Abstract
Rimicaris exoculata shrimps from hydrothermal vent ecosystems are known to host dense epibiotic communities inside their enlarged heads and digestive systems. Conversely, other shrimps from the family, described as opportunistic feeders have received less attention. We examined the nutrition and bacterial communities colonising ‘head’ chambers and digestive systems of three other alvinocaridids—Rimicaris variabilis, Nautilocaris saintlaurentae and Manuscaris sp.—using a combination of electron microscopy, stable isotopes and sequencing approaches. Our observations inside ‘head’ cavities and on mouthparts showed only a really low coverage of bacterial epibionts. In addition, no clear correlation between isotopic ratios and relative abundance of epibionts on mouthparts could be established among shrimp individuals. Altogether, these results suggest that none of these alvinocaridids rely on chemosynthetic epibionts as their main source of nutrition. Our analyses also revealed a substantial presence of several Firmicutes and Deferribacterota lineages within the foreguts and midguts of these shrimps, which closest known lineages were systematically digestive symbionts associated with alvinocaridids, and more broadly for Firmicutes from digestive systems of other crustaceans from marine and terrestrial ecosystems. Overall, our study opens new perspectives not only about chemosynthetic symbioses of vent shrimps but more largely about digestive microbiomes with potential ancient and evolutionarily conserved bacterial partnerships among crustaceans.
This work examines the nutrition and diversity or bacterial communities colonising ‘head’ chambers and digestive systems of shrimp endemic to hydrothermal vents of the South West Pacific. Our results suggest that, unlike relatives from the Atlantic Ocean, none of these shrimps rely on chemosynthetic epibionts as their main source of nutrition. On the other hand, vent shrimps from the South West Pacific exhibit conserved bacterial lineages within their digestive microbiomes similar to those of other vent shrimps and more largely, other crustaceans from marine and terrestrial ecosystems suggesting a potential ancient and evolutionarily conserved host‐symbiont partnerships among crustaceans.

INTRODUCTION
Microbial symbioses are a ubiquitous phenomenon in nature, expanding physiological capabilities and ecological niches of organisms (McFall‐Ngai et al., 2013). In many places, these associations constitute the structural base of ecosystems such as in hydrothermal vents. There, chemosynthetic symbioses with microorganisms using the chemical energy arising from vent fluid emissions are found in all invertebrates, establishing the foundation of lush faunal assemblages (Dubilier et al., 2008; Sogin et al., 2020).
Among them, Rimicaris exoculata and Rimicaris kairei form large aggregations of thousands of individuals, gathered in the close vicinity of fluid emissions respectively in the Mid Atlantic Ridge and the Central Indian Ridge (Zbinden & Cambon‐Bonavita, 2020). These shrimps host a complex community of cocci, rod‐shaped and filamentous epibionts on the inner side of their enlarged cephalothorax, that is, the branchiostegite, and on setae covering the surface of their hypertrophied mouthparts (Methou et al., 2022b; Petersen et al., 2010; Zbinden et al., 2004). These communities comprise a wide diversity of chemosynthetic partners including Campylobacterota, α‐, γ‐ and ζ‐Proteobacteria as well as Desulfobacterota among others (Cambon‐Bonavita et al., 2021; Guri et al., 2012; Jan et al., 2014; Jiang et al., 2020; Methou et al., 2022b; Petersen et al., 2010; Zbinden et al., 2008), from which their hosts derive most of their nutrition (Gebruk et al., 2000; Methou et al., 2020; Polz et al., 1998; Van Dover, 2002) through direct transtegumental transfer of organic compounds (Ponsard et al., 2013). This diversity of bacterial partners reflects a diversity of metabolisms based on a wide range of energy sources (Cambon‐Bonavita et al., 2021; Jan et al., 2014; Jiang et al., 2020) enabling these animals to thrive in vent fields with contrasting profiles of fluid chemistries.
Besides, R. exoculata and R. kairei shrimps harbour another community of resident symbionts within their digestive system (Aubé et al., 2022; Durand et al., 2010, 2015; Guéganton et al., 2022; Qi et al., 2022; Zbinden & Cambon‐Bonavita, 2003). In R. exoculata this digestive symbiosis exhibits a clear partitioning between organs with several lineages of Firmicutes affiliated to Mycoplasmatales located in the foregut (oesophagus and stomach) and Firmicutes from the Clostridia class as well as Candidatus Rimicarispirillum which are long thin Deferribacterota inserted between microvilli in their midgut (Aubé et al., 2022; Guéganton et al., 2022). Unlike chemoautotrophic epibionts from the cephalothoracic cavity, these symbionts are heterotrophic and were hypothesised to complement their host diet and participate in their immunity (Aubé et al., 2022). To date, other bacterial lineages often found in the digestive microbiome of R. exoculata such as Campylobacterota and Gammaproteobacteria (Durand et al., 2010, 2015) were only observed as transient rod‐shaped and cocci cells in its alimentary bolus (Guéganton et al., 2022).
These symbiotic communities of the cephalothoracic cavity and the foregut are renewed alongside their host exoskeleton at each moult, whereas those from the midgut are maintained throughout their adult life (Corbari et al., 2008; Guri et al., 2012). The constant renewal of their microhabitat coupled with an absence of similar or closely related lineages in the surrounding environment of their host, question the transmission pathways of the Mycoplasmatales located in the foregut (Durand et al., 2015). Similarly, the lack of geographic clustering of Deferribacterota symbionts in the midgut of R. exoculata, which are also absent from the environment, suggests a maternal inheritance (Durand et al., 2015). However, these lineages were never detected on their egg broods during the entire embryonic development (Guri et al., 2012; Methou et al., 2019).
Apart from R. exoculata and R. kairei, symbioses have been found in two other alvinocaridid species, R. hybisae from the Mid Cayman Rise and R. chacei from the Mid‐Atlantic Ridge, which however display different trophic relations towards their symbiosis (Apremont et al., 2018; Assié, 2016; Nye et al., 2012). R. chacei shrimps lack an hypertrophied cephalothorax and are only partially dependent on their chemosynthetic symbiosis, with a mixed diet of symbiotrophy, bacterivory and scavenging (Gebruk et al., 2000; Methou et al., 2020). Their digestive system also hosts similar symbiotic communities than for R. exoculata with the same partitioning among foreguts and midguts (Apremont et al., 2018; Guéganton et al., 2022). On the other hand, R. hybisae shows more similarity with the ecology of R. exoculata and R. kairei, forming dense aggregates around chimneys and with an enlarged cephalothorax heavily colonised by epibionts (Nye et al., 2012; Streit et al., 2015). However, recent evidence from gut contents and isotopic compositions of R. hybisae individuals distributed at the vent site periphery suggest they might have retained an ability to feed on other sources, including facultative carnivory (Versteegh et al., 2022), in addition to chemosynthetic bacterial sources (Streit et al., 2015).
In other alvinocaridids, nutritional strategies have been hypothesised to be mostly opportunistic and scavenging, with the use of several food sources including bacterial mats, detritus or the predation of small invertebrates (Gebruk et al., 2000; Stevens et al., 2008; Suh et al., 2022; Van Audenhaege et al., 2019). Based on stable isotope compositions, it was suggested that Rimicaris variabilis and Manuscaris sp. from the Manus Basin could either be conventional grazers/scavengers or feed on episymbiotic autotrophic bacteria in a similar fashion to R. exoculata (Van Audenhaege et al., 2019). Yet, no extensive study has investigated the microbial communities from their branchiostegites, mouthparts or digestive system so far.
Our study explores the bacterial communities colonising cephalothoracic cavities and digestive systems of three alvinocaridid species from hydrothermal vents of South West Pacific basins—Rimicaris variabilis, Nautilocaris saintlaurentae and Manuscaris sp.—as well as their nutrition, using a combination of electron microscopy, multiple stable isotopes and sequencing approaches. Our aim was to examine symbiotic relationships across alvinocaridid species with distinct ecologies to better understand the role and evolution of these symbioses. We address the following questions: (1) Do alvinocaridid species described as opportunistic feeders host symbiotic communities in their cephalothoracic cavity and/or their digestive system? (2) Do these potential symbiotic communities comprise similar or related bacterial lineages to epibionts of other Rimicaris species? (3) Can these alvinocaridid species rely, at least partially, on chemosynthetic epibionts for their nutrition?
EXPERIMENTAL PROCEDURES
Field sampling
Alvinocaridid shrimps were collected during the Futuna3 2012 and CHUBACARC 2019 oceanographic expeditions on board the R/V L'Atalante using a suction sampler manipulated by the HOV Nautile and the ROV Victor 6000, respectively. A total of 81 Rimicaris variabilis individuals were sampled from eight hydrothermal vent fields: Pacmanus and Susu Knolls in the Manus basin, La Scala in the Woodlark basin, Phoenix in the North Fiji basin, Fatu Kapa in the Futuna volcanic arc and Mangatolo, ABE and Tow Cam in the Lau basin (Figure 1A–C). In addition, 25 Nautilocaris saintlaurentae individuals were also sampled at the Phoenix, Fatu Kapa and Tow Cam vent fields, as well as one Manuscaris sp. at the Pacmanus vent field. Specimens were identified morphologically and confirmed by genetic barcoding of their COI gene with specific primers for alvinocarids, using the protocol from (Methou et al., 2020). All sequences have been deposited in GenBank under accession numbers OQ363903—OQ364004 (see Table S1 for sampling summary with associated individual ID). The 16S rRNA dataset is available in the NCBI SRA repository (submission identifier SUB12697284 and BioProject identifier PRJNA932596).
