Abstract
Biological pathways between alcohol consumption and alcohol liver disease (ALD) are not fully understood. We selected genes with known effect on (1) alcohol consumption, (2) liver function, and (3) gene expression. Expression of the orthologs of these genes in Caenorhabditis elegans and Drosophila melanogaster was suppressed using mutations and/or RNA interference (RNAi). In humans, association analysis, pathway analysis, and Mendelian randomization analysis were performed to identify metabolic changes due to alcohol consumption. In C. elegans, we found a reduction in locomotion rate after exposure to ethanol for RNAi knockdown of ACTR1B and MAPT. In Drosophila, we observed (1) a change in sedative effect of ethanol for RNAi knockdown of WDPCP, TENM2, GPN1, ARPC1B, and SCN8A, (2) a reduction in ethanol consumption for RNAi knockdown of TENM2, (3) a reduction in triradylglycerols (TAG) levels for RNAi knockdown of WDPCP, TENM2, and GPN1. In human, we observed (1) a link between alcohol consumption and several metabolites including TAG, (2) an enrichment of the candidate (alcohol-associated) metabolites within the linoleic acid (LNA) and alpha-linolenic acid (ALA) metabolism pathways, (3) a causal link between gene expression of WDPCP to liver fibrosis and liver cirrhosis. Our results imply that WDPCP might be involved in ALD.
Subject terms: Genetics, Biomarkers, Diseases, Health care, Molecular medicine
Introduction
Alcohol consumption is a major public health concern and is responsible for over 5% of the global burden of disease1. It has been known for a long time that excessive drinking leads to a range of molecular changes including acceleration of hepatic lipogenesis which leads to liver pathologies, such as liver fibrosis and liver cirrhosis commonly known as alcoholic liver disease (ALD). Although some of the known alcohol behavior genes such as ADH and CYP2E1 genes have a known role in hepatic lipogenesis2, the full picture of the biological pathways and molecular changes occurring as a result of alcohol consumption that leads to hepatic lipogenesis is not fully understood.
Advances in omics such as genomics and metabolomics within the last two decades have resulted in a boost in our understanding of the mechanism of diseases through agnostic approaches such as genome-wide association studies (GWAS) that revealed numerous genetic loci linked to complex diseases. Recently, we have applied GWAS to identify genetic variants in the form of single nucleotide polymorphisms (SNPs) that are associated with alcohol consumption3 as well as with circulating liver enzymes4 in the European populations. Some of the identified alcohol genes (e.g., ADH, KLB, DRD2) have been investigated for a better understanding of their involvement in alcohol consumption and health consequences such as hepatic lipogenesis. However, the biological effect of most of the identified alcohol-associated genes remains to be elucidated.
In this study, we aimed to shed light on the biological pathways and molecular changes linking alcohol consumption and liver pathologies. We first investigated molecular consequences of alcohol consumption and the pathways involved at metabolic level using omics approaches. Subsequently in search for common pathways between alcohol consumption and liver pathologies, we identified candidate genes with effect on both alcohol consumption and liver function and then investigated the biological effect of candidate genes in model organism i.e., ethanol-exposed C. elegans and Drosophila to generate knowledge that could ultimately be used to better understand alcohol related behavior and hepatic lipogenesis. We finally returned to use data from humans to further validate the most plausible candidate genes with evidence of potential involvement in lipogenesis.
Methods
Population
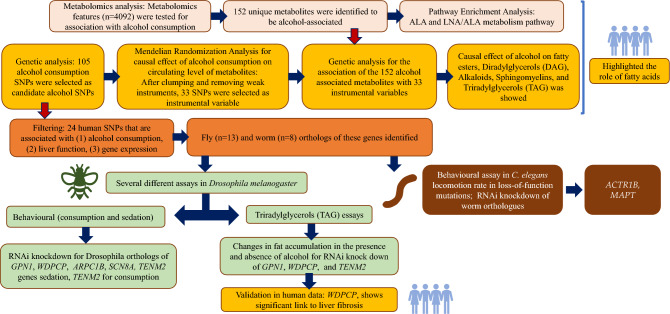
In the current study, we followed a multi-stage approach using population-based studies and model organisms to better understand pathways involved in alcohol consumption and its health consequences (Fig. 1). We used data from the Airwave Health Monitoring Study5, an occupational cohort of 53,116 police officers and staff ages 18 years and over across the UK (Supplementary Table 1). The Airwave Health Monitoring Study was approved by the UK National Research Ethics Service (NRES) North West—Haydock Regional Ethics Committee (REC reference: NRES/19/NW/0054; IRAS ID: 259978). The North-West Haydock REC approved the study protocol and all study documentation, and prior individual informed consent was obtained from each of the study participants. All methods were carried out in accordance with relevant guidelines and regulations. Participants were informed about the study and provided informed consent. Detailed information about the Airwave population, metabolic assays, data processing, metabolite annotation as well as genotyping and imputation is included in the “Supplementary Methods”.
Figure 1.
Overview of the study design and the findings.
Alcohol consumption
Alcohol consumption during the last seven days was assessed through a self-reported questionnaire in the Airwave study. The participants were asked to quantify (1) the number of glasses (small/125 ml) of red wine, white wine/champagne, fortified wine (includes sherry, port and vermouth), (2) the number of pub measures of spirits/liqueurs (includes whisky, gin, rum, vodka and brandy), and (3) the number of pints of beer or cider (include bitter, lager, stout, ale and Guinness) consumed in the last seven days. We considered the density of alcohol to be 0.79 g/ml at room temperature and calculated the total amount of alcohol consumed by summing up the grams of alcohol consumed from each category of alcoholic drink.
Analysis of the metabolome
We analyzed the association between alcohol consumption and circulating metabolites within the Airwave sample to identify candidate metabolites associated with alcohol consumption within the Airwave sample (alcohol-associated metabolites). Metabolomics features (n = 4092) were obtained from data acquired by the National Phenome Centre (NPC) using liquid-chromatography/mass spectrometry), covering a wide range of hydrophilic and lipid metabolite classes. We excluded unannotated features leaving 986 annotated metabolites for the analysis and tested the association of these metabolites with alcohol consumption. Subsequently, to assess if the resulting alcohol-associated metabolites were additionally involved in known metabolic pathways, we performed pathway enrichment analysis using all annotated alcohol-associated metabolomics features. To this end, Kyoto Encyclopedia of Genes and Genomes (KEGG)6 and The Small Molecule Pathway Database (SMPDB)7 were used and the statistical significance level was claimed using a P-value calculated using false discovery rate (Pfdr).
Genotyping and imputation
In the Airwave study5, genotyping was conducted using Illumina Infinium HumanExome-12v1-1 BeadChip Array. QC steps were performed to remove any samples with high missingness (> 3%) or outlier heterozygosity rates (> 3SD from the batch mean), duplicates (ID-based or genotype-based) or presented with high degree of relatedness. Markers with genotype call rate < 98%, significant deviation from Hardy–Weinberg equilibrium (P < 1 × 10–5), and low MAF (< 1%) were removed. Genotype data was imputed with 1000 Genomes phase 3 reference panel.
GWAS on metabolomics
We performed genome-wide association studies (GWAS) in the Airwave Study to obtain genetic association estimates for alcohol-related SNPs and selected metabolomic features. Untargeted Mass-Spectrometry (MS) was performed in heparin plasma samples of the Airwave Study, producing three separate datasets, namely hydrophilic interaction liquid chromatography (HILIC) positive (HPOS), lipid positive (LPOS), and lipid negative (LNEG). For each metabolomic dataset, we used principal component analysis (PCA) to identify outlier samples and exclude them. The data was residualized using the first ten principal components to account for population stratification. We transformed the data into z-scores, performed individually for each metabolomic feature, using median and median absolute deviations (MAD). Values that were more than 5 MAD from the median were removed and imputed using k-nearest neighbors’ imputation8. GWAS were conducted for each metabolomic feature with adjustment for age and sex (N = 1970). Due to the high number of metabolomic features, GWAS were performed using the high-dimensional association analyses (HASE) framework, which applies matrix operation and removes redundant calculation in high-dimensional association analyses to improve computational efficiency9.
