Abstract
Raw sewage samples from an area where hepatitis E is not endemic (Barcelona, Spain) were analyzed by reverse transcriptase-PCR followed by nested PCR. One of the 37 tested samples showed a positive result for hepatitis E virus (HEV). The detected strain was amplified by inoculation into rhesus monkeys, and the course of the infection was studied by analyzing serological and biochemical parameters and by monitoring the presence of HEV in serum and feces. Fecal suspensions from the rhesus monkeys were used as the source of viral particles for sequence analysis. Eighty percent of the genome of the isolated strain, named BCN, was sequenced and found to be phylogenetically related to Asian (Indian) strains, with a 98% nucleotide identity with an isolate from Madras, India. Since this was a single isolation we cannot conclude that HEV is regularly present in the sewage. However, the finding of viable HEV in sewage has implications for contamination of the environment and shellfish by HEV and must be considered in the diagnosis of viral hepatitis in regions of nonendemic hepatitis.
Hepatitis E virus (HEV) is an important enterically transmitted pathogen in many areas of Asia, Africa, and the Middle East, where it causes substantial morbidity and mortality, particularly among pregnant women (2). In industrialized countries where HEV is not endemic, infections occur occasionally as sporadic cases, most of which are imported, although seroepidemiological studies in these areas have indicated a 1 to 5% prevalence of antibody to HEV (anti-HEV) (13) even in the absence of clinical cases. Possible explanations for such a high prevalence of anti-HEV include the detection of antibodies cross-reactive with HEV, the presence of avirulent HEV strains (2), and a low level of exposure to HEV, resulting in a high subclinical attack rate in industrialized countries.
The reverse transcriptase-PCR (RT-PCR) technique has been used to sequence the genomes of several HEV strains (9, 16, 18) and to detect HEV genomes in clinical and experimental samples (16, 21). Sensitive and specific assays based on PCR have also been developed for the detection of other human viruses in raw sewage and environmental samples (8, 10, 15). We used RT-PCR specific for HEV to analyze raw sewage at the entry to a sewage treatment and purification plant in the city of Barcelona, Spain, an area in which HEV is not endemic. An HEV strain was recovered from the sewage stream and characterized.
MATERIALS AND METHODS
Sewage samples.
Thirty-seven samples of raw sewage with a mean value of 1.7 × 106 fecal coliform bacteria/100 ml were collected from the sewage network of Barcelona, Spain, from 1994 to 1998. The sewage samples were collected in the following periods: 12 samples (taken monthly) from September 1994 to August 1995; 3 samples in February, March, and September 1996; six samples in the summer of 1997; and 16 samples between September 1997 and February 1998. Each sample was collected in a sterile 500-ml polyethylene container, kept at 4°C for less than 8 h until the viral particles were concentrated in phosphate-buffered saline (PBS), and stored frozen at −80°C.
Viruses and cells.
Fecal suspensions containing the HEV SAR-55 strain (10% in PBS, pH 7.3), whose titers were determined by infectivity in cynomolgus monkeys (Macaca fascicularis) (21), were used as a positive control for the PCR analysis. Poliovirus type 1 (LSc 2ab strain) was propagated in Buffalo green monkey kidney cells growing in Eagle minimal essential medium (Auto-pow; ICN Biomedicals Inc.) containing 5% fetal bovine serum.
Concentration of viral particles and nucleic acid extraction.
The method applied for the recovery of viral particles and the nucleic acid extraction was chosen on the basis of previous studies (8, 15). Briefly, 40 ml of sewage sample was ultracentrifuged (229,600 × g for 1 h at 4°C) to pellet all the viral particles together with any suspended material. The sediment was resuspended by mixing it with 4 ml of 0.25 N glycine buffer, pH 9.5, on ice for 30 min, and the suspended solids were removed by centrifugation (12,000 × g for 15 min). The viruses in the supernatant were pelleted by ultracentrifugation (229,600 × g for 1 h at 4°C), resuspended in 0.1 ml of PBS, and stored at −80°C. In previous studies, the efficiency of recovery of the poliovirus type 1 LSc 2ab strain was 70% of the PFU seeded into sewage samples (15).
Viral nucleic acids were extracted with guanidinium thiocyanate and adsorption of the nucleic acids to silica particles (5).
