Abstract
The emergence and worldwide spread of SARS-CoV-2 during the COVID-19 pandemic necessitated the adaptation and rapid deployment of viral WGS and analysis techniques that had been previously applied on a more limited basis to other viral pathogens, such as HIV and influenza viruses. The need for WGS was driven in part by the low mutation rate of SARS-CoV-2, which necessitated measuring variation along the entire genome sequence to effectively differentiate lineages and characterize viral evolution. Several WGS approaches designed to maximize throughput and accuracy were quickly adopted by surveillance labs around the world. These broad-based SARS-CoV-2 genomic sequencing efforts revealed ongoing evolution of the virus, highlighted by the successive emergence of new viral variants throughout the course of the pandemic. These genomic insights were instrumental in characterizing the effects of viral mutations on transmissibility, immune escape and viral tropism, which in turn helped guide public health policy, the use of monoclonal antibody therapeutics and vaccine development strategies. As the use of direct-acting antivirals for the treatment of COVID-19 became more widespread, the potential for emergence of antiviral resistance has driven ongoing efforts to delineate resistance mutations and to monitor global sequence databases for their emergence. Given the critical role of viral genomics in the international effort to combat the COVID-19 pandemic, coordinated efforts should be made to expand global genomic surveillance capacity and infrastructure towards the anticipation and prevention of future pandemics.
SARS-CoV-2 evolution
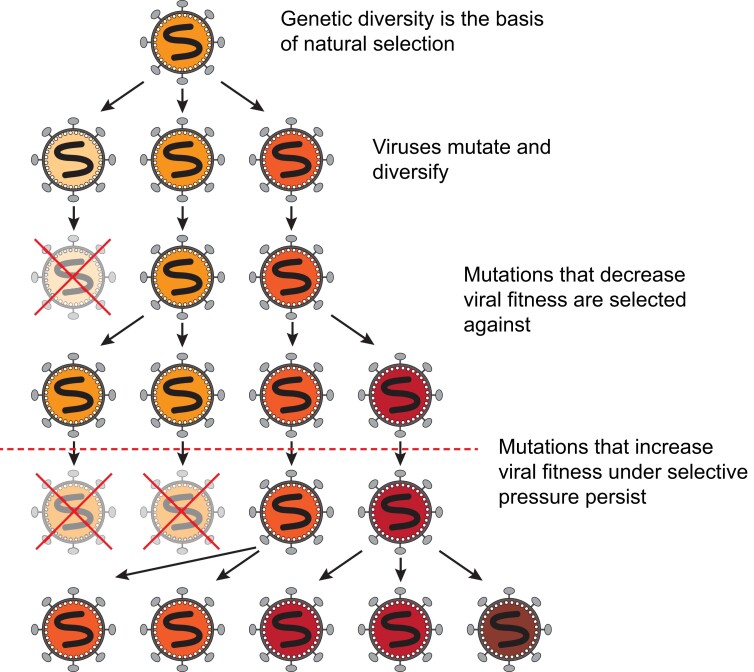
Viruses, like all organisms, face driving evolutionary forces that include mutation, natural selection and random genetic drift, resulting in a continuous process of evolution (Figure 1).1,2 Due to the unique properties of viruses, and especially those that replicate via an RNA-dependent RNA polymerase, these evolutionary forces operate in essentially different ways. A key feature for their distinct evolution is that they have some of the highest mutational frequencies found in nature.3–5 Such high mutation rates are achieved through their small genome sizes, rapid rates of viral replication and progeny production, error-prone viral polymerases, host enzyme-induced mutations [i.e. by APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like) or ADAR (adenosine deaminase acting on RNA) enzymes], and recombination during concurrent infections.3,6,7 As such, RNA viruses exist as ‘clouds’ of related genotypes, termed quasispecies, where a large proportion of progeny is expected to have one or more mutations relative to the consensus or average genome.8,9 This unique variability constitutes the basis of the evolutionary success for RNA viruses by allowing them to rapidly adapt via selection to changing environments or conditions.1,8,10 Moreover, this exceptional viral heterogeneity allows these viruses to tolerate the extreme population bottlenecks inherent to their life cycles—e.g. transmission, translocation to different anatomical compartments, immune evasion and therapeutic escape.2,11,12 The high genetic variability compensates for any intrinsic loss of fitness derived from successive population size losses where stochastic forces (genetic drift) and strong purifying selection could otherwise lead to viral extinction.9,13–15 Therefore, by means of a staggering capacity to adapt while maintaining population structure, viruses are very successful at evading immune responses and/or pharmacological challenges.
Figure 1.
Viral genetic diversity is the basis of natural selection and viral evolution. All viruses mutate over time. Mutations that are disadvantageous or that come at a cost to viral fitness are selected against. Mutations that confer a fitness advantage, especially in the presence of a selective pressure (represented by the horizontal dotted line), will be more likely to persist in future generations. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
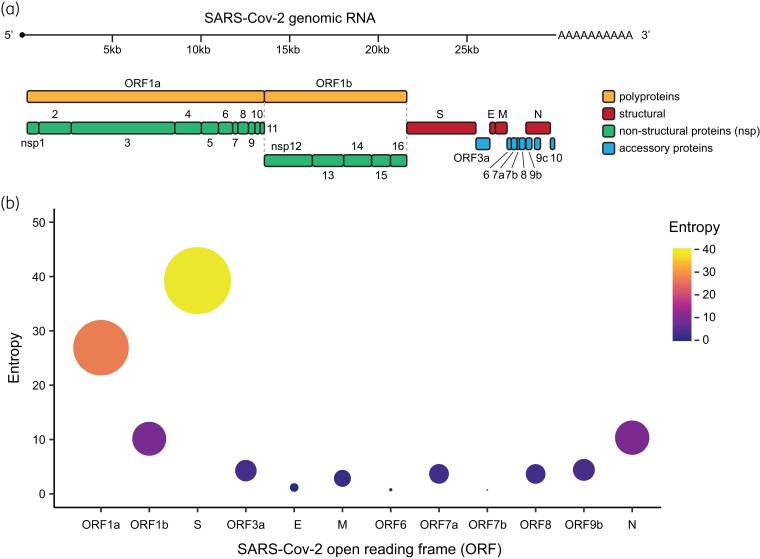
The importance of RNA viruses’ genetic variability in infections underscores the importance of studying viral evolution and the dynamics of viral diversity. This was clearly demonstrated during the SARS-CoV-2 pandemic, in which molecular surveillance and the study of viral population dynamics have been crucial for tracking epidemiological patterns and defining significant viral characteristics that are crucial for understanding essential viral properties.16,17 Whereas sequencing and analysing selected genes or genome regions can be sufficient to capture the variability of viruses with higher mutation rates such as HIV-1 (inter-host substitution rate of ∼5 × 10−3 substitutions/site/year)18 or influenza viruses (∼4 × 10−3 substitutions/site/year),19 the relatively large genome size and low mutation rate of SARS-CoV-2 (estimated substitution rate of ∼8 × 10−4 substitutions/site/year)20–22 means that epidemiologically and evolutionarily distinct viruses may differ by only a few nucleotide changes across the entire nearly 30 kb genome (Figure 2a).23 Additionally, selective processes operate very differently amongst distinct viral genomic regions leading to an uneven distribution of mutations throughout the SARS-CoV-2 genome (Figure 2b). This can render some low-diversity genes uninformative for defining evolutionary relationships. Thus, sequencing of the entire viral genome is necessary to effectively assess within- and between-host genetic variability in SARS-CoV-2. This has led to an unprecedented worldwide effort for SARS-CoV-2 WGS, resulting in the largest dataset of complete viral genome sequences for a species in the history of science, with more than 15.2 million SARS-CoV-2 genomes deposited in the GISAID (Global Initiative on Sharing All Influenza Data) public sequence database in just over 3 years.
