Abstract
Restriction fragment length polymorphism (RFLP) analysis of isolates of Cryptosporidium parvum has revealed two subgroups, termed H and C. The limited resolution of the RFLP method precludes an in-depth study of the genetic structure of C. parvum populations. Published C. parvum restriction polymorphisms lie within protein-coding regions known to be more homogeneous than noncoding sequences. To better assess the degrees of heterogeneity between and within C. parvum isolates, sequence polymorphism in the β-tubulin intron, the only C. parvum intron described to date, was investigated. In contrast to the two genotypes distinguished by multilocus RFLP, several alleles were detected by sequence and RFLP analysis of the β-tubulin intron and adjacent exon 2. Isolates carrying different β-tubulin alleles were found. Significantly, one of the β-tubulin alleles present in two geographically unrelated isolates combined features of C- and H-type isolates, suggesting that it might have arisen from a recombination event. A comparison of multiple samples of a calf-propagated laboratory isolate showed that the ratio of different β-tubulin alleles fluctuated during serial passage.
Cryptosporidium parvum is an enteric protozoan parasite of clinical and veterinary importance (7, 12). Phenotypic and genotypic characterizations of isolates obtained from human and animal hosts have demonstrated the occurrence of two subgroups (2, 3, 5, 6, 9, 10, 11, 14, 15). Restriction fragment length polymorphism (RFLP) analysis has been applied in several laboratories to characterize different isolates and assess the occurrence of C. parvum genotypes in human and animal hosts (1, 3, 6, 11, 15). These studies identified a group of isolates with identical RFLP genotypes which are found in humans only (termed H) and a second group which infects both humans and calves (termed C) (15). This finding has led some investigators to propose the existence of two transmission cycles, one exclusively anthroponotic and the other involving both human and animal hosts (2, 11).
In order to overcome the limited resolution achieved with RFLP, sequence polymorphism in the 84-bp intron within the β-tubulin gene (4) was investigated. The choice of this sequence was based on previous observations of extensive sequence polymorphism within another C. parvum intergenic region, the ribosomal internal transcribed spacer 1 (5, 8). The advantage of the β-tubulin intron over internal transcribed spacer 1 is that the β-tubulin gene is presumed to be single copy (4, 7a, 10a), whereas multiple and heterogenous copies of the ribosomal transcription unit are dispersed in the genome of C. parvum (8). This unusual feature of the rRNA loci makes them less suitable for genetic fingerprinting because of the difficulty in distinguishing between intragenomic heterogeneity and genotypically mixed samples.
Here we report on the identification of several β-tubulin alleles in C. parvum and on the correlation of this polymorphism with previously described RFLP and PCR markers. In contrast to previously published genetic markers in C. parvum, RFLP analysis of the β-tubulin locus identified several alleles within both RFLP subgroups.
MATERIALS AND METHODS
C. parvum isolates.
The host origin given for each isolate identifies the host from which the oocysts were recovered. Human isolates passaged through calves are listed under bovine isolates. This group includes isolates GCH1, CISD, Peru1, and Peru2. Isolates recovered directly from humans or primates are listed under the corresponding subheading.
(i) Human.
Isolates 2066K, 0541L, 0676I, and 0583K (15) were obtained from human immunodeficiency virus-positive individuals enrolled in a phase II/III study of nitazoxanide for cryptosporidiosis conducted by the National Institute of Allergy and Infectious Diseases AIDS Clinical Trials Group (ACTG 336). Isolate F16 was from the United Kingdom. OH was obtained from an immunocompetent individual from Ohio.
(ii) Bovine.
Isolate GCH1 originated from an AIDS patient and was maintained in calves at Tufts University School of Veterinary Medicine for over 6 years (13). Isolate Peru originated from a Peruvian patient and was passaged through calves at the University of Arizona. Peru1 and Peru2 designate two subsequent passages. Isolate UCP is believed to be of bovine origin and has been maintained in calves for over 8 years at ImmuCell Corp., Portland, Maine. Isolate CISD was originally isolated from a human immunodeficiency virus-negative patient and passaged through a calf. ICP is a bovine isolate from Idaho. TAMU was originally isolated from a foal, accidentally transmitted to a human, and thereafter maintained in calves.