We used the results from association analyses between the alcohol-associated metabolites and the known alcohol-associated SNPs to further perform a causal inference analysis on these results using the inverse variance weighted two-sample Mendelian randomization (MR) analysis10. The aim of the MR analysis was to identify changes in circulating metabolites caused by alcohol consumption. MR is a causal assessment method used in observational studies to mimic randomized controlled trials (RCTs) by taking advantage of the random assortment of alleles at conception. MR uses instrumental variables (i.e., genotype status), that are robustly associated with an exposure of interest, as a natural randomization tool occurring at conception11. The detailed methods for selection of alcohol consumption instruments have been described previously10,12. Briefly, we obtained genetic association statistics (β values) for 105 alcohol-associated SNPs3,13 from Liu and colleagues13. Clumping and removing weak instruments as described previously10,12 ensured the most robust instruments were used for the MR analysis (n = 33).
Gene selection for model organisms
We selected a list of 105 alcohol-related SNPs from recently conducted GWAS of alcohol consumption3,13. SNPs were selected if they presented a P-value lower than a GWAS significance threshold of 5 × 10−8 in their association with alcohol consumption. As we were interested in finding pathways involved in hepatic lipogenesis as a result of alcohol consumption, we sought candidate SNPs that showed strong effect with both alcohol consumption and liver function, using our recently published GWAS of circulating liver enzymes4. To account for multiple testing, a corrected P-value threshold of < 0.00048 was used for the association with liver enzymes. This P-value threshold corresponds to a nominal P-value (0.05) that has been adjusted for the number of alcohol-related SNPs (n = 105) using the Bonferroni method14. SNPs (n = 43; Supplementary Table 2) that were associated with at least one of the three liver enzymes alanine transaminase (ALT), alkaline phosphatase (ALP), and gamma-glutamyl transferase (GGT) were shortlisted to assess their link to gene expression using the GTEx database15. We eventually selected SNPs (n = 24) that demonstrated evidence of a statistically significant effect on gene expression of their nearest genes (a cis-eQTL effect; Table 1).
Table 1.
Overview of the genetic variants with effect on alcohol consumption, liver enzymes and gene expression.
| Alcohol SNP * | Annotated gene | eQTL tissue | eQTL effect size§ (effect allele) | eQTL P value† | Drosophila ortholog‡ | FlyBase score out of 15§ | C. elegans ortholog (score)‡ | Drosophila strains | C. elegans strain |
|---|---|---|---|---|---|---|---|---|---|
| rs823114 | NUCKS1 | Thyroid | − 0.16 (A) | 5.80 × 10–18 | – | – | – | – | – |
| rs1260326 | GCKR | Thyroid | 0.22 (C) | 1.80 × 10–08 | – | – | – | – | – |
| rs2178197 | GPN1 | Brain-Cerebellar Hemisphere | − 0.31 (G) | 6.50 × 10–08 | cg3704 | 13 | gop-2 (9) ¶ | CG3704 (55,294) | RG5036 gop-2 (gk5528), |
| rs13032049 | WDPCP | Adipose-Subcutaneous | − 0.16 (G) | 3.00 × 10–04 | frtz | 13 | – | Frz (55,649) | – |
| rs11692435 | ACTR1B | Thyroid | 0.89 (A) | 1.50 × 10–80 | arp1 | 12 | arp-1 (8) ¶ | Lethal, not studied | FX4735 arp-1 (tm4735) |
| rs12646808 | MSANTD1 | Thyroid | − 0.25 (T) | 1.30 × 10–09 | cg18766 | 8 | – | – | – |
| rs11940694 | KLB | Liver | 0.22 (A) | 4.00 × 10–05 | cg9701 | 6 | klo-1 (5), klo-2 (5) | – | Not studied |
| rs1229984 | ADH1B | Esophagus-Gastroesophageal Junction | − 1.4 (C) | 1.90 × 10–16 | fdh | 7 | adh-5 (5) | – | Not studied |
| rs10078588 | TENM2 | Thyroid | 0.29 (A) | 2.80 × 10–15 | ten-m | 13 | ten-1 (10) ¶ | TenM (29,390) | VC518 ten-1 (ok641), |
| rs34060476 | MLXIPL | Pancreas | 0.51 (G) | 4.50 × 10–18 | mondo | 12 | mml-1 (9) | Mondo (27,059) | RB954 mml-1 (ok849) |
| rs10249167 | ARPC1B | Thyroid | 0.3 (G) | 1.80 × 10–25 | arpc1 | 13 | arx-3 (10) ¶ | Arpc1 (31,246) | VC3166 arx-3 (ok1122) |
| rs988748 | BDNF | Brain–Nucleus accumbens basal ganglia | 0.22 (G) | 9.70 × 10–06 | – | – | – | – | – |
| rs2071305 | MYBPC3 | Whole blood | − 0.13 (C) | 2.30 × 10–11 | hbs | 1 | – | – | – |
| rs7121986 | DRD2 | Esophagus–Muscularis | 0.26 (C) | 3.50 × 10–08 | dop2r | 9 | dop-2 (6), dop-3 (6) | – | Not studied |
| rs10876188 | SLC4A8 | Cells-Cultured fibroblasts | − 0.12 (T) | 1.80 × 10–07 | ndae1 | 12 | abts-1 (9) | Lethal, not studied | RB1381 abts-1 (ok1566) |
| rs7958704 | SCN8A | Nerve–Tibial | − 0.25 (C) | 2.90 × 10–10 | para | 12 | cca-1 (1), unc-77 (1), egl-19 (4) | Para (33,923) | JD21 cca-1 (ad1650) |
| rs12312693 | STAT6 | Brain–Frontal Cortex BA9 | 0.23 (C) | 8.00 × 10–05 | stat92e | 9 | sta-2 (4), sta-1 (7) | – | RB796 sta-1 (ok587) |
| rs11625650 | KIF26A | Whole blood | − 0.26 (A) | 9.10 × 10–12 | cg14535 | 7 | vab-8 (2) | – | NG2484 vab-8 (gm84) |
| rs1421085 | FTO | Muscle–Skeletal | 0.14 (C) | 7.50 × 10–08 | – | – | – | – | – |
| rs11648570 | PMFBP1 | Esophagus–Mucosa | − 0.29 (C) | 9.00 × 10–05 | cg12702 | 1 | – | – | – |
| rs3803800 | TNFSF13 | Brain–Cortex | − 0.2 (G) | 2.50 × 10–08 | egr | 3 | – | – | – |
| rs1053651 | TCAP | Muscle–Skeletal | − 0.11 (C) | 4.90 × 10–11 | – | – | – | – | – |
| rs1991556 | MAPT | Brain–Cerebellum | − 0.28 (A) | 1.80 × 10–06 | tau | 7 | ptl-1 (8) | – | RB809 ptl-1 (ok621) |
| rs4815364 | ACSS1 | Brain–Cerebellar Hemisphere | − 0.47 (A) | 1.70 × 10–12 | accoas | 6 | – | – | – |
eQTL expression quantitative trait loci.
*Alcohol SNPs associated with liver enzymes at replication P value < 0.000476. Liver enzyme GWAS summary statistics obtained from Pazoki et al. 2021.
†Normalized effect size and P-value obtained from GTEx portal.
‡Orthologs were extracted from FlyBase and WormBase. § number of FlyBase tools that support the gene-pair relationship out of a total number of tools (n = 15) that computed relationships between Homo sapiens and Drosophila melanogaster.
¶Genes for which mutations were homozygous lethal and as such, heterozygote mutations balanced by chromosomal translocations were instead analyzed.
