ABSTRACT
Golgi stacks are the basic structural units of the Golgi. Golgi stacks are separated from each other and scattered in the cytoplasm of Drosophila cells. Here, we report that the ARF-GEF inhibitor Brefeldin A (BFA) induces the formation of BFA bodies, which are aggregates of Golgi stacks, trans-Golgi networks and recycling endosomes. Recycling endosomes are located in the centers of BFA bodies, while Golgi stacks surround them on their trans sides. Live imaging of S2 cells revealed that Golgi stacks repeatedly merged and separated on their trans sides, and BFA caused successive merger by inhibiting separation, forming BFA bodies. S2 cells carrying genome-edited BFA-resistant mutant Sec71M717L did not form BFA bodies at high concentrations of BFA; S2 cells carrying genome-edited BFA-hypersensitive mutant Sec71F713Y produced BFA bodies at low concentrations of BFA. These results indicate that Sec71 is the sole BFA target for BFA body formation and controls Golgi stack separation. Finally, we showed that impairment of Sec71 in fly photoreceptors induces BFA body formation, with accumulation of both apical and basolateral cargoes, resulting in inhibition of polarized transport.
Keywords: Brefeldin A, Sec71, Golgi ribbon, Trans-Golgi network, Recycling endosome, Drosophila
Summary: Golgi stacks are scattered in the cytoplasm of Drosophila cells. Sec71 is required for separating Golgi stacks, as its impairment, for example, through brefeldin A (BFA) treatment, produces Golgi aggregations termed BFA bodies.
INTRODUCTION
The Golgi is a membrane-bound organelle that is central to the secretory pathway and functions in the modification and sorting of secretory proteins and lipids to multiple destinations within the cell (Klumperman, 2011; Papanikou and Glick, 2014). The basic structural unit of the Golgi is the Golgi stack, composed of multiple flattened cisternae, tubules and vesicles. In plant, Caenorhabditis elegans, and Drosophila cells, Golgi stacks are separated from each other and scattered in the cytoplasm (Gosavi and Gleeson, 2017; Wei and Seemann, 2017; Yadav and Linstedt, 2011). In contrast, in mammalian cells dozens of Golgi stacks are connected by lateral links to form a Golgi ribbon, which resides in the vicinity of the centrosome via microtubule-based motors. Neither the mechanisms underlying nor the functional significance of ribbon formation have been resolved (Saraste and Prydz, 2019).
Trans-Golgi networks (TGNs), located on the trans sides of Golgi stacks, were originally considered to be the sorting centers for newly synthesized proteins destined for distinct cellular locations (Kienzle and von Blume, 2014; Luini and Parashuraman, 2016). Recycling endosomes (REs) are perinuclear compartments through which endocytosed materials are trafficked before being recycled back to the plasma membrane (Mayor et al., 1993; Yamashiro and Maxfield, 1987). However, both the TGN and RE are considered hubs of the exocytotic and endocytic pathways (Goldenring, 2015; Hierro et al., 2015; Makaraci and Kim, 2018).
We recently reported that REs are attached to the trans sides of Golgi stacks both in Drosophila and microtubule-disrupted HeLa cells (Fujii et al., 2020). REs can exist in two distinct, yet interchangeable states, Golgi-associated REs (GA-REs) and free REs – both undergo repeated detachment and reattachment. Moreover, free REs themselves divide and fuse together repeatedly. Since two distinct states of the TGN, Golgi-associated (GA-TGN) and free, are well established in plants (Kang et al., 2011; Uemura et al., 2019, 2014; Viotti et al., 2010), we propose that the plant TGN might be an equivalent organelle to the animal RE (Fujii et al., 2020).
Brefeldin A (BFA) is a fungal toxin that affects exocytotic and endocytic membrane trafficking in eukaryotes by inhibiting guanine-nucleotide exchange factors that regulate ARF GTPases (Anders and Jürgens, 2008; Casanova, 2007; Peyroche et al., 1996; Shin and Nakayama, 2004). In most mammalian cells, except for canine MDCK cells, BFA prevents the binding of peripheral COPI proteins to Golgi membranes and causes intensive tubule extension from Golgi membranes and redistribution of Golgi-resident proteins into the ER (Cole et al., 1996; Donaldson et al., 1990; Lippincott-Schwartz and Liu, 2006; Lippincott-Schwartz et al., 1989; Orci et al., 1991; Sciaky et al., 1997). In addition to its effect on the early secretory pathway, BFA induces the fusion of TGN, endosomes and lysosomes, forming tubule networks emanating from the juxtanuclear region (Hunziker et al., 1991; Lippincott-Schwartz et al., 1991; Wood et al., 1991). This endosomal impact of BFA is observed even in MDCK cells and is partly induced by the inhibition of γ-adaptin [the γ subunit of adapter protein complex 1 (AP1γ)] binding to TGN (Futter et al., 1998; Hunziker et al., 1991; Ishizaki et al., 2008). Tobacco BY2 and yeast cells react to BFA in a manner similar to mammalian cells: Golgi disruption, redistribution of Golgi-resident proteins into the ER, and fusion of TGN/endosomes, which are separated from the early secretory compartments (Hicke et al., 1997; Ito et al., 2017; Langhans et al., 2011; Peyroche et al., 1996; Yasuhara and Shibaoka, 2000; Yasuhara et al., 1995). In Arabidopsis, BFA does not induce Golgi absorption into the ER, but instead the Golgi and TGN aggregate, forming ‘BFA compartments’ or ‘BFA bodies’ (Dragwidge et al., 2019; Geldner et al., 2003; Robinson et al., 2008; Uemura and Nakano, 2013). Interestingly, in Arabidopsis, TGN markers aggregate at the centers of BFA bodies, whereas Golgi markers localize to the periphery (Robinson et al., 2008; Uemura and Nakano, 2013).
In Drosophila, BFA inhibits cellularization (Sisson et al., 2000) and plasma membrane transport of Delta (Kondylis and Rabouille, 2003); Golgi stacks are not disrupted, rather they form clusters or aggregates (Kondylis and Rabouille, 2009; Kondylis et al., 2007; Xu et al., 2002). However, cis-trans polarity in Golgi aggregation and the effects of BFA on the TGN or REs have not been well investigated. Here, we report that TGN/REs repeatedly merge and separate, and that BFA inhibits TGN/RE separation by inactivating the function of Sec71, a Drosophila guanine-nucleotide exchange factor (GEF) for ARF GTPases, resulting in the formation of BFA bodies. TGN/RE aggregates are located at the centers of BFA bodies and Golgi stacks surround the aggregate on their trans sides. The Golgi/RE organization of BFA bodies in S2 cells resembles that of normal COS-1 cells, suggesting that the morphological and structural organization of Golgi/RE reflects the kinetics of TGN/RE merger and separation in cells.
RESULTS
BFA-induced aggregation of Golgi stacks maintains cis-trans polarity in S2 cells
To investigate the effects of BFA on Golgi stacks and the TGN/REs, we incubated S2 cells expressing the medial-Golgi marker mannosidase II::GFP (ManII::GFP) or monomeric Turquoise2::Rab11 (mTq2::Rab11) for 2 h with or without 50 μM BFA at 25°C, followed by immunostaining with anti-GM130 (a cis-Golgi marker) and anti-Rab6 (TGN marker) antibodies (Fig. 1A,B). Approximately 30% of BFA-treated S2 cells showed aggregation of Golgi stacks, TGN and REs. Both mTq2::Rab11 and Rab6 localized to the central domain of the aggregate, but there was a clear difference in the localization of mTq2::Rab11, which was observed only at the central core, whereas Rab6 localized more broadly. In untreated cells, Rab11 localized specifically to the RE, whereas Rab6 was broadly distributed from trans-Golgi to the RE. The arrangement of ManII, Rab6, and Rab11 in BFA-induced aggregates likely reflects that in normal cells. GM130 localized only to the periphery of the aggregate, while ManII::EGFP was located between GM130 and the peripheral edge of Rab6 (Fig. 1A). The trans-Golgi markers ST::EGFP and GalT::EGFP colocalized with Rab6, but displayed stronger signals in the periphery than in the central region (Fig. 1C,D). The TGN marker Golgin245 colocalized very well with Rab6, but more weakly in the most central part of the aggregate (Fig. 1D). These results indicate that cis-trans polarity of Golgi stacks is generally maintained in BFA-induced aggregates, and that the trans sides of Golgi stacks are facing the central core formed by TGN/REs (Fig. 1E). Therefore, we describe BFA-induced aggregates in S2 cells as BFA bodies since their organization resembles that described in plant cells, in which TGN forms the center and trans-Golgi markers are located in the periphery (Robinson et al., 2008; Uemura et al., 2014).
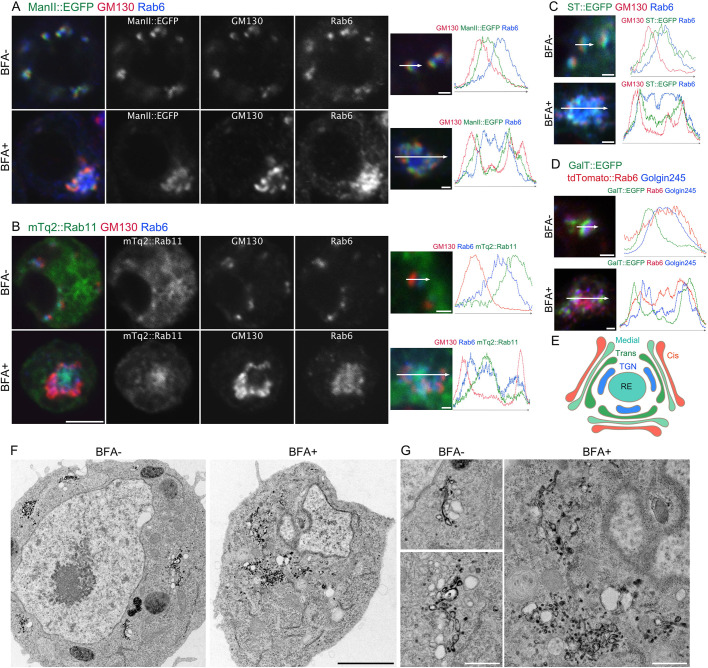
Fig. 1.
BFA induces the formation of BFA bodies. (A,B) Left, representative immunostaining of S2 cells expressing ManII::EGFP (A) or mTq2::Rab11 (B) (green) incubated without (upper panels) or with 50 μM BFA (lower panels) with anti-GM130 (red) and anti-Rab6 (blue) antibodies. Right panels, plots of signal intensities from image to the left. Signal intensities were measured along the 1.5 µm (upper) or 5 µm (lower) arrows shown in inset. Graphs show the overlap between channels. (C,D) Left pictures are immunostaining of S2 cells expressing ST::EGFP (green) (C) or GalT::EGFP (green) and tdTomato::Rab6 (red) (D) incubated without (upper panels) or with 50 μM BFA (lower panels) by anti-GM130 (red) and anti-Rab6 (blue) antibodies (C) or by anti-Golgin245 antibody (blue) (D). Right plots of signal intensities from image on the left. Signal intensity was measured along the 1.5 µm (upper) or 5 µm (lower) arrow in inset, graph shows the overlap between channels. (E) Schematic of the structure of a BFA body. (F,G) Electron micrographs of S2 cells expressing GalT::APEX2::EGFP without or with 50 μM BFA. GalT::APEX2::EGFP was visualized by osmium-enhanced DAB-depositions. Scale bars: 5 μm (A,B, left panels), 1 μm (A,B, right panels, C,D), 2 μm (F), and 500 nm (G).
Additionally, we investigated the membrane structure of BFA bodies by electron microscopy (EM) and a genetic EM-tag, APEX2 (Martell et al., 2017). Previously, we showed that DAB deposition generated by GalT::APEX2::EGFP is limited to the lumen of trans-Golgi cisternae in S2 cells (Fujii et al., 2020). In S2 cells in the absence of BFA, Golgi stacks with DAB deposition in trans cisternae were scattered in the cytoplasm; however, in S2 cells incubated for 2 h with BFA, Golgi stacks were not clear, but cisternae and tubules with DAB deposition were gathered in one place (Fig. 1F,G), where cisternae and tubule-networks likely represent BFA bodies and resemble those in plants (Geldner et al., 2003; Robinson et al., 2008).
As Golgi stacks in mammalian cells are gathered near centrosomes via microtubules, we determined whether microtubules are necessary for BFA body formation in S2 cells. Most Drosophila cells, including S2 cells, have no functional centrosomes during interphase (Rogers et al., 2008; Rusan and Rogers, 2009); however, S2 cells assemble functional centrosomes during mitosis. Thus, we first compared the positions of BFA bodies and microtubule asters spreading from functional centrosomes at early prophase, and found that BFA bodies were not located near microtubule asters (Fig. S2A, upper panels). We also closely investigated the positioning of the BFA body and microtubule lattice at interphase (Fig. S2A, panels in middle row). Although some tdTomato::Rab6-positive tubules extended along with microtubules outside the BFA body (Fig. S2A, panels in middle row, arrow), the BFA body itself was not tightly associated with microtubules. We further examined the effect of the microtubule-polymerization inhibitor colchicine on BFA body formation, and found that BFA bodies could be formed in the absence of microtubules (Fig. S2A, lower panels). Interestingly, there were small fragmented microtubules near Golgi stacks and other places following colchicine treatment that might correspond to previously reported microtubule nucleation sites on Golgi stacks or unidentified sites throughout the cytoplasm (Rogers et al., 2008; Rusan and Rogers, 2009). These results indicate that microtubules are not necessary for BFA body formation.
