Abstract
Background
Despite remarkable benefits have been provided by immune checkpoint inhibitors in gastric cancer (GC), predictions of treatment response and prognosis remain unsatisfactory, making identifying biomarkers desirable. The aim of this study was to develop and validate a CT imaging biomarker to predict the immunotherapy response in patients with GC and investigate the associated immune infiltration patterns.
Methods
This retrospective study included 294 GC patients who received anti-PD-1/PD-L1 immunotherapy from three independent medical centers between January 2017 and April 2022. A radiomics score (RS) was developed from the intratumoral and peritumoral features on pretreatment CT images to predict immunotherapy-related progression-free survival (irPFS). The performance of the RS was evaluated by the area under the time-dependent receiver operating characteristic curve (AUC). Multivariable Cox regression analysis was performed to construct predictive nomogram of irPFS. The C-index was used to determine the performance of the nomogram. Bulk RNA sequencing of tumors from 42 patients in The Cancer Genome Atlas was used to investigate the RS-associated immune infiltration patterns.
Results
Overall, 89 of 294 patients (median age, 57 years (IQR 48–66 years); 171 males) had an objective response to immunotherapy. The RS included 13 CT features that yielded AUCs of 12-month irPFS of 0.787, 0.810 and 0.785 in the training, internal validation, and external validation 1 cohorts, respectively, and an AUC of 24-month irPFS of 0.805 in the external validation 2 cohort. Patients with low RS had longer irPFS in each cohort (p<0.05). Multivariable Cox regression analyses showed RS is an independent prognostic factor of irPFS. The nomogram that integrated the RS and clinical characteristics showed improved performance in predicting irPFS, with C-index of 0.687–0.778 in the training and validation cohorts. The CT imaging biomarker was associated with M1 macrophage infiltration.
Conclusion
The findings of this prognostic study suggest that the non-invasive CT imaging biomarker can effectively predict immunotherapy outcomes in patients with GC and is associated with innate immune signaling, which can serve as a potential tool for individual treatment decisions.
Keywords: Immunotherapy; Biomarkers, Tumor
WHAT IS ALREADY KNOWN ON THIS TOPIC
Current immunotherapy-related biomarkers such as microsatellite instability (MSI), Epstein-Barr virus (EBV), combined positive score (CPS) of PD-L1 expression, and tumor mutation burden, are limited for predictions of personalized treatment response and prognosis in gastric cancer (GC). Radiomics has shown promising results in predicting the response to immunotherapy. However, there is no radiomics biomarker with underlying biological explanations to predict immunotherapy response in GC.
WHAT THIS STUDY ADDS
To the best of our knowledge, this is the first multicenter study using intratumoral and peritumoral features on pretreatment CT images with biological explanations for the prediction of immunotherapy response in GC. The non-invasive imaging biomarker constructed in this study outperformed currently used biomarkers such as MSI status, EBV, and CPS in predicting immunotherapy response, and is associated with innate immune signaling.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study showed that the non-invasive CT imaging biomarker has a better predictive value in the prediction of immunotherapy response and prognosis in GC than the current TNM stage and molecular biomarkers that associated with immunotherapy. Moreover, radiogenomics analysis showed that innate immune signaling is associated with the imaging biomarker. This non-invasive method can serve as a supplement to current immunotherapy treatment decisions with considerable socioeconomic effects since CT examination is a routine diagnostic approach in the clinical practice of GC.
Introduction
Gastric cancer (GC) is a highly prevalent malignancy, ranking as the fourth most common cancer-related disease worldwide.1 Despite advances in surgical techniques, chemotherapy, and targeted therapy, the prognosis for patients with advanced GC remains poor.2 Recently, the advent of immune checkpoint inhibitors (ICIs) has revolutionized the treatment of GC, with ICIs now approved as the first-line regimen for GC.3 However, while the success of ICIs in treating GC is remarkable, response rates to immunotherapy remain variable and limited.4 5 Therefore, there is a pressing need to identify biomarkers that can predict treatment response and guide the development of personalized immunotherapy strategies for GC patients.
Presently, microsatellite instability (MSI), Epstein-Barr virus (EBV), combined positive score (CPS) of PD-L1 expression, and tumor mutation burden (TMB) are used as biomarkers to identify GC patients who are likely to benefit from anti-PD-L1/PD-1 immunotherapy.6 7 However, studies have demonstrated that these biomarkers are less predictive of personalized treatment response and immunotherapy related prognosis, and have even resulted in controversial outcomes.8–10 Additionally, unselected GC patients often experience treatment failures and immune-related adverse events, which can cause severe and potentially life-threatening complications.11 12 Therefore, identifying novel and robust biomarkers is crucial for predicting immunotherapy response and developing personalized immunotherapy strategies for GC patients.
Radiomics is a promising non-invasive approach that extracts quantitative features from medical images to investigate underlying information about tumor biology.13 To date, radiomics has been regarded as a digital biopsy to evaluate the tumor microenvironment because the extracted features provide information on tumor heterogeneity and deeper characteristics of the molecular and genetic differences in the tumor microenvironment.14 15 Since the association between radiomics and immune microenvironment has been first evaluated in 2017, an increasing number of studies have confirmed the predictive power of imaging biomarkers in many cancer types, particularly in predicting immunotherapy response.16–18 Therefore, imaging biomarkers may outperform current biomarkers such as MSI/EBV/CPS/TMB in the prediction of immunotherapy response in GC. Despite some studies investigating the relationship between radiomics and immunotherapy response in GC, challenges remain, including limited sample size and unclear biological explanations.19 20
In this study, we aimed to develop and validate a non-invasive imaging biomarker for predicting immunotherapy response in GC using intratumoral and peritumoral features on CT images of 294 patients from three independent medical centers. Additionally, we investigated the underlying biological explanations of this biomarker using bulk RNA sequencing of tumors from 42 patients in The Cancer Genome Atlas (TCGA).
Materials and methods
Study patients
The study design is shown in figure 1. This retrospective study included four cohorts from three academic medical centers in China. The major inclusion and exclusion criteria are listed in online supplemental methods. A total of 294 patients with GC were enrolled in this study (online supplemental figure S1). The training cohort (n=150) and internal validation cohort (n=50) were collected from Nanfang Hospital of Southern Medical University (Guangzhou, China) between January 2018 and December 2020, and between January 2021 and April 2022, respectively. External validation cohort 1 (n=64) was collected from Guangdong Provincial Hospital of Chinese Medicine (Guangzhou, China) between January 2019 and April 2022. External validation cohort 2 was collected from Peking University Cancer Hospital (Beijing, China) between January 2017 and December 2020. Clinicopathological data including age, gender, tumor differentiation, size, tumor location, and TNM stage (eighth AJCC edition) were collected. HER2 status was determined using IHC analysis and amplification assay by fluorescence in situ hybridization. The mismatch repair (MMR) status was determined using IHC analysis of the expression of MLH1, MSH2, MSH6, and PMS2. EBV status was determined using in situ hybridization with probes against Epstein-Barr encoded RNA 1 (EBER1). A CPS≥1 was regarded as positive. In addition, 42 patients with GC from TCGA databases with baseline CT and RNA sequencing data available were collected as a genomics group for analysis of immune infiltration.