Figure 2.
Different SARS-CoV-2 ORFs have accumulated different amounts of variability. (a) Schematic of the SARS-CoV-2 genome and encoded proteins, separated broadly in the figure by function. (b) Entropy (a measure of genetic variability) in each SARS-CoV-2 ORF. Dot size and colour reflect the degree of entropy accumulated in the viral population since its emergence. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Methods and evolution of SARS-CoV-2 WGS
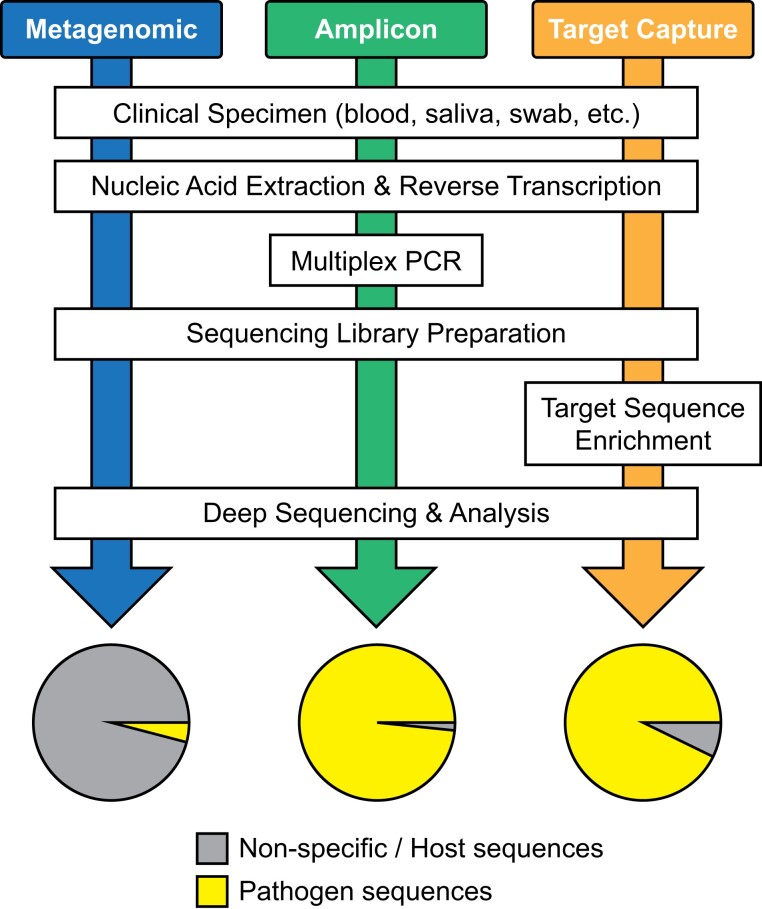
Multiple methods have been applied and adapted for sequencing the entire genome of SARS-CoV-2 (Figure 3). The first viral genome sequences were generated from respiratory specimens of workers at a seafood wholesale market in Wuhan, China, who had been hospitalized for severe respiratory distress syndromes in late December 2019.24,25 These earliest sequences were assembled from meta-transcriptomic sequencing of total RNA extracted from the patient specimens. The novelty of the newly emerged pathogen greatly limited the techniques available for isolating its genome sequence from the background of host and commensal microbe genomes, thus necessitating a broad, unbiased approach to sequencing. As COVID-19 spread and case numbers increased worldwide, the high cost and low specimen throughput of metagenomic sequencing approaches limited their broader application to genomic surveillance.
Figure 3.
Workflow diagram comparing metagenomic, amplicon-based and target capture strategies for pathogen genomics. The pie charts at the end depict the theoretical yield of pathogen sequence versus host or non-specific background. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Soon after the first SARS-CoV-2 genome sequences were made available, multiplex PCR primer sets producing tiled amplicons across the entire genome were designed to amplify the complete genome sequence directly from patient specimens. This tiled amplicon approach greatly minimized the amount of contaminating host or other nucleic acids from patient specimens and allowed viral genomes to be sequenced in greater volumes and at lower cost. One of the earliest amplicon sequencing protocols was developed in late January 2020 by the ARTIC network for molecular epidemiology for outbreak response (https://artic.network/ncov-2019) and was derived from similar approaches previously developed for sequencing Ebola and Zika viruses.26 The ARTIC primer set is comprised of 98–99 primer pairs that yield amplicons averaging roughly 350 bp in length. The primer sequences have been modified and adapted throughout the pandemic as mutations in certain primer binding sites have emerged in the genomes of new viral variants.27–29 The ARTIC primer scheme was widely adopted among labs performing SARS-CoV-2 sequencing for research and surveillance, and it has been incorporated into several commercial sequencing kits. A similar tiled amplicon scheme, the ‘midnight’ primer set consisting of 29 primer pairs producing roughly 1200 bp amplicons, was also developed and widely adopted.30,31 The lower number of primer binding sites in the midnight scheme reduces the relative risk of primer site mutation, theoretically necessitating fewer primer updates to respond to viral variant emergence.
Target capture approaches represent a third method applied to SARS-CoV-2 genomic sequencing. This technique employs DNA probes complementary to the viral sequence that selectively hybridize with SARS-CoV-2 genomic material and enrich it from the background host and commensal bacterial DNA. By incorporating longer probe sequences, approximately 120 bp each, and/or multiple overlapping probe sequences, these assays are more tolerant of viral mutation than amplicon-based approaches. Furthermore, the use of larger probe sets that also include sequences targeting other respiratory viruses allows this method to detect co-infections in COVID-19. Several target capture assays have been developed for SARS-CoV-232–34 and have been incorporated into some commercial kits. Once genomic libraries have been prepared from a patient or other samples using one of the above approaches, sequencing is performed on any one of multiple next-generation sequencing (NGS) platforms. As ongoing transmission of SARS-CoV-2 continues to drive evolution of the virus in response to natural and vaccine immunity, therapeutics and co-infections, diverse and adaptable genome sequencing approaches remain vital to effective global surveillance efforts.
Early diversification and the Spike D614G mutation
During the very early stages of the pandemic, analysis of the first SARS-CoV-2 sequences showed fairly homogeneous viral populations circulating globally.20,21,35 This picture dramatically changed around April 2020, when a distinct lineage of the virus rapidly expanded in Europe and the USA leading to a global genetic sweep.36,37 This new viral lineage was characterized by substitution of aspartic acid (D) with glycine (G) at position 614 in the Spike protein (S). S:D614G was rare in March 2020 but could be detected in over 74% of all published sequences by June 2020 (Figure 4). Viruses harbouring the S:D614G mutation rapidly replaced all other lineages circulating at the time, indicative of the significant fitness benefit afforded by the mutation. Since 2020, this mutation has become fixed in the SARS-CoV-2 genome and all subsequent viral lineages have maintained S:G614.
Figure 4.