Impairment of Sec71 function induces BFA body formation in S2 cells
From numerous studies on yeast, human, and plant cells, it is well known that BFA targets ARF-GEFs belonging to the Sec7/BIG and Gea/GBF families (Jackson, 2018; Peyroche et al., 1996; Sata et al., 1998; Wright et al., 2014). In the Drosophila genome, there is only one member of the Sec7/BIG family ARF-GEFs, namely, Sec71 (Cox et al., 2004), and a single member of the Gea/GBF family, ARF-GEF, namely Garz (Cox et al., 2004; Wang et al., 2012). Thus, we investigated whether impairment of Sec71 or Garz function caused BFA body formation. First, we examined the localization of overexpressed wild-type V5::Sec71 and V5::Garz, as well as their effects on Golgi organization. As reported previously (Armbruster and Luschnig, 2012; Christis and Munro, 2012; Wang et al., 2012, 2017), V5::Sec71 localized at the trans sides, while V5::Garz localized at the cis sides of Golgi stacks, with neither influencing their organization (Fig. 2A,B, upper panels, 2D,E, and Fig. S2C,D, upper panels, Fig. S2E,G). Next, we investigated the effects of V5::Sec71E677K or V5::GarzE740K expression. These mutants have been reported to impair GEF activity and function in a dominant-negative manner (Armbruster and Luschnig, 2012; Wang et al., 2017). The expression of V5::Sec71E677K induced Golgi stacks to aggregate (Fig. 2A, lower panels), with Rab6 localized broadly to the interior and GM130 peripherally (Fig. 2F). GalT::mTq2 signals were dominant in peripheral foci, but also were found in the interiors of aggregates (Fig. S2C, lower panels, Fig. S2F). These distributions were similar to those observed in BFA bodies in S2 cells (Fig. 1A–C). In these aggregates, V5::Sec71E677K was located at the center (Fig. 2A, lower panels, Fig. 2F and Fig. S2C, lower panels, Fig. S2F). In contrast, the expression of V5::GarzE740K caused the diffusion of Rab6 and GalT::mTq2, although GM130 foci were still visible and colocalized with V5::GarzE740K foci, as discussed below (Fig. 2B, Fig. S2D, lower panels). Similar to what is seen upon V5::Sec71E677K expression, double-stranded RNA knockdown of Sec71 resulted in Golgi aggregation with a radial polarity similar to that of BFA bodies – a cis-Golgi marker GM130 and a medial-Golgi marker p120 (also known as Glg1) (Yamamoto-Hino et al., 2012) localized at the periphery, with TGN markers Rab6 and Golgin245 located at the central region (Fig. 2C, panels in middle row, Fig. 2G–I and Fig. S2B panels in middle row). The RE marker mTq2::Rab11 also localized to the central region (Fig. 2I). In contrast, double-stranded RNA knockdown of Garz induced the diffusion of p120, Rab6 and Golgin245, but not of GM130 (Fig. 2C, lower panels; Fig. S2B, lower panels). These results confirmed that these phenotypes were caused by the impairment of Sec71 or Garz.
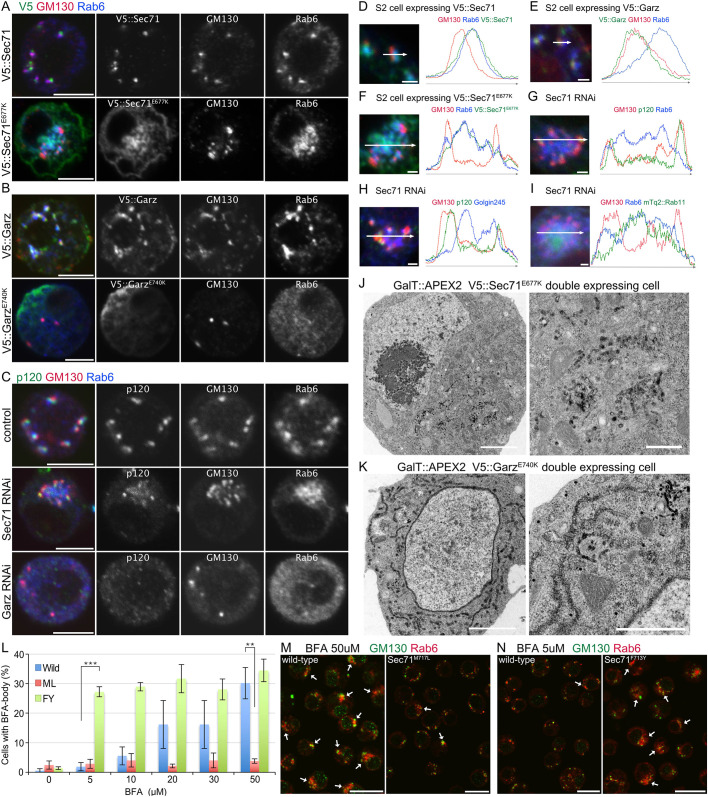
Fig. 2.
Sec71 is the only BFA target in BFA body formation in S2 cells. (A) Immunostaining of cells expressing V5::Sec71 (upper panels) or V5::Sec71E677K (lower panels) by anti-V5 (green), anti-GM130 (red), and anti-Rab6 (blue) antibodies. (B) Immunostaining of cells expressing V5::Garz (upper panels) or V5::GarzE740K (lower panels) by anti-V5 (green), anti-GM130 (red), and anti-Rab6 (blue) antibodies. (C) Immunostaining of cells transfected with double-stranded RNA against Sec71 (panels in middle row) or Garz (lower panels) or not transfected (upper panels) by anti-p120 (green), anti-GM130 (red), and anti-Rab6 (blue) antibodies. (D–I) Left, immunostaining of cells expressing V5::Sec71 (D), V5::Garz (E), V5::Sec71E677K (F), mTq2::Rab11 (green) (I), or transfected with double-stranded RNA against Sec71 (G–I). Anti-V5 (green), anti-GM130 (red), and anti-Rab6 (blue) antibodies (D–F). Anti-p120 (green), anti-GM130 (red), and anti-Rab6 (blue) antibodies (G). Anti-p120 (green), anti-GM130 (red), and anti-Golgin245 (blue) antibodies (H). Anti-GM130 (red) and anti-Rab6 (blue) antibodies (I). Right, plots of signal intensities from images on the left. Signal intensities were measured along the 1.5 µm (D,E) and 5 µm (F–I) arrows shown in insets. Graphs show the overlap between channels. (J,K) Electron micrographs of cells co-expressing GalT::APEX2::EGFP and V5::Sec71E677K (J) or V5::GarzE740K (K). GalT::APEX2::EGFP was visualized by osmium-enhanced DAB-deposition. (L) Percentages of wild-type, Sec71M717L and Sec71F713Y S2 cells with BFA bodies after 2 h of incubation with 0, 5, 10, 20, 30 and 50 μM BFA. BFA bodies were defined as focused Rab6 staining surrounded by GM130 staining. Results are mean±s.d. for n>250 in three independent experiments. (M) Immunostaining of wild-type cells (left) and genome-edited cells expressing BFA-resistant mutant Sec71M717L (right) treated with 50 μM BFA by anti-GM130 (green) and anti-Rab6 (red) antibodies. (N) Immunostaining of wild-type cells (left) and genome-edited cells with BFA-hypersensitive mutant Sec71F713Y (right) treated with 5 μM BFA with anti-GM130 (green) and anti-Rab6 (red) antibodies. Scale bars: 5 μm (A–C), 1 μm (D–I), 2 μm (J,K, left), 500 nm (J,K, right), 20 μm (M,N). **P<0.01, ***P<0.001 (unpaired two-tailed Student's t-test).
We investigated GalT::APEX2::EGFP-positive membrane structures in S2 cells expressing V5::Sec71E677K or V5::GarzE740K by electron microscopy. In V5::Sec71E677K-expressing S2 cells, the cisternae and tubules with DAB deposition gathered in one place (Fig. 2J). This phenotype resembles BFA-treated S2 cells (Fig. 1F,G), whereas in V5::GarzE740K-expressing S2 cells, ER and vesicles with DAB deposition were amplified, with no cytologically recognizable Golgi stacks (Fig. 2K). These results indicate that Sec71 loss-of-function phenocopies BFA treatment and results in structures similar to BFA bodies.
Sec71 is the sole BFA target in BFA body formation in S2 cells
BFA binds to the Sec7 domain of ARF-GEFs. Biochemical experiments, crystal structure analysis, and sequence alignment of BFA-sensitive and -insensitive ARF-GEFs revealed that the critical amino-acid residues in the Sec7 domain responsible for BFA binding; Y190, S191, M194, T197 and V204 in the Sec7 domain inhibit GDP/GTP exchange activity of ARF-GEF (Geldner et al., 2003; Peyroche et al., 1999; Renault et al., 2003; Viaud et al., 2007; Zeeh et al., 2006). Sec71 contains all of these residues except Y190 (F713 in Sec71). We edited the genome sequence of Sec71 by CRISPR/Cas9-mediated knock-in, and generated two types of S2 cells with Sec71 point mutations, Sec71M717L, which lacks the conserved M194 in the Sec7 domain and is expected to be BFA resistant, and Sec71F713Y, which gains the consensus sequence of Y190 on the Sec7 domain, which is expected to be more sensitive to BFA than the wild-type protein (Peyroche et al., 1999), and is therefore referred to as hypersensitive.
We investigated the sensitivity to BFA of these two mutant and wild-type S2 cells (Fig. 2L–N). S2 cells with Sec71M717L did not respond to BFA even at a high concentration (50 μM) – BFA bodies were formed in 3.8±0.73% of Sec71M717L S2 cells and 30.2±5.3% of wild-type S2 cells (mean±s.d.; Fig. 2L,M). In contrast, S2 cells with Sec71F713Y responded to BFA at a low concentration (5 μM) – BFA bodies were formed in 27.2±1.7% of Sec71F713Y S2 cells and 1.9±1.4% of wild-type S2 cells (Fig. 2L,N). Thus, substitutions of one amino acid in Sec71 alone can dramatically change the BFA sensitivity of S2 cells. Notably, the proportion of Sec71F713Y S2 cells with BFA bodies did not dramatically increase with increasing BFA doses from 5 μM (27.2±1.7%) to 50 μM (34.4±3.8%) (Fig. 2L). Moreover, wild-type S2 cells with BFA bodies reached a similar proportion at 50 μM (30.2±5.3%) (Fig. 2L). These results indicate that the effect of BFA is likely to reach a maximum at 5 μM for cells with Sec71F713Y mutant and 50 μM for the wild-type Sec71. These results indicate that impairment of Sec71 is necessary and sufficient for BFA body formation. Thus, Sec71 is the only BFA target responsible for BFA body formation.
Sec71 localized to the center of BFA bodies
We examined the localization of endogenous Sec71 using an anti-Sec71 antibody (Wang et al., 2017). As previously reported (Christis and Munro, 2012; Wang et al., 2017), wild-type Sec71 colocalized with the TGN markers Golgin245 and tdTomato::Rab6, and also localized between the cis-Golgi marker GM130 and the RE marker Rab11, indicating that endogenous Sec71 is on the TGN (Fig. 3A,C,E, upper panels and plots). In wild-type BFA bodies, Sec71 was extensively concentrated at centers (Fig. 3A,C,E, lower panels and plots). Rab11 and tdTomato::Rab6 also localized to the centers of BFA bodies but more broadly than Sec71 (Fig. 3C,E, lower panels and plot). Golgin245 was mostly found between GM130 and Sec71 (Fig. 3A, lower panels and plots). As shown above, the polarity of Golgi stacks is largely maintained in BFA bodies, as cis outwards and trans inwards (Fig. 1). These results indicate that BFA uncouples Sec71 from two other TGN markers, Rab6 and Golgin245, and induces Sec71 aggregation, resulting in BFA body formation.
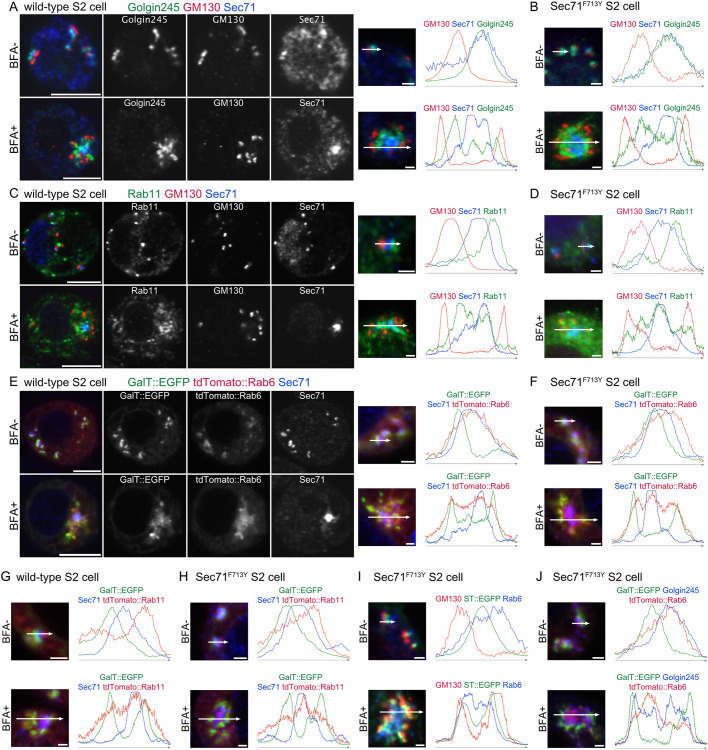
Fig. 3.