Ortholog selection
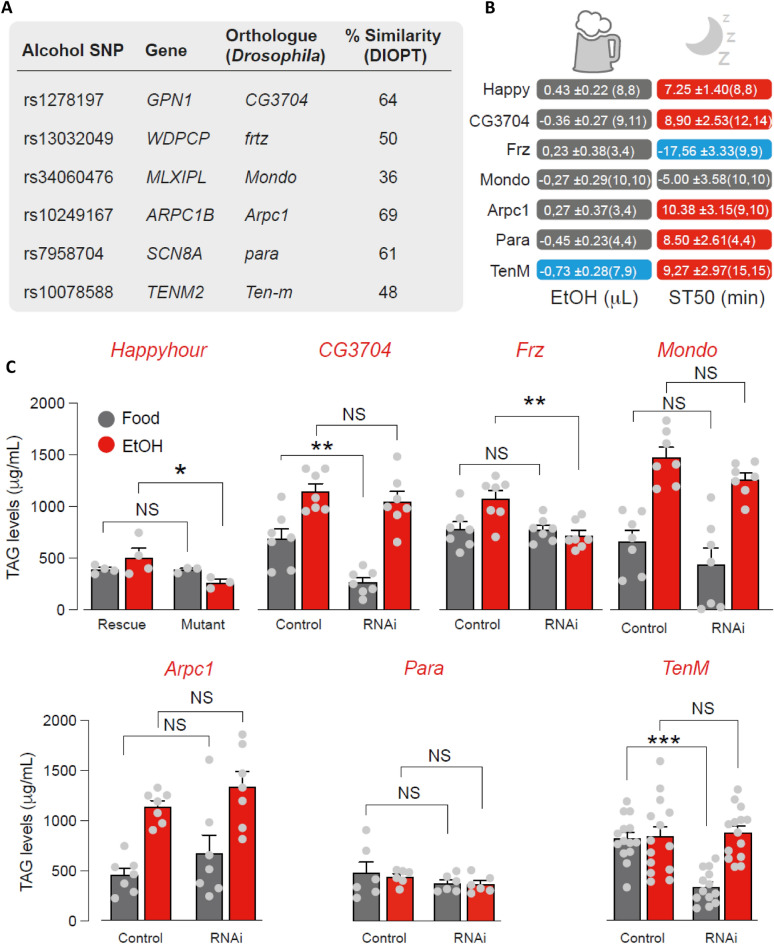
The 24 SNPs with cis-eQTL effect with their annotated genes were matched to their orthologs in Drosophila using FlyBase (DIOPT online tool version 8.5/9.0; beta; http://www.flyrnai.org/diopt). To ensure the selection of the most credible orthologs, we used scores calculated in FlyBase. This database provides a number of approaches that support the gene-pair relationship out of a total number of tools that computed relationships between Homo sapiens and Drosophila. Genes with a score of > 12 were shortlisted for further analysis in Drosophila. Nineteen Drosophila orthologs were identified of which eight had a score ≥ 12 including ARPC1B (arpc1), ACTR1B (arp1), GPN1 (CG3704), WDPCP (frz), MLXIPL (mondo), SLC4A8 (ndae1), SCN8A (para), and TENM2 (Ten-m).
To identify potential worm orthologs, the SNPs with cis-eQTL effect with their annotated genes were sought for their worm orthologs within WormBase (https://wormbase.org/). BLASTp analysis results of human protein sequence against C. elegans protein database were obtained in WormBase version WS280 using data from C. elegans Sequencing Consortium genome project (PRJNA13758). The human protein sequence for the protein encoded by the genes under this study was extracted from UniProt (https://www.uniprot.org/). Worm genes with the best alignment with the human protein sequence indicated by an Expect value (E value) < 1 × 10–5 and the highest BLASTp score (bits) were moved forward. The E value represents the number of alignments that could be found in similarity to the protein sequence by chance. Using WormBase, 13 worm orthologs were identified including ACTR1B (arp-1), ARPC1B (arx-3), GPN1 (gop-2), MLXIPL (mml-1), STAT6 (sta-1), TENM2 (ten-1), KIF26A (vab-8), SLC4A8 (abts-1), MAPT (ptl-1) and SCN8A (cca-1). We excluded KLB (klo-1), DRD2 (dop-2), and ADH1B (adh-5) from the C. elegans experiments as similar studies investigating these genes already exist16–18.
We did not identify any fly or worm orthologs for NUCKS1, GCKR, BDNF, FTO, TCAP. Additionally, no worm ortholog was found for WDPCP, MSANTD1, MYBPC3, PMFBP1, TNFSF13, and ACSS1.
Drosophila
Genetics and Drosophila melanogaster strains
All Drosophila stocks and crosses were maintained on standard cornmeal agar media at 25 °C on 12/12 h light/dark cycles. The following strains were used as positive control: wberlin; hppy17–51; + (hppy mutant) and wberlin; hppy17–51; hppy III (hppy mutant with a rescue genomic construct) (gifts from Prof. Ulrike Heberlein, Janelia Research Campus, Virginia, USA), RNAi lines were obtained from Bloomington Drosophila Stock Center (gene CG3704 (BDSC no: 55294), Frz (BDSC no: 55649), Mondo (BDSC no: 27059), Arpc1 (BDSC no: 31246), Para (BDSC no: 33923) and TenM (BDSC no: 29390)). All lines used were backcrossed to w1118 or [v]w1118 (RNAi lines). The expression of all RNAi constructs was driven by the ubiquitous driver, daGal4, which drives expression throughout development from embryonic to adult stage in all tissues. All the experiments on adult flies were performed using males.
Drosophila ethanol consumption assay
The CApillary FEeder (CAFE) assay19 was used to measure ethanol consumption. Eight male flies were placed into an experimental vial (8 cm height, 3.3 cm diameter) containing 6 microcapillary tubes (BRAND® disposable BLAUBRAND® micropipettes, intraMark, BR708707, with 1 µl marks), each containing 5 µl of liquid food. Liquid food was prepared by dissolving 50 mg of yeast granules in 1 ml of boiling water by vortexing, followed by brief centrifugation. Then, 40 mg of sucrose (Sigma‒Aldrich, 84097) was added to 800 µl of the dissolved yeast mixture, followed by vortexing. The microcapillary tubes were filled with liquid food up to the 5 µl mark. Ethanol food consists of normal food supplemented with 15% ethanol. Each experiment consisted of 5 experimental vials per genotype with each vial containing normal food (3 capillaries) and ethanol food (3 capillaries). The flies were acclimatized in the experimental vial without any food for 2 h prior to the start of the experiment. This step was also used to incentivise the flies to eat once the food was introduced. The experimental vials were placed in a plastic box with a cover to control humidity. The flies were allowed to feed for 19 h, after which the amount consumed (in mm) was measured with a digital calliper (Dasqua Bluetooth Digital Calliper 12″/300 mm, 24108120). The total amount of food consumed was calculated using the formula:
Drosophila ethanol sedation assay
Fly sedation assay was performed as previously described20. Briefly, 8 flies were transferred to a 25 mm x 95 mm transparent plastic vial in between two cotton plugs. A piece of cotton plug at the base of the vial served as a stable surface to observe the flies and another plug was used to cap the vial and deliver the ethanol. 500ul of 100% ethanol was added to the side of the cotton plug facing the flies. Sedation was observed manually as ST50, which is the time in minutes it takes for 50% of the flies in a sample vial to become sedated. Sedation events are recorded when the flies become inactive and lay on their backs for over 10 s.
Drosophila triradylglycerols (TAG) measurements
To assess the role of the candidate genes in lipogenesis, we assessed the effect of RNAi knockdown of the selected genes on TAG levels in Drosophila. Eight male flies of the indicated genotypes were placed into an experimental vial as described in the ethanol consumption assay, with all 6 microcapillary tubes filled with normal food (5% sucrose + 5% yeast) or ethanol food (normal food + 15% ethanol), for 2 days. TAGs were assessed through colorimetric assays using 96-well microtiter plates and an Infinite M200Pro multifunction reader (TECAN). The assays were performed as previously described21. Briefly, flies were homogenized in 110 µl of PBS + 0.05% Tween 20 (PBST) for 2 min on ice and immediately incubated at 70 °C for 10 min to inactivate endogenous enzymatic activity. A 35 µl fly homogenate sample and a glycerol standard (Sigma, no. G7793) were incubated together with either 35 µl of PBST (for free glycerol measurements) or 35 µl of TAG reagent (Sigma, no. T2449, for TAG measurements) at 37 °C for 60 min. After 3 min of centrifugation at full speed, 30 µl of each sample was transferred into a clear-bottom plate (two technical replicates per biological sample) together with 100 µl of free glycerol reagent (Sigma‒Aldrich, F6428) and incubated at 37 °C for 5 min. TAG absorbance was divided by the protein concentration of the respective sample, which was measured by Bradford assay (Sigma‒Aldrich, B6916).
