRNP K-OE and control cells, which indicated that hnRNP K extensively regulates gene transcription (Supplemental Figure S1(B)). The difference between the number of upregulated and downregulated genes indicates that hnRNP K may have a preferential regulation in upregulating gene expression. Detailed expression levels of the DEGs were presented in Supplemental Table S2. The hierarchical clustering of all DEGs was constructed and revealed the obvious expression change between the two datasets (Figure 2(A)).
Figure 2.
Changes of gene expression analysis responding to hnRNP K-OE. (A) Heat map of all 2405 DEGs identified in this study. FPKM values are log2-transformed and then median-centered by each gene. (B-C) The top 10 GO biological process terms of genes upregulated (B) and downregulated (C) by hnRNP K-OE. (A color version of this figure is available in the online journal.)
To investigate the enriched functions of these DEGs, 2405 DEGs were analyzed based on GO and KEGG pathway enrichment analysis. As a result, 632 up DEGs and 311 down DEGs were classified into 35 GO terms and 30 GO terms, respectively. Upregulated DEGs in hnRNP K-OE cells were chiefly enriched in cancer-associated pathways, including inflammatory response, transmembrane transport, lipoprotein metabolic process, and extracellular matrix organization (Figure 2(B)). The inflammatory response (corrected P value = 0.0043), which was the most enriched upregulated term in biological process, contains 23 inflammation-related genes, including CCL5, AOC3, NOS2, BCL6, FOS, IL17F, ADAM8, UCN, and CD14/180. The downregulated genes were mostly related to transport, positive regulation of angiogenesis, positive regulation of sequence-specific DNA binding transcription factors, cell adhesion, and cell–cell adhesion (Figure 2(C)). According to the results of KEGG analysis, DEGs were enriched in pathways, including rheumatoid arthritis, amoebiasis, ATP-binding cassette (ABC) transporters, peroxisome proliferator-activated receptor (PPAR) signaling pathway, transcriptional misregulation in cancer and legionellosis (Supplemental Figure S1(C) and (D)). It is notable that the most enriched upregulated annotations in both GO and KEGG analyses were associated with inflammation. The findings above indicate that hnRNP K can extensively affect the expression levels of inflammation-associated genes.
HnRNP K extensively regulated alternative splicing profile in HeLa cells
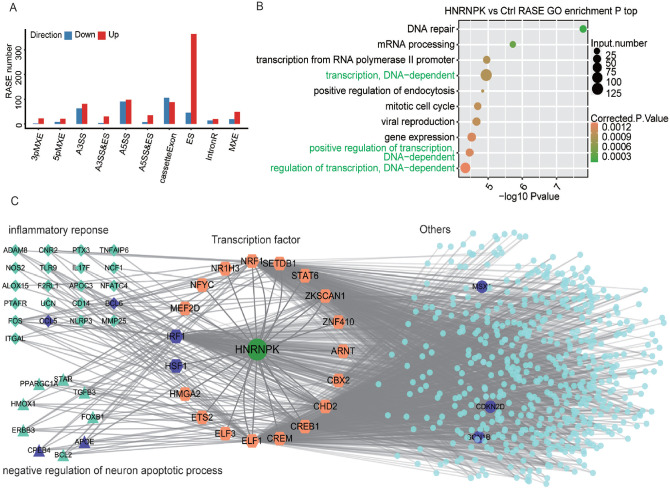
The impact of hnRNP K on alternative splicing (AS) in HeLa cells was explored according to the transcriptome data. From the aligning result, nearly 51% are junction reads, including 162,918 known junction reads and 221,547 novel junction reads. A stringent threshold of P value ⩽ 0.05 and changed T-value ⩾ 0.2 was set as the criteria to identify RASEs with a high confidence; A5SS, A3SS, and ES were the three most prevalent AS events induced by hnRNP K; and exon skipping (ES) events were extremely induced in hnRNP K-OE samples (Figure 3(A)). The results showed that hnRNP K extensively modulated ASEs in HeLa cells.
Figure 3.
HnRNP K extensively changed ASEs in HeLa cells. (A) Categories of alternative splicing events regulated by hnRNP K. (B) The top 10 GO biological process pathways of regulated alternative splicing genes. (C) HnRNP K-TFs-targets regulation network in HeLa cells. Hexagons represent transcription factors. Squares represent inflammatory response genes and triangles represent apoptotic-related genes. (A color version of this figure is available in the online journal.)
HnRNP K modulates gene expression by regulating alternative splicing of transcription factors
To discover the functions of alternative splicing genes regulated by hnRNP K, GO (Figure 3(B)) and KEGG (Supplemental Figure S2(A)) functional clustering analyses were conducted. The results demonstrated that these genes were highly enriched in transcription-associated pathways, in which large number of transcription factors (TFs) were included. We hypothesized that overexpression of hnRNP K might modulate gene expression by altering the AS of TFs.