(iii) Primate.
PC1 originated directly from a captive macaque at the New England Regional Primate Center, Southboro, Mass.
Oocyst purification and DNA extraction.
Oocysts were purified from C. parvum-positive stool by using a salt flotation step followed by centrifugation on a Nycodenz (Sigma) step gradient. Briefly, stool from which coarse debris had been removed by filtration through gauze or low-speed centrifugation (500 × g) was mixed with 2 volumes of saturated NaCl solution and centrifuged at 1,000 × g for 15 min. Oocysts were recovered from the supernatant diluted with 3 parts of water by centrifugation at 4,000 × g for 15 min. Oocysts were resuspended in a small volume of water and further purified by sedimentation at 100,000 × g on a 15%-30% (wt/vol) Nycodenz step gradient in phosphate-buffered saline for 1 h. DNA for genetic analysis was extracted from purified oocysts or, alternatively, directly from stool following an overnight incubation in 0.2% sodium dodecyl sulfate–200 μg of proteinase K per ml in water at 45°C as described previously (5).
PCR amplification and RFLP and sequence analyses of the β-tubulin gene.
A 538-bp fragment of the C. parvum β-tubulin gene (GenBank accession no. Y12615) spanning an 81- to 87-bp intron was amplified with sense primer btub5 (5′GATTGGTGCTAAATTCTGGG3′), located at positions 165 to 184, and antisense primer btub2 (5′GTCTGCAAAATACGATCTGG3′), at positions 708 to 689 (numbering according to GenBank accession no. Y12615). In some experiments btub4 (5′CCTGATCCTGTACCACCTCC3′; positions 648 to 629) was used instead of btub2. The GenBank sequence under accession no. Y12615 contains a 6-bp (ACTGGT) duplication at the 5′ end of exon 2, which was not seen in any of the samples sequenced here and was assumed to be an artifact. This resulted in a 6-bp discrepancy between size estimates derived from the GenBank entry and our sequences. Amplifications were performed with Taq DNA polymerase and 35 cycles of 94°C for 50 s, 52°C for 1 min, and 72°C for 50 s, with a 5-min extension at 72°C. For RFLP analysis, PCR products were digested with restriction enzyme Tsp509I (New England Biolabs, Beverly, Mass.) in PCR buffer (10 mM Tris-HCl [pH 9], 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2) for 30 to 60 min at 65°C.
PCR products were cloned in the pCRII vector (Invitrogen, Carlsbad, Calif.). Recombinant plasmids were purified from color-selected transformed Escherichia coli colonies and sequenced at the Tufts University School of Medicine core facility. The following plasmids were sequenced on both strands: icp-1, tamu-2, tamu-5, 0541L-5, 0541L-6, 0541L-8, 2066K-5, and 0583K-8. The remaining clones were sequenced on one strand.
RESULTS
RFLP analysis.
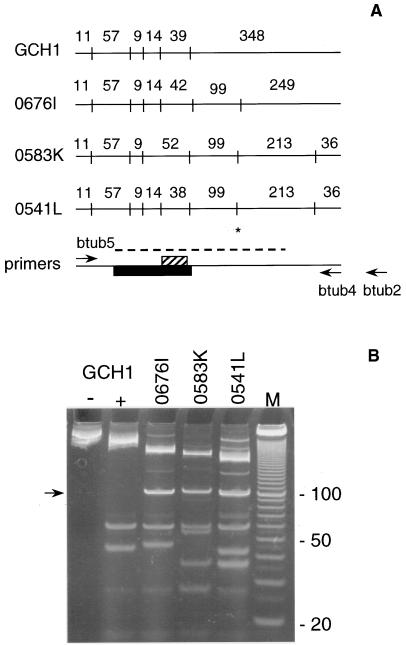
RFLP analysis was performed on a 478-bp PCR fragment of the β-tubulin gene defined by primers btub5 and btub4 (Fig. 1A). This amplicon includes the 3′ portion of exon 1, the intron, and the 5′ portion of exon 2. For RFLP analysis, the enzyme Tsp509I (/AATT) was used because of the high frequency with which this enzyme was expected to cut the AT-rich sequences typically found in C. parvum. Seven Tsp509I sites were present within this region, three of which were polymorphic (Fig. 1A). Two of these restriction polymorphisms, located at positions 399 and 613 within exon 2, segregated, with some exceptions, with the H and C genotypes. The allele with an intact Tsp509I site at position 399 (defined as Tsp509I+) was present in all subgroup H isolates. However, the reverse was not consistently found, as seen in isolate 0676I, which was classified as type C according to previous RFLP typing (15) yet was Tsp509I+. On the other hand, the β-tubulin allele which lacks the Tsp509I site at position 399 (Tsp509I−) was found in a subset of human isolates and in all bovine isolates. These are isolates which were classified as genotype C (15). Most samples could unambiguously be classified as Tsp509I+ or Tsp509I− because of the presence or absence of a diagnostic 99-bp restriction fragment. However, a number of isolates displayed mixed profiles, suggesting that both Tsp509I+ and Tsp509I− alleles were present in subgroups C and H (not shown).