Sec71 is localized to the center of the BFA body. Immunostaining and plots of signal intensities from wild-type (A,C,E,G) and Sec71F713Y S2 cells (B,D,F,H–J). Right, plots of signal intensities from images on the left. Signal intensities were measured along the 1.5 µm (upper panels) and 5 µm (lower panels) arrows shown in insets. In all panels, cells not treated with BFA are shown in the upper row and BFA-treated cells are shown in the lower row. 50 μM and 25 μM BFA was used for the wild-type cells and Sec71F713Y S2 cells, respectively. (A,B) Immunostaining with anti-Golgin245 (green), anti-GM130 (red), and anti-Sec71 (blue) antibodies. (C,D) Immunostaining with anti-Rab11 (green), anti-GM130 (red), and anti-Sec71 (blue) antibodies. (E,F) S2 cells expressing GalT::EGFP (green) and tdTomato::Rab6 (red) immunostained with anti-Sec71 (blue) antibody. (G,H) S2 cells expressing GalT::EGFP (green) and tdTomato::Rab11 (red) immunostained with anti-Sec71 (blue) antibody. (I) S2 cells expressing ST::EGFP (green) immunostained with anti-GM130 (red) and anti-Rab6 (blue) antibodies. (J) S2 cells expressing GalT::EGFP (green) and tdTomato::Rab6 (red) immunostained with anti-Golgin245 (blue) antibody. Scale bars: 5 μm (A,C,E, left panels), 1 μm (A,C,E, right panels, B,D,F,G–J).
To investigate whether Sec71F713Y and wild-type Sec71 behave in a similar manner, except for their sensitivities to BFA, we compared the distributions of Sec71 and Golgi/RE markers in both untreated and BFA-treated Sec71F713Y cells (Fig. 3B,D,F,H–J) with those in untreated and BFA-treated wild-type cells (Fig. 3A,C,E,G; Fig. 1C,D). The distributions of GM130, Golgin245, Rab11, GalT::EGFP, tdTomato::Rab6, tdTomato::Rab11, ST::EGFP and Sec71 in Sec71F713Y cells were similar to those in wild-type control cells. The sizes and shapes of Sec71F713Y BFA bodies were not obviously different from those of wild-type BFA bodies.
Live imaging of BFA body formation in S2 cells
To understand the mechanism of BFA body formation, we used confocal microscopy. To avoid using high BFA concentrations, we used BFA-hypersensitive Sec71F713Y S2 cells coexpressing trans-Golgi (GalT::EGFP) and TGN markers (tdTomato::Rab6) (Fig. 4A,B; Movies 1–3). In BFA-untreated cells, Golgi stacks move slowly but constantly, sometimes hitting each other and occasionally connecting by the merger of their TGNs (Fig. 4A, arrows; Movie 1). Separation of these TGN-connected Golgi stacks was also often observed (Fig. 4A, arrowheads; Movie 1). The mergers and separations of TGNs were confirmed in the XY and XZ projections of 3D time-lapse observations (Movies 2 and 3).
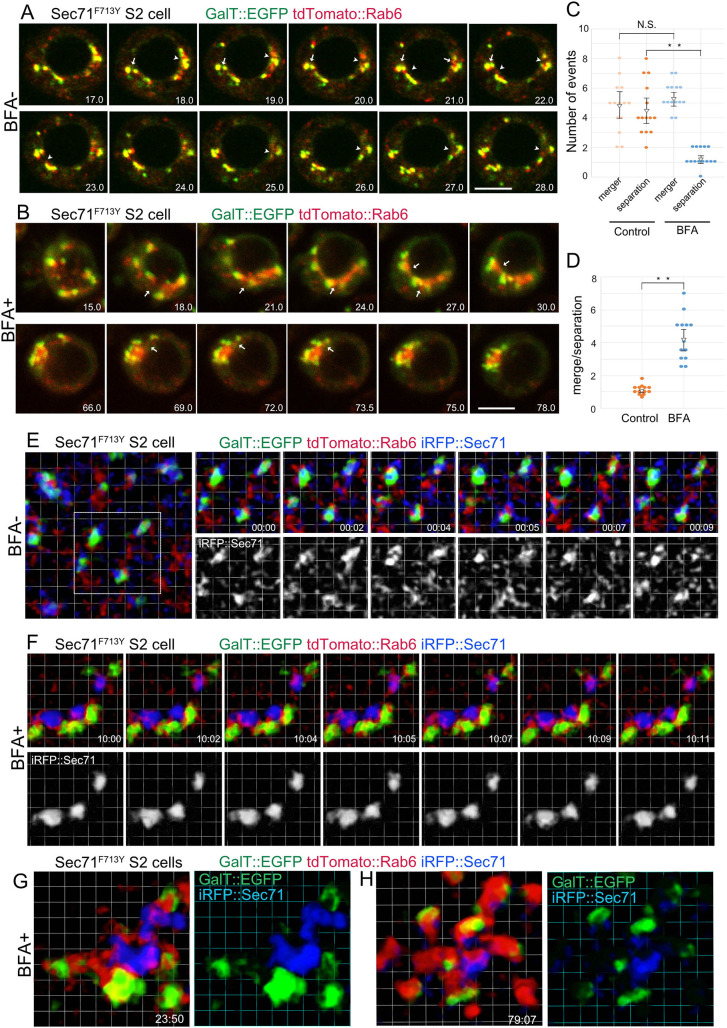
Fig. 4.
Live imaging of BFA body formation. (A,B) Frames from time-lapse movies of BFA-hypersensitive S2 cells carrying the Sec71F713Y substitution, and also expressing GalT::EGFP (green) and tdTomato::Rab6 (red) with (B) or without 25 μM BFA treatment (A) observed by confocal microscopy. Numbers at right bottom corners indicate the time (min) after BFA addition (B) or start point of time-lapse observation (A). Arrows or arrowheads indicate mergers or separations of TGN of two or more Golgi stacks. (C) Quantifications of mergers or separations of TGNs in untreated (left) and BFA-treated S2 cells (right). (D) Ratios of mergers to separations of TGNs in untreated (left) and BFA-treated (right) S2 cells carrying the Sec71F713Y substitution. Error bars show mean±s.d. (E–H) Frames of time-lapse movies of S2 cells carrying the BFA-hypersensitive Sec71F713Y substitution, also expressing GalT::EGFP (green), tdTomato::Rab6 (red) and iRFP::Sec71 (blue) with (F–H) or without 25 μM BFA treatment (E) observed by SCLIM. Numbers at the right bottom corners indicate the time (min:sec) after BFA addition (F–H) or start point of time-lapse observation (E). Scale bars: 5 μm (A,B); Grid: 1 μm (E–H). **P<0.01, N.S., not significant (unpaired two-tailed Student's t-test).
The merger and separation of TGNs was constantly repeated in BFA-untreated cells – 4.86±1.81 TGN merger and 4.57±1.72 TGN separation events were observed in five optical sections at a 0.5-µm interval of a cell within 60 min (mean±s.d.; Fig. 4C). After BFA addition, TGN mergers were similar to those of BFA untreated cells, but TGN separation was limited; 5.43±0.90 TGN mergers and 1.29±0.59 TGN separations occurred in the same conditions (Fig. 4C). The ratio of TGN mergers to TGN separations per cell increased from 1.09±0.28 to 4.23±1.34 upon BFA addition (Fig. 4C). As a result, Golgi stacks developed aggregates connected via TGNs at their cores (Fig. 4B; Movies 1 and 3). These observations indicate that BFA inhibits the separation of TGN, leading to the formation of BFA bodies.
To understand how BFA affects Sec71 dynamics, Sec71 movement before and after BFA addition was examined by super-resolution confocal live imaging microscopy (SCLIM). Before BFA treatment, iRFP::Sec71 fluorescence was relatively strong on the trans sides of GalT::EGFP-positive trans-Golgi membranes; however, the shapes of the iRFP-positive structures were unstable and most of the iRFP signal appeared hazy in the cytoplasm (Fig. 4E; Movie 4).
Minutes after the addition of BFA, as the dispersed iRFP signals decreased, iRFP::Sec71 became concentrated to form stable globular structures, locating near to GalT::EGFP-positive trans-Golgi membranes but not in direct contact. Both trans-Golgi and Sec71-positive structures were accompanied by tdTomato::Rab6, which often filled the space between them (Fig. 4F; Movie 4). At 24 and 79 min (Fig. 4G,H) after BFA addition, cells had aggregates of iRFP::Sec71 that were surrounded by GalT::EGFP-positive trans-Golgi membranes connected via tdTomato::Rab6 (Fig. 4G,H; Movie 4). These results indicate that BFA inhibits the dynamic behavior of Sec71, which results in the aggregation of Sec71-positive structures and Golgi stacks. This is consistent with the mechanism of action of BFA, which inhibits the turnover of ARF GTPases by stabilizing the complex of GDP-ARF–ARF-GEF, in the case of Drosophila, Sec71 on the TGN.
Sec71 exclusively localizes to TGN in fly photoreceptors
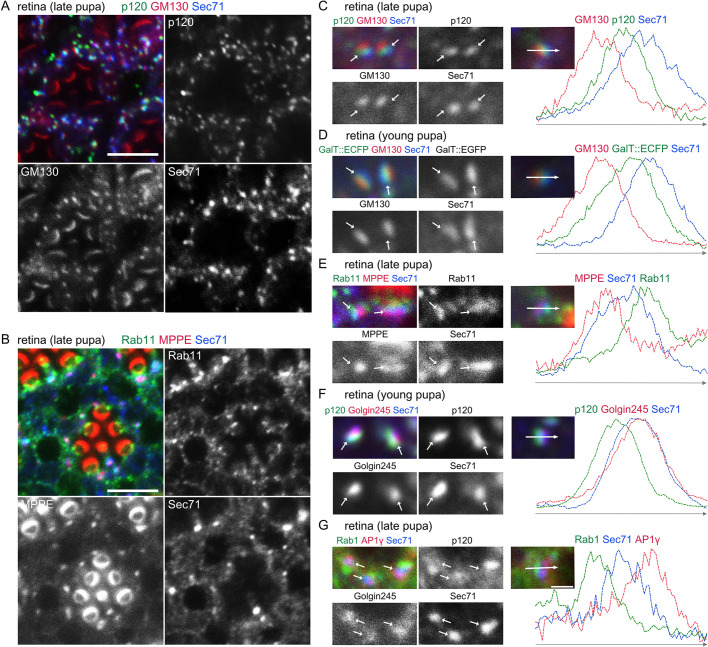
To understand the function of Sec71 in fly photoreceptors, we first examined the localization of endogenous Sec71. In contrast to Rab11, which is located not only on the trans sides of Golgi stacks, but also on post-Golgi vesicles at the base of the rhabdomeres (Otsuka et al., 2019; Satoh et al., 2005), Sec71 associated exclusively with the Golgi stack/GA-RE complex (Fig. 5A,B). Detailed analysis indicated that Sec71 localization within the Golgi stack/GA-RE complex was on the TGN (Fig. 5C–G), that is, the trans side of the cis-Golgi marker Rab1 (Fig. 5G), the medial-Golgi markers p120 and metallophosphoesterase (MPPE) (Fig. 5C,E,F) (Chen et al., 2005) and the trans-Golgi marker GalT::ECFP (Fig. 5D), and cis-side of the RE marker, Rab11 (Fig. 5E). Sec71 strongly colocalized with the TGN marker Golgin245 (Fig. 5F), but more to the cis side of another TGN marker, AP1γ (Fig. 5G) (Hirst et al., 2009). These results suggest that Sec71 mainly functions on the TGN of photoreceptors.
Fig. 5.
Sec71 exclusively localizes to TGN in photoreceptors. Immunostaining of retinas dissected from wild-type young and late-pupal flies with (A) anti-p120 (green), anti-GM130 (red) and anti-Sec71 (blue) antibodies; (B) anti-Rab11 (green), a medial-Golgi marker, anti-MPPE (red), and anti-Sec71 (blue) antibodies. Anti-MPPE antibody stains not only the medial Golgi but also the tips of the rhabdomeres. It is not known whether the latter staining represents genuine MPPE localization. (C–G) Left, immunostaining of retinas by the indicated antibodies. GalT::ECFP was expressed in D. Right, plots of signal intensities from images to the left. Signal intensities were measured along the 1.5 µm arrow in the insets; graphs show the overlap between channels. Scale bars: 5 μm (A,B), 1 μm (C–G).