To prove the abovementioned hypothesis, we extracted 68 hnRNP K–regulated TFs enriched in regulation of transcription, transcription, or DNA-dependent pathway. Based on TRRUST and ENCODE Transcription Factor Targets dataset, the DNA binding motifs of 19 out of 68 TFs were presented in the promoter regions of 573 DEGs induced by hnRNP K. GO analysis showed that these 573 DEGs were mainly involved in inflammatory response, extracellular matrix organization pathways and negative regulation of neuron apoptotic process (Figure 3(B)), which was highly similar to that in Figure 2(B). The results confirmed the hypothesis that the hnRNP K–regulated TF alternative splicing plays an important role in establishing the regulation network of hnRNP K (Figure 3(C)).
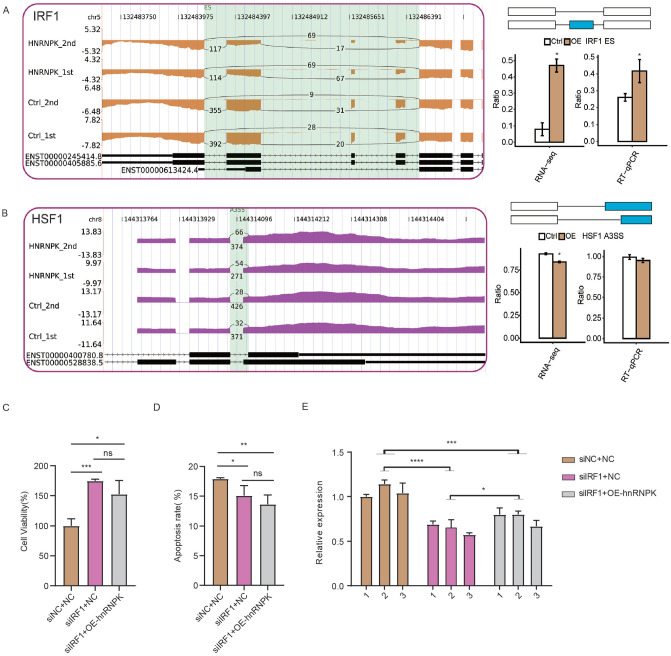
Validation of hnRNP K-IRF1-CCL5 axis in HeLa cells
Among the hnRNP K-TF-inflammatory gene regulatory network, we were particularly interested in the hnRNP K-IRF1-CCL5 axis. Inflammatory mediator CCL5 was found to play key roles in cancer progression, while IRF1 encodes an activator of immune response genes.42,43 Previous study revealed that IRF1 binds to a TTTTC motif-containing mouse CCL5 promoter. 44 To confirm our finding that hnRNP K may regulate inflammatory response and tumorigenesis via IRF1-CCL5 pathway, we performed RT-qPCR to further study their expression relationship. The ASEs regulated by hnRNP K in IRF1, HSF1, HDAC10 and BRD4 were verified by RT-qPCR experiment, which confirmed the RNA-seq results (Figure 4(A), (B) and S2(C) and (D)), except that HSF1 showed no significant difference (Figure 4(B)). We then silenced IRF1 expression by small interfering RNA (siIRF1) in HeLa cells to explore its influence on cellular phenotypes and the expression level of CCL5. SiIRF1 significantly increased proliferation rate and decreased apoptotic level of HeLa cells (Figure 4(C) and (D)), consistent with its known functions in cancer. 45 Under hnRNP K-OE condition, siIRF1 could also reverse maintain the increased proliferation rate and decreased apoptotic level (Figure 4(C) and (D)). Furthermore, we checked the expression level of CCL5 and found siIRF1 significantly reduced CCL5 expression level under hnRNP K-OE and normal conditions (Figure 4(E)). These results indicate that IRF1 regulates cellular phenotype of HeLa cells and could rescue CCL5 expression in hnRNP K-OE cells.
Figure 4.
Validation of hnRNP K-IRF1-CCL5 axis in HeLa cells. IGV-sashimi plots (left panel) and RT-qPCR validation (right panel) showed ASEs and the changed ratio of transcription factors, including (A) IRF1 and (B) HSF1. The schematic diagrams present the structures of AS (right panel, top) and Model (right panel, middle). Boxes denote the exon sequences. The horizontal line denotes the intron sequences. (C) Bar plot showing the cell proliferation rate in three groups. (D) Bar plot showing the cell apoptosis rate in three groups. (E) Bar plot showing the expression level of CCL5. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; Student’s t-test for A-D; two-way ANOVA test for E. (A color version of this figure is available in the online journal.)