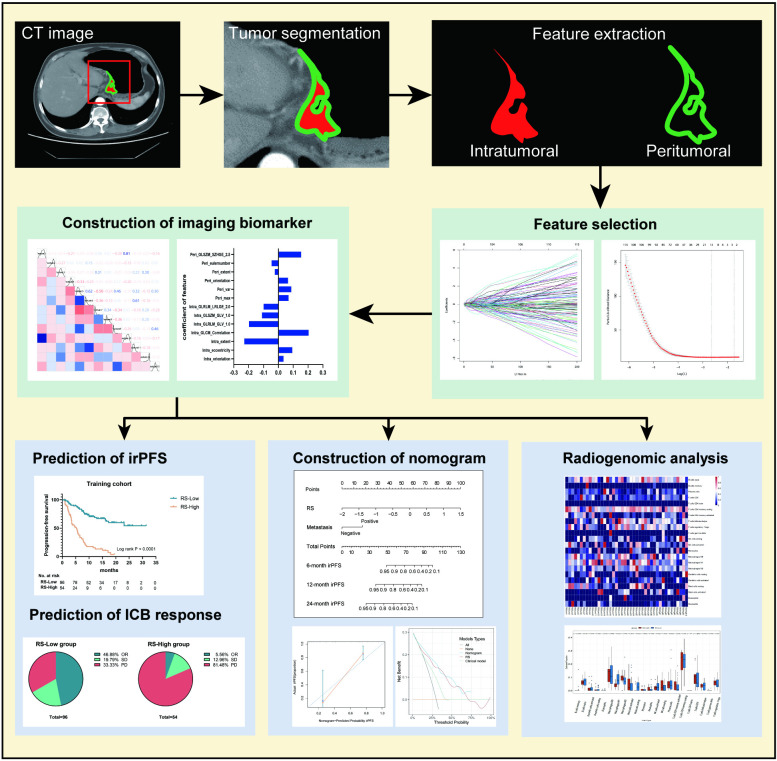
Figure 1.
Flow chart of the study steps. Tumor identification and segmentation were performed on the largest slice of the primary tumor CT image. Intratumoral and peritumoral textural features were extracted using MATLAB (R2017a). The LASSO Cox regression algorithm and Pearson correlation analysis were used to select the most predictive features of irPFS and construct the imaging biomarker. Prognostic performances of imaging biomarker were evaluated by predictions of immunotherapy response. A nomogram of irPFS was constructed by imaging biomarker and clinical characteristics to assess the incremental value in treatment decisions. Underlying immune infiltration of the imaging biomarker was investigated using radiogenomic analysis. ICB, immune checkpoint blockade; irPFS, immunotherapy-related disease-free survival; LASSO, least absolute shrinkage and selection operator; OR, objective response; PD, progressive disease; RS, radiomics score; SD, stable disease.
jitc-2023-007807supp001.pdf (1.1MB, pdf)
Immunotherapy responses were defined as complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD) according to irRECIST criteria.21 Objective response (OR) was defined for patients who achieved either CR or PR. Immunotherapy-related progression-free survival (irPFS) was defined as the time from anti-PD-1/PD-L1 immunotherapy initiation to tumor progression or death from any cause. Immunotherapy-related overall survival (irOS) was defined as the time from anti-PD-1/PD-L1 immunotherapy initiation to death from any cause.
Image acquisition, processing, and feature extraction
All patients underwent contrast-enhanced abdominal CT scans prior to immunotherapy. Portal venous phase images were retrieved from the picture archiving and communication system (Carestream Canada). Detailed acquisition is shown in online supplemental methods.
The tumor region on CT images was manually segmented by two radiologists (CC and YQ with 12 and 11 years of clinical experience in abdominal CT interpretation, respectively) using ITK-SNAP software (V.3.8.0). This study also extracted the information of the peritumor region by delineating a peripheral ring surrounding the primary tumor region, which automatically dilated the tumor boundaries by 2 mm on the outside and shrunk the tumor boundaries by 1 mm on the inside (a ring with a thickness of 3 mm). All CT images were processed following the Image Biomarker Standardization Initiative guidelines.22 Intraclass correlation coefficient (ICC) was calculated to evaluate intraobserver and interobserver reproducibility. In this study, a total of 584 quantitative features of each region of interest were extracted followed by z-score normalization. The detailed extraction process is summarized in online supplemental methods.
Development of radiomics imaging biomarker to predict immunotherapy response
In this study, we used the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm with 10-fold cross-validation and Pearson correlation analysis to identify the most predictive features of irPFS, which were then constructed into a radiomics score (RS) with their nonzero coefficients. The cut-off value of the RS was generated on the basis of the association with irPFS in the training cohort using X-tile software (V.3.6.1, Yale University). Patients in the training and validation cohorts were divided into RS-Low groups and RS-High groups according to the cut-off value.
Assessment of the predictive performance of imaging biomarker
Time-dependent receiver operating characteristic (ROC) curve analysis was applied to evaluate the predictive accuracies of RS and clinical variables. The area under the curves (AUCs) of 12-month and 24-month irPFS among the RS and clinical variables were used to compare the prediction performance. Kaplan-Meier analysis with the log-rank test was used to assess the irPFS and irOS of the defined RS groups to distinguish the prognostic value of the radiomics image biomarker.
Univariate Cox regression analysis was used to determine the prognostic values of the RS and clinicopathological variables. Variables that reached statistical significance (p<0.05) were then enrolled in a multivariate Cox regression. Moreover, variables that remained significant in the multivariate Cox analysis were integrated into nomogram construction for predicting irPFS and irOS to improve the predictive power of the radiomics image biomarker. Harrell’s concordance index (C-index) was used to quantify the relative improvement in the prediction accuracy of the prognostic nomogram. Calibration curves and decision curve analysis (DCA) were used to assess the performance and clinical utility.
Radiotranscriptomic analysis
In this study, 42 patients in the genomics group from TCGA were stratified into RS-low and RS-High groups according to the cut-off value of RS. CIBERSORT and Gene Set Enrichment Analysis (GSEA) analyses were employed to investigate the tumor infiltrating immune cells and underlying pathways between different RS groups.23 For this analysis, normalized mRNA expression values in the TPM (transcript per million) matrix were used as input for CIBERSORT. Immune cell types and normalized enrichment score (NES) with p values<0.05 were then used to annotate the imaging biomarker.
Statistical analysis
All statistical analyses were performed by using SPSS (V.22.0, IBM), GraphPad Prism (V.8), and R software (V.4.0.2). Categorical variables were analyzed using the χ2 test or Fisher’s exact test. Continuous variables were analyzed using a two-tailed t test (unpaired) or Mann-Whitney test. Survival curves were generated according to the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate analyses for survival were performed with the Cox proportional hazard model. Detailed algorithms used for statistical analysis are described in online supplemental materials. A two-sided p<0.05 was considered statistically significant.