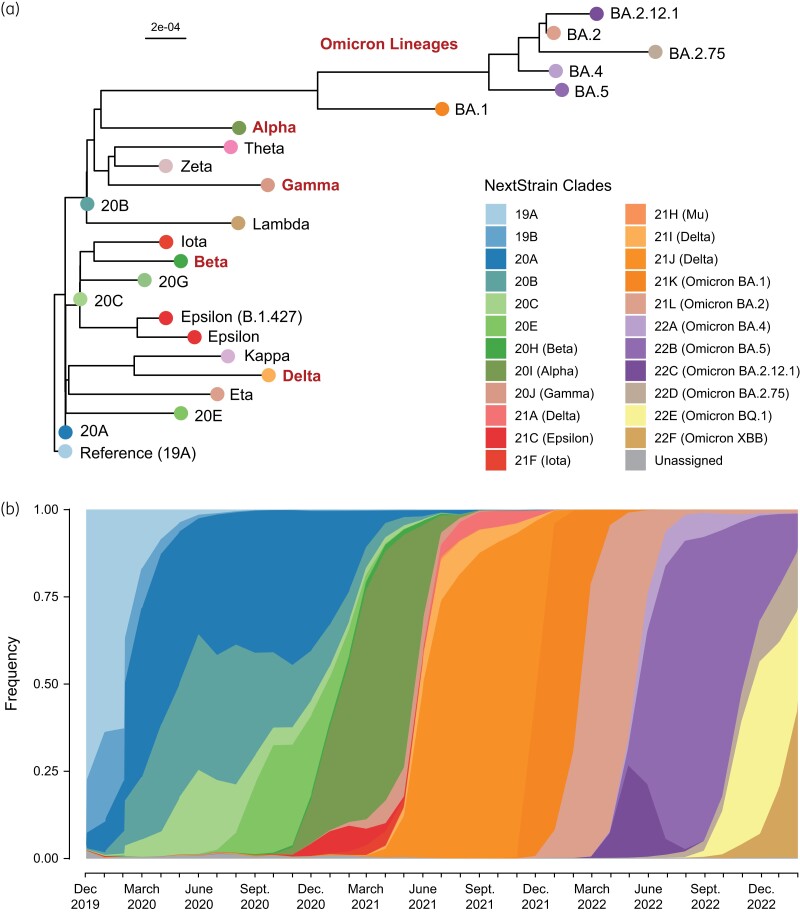
The ongoing evolution of SARS-CoV-2. (a) Phylogenetic tree of representative consensus sequences for globally prominent SARS-CoV-2 clades and variants of concern (Alpha, Beta, Gamma, Delta, and Omicron). (b) Cumulative frequency plot of globally prominent SARS-CoV-2 clades and variants of concern from December 2019 to February 2023. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The SARS-CoV-2 S protein mediates host cell entry by catalysing fusion of the virus envelope lipid bilayer with the target cell membrane.38 Because S decorates the virion surface, it is also a major antigen and induces neutralizing antibody responses.39 Therefore, S has served as a primary target for the development of diagnostics, therapeutics and—most importantly—vaccines. Consequently, this protein is subjected to strong selective processes, making it the most variable region in the SARS-CoV-2 genome (Figure 2b). Several studies performed on the S:D614G mutation demonstrated that this mutation facilitated enhanced cell entry through stabilization of the S subunits and through enhanced sampling of the open confirmation of the protein that enabled an interaction between the receptor binding domain (RBD) and the host receptor, ACE2.40,41 Subsequent studies confirmed that S:D614G conferred a clear biological fitness advantage in vitro, enhanced transmission in vivo, and resulted in higher viral loads in patient upper airways.37,42 S:D614G was the first naturally selected mutation in SARS-CoV-2 to be observed at the population level. Although the term had not yet been defined, this new lineage was the first SARS-CoV-2 ‘variant of concern’ (VOC) and was likely essential for the global expansion and consolidation of the pandemic.
Emergence of VOCs
In December 2020, the COVID-19 Genomics Consortium UK (CoG-UK) and Public Health England (PHE) reported the rapid expansion of a phylogenetically distinct lineage of SARS-CoV-2, termed B.1.1.7, that carried 17 lineage-defining non-synonymous mutations and deletions, 8 of which occurred in Spike alone (Figure 4a).43,44 Predicted to have emerged in September 2020, lineage B.1.1.7 accounted for over half of all cases in the UK by early December, becoming the first official SARS-CoV-2 VOC to be tracked by PHE.45 VOCs are defined by the WHO as variants with increasing relative prevalence and case counts that are associated with an increase in transmissibility, disease severity and/or therapeutic/immune evasion.46 The B.1.1.7 lineage quickly spread across the globe, accounting for a majority of cases worldwide by the spring of 2021 (Figure 4b). The WHO began labelling VOCs with letters of the Greek alphabet in May 2021, naming lineage B.1.1.7 the ‘Alpha’ variant.47 This coincided with the emergence of several VOCs in geographically disparate regions of the globe in early to mid-2021, including the Beta variant in South Africa (lineage B.1.351), the Delta variant in India (lineage B.1.617.2), and the Gamma variant in Brazil (lineage P.1), among others (Figure 4b).48–51
Despite being phylogenetically distinct (Figure 4a), some of the new VOCs shared mutations in Spike, including S:N501Y (Alpha, Beta and Gamma), S:E484K (Beta and Gamma) and S:K417N (Beta and Delta), suggestive of convergent evolution.52 Laboratory studies using non-infectious virus-like particles engineered to express Spike proteins with these different mutations (i.e. pseudotyped virus particles) demonstrated that the S:N501Y and S:E484K mutations increased binding efficiency to the host ACE2 receptor, facilitating cell entry.53–56 Studies in animal models and epidemiological studies of these variants in human populations demonstrated that these same mutations contributed to enhanced transmission, facilitating their rapid spread.57 The S:E484K and S:K417N mutations were furthermore found to contribute to immune resistance against certain classes of neutralizing antibodies.58–60
The emergence of VOCs harbouring changes in the Spike protein resulted in changes in the efficacy of monoclonal antibody therapeutics due to changes in their neutralizing capacity.61–63 For example, the S:E484K mutation (Beta and Gamma) and the S:T452R mutation (Delta) in the epitope region of Class II neutralizing antibodies conferred resistance to several monoclonal antibody therapeutics, including bamlanivimab.60,64–66 Indeed, several Spike mutations associated with VOCs that have been associated with immune escape have been observed to arise intra-host after monoclonal antibody administration,67 especially in immunocompromised hosts with persistent infection.68–71 This has led to the theory that persistent infection in immunocompromised or immunosuppressed hosts may have enabled the rapid evolution of SARS-CoV-2 and the simultaneous emergence of several distinct VOCs across the globe, though this is still under investigation.72
The co-circulation of many different VOCs with different monoclonal antibody therapeutic sensitivities in early 2021 increased the salience of viral genotypic information to optimize clinical care and resulted in a major investment in SARS-CoV-2 genomic surveillance.73 Given that timely assessment of the SARS-CoV-2 genotype in patients is not clinically tenable in most healthcare systems, the choice of monoclonal therapeutic agent to administer to COVID-19 patients has been determined based on the regional frequency of a specific variant.74 Whereas variant distributions have largely been assessed by WGS, a number of cheaper, PCR-based alternatives for rapid SARS-CoV-2 genotyping were developed in response to this need.75,76 These assays rely on PCR primer-probe sets that span one or more sequence mismatches between variants to enable specific amplification and detection of different VOCs. For example, a two amino acid deletion in Spike at positions 69 and 70 in the Alpha variant resulted in a loss of S gene amplification in a widely used multiplexed diagnostic assay (Thermopath TaqPath).43 This ‘S gene target failure’ (SGTF) became a reliable indicator of this deletion and a proxy for Alpha variant detection during early 2021. As this deletion has arisen independently in multiple lineages, only WGS can be used to unambiguously assign SARS-CoV-2 lineages, but PCR-based genotyping methods have been useful for powering large-scale clinical and translational research studies.57,77,78
Global predominance of the Omicron variants
By late fall of 2021, the Delta VOC was predominant worldwide and represented over 95% of all SARS-CoV-2 sequences (Figure 4b). However, in November 2021, a rapid increase in COVID-19 cases in Gauteng province, South Africa, coupled with a sudden increase in SGTF frequency prompted a rapid genomic surveillance response.79 These efforts uncovered a new, highly divergent SARS-CoV-2 lineage, B.1.1.529 (Figure 4a), which was designated as the Omicron VOC by the WHO only days afterwards.80 This highly divergent variant contained over 30 non-synonymous changes and deletions in Spike, including 15 in the Spike protein RBD alone.79 The Omicron VOC spread rapidly, causing massive surges in cases counts and hospitalizations in late 2021 and early 2022.81–83 By February 2022, Omicron had displaced Delta as the predominant VOC worldwide (Figure 4b).