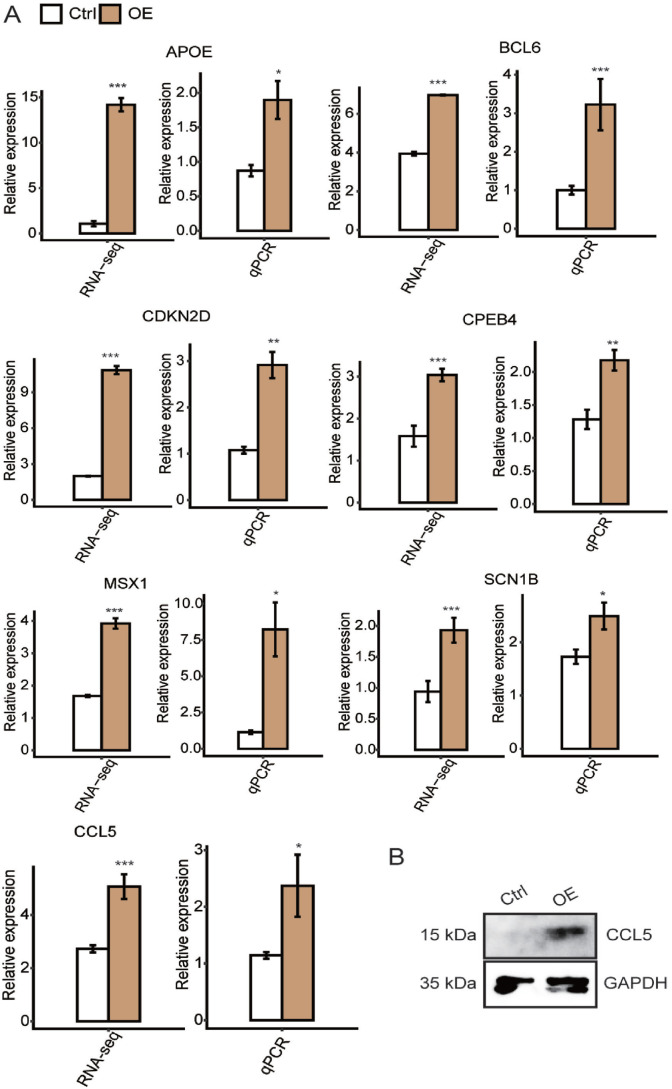
Other than CCL5, six other hnRNP K–upregulated DEGs (CPEB4, MSX1, BCL6, APOE, SCN1B, and CDKN2D) from inflammatory response and negative regulation of neuron apoptosis pathways, which are important biological process during tumorigenesis, were chosen for the purpose of qPCR validation. The results were shown in Figure 5(A), in which all seven genes were significantly increased along with hnRNP K overexpression. Please be noted that the total RNA template used for this validation experiments was the same as in RNA-seq experiments. HnRNP K–induced expression of CCL5 was also validated in WB experiments (Figure 5(B)).
Figure 5.
Validation of hnRNP K–regulated genes by qPCR and Western blot. (A) Bar plot showing the expression levels of seven genes with hnRNP K-OE and control vector in HeLa cells. (B) Validating the effect of hnRNP K–regulated gene CCL5 by conducting Western blot experiment in HeLa cells. (A color version of this figure is available in the online journal.)
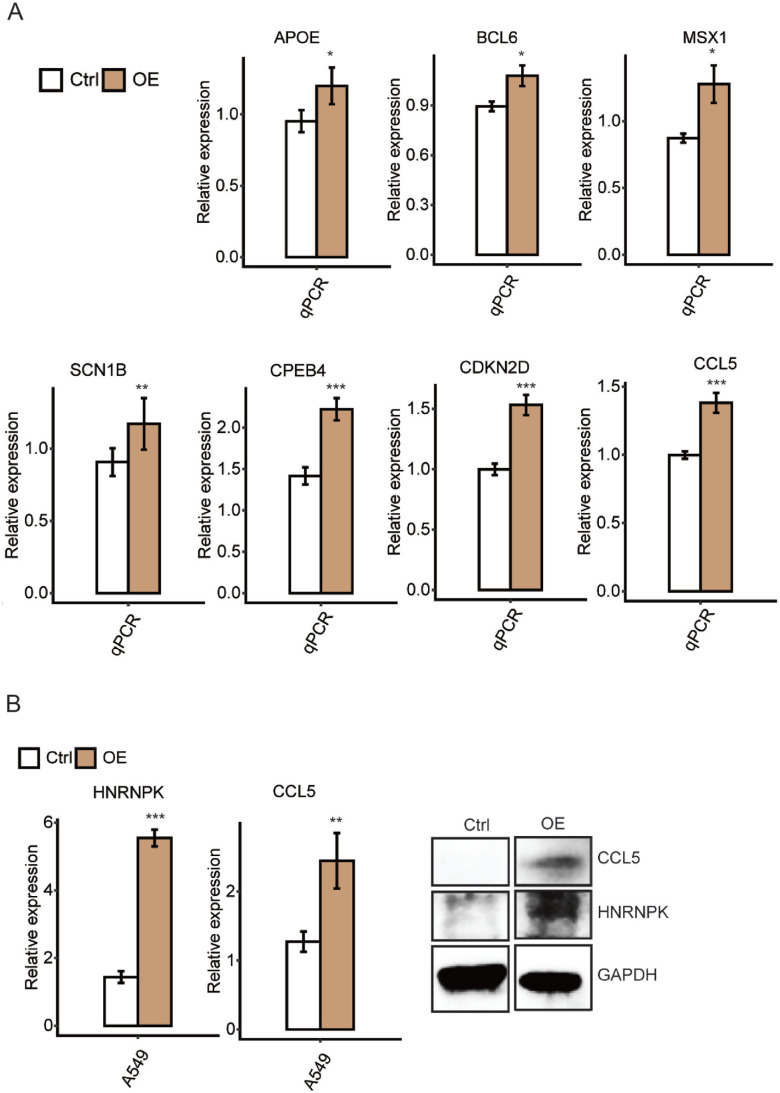
To further validate the above results, we performed a new set experiment from cell culture and plasmid transfection in triplicates. Using the total RNA from this new set of experiments, we conducted RT-qPCR experiment to assess the expression levels of the same seven genes, which demonstrated highly consistent results with the previous experiments (Figure 6(A)).