FIG. 1.
(A) Restriction maps of four β-tubulin alleles of C. parvum. The fragment sizes are indicated for each allele. The approximate locations of the Tsp509I sites (vertical lines), the intron (black box), and the T repeat (hatched box) were derived from multiple, aligned sequences. The PCR fragment is 478 bp long in GCH1. The codes for the isolates from which these sequences were amplified are shown at the left. Arrows show the approximate locations of the PCR primers used in this study. btub2 is located 60 bp downstream of btub4 and, together with primer btub5, amplifies a 538-bp fragment. The location of the restriction site defining the Tsp509I+ and Tsp509I− alleles is marked (*). The dashed line indicates the approximate range of the sequence included in the alignment shown in Fig. 2. (B) Tsp509I restriction profiles of isolates shown in panel A. GCH1 lanes − and + designate unrestricted and restricted btub5-btub4 PCR products, respectively. The arrow indicates the 99-bp fragment mentioned in the text. Lane M, 100-bp DNA ladder. The sizes (in base pairs) of three bands are shown at the right.
The seven Tsp509I sites found within the β-tubulin PCR fragment occurred in different combinations in human isolates, which gave rise to different restriction profiles (Fig. 1B). Contributing to the heterogeneity among profiles was the T repeat of variable length located within a 38- to 52-bp restriction fragment (Fig. 1A). The presence of the Tsp509I site at position 399 in exon 2 resulted in a 99-bp restriction fragment, typical of genotype H.
Sequence analysis.
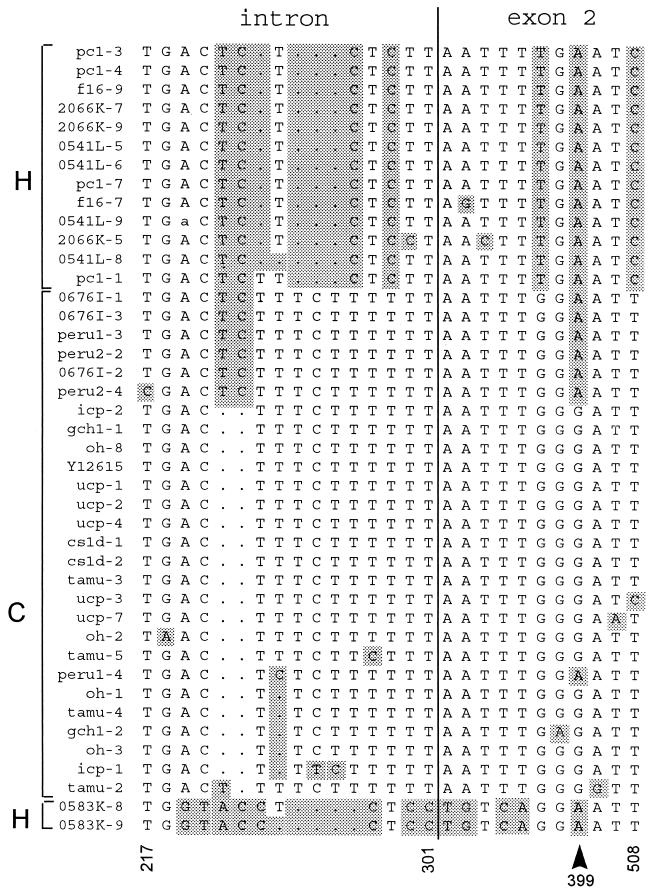
Sequence analysis of the intron and adjacent exon 2 was performed to obtain a more detailed view of heterogeneity at this locus. PCR products amplified from C. parvum DNA originating from human and animal infections were cloned, and multiple, randomly picked recombinant plasmids were sequenced. The alignment of 42 β-tubulin sequences ranging from 281 to 287 bp in length confirmed the expected high level of sequence polymorphism within the intron. Sixteen of 87 intron positions (18%) were polymorphic. In contrast, 11 of 200 positions (6%) within exon 2 were polymorphic (Fig. 2). The existence of an intron within this amplicon was confirmed by reverse transcription-PCR amplification of oocyst RNA (not shown).