C. elegans
Nematode strains and culture
All C. elegans strains were cultured on nematode growth medium (NGM) agar plates at 20 °C using Escherichia coli OP50 as a food source. For the wildtype worms, Bristol N2 strain was used. Genes for which mutations were homozygous lethal (arp-1, arx-3, gop-2 and ten-1), heterozygote mutations balanced by chromosomal translocations were instead analyzed. Loss-of-function mutations were not backcrossed into Bristol N2, but instead initially screened for phenotypic differences with those differences validated by RNAi.
Nematode RNA interference (RNAi)
RNAi experiments were performed on the NL2099 rrf-3 (pk1426) strain as previously described16,22 (Supplementary Table 3). RNAi was achieved by feeding23 using the ORFeome based RNAi library24. In brief, HT115 RNAi bacterial clones were initially cultured in LB media with 100 µg/ml ampicillin and subsequently spotted in three 50 µl drops on 60 mm diameter NGM plates containing 1 mM isopropyl β-1-thiogalactopyranoside (IPTG) and 25 µg/ml carbenicillin. Plates were left to dry for 4–7 days before seeding to improve RNAi efficiency. Following seeding, five L3–L4 worms were added to each RNAi plate and cultured at 20 °C until the F1 generation reached adulthood. Ethanol experiments were performed and analyzed as described above and compared to worms fed with an empty RNAi feeding vector.
Nematode behavioural assays
All ethanol experiments were performed at 20 °C in a temperature-controlled room as previously described16,22. Behavioural assays were conducted on young adult hermaphrodites selected from sparsely populated NGM plates. Nematodes with loss-of-function mutations in worm orthologues of ACTR1B (arp-1), ARPC1B (arx-3), GPN1 (gop-2), MLXIPL (mml-1), STAT6 (sta-1), TENM2 (ten-1), KIF26A (vab-8), SLC4A8 (abts-1), MAPT (ptl-1) and SCN8A (cca-1) were acutely exposed to ethanol and the resultant effect on rate of locomotion (thrashes per minute) was quantified in Dent’s solution (140 mM NaCl, 6 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, pH 7.4 with bovine serum albumin at 0.1 mg/ml) by measuring thrashes per minute (one thrash defined as one complete movement from maximum to minimum amplitude and back) following 10 min exposure to the drug. Ethanol was mixed with Dent’s solution at a concentration of 400 mM, which has previously been shown to produce a ~ 70% reduction in locomotion rate in wild-type worms22,25.
Bioinformatics
Secondary analysis
To gain a better insight into the biological pathways involved in the link between alcohol consumption and liver damage, we used the genetic variants within the genes highlighted by our model organism experiments and performed a series of secondary analyses using human data. We explored publicly available data from the UK Biobank deposited in the Edinburgh Gene Atlas26 using Phewas (Phenome-wide association analysis) databases to obtain association results between the genetic variants and 778 traits. We additionally used the genes highlighted by our model organism experiments to assess the causal effect of gene expression on liver conditions. Within these genes, the SNPs that have been identified to have a cis-eQTL effect within the previously published studies were selected and used as MR instrument against liver conditions within the twosampleMR package in R.
Statistical analyses
Within the Airwave study sample, we performed a linear regression to study the association of alcohol consumption with each of the metabolomic features (Metabolome-wide association study; MWAS). We adjusted the statistical analysis for age, sex, smoking status, and salary class. To account for multiple testing and the high degree of correlation in metabolomics datasets, we used a permutation-based method to estimate the significance level of the associations27,28. For each metabolomics platform, a P-value threshold equivalent to adjusting to a 5% Family-Wise Error Rate (i.e., Bonferroni method) was computed. A series of hypergeometric tests implemented in the R package MetaboAnalystR29 was used for pathway enrichment analysis where an FDR threshold of 0.05 was used as a significance threshold for each of the metabolomics platforms. To obtain an estimation for the association of known alcohol SNPs with our alcohol-associated metabolites to be used in the MR analysis, we performed linear regression analysis within the Airwave sample (see “Supplementary Methods” for details of the GWAS on metabolomics). Linear analyses between SNPs and metabolites were conducted for each metabolomic feature with adjustment for age, sex, and genetic principal components within a subsample of Airwave that included participants with both genetic and metabolite data (N = 1970). In C. elegans, locomotion rate was presented normalized as a percentage of the mean thrashing rate of untreated worms measured each day. All worm data were expressed as mean ± SE with an N = 30 individual worms. Locomotion rate significance was assessed by one-way analysis of variance (ANOVA) with post-hoc Tukey test for multiple comparisons. Statistical analyses of the Drosophila experimental data were performed using GraphPad Prism (www.graphpad.com). Drosophila data were presented as the mean values, and the error bars indicate ± SD.
Results
To understand biological effects of alcohol consumption in human population, we investigated the circulating metabolites within the Airwave study sample using an agnostic approach which revealed the association of 152 unique metabolites with alcohol consumption (Supplementary Data). Using MR approach to examine the causality of the above associations, we identified a possible causal association (Table 2) of alcohol consumption on changing circulating level for several lipid metabolites (TAGs, Diradylglycerols, Glycerophosphocholines, Sphingolipids), and an alkaloid (piperine). The most statistically significant causal association was observed with a triacylglycerol TG 60:2 (β = 1.24; 95% CI 0.52,1.95; P-value = 0.002). Pathway analysis on the 152 unique alcohol-metabolite associations showed that the linoleic acid (LNA) and alpha linolenic acid (ALA) metabolism pathway (LNA/ALA) within the Small Molecule Pathway Database (SMPDB) was enriched with alcohol-associated metabolomic features (Pfdr = 5.67 × 10–3). These features were annotated to Tetracosapentaenoic acid (24:5n-3; β = 0.01; 95% CI 0.008,0.012; P-value = 4.2 × 10–12), Eicosapentaenoic acid (β = 0.009; 95% CI 0.007,0.011; P-value = 2.4 × 10–12), Stearidonic acid (β = 0.008; 95% CI 0.006,0.01; P-value = 2.2 × 10–9), Arachidonic acid (β = 0.007; 95% CI 0.005,0.009; P-value = 9.7 × 10–8) and Adrenic acid (β = 0.01; 95% CI 0.008,0.012; P-value = 9.7 × 10–8).
Table 2.