Figure 6.
(A) A new set of experiment validating hnRNP K–regulated DEGs shown in Figure 4(A) in triplicate. (B) Validating the effect of hnRNP K–regulated gene CCL5 by qPCR and Western blot in A549 cells. *P < 0.05; **P < 0.01; ***P < 0.001. (A color version of this figure is available in the online journal.)
HnRNP K regulates expression of CCL5 in A549 cells
To confirm our findings that CCL5 is an important target of hnRNP K, a plasmid with hnRNP K overexpression was transfected into A549 cells. The expression of CCL5 and hnRNP K was validated by RT-qPCR and WB (Figure 6(B)). Our results demonstrated the successful overexpression of hnRNP K in A549 cells. In addition, overexpression of hnRNP K significantly induced the expression of CCL5, conforming to the results from HeLa cells.
Discussion
HnRNP K is a multifunctional protein implicated in RNA metabolism, RNA stability and translation, RNA splicing, and DNA transcription. Various lines of evidences have confirmed that aberrant expression of hnRNP K is contributing to tumor proliferation. For example, hnRNP K promotes cell proliferation in lung cancer, 19 bladder cancer, 46 and colon cancer. 26 In accordance with previous studies, results in our study revealed that hnRNP K overexpression extensively increased the proliferation of HeLa cells.
According to previous studies, hnRNP K was elevated in bladder cancer, which regulates expression of several genes to promote bladder cell proliferation, chemoresistance to cisplatin, and apoptosis inhibition. 46 Gao et al. reported that hnRNP K has the ability to increase tumor metastasis by regulating the expression of genes with cell motility, angiogenesis, and extracellular matrix functions. HnRNP K is also overexpressed and correlated with mutant p53 in pancreatic carcinoma. 47 Similarly, hnRNP K promotes the progression of prostate cancer by regulating the androgen receptor (AR) translational apparatus and its altered pattern of expression has potential values for diagnosis and prognosis in prostate cancer. 23 Hence, we adopted a high-throughput transcriptomic RNA-seq approach to reveal how hnRNP K affects tumor cells in transcriptome. Hybridization-based approaches can only be used to detect transcripts corresponding to sequences that already exist, whereas RNA-seq approaches have no upper limit on quantification. 48 In our study, transcriptome analysis of hnRNP K overexpression and control cells was performed, and a total of 2405 DEGs and 1052 RASGs were detected, suggesting that the activation of hnRNP K has a wide range of effects.
DEGs regulated by hnRNP K were unbiasedly analyzed and were shown to be enriched in multiple KEGG pathways and GO functional pathways. Previous study demonstrated that inflammation is involved in cancer progression, involving aberrant tissue repair, genotoxicity, proliferation, invasion, and migration. 49 In this study, hnRNP K upregulates expression of CCL5 and BCL6 among inflammatory response. As one of the chemokine superfamily members, CCL5 is well recognized in immune environment, in which it induces leukocyte-directed motility. 50 The expression of CCL5 is induced by inflammatory insult, whereas blocked when inflammation subsides. CCL5 acts mainly in inflammatory reactions, which are confirmed to be highly expressed by numerous tumor cells and exert certain pathogenic influences on malignant tumors and metastases.51 –53 According to previous studies, the level of CCL5 was found markedly increased in tissue and plasma of patients with CESC. 54 Besides, our hypothesis that hnRNP K regulates the expression of CCL5 was confirmed in A549 cells (Figure 6(B)). BCL6 can interact with large numbers of genes directly or indirectly and it is mainly required for germinal centers formation, which are necessary to produce an effective humoral immune response. 55 BCL6 also plays an important role in breast cancer, 56 ovarian cancer, 57 and non-small-cell lung cancer progression. 58 Above all, this study indicates that hnRNP K might contribute greatly to cancer progression by activating the inflammatory oncogenes. Meanwhile, it is of necessity to validate the functions of hnRNP K in CESC as well as other cancers by testing its expression level in tumor tissues, analyzing its influence on cell condition, and exploring the underlying mechanisms.
