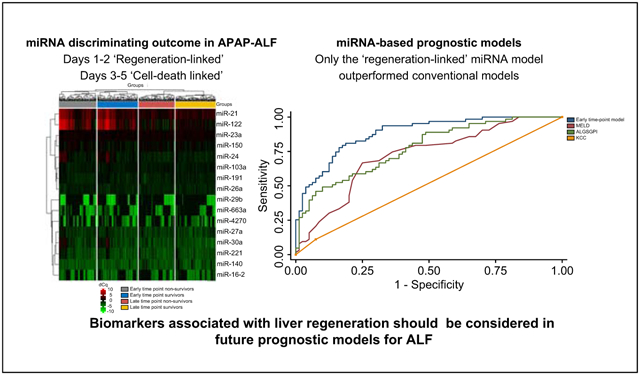
Abstract
Background & Aims:
Acetaminophen (APAP)-induced acute liver failure (ALF) remains the most common cause of ALF in the Western world. Conventional prognostic models, utilising markers of liver injury and organ failure, lack sensitivity for mortality prediction. We previously identified a microRNA signature that is associated with successful regeneration post-auxiliary liver transplant and with recovery from APAP-ALF. Herein, we aimed to use this microRNA signature to develop outcome prediction models for APAP-ALF.
Methods:
We undertook a nested, case-control study using serum samples from 194 patients with APAP-ALF enrolled in the US ALF Study Group registry (1998–2014) at early (day 1–2) and late (day 3–5) time-points. A microRNA qPCR panel of 22 microRNAs was utilised to assess microRNA expression at both time-points. Multiple logistic regression was used to develop models which were compared to conventional prognostic models using the DeLong method.
Results:
Individual microRNAs confer limited prognostic value when utilised in isolation. However, incorporating them within microRNA-based outcome prediction models increases their clinical utility. Our early time-point model (AUC = 0.78, 95% CI 0.71–0.84) contained a microRNA signature associated with liver regeneration and our late time-point model (AUC = 0.83, 95% CI 0.76–0.89) contained a microRNA signature associated with cell-death. Both models were enhanced when combined with model for end-stage liver disease (MELD) score and vasopressor use and both outperformed the King’s College criteria. The early time-point model combined with clinical parameters outperformed the ALF Study Group prognostic index and the MELD score.
Conclusions:
Our findings demonstrate that a regeneration-linked microRNA signature combined with readily available clinical parameters can outperform existing prognostic models for ALF in identifying patients with poor prognosis who may benefit from transplantation.
Lay summary:
While acute liver failure can be reversible, some patients will die without a liver transplant. We show that blood test markers that measure the potential for liver recovery may help improve identification of patients unlikely to survive acute liver failure who may benefit from a liver transplant.
Keywords: Regeneration, cell-death, outcome prediction, biomarker
Graphical abstract
Introduction
Acute liver failure (ALF) remains a rare, potentially reversible, life-threatening disorder. The syndrome is characterised by the development of hepatic encephalopathy (HE) and synthetic dysfunction within 26 weeks of symptom onset.1 Acetaminophen (APAP) is the most common aetiology of ALF in the Western world.2,3 Early clinical prognostication is vital in determining the need for life-saving emergent liver transplantation (LTx).
Several outcome prediction models aid clinicians in determining the risk of patient mortality without LTx including surrogate markers of liver injury and extrahepatic organ failure.4–6 The King’s College criteria (KCC) is the most commonly used model worldwide.4 However, a recent meta-analysis demonstrated that the KCC lacked sensitivity in predicting mortality in patients with APAP-ALF.7 Whilst this may partially represent improvements in the standard of non-transplant care in the 30 years since the development of this model, more recent models also have limitations in performance.5,7 There is a need to develop novel biomarkers that better discriminate outcome in ALF. Furthermore, the classical biomarkers used in existing models focus on severity of liver injury, give little insight on mechanisms mediating recovery and do not elucidate potential targets for therapeutic interventions in the future.
MicroRNA (miRNA) have been a focus of interest as biomarkers in multiple conditions including ALF.8–10 However, there has been limited progress in the development of prognostic miRNA-based biomarkers in ALF, reflecting a lack of standardised methodology in quantifying miRNA expression and the lack of utility of a single miRNA in this setting. Defining the impact of an individual miRNA is challenging as a single miRNA may regulate multiple genes and a single gene may be regulated by multiple miRNAs.10 Animal models of ALF have demonstrated differing miRNA expression depending on time after liver injury which may also impact on the performance of miRNA sampled at non-standardised time-points.11 A standardised methodological approach focusing on miRNA signatures over time may potentially improve the performance of miRNA as clinically tractable biomarkers.