Overview of the causal effect of alcohol on circulating metabolites using the inverse variance weighted two-sample Mendelian randomization multiple instrument method.
| Metabolite | Name | Main class | Beta (95% CI) | P-value |
|---|---|---|---|---|
| SLPOS_457.3339_0.7190 | CAR DC18:1 | Fatty esters | -1.13 (-2.14, -0.11) | 0.036 |
| SLPOS_579.5352_8.2395 | DG 34:0 | DAG | 0.91 (0.05,1.77) | 0.047 |
| SLPOS_606.5548_8.3326 | DG 36:1 | DAG | 0.92 (0.07,1.77) | 0.041 |
| SLPOS_745.5603_5.7424 | PC 33:2 | Glycerophosphocholines | 0.78 (0.07,1.5) | 0.040 |
| SLPOS_629.5424_6.9217 | PE 38:4 | Glycerophosphoethanolamines | 1.2 (0.15,2.25) | 0.032 |
| SHPOS_625.5174_2.8914 | PE 38:5 | Glycerophosphoethanolamines | 1.1 (0.1,2.1) | 0.039 |
| SLPOS_201.0516_0.6580 | Piperine | Alkaloids | 1.16 (0.1,2.22) | 0.040 |
| SHPOS_807.6348_4.4974 | SM 40:2;O2 | Sphingomyelins | 1.06 (0.15,1.96) | 0.028 |
| SHPOS_827.7093_0.5837 | TG 48:1 | TAG | 0.95 (0.13,1.77) | 0.030 |
| SLPOS_898.7965_11.2257 | TG 53:1 | TAG | 0.93 (0.1,1.75) | 0.036 |
| SLPOS_888.8079_10.7574 | TG 53:3 | TAG | 0.96 (0.06,1.85) | 0.045 |
| SLPOS_607.5599_11.1277 | TG 54:2 | TAG | 0.95 (0.19,1.71) | 0.019 |
| SLPOS_1027.7392_10.9051 | TG 54:3 | TAG | 1.06 (0.21,1.91) | 0.020 |
| SLPOS_984.8508_11.7812 | TG 58:1 | TAG | 1.12 (0.44,1.81) | 0.003 |
| SLPOS_687.6307_11.5996 | TG 58:2 | TAG | 1.08 (0.27,1.9) | 0.014 |
| SLPOS_689.6461_11.7895 | TG 60:2 | TAG | 1.24 (0.52,1.95) | 0.002 |
Effect estimates and 95% CI is given for the inverse variance weighted method of Mendelian randomization.
CI confidence interval, DAG diradylglycerols, TAG triradylglycerols.
We used Drosophila hppy mutant, described to have an increased resistance to ethanol sedation30, as positive control in all fly experiments. Drosophila with knock-down of ARPC1B (arpc1), GPN1 (CG3704), WDPCP (frz), MLXIPL (mondo), SCN8A (para), and TENM2 (Ten-m) together with hppy mutant were exposed to food supplemented with 15% Ethanol in a CAFE assay. A significant difference (Fig. 2A,B) was observed between ethanol consumed (μL) by flies with RNAi knockdown of TENM2 (Ten-m). Following exposure to ethanol vapour, the effect on Sedation Time 50% (ST50; minute) was quantified and we observed that in comparison to control (hppy), RNAi knockdown of fly orthologues of WDPCP (frz) showed a faster rate of sedation whilst TENM2 (Ten-m), GPN1 (CG3704), ARPC1B (arpc1) and SCN8A (para) showed a slower rate of sedation indicated by a higher ST50. WDPCP (frz), TENM2 (Ten-m) and GPN1 (CG3704) knockdown flies showed reduced TAG levels (Fig. 2C).
Figure 2.
Analysis of alcohol intake and sedation in adult flies. (A) Mapping Drosophila orthologues of human genes involved in ethanol consumption. Similarity of protein alignment based on the DRSC integrative ortholog prediction tool (DIOPT). (B) Analysis of ethanol intake (left column) and sedation (right column) in the Drosophila RNAi lines. The numerical values show the difference between means ± standard error of mean (RNAi–Control). Red or blue correspond to the measurements that were significantly increased or decreased, respectively (P ≤ 0.05) whereas grey indicates a significance higher than 0.05. The statistical significance was determined using an unpaired t-test. Values in parenthesis are the number of biological replicates for respectively, the control and the RNAi line. (C) Analysis of TAG levels in adult flies fed either with normal or ethanol-containing food (mean ± standard error of mean; asterisks, 2-way ANOVA with Tukey's multiple comparisons test). The number of biological replicates per experimental variable (n) is indicated in either the respective figure or figure legend. No sample was excluded from the analysis unless otherwise stated. Blinding was not performed. Normality was assessed before deciding on which parametric or non-parametric test to use for inferential statistics. Statistical significance is indicated as * for P < 0.05, ** for P < 0.01, *** for P < 0.001, **** for P < 0.0001 and NS for P ≥ 0.05.
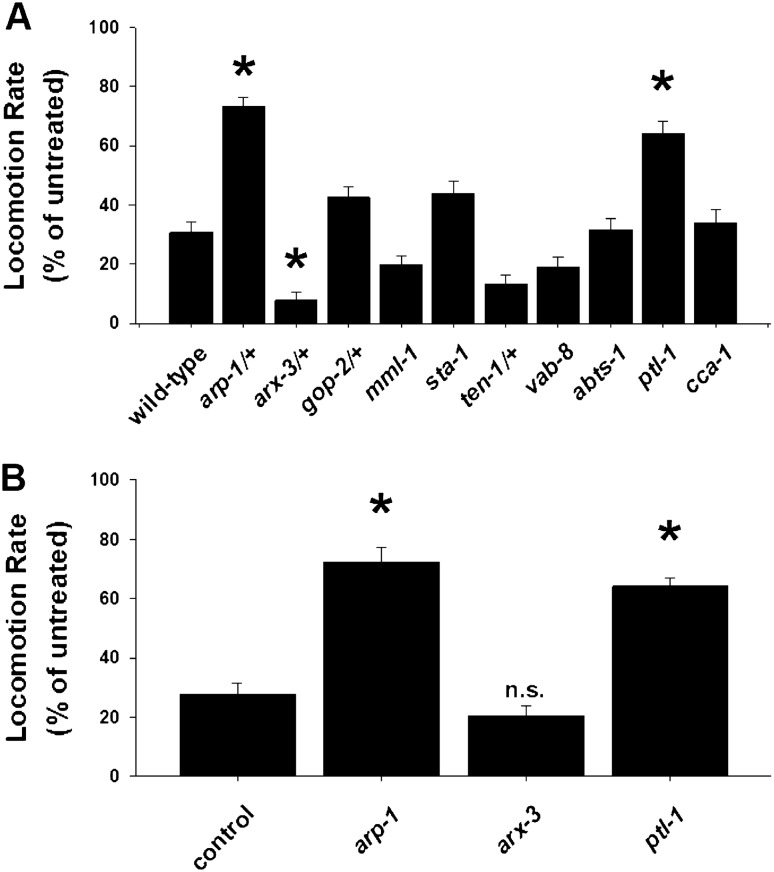
C. elegans loss-of-function mutants of TENM2 (ten-1), KIF26A (vab-8), SLC4A8 (abts-1) and SCN8A (cca-1) showed significant differences in basal locomotion rate (Supplementary Fig. S1A). After acute exposure to ethanol in C. elegance loss-of-function mutants, significant differences were identified in normalized locomotion rate of ACTR1B (arp-1), ARPC1B (arx-3) and MAPT (ptl-1) in comparison with Bristol N2 wild-type worms (Fig. 3A). RNAi knockdown of these genes confirmed that in comparison to controls, RNAi knockdown of worm orthologues of ACTR1B (arp-1) and MAPT (ptl-1) and ARPC1B (arx-3) did not have any effect on basal locomotion rate (Supplementary Fig. S1B) but RNAi knockdown of ACTR1B (arp-1) and MAPT (ptl-1) phenocopied the loss-of-function mutations after exposure to ethanol (Fig. 3B).
Figure 3.
Quantification of alcohol phenotypes for C. elegans genes. (A) Nematodes with loss-of-function mutations in worm orthologues of ACTR1B (arp-1/ +), ARPC1B (arx-3/ +), GPN1 (gop-2/ +), MLXIPL (mml-1), STAT6 (sta-1), TENM2 (ten-1/ +), KIF26A (vab-8), SLC4A8 (abts-1), MAPT (ptl-1) and SCN8A (cca-1) were acutely exposed to ethanol and the resultant effect on rate of locomotion (thrashes per minute) was quantified. In comparison with Bristol N2 wild-type worms, significant differences were identified for arp-1, arx-3 and ptl-1. Data is presented normalized to locomotion rate of untreated worms. *P < 0.01. (B) RNAi confirmation of positively identified genes involved in alcohol phenotypes. In comparison to controls, RNAi knockdown of worm orthologues of ACTR1B (arp-1) and MAPT (ptl-1) phenocopied the loss-of-function mutations, whereas ARPC1B (arx-3) knockdown had no effect. *P < 0.01. n.s., not significant.