The extensive prevalence of AS suggests the importance of genomic complexity-regulated biological process. As an important RBP, hnRNP K is found to mediate the AS pattern of many genes. Tyson-Capper and Gautrey 59 described hnRNP K as a splicing enhancer which promotes inclusion exon 2 Mcl-1 and subsequently produces the antiapoptotic Mcl-1L. HnRNP K could regulate alternative splicing of MRPL33 and then promote production of splice variant with the third exon inclusion in colorectal cancer cells. 21 Other than mediating splicing via direct binding with pre-mRNA, hnRNP K also activated the expression of the splicing regulator SRSF1 and then induced splicing activity. 22 We found that hnRNP K overexpression significantly changes the AS of numbers of genes in HeLa cells according to previous studies. A panel of those genes is enriched in transcription or regulation of transcription and in DNA-dependent pathways, including IRF1 and HSF1, which are well-known transcription factors. Belluti suggested that alternative splicing of cancer-related transcription factors can generate isoforms that promote cell proliferation, differentiation block, and apoptosis resistance. 60 IRF1 encodes a protein serving as an activator of genes implicated in both innate and acquired immune response. It was reported that IRF1 was implicated in CESC through STAT3/IRF1 pathway. 61 Lee et al. 62 discovered IRF1 variants with the exclusion of exons 7, 8, or 9, a crucial mechanism for negatively regulating IRF1 in CESC. Based on transcription factor and binding motif analysis, IRF1 was found to bind to the promoter of several genes, including CCL5 and BCL6, which are important in inflammatory response (Figure 3(C)). This result is consistent with the finding of Liu et al. 44 who have identified IRF1 bound to mouse CCL5 promoter directly with a TTTTC motif at −147 to −143 and provided a novel defense mechanism against tumor development. We validated IRF1 could affect cellular phenotypes and rescue CCL5 expression under hnRNP K-OE condition in HeLa cells; we speculate that hnRNP K indirectly modulates the expression level of genes implicated in inflammatory responses via regulating AS of TFs.
Except for IRF1, the other three transcription factors also play essential roles in inflammation process via modulating gene expression. For example, HSF1 activates the inhibition of NF-kB cytokine production mediated by hypercapnia 63 or exhibits anti-inflammatory functions in hepatocytes induced by zerumbone. 64 IL-6 was induced by oxidant stress. CXCL8 was remarkably inhibited by BRD4 restriction, indicating its essential function in regulating inflammatory genes. 65 HDAC10 was reported to alleviate inflammation after intracerebral hemorrhage 66 or promote IL-1β-mediated inflammatory activation of mesenchymal stem cells derived from synovium. 67 In addition, previous studies have shown that HSF1, BRD4, and HDAC10 were implicated in CESC,68 –70 supporting our hypothesis that hnRNP K might regulate expression of genes involved in inflammation via modulating alternative splicing of transcription factors. Nevertheless, the splicing isoforms of these genes and their exact functions on CESC remain to be disclosed. Another interesting point that needs to be further investigated is whether human papillomavirus (HPV) regulates hnRNP K expression. It has been reported hnRNP K inhibited the protein level of HPV-16 L2 mRNA in vitro, 71 while no other study reported the relationship between HPV and hnRNP K in cervical cancer.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221110649 for HnRNP K regulates inflammatory gene expression by mediating splicing pattern of transcriptional factors by Siyi Liu, Yong Duan, Ran You, Dong Chen and Jinhai Tan in Experimental Biology and Medicine
Footnotes
Authors’ Contributions: JT and SL devised the study. YD and RY conducted the experiments. SL and DC collected and analyzed the dada. SL prepared this manuscript. JT oversaw and revised the final manuscript. All authors have read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Health commission of Hubei Province scientific research project (WJ2019H011) and Zhongnan Hospital of Wuhan University Medical Science and Technology Innovation Platform project (PTXM2020033).
Data Availability: The raw reads of RNA-seq data have been deposited in NCBIs Gene Expression Omnibus (GEO) with accession number GSE148385.