FIGURE 1.

(A) Alvinocaridids shrimps on the wall of an active vent chimney at Pacmanus (Manus basin). (B) Alvinocaridids shrimps around assemblages of barnacles and at La Scala (Woodlark Basin). (C) Sampling localities of alvinocaridid shrimps from Southwest Pacific basins. Colour dots depict hydrothermal vent field locations. Shapes depict shrimp species collected at a given sampling field.
Upon their recovery on board, dissected parts or whole shrimps were stored frozen at −80°C before processing for stable isotopes or 16S rRNA metabarcoding. Specimens were dissected directly on board or in a shore‐based laboratory under sterile conditions to retrieve their anatomical parts: scaphognathites and exopodites mouthparts as well as branchiostegites from the cephalothoracic cavity, foreguts and midguts of their digestive system and pieces of abdominal muscles. Pieces from cephalothoracic cavities and mouthparts were also fixed in a 2.5% glutaraldehyde filtrated seawater solution for 16 h at 4°C, rinsed and then stored at 4°C in filtrated seawater with 0.44 g/L of NaN3 at pH 7.4 until use for scanning electron microscopy (SEM) observations.
Scanning electron microscopy
Dissected mouthparts and branchiostegites were dehydrated with an ethanol series (25%, 50%, 75% and 100% ethanol) and then for 5 h in a critical point dryer CPD 020 (Balzers Union, Balzers, Liechtenstein). Samples were then gold‐coated with an SCD 040 (Balzers Union). Observations and imaging were performed using a Quanta 200 microscope (FEI‐Thermo Fisher, Hillsboro, OR, USA).
Stable isotope analysis
The abdominal muscle of alvinocaridid shrimps was oven‐dried to constant mass at 50°C (>48 h) and ground into a homogeneous powder using a mortar and pestle. Measurements of stable isotope ratio were performed by continuous flow–elemental analysis–isotope ratio mass spectrometry (CF‐EA‐IRMS) at University of Liège (Belgium), using a vario MICRO cube C‐N‐S elemental analyser (Elementar Analysensysteme GMBH, Hanau, Germany) coupled to an IsoPrime100 isotope ratio mass spectrometer (Isoprime, Cheadle, United Kingdom). Isotopic ratios were expressed in ‰ using the widespread δ notation (Coplen, 2011) relative to the international references: Vienna Pee Dee Belemnite (for carbon), atmospheric air (for nitrogen) and Vienna Canyon Diablo Troilite (for sulphur). Primary analytical standards used for these analyses were the following: Sucrose (IAEA‐C‐6; δ13C = −10.8 ± 0.5‰; mean ± SD), ammonium sulphate (IAEA‐N‐2; δ15N = 20.4 ± 0.1‰; mean ± SD) and silver sulphide (IAEA‐S‐2; δ34S = 22.6 ± 0.1‰; mean ± SD). A secondary analytical standard, Sulfanilic acid (Sigma‐Aldrich; δ13C = −25.6 ± 0.4‰; δ15N = −0.13 ± 0.4‰; δ34S = 5.9 ± 0.5‰; means ± SD) was also used as well as an internal laboratory standard (seabass muscle). These standards were analysed interspersed among samples with one replicate of each standard every 15 analyses. Standard deviations on multi‐batch replicate measurements of secondary and internal laboratory standards were 0.2‰ for δ13C and δ15N and 0.4‰ for δ34S.
SIBER (Stable Isotope Bayesian Ellipses in R; Jackson et al., 2011) was used to explore ecological niches in an R 4.2.1 statistical environment (R Core Team, 2020). Two separate sets of standard ellipses were constructed: one with δ13C and δ15N data and another with δ13C and δ34S data. Areas of these ellipses were also estimated using the Bayesian model (SEAB) with direct intergroup pairwise comparisons of SEAB. The model solutions were presented using credibility intervals of probability density function distributions. Areas of all ellipses were also estimated using the SEAc correction for small sample sizes, as outlined in (Jackson et al., 2011).
DNA extraction and sequencing
Twenty‐three Rimicaris variabilis, six Nautilocaris saintlaurentae and one Manuscaris sp. specimens were used for DNA extraction of their mouthparts, foreguts and midguts, as well as 17 additional R. variabilis and two additional N. saintlaurentae for mouthparts only, using the Nucleospin® Soil Kit (Macherey‐Nagel, Germany) following manufacturer's instructions. Three blanks (i.e., a negative DNA extraction control) were also performed in parallel with DNA extractions of shrimp specimens.
For sequencing of the V3‐V4 variable region of 16S rRNA (Fadrosh et al., 2014) using Illumina's MiSeq technology, libraries were prepared using two successive PCR steps. (a) PCR1: samples were amplified in triplicate using the 341/785 primers (Herlemann et al., 2011) to generate a 450 bp fragment. The P5 and P7 Illumina primers were included in the 5′ part of the 341 forward and 785 reverse primers, with Truseq read 1 (CTTTCCCTACACGACGC TCTTCCGATCT) and Truseq read 2 Illumina adapters (GGAGTTCAGACGTGTGCTCTTCCGATCT) respectively. PCR1 amplifications were performed in a final volume of 50 μL using 1 or 2 μL of DNA, 1.25 U of TaqCore polymerase (MP Biomedicals), standard Buffer with final 1.5 mM MgCl2, 0.5 mM of each dNTP and 0.2 μM of each primer under the following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 53°C for 30 s and 72°C for 1 min, and a final elongation step at 72°C for 6 min. (b) PCR2: the three PCR1 replicates of each sample were then pooled and sent to the GenoToul platform (GeT‐BioPuce, INSA, Toulouse, France). Amplicons were first purified and dosed. Then they were used as templates for the PCR2 to which are added Illumina‐tailed primers targeting the half of Illumina adapters P5 and P7 used in the first PCR and a unique index per sample. After purification, all amplicons were pooled in equimolar concentrations to be sequenced on an Illumina MiSeq system using paired‐end sequencing with a V3 kit (300 bp × 2).
Metabarcoding analysis
A total of 16,817,628 raw reads across 109 samples, averaging 150,157 reads per sample, were analysed using the DADA2 pipeline (Callahan et al., 2016) in an R 4.2.1 statistical environment (R Core Team, 2020). Sequences were truncated to 250 bp for forward reads and to 240 bp for reverse reads based on the average quality scores. Additionally, reads displaying ‘N’, a quality score below 2, and/or more than 2 expected errors were discarded. The error model was trained using 1,000,000 sequences before denoising, and chimeric sequences were removed based on a consensus approach before the paired ends were assembled. Contaminants were removed using blank controls with the MicroDecon R package (McKnight et al., 2019b).
The final data set contained 10,635,660 reads, with an average of 94,961 sequences per sample after quality filtering. Representative sequences were classified into taxonomic groups using the SILVA 138 database (Quast et al., 2013). Additional filtering on abundance was conducted at a threshold of 0.01% (Bokulich et al., 2013) to remove sequences containing non‐biologically relevant amplicon sequence variants (ASVs) (Breusing et al., 2022). ASVs affiliated with mitochondria sequences of alvinocaridid shrimps were also manually removed from the data set.
Visualisation and statistical analyses of 16S rRNA bacterial diversity were performed using the Phyloseq (v. 1.4.0) (McMurdie & Holmes, 2013) and vegan (v. 2.6.2) (Oksanen et al., 2008) R packages. Alpha diversity across the 109 samples was explored with ASV number for richness and Inverse Simpson Index for evenness. Differences in richness and evenness among categories (hosting organs, vent fields, alvinocaridid host species) were compared with Kruskal–Wallis tests followed by Dunn post‐hoc tests. For Beta diversity, the dataset was normalised via proportions (McKnight et al., 2019a) with the ‘transform_sample_counts’ function (Phyloseq R package) and analysed using Bray–Curtis distance matrices with the ‘distance’ function (Phyloseq R package). Homogeneity between categories was tested with the ‘betadisper’ function (vegan R package), and significant differences between categories were tested by permutational analysis of variance (PERMANOVA; 999 permutations) with the ‘adonis2’ function (vegan R package). Constrained ordinations with stable isotopes ratios for each hosting organs were achieved by canonical analyses on the principal coordinates (CAP) using the ‘ordinate’ (Phyloseq R package) and ‘scores’ (vegan R package) functions.