FIG. 2.
Sequence alignment of β-tubulin sequences from 14 C. parvum isolates. The alignment was generated with the PileUp program (Wisconsin Sequence Analysis Package). Isolate codes are shown on the left. The number following the hyphen designates the plasmid number. The multilocus RFLP genotype (15) is indicated leftmost. Only the polymorphic positions are shown. Changes from the sequence under GenBank accession no. Y12615 are shaded. A 6-bp duplication in Y12615 presumed to be artifactual was omitted (see text). Selected positions are numbered below the sequences according to accession no. Y12615. The intron-exon boundary is indicated with a vertical line. The arrowhead at position 399 indicates the 5′-most nucleotide of a polymorphic Tsp509I site in exon 2. Clones icp-1, tamu-2, tamu-5, 0541L-5, 0541L-6, 0541L-8, 2066K-5, and 0583K-8 were sequenced on both strands. All others were sequenced on one strand. A sequence ambiguity in 0541L-9 is indicated in lowercase.
The β-tubulin sequence alignment defined four groups of β-tubulin alleles. Two of these matched the genotypes H and C described previously (6, 15). A third group was comprised of isolates 0676I, Peru2, and one sequence of Peru1. On the basis of RFLP analysis, these isolates were assigned to subgroup C. Lastly, two clones from human isolate 0583K constituted a fourth group. Two β-tubulin sequences originating from this isolate revealed numerous differences from the other alleles.
The comparison of multiple clones from individual isolates revealed intraisolate heterogeneity. For example, in isolate Peru, three clones grouped with the 0676I sequences and one was related to the sequences found in other C isolates. In contrast, there was a high degree of homogeneity among C isolates, regardless of their human or bovine source.
The relationship between the C, H, and 0676I-Peru sequences suggested that the last group arose from multiple recombination events between the H and C sequences. This assumption was based on the observation that the 0676I-Peru sequences alternately share polymorphic nucleotides with the H and the C groups. For example, this is apparent within the intron, where the 0676I-Peru group shares a polymorphic T and C with the H group and then six positions with the C group (Fig. 2). This recombinant sequence could have arisen from one crossover event in the intron and two in the exon, flanking position 399.
Variable ratio of β-tubulin alleles in a calf-propagated isolate.
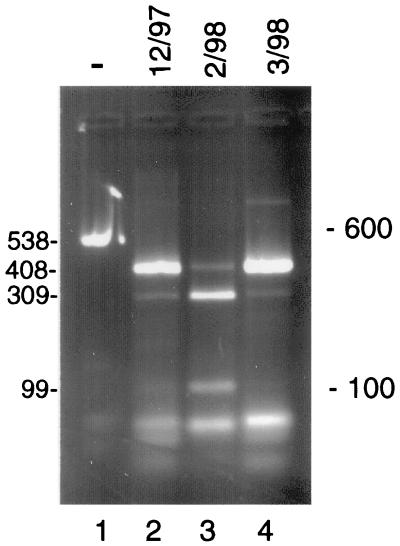
RFLP analysis of β-tubulin PCR products amplified from different calf passages of isolate GCH1 revealed a changing ratio of Tsp509I+ and Tsp509I− alleles. This resulted in restriction profiles with variable ratios of uncut (408-bp) to cut (309- and 99-bp) fragments. This is illustrated by three calf passages of isolate GCH1 collected between December 1997 and March 1998 (Fig. 3). The December and March samples showed a predominance of Tsp509I− DNA, while the sample from February showed the opposite pattern. In contrast to the β-tubulin profile, no changes were seen in the calf-propagated samples with other RFLP markers. All GCH1 samples consistently displayed genotype C at these loci (not shown). Similar changes in the ratio of β-tubulin alleles were made on GCH1 passaged in mice (not shown).