Rh1 and Na+K+-ATPase accumulated in Golgi aggregates in Sec71-deficient photoreceptors
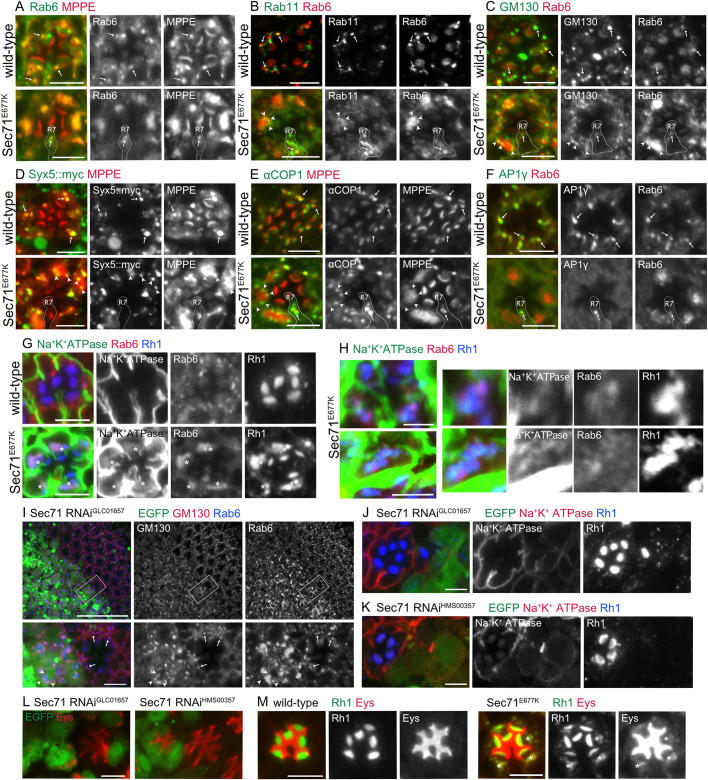
We next investigated the effects of Sec71 impairment in fly photoreceptors. We first expressed dominant-negative Sec71 (Sec71E677K) by means of an Rh1-Gal4 driver, which induces the expression in R1-6 peripheral photoreceptors from late pupal stages. In Sec71E677K-expressing late pupal photoreceptors, the TGN marker Rab6 and the medial-Golgi marker MPPE colocalized on the entire Golgi aggregate (Fig. 6A). Most of Rab11, an RE marker, seemed to be diffused, but some staining remained in the Golgi aggregate (Fig. 6B). The cis-Golgi markers GM130 and Syntaxin5::myc (Syx5::myc) (Norgate et al., 2010) localized to the peripheries of aggregates (Fig. 6C,D). As ARF-GEFs recruit coat proteins to Golgi stacks, we investigated the localization of a subunit of COPI, αCOP (Kitazawa et al., 2012) and AP1γ (Hirst et al., 2009). Both αCOP and AP1γ localized on Golgi stacks in wild-type photoreceptors (Fig. 6E,F, upper panels). However, AP1γ was completely diffuse in Sec71E677K-expressing R1–6 peripheral photoreceptors, although αCOP localized peripherally in Golgi aggregates, similar to other cis-Golgi markers (Fig. 6E,F, lower panels). Notably, punctate staining of AP1γ was found in R7 cells, which do not express Sec71E677K. On the other hand, in GarzE740K expressing photoreceptors, Rab6, MPPE, αCOP, and AP1γ all were diffuse (Fig. S3A–D). The diffuse Rab6 and MPPE colocalized with ER markers Calnexin99A (Cnx) (Rosenbaum et al., 2006) and ER-membrane complex subunit 3 (EMC3) (Satoh et al., 2015), respectively (Fig. S3E,F). GarzE740K likely induced redistribution of Golgi resident proteins into the ER. Interestingly, GM130 staining still remained punctate; we discussed this issue below. Thus, the phenotypes of photoreceptors expressing Sec71E677K and GarzE740K correspond to those of S2 cells under Sec71 and Garz impairment, respectively.
Fig. 6.
Basolateral and rhabdomere transport is inhibited in Sec71E677K-expressing photoreceptors. (A–G) Immunostaining of retinas from Rh1-Gal4/+ (upper panels) and Rh1-Gal4/UAS- Sec71E677K (lower panels) late-pupal flies. Sec71E677K is expressed in the R1–6 peripheral photoreceptors (A–C,E–G). Immunostaining of retinas from Rh1-Gal4/UAS-Syx5::myc (upper panels) and Rh1-Gal4/UAS-Syx5::myc, UAS- Sec71E677K (lower panels) late-pupal flies. Sec71E677K is expressed in the R1–6 peripheral photoreceptors (D). Anti-Rab6 (green) and anti-MPPE (red) antibodies (A). Anti-Rab11 (green) and anti-Rab6 (red) antibodies (B). Anti-GM130 (green) and anti-Rab6 (red) antibodies (C). Anti-myc (green) and anti-MPPE (red) antibodies (D). Anti-αCOPI (green) and anti-MPPE (red) antibodies (E). Anti-AP1γ (green) and anti-Rab6 (red) antibodies (F). Anti-Na+K+-ATPase-α (green), anti-Rab6 (red), and anti-Rh1 (blue) antibodies (G). Arrows indicate Golgi stacks in the wild-type cells, and arrowheads indicate the foci of cis-Golgi makers surrounding the medial and late Golgi aggregates. Cytoplasmic accumulation of Rh1 is indicated by asterisks. (H) Immunostaining of retinas from Rh1-Gal4/UAS-Sec71E677K late-pupal flies using anti-Na+K+-ATPase-α (green), anti-Rab6 (red), and anti-Rh1 (blue) antibodies. (I–L) Immunostaining of retinas dissected from Sec71-RNAiGLC01657 (I,J,L, left) or Sec71RNAiHMS00357 (K,L, right) mosaic retinas. GFP marks Sec71-RNAi-expressing cells. Anti-GM130 (red) and anti-Rab6 (blue) antibodies (I). Anti-Na+K+-ATPase-α (red) and anti-Rh1 (blue) antibodies (J,K). Anti-Eys antibody (red) (L), Arrows indicate Golgi stacks in the wild-type cells, and arrowheads indicate the foci of cis-Golgi makers surrounding Golgi aggregates. (M) Immunostaining of retinas from Rh1-Gal4/+ (left) and Rh1-Gal4/UAS- Sec71E677K (right) late-pupal flies. Sec71E677K is expressed in the R1–6 peripheral photoreceptors. Anti-Rh1 (green) and anti-Eys (red) antibodies. Cytoplasmic accumulation of Rh1 and Eys is indicated by asterisks. Scale bars: 5 μm (A–G), 2 μm (H), 50 μm (I, upper), 5 μm (I, lower, J–M).
Recently, RNAi-based screening identified Sec71 as an important factor for apical trafficking in fly photoreceptors (Laffafian and Tepass, 2019). Moreover, Sec71 was shown to be essential not only for apical transport, including Rh1 and Eys, but also for basolateral transport of Na+K+-ATPase. Similarly, we found that Rh1 accumulation in the rhabdomere was severely inhibited by Sec71E677K expression induced by Rh1-Gal4 (Fig. 6G). However, the defect in Na+K+-ATPase transport was limited in these photoreceptors (Fig. 6G), as Rh1-Gal4 expression probably occurs too late to sufficiently inhibit Na+K+-ATPase transport. Interestingly, we found accumulation of robust Rh1 and some Na+K+-ATPase in Golgi aggregates (Fig. 6G,H). The staining of Rh1 and Na+K+-ATPase partially overlapped but clearly separated within Golgi aggregates, suggesting that sorting of these proteins might be done without Sec71 (Fig. 6H).
To verify the function of Sec71 in fly photoreceptors, we attempted to create mosaic eyes containing Sec71-deficient clones using a strong mutant allele, Sec71ex11, using the FRT-FLP system. However, we failed to obtain Sec71ex11 homozygous clones, suggesting the lethality of Sec71 deficiency. Thus, we knocked down Sec71 using RNAi. To make mosaic retinas, we used coin-FLP-Gal4 (Bosch et al., 2015) with eyeless-FLP, and expressed Sec71-RNAiGLC01657 or Sec71-RNAiHMS00357 from early eye development. These two RNAi lines target different parts of the Sec71 mRNA (see Flybase). Sec71-RNAiGLC01657-expressing photoreceptors had Rab6-positive aggregates surrounded by GM130-positive foci (Fig. 6I). The transport of Rh1 and Na+K+-ATPase was severely inhibited (Fig. 6J). Sec71-RNAiHMS00357-expressing photoreceptors also showed severe Rh1 and Na+K+-ATPase defects (Fig. 6K). These phenotypes of photoreceptors expressing Sec71-RNAi are consistent with those of cells expressing Sec71E677K. Unlike ommatidia expressing Sec71E677K, the inter-rhabdomeral space (IRS) of Sec71-RNAi-expressing ommatidia was not fully open (Fig. 6I–K). As the IRS is formed by the secretion of Eys from the apical membrane of photoreceptors (Husain et al., 2006; Laffafian and Tepass, 2019), we investigated the localization of Eys (Fig. 6L). It accumulated in the cytoplasm of the Sec71-RNAiGLC01657-expressing photoreceptors, with IRSs that were small or difficult to recognize. This difference in phenotypes between cells expressing Sec71E677K and cells expressing Sec71-RNAi could be explained by differences in the onset of Sec71 deficiency, such that Rh1Gal4-driven Sec71E677K inhibits Sec71 function starting from the late pupal stage, whereas coin-FLP-Gal4-driven Sec71-RNAi knockdown starts from early in eye development. Eys occasionally accumulated in the cytoplasm with Rh1 in cells expressing Sec71E677K (Fig. 6M), supporting this hypothesis. These results indicate that Sec71-deficiency inhibits anterograde transport to the apical and basolateral membrane as well as the secretion to the IRS.
Tubule network and vesicle accumulation in Sec71-deficient photoreceptors
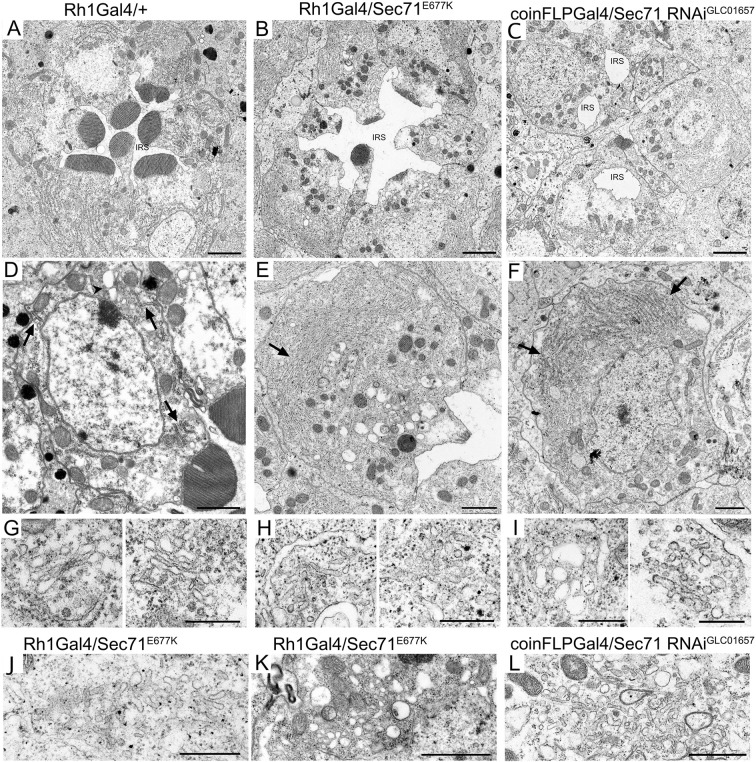
To investigate membrane structure in Sec71-deficient photoreceptors, we observed thin sections of Rh1-Gal4/+, Rh1-Gal4/UAS-Sec71E677K, and coin-FLP-Gal4/UAS-Sec71-RNAiGLC01657 pupal photoreceptors using electron microscopy (Fig. 7). In wild-type photoreceptors, the rhabdomeres were round-shaped (Fig. 7A,D), but in Sec71-deficient photoreceptors they were not round, as well as being rather smaller and thinner than in wild-type cells (Fig. 7B,C,E,F). Several ER membrane sheets and Golgi stacks were observed in typical thin sections of wild-type photoreceptors (Fig. 7D, arrows, arrowhead). However, in Sec71E677K-expressing photoreceptors, the ER membrane was amplified and Golgi stacks were not observed; instead, tubule networks and vesicles were often observed (Fig. 7E, arrow, 7H,J,K). The amplification of ER membrane and tubule networks was also observed in Sec71-RNAi photoreceptors (Fig. 7F, arrows, 7I,L). The accumulation of cytoplasmic Rh1 and Na+K+-ATPase likely localizes on these tubule networks and vesicles, which are most probably Golgi aggregates. Consistent with our confocal microscopy observations, the IRS of Sec71-RNAi ommatidia was found to be quite small and fragmented (Fig. 7C,F).
Fig. 7.