Fortunately, early detection and reporting of the Omicron variant enabled a rapid research and public health response, such that healthcare systems were able to adjust their clinical care practices in real time as the Omicron VOC emerged in their regions. Most notably, the significant antigenic shift in Spike dramatically curtailed the effectiveness of most approved monoclonal therapeutics, including all approved Class I and Class II antibodies at the time.84 The Class III antibodies sotrovimab and bebtelovimab, which targeted relatively conserved regions of the RBD away from the receptor-binding motif, were the only two approved monoclonal antibody therapeutics with sustained efficacy against the Omicron VOC, though even these lost efficacy as the variant continued to diversify.62,84–86
The large number of mutations in the antigenic regions of Spike reduced the efficacy of certain monoclonal antibodies used as therapeutics, but were also sufficient to circumvent polyclonal antibody responses induced by vaccination and/or prior infection.87 During the very first wave of the Omicron VOC in South Africa, it was found that fully vaccinated individuals were becoming infected at higher rates than previously seen.79 Indeed, one of the first studies to monitor vaccine effectiveness against Omicron found that two doses of the BNT162b2 mRNA vaccine produced by Pfizer and BioNTech provided only 33% efficacy against ‘breakthrough’ infection (compared with over 70% for Delta) and provided only 70% efficacy against hospitalization (compared with over 90% for Delta).88 Follow-up studies in several countries confirmed these results, but also found that administration of a third-dose booster shot significantly increased neutralizing antibody titre and vaccine efficacy against Omicron.89,90 Parallel studies at several sites likewise reported a drop in the effectiveness of prior infection in protecting against Omicron reinfection.91
Although this increased risk of vaccine breakthrough infections and reinfections resulted in a massive increase in Omicron cases, hospitalizations and deaths due to COVID-19 did not increase proportionally.92–94 In a study of over 1 million adults who tested positive for SARS-CoV-2 in the UK in December 2021, death certificate records verified that the risk of COVID-19-related death was 66% lower in people infected with Omicron compared with the Delta VOC, even when controlling for vaccination and prior infection.95 Several similar studies likewise found that the Omicron variant was associated with less severe disease and better patient outcomes.96,97 This, combined with a different disease presentation featuring increased likelihood of sore throat and decreased likelihood of loss of smell compared with other variants,98 suggests that the underlying disease pathogenesis of Omicron may be different from prior variants. It has been suggested that these changes may be due to the Omicron variant’s altered viral tropism due to use of an alternative cell entry pathway favouring the upper respiratory tract, but these studies are still ongoing.99,100
Since the emergence and global spread of the Omicron VOC, the virus has continued to diversify, with several successive sublineages outcompeting one another over time (BA.1, BA.2, BA.4, BA.5, XBB.1.5, etc.). Each of these sublineages has been found to have slightly different fitness advantages, with different immune escape mutations and neutralizing antibody resistance profiles, but all descend from the same parental lineage.101 As such, each sublineage has been classified as a ‘variant of interest’ (VOI) by the WHO, but none has been classified as a new VOC or issued a new Greek alphabet moniker.102 To date, no named VOC (Alpha, Beta, Delta, Gamma, Omicron) has descended from another VOC (Figure 4a), though the current global dominance of the Omicron sublineages makes it more likely that future variants will be derived from this genetic background.
Viral persistence and antiviral resistance
Intra-host longitudinal viral genomics studies have described the emergence of viral mutations in patients with COVID-19 over the course of a single infection. This viral evolution is most readily observed in the context of prolonged or persistent infections in immunocompromised hosts who have impaired viral clearance. For example, we reported the emergence of S mutations associated with immune escape in two immunosuppressed patients with persistent SARS-CoV-2 infection with detectable viral loads and intermittent symptoms over 106–138 days.70 In addition to increasing intra-host viral diversity over time, immune escape mutations at S:E484 and S:241–243del (characteristic of the Beta and Gamma VOCs) independently emerged in both individuals.103 Other studies have similarly described the development of immune escape mutations in SARS-CoV-2 during persistent infections, which is thought to be one source for the emergence of new variants with enhanced infection and/or immune evasion potential.104–107
Diversification in the SARS-CoV-2 genome at drug target sites could lead to the emergence of antiviral resistance, particularly in cases where there is persistent viral replication in hosts undergoing treatment with direct-acting antivirals such as remdesivir, molnupiravir or nirmatrelvir/ritonavir. Remdesivir is a nucleotide analogue incorporated by the viral RNA-dependent RNA polymerase (RdRp) into nascent viral transcripts resulting in premature chain termination and inhibition of viral replication.108,109 Mutations in nsp12 in ORF1b, which encodes the RdRp, have been reported in in vitro selection experiments (nsp12:V166L) and have been detected in patients being treated with remdesivir (nsp12:E802D and nsp12:V792I).110–112 Although remdesivir has been approved for the treatment of SARS-CoV-2 since May 2020 in the USA and many other countries, the frequency of these or other potentially resistance-associated mutations remains very low in global sequence databases.113 The frequency of 12 known or potential remdesivir resistance mutations catalogued by the Stanford Antiviral and Resistance Database (CoV-RDB) is at most 0.07% among sequenced SARS-CoV-2 genomes.114,115 Several of these mutations were found to come at a significant replicative fitness cost, which may explain their limited spread, though it is also possible that remdesivir does not elicit strong selective pressure in vivo.