Results
Clinicopathological characteristics
This study included 294 GC patients (171 male, 123 female) with a median age of 57 years (IQR, 48–66 years) from three independent medical centers (table 1 and online supplemental figure S1). The training cohort consisted of 150 patients, while the internal validation, external validation cohort 1, and cohort 2 consisted of 50, 64, and 30 patients, respectively (table 1). Most of the patients (n=272) were diagnosed at stages III and IV (table 1). The clinicopathological characteristics of the training and validation cohorts according to the RS groups are summarized in online supplemental table S1–4.
Table 1.
Characteristics of patients with GC in each cohort
| Variables | Training cohort, n=150 | Internal validation cohort, n=50 | External validation cohort 1, n=64 | External validation cohort 2, n=30 |
| n (%) | n (%) | n (%) | n (%) | |
| Median age (IQR) | 54 (46–64) | 62 (51–67) | 60 (48–66) | 65 (49–71) |
| Gender | ||||
| Male | 86 (57.3) | 29 (58.0) | 34 (53.1) | 22 (73.3) |
| Female | 64 (42.7) | 21 (42.0) | 30 (46.9) | 8 (26.7) |
| Differentiation | ||||
| Well or moderate | 19 (12.7) | 7 (14.0) | 7 (10.9) | 6 (20.0) |
| Poor or undifferentiated | 131 (87.3) | 43 (86.0) | 57 (89.1) | 24 (80.0) |
| Size | \ | |||
| ≥4 cm | 96 (64.0) | 30 (60.0) | 56 (87.5) | |
| <4 cm | 54 (36.0) | 20 (40.0) | 8 (12.5) | |
| Location | \ | |||
| Cardia | 36 (24.0) | 7 (14.0) | 8 (12.5) | |
| Body | 44 (29.3) | 17 (34.0) | 21 (32.8) | |
| Antrum | 62 (41.4) | 24 (48.0) | 28 (43.7) | |
| Whole | 8 (5.3) | 2 (4.0) | 3 (4.7) | |
| Remnant | 0 | 0 | 4 (6.3) | |
| TNM stage | ||||
| I | 1 (0.7) | 1 (2.0) | 0 | 1 (3.3) |
| II | 14 (9.3) | 4 (8.0) | 0 | 1 (3.3) |
| III | 43 (28.7) | 10 (20.0) | 14 (21.9) | 10 (33.4)) |
| IV | 92 (61.3) | 35 (70.0) | 50 (78.1) | 18 (60.0) |
| HER-2 status | ||||
| Positive | 22 (14.7) | 8 (16.0) | 7 (10.9) | 1 (3.3) |
| Negative | 119 (79.3) | 38 (76.0) | 34 (53.1) | 25 (83.4) |
| Unknown | 9 (6.0) | 4 (8.0) | 23 (36.0) | 4 (13.3) |
| MSI status | ||||
| MSI-H/MMR deficient | 7 (4.7) | 5 (10.0) | 5 (7.8) | 8 (26.7) |
| MSS/MMR proficient | 128 (85.3) | 41 (82.0) | 32 (50.0) | 18 (60.0) |
| Unknown | 15 (10.0) | 4 (8.0) | 27 (42.2) | 4 (13.3) |
| CPS status | ||||
| ≥1 | 120 (80.0) | 40 (80.0) | 11 (17.2) | 13 (43.3) |
| <1 | 25 (16.7) | 5 (10.0) | 13 (20.3) | 8 (26.7) |
| Unknown | 5 (3.3) | 5 (10.0) | 40 (62.5) | 9 (30.0) |
| EBV status | \ | |||
| Positive | 8 (5.3) | 2 (4.0) | 3 (10.0) | |
| Negative | 96 (64.0) | 30 (60.0) | 25 (83.3) | |
| Unknown | 46 (30.7) | 18 (36.0) | 2 (6.7) | |
| Immunotherapy response | ||||
| CR | 19 (12.7) | 7 (14.0) | 1 (1.6) | 0 |
| PR | 29 (19.3) | 14 (28.0) | 11 (17.2) | 8 (26.7) |
| SD | 26 (17.3) | 6 (12.0) | 25 (39.0) | 7 (23.3) |
| PD | 76 (50.7) | 23 (46.0) | 27 (42.2) | 15 (50.0) |
| Drugs | ||||
| Anti-PD 1 | 150 (100) | 50 (100) | 64 (100) | 22 (73.3) |
| Anti-PD-L1 | 0 (0) | 0 (0) | 0 (0) | 8 (26.7) |
| Treatment | ||||
| Anti-PD 1(L1) only | 5 (3.3) | 3 (6.0) | 1 (1.6) | 13 (43.3) |
| PD-1(L1) and chemo | 145 (96.7) | 47 (94.0) | 63 (98.4) | 17 (56.7) |
| Treatment line | ||||
| 1 | 88 (58.7) | 32 (64.0) | 34 (53.1) | 19 (63.3) |
| 2 | 34 (22.7) | 10 (20.0) | 24 (37.5) | 7 (23.4) |
| 3 or higher | 28 (18.6) | 8 (16.0) | 6 (9.4) | 4 (13.3) |
| Median irPFS (range, month) | 7.87 (0.70–31.37) | 8.78 (1.20–25.47) | 4.45 (0.53–16.2) | 11.28 (2.20–41.93) |
| Median irOS (range, month) | 12.72 (0.87–32.63) | 11.60 (1.43–25.47) | 5.23 (0.53–16.2) | 30.40 (8.10–76.63) |
CPS, combined positive score; CR, complete response; EBV, Epstein-Barr virus; GC, gastric cancer; HER2, human epidermal growth factor receptor 2; irOS, immunotherapy-related overall survival; irPFS, immunotherapy-related progression-free survival; MMR, mismatch repair; MSI, microsatellite instability; MSS, microsatelite instability-stable; PD, progressive disease; PR, partial response; SD, stable disease.
The OR rates of the training cohort, internal validation cohort, and external validation cohorts 1 and 2 were 32% (n=48), 42% (n=21), 18.8% (n=12), and 26.7% (n=8), respectively. The median irPFS of patients in the training cohort, internal validation cohort, and external validation cohorts 1 and 2 were 7.87 (range 0.70–31.37) months, 8.78 (1.20–25.47) months, 4.45 (0.53–16.2) months and 11.28 (2.20–41.93) months, respectively. Moreover, the median irOS of patients was 12.72 (range 0.87–32.63) months, 11.60 (1.43–25.47) months, 5.23 (0.53–16.2) months, and 30.40 (8.10–76.63) months in the training cohort, internal validation cohort, and external validation cohorts 1 and 2, respectively.