Secondary analysis
The Phewas analysis (Table 3) on the three genetic variants within WDPCP, TENM2, and GPN1 showed a link between rs10078588 (TENM2) and food and liquid intake (beef, oily fish, fresh fruit, bread, alcohol). rs13032049 (WDPCP) was linked to salt intake and smoking. rs2178197 (GPN1) was linked to hypertension, and hematologic traits. All three SNPs showed association with disorders of lipoprotein metabolism and other lipidaemias (ICD code E78) within the UK Biobank (Table 4). We finally performed a Mendelian randomization analysis on the expression of WDPCP gene (ENSG00000115507) and liver conditions using cis-eQTL data (Table 5) and identified a link between expression of WDPCP with liver fibrosis and cirrhosis (β = − 0.20; 95% CI − 0.39, − 0.01; P-value = 0.04) as well as liver and bile duct cancer (β = 0.0003; 95% CI 3.27 × 10–05, 5.85 × 10–04; P-value = 0.02). We also observed a suggestive link with the changes in liver fatty acid-binding protein (Table 5).
Table 3.
Overview of the significant associations between SNPs in WDPCP, TENM2, and GPN1 with Phewas traits within the UK Biobank Edinburgh Gene Atlas.
| Trait | Beta | Z value | P-value |
|---|---|---|---|
| rs10078588-A (TENM2) | |||
| Bread intake, number of slices/week | 0.09 | 5.66 | 7.57 × 10–9 |
| Alcohol weekly intake frequency (high to low frequency) | − 0.02 | 5.21 | 9.63 × 10–8 |
| Oily fish intake, number of times/week | − 0.01 | 4.53 | 2.88 × 10–6 |
| Beef intake, number of intake times/week | 0.01 | 4.45 | 4.34 × 10–6 |
| Fresh fruit intake, number of pieces/day | − 0.01 | 4.44 | 4.45 × 10–6 |
| rs13032049-G (WDPCP) | |||
| Salt added to food, frequency | 0.01 | 5.01 | 2.77 × 10–7 |
| Smoking status, current vs. previous and never | 0.01 | 4.12 | 1.88 × 10–5 |
| rs2178197-G (GPN1) | |||
| Hypertension | − 3.60 × 10–3 | − 4.26 | 1.02 × 10–5 |
| Mean sphered cell volume, fl | 0.04 | − 4.98 | 3.15 × 10–7 |
| Red blood cell (erythrocyte) count, × 1012 cells/L | − 3.30 × 10–3 | − 5.41 | 3.21 × 10–8 |
| Number of treatments/medications taken | − 0.02 | − 4.64 | 1.71 × 10–6 |
| Monocyte percentage, % | 0.02 | − 4.97 | 3.39 × 10–7 |
| Number of operations, self-reported | − 0.01 | − 4.33 | 7.58 × 10–6 |
| Mean reticulocyte volume, fL | 0.06 | − 4.39 | 5.66 × 10–6 |
| High light scatter reticulocyte percentage, % | − 1.60 × 10–3 | − 4.37 | 6.32 × 10–6 |
| Reticulocyte count, × 1012 cells/L | − 2.00 × 10–4 | − 5.15 | 1.33 × 10–7 |
| Reticulocyte percentage, % | − 4.20 × 10–3 | − 4.09 | 2.12 × 10–5 |
| Lymphocyte percentage, % | 0.06 | − 4.49 | 3.59 × 10–6 |
| Neutrophil percentage, % | − 0.07 | − 4.48 | 3.81 × 10–6 |
| High light scatter reticulocyte count, × 1012 cells/L | − 1.00 × 10–4 | − 5.39 | 3.50 × 10–8 |
Significant associations are depicted in bold. Statistical significance level P-value = 2.14 × 10–5 was considered equivalent to a P-value adjusted for multiple testing for the analysis of 3 SNPs and 778 phenotypes within the Gene Atlas. The associations are provided per copy of the allele that increases alcohol intake.
Table 4.
Overview of the association of SNPs in WDPCP, TENM2, and GPN1 with disorders of lipoprotein metabolism and other lipidaemias (ICD code E78) within the UK Biobank Edinburgh Gene Atlas.
| SNP | chromosome | Base pair location | Effect allele | Beta (effect estimate) | 95% CI lower bound | 95% CI upper bound | P-value | MAF | HWE |
|---|---|---|---|---|---|---|---|---|---|
| rs10078588 | 5 (TENM2) | 166,816,176 | A | − 0.001 | − 2.49 × 10–04 | − 2.44 × 10–03 | 0.02 | 0.47 | 0.5 |
| rs13032049 | 2 (WDPCP) | 63,581,507 | G | − 0.001 | − 1.59–04 | − 2.60 × 10–03 | 0.03 | 0.28 | 0.51 |
| rs2178197 | 2 (GPN1) | 27,860,551 | G | − 0.001 | − 3.36 × 10–04 | − 2.55 × 10–03 | 0.01 | 0.42 | 0.21 |
Results obtained from the Edinburgh Gene Atlas.
CI confidence interval, SNP single nucleotide polymorphism, MAF minor allele frequency, HWE Hardy Weinberg Equilibrium.
Table 5.
Overview of the significant results from inverse variance weighted Mendelian randomization analysis for the effect of gene expression of ENSG00000115507 on liver traits using the MRC IEU OpenGWAS data infrastructure51.
| Liver trait | Number of SNPs | Effect estimate | Standard error | P value |
|---|---|---|---|---|
| Fatty acid-binding protein, liver | 6 | − 0.10 | 0.05 | 0.06 |
| Liver enzyme levels (alanine transaminase) | 6 | − 0.002 | 0.001 | 0.2 |
| Fibrosis and cirrhosis of liver | 6 | − 0.20 | 0.1 | 0.04 |
| Liver & bile duct cancer | 6 | 0.0003 | 0.0001 | 0.03 |
Discussion
In this study we used data from humans, C. elegans, and Drosophila and identified a link between genes implicated in alcohol consumption and lipid metabolism. We identified that alcohol consumption changes the metabolites within linoleic acid (LNA) and alpha linolenic acid (ALA) metabolism pathway (LNA/ALA) and demonstrates causal effect on changes in several lipid metabolites. We highlighted that change of function of the genes implicated in alcohol consumption leads to changes in ethanol consumption, sedation after exposure to ethanol vapor, and changes in accumulation of fat in Drosophila as well as changes in locomotion rate after exposure to ethanol in C. elegans. Our results demonstrate that three alcohol-implicated genes namely WDPCP (frz), TENM2 (Ten-m), and GPN1 (CG3704) might be involved in fat accumulation.
In our study, we identified several metabolites to be causally altered due to alcohol consumption and identified the LNA/ALA pathway involved in alcohol consumption. A metabolite, eicosapentaenoic acid that is synthesized from linolenic acid, has previously been associated with altered acute behavioral responses to alcohol in C. elegans (altered locomotion)31, and mice32. In addition in humans, long-chain polyunsaturated fatty acids are shown to be associated with alcohol sensitivity33 and genes that are essential to generate ω-3 long-chain polyunsaturated fatty acids are shown to be associated with alcohol-related phenotypes34. Previous studies also showed associations between lipid metabolites and alcohol consumption35. We previously showed alteration of high-density lipoprotein to be associated with alcohol consumption within the UK Biobank10. The metabolomics part of the current study is a step forward in that firstly it uses causal inference tools (MR method) to demonstrate the causality of the association between lipid metabolites specifically long chain fatty acids with alcohol consumption. Secondly, by performing a pathway analysis, we were able to pinpoint a specific LNA/ALA pathway in alcohol consumption.
In our study, we used alcohol-associated genetic variants to explore (1) the alcohol-induced biological changes in human metabolites and (2) alcohol-induced biological effect of genes annotated to alcohol-associated genetic variants in C. elegans and Drosophila melanogaster. Of the alcohol-implicated genes that we investigated in C. elegans, ACTR1B (arp-1) and MAPT (ptl-1), show significant effects on the worms’ locomotion upon acute exposure to ethanol. In addition, TENM2 (Ten-m) shows significant effects on ethanol consumption in Drosophila and apart from MLXIPL (mondo), RNAi lines for all the genes investigated in Drosophila change the time to sedation from ethanol.