RESULTS
Scanning electron microscopy observations
Observation of Rimicaris variabilis branchiostegites under Scanning Electron Microscopy (SEM) showed that the inner part of their cephalothoracic cavities was mostly devoid of bacterial colonisation (Figure 2A,B). Conversely, a more abundant bacterial colonisation was observed on the external surface of R. variabilis cephalothorax, in particular on setae aligned along their ventral side, which were covered by thick and thin filamentous bacteria (Figure 2C). In 7 out of the 12 R. variabilis individuals observed, single‐layered mats of rod‐shaped bacteria were found on their cephalothorax inner surfaces, either on the anterior part facing mouthparts or on the posterior part facing the gills (Figure 2D). In some instances (2 out of 12 individuals), small spots of filamentous bacteria were localised on the most anterior part of the branchiostegites.
FIGURE 2.

Scanning Electron Microscopy (SEM) observations of microbial communities on the surface of Rimicaris variabilis branchiostegites and mouthparts. (A) Overview of the branchiostegite inner side. scale = 1 mm. (B) Enlargement of the branchiostegite inner side devoid of bacterial colonisation; scale = 500 μm. (C) Filamentous bacteria colonising the ventral setae along the external side of the branchiostegite. scale = 100 μm. (D) Single‐layered bacterial mats colonising inner side of R. variabilis branchiostegite. scale = 10 μm. (E) Overview of a scaphognathite dorsal side. scale = 1.5 mm. (F) Dense aggregations of filamentous bacteria covering plumose setae of the scaphognathite margin. scale = 200 μm. (G) Filamentous bacteria colonising the scaphognathite surface. scale = 50 μm. (H) Small cocci and rod‐shaped bacteria colonising the scaphognathite surface. scale = 50 μm.
Bacterial colonisation was more widespread on R. variabilis mouthparts, although remaining limited to particular areas (Figure 2E). Dense aggregations of thick and thin filamentous bacteria‐covered plumose setae distributed along scaphognathite and exopodite margins (Figure 2F). The dorsal and ventral surfaces of these two mouthparts lacked bacteriophore setae (Figure 2E) and were generally only colonised by small cocci and rod‐shaped bacteria on most of their surface (Figure 2G). In 2 out of 10 R. variabilis mouthparts observed, ventral and dorsal surfaces of scaphognathites were colonised by filamentous bacteria, but to a lesser extent than marginal setae (Figure 2H).
Observations of Nautilocaris saintlaurentae and Manuscaris sp. branchiostegites under SEM revealed similar patterns of bacterial colonisation compared to R. variabilis with most of the inner parts of their cephalothoracic cavities devoid of bacteria (Figure 3A) or covered by single layered mats of rod‐shaped bacteria (Figure 3B). As for R. variabilis, a few regionalized spots of filamentous bacteria were also present on the most anterior part of the Manuscaris sp. branchiostegite, close to the cephalothorax opening (Figure 3C). Bacterial colonisation on mouthparts of these two species was also mostly limited to marginal setae covered by filamentous bacteria (Figure 3D), with only mono layers of cocci and rod‐shaped bacteria on the surfaces of their scaphognathites and exopodites.
FIGURE 3.

Scanning Electron Microscopy (SEM) observations of microbial communities on the surface of Nautilocaris saintlaurentae and Manuscaris sp. branchiostegites and mouthparts. (A) Overview (composite image) of the inner side of N. saintlaurentae branchiostegite. scale = 1 mm. (B) Single‐layered bacterial mats colonising the inner side of Manuscaris sp. branchiostegite. scale = 50 μm. (C) Spot of filamentous bacteria colonising the most anterior part of the Manuscaris sp. branchiostegite. scale = 50 μm. (D) Dense aggregations of filamentous bacteria covering plumose setae of N. saintlaurentae scaphognathite margin. scale = 50 μm.
Stable isotopes analysis
Rimicaris variabilis and Nautilocaris saintlaurentae populations showed limited variations in δ13C among vent fields (Kruskal–Wallis, p < 0.001; Figure S1A), with only significantly lower δ13C values for R. variabilis from Susu Knolls compared to those from Tow Cam, Pacmanus and La Scala (Dunn tests, p < 0.001; see Table S2 for detailed p values). Slight variations in δ15N among vent fields could be observed as well (Kruskal–Wallis, p < 0.001; Figure S1B) with higher δ15N values in R. variabilis from Pacmanus and La Scala compared to those from ABE, Fatu Kapa and Susu Knolls (Dunn tests, p < 0.001). Significant differences in δ34S of Rimicaris variabilis were also found among vent fields (Kruskal–Wallis, p < 0.001; Figure S1C) with a trend of 34S‐depletion in shrimps from vent fields of the most eastern basins—Manus and Woodlark—compared to shrimp populations from more western basins—North Fiji and Lau—(Dunn tests, p < 0.001). At Fatu Kapa, δ13C, δ15N and δ34S of R. variabilis and N. saintlaurentae were similar between the two species (Dunn tests, p > 0.001).
SIBER analysis confirmed that carbon and sulphur isotopic niches of alvinocaridids from Manus and Woodlark basins were clearly separated from those of North Fiji and Lau populations (Figure 4A). However, the same trend was not observed for carbon and nitrogen isotopic niches (Figure 4B) with some overlap between R. variabilis from Pacmanus and Fatu Kapa (1.19‰2, i.e., 15.3% of the smallest ellipse area), from Tow Cam and Pacmanus (0.74‰2, i.e., 28.9% of the smallest ellipse area) or from Tow Cam and La Scala (0.31‰2, i.e., 12.1% of the smallest ellipse area). In general, the limited overlap was observed between R. variabilis ellipses from vent fields within the same basin with no overlap between the two Manus vent fields and small overlaps between vent fields from the Lau basin (22.4% of the smallest ellipse areas at most), except for carbon and sulphur ellipses of ABE and Fatu Kapa which were strongly overlapping (4.55‰2, i.e., 69.12% of the smallest ellipse area). Between Phoenix and Fatu Kapa, carbon and sulphur ellipses of N. saintlaurentae overlapped only by 0.64‰2 (i.e., 6.5% of the smallest ellipse area) but strongly overlapped for carbon and nitrogen ones (3.45 ‰2, i.e., 62.4% of the smallest ellipse area). At Fatu Kapa, ellipses of R. variabilis and N. saintlaurentae overlapped clearly, in particular for carbon and sulphur ellipses (6.7‰2, i.e., 57.7% of the smallest ellipse area) but also for carbon and nitrogen ellipses (2.02‰2, i.e., 36.6% of the smallest ellipse area).
FIGURE 4.

Isotopic niches of alvinocaridid shrimps from southwest Pacific basins. (A) Carbon and sulphur isotopic niches. (B) Carbon and nitrogen isotopic niches. (A,B) Each dot corresponds to the isotopic ratios of a shrimp individual; colours depict hydrothermal vent field locations and shapes depict different alvinocaridid species. (C) Model‐estimated bivariate standard area (SEAB) for carbon and sulphur ellipses. (D) Model‐estimated bivariate standard area (SEAB) for carbon and sulphur ellipses. (C,D) Boxes in dark grey, medium grey and light grey correspond, respectively, to the 50%, 75% and 95% credibility intervals of probability density function distributions of the model solutions, and black dots are the modes of these distributions. Red dots are the standard ellipse areas computed using a frequentist algorithm adapted for small sample sizes (SEAC).
Areas of the standard ellipses associated with each shrimp species and vent field populations varied widely (Figure 4C,D), with SEAc values ranging from 1.85‰2 (carbon and nitrogen ellipse of R. variabilis from Susu Knolls) to 46.24‰2 (carbon and sulphur ellipse of R. variabilis from Pacmanus). Overall, R. variabilis from Pacmanus had the widest isotopic niches (Figure 4C,D), with larger niches than any other shrimp population in nearly all model solutions (>99.99% of model solutions for both carbon and sulphur niches and carbon and nitrogen niches) except for carbon and sulphur niches of R. variabilis from La Scala (only 72.09% of model solutions). These broad isotopic niches in some vent fields seemed to result in part from spatial variations, with in general, more similar and clustered isotopic ratios in individuals collected from the same sampling point, particularly at La Scala (Figure S2C). Differences in niche sizes between alvinocaridid species at Fatu Kapa were not well supported by the model, with larger carbon and nitrogen niches for R. variabilis in 82.39% of model solutions and larger carbon and sulphur niches in 59.06% of model solutions.
16S rRNA metabarcoding analysis
Read distributions among samples following processing and filtering steps varied from 27,759 reads (FU3‐Cr9‐Sc) to 153,987 reads (CHUB‐Cr53‐Sc) resulting in a maximum of 5.5‐fold difference among samples. Alpha Diversity analyses revealed slight variations in ASV richness among host organs (Kruskal–Wallis, H = 14.28, p < 0.001) or among alvinocaridid species (Kruskal–Wallis, H = 6.84, p < 0.05) with a significantly higher number of ASVs in stomach compared to mouthparts communities (Dunn's Multiple Comparison Test, p < 0.01) and a slightly higher number of ASVs in Rimicaris variabilis compared to Nautilocaris saintlaurentae communities (Dunn's Multiple Comparison Test, p < 0.05). In contrast, ASV richness was similar among back‐arc basins (Kruskal–Wallis, H = 7.08, p > 0.05) or among vent fields (Kruskal–Wallis, H = 7.58, p > 0.05). Similarly, Inverse Simpson values did not indicate any variations of evenness among organs, shrimp species, regions or vent fields (Kruskal–Wallis tests, p > 0.05).