Nirmatrelvir, the active component of nirmatrelvir/ritonavir, is an oral peptidomimetic that employs a nitrile electrophilic functional group, or ‘warhead’, to covalently bind the catalytic cysteine residue in the active site of the 3CL main protease (Mpro) required for processing and expression of critical viral proteins during replication.116,117 Protease mutations have been described in in vitro selection experiments, including Mpro:E166A and Mpro:L167F, which each confer low-level resistance to nirmatrelvir, as well as the triple mutant Mpro:L50F/E166A/L167F, which was 6 to 72 times more resistant to nirmatrelvir in biochemical assays.118 Notably, these substitutions were also associated with a significant reduction in Mpro enzymatic activity, implying a steep evolutionary fitness cost. To date, no single nucleotide mutations in Mpro have been described that both confer high-level resistance to nirmatrelvir/ritonavir and preserve enzyme function, suggesting that clinically significant resistance to nirmatrelvir/ritonavir likely requires multiple compensatory mutations. 119,120
Molnupiravir is the oral prodrug of β-d-N(4)-hydroxycytidine, a ribonucleoside recognized by RdRp (nsp12/ORF1b) that acts by inducing lethal mutagenesis, most often through transition mutations (G→A or C→U) made during viral replication.121 An examination of the global population structure of SARS-CoV-2 genome sequences revealed long phylogenetic branches enriched for transition mutations that occurred almost exclusively after 2022 when molnupiravir treatment was introduced, and that were also localized to countries and age groups in which molnupiravir was more heavily prescribed.122 Molnupiravir is thought to have a high barrier to resistance, and to date no studies have reported numerically significant, lineage-fixed mutations with clinical molnupiravir resistance.123 However, given its limited clinical performance and the inherent risk of inducing mutagenesis towards accelerated adaptive evolution, clinical usage of this agent over other alternative therapeutics is under question.124,125
Lessons learned and future considerations
Global efforts for genomic sequencing and surveillance during the COVID-19 pandemic have been historic in their scope and volume. Previous pathogen sequencing efforts played an important role in informing and supporting this global effort. After the first HIV genome sequences were published in 1985, sequencing was applied to characterizing antiviral resistance mutations, tracking potential risk networks for public health interventions, and studying global and within-host evolutionary patterns in HIV.126–130 Similarly, genomic surveillance of influenza viruses has been increasing in the last two decades with the goals of tracking the emergence of new viral strains, characterizing outbreaks within institutions or communities, and guiding vaccine development.131–134 Genome sequence databases, such as those maintained by the NCBI or GISAID, provided ready infrastructure able to be quickly adapted to the needs of SARS-CoV-2 data sharing.135 Similarly, sequencing techniques and analysis tools developed for the study of other viruses were rapidly adapted to the public health, clinical and research needs of the COVID-19 pandemic, underscoring the value of robust pathogen genomics expertise in responding to new pandemic threats.
With increasing natural and vaccine-based immunity and waning COVID-19 case counts worldwide, sequencing programmes and infrastructure that were implemented or scaled up to meet the demands of SARS-CoV-2 surveillance and study are being transitioned away from national-level efforts into more state or regional public health programmes or are being dismantled entirely. The challenge facing sequencing programmes in a period of contracting resources is in maintaining the appropriate balance of surveillance of persistent infectious public health threats and specimen banking capacity to quickly respond to outbreaks or pandemics with timely and effective pathogen sequencing. Unfortunately, despite the shared experience of the scientific community over the last 3 years and the improvements in genomic surveillance capacity and expertise gained over this time, several structural limitations that predated the COVID-19 pandemic remain largely in place. These include limited communication and coordination among public health, academic and private industry entities, lack of clearly defined central direction of surveillance efforts, and important patient privacy protections that can also present significant barriers to effective data sharing and interpretation. These limitations each serve to increase costs and reduce the effectiveness of pathogen genomic surveillance efforts for tracking, treating or preventing the next pandemic. Ongoing and future initiatives to improve data sharing, cross-centre communication and funding in this area are greatly needed.
Finally, the importance of boosting global contributions to pandemic surveillance cannot be overstated. The great majority of SARS-CoV-2 genomic sequencing has been performed in high-income countries, primarily in North America and Europe, with considerable disparities in sequencing volume from low-income countries around the world.136 To ensure rapid detection of the next pandemic threat and the greatest possible lead-time for developing and approving therapeutics and vaccines, a comprehensive global sequence-based surveillance network is needed. This will require building infrastructure and expertise for pathogen genomic sequencing not just in high-income regions, but also in locations at comparatively higher risk of zoonotic or environmental infection spillover events. These often include low-income countries and regions with higher amounts of human/wildlife interactions, greater population densities, or inadequate sanitation or healthcare resources.137 Investing in global genomic sequencing resources and training will allow for faster, more representative sampling and data availability for earlier detection of new pandemic threats, more timely and effective therapeutics development, and better targeted public health responses in affected countries and globally.
Contributor Information
Ramon Lorenzo-Redondo, Department of Medicine, Division of Infectious Diseases, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA; Center for Pathogen Genomics and Microbial Evolution, Northwestern University Havey Institute for Global Health, Chicago, IL 60611, USA.
Alexandre Machado de Sant’Anna Carvalho, Department of Medicine, Division of Infectious Diseases, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA; Center for Pathogen Genomics and Microbial Evolution, Northwestern University Havey Institute for Global Health, Chicago, IL 60611, USA.
Judd F Hultquist, Department of Medicine, Division of Infectious Diseases, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA; Center for Pathogen Genomics and Microbial Evolution, Northwestern University Havey Institute for Global Health, Chicago, IL 60611, USA.
Egon A Ozer, Department of Medicine, Division of Infectious Diseases, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA; Center for Pathogen Genomics and Microbial Evolution, Northwestern University Havey Institute for Global Health, Chicago, IL 60611, USA.
Funding
Funding for this work was provided by: a Dixon Translational Research Grant made possible by the generous support of the Dixon Family Foundation (E.A.O. and J.F.H.); two COVID-19 Supplemental Research awards from the National Institutes of Health’s (NIH’s) National Center for Advancing Translational Sciences (NCATS; UL1 TR001422—J.F.H., and UL1 TR002389—J.F.H., E.A.O., R.L.-R.); a supplement to the Northwestern University Cancer Center (P30 CA060553—J.F.H.); the NIH-supported Third Coast CFAR (P30 AI117943—R.L.-R., J.F.H.); NIH grant R21 AI163912 (J.F.H.); NIH grant U19 AI135964 (E.A.O.); NIH grant U19 AI171110 (J.F.H., E.A.O., R.L.-R.); and through a generous contribution from the Walder Foundation’s Chicago Coronavirus Assessment Network (Chicago CAN) Initiative (J.F.H., E.A.O., R.L.-R.). The funding sources had no role in the study design, data collection, analysis, interpretation or writing of the report. This paper was published as part of a supplement financially supported by an educational grant from Roche Molecular Systems.
Transparency declarations
J.F.H. has received research support, paid to Northwestern University, from Gilead Sciences and is a paid consultant for Merck. All other authors declare no conflicts of interest.
Author contributions
Conceptualization, R.L.-R., E.A.O, J.F.-H.; formal analysis, R.L.-R.; resources, R.L.-R., E.A.O., J.F.H.; writing—original draft, A.M.S.C., R.L.-R., E.A.O, J.F.H.; writing—review and editing, A.M.S.C., R.L.-R., E.A.O, J.F.H.; visualization, R.L.-R., E.A.O, J.F.H.; supervision, R.L.-R., E.A.O, J.F.H.; project administration, R.L.-R., E.A.O, J.F.H.; funding acquisition, R.L.-R., E.A.O, J.F.H.