FIG. 3.
Variable β-tubulin restriction profiles in GCH1 oocysts from three calf passages. A 538-bp fragment was PCR amplified with primers btub5 and btub2 from three samples of GCH1 oocyst DNA collected from calf passages at the dates shown (lanes 2 to 4). The difference in restriction profiles was caused by variable ratios of β-tubulin alleles with or without a Tsp509I site at position 399. The sizes of the uncut PCR product (lane 1) and the relevant restriction fragments are shown (in base pairs) at the left. The length of the uncut PCR product (538 bp) and digests (408 bp or 309 and 99 bp) is not additive due to the presence of several unresolved small fragments totalling 129 to 133 bp (Fig. 1A). The positions of 600- and 100-bp size markers are indicated on the right.
In view of the unexpected variability of the β-tubulin restriction profiles, a control experiment was performed to rule out possible artifacts. First, we ruled out incomplete digestion with Tsp509I, since the full-length, 538-bp PCR product (Fig. 3, lane 1) was not visible in the digests (lanes 2 to 4). Second, in order to confirm that the mixed β-tubulin RFLP profiles originated from genotypically mixed C. parvum DNA, the following control experiment was performed. PCR products were amplified from DNA samples showing an approximately equal ratio of Tsp509I+ and Tsp509I− DNA and cloned into a plasmid vector. Recombinant plasmids were subjected individually to PCR amplification with primers btub5 and btub2 and were RFLP typed with Tsp509I. Among nine plasmids analyzed in this manner, five were Tsp509I− and four were Tsp509I+ (not shown). As to be expected from cloned sequences, no mixed patterns were observed. This observation supports our interpretation that mixed Tsp509I restriction profiles originated from mixed DNA templates. Mixed β-tubulin RFLP profiles were also found in some human isolates of genotype H, indicating that isolates classified as genotype C or H may carry different β-tubulin alleles.
DISCUSSION
The sequence alignment and the RFLP analysis of the C. parvum β-tubulin intron and 5′ portion of exon 2 were consistent with the existence of two main subgroups in this species. In addition, this analysis demonstrated the presence of additional polymorphisms within each subgroup and revealed two new subgroups, one of them showing significant divergence. The occurrence of additional heterogeneity is consistent with earlier genotypic and phenotypic observations. Specifically, sequence heterogeneity was observed among genotype H (genotype 1) isolates at the TRAP-C2 locus (10) and among different PCR clones amplified from the poly(T) locus (14). Differences in virulence in tissue culture, as defined by the release of lactate dehydrogenase and decrease in transmonolayer resistance, were also consistent with heterogeneity within genetic subgroups (15).
Both newly identified subgroups are of interest, the 0676I-Peru group because it shows sequence patterns suggestive of a recombination between alleles found in the H and C subgroups and the 05483K sequences for showing considerable divergence from other β-tubulin alleles. Sequences suggestive of recombination between the H and C genotypes were unexpected, since the two subgroups are assumed to be reproductively isolated. This view is based on the absence of recombinant isolates as defined by multilocus RFLP analysis. With respect to the unusual 05483K β-tubulin sequence, a unique restriction polymorphism was found in the poly(T) locus of this isolate (14a), a further indication that isolate 05483K possesses a distinct genotype.
The high frequency of mixed β-tubulin genotypes contrasts with observations on other RFLP markers, which rarely display mixed profiles (6). We assume that a certain proportion of natural populations of C. parvum are genotypically mixed and that the examination of other highly variable loci will reveal additional mixed profiles.