ORCID iD: Jinhai Tan  https://orcid.org/0000-0001-7644-2970
https://orcid.org/0000-0001-7644-2970
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: Cancer J Clin 2011;61:69–90 [DOI] [PubMed] [Google Scholar]
- 2. Sankaranarayanan R, Esmy PO, Rajkumar R, Muwonge R, Swaminathan R, Shanthakumari S, Fayette J-M, Cherian J. Effect of visual screening on cervical cancer incidence and mortality in Tamil Nadu, India: a cluster-randomised trial. Lancet 2007;370:398–406 [DOI] [PubMed] [Google Scholar]
- 3. Sankaranarayanan R. HPV vaccination: the promise & problems. Indian J Med Res 2009;130:322–6 [PubMed] [Google Scholar]
- 4. Ronco G, Giorgi-Rossi P, Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, Ghiringhello B, Girlando S, Gillio-Tos A, De Marco L, Naldoni C, Pierotti P, Rizzolo R, Schincaglia P, Zorzi M, Zappa M, Segnan N, Cuzick J, New Technologies for Cervical Cancer screening (NTCC) Working Group. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol 2010;11:249–57 [DOI] [PubMed] [Google Scholar]
- 5. Ronco G, Dillner J, Elfstrom KM, Tunesi S, Snijders PJ, Arbyn M, Kitchener H, Segnan N, Gilham C, Giorgi-Rossi P, Berkhof J, Peto J, Meijer CJ. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet (London, England) 2014;383:524–32 [DOI] [PubMed] [Google Scholar]
- 6. Angioli R, Plotti F, Luvero D, Aloisi A, Guzzo F, Capriglione S, Terranova C, De Cicco Nardone C, Benedetti-Panici P. Feasibility and safety of carboplatin plus paclitaxel as neoadjuvant chemotherapy for locally advanced cervical cancer: a pilot study. Tumour Biol 2014;35:2741–6 [DOI] [PubMed] [Google Scholar]
- 7. Organista-Nava J, Gomez-Gomez Y, Gariglio P. Embryonic stem cell-specific signature in cervical cancer. Tumour Biol 2014;35:1727–38 [DOI] [PubMed] [Google Scholar]
- 8. Fang M, Kan Y, Dong D, Yu T, Zhao N, Jiang W, Zhong L, Hu C, Luo Y, Tian J. Multi-habitat based radiomics for the prediction of treatment response to concurrent chemotherapy and radiation therapy in locally advanced cervical cancer. Front Oncol 2020;10:563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vergote I, Magrina JF, Zanagnolo V, Magtibay PM, Butler K, Gil-Moreno A, Feijoo B-D, Kimmig R, Canis M, Bourdel N. The LACC trial and minimally invasive surgery in cervical cancer. J Min Inv Gynecol 2020;27:462–3 [DOI] [PubMed] [Google Scholar]
- 10. Boldrini L, Piras A, Chiloiro G, Autorino R, Cellini F, Cusumano D, Fionda B, D’Aviero A, Campitelli M, Marazzi F, Balducci M, Valentini V, Gambacorta MA. Low Tesla magnetic resonance guided radiotherapy for locally advanced cervical cancer: first clinical experience. Tumori 2020;106:497–505 [DOI] [PubMed] [Google Scholar]
- 11. Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer 2017;3:506–28 [DOI] [PubMed] [Google Scholar]
- 12. Nyati KK, Zaman MM, Sharma P, Kishimoto T. Arid5a, an RNA-binding protein in immune regulation: RNA stability, inflammation, and autoimmunity. Trends Immunol 2020;41:255–68 [DOI] [PubMed] [Google Scholar]
- 13. Chou HL, Tian L, Kumamaru T, Hamada S, Okita TW. Multifunctional RNA binding protein OsTudor-SN in storage protein mRNA transport and localization. Plant Physiol 2017;175:1608–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu X, Harvey SE, Zheng R, Lyu J, Grzeskowiak CL, Powell E, Piwnica-Worms H, Scott KL, Cheng C. The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing. Nat Commun 2020;11:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, Cao X. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci 2019; 116:976–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet 2008;24:416–25 [DOI] [PubMed] [Google Scholar]
- 17. Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett 1997;403:113–5 [DOI] [PubMed] [Google Scholar]
- 18. Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol 1996;16:2350–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallardo M, Hornbaker MJ, Zhang X, Hu P, Bueso-Ramos C, Post SM. Aberrant hnRNP K expression: all roads lead to cancer. Cell Cycle (Georgetown, Tex) 2016;15:1552–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pelisch F, Pozzi B, Risso G, Muñoz MJ, Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J Biol Chem 2012;287:30789–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu L, Luo C, Luo Y, Chen L, Liu Y, Wang Y, Han J, Zhang Y, Wei N, Xie Z. MRPL33 and its splicing regulator hnRNPK are required for mitochondria function and implicated in tumor progression. Oncogene 2018;37:86–94 [DOI] [PubMed] [Google Scholar]
- 22. Peng WZ, Liu JX, Li CF, Ma R, Jie JZ. hnRNPK promotes gastric tumorigenesis through regulating CD44E alternative splicing. Cancer Cell Int 2019;19:335–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barboro P, Repaci E, Rubagotti A, Salvi S, Boccardo S, Spina B, Truini M, Introini C, Puppo P, Ferrari N, Carmignani G, Boccardo F, Balbi C. Heterogeneous nuclear ribonucleoprotein K: altered pattern of expression associated with diagnosis and prognosis of prostate cancer. Br J Cancer 2009;100:1608–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benelli R, Monteghirfo S, Balbi C, Barboro P, Ferrari N. Novel antivascular efficacy of metronomic docetaxel therapy in prostate cancer: hnRNP K as a player. Int J Cancer 2009;124:2989–96 [DOI] [PubMed] [Google Scholar]