Based on PERMANOVA analyses, bacterial community composition was significantly influenced mostly by geography (i.e., among vent field; F = 4.76, R 2 = 0.224, p < 0.001) but also by hosting organs (F = 5.94, R 2 = 0.079, p < 0.001) or host species (F = 3.22, R 2 = 0.043, p < 0.001). However, homogeneity of variances among vent fields (betadisper; F = 3.349, p < 0.01) or shrimp species (betadisper: F = 16.409, p < 0.001) was not met. Moreover, R. variabilis and N. saintlaurentae communities composition from Fatu Kapa and Phoenix taken alone—a balanced dataset (betadisper: F = 0.483, p > 0.05)—did not significantly differ between the two species (PERMANOVA: F = 1.742, R 2 = 0.052, p > 0.05).
Canonical analysis on the principal coordinates (CAP) supported a correlation between stable isotopic composition of abdominal muscles and bacterial communities of alvinocaridid mouthparts (ANOVA‐like: F = 3.027, R 2 = 0.206, p < 0.001; Figure 5A) with a significant contribution of δ15N (F = 3.025, p < 0.01) and δ34S (F = 4.556, p < 0.001), but not δ13C (F = 1.5002, p > 0.05). RDA models showed similar results for bacterial communities of alvinocaridid foreguts (ANOVA‐like: F = 1.581, R 2 = 0.182, p < 0.01; Figure 5A) and alvinocaridid midguts (ANOVA‐like: F = 1.762, R 2 = 0.203, p < 0.001; Figure 5C) with a significant contribution for δ34S but not or only slightly for δ13C on midguts and not for δ15N (see Table S3). PERMANOVA analyses further confirmed that community compositions of each organ were mostly influenced by δ34S variations with only a significant influence of δ15N for bacterial communities of mouthparts and a slight effect of δ13C for bacterial communities of midguts (PERMANOVA tests; see Table S4 for detailed values).
FIGURE 5.

Constrained ordinations of 16S rRNA bacterial diversity by stable isotope ratios using canonical analysis on the principal coordinates (CAP) for each hosting organs. (A) Mouthparts bacterial communities; (B) Foreguts bacterial communities; (C) Midguts bacterial communities. Results from ANOVA‐like permutation tests for CAP are displayed on each plot panel. Stable isotopic ratios that significantly contributed to CAP results are marked with an asterisk (p < 0.01, see Table S3). Points are coloured by hydrothermal vent field locations with shapes depicting distinct alvinocaridid species.
The composition of microbial communities on mouthparts was largely dominated by Campylobacterota ASVs (80.5% of mean relative abundance) followed by Proteobacteria ASVs (15.4%) (Figure 6A). They also included lower proportions of Bacteroidota ASVs (1.9%) and Firmicutes ASVs (1.1%); (Figure 6A). Campylobacterota ASVs also dominated microbial communities of foreguts (66.4%) and midguts (43.1%), but other groups had much higher relative abundances than on mouthparts, in particular, Firmicutes (11.7% in foreguts and 27.6% in midguts, respectively) but also Verrucomicrobiota (3.2% in foreguts and 2.9% in midguts respectively) and Bacteroidota to a lower extent (3.9% in foreguts and 7.7% in midguts respectively) (Figure 6B,C). In contrast, lower relative abundances of Proteobacteria were retrieved both in foreguts (8.5%) and in midguts (7.4%). Substantial proportions of Desulfobacterota (2.8%) and Patescibacteria (1.1%) were also found in foreguts; (Figure 6B) and of Fusobacteriota in midguts (2.5%); (Figure 6C). Large proportions of bacterial sequences, not affiliated to any particular phylum according to the Silva 138 database, were observed as well in foreguts (1.1%) and in midguts (7.7%).
FIGURE 6.
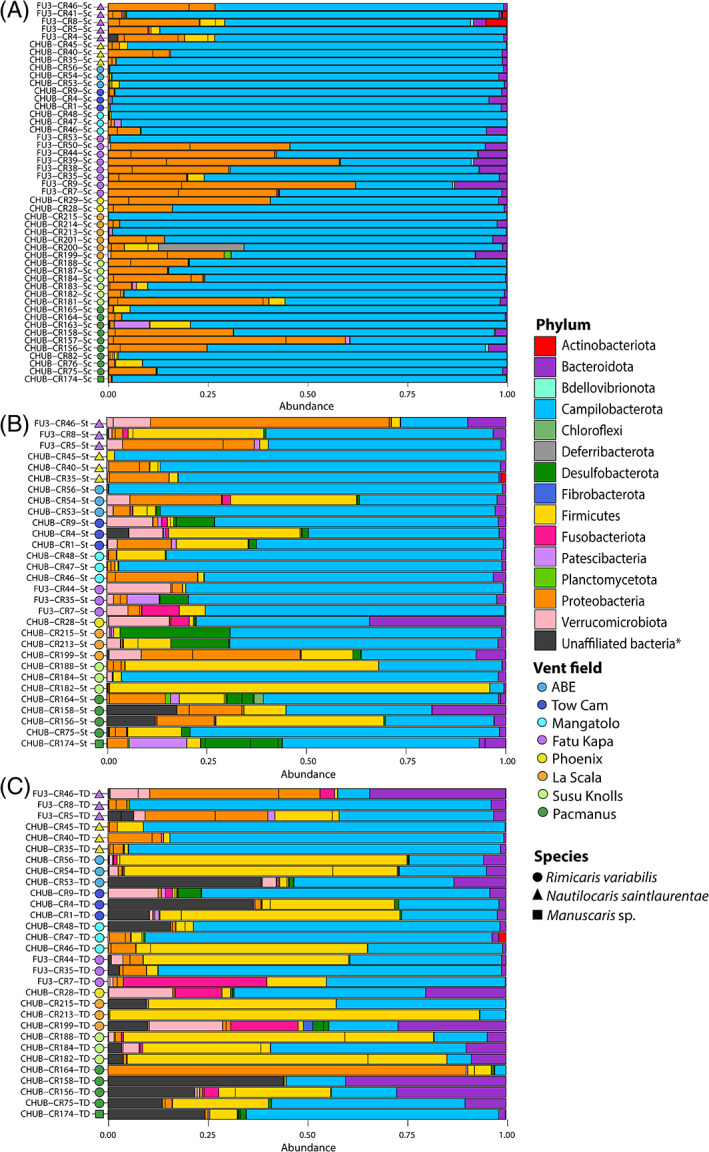
Relative abundances of 16S rRNA gene sequence reads from bacterial communities associated with southwest Pacific alvinocaridids according to their classification at the phylum level (Silva 138 database). *Bacteria unaffiliated at the phylum level according to the Silva 138 database include also some ASVs belonging to the Deferribacterota phylum (see Figure 7). (A) Mouthparts bacterial communities; (B) Foreguts bacterial communities; (C) Midguts bacterial communities.
Best BLAST hits of these unaffiliated ASVs with the NCBI database revealed two main bacterial lineages, the most abundant lineage showing clear sequence similarity with R. chacei and R. exoculata Deferribacterota gut symbionts (98.1% for both ASVs) whereas the other corresponded to an unknown bacterial lineage similar to a symbiont retrieved from a cuttlefish nidamental gland (97.9% and 98%); (Figure 7A and Table S5). Phylogenetic reconstruction of these unaffiliated ASVs agglomerated by phylogenetic similarity together with ASVs affiliated to Deferribacterota (ASV9011 and ASV9059) confirmed their similarity with the ‘unaffiliated’ Deferribacterota ASVs (ASV9038 and ASV9045) (Figure 7A). The Deferribacterota ASV9011 and ASV9059 were identical to a gut symbiont of R. chacei (Table S5) but were only found in the mouthpart of one R. variabilis individual (CHUB‐Cr200‐Sc). On the other, the Deferribacterota ASV9038 and ASV9045 were retrieved within each hosting organ and each alvinocaridid species (Figure 7A), although these were notably absent from all organs of seven individuals (CHUB‐Cr28, CHUB‐Cr40, CHUB‐Cr45, CHUB‐Cr47, CHUB‐Cr164, FU3‐Cr7 and FU3‐Cr44).
FIGURE 7.

Phylogenetic trees of (A) Deferribacterota and unaffiliated bacterial ASVs and (B) Firmicutes ASVs agglomerated by similarity (h = 0.1). Panel A: Bacterial ASVs without phylum affiliation with the Silva 138 database are marked with an asterisk. All panels: Trees were constructed with the Maximum Likelihood method, based on the General Time Reversible model with Gamma distribution and allowing for some sites to be invariable (GTR + I + G). Prop: Relative abundance of the ASV among total sequence reads within the dataset. %Sim: % of similarity with the ASV best BLAST hit (R. exoculata or R. chacei digestive epibiont; details on Table S5). Each dot represents the occurrence of the lineage in an individual with shapes depicting alvinocaridid species (circle: R. variabilis; triangle: N. saintlaurentae; square: Manuscaris sp.) and colours depicting the hosting organ (red: mouthpart; green: stomach; blue: digestive tube).