WDPCP which was first characterized in Drosophila36 is known to be involved in cell polarity and ciliogenesis37,38. In humans, mutations in WDPCP gene cause Bardet-Biedl Syndrome39 presenting with a variety of symptoms in different organs including obesity, blindness, and polydactyly. WDPCP gene is widely expressed known to be involved in hedgehog signaling40. Recently, Liu and colleagues13 identified that a G allele in rs13032049 within the WDPCP gene was associated with an increased consumption of alcohol. The G allele in rs13032049 shows a strong association with a lower expression of the WDPCP gene. We found the SNP to be strongly associated with increased liver enzyme GGT and behavioral traits such as smoking and adding salt to food. In our Drosophila experiments, the observed effect of WDPCP (frz) knockdown on the changed TAG levels occurred under exposure to ethanol and followed similar patterns as hppy mutants. This implies that minimizing alcohol consumption could reduce fat accumulation and thus could potentially reduce the risk of hepatic lipogenesis. In our further MR analysis, we used publicly available databases derived from GWAS in human population and showed a link between the gene expression of WDPCP and liver fibrosis and liver cirrhosis. The analysis also showed a suggestive link with the liver fatty acid binding protein that is involved in the metabolism of lipids41. This evidence suggests that WDPCP might be an important gene involved in the pathway between alcohol consumption, accumulation of fat and liver fibrosis. This could have public health implications in terms of the identification of high-risk groups and targeting preventive measures as well as drug development. More studies in vivo and in vitro are needed to focus on WDPCP and provide more details on its role in lipid metabolism and liver pathologies.
Changes in TAG levels of Drosophila occurred with exposure to normal food rather than ethanol for RNAi knockdown of TENM2 (Ten-m) and GPN1 (CG3704) indicating that loss of function of these genes could have a direct role in the accumulation of fat in liver independent of exposure to ethanol. RNAi knockdown of both genes shows increased tolerance to the sedative effect of ethanol which could justify the effect of these genes on a more frequent alcohol consumption in humans, possibly due to alcohol tolerance. GPN1 is located on chromosome 2p23.3 and the encoded protein is implicated in the regulation of TGFβ superfamily signaling42 that is demonstrated to play a role in obesity43, accumulation of fat in the liver42,44 as well as regulation of ADH1 gene that enhances alcohol-induced liver damage and lipid metabolism45. The existing evidence alongside our findings on the role of GPN1 in alcohol consumption and lipid metabolism in Drosophila implies that GPN1 might play a role upstream of TGFβ in the regulation of the metabolism of alcohol and lipids. Further studies are needed to highlight the relationship between GPN1 and TGFβ in alcohol consumption and alcohol-induced liver damage.
TENM2 (Ten-m) is located on chromosome 5q34, and the encoded protein is involved in cell adhesion46. TENM2 is found to be highly enriched in white adipocyte progenitor cells47. TENM2 deficiency in human fat cells leads to expression of UCP1, the primary marker of brown adipose tissue48. Genetic variants in TENM2 have shown to be linked to obesity49. Our secondary analyses confirmed an association between the genetic variant in TENM2 and excess of food and liquid intake which suggests the link between TENM2, and alcohol consumption could also be due to systematic increase in consumption of all food and beverages rather than alcohol alone. The evidence in this study alongside the existing literature highlights that the observed effect of TENM2 (Ten-m) RNAi knockdown on the changes in TAG levels in Drosophila could potentially be related to biological pathways implicated in adipose tissue rather than pure liver-related pathways.
One strength of our study is in that we performed our analyses in human and two different model organisms, allowing for a more comprehensive insight into biological mechanisms involved in the function of alcohol consumption genes under different biological scenarios. A second strength of this study is in the use of RNAi technique which provides insight into the function of genes and what biological manifestation they would have when exposed to ethanol. The third strength of our study is in the use of CAFE assay that allows for the investigation of food and alcohol consumption in Drosophila in a more controlled environment. In our CAFE assay, each experimental box per genotype contained both normal food and ethanol food (food supplemented with 15% ethanol) providing the insects with a choice. Another strength of our study is that in our TAG levels experiments, we made our conclusions based on the comparisons between flies (RNAi vs. control) that were exposed to identical food and environmental conditions which increases the robustness of our conclusions. Finally, we combined the results with human studies to get better insight into the link between alcohol consumption and lipid metabolites.
Alcohol consumption in Airwave participants was calculated based on self-reported data which could have affected the precision of the alcohol consumed due to recall bias. To reduce this limitation, the duration of recall was limited to the last 7 days in the Airwave study. We should acknowledge that the concentration of alcohol within alcoholic drinks is not standard50, and our calculation of alcohol consumed could be affected by these variations. Although the genetic variants used for our investigations were originally found in human studies and we also performed a metabolomics analysis between alcohol consumption and circulating metabolites in the human population, the main part of the study was performed in model organisms and the results of this study might not directly generalisable to patients and the public without performing further population studies.
Conclusion
We found that alcohol-associated genes may be involved in the metabolism of lipids. Our study highlights three genes (WDPCP, TENM2, and GPN1) that may be involved in the accumulation of lipids. Of these genes, WDPCP exhibits its effects on lipid accumulation in Drosophila with exposure to ethanol. The gene expression of WDPCP in the human population supports a link to liver fibrosis. Further studies are necessary to investigate the role of this gene in ALD.
Supplementary Information
Acknowledgements
R.P. was supported by Rutherford Fund fellowship from the Medical Research Council (MR/R026505/1 and MR/R026505/2). B.A., X.J., and F.O. were supported by Rutherford Fund from Medical Research Council MR/R026505/2. R.M. was funded by the President’s PhD Scholarship from Imperial College London. PE is Director of the MRC Centre for Environment and Health and acknowledges support from the Medical Research Council (MR/S019669/1). PE also acknowledges support from the UK Dementia Research Institute, Imperial College London (UKDRI-5001), Health Data Research UK London (HDRUK-1004231) and the British Heart Foundation Imperial College London Centre for Research Excellence (BHF-RE/18/4/34215). The Airwave Health Monitoring Study was funded by the UK Home Office (780- TETRA, 2003-2018) and is currently funded by the MRC and ESRC (MR/R023484/1) with additional support from the NIHR Imperial College Biomedical Research Centre in collaboration with Imperial College NHS Healthcare Trust. R.C.P is supported by the UK Dementia Research Institute (UKDRI-5001), which receives its funding from UK DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK. Work in LMM’s laboratory is supported by the UK Medical Research Council, intramural project MC_UU_00025/3 (RG94521). The views expressed are those of the authors and not necessarily those of the sponsors. We thank Prof. Ulrike Heberlein, (Janelia Research Campus, Virginia, USA) for generously providing us the hppy17-51 fly lines. This research was funded, in whole or in part, by the Medical Research Council (MR/R026505/1 and MR/R026505/2). A CC BY or equivalent licence is applied to the Author Accepted Manuscript (AAM) arising from this submission, in accordance with the grant’s open access conditions.
Author contributions
R.P. contributed to study design, supervision, leading the project, and data interpretation. X.J. performed the initial analysis including SNP selection and shortlisting for the experiments. R.C.P performed the metabolome wide association analysis within the Airwave study, R.M. performed metabolomics GWAS within the Airwave study. F.O. performed the pathway enrichment analysis. B.A. performed the Drosophila experiments. S.H.Y.L. and L.M.M. supervised the Drosophila experiments. J.B. conducted the C. elegans experiments. A.B. provided scientific input and supervision regarding data metabolomics and GWAS analysis within the Airwave study. I.T. and P.E. acquired the Airwave data. F.O. and R.P. drafted the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Felix O’Farrell, Benjamin Aleyakpo, Rima Mustafa, Xiyun Jiang and Rui Climaco Pinto.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-47371-7.