- 25. Ciarlo M, Benelli R, Barbieri O, Minghelli S, Barboro P, Balbi C, Ferrari N. Regulation of neuroendocrine differentiation by AKT/hnRNPK/AR/beta-catenin signaling in prostate cancer cells. Int J Cancer 2012;131: 582–90 [DOI] [PubMed] [Google Scholar]
- 26. Carpenter B, McKay M, Dundas SR, Lawrie LC, Telfer C, Murray GI. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br J Cancer 2006;95:921–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Otoshi T, Tanaka T, Morimoto K, Nakatani T. Cytoplasmic accumulation of heterogeneous nuclear ribonucleoprotein K strongly promotes tumor invasion in renal cell carcinoma cells. PLoS ONE 2015;10: e0145769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chung IC, Chen LC, Chung AK, Chao M, Huang HY, Hsueh C, Tsang NM, Chang KP, Liang Y, Li HP, Chang YS. Matrix metalloproteinase 12 is induced by heterogeneous nuclear ribonucleoprotein K and promotes migration and invasion in nasopharyngeal carcinoma. BMC Cancer 2014;14:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang L, Feng J, Kong S, Wu M, Xi Z, Zhang B, Fu W, Lao Y, Tan H, Xu H. Nujiangexathone A, a novel compound from Garcinia nujiangensis, suppresses cervical cancer growth by targeting hnRNPK. Cancer Lett 2016;380:447–56 [DOI] [PubMed] [Google Scholar]
- 30. Lee WJ, Shin CH, Ji H, Jeong SD, Park MS, Won HH, Pandey PR, Tsitsipatis D, Gorospe M, Kim HH. hnRNPK-regulated LINC00263 promotes malignant phenotypes through miR-147a/CAPN2. Cell Death Dis 2021;12:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song Ba Y, Wang Ma F, Wei Ma Y, Chen Ba D, Deng Ba G. ATP5A1 participates in transcriptional and posttranscriptional regulation of cancer-associated genes by modulating their expression and alternative splicing profiles in HeLa Cells. Technol Cancer Res Treat 2021; 20:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jobke B, Milovanovic P, Amling M, Busse B. Bisphosphonate-osteoclasts: changes in osteoclast morphology and function induced by antiresorptive nitrogen-containing bisphosphonate treatment in osteoporosis patients. Bone 2014;59:37–43 [DOI] [PubMed] [Google Scholar]
- 33. Plumb JA, Milroy R, Kaye S. Effects of the pH dependence of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide-formazan absorption on chemosensitivity determined by a novel tetrazolium-based assay. Cancer Res 1989;49:4435–40 [PubMed] [Google Scholar]
- 34. Yang X, Zhan P, Feng S, Ji H, Tian W, Wang M, Cheng C, Song B. SRSF6 regulates alternative splicing of genes involved in DNA damage response and DNA repair in HeLa cells. Oncol Rep 2020;44:1851–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y, Zhang L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech 2017; 1860:911–21 [DOI] [PubMed] [Google Scholar]
- 39. Liu J, Li C, Wang J, Xu D, Wang H, Wang T, Li L, Li H, Nan P, Zhang J, Wang Y, Huang C, Chen D, Zhang Y, Wen T, Zhan Q, Ma F, Qian H. Chromatin modifier MTA1 regulates mitotic transition and tumorigenesis by orchestrating mitotic mRNA processing. Nat Commun 2020; 11:4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li RZ, Hou J, Wei Y, Luo X, Ye Y, Zhang Y. hnRNPDL extensively regulates transcription and alternative splicing. Gene 2019;687:125–34. [DOI] [PubMed] [Google Scholar]
- 41. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L.KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm 2014;2014:292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Forero A, Ozarkar S, Li H, Lee CH, Hemann EA, Nadjsombati MS, Hendricks MR, So L, Green R, Roy CN. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity 2019;51:451e6–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Guan X, Ma X. Interferon regulatory factor 1 is an essential and direct transcriptional activator for interferon γ-induced RANTES/CCl5 expression in macrophages. J Biol Chem 2005;280:24347–55 [DOI] [PubMed] [Google Scholar]
- 45. Romeo G, Fiorucci G, Chiantore MV, Percario ZA, Vannucchi S, Affabris E. IRF-1 as a negative regulator of cell proliferation. J Interferon Cytokine Res 2002;22:39–47 [DOI] [PubMed] [Google Scholar]
- 46. Chen X, Gu P, Xie R, Han J, Liu H, Wang B, Xie W, Xie W, Zhong G, Chen C, Xie S, Jiang N, Lin T, Huang J. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med 2017;21:1266–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Renyuan Z, Reneé S, Nelson MA, Achyut B, Jiaqi S. Increased expression of the heterogeneous nuclear ribonucleoprotein K in pancreatic cancer and its association with the mutant p53. Int J Cancer 2010;126: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008;320:1344–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 2013;13:759–71 [DOI] [PubMed] [Google Scholar]