Sensitivity of the method for HEV.
Raw urban sewage samples or PBS, pH 7.4, was spiked with a standard stock of the SAR-55 strain of HEV (final concentration, 102 infectious doses/ml) (21) in order to evaluate the sensitivity of the method. The stability at 20°C of the HEV both in sewage water and in PBS was checked by comparison with that of poliovirus type 1 (final concentration, 105 infectious doses/ml). Tenfold serial dilutions of aliquots were analyzed by PCR immediately and at 1 and 2 months after the inoculation.
Primates and inoculation.
Two rhesus monkeys (Macaca mulatta) were each inoculated intravenously with 25 ml of sewage (5 ml/day). Blood and feces were collected weekly. Serum levels of the liver enzymes alanine aminotransferase, isocitrate dehydrogenase, and γ-glutamyl transpeptidase were assayed as previously described (18). Viral RNA was detected by RT-PCR.
Serologic test.
Anti-HEV was detected by a modified enzyme-linked immunosorbent assay protocol (19).
Enzymatic amplification.
For detection of the HEV genomes, 5-μl aliquots of the nucleic acids extracted were used for reverse transcription plus 1.5 mM MgCl2 and 1× PCR buffer II (Perkin-Elmer Roche, Inc.), which contained 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 0.01 M dithiothreitol, 200 μM (each) deoxynucleoside triphosphate, and 12.5 pmol of reverse detection primer HEVr473 (5′-CCTCGAAGCAGTAAGTGCGGTC-3′) in a total volume of 10 μl. The reaction mixture was incubated at 95°C for 5 min before the addition of 50 U of Moloney murine leukemia virus RT (Perkin-Elmer Roche, Inc.) and 10 U of RNase inhibitor (Perkin-Elmer Roche, Inc.). After 30 min at 42°C, the reaction mixture was again heated for 5 min at 95°C.
For a typical one-step reaction, 10 μl of the cDNA solution (corresponding to 2 ml of sewage sample, 5 μl of serum, or 5 μl of 10% fecal suspension in PBS, pH 7.4) was used. Amplification was carried out in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate, 12.5 pmol of direct detection primer HEV51 (5′-GGCTCCTGGCATCACTACTG-3′), and 2 U of AmpliTaq DNA polymerase (Perkin-Elmer Roche, Inc.). Thermal cycling of the amplification mixture was performed in a programmable heat block (Gene Amp PCR System 2400; Perkin-Elmer). In all PCR assays the first cycle of denaturation was carried out for 2 min at 94°C. The conditions for the amplification were denaturing at 92°C for 90 s, annealing at 55°C for 90 s, and extension at 72°C for 120 s.
The external primers were used in the first 30 cycles of amplification. Then 1 μl of the reaction mixture was added to a new batch of 50 μl of PCR mixture, containing 12.5 pmol each of the nested detection primers HEV101 (5′-ACTCTGCCCTTGCGAATGCT-3′) and HEVr383 (5′-TACCAACGCTGAACATCACG-3′), in a repeat 30-cycle amplification. The products were analyzed by agarose gel electrophoresis with ethidium bromide as a stain.
Sequencing and analysis of the viral genome.
The sequences of eight human HEV strains were aligned by using the CLUSTALW-1.7 program of the Genetics Computer Group (GCG) software package (version 9.1; University of Wisconsin Genetics Computer Group, Madison, Wis.). Based on this alignment, primers to amplify different segments along the full genome of HEV were designed with OLIGO-4.01s primer analysis software (National Biosciences, Inc.). Some primers were used for amplification by RT-PCR, followed by nested PCR with the same conditions as for the detection primers. Other regions were amplified by long RT-PCR followed by long nested PCR by using the SuperScript One-Step RT-PCR system (Gibco-BRL) with Elongase (Gibco-BRL) enzyme mix following the manufacturer’s instructions.