We have previously described a distinct hepatic miRNA signature associated with successful native liver regeneration following auxiliary LTx. Using an in vitro model, we demonstrated that this miRNA signature induces proliferation.12 We have subsequently demonstrated that the presence of this regeneration-linked signature in serum is associated with clinical recovery in both ALF and chronic liver disease.13
We hypothesised that the integration of miRNA signatures within outcome prediction models for APAP-ALF would improve prognostication. Herein, we aimed to develop miRNA-based outcome prediction models with superior performance to conventional models for the prediction of 21-day mortality from APAP-ALF.
Patients and methods
Study design
This study was designed as a nested case-control study of prospectively collected data and bio-samples from 194 patients enrolled in the US ALF Study Group (USALFSG) registry/bio-repository. Inclusion criteria for enrolment to the registry were evidence of ALF defined by an international normalised ratio (INR) >1.5 and HE within 26 weeks of hepatic insult. Patients with pre-existing chronic liver disease or alcohol-induced liver failure were excluded. For this study, patients were excluded if: i) they were younger than 18 years old; ii) they did not develop HE within 7 days of liver injury; iii) the aetiology of ALF was not felt to be APAP; or iv) they underwent LTx. The latter were excluded as selection for transplantation was not standardised across USALFSG sites and to mitigate the potential impact of patients who were transplanted who may have survived without LTx. Samples and clinical information were collected at 2 clinical time-points; early (day 1–2) and late (day 3–5). The primary outcome was 21-day survival.
We identified 928 APAP-ALF patients enrolled from 16 tertiary academic centres between January 1998-December 2014 who fulfilled inclusion/exclusion criteria. We were able to access paired time-point samples for 95 survivors and 83 non-survivors. To maintain the power of the study, further unpaired samples at both time-points were randomly selected for analysis by personnel not involved with the analysis of samples or statistical analysis of this manuscript. Final group numbers were: 96 early time-point survivors; 97 late time-point survivors; 92 early time-point non-survivors; and, 87 late time-point survivors.
This study was approved by the authors’ institutional review board/health research ethics board (REC number 12/LO/0167) and institutional review boards of all participating USALFSG enrolling sites and has been conducted according to the principles expressed in the 1975 Declaration of Helsinki. Given patients lacked capacity to provide informed written consent due to the nature of ALF, written assent was obtained from the next of kin from each patient. All centres implemented monitoring and therapeutics according to institutional standards of care. Reporting of the analysis of this study complies to the STROBE Guidelines for reporting case-control studies.14 All authors had access to the study data and reviewed and approved the final manuscript.
Clinical data
Demographic, clinical, laboratory and outcome data were recorded prospectively at both early and late time-points simultaneous to blood sampling as part of enrolment to the USALFSG registry. Data assessed in this study included age, sex, laboratory data (full blood count, creatinine, liver function tests, INR, ammonia, lactate and arterial pH), HE grade (as per West Haven Criteria15) and requirement for organ support (mechanical ventilation, vasopressors and renal replacement therapy). Prognostic scores including the KCC,4 model for end-stage liver disease (MELD) score16 and ALFSG prognostic index (PI)5 were calculated from these data at both time-points (see Table S1).
miRNA analysis
RNA extraction
Serum samples were thawed on ice and centrifuged at 3000 x g for 5 minutes in a 4°C microcentrifuge. An aliquot of 200 μl per sample was transferred to a FluidX tube and 60 μl of lysis solution Biofluids containing 1 μg carrier-RNA and RNA spike-in template mixture were added to each sample. Each sample was mixed for 1 min then incubated for 7 min at room temperature. Following this, 20 μl of protein precipitation solution BF was added. Total RNA was extracted from serum using miRCURY™ RNA isolation kit–biofluids (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol.17 The purified total RNA was eluted to a final volume of 50 μl.