Development of radiomics imaging biomarker to predict immunotherapy response
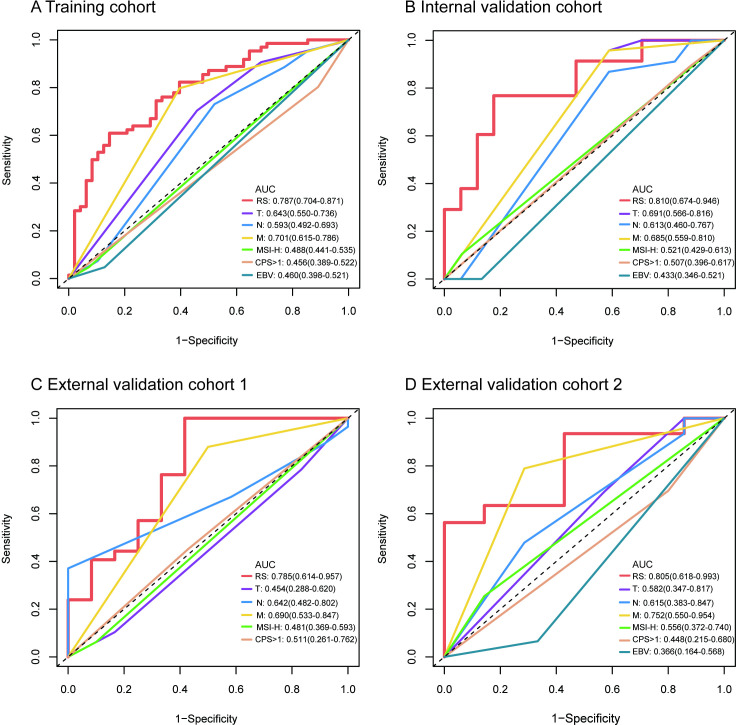
In this study, the intraobserver and interobserver ICCs were greater than 0.75, indicating high reproducibility of the radiomic features. Therefore, all outcomes were based on the measurements of the first radiologist. A total of 584 quantitative features (292 in the intratumoral area and 292 in the peritumoral area) were extracted and subjected to LASSO Cox regression analysis. Ultimately, seven intratumoral and six peritumoral features with non-zero coefficients were selected using the minimum criteria with a log (λ) value of −2.655414 for construction of the RS, which was calculated using a specific formula described in online supplemental figure S2. The contributions and correlations of each selected feature to irPFS are shown in online supplemental figure S3,S4. The RS was able to accurately predict immunotherapy response, as demonstrated by its strong association with irPFS. The predictive accuracies of RS and clinicopathological characteristics were assessed by time-dependent ROC analysis. The RS showed the highest AUCs of 12-month irPFS of 0.787 (95% CI 0.704 to 0.871), 0.810 (95% CI 0.674 to 0.948), and 0.785 (95% CI 0.614 to 0.957) in the training cohort, internal validation cohort, and external validation cohort 1, respectively (figure 2A–C). In external validation cohort 2, the RS still showed the highest AUC of 24-month irPFS of 0.805 (95% CI 0.618 to 0.993, figure 2D).
Figure 2.
Time-dependent ROC curves of the RS and clinical characteristics in the training, internal and external validation cohorts. (A) AUCs of 12-month irPFS of the RS and clinical characteristics in the training cohort. (B) AUCs of 12-month irPFS of the RS and clinical characteristics in the internal validation cohort. (C), AUCs of 12-month irPFS of the RS and clinical characteristics in external validation cohort 1. (D) AUCs of 24-month irPFS of the RS and clinical characteristics in external validation cohort 2. CPS: combined positive score of PD-L1 expression. AUC, area under the curve; CPS, Combined Positive Score; EBV, Epstein-Barr virus; irPFS, immunotherapy-related progression-free survival; M, distant metastasis; MSI-H, microsatellite instability-high; N, lymph node metastasis; ROC, receiver operating characteristic; RS, radiomics score; T, depth of tumor invasion.
Patients were divided into RS-High and RS-Low groups based on an optimum cut-off value of 0.22 generated by X-tile (online supplemental figure S5). The PD rates of the RS-High groups were significantly higher than those of the RS-Low groups in the training, internal, and external two cohorts (p<0.001 in the training cohort, p=0.040 in the internal validation cohort, and p=0.035 in external validation cohort 2, respectively). The RS was also highly predictive of patient survival, with the RS-Low groups exhibiting significantly longer median irPFS than the RS-High groups in all cohorts. Specifically, the median irPFS of the RS-Low groups were 10.57 (range 0.70–31.37) months, 10.97 (1.20–23.80) months, 4.77 (1.00–16.20) months, and 22.40 (3.07–41.93) months in the training cohort, internal validation cohort, external validation cohort 1, and external validation cohort 2, respectively, which were all longer than those of the RS-High groups (3.64 (0.87–19.47)), 8.07 (1.73–25.47), 3.73 (0.53–14.03), and 6.20 (2.20–22.27), respectively, online supplemental table S1-4).
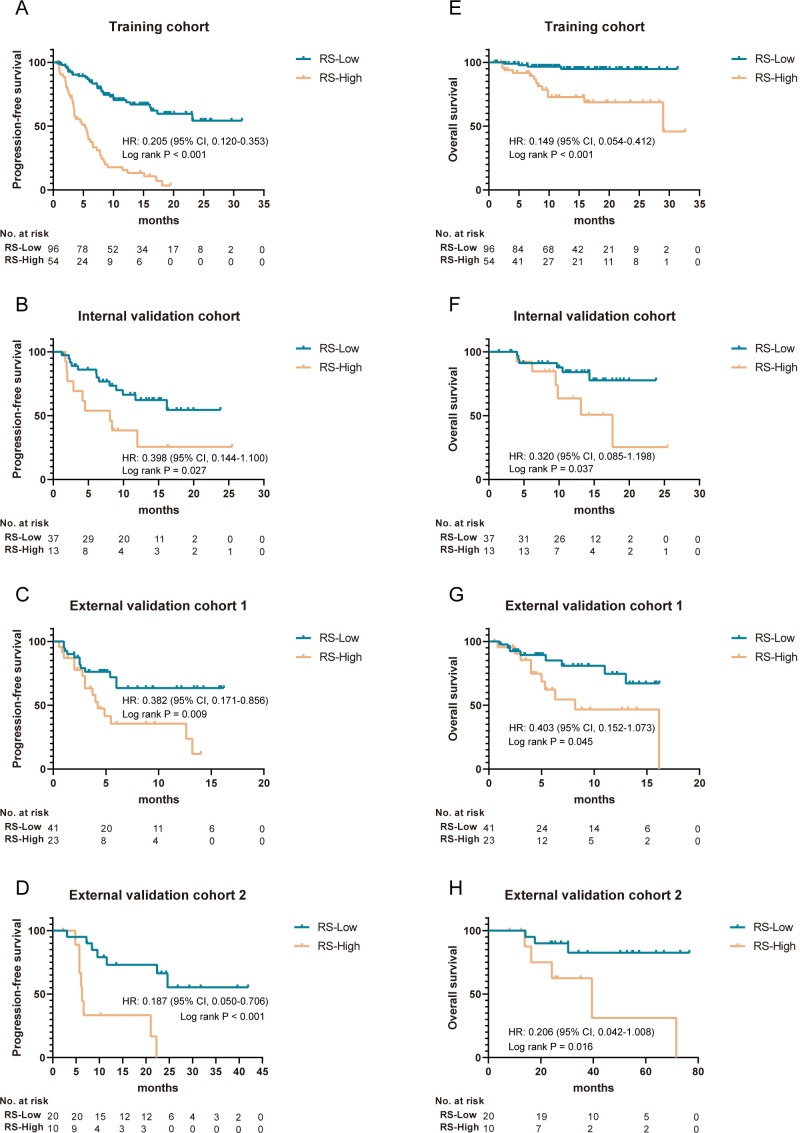
Prognostic value of the imaging biomarker for immunotherapy
We used Kaplan-Meier analysis with the log-rank test to distinguish the prognostic value of the radiomics image biomarker. The results showed that patients in the RS-Low group had a better 12-month irPFS rate and 12-month irOS rate than those in the RS-High group in the training cohort (68.7% and 94.8%, respectively, for the RS-Low group, and 13.3% and 72.7%, respectively, for the RS-High group, HR 0.205 (95% CI 0.120 to 0.353) and 0.149 (95% CI 0.054 to 0.412), p<0.001, figure 3A,E). This analysis was also performed in the internal validation cohort and confirmed that patients in the RS-Low group still had better irPFS and irOS (HR 0.398 (95% CI 0.144 to 1.100) and 0.320 (95% CI 0.085 to 1.198), respectively, p=0.027 and p=0.037, respectively, figure 3B,F).