Tubule networks develop in Sec71-deficient photoreceptors. Electron micrographs of photoreceptors from Rh1-Gal4/+ (A,D,G), Rh1-Gal4/UAS- Sec71E677K (B,E,H,J,K) and coinFLPGal4/UAS-Sec71RNAiGL01657 (C,F,I,L) late-pupal flies. (A–C) Electron micrographs of a single ommatidium. The IRS is small and fragmented in C. (D–F) Electron micrographs of single photoreceptors. Arrows show the ER membrane and the arrowhead shows a Golgi stack. (G–I) Electron micrographs of Golgi stacks or related organelles. (J–L) Electron micrographs of tubules and vesicles observed in Sec71-deficient cells. Scale bars: 2 μm (A–C), 1 μm (D–F), 500 nm (G–L).
These results indicate that structures of ER and Golgi stacks are strongly affected, and transport for both apical and basolateral membrane domains is inhibited in Sec71-deficient photoreceptors.
DISCUSSION
Impairment of Sec71 function results in TGN/RE aggregation, leading to formation of BFA bodies
We showed that the ARF-GEF inhibitor BFA induces the formation of BFA bodies in Drosophila cells (Fig. 1). Detailed analysis of BFA bodies using confocal microscopy indicates that aggregated TGN/RE is central and Golgi stacks are located at the periphery of BFA bodies (Fig. 1). We also showed that the sensitivity of S2 cells to BFA could be completely controlled by genome editing of a single ARF-GEF gene, Sec71; S2 cells carrying the BFA-resistant Sec71M717L mutation do not form BFA bodies even at a high BFA concentration, whereas S2 cells carrying BFA-hypersensitive Sec71F713Y form BFA bodies at low BFA concentration (Fig. 2L–N). These results indicate that Sec71 is the sole BFA target for BFA body formation in S2 cells. In addition, BFA-hypersensitive S2 cells provide an excellent model system to study the role of ARF-GEF and ARF1 in the TGN.
In normal S2 cells, Golgi stacks are moving constantly and often fuse together on their TGNs, and then separate spontaneously (Fig. 4A,C; Movies 1–3). We previously reported that REs are attached to the trans sides of Golgi stacks in both Drosophila and microtubule-disrupted HeLa cells (Fujii et al., 2020). Moreover, REs and Golgi stacks as well as free REs themselves undergo repeated detachment and reattachment. Although RE detachment and reattachment are much more rapid events than the TGN mergers and separations of Golgi stacks reported here, RE dynamics could be the basis of Golgi stack attachment to their TGNs. In BFA-treated S2 cells, TGN/RE separations were suppressed but TGN/RE-mergers were not affected, resulting in the formation of BFA bodies (Fig. 4B–D). Around the trans sides of Golgi stacks in normal S2 cells, Sec71 appears to be dynamic and unstable (Fig. 4E). BFA quickly stabilizes Sec71 on the TGN/RE and Sec71 finally localizes to the center of BFA bodies (Fig. 4F–H). Sec71 has been shown to accelerate the transport carrier formation at the TGN/RE through ARF1 activation (Christis and Munro, 2012; Wang et al., 2017). Together with the function of Sec71 already indicated or expected from the studies of homologs (Casanova, 2007; Ishizaki et al., 2008; McDonold and Fromme, 2014; Shin and Nakayama, 2004), we suggest that the deficiency of the transport carrier formation inhibits the separation of TGN/RE, resulting in BFA body formation. The simplified model in Fig. 8 shows that separations and mergers of TGN/RE lead to separation and aggregation of Golgi stacks, respectively, and BFA inhibits Sec71-mediated separations, resulting in BFA body formation.
Fig. 8.
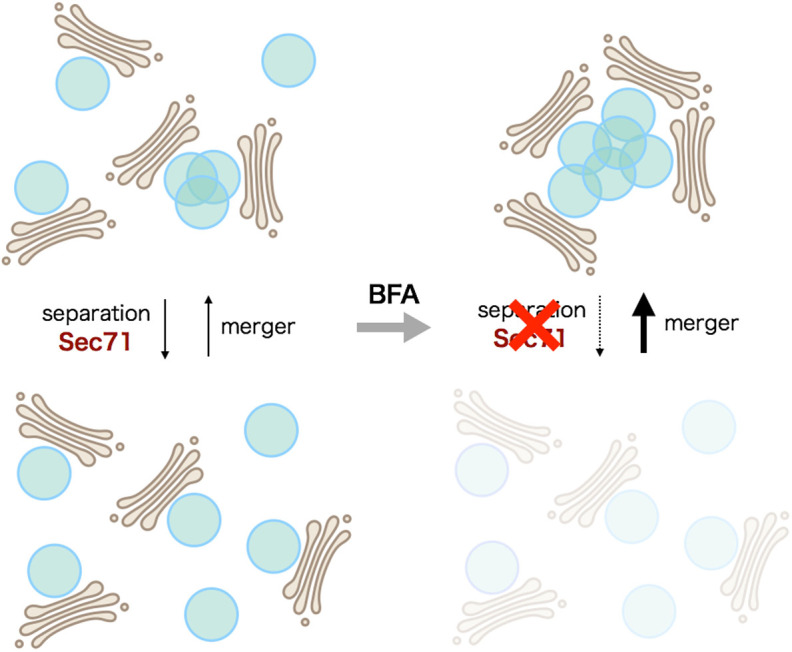
BFA body formation model. Golgi stacks and TGN/RE are shown in brown and blue, respectively. TGN/RE separations are driven by Sec71, and BFA inhibits Sec71 function. Without BFA, both separations and mergers of TGN/REs occur repeatedly, resulting in Golgi stack-associated TGN/RE being mostly separated in the steady state. After BFA addition, the separation of TGN/RE is greatly decreased, whereas mergers of TGN/RE occur normally, resulting in BFA body formation.
The appearance of BFA bodies in Drosophila S2 cells resembles that of Golgi stacks and REs in many COS-1 cells and some populations of HeLa and MDCK cells, with REs closely associated with the centrosome as one large aggregate and Golgi stacks surrounding the REs with their trans sides inward (Misaki et al., 2010). This suggests that one of the determinants of the cell-wide arrangement of Golgi stacks and RE is the kinetic balance between the merger and separation of TGN and REs.
The sole fly GBF1 ortholog, Garz, is insensitive to BFA
The most prominent effects of BFA on yeast, mammalian cells and tobacco BY2 cells are the breakup of the Golgi and redistribution of Golgi-resident proteins into the ER (Ito et al., 2012; Lippincott-Schwartz et al., 1989; Peyroche et al., 1996; Yasuhara and Shibaoka, 2000; Yasuhara et al., 1995). However, in S2, Arabidopsis and maize cells, Golgi-resident proteins were resistant to BFA (Fig. 1) (Baluška et al., 2002; Langhans et al., 2011; Uemura et al., 2014). The effects seen in many cell types can be explained by differences in the BFA sensitivity of GBF1 orthologs (Casanova, 2007; Peyroche et al., 2001, 1996; Robinson et al., 2008; Teh and Moore, 2007; Wright et al., 2014). For example, MDCK cells, in which the Golgi is not absorbed into the ER (Hunziker et al., 1991), have a substitution of one consensus amino acid residue of GBF1 responsible for BFA sensitivity in the Sec7 domain compared to other mammalian GBF1s, resulting in resistance to BFA (Verheije et al., 2008). In this report, we showed that redistribution of Golgi-resident proteins into the ER is not induced by BFA (Fig. 1) but induced by the impairment of Garz activity, as demonstrated by dominant-negative GarzE740K expression or knockdown by RNAi in S2 cells (Fig. 2B,C,K; Fig. S2B,D). As Garz lacks the consensus residues required for BFA binding (S191, T197 and V204 in the Sec7 domain), it must be insensitive to BFA. Thus, BFA does not induce breakup of the Golgi in S2 cells.
Neither the expression of dominant-negative GarzE740K nor RNAi knockdown in S2 cells or Drosophila photoreceptors redistributes GM130, a cis-Golgi localizing Golgin, to the ER. The remaining GM130 foci are reminiscent of a previously described structure, the Golgi-entry core compartment (GECCO), small foci containing a particular subset of cis-Golgi proteins formed in BFA-treated Tobacco BY2 cells (Ito et al., 2018, 2012). The GECCO was shown to be formed by COPII-independent anterograde transport from the ER, even in the presence of aggressive retrograde traffic induced by GBF1 deficiency. GM130 foci in BFA-treated mammalian cells have also been reported previously (Jiang et al., 2006; Walenta et al., 2001).
Sec71 is required for polarized transports towards both apical and basolateral domains
Sec71 is the only Drosophila ortholog of mammalian BIG1 and BIG2 (also known as ARFGEF1 and ARFGEF2, respectively), which are involved in post-Golgi vesicle formation by recruiting AP1 (Futter et al., 1998). As expected, impairment of Sec71 results in the diffusion of AP1γ in fly photoreceptors (Fig. 6F). We also showed that Golgi stacks and REs form Golgi aggregates, and that cis-Golgi markers localize in peripheral regions, similar to BFA bodies (Fig. 6A–F). Consistent with a previous report (Laffafian and Tepass, 2019), the lack of Sec71 inhibits the polarized transport of Rh1 and Na+K+-ATPase to the apical and basolateral domains (Fig. 6G,H,J,K,M). Interestingly, both Rh1 and Na+K+-ATPase accumulated in Golgi aggregates; however, the two membrane cargoes were segregated from each other, suggesting that sorting might occur even in cells without Sec71 activity (Fig. 6H). We previously showed that GPI, but not VSV-G, localizes to GA-RE, suggesting that segregation of these cargoes occurs at the interface between Golgi stacks and GA-RE (Fujii et al., 2020). Segregation might not require Sec71-mediated coat protein assembly. It is important to determine the exact timing of sorting and carrier formation in future studies.
MATERIALS AND METHODS
Construction of plasmids
For PCR amplification, the high-fidelity DNA-polymerase KOD plus Neo (TOYOBO, Osaka, Japan) was used, unless otherwise noted. Primers and other oligonucleotides are listed in Table S1. Sequences of plasmids used in this study are shown in Table S2.
The pMK-V5::Sec71 and pMK-V5::Garz plasmids were constructed from cDNAs of total RNA from w1118 third-instar larvae, produced by PCR using KOD FX Neo (TOYOBO, Osaka, Japan) with primers Sec71-GF1/Sec71-GR2 and Garz-GF1/Garz-GR1, respectively. DNA sequences of 18–20 bp homologous to the vector were added by 10 additional cycles of PCR with primers GL3-Sec71/Sec71-MK-Sp and GL3-garz/garz-MK-Sp, respectively. The DNA fragment encoding the V5 epitope was amplified from pMK-V5::pcs (Otsuka et al., 2019) with primers B-MK-V5 and GL3-R. Using NEBuilder HiFi DNA Assembly Master Mix (NEB, Ipswich, MA, USA), these fragments were assembled between the BamHI and SpeI sites of pMK33-CFH-BD to obtain pMK-V5::Sec71 and pMK-V5::garz. The clones were sequenced using the primers listed below, and one of each clone, without any mutations, was selected and used for subsequent experiments.
pMK-V5::Sec71E677K and pMK-V5::garzE740K were generated from pMK-V5::Sec71 and pMK-V5::garz using Gibson assembly-mediated site-directed mutagenesis. These V5-tagged genes were amplified with primers K-MT-V5 and MK-MT-Ap and transferred to KpnI-ApaI sites of pMT-puro to construct pMT-hyg-V5::Sec71, pMT-hyg-V5::Sec71E677K, pMT-hyg-V5::garz and pMT-hyg-V5::garzE740K.
The pMT-hyg-V5::Sec71, pMT-hyg-V5::Sec71M717L and pMT-hyg-V5::Sec71F713Y plasmids were constructed using Gibson assembly-mediated site-directed mutagenesis. The fluorescent protein iRFP713 came from piRFP (Addgene #31857). The coding region of Sec71M717L was amplified from pMT-hyg-V5::Sec71M717L with primers Asc-GL3 and MT-Mlu-short, then cloned into pMT-hyg-V5::iRFP713::myc to produce pMT-hyg-V5::iRFP713::Sec71.
To construct CMV-ManII::EGFP and CMV-ST::EGFP, DNAs encoding amino acid residues 1–116 of mouse mannosidase II (Uniprot ID: P27046) and 1–43 of human sialyl-transferase (Uniprot ID: P15907) were amplified from plasmids Str-KDEL_ManII-SBP-tagBFP (Addgene #65254) and Str-KDEL_ST-SBP-tagBFP (AddGene #65266), deposited by Franck Perez (Boncompain et al., 2012), with primers Sac-ManII/ManII-B and Sac-ST/ST-Sal, respectively. These fragments were digested with SacI/BamHI and SacI/SalI, respectively, and then inserted into EGFP-golgi, replacing the GalT gene.
To construct pMT-GalT::EGFP, pMT-ST::EGFP and pMT-ManII::EGFP, NheI/NotI-digested fragments of CMV-GalT::EGFP, CMV-ST::EGFP and CMV-ManII::EGFP were inserted between SpeI and NotI sites of pMT-puro (Addgene plasmid #17923; deposited by David Sabatini).