Phylogenetic reconstruction of Firmicutes ASVs agglomerated by phylogenetic similarity showed three main bacterial lineages in this phylum (Figure 7B) with three ASVs affiliated to the Candidatus Hepatoplasmata genus (class Bacilli), three ASVs affiliated to the Candidatus Bacilloplasma genus (class Bacilli) and one ASV affiliated to the Tyzzerella genus (class Clostridia). Best BLAST hits of these ASVs were always Rimicaris exoculata or Rimicaris chacei foreguts and midguts symbionts with sequence similarity comprised between 98.7% and 99.9% (Figure 7B and Table S5). Most Firmicutes ASVs were present within each hosting organs and among each alvinocaridid species except ASV1723 that was found within R. variabilis foreguts and midguts only and ASV1649 that was only within R. variabilis and N. saintlaurentae foreguts and midguts (Figure 7B).
DISCUSSION
Nutritional strategies of alvinocaridid shrimps from hydrothermal vents of the Southwest Pacific basins
Our observations on the inner side of the cephalothoracic cavities showed only a scarce coverage of bacterial epibionts for either Rimicaris variabilis, Nautilocaris saintlaurentae or Manuscaris shrimps (Figures 2 and 3). A slightly more developed colonisation was observed on their mouthparts with some filamentous bacteria although limited to particular areas, mostly the plumose setae on the mouthpart margins. This sharply contrasts with colonisation patterns seen not only in alvinocaridid species relying mostly on their cephalothoracic chemosymbiosis such as Rimicaris exoculata (Petersen et al., 2010; Zbinden et al., 2004; Zbinden & Cambon‐Bonavita, 2020) or Rimicaris kairei (Methou et al., 2022b), but also in those with a mixed diet, only partially dependent on this symbiosis like Rimicaris chacei (Apremont et al., 2018), and which all exhibit extensive colonisation of their cephalothoracic cavities by filamentous bacteria. Although earlier works stress out the importance of the moult cycle in R. exoculata symbiosis (Corbari et al., 2008), with a sparse colonisation within the cephalothoracic cavity in early moult stage individuals, it is unlikely that moult stages have introduced a bias in our observations for alvinocaridids from South West Pacific basins. Indeed, the inversely dense colonisation of ventral setae along the external face of their cephalothorax (Figure 2C) suggests that limited colonisation of their branchiostegites does not stem from a recent renewal of the exoskeleton but is found all along their moult cycle.
In addition, no clear correlation between the relative abundance of epibionts colonising their mouthparts and stable isotopes ratios of carbon could be established among alvinocaridid individuals analysed in our study (Figure 5). In hydrothermal vent ecosystems, these variations in δ13C are mostly attributed to the use of different carbon fixation pathways by chemosynthetic microorganisms, with depleted δ13C ratios for those using the CBB cycle (typically −36%–−30‰) and enriched δ13C ratios for rTCA‐fixed carbon sources (typically −15%–−10‰) (Hügler & Sievert, 2011; Portail et al., 2018). Both carbon fixation pathways can be found in chemosynthetic epibiont communities with Campylobacterota using the rTCA cycle and Proteobacteria using the CBB cycle (Cambon‐Bonavita et al., 2021; Jan et al., 2014; Jiang et al., 2020). However, the relationship between individual δ13C ratios and the relative abundance of rTCA‐ or CBB‐fixing bacterial lineages did not hold for our dataset. As an example, the R. variabilis individual exhibiting the highest relative abundance of Proteobacteria lineages within its mouthparts (FU3‐CR9; see Figure 5.) had a more enriched δ13C ratio (−16.6‰) compared to a R. variabilis individual from the same site (FU3‐CR53; −23.6‰) whose mouthparts were completely dominated by Campylobacterota lineages and Proteobacteria being almost absent. Collectively, these results from microscopic observations, bacterial diversity, and isotopic ratios all suggest that neither R. variabilis, N. saintlaurentae nor Manuscaris sp. rely on chemosynthetic epibionts as their main source of nutrition.
Aside from a chemosymbiotic diet, other feeding modes such as bacterial grazers, scavengers or detritivores have been proposed for alvinocaridids shrimps, including those from the Manus and North Fiji basins (Suh et al., 2022; Van Audenhaege et al., 2019). Our results are consistent with previous studies on R. variabilis (Suh et al., 2022; Van Audenhaege et al., 2019), showing large trophic niches with particularly variable isotopic composition for carbon. Still, with a maximum of 11.6‰ for δ34S, their feeding sources remain within the range of chemosynthetically derived organic matter with no clear input of photosynthetic origin (Erickson et al., 2009; Reid et al., 2013; Van Dover & Fry, 1994). A notable exception was the N. saintlaurentae sampled at the Phoenix site (North Fiji basin), with a clearly higher δ34S ratio, up to 15.6‰, pointing out a potential mixed diet for this species with a larger contribution of photosynthetic material. However, the small size of these individuals and the observation of red lipid storages during their dissection (Methou, personal observation) rather indicate the influence of an ontogenetic shift as seen in juveniles and subadults stages of alvinocaridid shrimps from the Mid Atlantic Ridge (Methou et al., 2020; Pond et al., 1997), Central Indian Ridge (Van Dover, 2002) or the Mariana Arc (Stevens et al., 2008). Overall, these results suggest a generalist behaviour with various potential chemosynthetic food sources at the species level but more specialised feeding habits at a local scale. This is supported by the more similar and clustered isotopic ratios of shrimp individuals from the same sampling point within a vent field, arguing for a relatively strong habitat fidelity (Figure S2A–D). Thus, although being potentially highly mobile animals, alvinocaridids from the southwest Pacific might remain faithful to the same assemblage of foundation species once they settled, or at least at the timescales integrated by stable isotopic compositions of their abdominal muscles. Nevertheless, the presence of large alvinocaridid assemblages on chimney outcrops (Figure 1A), outside of mussel beds, tubeworm bushes or hairy snail colonies, indicates that these shrimps do not solely rely on detritus of these foundational symbiotrophs but are also able to feed on other nutrition sources, such as bacterial mats, possibly. Thereby, both detritivory and bacterivory diets could coexist in R. variabilis and N. saintlaurentae, although with strong intraspecific variations among individuals.
Interestingly, no isotopic niche partitioning was observed between R. variabilis and N. saintlaurentae from Fatu Kapa suggesting similar diets for the two co‐occurring species (Figure 4). Since both were sometimes collected from the same assemblage, there was no clear indication of spatial segregation in distinct habitat either. To avoid competitive exclusion, niche theory predicts that sympatric species differ by their resource use and/or spatio‐temporal habitat distribution, particularly in the case of closely related species with similar morphological traits and/or limited resource availability (Hutchinson, 1957; Schoener, 1974). However, the high biological productivity of hydrothermal vent ecosystems coupled with their temporal instability at short time scales might not allow to overreach the carrying capacity of these environments on the resources used by these shrimps, enabling long‐term coexistence of similar species for the same food source. This contrasts with Rimicaris shrimps co‐occurring in high densities assemblages on the Mid Atlantic Ridge, for which clear spatial and trophic niche partitioning could be observed between R. exoculata and R. chacei (Methou et al., 2020; Methou et al., 2022a). In the case of these species relying on their chemosymbiosis, competition for food is interlinked with competition for a limited space—that is, the access to the vent fluid source—resulting ultimately in niche partitioning for the case of vent holobionts (Beinart et al., 2012; Methou et al., 2022a; Van Audenhaege et al., 2019). On the other hand, vent species with a distinct type of diet, such as alvinocaridids from the southwest Pacific might experience a more relaxed competition enabling co‐occurring species to occupy the same ecological niche.
Resident bacterial communities within the digestive system of alvinocaridid shrimps
Although bacterial coverage on mouthparts of R. variabilis, N. saintlaurentae and Manuscaris sp. was very low comparatively to Rimicaris species from the Atlantic or Indian Oceans, the composition of their epibiotic communities mirrors those previously observed in cephalothoracic cavities of the latter (Apremont et al., 2018; Cambon‐Bonavita et al., 2021; Guri et al., 2012; Jan et al., 2014; Methou et al., 2022b; Petersen et al., 2010; Zbinden et al., 2008). Thus, we found a similar phylogenetic diversity with a dominance of Campylobacterota, followed by several families of Proteobacteria—including α‐, γ‐ and ζ‐proteobacteria—as well as Bacteroidota epibionts.
In contrast, composition of their digestive communities differs, in part, from those of R. exoculata and R. chacei in the Mid Atlantic Ridge (Apremont et al., 2018; Aubé et al., 2022; Durand et al., 2010, 2015; Guéganton et al., 2022) or R. kairei in the Central Indian Ridge (Qi et al., 2022), which exhibited a clear partitioning of bacterial lineages between their digestive organs. Indeed, bacterial communities of their midguts were mainly composed of Deferribacterota and Firmicutes from the Clostridia class, whereas Firmicutes affiliated to Mycoplasmatales (class Bacilli) were dominant in foreguts (Aubé et al., 2022; Durand et al., 2010; Guéganton et al., 2022). These two phyla constitute resident communities within their respective hosting organs in R. exoculata and R. chacei, whereas other bacterial lineages such as Campylobacterota or Gammaproteobacteria were only observed in the alimentary bolus (Guéganton et al., 2022).