Figure 3.
Kaplan-Meier survival analysis of irPFS and irOS according to the RS in the training, internal, and external validation cohorts. (A–D) Kaplan-Meier survival curves of irPFS of different RS groups in the training, internal validation, external validation 1, and external validation 2 cohorts. (E–H) Kaplan-Meier survival curves of irOS of different RS groups in the training, internal validation, external validation 1, and external validation 2 cohorts. P values were calculated using the log-rank test. irOS, immunotherapy-related overall survival; irPFS, immunotherapy-related disease-free survival; RS, radiomics score.
Furthermore, the imaging biomarker was evaluated in two external validation cohorts, and similar results were obtained, indicating the prognostic value of the imaging biomarker in different populations (HR 0.382 (95% CI 0.171 to 0.856), p=0.009, for irPFS, and 0.403 (95% CI 0.152 to 1.073), p=0.045, for irOS in external validation cohort 1, and 0.187 (95% CI 0.050 to 0.706), p<0.001, for irPFS, and 0.206 (95% CI 0.042 to 1.008), p=0.016, for irOS in external cohort 2, respectively, figure 3C, D, G and H). Moreover, subgroup survival analyses defined by clinicopathological risk factors revealed significant differences in irPFS and irOS between patients in the RS-Low and RS-High groups (online supplemental figure S6,S7).
Univariate Cox regression analysis revealed that the radiomics imaging biomarker was a prognostic factor for irPFS (HR ranges 7.235–12.680, all p<0.01) and irOS (HR ranges 6.666–11.448, all p<0.05) in each cohort (online supplemental table S5-S8). Multivariate Cox regression analysis also showed that the radiomics imaging biomarker remained an independent prognostic factor for irPFS (HR ranges 6.993–9.108, all p<0.05) and irOS (HR ranges 6.666–11.448, all p<0.05) in each cohort (table 2).
Table 2.
Multivariate Cox regression analyses for immunotherapy related disease-free survival and overall survival in patients with gastric cancer in the training and validation cohorts
| Variables | irPFS | irOS | |||
| HR (95%CI) | P value | HR (95%CI) | P value | ||
| Training cohort | |||||
| RS | 9.108 (4.891 to 16.961) | <0.001 | 9.122 (2.559 to 32.516) | 0.001 | |
| Gender (male vs female) | 0.641 (0.400 to 1.028) | 0.065 | \ | \ | |
| Depth of invasion | 1.092 (0.779 to 1.531) | 0.609 | 2.584 (0.942 to 7.089) | 0.065 | |
| Distant metastasis | 3.111 (1.690 to 5.727) | <0.001 | \ | \ | |
| CPS status (≥1 vs <1) | 0.586 (0.329 to 1.044) | 0.070 | \ | \ | |
| Internal validation cohort | |||||
| RS | 6.993 (1.565 to 31.245) | 0.011 | 6.666 (1.447 to 30.712) | 0.015 | |
| Depth of invasion | 2.263 (0.537 to 9.541) | 0.266 | \ | \ | |
| Distant metastasis | 4.278 (0.477 to 38.378) | 0.194 | \ | \ | |
| External validation cohort 1 | |||||
| RS | 8.740 (2.558 to 29.856) | 0.001 | 11.448 (2.622 to 449.978) | 0.001 | |
| Age (years) (≥60 vs <60) | 0.769 (0.335 to 1.765) | 0.535 | \ | \ | |
| External validation cohort 2 | |||||
| RS | 7.572 (1.697 to 33.787) | 0.008 | 10.096 (1.566 to 65.110) | 0.015 | |
P values reported are two-tailed from Cox proportional hazard regression analyses.
CPS, Combined Positive Score; irOS, immunotherapy-related overall survival; irPFS, immunotherapy-related progression-free survival; RS, Radiomics Score.
Importantly, the classification of patients into different RS groups revealed significantly higher OR rates (46.88%, 51.35%, 19.51%, and 35.00% in the training cohort, internal validation cohort, external validation cohorts 1 and 2, respectively) in the RS-Low groups than in the RS-High groups (5.56%, 15.38%, 17.39%, and 10.00% respectively, figure 4A–H). The patients in the RS-High groups had higher PD rates than those in the RS-Low groups in the training and validation cohorts (figure 4A–H). These findings suggest that the radiomics imaging biomarker has a high prognostic value for immunotherapy and can be used as an effective tool for patient stratification and treatment planning.
Figure 4.
Proportion of different immunotherapy responses according to RS groups in the training, internal, and external validation cohorts. (A–D) Proportion of different immunotherapy responses of RS-Low groups in the training, internal validation, external validation 1, and external validation 2 cohorts. (E–H) Proportion of different immunotherapy responses of RS-High groups in the training, internal validation, external validation 1, and external validation 2 cohorts. OR, objective response; PD, progressive disease; RS, radiomics score; SD, stable disease.