References
- 1. Dolan PT, Whitfield ZJ, Andino R. Mapping the evolutionary potential of RNA viruses. Cell Host Microbe 2018; 23: 435–46. 10.1016/j.chom.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dolan PT, Whitfield ZJ, Andino R. Mechanisms and concepts in RNA virus population dynamics and evolution. Annu Rev Virol 2018; 5: 69–92. 10.1146/annurev-virology-101416-041718 [DOI] [PubMed] [Google Scholar]
- 3. Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol 2018; 16: e3000003. 10.1371/journal.pbio.3000003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lauring AS, Frydman J, Andino R. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 2013; 11: 327–36. 10.1038/nrmicro3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanjuan R. From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog 2012; 8: e1002685. 10.1371/journal.ppat.1002685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peck KM, Lauring AS. Complexities of viral mutation rates. J Virol 2018; 92: e01031-17. 10.1128/JVI.01031-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanjuan R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci 2016; 73: 4433–48. 10.1007/s00018-016-2299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev 2012; 76: 159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Domingo E, Perales C. Viral quasispecies. PLoS Genet 2019; 15: e1008271. 10.1371/journal.pgen.1008271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makeyev EV, Bamford DH. Evolutionary potential of an RNA virus. J Virol 2004; 78: 2114–20. 10.1128/JVI.78.4.2114-2120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zwart MP, Elena SF. Matters of size: genetic bottlenecks in virus infection and their potential impact on evolution. Annu Rev Virol 2015; 2: 161–79. 10.1146/annurev-virology-100114-055135 [DOI] [PubMed] [Google Scholar]
- 12. Gutierrez S, Michalakis Y, Blanc S. Virus population bottlenecks during within-host progression and host-to-host transmission. Curr Opin Virol 2012; 2: 546–55. 10.1016/j.coviro.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 13. Allman B, Koelle K, Weissman D. Heterogeneity in viral populations increases the rate of deleterious mutation accumulation. Genetics 2022; 222: iyac127. 10.1093/genetics/iyac127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lorenzo-Redondo R, Borderia AV, Lopez-Galindez C. Dynamics of in vitro fitness recovery of HIV-1. J Virol 2011; 85: 1861–70. 10.1128/JVI.01254-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elena SF, Sanjuán R. Virus evolution: insights from an experimental approach. Annu Rev Ecol Evol Syst 2007; 38: 27–52. 10.1146/annurev.ecolsys.38.091206.095637 [DOI] [Google Scholar]
- 16. Hill V, Ruis C, Bajaj Set al. Progress and challenges in virus genomic epidemiology. Trends Parasitol 2021; 37: 1038–49. 10.1016/j.pt.2021.08.007 [DOI] [PubMed] [Google Scholar]
- 17. Oude Munnink BB, Worp N, Nieuwenhuijse DFet al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med 2021; 27: 1518–24. 10.1038/s41591-021-01472-w [DOI] [PubMed] [Google Scholar]
- 18. Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev 2006; 8: 125–40. [PubMed] [Google Scholar]
- 19. Bedford T, Riley S, Barr IGet al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 2015; 523: 217–20. 10.1038/nature14460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Worobey M, Pekar J, Larsen BBet al. The emergence of SARS-CoV-2 in Europe and North America. Science 2020; 370: 564–70. 10.1126/science.abc8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gonzalez-Reiche AS, Hernandez MM, Sullivan MJet al. Introductions and early spread of SARS-CoV-2 in the New York city area. Science 2020; 369: 297–301. 10.1126/science.abc1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boni MF, Lemey P, Jiang Xet al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 2020; 5: 1408–17. 10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- 23. Rausch JW, Capoferri AA, Katusiime MGet al. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc Natl Acad Sci U S A 2020; 117: 24614–6. 10.1073/pnas.2017726117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu F, Zhao S, Yu Bet al. A new coronavirus associated with human respiratory disease in China. Nature 2020; 579: 265–9. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu N, Zhang D, Wang Wet al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727–33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quick J, Grubaugh ND, Pullan STet al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 2017; 12: 1261–76. 10.1038/nprot.2017.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis JJ, Long SW, Christensen PAet al. Analysis of the ARTIC version 3 and version 4 SARS-CoV-2 primers and their impact on the detection of the G142D amino acid substitution in the spike protein. Microbiol Spectr 2021; 9: e0180321. 10.1128/Spectrum.01803-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambisia AW, Mohammed KS, Makori TOet al. Optimization of the SARS-CoV-2 ARTIC network V4 primers and whole genome sequencing protocol. Front Med (Lausanne) 2022; 9: 836728. 10.3389/fmed.2022.836728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borcard L, Gempeler S, Terrazos Miani MAet al. Investigating the extent of primer dropout in SARS-CoV-2 genome sequences during the early circulation of Delta variants. Front Virol 2022; 2: 840952. 10.3389/fviro.2022.840952 [DOI] [Google Scholar]
- 30. Freed NE, Vlkova M, Faisal MBet al. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding. Biol Methods Protoc 2020; 5: bpaa014. 10.1093/biomethods/bpaa014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pembaur A, Sallard E, Weil PPet al. Simplified point-of-care full SARS-CoV-2 genome sequencing using nanopore technology. Microorganisms 2021; 9: 2598. 10.3390/microorganisms9122598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao M, Liu X, Ji Jet al. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med 2020; 12: 57. 10.1186/s13073-020-00751-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wen S, Sun C, Zheng Het al. High-coverage SARS-CoV-2 genome sequences acquired by target capture sequencing. J Med Virol 2020; 92: 2221–6. 10.1002/jmv.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagy-Szakal D, Couto-Rodriguez M, Wells HLet al. Targeted hybridization capture of SARS-CoV-2 and metagenomics enables genetic variant discovery and nasal microbiome insights. Microbiol Spectr 2021; 9: e0019721. 10.1128/Spectrum.00197-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fauver JR, Petrone ME, Hodcroft EBet al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell 2020; 181: 990–6.e5. 10.1016/j.cell.2020.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korber B, Fischer WM, Gnanakaran Set al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182: 812–27.e19. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorenzo-Redondo R, Nam HH, Roberts SCet al. A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways. EBioMedicine 2020; 62: 103112. 10.1016/j.ebiom.2020.103112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoffmann M, Kleine-Weber H, Schroeder Set al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–80.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J, Xiao T, Cai Yet al. Structure of SARS-CoV-2 spike protein. Curr Opin Virol 2021; 50: 173–82. 10.1016/j.coviro.2021.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Plante JA, Liu Y, Liu Jet al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021; 592: 116–21. 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J, Cai Y, Xiao Tet al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021; 372: 525–30. 10.1126/science.abf2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou B, Thao TTN, Hoffmann Det al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021; 592: 122–7. 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 43. Public Health England . Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01. Technical Briefing 3.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959360/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3.pdf
- 44. Rambaut A, Loman N, Pybus Oet al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological, 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- 45. Public Health England . Investigation of novel SARS-COV-2 variant: variant of concern 202012/01.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf
- 46. Technical Advisory Group on SARS-CoV-2 Virus Evolution . Historical working definitions and primary actions for SARS-CoV-2 variants. World Health Organization, 2023.https://www.who.int/publications/m/item/historical-working-definitions-and-primary-actions-for-sars-cov-2-variants
- 47. World Health Organization . Tracking SARS-CoV-2 variants.https://www.who.int/activities/tracking-SARS-CoV-2-variants