In the three species of alvinocaridids from the southwest Pacific, Deferribacterota affiliated to Candidatus Rimicarispirillum and several lineages of Firmicutes affiliated to Mycoplasmatales and Clostridia were found in both the midguts and foreguts communities of every shrimp individual, often constituting the dominant lineages within their community (Figure 5B,C). These Firmicutes and Deferribacterota were also found on the mouthparts of some individuals but in lower proportions than in their digestive systems (Figure 6A). This absence of partitioning among hosting organs in alvinocaridids from the Southwest Pacific is thus more similar to the case of the terrestrial isopod Armadillidium vulgare whose Firmicutes symbiont, Candidatus Hepatoplasma crinochetorum, occupying predominantly their hepatopancreas and caeca (Bouchon et al., 2016; Wang et al., 2004), was also found in other tissues, including their hindgut, nerve cord, gonads as well as their haemolymph and faeces (Dittmer et al., 2016).
It is surprising that only Deferribacterota ASVs identical to known sequences were correctly affiliated and not those with 98.1% similarity based on the Silva 138 database (Figure 7). Recent metagenomics analyses of R. exoculata gut symbiont showed low alignment coverages of these Deferribacterota symbionts with other known lineages within this phylum leading to the creation of a new genus Candidatus Rimicarispirillum within a new family Candidatus Microvillispirillaceae for their classification (Aubé et al., 2022). This low alignment coverage could explain their affiliation failure with the current reference database for 16S rRNA metabarcoding studies and highlights may be a need to revise the current classification for this phylum.
Our results also reveal that the closest known lineages of Deferribacterota and Firmicutes found within the foreguts and midguts of southwest Pacific alvinocaridids were systematically symbionts associated with R. exoculata or R. chacei (Figure 7.) supporting their vertical inheritance and an association maintained along the evolutionary history of these hydrothermal vent shrimps. More broadly in the case of Firmicutes, related lineages like Candidatus Hepatoplasma or Candidatus Bacilloplasma were also retrieved in digestive systems of several crustaceans such as terrestrial, intertidal or deep‐sea isopods (Bouchon et al., 2016; Dittmer et al., 2016; Eberl, 2010; Fraune & Zimmer, 2008; Wang et al., 2004, 2016), hadal amphipods (Cheng et al., 2019) or coastal crab and shrimp species (Chen et al., 2015; Zhang et al., 2014, 2016). These Candidatus Hepatoplasma symbionts exhibited a high level of specificity with their hosts in terrestrial isopods (Fraune & Zimmer, 2008). Taken together, these results would even suggest an ancient and evolutionarily conserved partnership in the crustacean subphylum.
Conversely, the presence of each of these Deferribacterota and Firmicutes lineages within the digestive system of distantly‐related alvinocaridids like R. variabilis and N. saintlaurentae but from the same geographic area (Figure 7.) is more congruent with a horizontal mode of transmission. Similarly, hepatopancreas of the co‐occurring intertidal isopods, Ligia pallasii and L. occidentalis, hosted the same lineage of Candidatus Hepatoplasma (Eberl, 2010). It has been suggested that inter‐moults and inter‐generational transmission of Mycoplasmatales and Deferribacteres symbionts could be achieved by trophallaxis among individuals or by ingestion of their old cuticle (Durand et al., 2015). In the light of our results, this reinfection must be possible not only among individuals from the same species but also among individuals from the same family.
CONCLUSION
Our study confirms that these opportunistic alvinocarids from the South West Pacific basins do not rely heavily on chemosymbiosis as an alternative or complementary part of their diet. Rather, they most likely feed on other food sources available at vent ecosystems, including bacterial mats, detritus or mucus discarded by the foundational symbiotroph species. On the other hand, their digestive microbiome, notably bacteria from the Firmicutes and Deferribacterota group, was highly conserved compared to other alvinocaridids but also more largely among crustaceans, suggesting overall a possible ancient and evolutionarily conserved bacterial partnership. However, the distribution of these Firmicutes and Deferribacterota lineages within the different organs of the digestive system differs from those of other alvinocaridids where they are mostly restricted to the foregut and the midgut respectively. A larger sampling comparing digestive microbiomes of different alvinocaridid species from several regions would be required to disentangle the respective influence of geography, host diet and host phylogeny of these associations.
AUTHOR CONTRIBUTIONS
Pierre Methou: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (equal). Valérie Cueff‐Gauchard: Data curation (supporting); formal analysis (equal); methodology (equal); resources (supporting); validation (supporting); visualization (equal); writing – review and editing (equal). Loïc N Michel: Data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); resources (equal); validation (equal); writing – review and editing (equal). Nicolas Gayet: Investigation (supporting); methodology (equal). Florence Pradillon: Data curation (equal); resources (equal); supervision (equal); validation (equal); writing – review and editing (equal). Marie‐Anne Cambon‐Bonavita: Funding acquisition (lead); project administration (lead); resources (equal); supervision (lead); validation (equal); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
The author declares no competing interests.
Supporting information
Figure S1. Isotopic ratios of alvinocaridids from Southwest Pacific basins for (A) carbon, (B) nitrogen and (C) sulphur.
Figure S2. Carbon and sulphur isotopic ratios of alvinocaridid shrimps highlighting sampling events at (A) the Pacmanus field; (B) Susu Knolls field; (C) La Scala field; and (D) Fatu Kapa field.
Table S1. Summary of alvinocaridid shrimp sampling and conducted analyses with individual GenBank Identifier.
Table S2. p values of Dunn tests used for isotopic ratio comparisons (δ13C, δ15N δ34S) among alvinocaridid species and vent fields.
Table S3. Results from ANOVA‐like permutation tests for RDA by term with 999 permutations for each hosting organs.
Table S4. Results from PERMANOVA tests of isotopic ratios with 999 permutations for each hosting organs.
Table S5. Best Blast hit results of Firmicutes ASVs from hosting organs of SW Pacific alvinocaridid shrimps
ACKNOWLEDGEMENTS
We thank the captain and crew of the French Research Vessel L'Atalante and the team in charge of the ROV Victor 6000 during the ChubacArc expedition (2019) (https://doi.org/10.17600/18001111) and the HOV Nautile during the Futuna 3 expedition (2012) (https://doi.org/10.17600/12010040). Faunal collections were conducted with the necessary authority permissions of the foreign countries. Permission for sampling in Exclusive Economic Zones (EEZ) was issued by the Papua New Guinea, The Republic of Fiji and the Kingdom of Tonga. We obtained the agreement to sample in Wallis et Futuna waters from the Haut Commissariat à la République in New Caledonia and the Préfecture in Wallis and Futuna. Research animals were invertebrate caridean shrimps and no live experiments with animals were conducted in this study. We are also grateful to the GenoToul platform (GeT‐BioPuce, INSA, Toulouse, France) for providing resources for DNA sequencing. Pierre Methou was supported by a JAMSTEC Young Research Fellow fellowship. Marie‐Anne Cambon‐Bonavita, Florence Pradillon, Nicolas Gayet and Valérie Cueff‐Gauchard are supported by Ifremer, REMIMA grant. The 2012 Futuna3 cruise was financed through a public/private consortium comprising the French government, Ifremer and industrial groups Eramet and Technip. Laboratory analyses were supported by the Ifremer REMIMA programme.
Methou, P. , Cueff‐Gauchard, V. , Michel, L.N. , Gayet, N. , Pradillon, F. & Cambon‐Bonavita, M.‐A. (2023) Symbioses of alvinocaridid shrimps from the South West Pacific: No chemosymbiotic diets but conserved gut microbiomes. Environmental Microbiology Reports, 15(6), 614–630. Available from: 10.1111/1758-2229.13201
DATA AVAILABILITY STATEMENT
All COI barcode sequences have been deposited in GenBank under accession numbers OQ363903 – OQ364004 (see Table S1 for sampling summary with associated individual ID). The 16S rRNA dataset is available in the NCBI SRA repository (submission identifier SUB12697284 and BioProject identifier PRJNA932596). All stable isotopes measurements are available in the DeepIso database (https://www.seanoe.org/data/00654/76595/).
REFERENCES
- Apremont, V. , Cambon‐Bonavita, M.‐A. , Cueff‐Gauchard, V. , François, D. , Pradillon, F. , Corbari, L. et al. (2018) Gill chamber and gut microbial communities of the hydrothermal shrimp Rimicaris chacei Williams and Rona 1986: a possible symbiosis. PLoS One, 13, e0206084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assié, A. (2016) Deep Se(a)quencing: A study of deep sea ectosymbioses using next generation sequencing.