As exemplified by the GCH1 β-tubulin RFLPs, the ratio between Tsp509I+ and Tsp509I− alleles is unstable. The reason for this phenomenon is unknown. Since changes were seen during serial passage in calves, it cannot be excluded that new β-tubulin alleles are imported with the calves prior to infection. In this context, it is interesting that in two calf-propagated isolates (GCH1 and Peru), a predominantly Tsp509I+ profile coincided with low infectivity in calves. As pointed out above, Tsp509I+ alleles are not frequently seen in calves. It is presently unclear whether different β-tubulin alleles have any phenotypic relevance or are linked to other phenotypically relevant traits. Given a sufficient number of isolates, it should be possible to address this question by assaying isolates bearing different β-tubulin alleles for virulence in tissue culture or in mice. Evidence against any phenotypic significance of the β-tubulin polymorphism is the fact that the majority of polymorphisms within exon 2 were silent. Substitutions changing the amino acid sequence were present in single clones only, suggesting that they may be polymerase or sequencing artifacts.
Although new information from previous studies of genotypic polymorphism in C. parvum was gained through sequence analysis, this approach is time-consuming and expensive. Based on the sequence information gained from this study, it is now possible to design PCR-based methods capable of directly discriminating between β-tubulin alleles. To this end, PCR primers flanking the T repeat located within the intron are being evaluated in parallel with the Tsp509I RFLP profiles for routine isolate characterization. The β-tubulin marker alone or together with microsatellite markers under development should provide faster and less expensive methods for genotyping C. parvum isolates. The present study validates the idea that untranslated regions are suitable targets for such applications. The analysis of hypervariable loci will facilitate the identification of clinically relevant markers and find application in epidemiological studies.
ACKNOWLEDGMENTS
This work was supported by USDA grant 94-371020914 and NIH Cooperative Agreement U019AI33384.
Our thanks go to Carl Fichtenbaum, Jeff Griffiths, and the NIH ACTG 336 team, patients, and site personnel for providing samples, to Heidi Scaltreto for technical assistance, and to Sylvie Le Blancq and Mike Piper for mapping the β-tubulin gene. Samples were kindly provided by Lucy Ward (Ohio State University), Marilyn Marshall (University of Arizona), Joe Crabb (ImmuCell, Portland, Maine), Furio Spano (University of Rome, Rome, Italy), Karen Snowden (Texas A&M University), Hal Stibbs (Waterborne, New Orleans, La.), and Keith Mansfield (New England Regional Primate Center, Southboro, Mass.).
REFERENCES
- 1.Awad-El-Kariem F M, Robinson H A, Dyson D A, Evans D, Wright S, Fox T M, McDonald V. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 2.Awad-El-Kariem F M, Robinson H A, Petry F, McDonald V, Evans D, Casemore D. Differentiation between human and animal isolates of Cryptosporidium parvum using molecular and biological markers. Parasitol Res. 1998;84:297–301. doi: 10.1007/s004360050399. [DOI] [PubMed] [Google Scholar]
- 3.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Toubas D, Gabriel-Pospisil F, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 4.Cacciò S, La Rosa G, Pozio E. The beta-tubulin gene of Cryptosporidium parvum. Mol Biochem Parasitol. 1997;89:307–311. doi: 10.1016/s0166-6851(97)00122-9. [DOI] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carraway M, Tzipori S, Widmer G. New restriction fragment length polymorphism marker in Cryptosporidium parvum identifies mixed parasite populations and genotypic instability in response to host change. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrant R L. Cryptosporidiosis: an emerging, highly infectious threat. Emerg Infect Dis. 1997;3:51–57. doi: 10.3201/eid0301.970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Le Blancq, S. (Columbia University). Personal communication.
- 8.Le Blancq S M, Khramtsov N, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 9.Morgan U M, Constantine C C, O’Donoghue P, Meloni B P, O’Brien P A, Thompson R C A. Molecular characterization of Cryptosporidium isolates from human and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 10.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence for two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Piper, M. (Medical Research Council, Cambridge, United Kingdom). Personal communication.
- 11.Spano F, Putignani L, McLaughlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;152:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 12.Tzipori S. Cryptosporidiosis in perspective. Adv Parasitol. 1988;27:63–130. doi: 10.1016/S0065-308X(08)60353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzipori S, Rand W, Griffiths J, Widmer G, Crabb J. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin Diagn Lab Immunol. 1994;1:450–463. doi: 10.1128/cdli.1.4.450-463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:224–241. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 14a.Widmer, G., L. Tchack, and S. Tzipori. Unpublished data.
- 15.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]