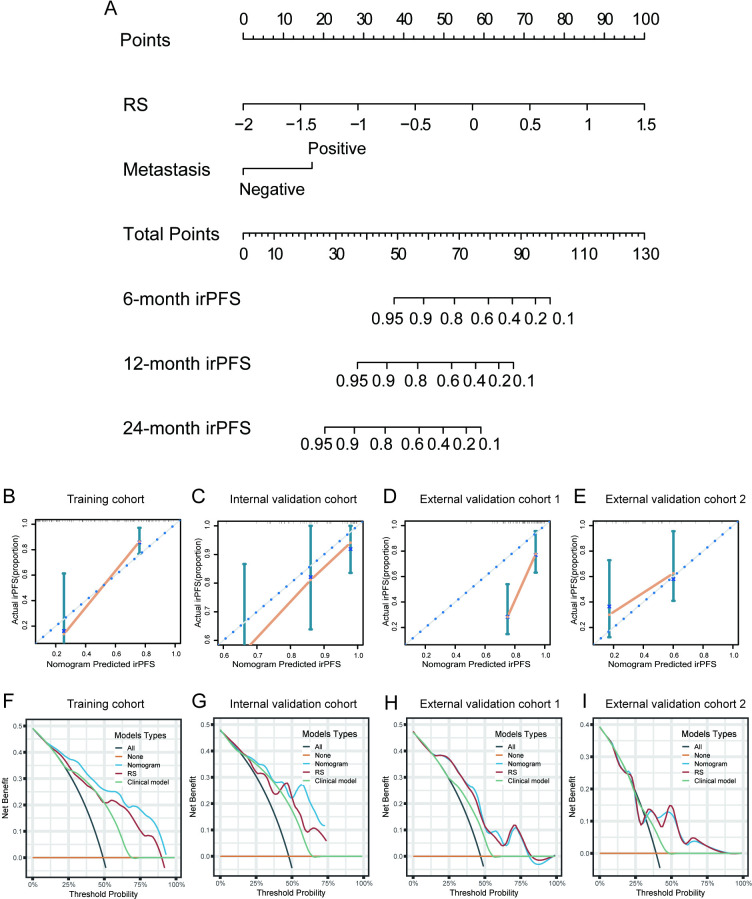
Assessment of the incremental value of the nomogram for irPFS
The RS and distant metastasis were found to be significantly associated with irPFS after performation of multivariate Cox regression analysis, which was integrated to construct the nomogram to improve the prediction accuracy of irPFS (p<0.01, table 2 and figure 5A). Nomogram of irOS was also constructed based on RS and distant metastasis (online supplemental figure S8). The calibration curves of the radiomics nomogram for irPFS showed good agreement between the estimated and actual observation in each cohort (figure 5B–E). DCA curves further confirmed the incremental value of the nomogram to irPFS, as it showed a better overall net benefit than the clinical model (distant metastasis) and RS across the majority of the range of reasonable threshold probabilities (figure 5F–I).
Figure 5.
Construction and validation the irPFS nomogram. (A), Nomogram for 6-month, 12-month, and 24-month irPFS in the training cohort based on RS and tumor metastasis. (B) Calibration curves of the nomogram in terms of agreement between predicted and observed 24-month irPFS in the training cohort. (C) Calibration curves of the nomogram in terms of agreement between predicted and observed 6-month irPFS in the internal validation cohort. (D) Calibration curves of the nomogram in terms of agreement between predicted and observed 6-month irPFS in the external validation cohort 1. (E) Calibration curves of the nomogram in terms of agreement between predicted and observed 24-month irPFS in the external validation cohort 2. (F–I) Decision curve analyses of the nomogram, RS, and clinical model for 12-month irPFS in the training, internal validation, external validation 1, and external validation 2 cohorts. irPFS, immunotherapy-related progression-free survival; RS, radiomics score.
The discrimination performance of the radiomics nomogram was evaluated using the C-index, which showed improved prediction of irPFS and irOS compared with RS and the clinical model in the training cohort (C-index for the radiomics nomogram: 0.778 (95% CI 0.729 to 0.827) and 0.779 (95% CI 0.689 to 0.870), respectively, table 3). Similar results were observed in the internal and external validation cohorts, further confirming the robustness of the nomogram in different populations (table 3).
Table 3.
Comparing the prediction power of the integrated nomogram with RS and clinical model in the training and validation cohorts
| Variable | irPFS | irOS | |
| C-Index (95% CI) | C-Index (95% CI) | ||
| Training cohort | |||
| Nomogram | 0.778 (0.729 to 0.827) | 0.779 (0.689 to 0.870) | |
| RS | 0.749 (0.694 to 0.804) | 0.767 (0.661 to 0.873) | |
| Clinical model | 0.643 (0.592 to 0.694) | 0.599 (0.491 to 0.707) | |
| Internal validation cohort | |||
| Nomogram | 0.767 (0.651 to 0.883) | 0.697 (0.538 to 0.856) | |
| RS | 0.714 (0.589 to 0.839) | 0.666 (0.501 to 0.831) | |
| Clinical model | 0.646 (0.558 to 0.734) | 0.597 (0.478 to 0.718) | |
| External Validation cohort 1 | |||
| Nomogram | 0.713 (0.617 to 0.809) | 0.714 (0.591 to 0.837) | |
| RS | 0.709 (0.609 to 0.809) | 0.712 (0.583 to 0.841) | |
| Clinical model | 0.556 (0.464 to 0.648) | 0.553 (0.445 to 0.661) | |
| External validation cohort 2 | |||
| Nomogram | 0.687 (0.550 to 0.824) | 0.752 (0.562 to 0.942) | |
| RS | 0.690 (0.557 to 0.823) | 0.752 (0.576 to 0.928) | |
| Clinical model | 0.579 (0.436 to 0.722) | 0.600 (0.426 to 0.744) |
irOS, immunotherapy-related overall survival; irPFS, immunotherapy-related progression-free survival; RS, Radiomics Score.
Immune infiltration patterns associated with the imaging biomarker
In this study, CIBERSORT and GSEA analyses were performed to analyze the immune cell infiltration and underlying pathways between the RS-Low and RS-High groups. The infiltration proportions of M1 macrophages and resting mast cells were identified to be significantly higher in the patients in the RS-Low group (figure 6B). GSEA showed that the RS-High group was significantly positively correlated with tumor promoting-associated pathways, such as MYC target signaling, E2F target signaling, cell cycle signaling, and DNA repair signaling (figure 6C). Moreover, metabolism-associated pathways involved in oxidative phosphorylation signaling, fatty acid metabolism signaling, and glycolysis signaling were significantly positively correlated with the RS-High group (figure 6C). However, tumor suppression associated pathways and immune activation-associated pathways, such as apoptosis signaling and inflammatory response signaling, were significantly negatively correlated with the RS-High group (figure 6C).
Figure 6.
Radiogenomic analysis of immune infiltration patterns associated with imaging biomarker. (A) Heatmap of the infiltrated immune cell scores between groups stratified by RS in TCGA samples. (B) Estimated proportions of 22 types of immune cells in different RS groups. (C) NES of the pathways significantly associated with the RS-High group based on the GSEA. P values were generated using the Kruskal or Mann-Whitney test. GSEA, gene set enrichment analysis; NES, Normalized Enrichment Score; RS, radiomics score; TCGA, The Cancer Genome Atlas.
Discussion
Immunotherapy has revolutionized cancer treatment, but identifying biomarkers to predict treatment outcomes remains a major challenge. The CT radiomics model has emerged as a promising method to predict treatment outcomes in many cancer types, such as melanoma and non-small cell lung cancer.18 24 25 However, there is a lack of development and validation of CT imaging biomarker with biological understanding to predict immunotherapy in GC. Here, we developed an imaging biomarker based on the intratumoral and peritumoral features of CT images to non-invasively predict the immunotherapy response of GC, with C-indices of 0.749, 0.714, 0.709, and 0.690 for the prediction of irPFS in the training cohort, internal validation cohort, and external validation cohorts 1 and 2, respectively. Multivariate Cox regression analysis identified that the RS was an independent prognostic factor of irPFS and irOS in each cohort, indicating the predictive value of the CT imaging biomarker. Moreover, the nomogram that integrated RS and clinical characteristics performed better than any single predictor in the prediction of irPFS, demonstrating the incremental value of CT imaging biomarker for the individualized evaluation of irPFS. Importantly, radiotranscriptomic analysis revealed that the infiltration of M1 macrophages and inflammatory response signaling were significantly associated with imaging biomarker, indicating that the current imaging biomarker may be associated with the innate immune signaling.