To construct pMT-GalT::EGFP-T2A-tdTomato::Rab11, the DNA fragment encoding GalT::EGFP was amplified from CMV-GalT::EGFP with primers msK-GalT-F and EGFP-dT2A and then with msK-GalT-F and dT2Aa. The DNA fragment coding tdTomato::Rab11 was amplified from pUAST-tdTomato-Rab11 WT (Addgene plasmid #53473; deposited by Matthew Scott), with primers dT2A-EGFP and Rab11-MT-Mlu, and then with dT2As and Rab11-MT-Mlu. To construct pMT-GalT::EGFP-T2A-tdTomato::Rab11, these two fragments were assembled between the KpnI and MluI sites of pMT-puro, using Gibson assembly. The plasmid pMT-GalT::EGFP-T2A-tdTomato::Rab6 was constructed similarly to the tdTomato-Rab6 fragment amplified from pMT-tdTomato::Rab6 with primers dT2A-EGFP and MT-Mlu-short.
S2 cell culture
Drosophila S2 cells were cultured at 25°C in Schneider's medium (Gibco) supplemented with 10% inactivated fetal bovine serum and penicillin-streptomycin, as described in the standard protocol at DRSC/TRiP Functional Genomics Resources, Harvard University (https://fgr.hms.harvard.edu/fly-cell-culture).
Knockdown by RNA interference in S2 cells
Predesigned RNAi templates, DRSC07193 for Garz and DRSC01893 for Sec71, were selected from the UP-TORR Database (Hu et al., 2013). The templates for RNA synthesis were amplified from w1118 genomic DNA using KOD FX Neo (TOYOBO, Osaka, Japan) with primers garz-DRSC07193-F/garz-DRSC07193-R for Garz, and Sec71-DRSC01893-F/Sec71-DRSC01893-R for Sec71, respectively. T7 RNA polymerase promoters were added to both ends using 10 additional rounds of PCR using KOD Plus Neo (TOYOBO) with primers garz-DRSC07193-T7F/garz-DRSC07193-T7R for Garz, and Sec71-DRSC01893-T7F/Sec71-DRSC01893-T7R for Sec71, respectively. From these templates, double-stranded RNA was synthesized using the T7 High Yield RNA Synthesis Kit (NEB, Ipswich, MA, USA) and purified using RNA Clean & Concentrator-25 (Zymo Research, Irvine, CA, USA). Aliquots of 0.5 ml of 30–50% confluent S2 cells were transfected with 1 µg of dsRNA using 3 µl of FuGeneHD (Promega, Madison, WI, USA). At 18–24 h after transfection, the medium was replaced. Cells were incubated for 4–5 days after transfection, then subjected to immunostaining as described below.
Establishment of genome-edited S2 cells with BFA-resistant mutant Sec71M717L or BFA-hypersensitive mutant Sec71F713Y
Designs of CRISPR and repair templates are shown in Fig. S1. A pair of CRISPR target sites was chosen to nest the 713Y and 717M mutations between them. In addition to the desired substitutions, silent mutations were introduced on the proto-spacers to make the successfully edited allele CRISPR/Cas9-resistant. Approximately 200 bp of genomic sequences were added for homology-dependent repair on both sides. The repair templates were synthesized as 500-bp double-stranded DNA (dsDNA) fragments (gBlocks Gene Fragments, IDT), and PCR amplified using primers Sec71-GF14 and Sec71-GR6.
Chemically synthesized Alt-R CRISPR-Cas9 crRNAs (IDT, Coralville, IA, USA) were annealed with Alt-R CRISPR-Cas9 tracrRNAs (IDT, Coralville, IA, USA) to form duplexes. Then, 6 pmol of the duplex was incubated with 6 pmol of Alt-R S.p. We used Cas9 Nuclease V3 (IDT, Coralville, IA, USA) as the ribonucleoprotein (RNP) complex, according to the manufacturer's instructions. An aliquot of 0.5 ml of S2+ cells was transfected with 6 pmol of RNP and 250 ng of dsDNA repair template, using 3 µl of Lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA). To screen for BFA-resistant cells, Sec71M717L-knock-in cells were subcultured in medium containing 50 µM BFA for 30 days.
Because the null allele of Sec71 showed strong lethality in the mosaic retina, a biallelic knockout of Sec71 was expected to be lethal in S2 cells. Therefore, an additional knockout after knock-in by the same pair of CRISPR/Cas9 was expected to eliminate unedited alleles and enrich for the genome-edited allele. To enrich for BFA-hypersensitive cells, the Sec71F713Y knock-in was repeated three times; thereafter, the cells were knocked out four times by transfecting the RNP without repair template. To evaluate the efficiency of genome editing, the 849-bp fragment of genomic DNA was amplified using Sec71-GF5 and Sec71-GR4 primers, then digested with AluI, which digests only the unedited allele. As a result, the Sec71M717L knock-in reached 100% of the allele, with Sec71F713Y knock-in cells estimated to be 100%. These genome-edited cells were transfected with pMT-GalT::EGFP-T2A-tdTomato::Rab6 or pMT-GalT::EGFP-T2A-tdTomato::Rab11, using FuGeneHD (Promega). Transformants were selected by culturing with 2 µg/ml puromycin. After establishing stable transformants, efficiencies of genome editing were reconfirmed by PCR and AluI digestion.
Immunostaining of S2 cells
Drosophila S2 cells were transiently transfected with pMT-ManII::EGFP, pMT-mTq2-Rab11, pMT-ST::EGFP, pMT-GalT::EGFP, pMK-V5-Sec71, or pMK-V5::Sec71E677K, pMK-V5-Garz, or pMK-V5::GarzE740K using FuGENE HD (Promega). Expression was induced by adding 1 mM CuSO4 at 2 days after transfection. For BFA treatment, the cells were incubated for 2 h with 50 μM BFA (Cayman Chemical Company, Ann Arbor, MI, USA). S2 cells previously transfected with pAc-mTq2-Rab11 or wild-type S2 cells were transfected with double-stranded RNA as described above. Cells were fixed in 4% paraformaldehyde in PBS for 5 min at room temperature, rinsed three times for 2 min each in PBS, treated for 5 min in PBS containing 0.1% Triton X-100, and three 2 min rinses in PBS. Cells were incubated for 30 min in 20% bovine serum in PBS for blocking. Cells were incubated for 2 h in primary antibodies with 5% bovine serum in PBS. After three rinses for 2 min each in PBS, cells were incubated for 2 h with secondary antibodies. Cells were rinsed three times for 2 min each in PBS and then mounted in 50% glycerol in PBS containing 0.25% n-propyl gallate to inhibit fading. Primary antibodies were: mouse anti-V5 monoclonal antibody 6F5 (1:150) (WAKO Chemical #CTN3094, Osaka, Japan), rabbit anti-GM130 (1:300) (Abcam #ab30637, Cambridge, UK), guinea pig anti-Rab6 (1:300) (Iwanami et al., 2016), goat anti-Golgin245 (1:300) (DSHB) (Riedel et al., 2016), anti-Sec71 (1:300) (Wang et al., 2017), anti-AP1γ (1:2000) (Hirst et al., 2009), rat anti-p120 (1:15) (a gift from Dr Satoshi Goto, Rikkyo University, Tokyo, Japan) (Yamamoto-Hino et al., 2012), and rat anti-Rab11 (1:200) (Otsuka et al., 2019). Secondary antibodies were anti-mouse, anti-rabbit, and anti-guinea pig-IgG antibodies labeled with Alexa Fluor 488, 568 and 647 (1:300) (Life Technologies, Carlsbad, CA, USA). Sample images were recorded using FV1000 (PlanApo N 60×1.42 NA objective lens; Olympus, Tokyo, Japan) or FV3000 (PLAPON60XOSC2 1.4 NA objective lens; Olympus, Tokyo, Japan) confocal microscopes. To minimize bleedthrough, each color in double- or triple-stained samples was imaged sequentially. Images were processed in accordance with the Guidelines for Proper Digital Image Handling using Fiji, Affinity photo, and/or Adobe Photoshop CS3 (Adobe, San Jose, CA, USA).
For plotting immunostaining intensity across the Golgi stacks of BFA bodies, lines were drawn through each Golgi stack or BFA body, and intensities were measured using Fiji software and plotted using PLOT2 (micw.org). Representative plots are presented.
To quantify the proportions of S2 cells possessing BFA bodies, untreated and BFA-treated S2 cells were immunostained with anti-Rab6 and anti-GM130 antibodies, then observed under a FV3000 microscope with a 60× objective lens. In these images, more than 250 cells for each condition were semi-automatically annotated for possessing BFA bodies or not; background staining by anti-Rab6 and anti-GM130 were used to define cells, with a concentrated Rab6 staining surrounded by GM130 staining scored as a BFA body. We performed three independent experiments for each condition, with means and standard deviations plotted in Fig. 2L.
Live imaging of S2 cells by confocal microscopy
BFA-hypersensitive S2 cells were transfected with pMT-GalT-EGFP-T2A-tdTomato-Rab6 and selected with 2 µg/ml puromycin for 2 weeks. Expression was induced by adding 0.5 mM CuSO4 for 1 day. Cells were attached to a µ-Slide 8-well chambered coverslip (ibidi, Martinsried, Germany) treated with poly-L-lysine (Merck KGaA, Darmstadt, Germany), and imaged on an FV3000 confocal microscope equipped with a PLAPON60XOSC2 1.4 NA objective lens (Olympus, Tokyo, Japan). For BFA treatment, the cells were incubated with 25 μM BFA (Cayman Chemical Company). For each series, Z-stacks of three slices at 0.5-µm intervals were taken every 1.5 min for 90 min. For quantification of the number of TGN merge and separation events, Z-stacks of 0.5 µm, apart from five slices, were taken every 1 min for 60 min. TGN mergers and separations were counted in 16 untreated and BFA-treated cells, respectively, and plotted per cell (Fig. 4C). The ratio of TGN-merger to TGN separation within the same cell is plotted in Fig. 4D. Time-lapse series and movies were processed using ImageJ.
To show side views of merger and separation events, Z-stacks of 49 slices at 0.25-µm intervals were taken every 20–60 s for 60 min. Volume-rendered images were generated using FluoRender.
Live imaging of S2 cells by SCLIM
BFA-hypersensitive S2 cells stably transformed by trans-Golgi (GalT::EGFP) and TGN markers (tdTomato::Rab6) (see above) were further transfected with iRFP::Sec71 and selected with 200 µg/ml hygromycin B for 2 weeks. BFA-hypersensitive S2 cells with GalT::EGFP, tdTomato::Rab6, and iRFP::Sec71 were inoculated on glass-based dishes (Iwaki, Shizuoka, Japan). Cells were cultured for a day in phenol red-free medium to reduce background fluorescence, before observation by super-resolution confocal live imaging microscopy (SCLIM) (Kurokawa et al., 2013, 2019). For BFA treatment, cells were incubated for 2 h with 25 μM BFA (Cayman Chemical Company). Z-stack images obtained by SCLIM were converted into 3D voxel data and processed by deconvolution with Volocity (Perkin Elmer, Waltham, MA, USA) using the theoretical point-spread function for spinning-disk confocal microscopy. Volume-rendered images were generated using Volocity or FluoRender, and time-lapse series and movies were processed using ImageJ.
Electron microscopy imaging of GalT::APEX2::EGFP
Drosophila S2 cells were transformed by pMT-hyg-GalT-APEX2-EGFP using FuGENE HD (Promega), then selected in 200 µg/ml hygromycin B for 3 weeks to establish a GalT-APEX2-EGFP stable transformant. Expression of GalT::APEX2::EGFP was induced by adding 0.5 mM CuSO4 for 1 day. For Fig. 1F,G, the cells were incubated for 2 h with or without 50 μM BFA (Cayman Chemical Company), then fixed in 2% glutaraldehyde, 2% paraformaldehyde and 2 mM CaCl2 in 0.1 mM cacodylate buffer (pH 7.4) for 1 h on ice. For Fig. 2H,I, S2 cells stably transformed with GalT-APEX2-EGFP were transiently transfected with pMK-V5::Sec71E677K or pMK-V5::GarzE740K using FuGENE HD (Promega), then expression of both GalT::APEX2::EGFP and V5::Sec71E677K/V5::GarzE740K was induced by adding 0.5 mM CuSO4 for 16–18 h. EM imaging of APEX2 was performed as described previously (Otsuka et al., 2019). The 70–90 nm sections were imaged using a JEM1400 transmission electron microscope (JEOL, Tokyo, Japan) operated at 80 kV; montage images were taken with a CCD camera system (JEOL, Tokyo, Japan).
Drosophila stocks
Flies were grown at 20–25°C in standard cornmeal-glucose-agar-yeast medium, either in the laboratory with room light or in a 12L/12D incubator. The following fly stocks were used: Rh1-Gal4 (Chihiro Hama, Kyoto Sangyo University, Japan), longGMR-Gal4 (Bloomington Stock No. 8605, Bloomington, IN, USA; indicated as BL8605 in the following stocks), coin-FLP-Gal4 with UAS-2xEGFP (BL58751), UAS-V5::Sec71E677K (Fengwei Yu, Temasek Life Sciences Laboratory, Singapore), UAS-Syx5::myc (Richaud Burke, Monash University, Australia), UAS-GalT::ECFP (Satoh et al., 2005), UAS-Sec71-RNAiGLC01657 (BL50539) and UAS-Sec71-RNAiHMS00357 (BL32366). The females of ey-FLP, UAS-Dicer (BL24644) with second or third balancers were crossed with the males with UAS-Sec71-RNAiGLC01657 or UAS-Sec71-RNAiHMS00357, respectively, and their male progeny were crossed to females with coin-FLP-Gal4 and UAS-2xEGFP to obtain mosaic retinas. The final cross for UAS-Sec71-RNAiHMS00357 to obtain mosaic retinas was maintained at 18°C; otherwise, RNAi clones died during early stages of eye development.