Nested products were purified either by electroelution, followed by phenol:chloroform:isoamyl-alcohol (25:24:1) extraction and precipitation with absolute ethanol and 0.3 M sodium acetate (pH 5.5), or with the QIAquick PCR purification kit (QIAGEN, Inc.). Both strands of the purified DNA were sequenced with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase FS (Perkin-Elmer, Applied Biosystems) following the manufacturer’s instructions. The results were checked with the ABI PRISM 377 automated sequencer (Perkin-Elmer, Applied Biosystems). The sequences were compared with the HEV sequences in the GenBank and EMBL (European Molecular Biology Library) databases by using the FASTA program of the GCG. Phylogenetic analysis of the sequenced regions was performed with the Sequencher package. GenBank accession numbers of the sequences of the HEV strains used for phylogenetic studies are shown in Table 1.
TABLE 1.
HEV strains used in the phylogenetic analysis
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession no. AF058679, AF058680, AF058681, AF061581, AF058683, AF058684, and AF058682.
RESULTS
Sewage analysis.
One of the 37 sewage samples (taken 31 August, 1995) was positive for HEV after nested PCR amplification. The HEV-positive sewage sample contained both enteroviruses and human adenoviruses, but hepatitis A viruses were not detected by RT-PCR. This sample did not show any other features distinguishable from those of other samples (14). Thirty-five of the 37 samples were also positive for human adenoviruses (data not shown).
Sensitivity of the method and HEV stability test.
In the seeded PBS samples HEV and poliovirus were detected by nested PCR at unchanged titer after incubation for 2 months at 20°C. Poliovirus was also found at unchanged titer after 2 months of incubation in sewage, but the level of HEV had fallen to 1% of its initial titer after 1 month in sewage and fell to undetectable levels after 2 months. Thus, the stability of HEV in sewage appeared to be less than that of poliovirus.
Biochemical, serological, and virological analyses of HEV infection in rhesus monkeys.
The HEV detected in the sewage was viable because both monkeys inoculated with the sewage became infected and excreted virus in the feces (data not shown). Fecal shedding began during the first or second week postinoculation and continued until the fifth week. Seroconversion to anti-HEV (immunoglobulin M and immunoglobulin G) occurred at week 5. Virus was not detected in the serum. The liver enzyme levels in serum remained normal except for one minor rise in alanine aminotransferase activity in one of the monkeys at week 5.
Molecular characterization of the Barcelona strain.
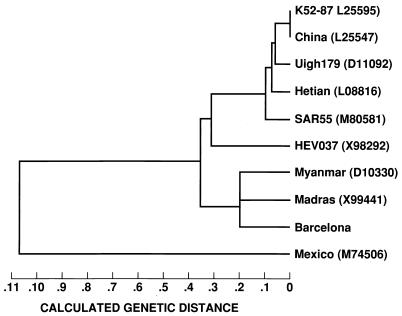
Fecal suspensions from the rhesus monkeys were used as the source of viral particles for the characterization of the BCN strain of HEV recovered from the sewage. We sequenced 580 nucleotides of the putative methyltransferase domain; 1,474 nucleotides that included the Y domain, the putative papain-like protease, and the polyproline hinge domain; 676 nucleotides of the putative helicase domain; two fragments of 276 and 637 nucleotides corresponding to the RNA-directed RNA polymerase; and a fragment that included 148 nucleotides of the 3′ end of ORF2 and 49 nucleotides of the 3′-noncoding region. The longest segment, 1,946 nucleotides, encompasses the 3′ end of ORF1, the full ORF3, and nearly all of ORF2. The 5,785 nucleotides sequenced represent 80% of the genome. Nucleotide sequence comparison with data from GenBank and EMBL and phylogenetic analysis revealed that HEV BCN is closely related to Indian strains, showing about 98% nucleotide identity with the Madras isolate (GenBank accession no. X99441) (Fig. 1), and shares a lower level of similarity (79%) with strains isolated in North Africa (Fig. 2).
FIG. 1.
Phylogenetic analysis using the longest of the sequenced segments (1,946 nucleotides) of the genome. GenBank accession numbers are indicated.