- 48. Tegally H, Wilkinson E, Giovanetti Met al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592: 438–43. 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- 49. Cherian S, Potdar V, Jadhav Set al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms 2021; 9: 1542. 10.3390/microorganisms9071542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mlcochova P, Kemp SA, Dhar MSet al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021; 599: 114–9. 10.1038/s41586-021-03944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Faria NR, Mellan TA, Whittaker Cet al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021; 372: 815–21. 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harvey WT, Carabelli AM, Jackson Bet al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021; 19: 409–24. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barton MI, MacGowan SA, Kutuzov MAet al. Effects of common mutations in the SARS-CoV-2 spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife 2021; 10: e70658. 10.7554/eLife.70658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tian F, Tong B, Sun Let al. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife 2021; 10: e69091. 10.7554/eLife.69091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim YJ, Jang US, Soh SMet al. The impact on infectivity and neutralization efficiency of SARS-CoV-2 lineage B.1.351 pseudovirus. Viruses 2021; 13: 633. 10.3390/v13040633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Starr TN, Greaney AJ, Hilton SKet al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020; 182: 1295–310.e20. 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Volz E, Mishra S, Chand Met al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021; 593: 266–9. 10.1038/s41586-021-03470-x [DOI] [PubMed] [Google Scholar]
- 58. Chen RE, Zhang X, Case JBet al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 2021; 27: 717–26. 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou D, Dejnirattisai W, Supasa Pet al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021; 184: 2348–61.e6. 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li Q, Nie J, Wu Jet al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 2021; 184: 2362–71.e9. 10.1016/j.cell.2021.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Corti D, Purcell LA, Snell Get al. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 2021; 184: 3086–108. 10.1016/j.cell.2021.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou H, Tada T, Dcosta BMet al. Neutralization of SARS-CoV-2 Omicron BA.2 by therapeutic monoclonal antibodies. bioRxiv 2022. 10.1101/2022.02.15.480166 [DOI] [PMC free article] [PubMed]
- 63. Cox M, Peacock TP, Harvey WTet al. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat Rev Microbiol 2023; 21: 112–24. 10.1038/s41579-022-00809-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilhelm A, Toptan T, Pallas Cet al. Antibody-mediated neutralization of authentic SARS-CoV-2 B.1.617 variants harboring L452R and T478K/E484Q. Viruses 2021; 13: 1693. 10.3390/v13091693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu H, Wei P, Zhang Qet al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to bamlanivimab in vitro. MAbs 2021; 13: 1919285. 10.1080/19420862.2021.1919285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Z, Schmidt F, Weisblum Yet al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021; 592: 616–22. 10.1038/s41586-021-03324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Choudhary MC, Chew KW, Deo Ret al. Emergence of SARS-CoV-2 escape mutations during bamlanivimab therapy in a phase II randomized clinical trial. Nat Microbiol 2022; 7: 1906–17. 10.1038/s41564-022-01254-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gupta A, Konnova A, Smet Met al. Host immunological responses facilitate development of SARS-CoV-2 mutations in patients receiving monoclonal antibody treatments. J Clin Invest 2023; 133: e166032. 10.1172/JCI166032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Destras G, Assaad S, Bal Aet al. Bamlanivimab as monotherapy in two immunocompromised patients with COVID-19. Lancet Microbe 2021; 2: e424. 10.1016/S2666-5247(21)00189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simons LM, Ozer EA, Gambut Set al. De novo emergence of SARS-CoV-2 spike mutations in immunosuppressed patients. Transpl Infect Dis 2022; 24: e13914. 10.1111/tid.13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choi B, Choudhary MC, Regan Jet al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383: 2291–3. 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weigang S, Fuchs J, Zimmer Get al. Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat Commun 2021; 12: 6405. 10.1038/s41467-021-26602-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen Z, Azman AS, Chen Xet al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet 2022; 54: 499–507. 10.1038/s41588-022-01033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cavazzoni P. Coronavirus (COVID-19) update: FDA limits use of certain monoclonal antibodies to treat COVID-19 due to the Omicron variant. Food and Drug Administration, 2022.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-limits-use-certain-monoclonal-antibodies-treat-covid-19-due-omicron
- 75. Wang H, Miller JA, Verghese Met al. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J Clin Microbiol 2021; 59: e0085921. 10.1128/JCM.00859-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vogels CBF, Breban MI, Ott IMet al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol 2021; 19: e3001236. 10.1371/journal.pbio.3001236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kidd M, Richter A, Best Aet al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis 2021; 223: 1666–70. 10.1093/infdis/jiab082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Galloway SE, Paul P, MacCannell DRet al. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep 2021; 70: 95–9. 10.15585/mmwr.mm7003e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Viana R, Moyo S, Amoako DGet al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. Nature 2022; 603: 679–86. 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. World Health Organization . Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern.https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 81. Paton RS, Overton CE, Ward T. The rapid replacement of the SARS-CoV-2 Delta variant by Omicron (B.1.1.529) in England. Sci Transl Med 2022; 14: eabo5395. 10.1126/scitranslmed.abo5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lambrou AS, Shirk P, Steele MKet al. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) variants—United States, June 2021-January 2022. MMWR Morb Mortal Wkly Rep 2022; 71: 206–11. 10.15585/mmwr.mm7106a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lundberg AL, Lorenzo-Redondo R, Ozer EAet al. Has Omicron changed the evolution of the pandemic? JMIR Public Health Surveill 2022; 8: e35763. 10.2196/35763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. VanBlargan LA, Errico JM, Halfmann PJet al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med 2022; 28: 490–5. 10.1038/s41591-021-01678-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhou H, Dcosta BM, Landau NRet al. Resistance of SARS-CoV-2 omicron BA.1 and BA.2 variants to vaccine-elicited sera and therapeutic monoclonal antibodies. Viruses 2022; 14: 1334. 10.3390/v14061334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bruel T, Hadjadj J, Maes Pet al. Serum neutralization of SARS-CoV-2 omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med 2022; 28: 1297–302. 10.1038/s41591-022-01792-5 [DOI] [PubMed] [Google Scholar]
- 87. Kuhlmann C, Mayer CK, Claassen Met al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 2022; 399: 625–6. 10.1016/S0140-6736(22)00090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Collie S, Champion J, Moultrie Het al. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N Engl J Med 2022; 386: 494–6. 10.1056/NEJMc2119270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thompson MG, Natarajan K, Irving SAet al. Effectiveness of a third dose of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and Omicron variant predominance—VISION network, 10 states, August 2021-January 2022. MMWR Morb Mortal Wkly Rep 2022; 71: 139–45. 10.15585/mmwr.mm7104e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Planas D, Saunders N, Maes Pet al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022; 602: 671–5. 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 91. Pulliam JRC, van Schalkwyk C, Govender Net al. Increased risk of SARS-CoV-2 reinfection associated with emergence of omicron in South Africa. Science 2022; 376: eabn4947. 10.1126/science.abn4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nyberg T, Ferguson NM, Nash SGet al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet 2022; 399: 1303–12. 10.1016/S0140-6736(22)00462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Iuliano AD, Brunkard JM, Boehmer TKet al. Trends in disease severity and health care utilization during the early omicron variant period compared with previous SARS-CoV-2 high transmission periods—United States, December 2020-January 2022. MMWR Morb Mortal Wkly Rep 2022; 71: 146–52. 10.15585/mmwr.mm7104e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wolter N, Jassat W, Walaza Set al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet 2022; 399: 437–46. 10.1016/S0140-6736(22)00017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ward IL, Bermingham C, Ayoubkhani Det al. Risk of COVID-19 related deaths for SARS-CoV-2 omicron (B.1.1.529) compared with delta (B.1.617.2): retrospective cohort study. BMJ 2022; 378: e070695. 