References
- 1.World Health, O . Global Status Report on Alcohol and Health 2018: Executive Summary. World Health Organization; 2018. [Google Scholar]
- 2.Osna NA, Donohue TM, Jr, Kharbanda KK. Alcoholic liver disease: Pathogenesis and current management. Alcohol Res. 2017;38:147–161. [PMC free article] [PubMed] [Google Scholar]
- 3.Evangelou E, et al. New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat. Hum. Behav. 2019;3:950–961. doi: 10.1038/s41562-019-0653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pazoki R, et al. Genetic analysis in European ancestry individuals identifies 517 loci associated with liver enzymes. Nat. Commun. 2021;12:2579. doi: 10.1038/s41467-021-22338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott P, et al. The Airwave Health Monitoring Study of police officers and staff in Great Britain: Rationale, design and methods. Environ. Res. 2014;134:280–285. doi: 10.1016/j.envres.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 6.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jewison T, et al. SMPDB 2.0: Big improvements to the small molecule pathway database. Nucleic Acids Res. 2014;42:D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastie T, T. R., Narasimhan, B. & Chu, G. impute: Imputation for Microarray Data (2020).
- 9.Roshchupkin GV, et al. HASE: Framework for efficient high-dimensional association analyses. Sci. Rep. 2016;6:36076. doi: 10.1038/srep36076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Farrell F, Jiang X, Aljifri S, Pazoki R. Molecular alterations caused by alcohol consumption in the UK biobank: A Mendelian randomisation study. Nutrients. 2022;14:2943. doi: 10.3390/nu14142943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith GD, Ebrahim S. 'Mendelian randomization': Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Anasanti MD, Drenos F, Blakemore AI, Pazoki R. Urinary sodium excretion enhances the effect of alcohol on blood pressure. Healthcare. 2022;10:1296. doi: 10.3390/healthcare10071296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, et al. Association studies of up to 12 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet. Epidemiol. 2008;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 15.The Genotype-Tissue Expression (GTEx) project. Nat Genet45, 580–585 (2013). [DOI] [PMC free article] [PubMed]
- 16.Thompson, A., et al. Functional validity, role, and implications of heavy alcohol consumption genetic loci. Science advances6, eaay5034 (2020). [DOI] [PMC free article] [PubMed]
- 17.Pandey P, Singh A, Kaur H, Ghosh-Roy A, Babu K. Increased dopaminergic neurotransmission results in ethanol dependent sedative behaviors in Caenorhabditis elegans. PLoS Genet. 2021;17:e1009346. doi: 10.1371/journal.pgen.1009346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alaimo JT, et al. Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcoholism. 2012;36:1840–1850. doi: 10.1111/j.1530-0277.2012.01799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diegelmann S, et al. The CApillary FEeder assay measures food intake in Drosophila melanogaster. J. Vis. Exp. 2017;121:15024. doi: 10.3791/55024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aleyakpo B, et al. G-protein αq gene expression plays a role in alcohol tolerance in Drosophila melanogaster. Brain Neurosci. Adv. 2019;3:2398212819883081. doi: 10.1177/2398212819883081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68:105–115. doi: 10.1016/j.ymeth.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JR, Rajamanoharan D, McCue HV, Rankin K, Barclay JW. Small heat shock proteins are novel common determinants of alcohol and nicotine sensitivity in Caenorhabditis elegans. Genetics. 2016;202:1013–1027. doi: 10.1534/genetics.115.185025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 24.Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies AG, et al. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/S0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- 26.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castagné R, et al. Improving visualization and interpretation of metabolome-wide association studies: An application in a population-based cohort using untargeted 1H NMR metabolic profiling. J. Proteome Res. 2017;16:3623–3633. doi: 10.1021/acs.jproteome.7b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chadeau-Hyam M, et al. Metabolic profiling and the metabolome-wide association study: Significance level for biomarker identification. J. Proteome Res. 2010;9:4620–4627. doi: 10.1021/pr1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong J, et al. MetaboAnalyst 40: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corl AB, et al. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 31.Raabe RC, Mathies LD, Davies AG, Bettinger JC. The omega-3 fatty acid eicosapentaenoic acid is required for normal alcohol response behaviors in C. elegans. PLoS ONE. 2014;9:e105999. doi: 10.1371/journal.pone.0105999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolstenholme JT, et al. Dietary omega-3 fatty acids differentially impact acute ethanol-responsive behaviors and ethanol consumption in DBA/2J versus C57BL/6J mice. Alcoholism. 2018;42:1476–1485. doi: 10.1111/acer.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards AC, et al. Long-chain ω-3 levels are associated with increased alcohol sensitivity in a population-based sample of adolescents. Alcoholism. 2019;43:2620–2626. doi: 10.1111/acer.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aliev F, Barr PB, Davies AG, Dick DM, Bettinger JC. Genes regulating levels of ω-3 long-chain polyunsaturated fatty acids are associated with alcohol use disorder and consumption, and broader externalizing behavior in humans. Alcoholism. 2022;46:1657–1664. doi: 10.1111/acer.14916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langenau J, Boeing H, Bergmann MM, Nöthlings U, Oluwagbemigun K. The association between alcohol consumption and serum metabolites and the modifying effect of smoking. Nutrients. 2019;11:2331. doi: 10.3390/nu11102331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collier S, Lee H, Burgess R, Adler P. The WD40 repeat protein fritz links cytoskeletal planar polarity to frizzled subcellular localization in the drosophila epidermis. Genetics. 2005;169:2035–2045. doi: 10.1534/genetics.104.033381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui C, et al. Wdpcp, a PCP protein required for ciliogenesis, regulates directional cell migration and cell polarity by direct modulation of the actin cytoskeleton. PLoS Biol. 2013;11:e1001720. doi: 10.1371/journal.pbio.1001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SK, et al. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur. J. Hum. Genet. 2013;21:8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langhans MT, et al. Wdpcp regulates cellular proliferation and differentiation in the developing limb via hedgehog signaling. BMC Dev. Biol. 2021;21:10. doi: 10.1186/s12861-021-00241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin GG, et al. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J. Biol. Chem. 2003;278:21429–21438. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 42.Taneja-Bageshwar S, Gumienny TL. Regulation of TGFβ superfamily signaling by two separable domains of glypican LON-2 in C. elegans. Worm. 2013;2:e23843. doi: 10.4161/worm.23843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, et al. Hepatocyte TGF-β signaling inhibiting WAT browning to promote NAFLD and obesity is associated with Let-7b-5p. Hepatol. Commun. 2022;6:1301–1321. doi: 10.1002/hep4.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang X, et al. Impaired reciprocal regulation between SIRT6 and TGF-β signaling in fatty liver. FASEB J. 2022;36:e22335. doi: 10.1096/fj.202101518R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciuclan L, et al. TGF-beta enhances alcohol dependent hepatocyte damage via down-regulation of alcohol dehydrogenase I. J. Hepatol. 2010;52:407–416. doi: 10.1016/j.jhep.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Boucard AA, Maxeiner S, Südhof TC. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: Regulation by alternative splicing. J. Biol. Chem. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tews D, et al. Comparative gene array analysis of progenitor cells from human paired deep neck and subcutaneous adipose tissue. Mol. Cell. Endocrinol. 2014;395:41–50. doi: 10.1016/j.mce.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 48.Tews D, et al. Teneurin-2 (TENM2) deficiency induces UCP1 expression in differentiating human fat cells. Mol. Cell. Endocrinol. 2017;443:106–113. doi: 10.1016/j.mce.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Scannell Bryan M, et al. Genome-wide association studies and heritability estimates of body mass index related phenotypes in bangladeshi adults. PLoS ONE. 2014;9:e105062. doi: 10.1371/journal.pone.0105062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferner RE, Chambers J. Alcohol intake: Measure for measure. BMJ. 2001;323:1439–1440. doi: 10.1136/bmj.323.7327.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elsworth B, et al. The MRC IEU OpenGWAS data infrastructure. Biorxiv. 2020;35:99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).