- Aubé, J. , Cambon‐Bonavita, M.‐A. , Velo‐Suárez, L. , Cueff‐Gauchard, V. , Lesongeur, F. , Guéganton, M. et al. (2022) A novel and dual digestive symbiosis scales up the nutrition and immune system of the holobiont Rimicaris exoculata. Microbiome, 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinart, R.A. , Sanders, J.G. , Faure, B. , Sylva, S.P. , Lee, R.W. , Becker, E.L. et al. (2012) Evidence for the role of endosymbionts in regional‐scale habitat partitioning by hydrothermal vent symbioses. Proceedings of the National Academy of Sciences, 109, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N.A. , Subramanian, S. , Faith, J.J. , Gevers, D. , Gordon, I. , Knight, R. et al. (2013) Quality‐filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods, 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon, D. , Zimmer, M. & Dittmer, J. (2016) The terrestrial isopod microbiome: an all‐in‐one toolbox for animal‐microbe interactions of ecological relevance. Frontiers in Microbiology, 7, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breusing, C. , Castel, J. , Yang, Y. , Broquet, T. , Sun, J. , Jollivet, D. et al. (2022) Global 16S rRNA diversity of provannid snail endosymbionts from Indo‐Pacific deep‐sea hydrothermal vents. Environmental Microbiology Reports, 14, 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J. , McMurdie, P.J. , Rosen, M.J. , Han, A.W. , Johnson, A.J.A. & Holmes, S.P. (2016) DADA2: high‐resolution sample inference from Illumina amplicon data. Nature Methods, 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambon‐Bonavita, M.‐A. , Aubé, J. , Cueff‐Gauchard, V. & Reveillaud, J. (2021) Niche partitioning in the Rimicaris exoculata holobiont: the case of the first symbiotic Zetaproteobacteria. Microbiome, 9, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Di, P. , Wang, H. , Li, B. , Pan, Y. , Yan, S. et al. (2015) Bacterial community associated with the intestinal tract of Chinese mitten crab (Eriocheir sinensis) farmed in Lake tai, China. PLoS One, 10, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. , Wang, Y. , Li, J. , Yan, G. & He, L. (2019) Comparative analysis of the gut microbial communities between two dominant amphipods from the challenger deep, Mariana Trench. Deep Sea Research Part I: Oceanographic Research Papers, 151, 103081. [Google Scholar]
- Coplen, T.B. (2011) Guidelines and recommended terms for expression of stable‐isotope‐ratio and gas‐ratio measurement results. Rapid Communications in Mass Spectrometry, 25, 2538–2560. [DOI] [PubMed] [Google Scholar]
- Corbari, L. , Zbinden, M. , Cambon‐Bonavita, M.A. , Gaill, F. & Compère, P. (2008) Bacterial symbionts and mineral deposits in the branchial chamber of the hydrothermal vent shrimp rimicaris exoculata: relationship to moult cycle. Aquatic Biology, 1, 225–238. [Google Scholar]
- Dittmer, J. , Lesobre, J. , Moumen, B. & Bouchon, D. (2016) Host origin and tissue microhabitat shaping the microbiota of the terrestrial isopod Armadillidium vulgare . FEMS Microbiology Ecology, 92, 1–15. [DOI] [PubMed] [Google Scholar]
- Dubilier, N. , Bergin, C. & Lott, C. (2008) Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nature Reviews Microbiology, 6, 725–740. [DOI] [PubMed] [Google Scholar]
- Durand, L. , Roumagnac, M. , Cueff‐Gauchard, V. , Jan, C. , Guri, M. , Tessier, C. et al. (2015) Biogeographical distribution of Rimicaris exoculata resident gut epibiont communities along the Mid‐Atlantic Ridge hydrothermal vent sites. FEMS Microbiology Ecology, 91, 1–15. [DOI] [PubMed] [Google Scholar]
- Durand, L. , Zbinden, M. , Cueff‐Gauchard, V. , Duperron, S. , Roussel, E.G. , Shillito, B. et al. (2010) Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiology Ecology, 71, 291–303. [DOI] [PubMed] [Google Scholar]
- Eberl, R. (2010) Sea‐land transitions in isopods: pattern of symbiont distribution in two species of intertidal isopods Ligia pallasii and Ligia occidentalis in the Eastern Pacific. Symbiosis, 51, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, K.L. , Macko, S.A. & Van Dover, C.L. (2009) Evidence for a chemoautotrophically based food web at inactive hydrothermal vents (Manus Basin). Deep‐Sea Research Part II: Topical Studies in Oceanography, 56, 1577–1585. [Google Scholar]
- Fadrosh, D.W. , Bing Ma, P.G. , Sengamalay, N. , Ott, S. , Brotman, R.M. , Ravel, J. et al. (2014) An improved dual‐indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeqplatform. Microbiome, 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune, S. & Zimmer, M. (2008) Host‐specificity of environmentally transmitted mycoplasma‐like isopod symbionts. Environmental Microbiology, 10, 2497–2504. [DOI] [PubMed] [Google Scholar]
- Gebruk, A.V. , Southward, E.C. , Kennedy, H. & Southward, A.J. (2000) Food sources, behaviour, and distribution of hydrothermal vent shrimps at the Mid‐Atlantic Ridge. Journal of the Marine Biological Association of the United Kingdom, 80, 485–499. [Google Scholar]
- Guéganton, M. , Rouxel, O. , Durand, L. , Cueff‐Gauchard, V. , Gayet, N. , Pradillon, F. et al. (2022) Anatomy and Symbiosis of the digestive system of the vent shrimps Rimicaris exoculata and Rimicaris chacei revealed through imaging approaches. Frontiers in Marine Science, 7, 454–464. [Google Scholar]
- Guri, M. , Durand, L. , Cueff‐Gauchard, V. , Zbinden, M. , Crassous, P. , Shillito, B. et al. (2012) Acquisition of epibiotic bacteria along the life cycle of the hydrothermal shrimp Rimicaris exoculata . The ISME Journal, 6, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann, D.P.R. , Labrenz, M. , Jürgens, K. , Bertilsson, S. , Waniek, J.J. & Andersson, A.F. (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal, 5, 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler, M. & Sievert, S.M. (2011) Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annual Review of Marine Science, 3, 261–289. [DOI] [PubMed] [Google Scholar]
- Hutchinson, G. (1957) Concluding remarks. Cold Spring Harbor Symposia on Quantitative Biology, 22, 66–77. [Google Scholar]
- Jackson, A.L. , Inger, R. , Parnell, A.C. & Bearhop, S. (2011) Comparing isotopic niche widths among and within communities: SIBER—stable isotope Bayesian ellipses in R. The Journal of Animal Ecology, 80, 595–602. [DOI] [PubMed] [Google Scholar]
- Jan, C. , Petersen, J.M. , Werner, J. , Teeling, H. , Huang, S. , Glöckner, F.O. et al. (2014) The gill chamber epibiosis of deep‐sea shrimp Rimicaris exoculata: an in‐depth metagenomic investigation and discovery of Zetaproteobacteria. Environmental Microbiology, 16, 2723–2738. [DOI] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X. , Dong, C. , Huang, Z. , Cambon‐Bonavita, M.‐A. , Alain, K. et al. (2020) “Candidatus Desulfobulbus rimicarensis,” an uncultivated Deltaproteobacterial Epibiont from the Deep‐Sea Hydrothermal Vent Shrimp Rimicaris exoculata . Applied and Environmental Microbiology, 86, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall‐Ngai, M. , Hadfield, M.G. , Bosch, T.C.G. , Carey, H.V. , Domazet‐Lošo, T. , Douglas, A.E. et al. (2013) Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences, 110, 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight, D.T. , Huerlimann, R. , Bower, D.S. , Schwarzkopf, L. , Alford, R.A. & Zenger, K.R. (2019a) Methods for normalizing microbiome data: an ecological perspective. Methods in Ecology and Evolution, 10, 389–400. [Google Scholar]
- McKnight, D.T. , Huerlimann, R. , Bower, D.S. , Schwarzkopf, L. , Alford, R.A. & Zenger, K.R. (2019b) microDecon: a highly accurate read‐subtraction tool for the post‐sequencing removal of contamination in metabarcoding studies. Environmental DNA, 1, 14–25. [Google Scholar]
- McMurdie, P.J. & Holmes, S. (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methou, P. , Hernández‐Ávila, I. , Aube, J. , Cueff‐Gauchard, V. , Gayet, N. , Amand, L. et al. (2019) Is it first the egg or the shrimp?—diversity and variation in microbial communities colonizing broods of the vent shrimp Rimicaris exoculata during embryonic development. Frontiers in Microbiology, 10, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methou, P. , Hernández‐Ávila, I. , Cathalot, C. , Cambon‐Bonavita, M.‐A. & Pradillon, F. (2022a) Population structure and environmental niches of Rimicaris shrimps from the Mid‐Atlantic Ridge. Marine Ecology Progress Series, 684, 1–20. [Google Scholar]
- Methou, P. , Hikosaka, M. , Chen, C. , Watanabe, H.K. , Miyamoto, N. , Makita, H. et al. (2022b) Symbiont community composition in Rimicaris kairei shrimps from Indian Ocean vents with notes on mineralogy. Appl Environ Microbiol, 88, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methou, P. , Michel, L.N. , Segonzac, M. , Cambon‐Bonavita, M.‐A. & Pradillon, F. (2020) Integrative taxonomy revisits the ontogeny and trophic niches of Rimicaris vent shrimps. Royal Society Open Science, 7, 200837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye, V. , Copley, J. & Plouviez, S. (2012) A new species of Rimicaris (Crustacea: Decapoda: Caridea: Alvinocarididae) from hydrothermal vent fields on the Mid‐Cayman Spreading Centre, Caribbean. Journal of the Marine Biological Association of the United Kingdom, 92, 1057–1072. [Google Scholar]
- Oksanen, J. , Kindt, R. , Legendre, P. , O'Hara, B. , Simpson, G.L. , Solymos, P.M. et al. (2008) The vegan package. Community Ecology Package, 1–190. [Google Scholar]
- Petersen, J.M. , Ramette, A. , Lott, C. , Cambon‐Bonavita, M.‐A. , Zbinden, M. & Dubilier, N. (2010) Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma‐ and epsilonproteobacteria at four Mid‐Atlantic Ridge hydrothermal vent fields. Environmental Microbiology, 12, 2204–2218. [DOI] [PubMed] [Google Scholar]
- Polz, M.F. , Robinson, J.J. , Cavanaugh, C.M. , Van Dover, C.L. , Van, C.L. & Van Dover, C.L. (1998) Trophic ecology of massive shrimp aggregations at a Mid‐Atlantic Ridge hydrothermal vent site. Limnology and Oceanography, 43, 1631–1638. [Google Scholar]
- Pond, D.W. , Segonzac, M. , Bell, M.V. , Dixon, D.R. , Fallick, A.E. , Sargent, J.R. et al. (1997) Lipid and lipid carbon stable isotope composition of the hydrothermal vent shrimp Mirocaris fortunata: evidence for nutritional dependence on photosynthetically fixed carbon. Marine Ecology Progress Series, 157, 221–231. [Google Scholar]
- Ponsard, J. , Cambon‐Bonavita, M.‐A. , Zbinden, M. , Lepoint, G. , Joassin, A. , Corbari, L. et al. (2013) Inorganic carbon fixation by chemosynthetic ectosymbionts and nutritional transfers to the hydrothermal vent host‐shrimp Rimicaris exoculata . The ISME Journal, 7, 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portail, M. , Brandily, C. , Cathalot, C. , Colaço, A. , Gélinas, Y. , Husson, B. et al. (2018) Food‐web complexity across hydrothermal vents on the Azores triple junction. Deep Sea Research Part I: Oceanographic Research Papers, 131, 101–120. [Google Scholar]
- Qi, L. , Lian, C.A. , Zhu, F.C. , Shi, M. & He, L.S. (2022) Comparative analysis of intestinal microflora between two developmental stages of Rimicaris kairei, a hydrothermal shrimp from the Central Indian Ridge. Frontiers in Microbiology, 12, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Research, 41, 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2020) R: A language and environment for statistical computing.