10.1136/bmj-2022-070695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mayr FB, Talisa VB, Castro ADet al. COVID-19 disease severity in US veterans infected during Omicron and Delta variant predominant periods. Nat Commun 2022; 13: 3647. 10.1038/s41467-022-31402-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sievers C, Zacher B, Ullrich Aet al. SARS-CoV-2 Omicron variants BA.1 and BA.2 both show similarly reduced disease severity of COVID-19 compared to Delta, Germany, 2021 to 2022. Euro Surveill 2022; 27: 2200396. 10.2807/1560-7917.ES.2022.27.22.2200396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Menni C, Valdes AM, Polidori Let al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID study. Lancet 2022; 399: 1618–24. 10.1016/S0140-6736(22)00327-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Meng B, Abdullahi A, Ferreira Iet al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022; 603: 706–14. 10.1038/s41586-022-04474-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhao H, Lu L, Peng Zet al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 2022; 11: 277–83. 10.1080/22221751.2021.2023329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cao Y, Yisimayi A, Jian Fet al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022; 608: 593–602. 10.1038/s41586-022-04980-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. World Health Organization . Statement on the update of WHO’s working definitions and tracking system for SARS-CoV-2 variants of concern and variants of interest. 2023. https://www.who.int/news/item/16-03-2023-statement-on-the-update-of-who-s-working-definitions-and-tracking-system-for-sars-cov-2-variants-of-concern-and-variants-of-interest [PMC free article] [PubMed]
- 103. Weisblum Y, Schmidt F, Zhang Fet al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020; 9: e61312. 10.7554/eLife.61312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wrobel AG, Benton DJ, Roustan Cet al. Evolution of the SARS-CoV-2 spike protein in the human host. Nat Commun 2022; 13: 1178. 10.1038/s41467-022-28768-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moelling K. Within-host and between-host evolution in SARS-CoV-2-new variant’s source. Viruses 2021; 13: 751. 10.3390/v13050751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cele S, Karim F, Lustig Get al. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 2022; 30: 154–62.e5. 10.1016/j.chom.2022.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Voloch CM, da Silva Francisco R Jr, de Almeida LGPet al. Intra-host evolution during SARS-CoV-2 prolonged infection. Virus Evol 2021; 7: veab078. 10.1093/ve/veab078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gordon CJ, Tchesnokov EP, Woolner Eet al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 2020; 295: 6785–97. 10.1074/jbc.RA120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tchesnokov EP, Gordon CJ, Woolner Eet al. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem 2020; 295: 16156–65. 10.1074/jbc.AC120.015720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Checkmahomed L, Carbonneau J, Du Pont Vet al. In vitro selection of remdesivir-resistant SARS-CoV-2 demonstrates high barrier to resistance. Antimicrob Agents Chemother 2022; 66: e0019822. 10.1128/aac.00198-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gandhi S, Klein J, Robertson AJet al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat Commun 2022; 13: 1547. 10.1038/s41467-022-29104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hogan JI, Duerr R, Dimartino Det al. Remdesivir resistance in transplant recipients with persistent coronavirus disease 2019. Clin Infect Dis 2023; 76: 342–5. 10.1093/cid/ciac769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Focosi D, Maggi F, McConnell Set al. Very low levels of remdesivir resistance in SARS-COV-2 genomes after 18 months of massive usage during the COVID19 pandemic: a GISAID exploratory analysis. Antiviral Res 2022; 198: 105247. 10.1016/j.antiviral.2022.105247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tzou PL, Tao K, Pond SLKet al. Coronavirus resistance database (CoV-RDB): SARS-CoV-2 susceptibility to monoclonal antibodies, convalescent plasma, and plasma from vaccinated persons. PLoS One 2022; 17: e0261045. 10.1371/journal.pone.0261045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Alisoltani A, Jaroszewski L, Godzik Aet al. Viralvar: a web tool for multilevel visualization of SARS-CoV-2 genomes. Viruses 2022; 14: 2714. 10.3390/v14122714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Owen DR, Allerton CMN, Anderson ASet al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 2021; 374: 1586–93. 10.1126/science.abl4784 [DOI] [PubMed] [Google Scholar]
- 117. Bai B, Arutyunova E, Khan MBet al. Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors. RSC Med Chem 2021; 12: 1722–30. 10.1039/D1MD00247C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jochmans D, Liu C, Donckers Ket al. The substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. mBio 2023; 14: e0281522. 10.1128/mbio.02815-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Iketani S, Mohri H, Culbertson Bet al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature 2023; 613: 558–64. 10.1038/s41586-022-05514-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hu Y, Lewandowski EM, Tan Het al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. ACS Cent Sci 2023; 9: 1658–69. 10.1021/acscentsci.3c00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gordon CJ, Tchesnokov EP, Schinazi RFet al. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem 2021; 297: 100770. 10.1016/j.jbc.2021.100770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sanderson T, Hisner R, Donovan-Banfield IAet al. Identification of a molnupiravir-associated mutational signature in SARS-CoV-2 sequencing databases. medRxiv 2023: 2023.01.26.23284998. 10.1101/2023.01.26.23284998 [DOI] [PMC free article] [PubMed]
- 123. Agostini ML, Pruijssers AJ, Chappell JDet al. Small-molecule antiviral beta-d-N(4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J Virol 2019; 93: e01348-19. 10.1128/JVI.01348-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Butler CC, Hobbs FDR, Gbinigie OAet al. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet 2023; 401: 281–93. 10.1016/S0140-6736(22)02597-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Huang P-Y, Liu T-H, Wu J-Yet al. Clinical efficacy and safety of molnupiravir for nonhospitalized and hospitalized patients with COVID-19: a systematic review and meta-analysis of randomized control trials. J Med Virol 2023; 95: e28621. 10.1002/jmv.28621 [DOI] [PubMed] [Google Scholar]
- 126. Wain-Hobson S, Sonigo P, Danos Oet al. Nucleotide sequence of the AIDS virus, LAV. Cell 1985; 40: 9–17. 10.1016/0092-8674(85)90303-4 [DOI] [PubMed] [Google Scholar]
- 127. Ratner L, Haseltine W, Patarca Ret al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 1985; 313: 277–84. 10.1038/313277a0 [DOI] [PubMed] [Google Scholar]
- 128. Callegaro A, Di Filippo E, Astuti Net al. Early clinical response and presence of viral resistant minority variants: a proof of concept study. J Int AIDS Soc 2014; 17: 19759. 10.7448/IAS.17.4.19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Swenson LC, Daumer M, Paredes R. Next-generation sequencing to assess HIV tropism. Curr Opin HIV AIDS 2012; 7: 478–85. 10.1097/COH.0b013e328356e9da [DOI] [PubMed] [Google Scholar]
- 130. Luk KC, Berg MG, Naccache SNet al. Utility of metagenomic next-generation sequencing for characterization of HIV and human pegivirus diversity. PLoS One 2015; 10: e0141723. 10.1371/journal.pone.0141723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ghedin E, Sengamalay NA, Shumway Met al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005; 437: 1162–6. 10.1038/nature04239 [DOI] [PubMed] [Google Scholar]
- 132. Briand FX, Schmitz A, Ogor Ket al. Emerging highly pathogenic H5 avian influenza viruses in France during winter 2015/16: phylogenetic analyses and markers for zoonotic potential. Euro Surveill 2017; 22: 30473. 10.2807/1560-7917.ES.2017.22.9.30473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Houlihan CF, Frampton D, Ferns RBet al. Use of whole-genome sequencing in the investigation of a nosocomial influenza virus outbreak. J Infect Dis 2018; 218: 1485–9. 10.1093/infdis/jiy335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. MacFadden DR, McGeer A, Athey Tet al. Use of genome sequencing to define institutional influenza outbreaks, Toronto, Ontario, Canada, 2014-15. Emerg Infect Dis 2018; 24: 492–7. 10.3201/eid2403.171499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill 2017; 22: 30494. 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ohlsen EC, Hawksworth AW, Wong Ket al. Determining gaps in publicly shared SARS-CoV-2 genomic surveillance data by analysis of global submissions. Emerg Infect Dis 2022; 28: S85–92. 10.3201/eid2813.220780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ling-Hu T, Rios-Guzman E, Lorenzo-Redondo Ret al. Challenges and opportunities for global genomic surveillance strategies in the COVID-19 era. Viruses 2022; 14: 2532. 10.3390/v14112532 [DOI] [PMC free article] [PubMed] [Google Scholar]