Immunostaining of fly retinas
Fixation and staining were performed as described previously (Fujii et al., 2020; Satoh and Ready, 2005). Primary antibodies were: rabbit anti-Rh1 (1:1000) (Satoh et al., 2005), mouse monoclonal anti-Na+K+-ATPase α subunit (1:300 ascites) (DSHB, IA, USA), rabbit anti-GM130 (1:300) (Abcam #ab30637, Cambridge, UK), rabbit anti-MPPE (1:1000) (a gift from Junhai Han, Southeast University, Nanjing, China), rat anti-p120 (1:15) (a gift from Dr Satoshi Goto) (Yamamoto-Hino et al., 2012), rabbit anti-Rab6 (1:300) (Iwanami et al., 2016), guinea pig anti-Rab6 (1:300) (Iwanami et al., 2016), guinea pig anti-Sec71 (1:500) (Wang et al., 2017), rabbit anti-AP1γ (1:2000) (Hirst et al., 2009), Goat anti-Golgin245 (1:500) (DSHB) (Riedel et al., 2016), mouse anti-myc (1:15) (DSHB), mouse anti-Eys (1:15) (DSHB), guinea pig anti-αCOP (1:150) (a gift from Dr Yoshihiro Inoue, Kyoto Sangyo University, Kyoto, Japan) (Kitazawa et al., 2012), rabbit anti-Cnx (1:300) (Satoh et al., 2015), rat anti-EMC3 (1:300) (Satoh et al., 2015), rat anti-Rab1 (1:250) (Otsuka et al., 2019), and rat anti-Rab11 (1:300) (Otsuka et al., 2019). Secondary antibodies were anti-mouse, anti-rabbit, anti-rat, anti-goat, and/or anti-guinea pig labeled with Alexa Fluor 488, 568 and 647 (1:300) (Life Technologies). Sample images were recorded using FV1000 (PlanApo N 60×1.42 NA objective lens; Olympus) or FV3000 (UPLSAPO60XS2 silicone-immersion 60x objective lens; Olympus) confocal microscopes. To minimize bleed-through, each signal in double- or triple-stained samples was imaged sequentially. Images were processed in accordance with the Guidelines for Proper Digital Image Handling using Fiji, Affinity photo, and/or Adobe Photoshop CS3.
Electron microscopy
Electron microscopy was performed as described previously (Satoh et al., 1997). Samples were observed under a JEM1400 EM (JEOL, Tokyo, Japan), and montages were prepared using a CCD camera system (JEOL). Phenotypes were investigated using sections at the depth at which multiple photoreceptor nuclei within ommatidia were observed.
Supplementary Material
Acknowledgements
We thank Dr Junhai Han (Southeast University, Nanjing, China), Dr Satoshi Goto (Rikkyo University, Tokyo, Japan), Dr Robert Burke (Monash University, Victoria, Australia), Dr Fengwei Yu (Temasek Life Sciences Laboratory, Singapore), and Dr Stefan Luschnig (University of Zurich, Zurich, Switzerland) for kindly providing reagents. We also thank the Bloomington Drosophila Stock Center and Drosophila Genomics and Genetic Resources. This work was made possible in part by software funded by the NIH: FluoRender: Visualization-based and Interactive Analysis for Multichannel Microscopy Data, 1 R01 EB023947, and National Institute of General Medical Sciences of the National Institutes of Health grant P41 GM103545.
Footnotes
Author contributions
Conceptualization: T.S., A.K.S.; Methodology: S.F., K.K., T.S., A.K.S.; Formal analysis: S.F.; Investigation: S.F., K.K., T.T., R.I., A.T., T.S.; Data curation: S.F., T.S., A.K.S.; Writing - original draft: T.S., A.K.S.; Writing - review & editing: K.K., A.N.; Supervision: A.N., T.S., A.K.S.; Project administration: T.S., A.K.S.; Funding acquisition: K.K., A.N., T.S., A.K.S.
Funding
This work was supported by Precursory Research for Embryonic Science and Technology grant 25-J-J4215, Japan Society for the Promotion of Science, KAKENHI grant 15K07050, Sumitomo Foundation for Basic Science Research Projects, Astellas Foundation for Research on Metabolic Disorders, and Female Researcher Joint Research Grant from Hiroshima University to A.S., KAKENHI grant 19K06566 to T.S., and KAKENHI grants 25221103, 17H06420, and 18H05275 to K.K. and A.N.
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.245571.reviewer-comments.pdf
References
- Anders, N. and Jürgens, G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65, 3433-3445. 10.1007/s00018-008-8227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster, K. and Luschnig, S. (2012). The Drosophila Sec7 domain guanine nucleotide exchange factor protein Gartenzwerg localizes at the cis-Golgi and is essential for epithelial tube expansion. J. Cell Sci. 125, 1318-1328. 10.1242/jcs.096263 [DOI] [PubMed] [Google Scholar]
- Baluška, F., Hlavacka, A., Šamaj, J., Palme, K., Robinson, D. G., Matoh, T., McCurdy, D. W., Menzel, D. and Volkmann, D. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol. 130, 422-431. 10.1104/pp.007526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain, G., Divoux, S., Gareil, N., de Forges, H., Lescure, A., Latreche, L., Mercanti, V., Jollivet, F., Raposo, G. and Perez, F. (2012). Synchronization of secretory protein traffic in populations of cells. Nat. Methods 9, 493-498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Bosch, J. A., Tran, N. H. and Hariharan, I. K. (2015). CoinFLP: a system for efficient mosaic screening and for visualizing clonal boundaries in Drosophila. Development 142, 597-606. 10.1242/dev.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova, J. E. (2007). Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 8, 1476-1485. 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- Chen, J., Call, G. B., Beyer, E., Bui, C., Cespedes, A., Chan, A., Chan, J., Chan, S., Chhabra, A., Dang, P., et al. (2005). Discovery-based science education: functional genomic dissection in Drosophila by undergraduate researchers. PLoS Biol. 3, e59. 10.1371/journal.pbio.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christis, C. and Munro, S. (2012). The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J. Cell Biol. 196, 327-335. 10.1083/jcb.201107115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, N. B., Smith, C. L., Sciaky, N., Terasaki, M., Edidin, M. and Lippincott-Schwartz, J. (1996). Diffusional mobility of Golgi proteins in membranes of living cells. Science 273, 797-801. 10.1126/science.273.5276.797 [DOI] [PubMed] [Google Scholar]
- Cox, R., Mason-Gamer, R. J., Jackson, C. L. and Segev, N. (2004). Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15, 1487-1505. 10.1091/mbc.e03-06-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, J. G., Lippincott-Schwartz, J., Bloom, G. S., Kreis, T. E. and Klausner, R. D. (1990). Dissociation of a 110-kD peripheral membrane protein from the Golgi apparatus is an early event in brefeldin A action. J. Cell Biol. 111, 2295-2306. 10.1083/jcb.111.6.2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragwidge, J. M., Scholl, S., Schumacher, K. and Gendall, A. R. (2019). NHX-type Na+(K +)/H+antiporters are required for TGN/EE trafficking and endosomal ion homeostasis in Arabidopsis thaliana. J. Cell Sci. 132, jcs226472. 10.1242/jcs.226472 [DOI] [PubMed] [Google Scholar]
- Fujii, S., Kurokawa, K., Inaba, R., Hiramatsu, N., Tago, T., Nakamura, Y., Nakano, A., Satoh, T. and Satoh, A. K. (2020). Recycling endosomes attach to the trans-side of Golgi stacks in Drosophila and mammalian cells. J. Cell Sci. 133, jcs236935. 10.1242/jcs.236935 [DOI] [PubMed] [Google Scholar]
- Futter, C. E., Gibson, A., Allchin, E. H., Maxwell, S., Ruddock, L. J., Odorizzi, G., Domingo, D., Trowbridge, I. S. and Hopkins, C. R. (1998). In polarized MDCK cells basolateral vesicles arise from clathrin-γ-adaptin-coated domains on endosomal tubules. J. Cell Biol. 141, 611-623. 10.1083/jcb.141.3.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A. and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219-230. 10.1016/S0092-8674(03)00003-5 [DOI] [PubMed] [Google Scholar]
- Goldenring, J. R. (2015). Recycling endosomes. Curr. Opin. Cell Biol. 35, 117-122. 10.1016/j.ceb.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosavi, P. and Gleeson, P. A. (2017). The function of the golgi ribbon structure - an enduring mystery unfolds! BioEssays 39, 1700063. 10.1002/bies.201700063 [DOI] [PubMed] [Google Scholar]
- Hicke, L., Zanolari, B., Pypaert, M., Rohrer, J. and Riezman, H. (1997). Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol. Biol. Cell 8, 13-31. 10.1091/mbc.8.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro, A., Gershlick, D. C., Rojas, A. L. and Bonifacino, J. S. (2015). Formation of tubulovesicular carriers from endosomes and their fusion to the trans-Golgi network. Int. Rev. Cell Mol. Biol. 318, 159-202. 10.1016/bs.ircmb.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Hirst, J., Sahlender, D. A., Choma, M., Sinka, R., Harbour, M. E., Parkinson, M. and Robinson, M. S. (2009). Spatial and functional relationship of GGAs and AP-1 in Drosophila and HeLa cells. Traffic 10, 1696-1710. 10.1111/j.1600-0854.2009.00983.x [DOI] [PubMed] [Google Scholar]
- Hu, Y., Roesel, C., Flockhart, I., Perkins, L., Perrimon, N. and Mohr, S. E. (2013). UP-TORR: online tool for accurate and Up-to-Date annotation of RNAi Reagents. Genetics 195, 37-45. 10.1534/genetics.113.151340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker, W., Whitney, J. A. and Mellman, I. (1991). Selective inhibition of transcytosis by brefeldin A in MDCK cells. Cell 67, 617-627. 10.1016/0092-8674(91)90535-7 [DOI] [PubMed] [Google Scholar]
- Husain, N., Pellikka, M., Hong, H., Klimentova, T., Choe, K.-M., Clandinin, T. R. and Tepass, U. (2006). The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev. Cell 11, 483-493. 10.1016/j.devcel.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Ishizaki, R., Shin, H.-W., Mitsuhashi, H. and Nakayama, K. (2008). Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol. Biol. Cell 19, 2650-2660. 10.1091/mbc.e07-10-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y., Uemura, T., Shoda, K., Fujimoto, M., Ueda, T. and Nakano, A. (2012). cis-Golgi proteins accumulate near the ER exit sites and act as the scaffold for Golgi regeneration after brefeldin A treatment in tobacco BY-2 cells. Mol. Biol. Cell 23, 3203-3214. 10.1091/mbc.e12-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y., Toyooka, K., Fujimoto, M., Ueda, T., Uemura, T. and Nakano, A. (2017). The trans-Golgi Network and the Golgi Stacks Behave Independently During Regeneration After Brefeldin A Treatment in Tobacco BY-2 Cells. Plant Cell Physiol. 58, 811-821. 10.1093/pcp/pcx028 [DOI] [PubMed] [Google Scholar]
- Ito, Y., Uemura, T. and Nakano, A. (2018). The Golgi entry core compartment functions as a COPII-independent scaffold for ER-to-Golgi transport in plant cells. J. Cell Sci. 131, jcs203893. 10.1242/jcs.203893 [DOI] [PubMed] [Google Scholar]
- Iwanami, N., Nakamura, Y., Satoh, T., Liu, Z. and Satoh, A. K. (2016). Rab6 is required for multiple apical transport pathways but not the basolateral transport pathway in Drosophila photoreceptors. PLoS Genet. 12, e1005828. 10.1371/journal.pgen.1005828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C. L. (2018). Activators and effectors of the small G protein Arf1 in regulation of Golgi dynamics during the cell division cycle. Front. Cell Dev. Biol. 6, 29. 10.3389/fcell.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S., Rhee, S. W., Gleeson, P. A. and Storrie, B. (2006). Capacity of the Golgi apparatus for cargo transport prior to complete assembly. Mol. Biol. Cell 17, 4105-4117. 10.1091/mbc.e05-12-1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.-H., Nielsen, E., Preuss, M. L., Mastronarde, D. and Staehelin, L. A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12, 313-329. 10.1111/j.1600-0854.2010.01146.x [DOI] [PubMed] [Google Scholar]
- Kienzle, C. and von Blume, J. (2014). Secretory cargo sorting at the trans-Golgi network. Trends Cell Biol. 24, 584-593. 10.1016/j.tcb.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Kitazawa, D., Yamaguchi, M., Mori, H. and Inoue, Y. H. (2012). COPI-mediated membrane trafficking is required for cytokinesis in Drosophila male meiotic divisions. J. Cell Sci. 125, 3649-3660. 10.1242/jcs.103317 [DOI] [PubMed] [Google Scholar]
- Klumperman, J. (2011). Architecture of the mammalian Golgi. Cold Spring Harb. Perspect. Biol. 3, a005181. 10.1101/cshperspect.a005181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis, V. and Rabouille, C. (2003). A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell Biol. 162, 185-198. 10.1083/jcb.200301136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis, V. and Rabouille, C. (2009). The Golgi apparatus: lessons from Drosophila. FEBS Lett. 583, 3827-3838. 10.1016/j.febslet.2009.09.048 [DOI] [PubMed] [Google Scholar]