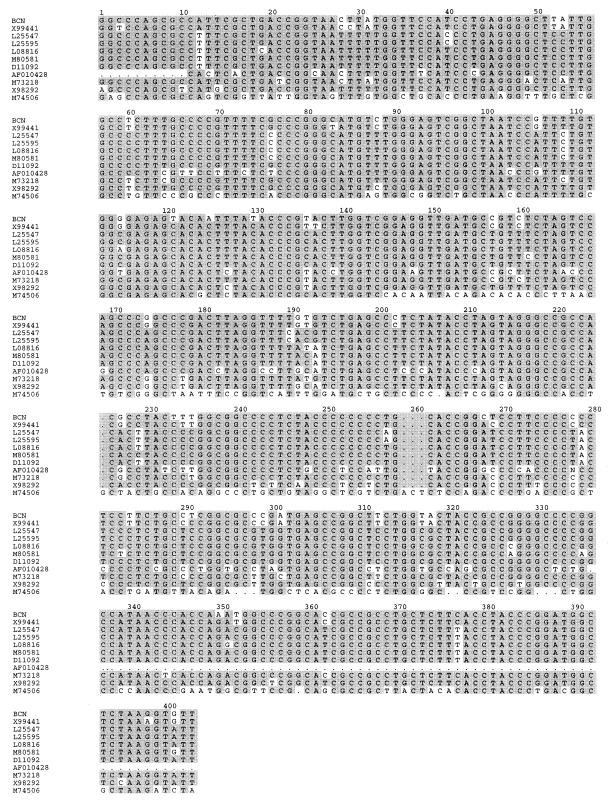
FIG. 2.
Sequence alignment of the hypervariable regions of the BCN strain of HEV and other HEV strains. GenBank accession numbers are indicated on the left. Shaded areas indicate sequence identities.
DISCUSSION
We were able to detect and isolate an infectious strain of HEV from the influent of a wastewater treatment plant that services Barcelona, Spain, and receives inflow of 670,000 m3/day. The differences between the nucleotide sequence of the detected HEV genome and that of the strain used as a positive control, and the results of the infectivity assays, rule out the possibility of virus detection due to cross-contamination in the laboratory.
The presence of HEV strains in areas where hepatitis E is not endemic was suggested following descriptions of sporadic HEV cases and a higher-than-expected prevalence of anti-HEV among blood donors in those countries (11, 12). In Spain, sporadic cases of hepatitis E were reported to be related to the consumption of shellfish grown in sewage-polluted areas (3, 17), and cases of community-acquired hepatitis E have been described among previously healthy subjects and patients on hemodialysis (6). Since over a 4-year period we obtained only one HEV-positive sample from the many sewage samples we tested, the virus may only rarely be present in sewage in high enough concentrations to be detected.
The BCN strain of HEV showed a typical course of infection in the infected animals (19, 20) except that they did not develop hepatitis. The sequence of the BCN strain is phylogenetically related to Asian (Indian) strains, with a 98% identity with the Madras strain, 92% identity with SAR-55 strain (from Pakistan), and only 79% identity with the known sequences of Moroccan or Algerian strains of HEV.
The stability of the HEV SAR-55 strain in sewage was lower than that of poliovirus 1 but sufficient to allow detection after 1 month at 20°C. The recovery of infectious HEV from sewage raises the question of the possibility of contamination of the environment and shellfish with HEV.
ACKNOWLEDGMENTS
This work was supported by grant GRQ 94-1073 and research grant Ajut a la Recerca 22.02.441.01-6(95) from the Generalitat de Catalunya. S. P. is a fellow of the Generalitat de Catalunya.
We thank Serveis Científic-Tècnics of the University of Barcelona for their help in the sequencing of the PCR products. Excellent animal care was provided by the staff of Bioqual (Gaithersburg, Md.).