Recently, radiomics, which can convert images into statically quantifiable and interpretable data, has been widely applied in the prediction of tumor heterogeneity, distant metastasis, pathological response, and survival in many cancer types.15 26–28 For a long time, traditional radiomics studies have tried to extract the most predictive information from the intratumoral region of medical images, as the tumor region contains the most meaningful part of the view. Since the tumor immune microenvironment (TIME) has been proven to modulate antitumor immune response, peritumoral regions of informative images, which contain various components for understanding tumor behavior, are increasingly appreciated by researchers around the world.29 30 A previous study also found that features from peritumoral regions of CT images can be used for evaluating the TIME and were associated with immunotherapy response in GC.19 In the present study, the RS that developed for the prediction of immunotherapy response was constructed by seven intratumoral and six peritumoral features. Intratumoral features of Intra_extent, Intra_GLCM_Corrlation, and Intra_GLRLM_GLV_1.0 were found to be the top three important features of RS. Moreover, we found that the grey-level size zone matrix features from the peritumoral region ranked as the fourth contribution among these 13 selected features to irPFS, indicating the important value of peritumoral features of CT images to assess immunotherapy response in GC.
Currently, clinical predictors for immunotherapy of GC are limited to biomarkers such as CPS, MSI/MMR status, EVB status and TMB status. However, an increasing number of studies and clinical trials have pointed out limitations regarding application of the above biomarkers in individual. The KEYNOTE-061 trial reported that immunotherapy did not improve overall survival for advanced gastric or gastroesophageal junction cancer with a PD-L1 CPS of 1 or higher, while another clinical trial reported that patients with a PD-L1 CPS less than 1 can also benefit from immunotherapy, indicating that CPS is not adequate to select which patients would benefit from immunotherapy.8 31 On the other hand, although EBV and MSI were considered treatment predictors, patients with positive status comprised only approximately 4% of the GC patients, missing a large proportion of patients who may benefit from immunotherapy.9 32 Although these biomarkers are valuable for immunotherapy, they reflect a small part of the complex heterogeneity of GC. Therefore, these biomarkers themselves showed limited predictive power in guiding immunotherapy. Importantly, in this mutilcenter study, most of the patients received immunotherapy following the above biomarkers, but the real OR rate was still low, and most of the patients were ultimately found to have PD. Moreover, biomarkers of CPS, MSI, and EBV were found to be less associated with immunotherapy response, indicating that these biomarkers may not be impressive for treatment decisions in regular clinical treatment. The CT imaging biomarker outperformed any single clinical characteristic and currently used biomarkers in the training and internal validation cohorts, and the stable performance in the two external validation cohorts suggests that the radiomics approach based on CT images could function as a useful treatment guide for the response to immunotherapy in GC.
Biological meaning is essential for the application of radiomics models in different datasets to ensure reproducibility and intensity.33 For years, researchers around the world have tried to investigate the relationship between radiomics features and their involved biological processes in different prediction models.34–36 In this study, we used bulk RNA sequencing of GC from TCGA to reveal the underlying immune infiltration and found that the CT imaging biomarker was associated with M1 macrophage infiltration and inflammatory response signaling, suggesting that the defects of innate immune system may account for the worse irPFS in GC.37 38 Given this association, there is potential merit in combining immunotherapy with treatments that target innate immune pathways, which could potentially amplify the overall immune response against the tumor, benefiting patients with a high RS. However, the genomic samples used in the present study were limited as there were only 42 patients with both RNA sequencing and CT image data. Therefore, a large sample of genomic or immunohistochemistry profiling may help to investigate the deeper characterization and understanding of tumor biology.
Despite the promising results, there are still some limitations in the present study. The first limitation is the retrospective nature of data collection. Although we have tried to use four cohorts from three medical centers across southern and northern of China, prospective studies with larger samples are still needed for further validation. Second, the CT images were obtained from different scanners and medical centers. To minimize the parameter differences, data standardization before model construction was performed. Finally, although we found that the CT imaging biomarker may be associated with the innate immune system, the study population is non-Asian, which may affect the results, and the samples for clarifying the biological meaning of radiomics features are limited. Therefore, larger samples of bulk RNA sequencing or single-cell RNA sequencing of Asian populations are necessary in future studies to validate the findings of the present study.
Conclusion
In conclusion, this study demonstrates that the CT imaging biomarker based on radiomics features can non-invasively and effectively predict the immunotherapy response and outcomes in GC and can serve as a potential tool for individual treatment decision. Importantly, the imaging biomarker is associated with the dysregulation of innate immunity.
Acknowledgments
We acknowledge the TCGA database and GSEA tool for providing their platforms and contributors for uploading their meaningful datasets.
Footnotes
Contributors: YJ, GL, and WH conceived and designed the study. WH, WX, LT, QY, CC, ZS, TZ, HF, ZH, ZZ, YZ, HD, LS, and WW collected and assembled the data; WH, KZ, YJ, QY, CC, YX, XL, and LY performed the data analysis and interpretation; WH, YJ, and GL wrote the manuscript. YJ is responsible for the overall content as guarantor. All authors have read and approved the final manuscript.