- Reid, W.D.K. , Sweeting, C.J. , Wigham, B.D. , Zwirglmaier, K. , Hawkes, J.A. , McGill, R.A.R. et al. (2013) Spatial differences in east scotia ridge hydrothermal vent food webs: influences of chemistry, microbiology and predation on trophodynamics. PLoS One, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoener, T.W. (1974) Resouce partitioning in ecological communities: research on how similar species divide resources helps. Science, 185, 27–39. [DOI] [PubMed] [Google Scholar]
- Sogin, E.M. , Leisch, N. & Dubilier, N. (2020) Chemosynthetic symbioses. Current Biology, 30, R1137–R1142. [DOI] [PubMed] [Google Scholar]
- Stevens, C.J. , Limén, H. , Pond, D.W. , Gélinas, Y. & Juniper, S.K. (2008) Ontogenetic shifts in the trophic ecology of two alvinocaridid shrimp species at hydrothermal vents on the Mariana Arc, western Pacific Ocean. Marine Ecology Progress Series, 356, 225–237. [Google Scholar]
- Streit, K. , Bennett, S.A. , Van Dover, C.L. & Coleman, M. (2015) Sources of organic carbon for Rimicaris hybisae: tracing individual fatty acids at two hydrothermal vent fields in the mid‐Cayman rise. Deep Sea Research Part I: Oceanographic Research Papers, 100, 13–20. [Google Scholar]
- Suh, Y.J. , Kim, M. , Lee, W.‐K. , Yoon, H. , Moon, I. , Jung, J. et al. (2022) Niche partitioning of hydrothermal vent fauna in the North Fiji Basin, Southwest Pacific inferred from stable isotopes. Marine Biology, 169, 1–15. [Google Scholar]
- Van Audenhaege, L. , Fariñas‐Bermejo, A. , Schultz, T. & Lee Van Dover, C. (2019) An environmental baseline for food webs at deep‐sea hydrothermal vents in Manus Basin (Papua New Guinea). Deep Sea Research Part I: Oceanographic Research Papers, 148, 88–99. [Google Scholar]
- Van Dover, C.L. (2002) Trophic relationships among invertebrates at the Kairei hydrothermal vent field (Central Indian Ridge). Marine Biology, 141, 761–772. [Google Scholar]
- Van Dover, C.L. & Fry, B. (1994) Microorganisms as food resources at deep‐sea hydrothermal vents. Limnology and Oceanography, 39, 51–57. [Google Scholar]
- Versteegh, E.A.A. , Van Dover, C.L. , Van Audenhaege, L. & Coleman, M. (2022) Multiple nutritional strategies of hydrothermal vent shrimp (Rimicaris hybisae) assemblages at the mid‐Cayman rise. Deep Sea Research Part I: Oceanographic Research Papers, 192, 103915. [Google Scholar]
- Wang, Y. , Huang, J.M. , Wang, S.L. , Gao, Z.M. , Zhang, A.Q. , Danchin, A. et al. (2016) Genomic characterization of symbiotic mycoplasmas from the stomach of deep‐sea isopod bathynomus sp. Environmental Microbiology, 18, 2646–2659. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Stingl, U. , Anton‐erxleben, F. , Geisler, S. , Brune, A. & Zimmer, M. (2004) “Candidatus Hepatoplasma crinochetorum,” a new, stalk‐forming lineage of Mollicutes colonizing the midgut glands of a terrestrial isopod. Applied and Environmental Microbiology, 70, 6166–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden, M. & Cambon‐Bonavita, M. (2020) Biology and ecology of Rimicaris exoculata, a symbiotic shrimp from deep‐sea hydrothermal vents. Marine Ecology Progress Series, 652, 187–222. [Google Scholar]
- Zbinden, M. & Cambon‐Bonavita, M.A. (2003) Occurrence of Deferribacterales and Entomoplasmatales in the deep‐sea Alvinocarid shrimp Rimicaris exoculata gut. FEMS Microbiology Ecology, 46, 23–30. [DOI] [PubMed] [Google Scholar]
- Zbinden, M. , Le Bris, N. , Gaill, F. & Compère, P. (2004) Distribution of bacteria and associated minerals in the gill chamber of the vent shrimp Rimicaris exoculata and related biogeochemical processes. Marine Ecology Progress Series, 284, 237–251. [Google Scholar]
- Zbinden, M. , Shillito, B. , Le Bris, N. , de Villardi de Montlaur, C. , Roussel, E. , Guyot, F. et al. (2008) New insigths on the metabolic diversity among the epibiotic microbial communitiy of the hydrothermal shrimp Rimicaris exoculata. Journal of Experimental Marine Biology and Ecology, 359, 131–140. [Google Scholar]
- Zhang, M. , Sun, Y. , Chen, K. , Yu, N. , Zhou, Z. , Chen, L. et al. (2014) Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture, 434, 449–455. [Google Scholar]
- Zhang, M. , Sun, Y. , Chen, L. , Cai, C. , Qiao, F. , Du, Z. et al. (2016) Symbiotic bacteria in gills and guts of Chinese mitten crab (Eriocheir sinensis) differ from the free‐living bacteria in water. PLoS One, 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Isotopic ratios of alvinocaridids from Southwest Pacific basins for (A) carbon, (B) nitrogen and (C) sulphur.
Figure S2. Carbon and sulphur isotopic ratios of alvinocaridid shrimps highlighting sampling events at (A) the Pacmanus field; (B) Susu Knolls field; (C) La Scala field; and (D) Fatu Kapa field.
Table S1. Summary of alvinocaridid shrimp sampling and conducted analyses with individual GenBank Identifier.
Table S2. p values of Dunn tests used for isotopic ratio comparisons (δ13C, δ15N δ34S) among alvinocaridid species and vent fields.
Table S3. Results from ANOVA‐like permutation tests for RDA by term with 999 permutations for each hosting organs.
Table S4. Results from PERMANOVA tests of isotopic ratios with 999 permutations for each hosting organs.
Table S5. Best Blast hit results of Firmicutes ASVs from hosting organs of SW Pacific alvinocaridid shrimps
Data Availability Statement
All COI barcode sequences have been deposited in GenBank under accession numbers OQ363903 – OQ364004 (see Table S1 for sampling summary with associated individual ID). The 16S rRNA dataset is available in the NCBI SRA repository (submission identifier SUB12697284 and BioProject identifier PRJNA932596). All stable isotopes measurements are available in the DeepIso database (https://www.seanoe.org/data/00654/76595/).