REFERENCES
- 1.Aye T T, Uchida T, Ma X Z, Iida F, Shikata T, Zhuang H, Win K M. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balayan M S. Epidemiology of hepatitis E virus infection. J Viral Hepatitis. 1977;4:155–165. doi: 10.1046/j.1365-2893.1997.00145.x. [DOI] [PubMed] [Google Scholar]
- 3.Balayan M S. Hepatitis E virus infection in Europe: regional situation regarding laboratory diagnosis and epidemiology. Clin Diagn Virol. 1993;1:1–19. doi: 10.1016/0928-0197(93)90027-3. [DOI] [PubMed] [Google Scholar]
- 4.Bi S L, Purdy M A, McCaustland K A, Margolis H S, Bradley D W. The sequence of hepatitis E virus isolated directly from a single source during an outbreak in China. Virus Res. 1993;28:233–247. doi: 10.1016/0168-1702(93)90024-h. [DOI] [PubMed] [Google Scholar]
- 5.Boom R, Sol C J A, Salimans M M M, Jansen C J, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buti M, Jardi R, Cotrina M, Rodriguez-Frias F, Troonen H, Viladomiu L, Esteban J I, Esteban R, Guardia J. Hepatitis E virus infection in acute hepatitis in Spain. J Virol Methods. 1995;55:49–54. doi: 10.1016/0166-0934(95)00044-u. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee R, Tsarev S, Pillot J, Coursaget P, Emerson S U, Purcell R H. African strains of hepatitis E virus that are distinct from Asian strains. J Med Virol. 1997;53:139–144. [PubMed] [Google Scholar]
- 8.Girones R, Puig M, Allard A, Lucena F, Wadell G, Jofre J. Detection of adenovirus and enterovirus by PCR amplification in polluted waters. Water Sci Technol. 1995;31:351–357. [Google Scholar]
- 9.Huang C C, Nguyen D, Fernandez J, Yun K Y, Fry K E, Bradley D W, Tam A W, Reyes G R. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- 10.Jothikumar N, Aparna K, Kamatchiammal S, Paulmurugan R, Saravanadevi S, Khanna P. Detection of hepatitis E virus in raw and treated wastewater with the polymerase chain reaction. Appl Environ Microbiol. 1993;59:2558–2562. doi: 10.1128/aem.59.8.2558-2562.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khuroo M S, Teli M, Skidmore S, Sofi M A, Khuroo M I. Incidence and severity of viral hepatitis in pregnancy. Am J Med. 1981;70:252–255. doi: 10.1016/0002-9343(81)90758-0. [DOI] [PubMed] [Google Scholar]
- 12.Mast E E, Krawczynski K. Hepatitis E: an overview. Annu Rev Med. 1996;47:257–266. doi: 10.1146/annurev.med.47.1.257. [DOI] [PubMed] [Google Scholar]
- 13.Paul D A, Knigge M F, Ritter A, Gutierrez R, Pilot-Matias T, Chau K H, Dawson G J. Determination of Hepatitis E virus seroprevalence by using recombinant fusion proteins and synthetic peptides. J Infect Dis. 1994;169:801–806. doi: 10.1093/infdis/169.4.801. [DOI] [PubMed] [Google Scholar]
- 14.Pina S, Puig M, Lucena F, Jofre J, Girones R. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol. 1998;64:3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig M, Jofre J, Lucena F, Allard A, Wadell G, Girones R. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl Environ Microbiol. 1994;60:2963–2970. doi: 10.1128/aem.60.8.2963-2970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam A W, Smith M M, Guerra M E, Huang C C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torne J, Miralles R, Tomas S, Saballs P. Thyphoid fever and acute non-A non-B hepatitis after shellfish consumption. Eur J Clin Microbiol Infect Dis. 1988;7:581–582. doi: 10.1007/BF01962622. [DOI] [PubMed] [Google Scholar]
- 18.Tsarev S A, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsarev S A, Emerson S U, Tsareva T S, Yarbough P O, Lewis M, Govindarajan S, Reyes G R, Shapiro M, Purcell R H. Variation in course of hepatitis E in experimentally infected cynomolgus monkeys. J Infect Dis. 1993;167:1302–1306. doi: 10.1093/infdis/167.6.1302. [DOI] [PubMed] [Google Scholar]
- 20.Tsarev S A, Tsareva T S, Emerson S U, Kapikian A Z, Ticehurst J, London W, Purcell R H. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J Infect Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- 21.Tsarev S A, Tsareva T S, Emerson S U, Yarbough P O, Legters L J, Moskal T, Purcell R H. Infectivity titration of a prototype strain of hepatitis E virus in cynomolgus monkeys. J Med Virol. 1994;43:135–142. doi: 10.1002/jmv.1890430207. [DOI] [PubMed] [Google Scholar]
- 22.Yin S R, Purcell R H, Emerson S U. A new Chinese isolate of hepatitis E virus: comparison with strains recovered from different geographical regions. Virus Genes. 1994;9:23–32. doi: 10.1007/BF01703432. [DOI] [PubMed] [Google Scholar]