Funding: This work was supported by grants from Project funded by China Postdoctoral Science Foundation (grant number: 2023M731567), Guangdong Provincial Key Laboratory of Precision Medicine for Gastrointestinal Cancer (grant number: 2020B121201004), the Key-Area Research and Development Program of Guangdong Province (grant number: 2021B0101420005), the Guangdong Provincial Major Talents Project (grant number: No.2019JC05Y361), and National Natural Science Foundation of China (grant number: 81872013, 82102156, 82302301).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data generated in this study are available from the corresponding author on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Institutional Review Board of Nanfang Hospital of Southern Medical University (NFEC2023375), Guangdong Provincial Hospital of Chinese Medicine (BF2019-066), and Peking University Cancer Hospital (2020KT08). Informed consent was waived since this was a retrospective study.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635–48. 10.1016/S0140-6736(20)31288-5 [DOI] [PubMed] [Google Scholar]
- 3.Smyth EC, Gambardella V, Cervantes A, et al. Checkpoint inhibitors for gastroesophageal cancers: Dissecting heterogeneity to better understand their role in first-line and adjuvant therapy. Ann Oncol 2021;32:590–9. 10.1016/j.annonc.2021.02.004 [DOI] [PubMed] [Google Scholar]
- 4.Moehler M, Shitara K, Garrido M, et al. Lba6_Pr Nivolumab (Nivo) plus chemotherapy (Chemo) versus Chemo as first-line (1L) treatment for advanced gastric cancer/gastroesophageal junction cancer (GC/GEJC)/Esophageal adenocarcinoma (EAC): first results of the Checkmate 649 study. Annals of Oncology 2020;31:S1191. 10.1016/j.annonc.2020.08.2296 [DOI] [Google Scholar]
- 5.Salas-Benito D, Pérez-Gracia JL, Ponz-Sarvisé M, et al. Paradigms on Immunotherapy combinations with chemotherapy. Cancer Discov 2021;11:1353–67. 10.1158/2159-8290.CD-20-1312 [DOI] [PubMed] [Google Scholar]
- 6.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for Checkpoint inhibitor Immunotherapy. Nat Rev Cancer 2019;19:133–50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGrail DJ, Pilié PG, Rashid NU, et al. High tumor Mutation burden fails to predict immune Checkpoint blockade response across all cancer types. Ann Oncol 2021;32:661–72. 10.1016/j.annonc.2021.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Zhou Z-W, Lu J, et al. PD-L1P146R is Prognostic and a negative Predictor of response to Immunotherapy in gastric cancer. Mol Ther 2022;30:621–31. 10.1016/j.ymthe.2021.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Xiu J, Farrell A, et al. Mutational analysis of Microsatellite-stable gastrointestinal cancer with high tumour mutational burden: a retrospective cohort study. The Lancet Oncology 2023;24:151–61. 10.1016/S1470-2045(22)00783-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishino M, Ramaiya NH, Hatabu H, et al. Monitoring immune-Checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol 2017;14:655–68. 10.1038/nrclinonc.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda K, Shoji H, Nagashima K, et al. Correlation between immune-related adverse events and prognosis in patients with gastric cancer treated with Nivolumab. BMC Cancer 2019;19:974. 10.1186/s12885-019-6150-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shitara K, Van Cutsem E, Bang Y-J, et al. Efficacy and safety of Pembrolizumab or Pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: the KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol 2020;6:1571–80. 10.1001/jamaoncol.2020.3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749–62. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 15.Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (Radiomics) in oncology. Ann Oncol 2017;28:1191–206. 10.1093/annonc/mdx034 [DOI] [PubMed] [Google Scholar]
- 16.Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of Radiomic phenotypes in lung cancer. Elife 2017;6:e23421. 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schniering J, Maciukiewicz M, Gabrys HS, et al. Computed tomography-based Radiomics Decodes Prognostic and molecular differences in interstitial lung disease related to systemic sclerosis. Eur Respir J 2022;59:2004503. 10.1183/13993003.04503-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer Immunotherapy using noninvasive Radiomic biomarkers. Ann Oncol 2019;30:998–1004. 10.1093/annonc/mdz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Jiang Y, Xiong W, et al. Noninvasive imaging of the tumor immune Microenvironment correlates with response to Immunotherapy in gastric cancer. Nat Commun 2022;13:5095. 10.1038/s41467-022-32816-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Z, Huang A, Wang L, et al. A Radiomics model predicts the response of patients with advanced gastric cancer to PD-1 inhibitor treatment. aging (Albany NY). Aging (Albany NY) 2022;14:907–22. 10.18632/aging.203850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino M, Giobbie-Hurder A, Gargano M, et al. Developing a common language for tumor response to Immunotherapy: immune-related response criteria using Unidimensional measurements. Clin Cancer Res 2013;19:3936–43. 10.1158/1078-0432.CCR-13-0895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative Radiomics for high-throughput image-based Phenotyping. Radiology 2020;295:328–38. 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with Digital Cytometry. Nat Biotechnol 2019;37:773–82. 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun R, Limkin EJ, Vakalopoulou M, et al. A Radiomics approach to assess tumour-infiltrating Cd8 cells and response to anti-PD-1 or anti-PD-L1 Immunotherapy: an imaging biomarker, retrospective Multicohort study. Lancet Oncol 2018;19:1180–91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 25.Sun R, Henry T, Laville A, et al. Imaging approaches and Radiomics: toward a new era of Ultraprecision Radioimmunotherapy. J Immunother Cancer 2022;10:e004848. 10.1136/jitc-2022-004848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dercle L, McGale J, Sun S, et al. Artificial intelligence and Radiomics: fundamentals, applications, and challenges in Immunotherapy. J Immunother Cancer 2022;10:e005292. 10.1136/jitc-2022-005292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Zhou K, Jiang Y, et al. Radiomics Nomogram for prediction of peritoneal metastasis in patients with gastric cancer. Front Oncol 2020;10:1416. 10.3389/fonc.2020.01416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Chen C, Xie J, et al. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine 2018;36:171–82. 10.1016/j.ebiom.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Feng S, Wei J, et al. Pretreatment prediction of Immunoscore in hepatocellular cancer: a Radiomics-based clinical model based on GD-EOB-DTPA-enhanced MRI imaging. Eur Radiol 2019;29:4177–87. 10.1007/s00330-018-5986-x [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Wang H, Wu J, et al. Noninvasive imaging evaluation of tumor immune Microenvironment to predict outcomes in gastric cancer. Ann Oncol 2020;31:760–8. 10.1016/j.annonc.2020.03.295 [DOI] [PubMed] [Google Scholar]
- 31.Shitara K, Özgüroğlu M, Bang Y-J, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or Gastro-Oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. The Lancet 2018;392:123–33. 10.1016/S0140-6736(18)31257-1 [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Liu Z, Zeng B, et al. Epstein-Barr virus-associated gastric cancer: A distinct subtype. Cancer Letters 2020;495:191–9. 10.1016/j.canlet.2020.09.019 [DOI] [PubMed] [Google Scholar]
- 33.Tomaszewski MR, Gillies RJ. The biological meaning of Radiomic features. Radiology 2021;299:505–16.:E256. 10.1148/radiol.2021219005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z, Li H, Liu Q, et al. CT Radiomics to predict Macrotrabecular-massive subtype and immune status in hepatocellular carcinoma. Radiology 2023;307:e221291. 10.1148/radiol.221291 [DOI] [PubMed] [Google Scholar]
- 35.Zhou M, Leung A, Echegaray S, et al. Non-small cell lung cancer Radiogenomics map identifies relationships between molecular and imaging phenotypes with Prognostic implications. Radiology 2018;286:307–15. 10.1148/radiol.2017161845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer S, Tahoun M, Klaan B, et al. A Radiogenomic approach for decoding molecular mechanisms underlying tumor progression in prostate cancer. Cancers (Basel) 2019;11:1293. 10.3390/cancers11091293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf NK, Kissiov DU, Raulet DH. Roles of natural killer cells in immunity to cancer, and applications to Immunotherapy. Nat Rev Immunol 2023;23:90–105. 10.1038/s41577-022-00732-1 [DOI] [PubMed] [Google Scholar]
- 38.Sharma MD, Pacholczyk R, Shi H, et al. Inhibition of the BTK-IDO-mTOR axis promotes differentiation of monocyte-lineage Dendritic cells and enhances anti-tumor T cell immunity. Immunity 2021;54:2354–71. 10.1016/j.immuni.2021.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007807supp001.pdf (1.1MB, pdf)
Data Availability Statement
Data are available on reasonable request. All data generated in this study are available from the corresponding author on reasonable request.