- 50. Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett 2008;267:271–85 [DOI] [PubMed] [Google Scholar]
- 51. Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol 2006;7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Christopherson K, 2nd, Hromas R. Chemokine regulation of normal and pathologic immune responses. Stem Cells 2001;19:388–96 [DOI] [PubMed] [Google Scholar]
- 53. Mackay CR. Chemokines: immunology’s high impact factors. Nat Immunol 2001;2:95–101 [DOI] [PubMed] [Google Scholar]
- 54. Niwa Y, Akamatsu H, Niwa H, Sumi H, Ozaki Y, Abe A. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clin Cancer Res 2001;7:285–9 [PubMed] [Google Scholar]
- 55. Leeman-Neill RJ, Bhagat G. BCL6 as a therapeutic target for lymphoma. Expert Opin Ther Targets 2018;22:143–52 [DOI] [PubMed] [Google Scholar]
- 56. Ang L, Zheng L, Wang J, Huang J, Hu HG, Zou Q, Zhao Y, Liu QM, Zhao M, Wu ZS. Expression of and correlation between BCL6 and ZEB family members in patients with breast cancer. Exp Ther Med 2017; 14:3985–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu L, Feng H, Jin S, Tan M, Gao S, Zhuang H, Hu Z, Wang H, Song Z, Lin B. High expressions of BCL6 and Lewis y antigen are correlated with high tumor burden and poor prognosis in epithelial ovarian cancer. Tumour Biol 2017;39:1–12 [DOI] [PubMed] [Google Scholar]
- 58. Deb D, Rajaram S, Larsen JE, Dospoy PD, Marullo R, Li LS, Avila K, Xue F, Cerchietti L, Minna JD, Altschuler SJ, Wu LF. Combination therapy targeting BCL6 and phospho-STAT3 defeats intratumor heterogeneity in a subset of non-small cell lung cancers. Cancer Res 2017;77:3070–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tyson-Capper A, Gautrey H. Regulation of Mcl-1 alternative splicing by hnRNP F, H1 and K in breast cancer cells. RNA Biol 2018;15: 1448–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belluti S, Rigillo G, Imbriano C. Transcription factors in cancer: when alternative splicing determines opposite cell fates. Cells 2020;9:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walch-Rückheim B, Pahne-Zeppenfeld J, Fischbach J, Wickenhauser C, Horn LC, Tharun L, Büttner R, Mallmann P, Stern P, Kim Y-J. STAT3/IRF1 pathway activation sensitizes cervical cancer cells to chemotherapeutic drugs. Cancer Res 2016;76:3872–83 [DOI] [PubMed] [Google Scholar]
- 62. Lee E-J, Jo M, Park J, Zhang W, Lee J-H. Alternative splicing variants of IRF-1 lacking exons 7, 8, and 9 in cervical cancer. Biochem Biophys Res Commun 2006;347:882–8 [DOI] [PubMed] [Google Scholar]
- 63. Lu Z, Casalino-Matsuda SM, Nair A, Buchbinder A, Budinger GRS, Sporn PHS, Gates KL. A role for heat shock factor 1 in hypercapnia-induced inhibition of inflammatory cytokine expression. FASEB J 2018; 32:3614–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Igarashi Y, Ohnishi K, Irie K, Murakami A. Possible contribution of zerumbone-induced proteo-stress to its anti-inflammatory functions via the activation of heat shock factor 1. PLoS ONE 2016;11:e0161282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khan YM, Kirkham P, Barnes PJ, Adcock IM. Brd4 is essential for IL-1β-induced inflammation in human airway epithelial cells. PLoS ONE 2014;9:e95051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang L, Zheng S, Zhang L, Xiao H, Gan H, Chen H, Zhai X, Liang P, Zhao J, Li Y. Histone deacetylation 10 alleviates inflammation after intracerebral hemorrhage via the PTPN22/NLRP3 pathway in rats. Neuroscience 2020;432:247–59 [DOI] [PubMed] [Google Scholar]
- 67. Liao W, Sun J, Liu W, Li W, Jia J, Ou F, Su K, Zheng Y, Zhang Z, Sun Y. HDAC10 upregulation contributes to interleukin 1beta-mediated inflammatory activation of synovium-derived mesenchymal stem cells in temporomandibular joint. J Cell Physiol 2019;234:12646–62 [DOI] [PubMed] [Google Scholar]
- 68. Rataj O, Haedicke Jarboui -J, Stubenrauch F, Iftner T. Brd4 inhibition suppresses HPV16 E6 expression and enhances chemoresponse: a potential new target in cervical cancer therapy. Int J Cancer 2019;144:2330–8 [DOI] [PubMed] [Google Scholar]
- 69. Song C, Zhu S, Wu C, Kang J. Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J Biol Chem 2013;288:28021–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Das S, Bhattacharyya NP. Heat shock factor 1 regulates hsa-miR-432 expression in human cervical cancer cell line. Biochem Biophys Res Commun 2014;453:461–6 [DOI] [PubMed] [Google Scholar]
- 71. Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem 1998;273:22648–56 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221110649 for HnRNP K regulates inflammatory gene expression by mediating splicing pattern of transcriptional factors by Siyi Liu, Yong Duan, Ran You, Dong Chen and Jinhai Tan in Experimental Biology and Medicine