- Kondylis, V., van Nispen tot Pannerden, H. E., Herpers, B., Friggi-Grelin, F. and Rabouille, C. (2007). The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev. Cell 12, 901-915. 10.1016/j.devcel.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Kurokawa, K., Ishii, M., Suda, Y., Ichihara, A. and Nakano, A. (2013). Live cell visualization of Golgi membrane dynamics by super-resolution confocal live imaging microscopy. Methods Cell Biol. 118, 235-242. 10.1016/B978-0-12-417164-0.00014-8 [DOI] [PubMed] [Google Scholar]
- Kurokawa, K., Osakada, H., Kojidani, T., Waga, M., Suda, Y., Asakawa, H., Haraguchi, T. and Nakano, A. (2019). Visualization of secretory cargo transport within the Golgi apparatus. J. Cell Biol. 218, 1602-1618. 10.1083/jcb.201807194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffafian, A. and Tepass, U. (2019). Identification of genes required for apical protein trafficking in Drosophila photoreceptor cells. G3 (Bethesda) 9, 4007-4017. 10.1534/g3.119.400635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans, M., Förster, S., Helmchen, G. and Robinson, D. G. (2011). Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. J. Exp. Bot. 62, 2949-2957. 10.1093/jxb/err007 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J. and Liu, W. (2006). Insights into COPI coat assembly and function in living cells. Trends Cell Biol. 16, e1-e4. 10.1016/j.tcb.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S. and Klausner, R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801-813. 10.1016/0092-8674(89)90685-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L., Tipper, C., Amherdt, M., Orci, L. and Klausner, R. D. (1991). Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67, 601-616. 10.1016/0092-8674(91)90534-6 [DOI] [PubMed] [Google Scholar]
- Luini, A. and Parashuraman, S. (2016). Signaling at the Golgi: sensing and controlling the membrane fluxes. Curr. Opin. Cell Biol. 39, 37-42. 10.1016/j.ceb.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Makaraci, P. and Kim, K. (2018). trans-Golgi network-bound cargo traffic. Eur. J. Cell Biol. 97, 137-149. 10.1016/j.ejcb.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Martell, J. D., Deerinck, T. J., Lam, S. S., Ellisman, M. H. and Ting, A. Y. (2017). Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat. Protoc. 12, 1792-1816. 10.1038/nprot.2017.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, S., Presley, J. F. and Maxfield, F. R. (1993). Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J. Cell Biol. 121, 1257-1269. 10.1083/jcb.121.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonold, C. M. and Fromme, J. C. (2014). Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-golgi network. Dev. Cell 30, 759-767. 10.1016/j.devcel.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaki, R., Morimatsu, M., Uemura, T., Waguri, S., Miyoshi, E., Taniguchi, N., Matsuda, M. and Taguchi, T. (2010). Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J. Cell Biol. 191, 23-29. 10.1083/jcb.200911143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgate, M., Southon, A., Greenough, M., Cater, M., Farlow, A., Batterham, P., Bush, A. I., Subramaniam, V. N., Burke, R. and Camakaris, J. (2010). Syntaxin 5 is required for copper homeostasis in Drosophila and mammals. PLoS ONE 5, e14303. 10.1371/journal.pone.0014303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci, L., Tagaya, M., Amherdt, M., Perrelet, A., Donaldson, J. G., Lippincott-Schwartz, J., Klausner, R. D. and Rothman, J. E. (1991). Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell 64, 1183-1195. 10.1016/0092-8674(91)90273-2 [DOI] [PubMed] [Google Scholar]
- Otsuka, Y., Satoh, T., Nakayama, N., Inaba, R., Yamashita, H. and Satoh, A. K. (2019). Parcas is the predominant Rab11-GEF for rhodopsin transport in Drosophila photoreceptors. J. Cell Sci. 132, jcs231431. 10.1242/jcs.231431 [DOI] [PubMed] [Google Scholar]
- Papanikou, E. and Glick, B. S. (2014). Golgi compartmentation and identity. Curr. Opin. Cell Biol. 29, 74-81. 10.1016/j.ceb.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J. and Jackson, C. L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285. 10.1016/S1097-2765(00)80455-4 [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S. and Jackson, C. L. (1996). Nucleotide exchange on ARF mediated by yeast Geal protein. Nature 384, 479-481. 10.1038/384479a0 [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Courbeyrette, R., Rambourg, A. and Jackson, C. L. (2001). The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 114, 2241-2253. [DOI] [PubMed] [Google Scholar]
- Renault, L., Guibert, B. and Cherfils, J. (2003). Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426, 525-530. 10.1038/nature02197 [DOI] [PubMed] [Google Scholar]
- Riedel, F., Gillingham, A. K., Rosa-Ferreira, C., Galindo, A. and Munro, S. (2016). An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol Open 5, 987-992. 10.1242/bio.018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D. G., Langhans, M., Saint-Jore-Dupas, C. and Hawes, C. (2008). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13, 405-408. 10.1016/j.tplants.2008.05.010 [DOI] [PubMed] [Google Scholar]
- Rogers, G. C., Rusan, N. M., Peifer, M. and Rogers, S. L. (2008). A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell 19, 3163-3178. 10.1091/mbc.e07-10-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, E. E., Hardie, R. C. and Colley, N. J. (2006). Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron 49, 229-241. 10.1016/j.neuron.2005.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan, N. M. and Rogers, G. C. (2009). Centrosome function: sometimes less is more. Traffic 10, 472-481. 10.1111/j.1600-0854.2009.00880.x [DOI] [PubMed] [Google Scholar]
- Saraste, J. and Prydz, K. (2019). A new look at the functional organization of the Golgi ribbon. Front. Cell Dev. Biol. 7, 171. 10.3389/fcell.2019.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata, M., Donaldson, J. G., Moss, J. and Vaughan, M. (1998). Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc. Natl. Acad. Sci. USA 95, 4204-4208. 10.1073/pnas.95.8.4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, A. K. and Ready, D. F. (2005). Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Curr. Biol. 15, 1722-1733. 10.1016/j.cub.2005.08.064 [DOI] [PubMed] [Google Scholar]
- Satoh, A., Tokunaga, F., Kawamura, S. and Ozaki, K. (1997). In situ inhibition of vesicle transport and protein processing in the dominant negative Rab1 mutant of Drosophila. J. Cell Sci. 110, 2943-2953. [DOI] [PubMed] [Google Scholar]
- Satoh, A. K., O'Tousa, J. E., Ozaki, K. and Ready, D. F. (2005). Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132, 1487-1497. 10.1242/dev.01704 [DOI] [PubMed] [Google Scholar]
- Satoh, T., Ohba, A., Liu, Z., Inagaki, T. and Satoh, A. K. (2015). dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. eLife 4, e06306. 10.7554/eLife.06306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaky, N., Presley, J., Smith, C., Zaal, K. J. M., Cole, N., Moreira, J. E., Terasaki, M., Siggia, E. and Lippincott-Schwartz, J. (1997). Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139, 1137-1155. 10.1083/jcb.139.5.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H.-W. and Nakayama, K. (2004). Guanine nucleotide-exchange factors for arf GTPases: their diverse functions in membrane traffic. J. Biochem. 136, 761-767. 10.1093/jb/mvh185 [DOI] [PubMed] [Google Scholar]
- Sisson, J. C., Field, C., Ventura, R., Royou, A. and Sullivan, W. (2000). Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 151, 905-918. 10.1083/jcb.151.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh, O.-K. and Moore, I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493-496. 10.1038/nature06023 [DOI] [PubMed] [Google Scholar]
- Uemura, T. and Nakano, A. (2013). Plant TGNs: dynamics and physiological functions. Histochem. Cell Biol. 140, 341-345. 10.1007/s00418-013-1116-7 [DOI] [PubMed] [Google Scholar]
- Uemura, T., Suda, Y., Ueda, T. and Nakano, A. (2014). Dynamic behavior of the trans-golgi network in root tissues of Arabidopsis revealed by super-resolution live imaging. Plant Cell Physiol. 55, 694-703. 10.1093/pcp/pcu010 [DOI] [PubMed] [Google Scholar]
- Uemura, T., Nakano, R. T., Takagi, J., Wang, Y., Kramer, K., Finkemeier, I., Nakagami, H., Tsuda, K., Ueda, T., Schulze-Lefert, P., et al. (2019). A golgi-released subpopulation of the trans-golgi network mediates protein secretion in arabidopsis. Plant Physiol. 179, 519-532. 10.1104/pp.18.01228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije, M. H., Raaben, M., Mari, M., Te Lintelo, E. G., Reggiori, F., van Kuppeveld, F. J. M., Rottier, P. J. M. and de Haan, C. A. M. (2008). Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4, e1000088. 10.1371/journal.ppat.1000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud, J., Zeghouf, M., Barelli, H., Zeeh, J.-C., Padilla, A., Guibert, B., Chardin, P., Royer, C. A., Cherfils, J. and Chavanieu, A. (2007). Structure-based discovery of an inhibitor of Arf activation by Sec7 domains through targeting of protein-protein complexes. Proc. Natl. Acad. Sci. USA 104, 10370-10375. 10.1073/pnas.0700773104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti, C., Bubeck, J., Stierhof, Y.-D., Krebs, M., Langhans, M., van den Berg, W., van Dongen, W., Richter, S., Geldner, N., Takano, J., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22, 1344-1357. 10.1105/tpc.109.072637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta, J. H., Didier, A. J., Liu, X. and Krämer, H. (2001). The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152, 923-934. 10.1083/jcb.152.5.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Meyer, H., Ochoa-Espinosa, A., Buchwald, U., Onel, S., Altenhein, B., Heinisch, J. J., Affolter, M. and Paululat, A. (2012). GBF1 (Gartenzwerg)-dependent secretion is required for Drosophila tubulogenesis. J. Cell Sci. 125, 461-472. 10.1242/jcs.092551 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Zhang, H., Shi, M., Liou, Y.-C., Lu, L. and Yu, F. (2017). Sec71 functions as a GEF for the small GTPase Arf1 to govern dendrite pruning of Drosophila sensory neurons. Development 144, 1851-1862. 10.1242/dev.146175 [DOI] [PubMed] [Google Scholar]
- Wei, J.-H. and Seemann, J. (2017). Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr. Opin. Cell Biol. 47, 43-51. 10.1016/j.ceb.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, S. A., Park, J. E. and Brown, W. J. (1991). Brefeldin A causes a microtubule-mediated fusion of the trans-Golgi network and early endosomes. Cell 67, 591-600. 10.1016/0092-8674(91)90533-5 [DOI] [PubMed] [Google Scholar]
- Wright, J., Kahn, R. A. and Sztul, E. (2014). Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell. Mol. Life Sci. 71, 3419-3438. 10.1007/s00018-014-1602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Boulianne, G. L. and Trimble, W. S. (2002). Drosophila syntaxin 16 is a Q-SNARE implicated in Golgi dynamics. J. Cell Sci. 115, 4447-4455. 10.1242/jcs.00139 [DOI] [PubMed] [Google Scholar]
- Yadav, S. and Linstedt, A. D. (2011). Golgi positioning. Cold Spring Harb. Perspect. Biol. 3, a005322. 10.1101/cshperspect.a005322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Hino, M., Abe, M., Shibano, T., Setoguchi, Y., Awano, W., Ueda, R., Okano, H. and Goto, S. (2012). Cisterna-specific localization of glycosylation-related proteins to the Golgi apparatus. Cell Struct. Funct. 37, 55-63. 10.1247/csf.11037 [DOI] [PubMed] [Google Scholar]
- Yamashiro, D. J. and Maxfield, F. R. (1987). Acidification of morphologically distinct endosomes in mutant and wild-type Chinese hamster ovary cells. J. Cell Biol. 105, 2723-2733. 10.1083/jcb.105.6.2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara, H. and Shibaoka, H. (2000). Inhibition of cell-plate formation by brefeldin A inhibited the depolymerization of microtubules in the central region of the phragmoplast. Plant Cell Physiol. 41, 300-310. 10.1093/pcp/41.3.300 [DOI] [PubMed] [Google Scholar]
- Yasuhara, H., Sonobe, S. and Shibaoka, H. (1995). Effects of brefeldin A on the formation of the cell plate in tobacco BY-2 cells. Eur. J. Cell Biol. 66, 274-281. [PubMed] [Google Scholar]
- Zeeh, J.-C., Zeghouf, M., Grauffel, C., Guibert, B., Martin, E., Dejaegere, A. and Cherfils, J. (2006). Dual specificity of the interfacial inhibitor brefeldin a for arf proteins and sec7 domains. J. Biol. Chem. 281, 11805-11814. 10.1074/jbc.M600149200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.