miRNA real-time quantitative PCR
20 μl of RNA was reverse transcribed using the miRCURY™ LNA RT kit (Qiagen, Hilden Germany). Complementary DNA (cDNA) was diluted 50x and assayed in 10 μl PCRs according to the manufacturer’s protocol.17 22 miRNAs were selected for analysis based on our previous study findings,12,13 review of the literature for association with regeneration and cell-death,8,9,18–20 and to allow for normalisation (Table S2). Also included within this panel were; RNA spike-ins (UniSp2 and 4) to ensure RNA isolation efficiency, a cDNA control marker (UniSp6) to assess reverse transcription (RT) and a DNA spike-in (UniSp3) to assess PCR efficiency. Each miRNA was assayed once by quantitative PCR (qPCR) on the miRNA Ready-to-Use PCR Custom Panel (Qiagen, Hilden Germany) using the miRCURY™ LNA SYBR Green master mix. Negative controls excluding the template from the RT reaction were performed and profiled in comparison to the samples. Amplification was performed in a LightCycler® 480 Real-Time PCR System (Roche, Basel, Switzerland) in 384 well plates. Amplification curves were analysed using the Roche LC software (Roche, Basel, Switzerland), both for determination of quantification cycle (Cq) (by the second derivative method) and melting curve analysis.
Methods of assessment and control of haemolysis and miRNA data analysis are described in the supplementary methods.
Statistical analysis
Comparisons were made between survivors and non-survivors at both time-points. Continuous demographic, clinical and laboratory variables were analysed for normality using the D’Agostino and Pearson test. Normally distributed data were analysed using t tests with results reported as mean (SD) and non-normally distributed data were analysed using Mann-Whitney U tests with results reported as median (IQR). Categorical data were analysed by Fisher’s exact test and results reported as number (%).
After natural logarithmic transformation for normalisation (supplementary methods), all miRNA were compared between survivors and non-survivors using t tests. If a p value <0.05 were achieved, receiver-operating characteristic curve analysis was performed and reported as AUC (95% CI). To increase the utility of less prevalently expressed miRNA, those detected in less than 85% of samples were treated as categorical variables (detected(D)=1, not detected(ND)=0) and analysed using Fisher’s exact tests and reported as an odds ratio (OR) (95% CI) for 21-day mortality.
Multiple logistic regression was used to develop miRNA models to predict 21-day mortality at early and late time-points (supplementary methods). Complete case analysis was used, excluding individuals with missing data. Results were recorded as ß estimate, OR with 95% CI and p values. Sensitivity analyses were performed for grade of encephalopathy at the early time-point and time to death at both time-points.
MetaCore™ pathway analysis (GeneGo Inc, Michigan, USA) was used to identify biological processes associated with the miRNA signature expressed within each model (supplementary methods). These models were adjusted for liver injury and critical illness by developing further multiple logistic regression models including MELD score and vasopressor use. A threshold value was determined for each model by Youden’s Index to predict 21-day mortality. All models (with and without threshold values) were compared to the KCC, MELD score and ALFSGPI using DeLong method in paired samples only.21 Correction for multiple comparisons was performed using the Benjamini-Hochberg procedure22 with a false discovery rate set at 0.05. All univariate and multivariate analyses were performed using Prism V8.4.2 (GraphPad, San Diego, USA) or Stata 16.0 (StataCorp, Texas, USA).
Results
Comparison of clinical data for survivors and non-survivors
Clinical, demographic and laboratory data at both time-points are shown in Table 1. At the early time-point, data were provided for 96 survivors and 92 non-survivors. At the late time-point data were provided for 97 survivors and 87 non-survivors.
Table 1.
Comparison of demographic, biochemical and laboratory data between survivors and non-survivors at both early and late time-points.
| Survivors (n = 96) | Non-survivors (n = 92) | ||||
|---|---|---|---|---|---|
| Early time-point (n = 188) | n | Number (%) or median (IQR) | n | Number (%) or median (IQR) | p value |
| Age | 96 | 35 (28–42) | 92 | 40 (30–48) | 0.06 |
| Sex (female) | 96 | 73 (75%) | 92 | 72 (78%) | 0.64 |
| Listed for transplant | 96 | 15 (16%) | 92 | 23 (25%) | 0.15 |
| Median time to death (days) | NA | NA | 92 | 6 (3–9) | |
| Biochemistry | |||||
| Bilirubin (mg/dl) | 95 | 4.1 (2.5–5.8) | 92 | 5.1 (3.5–7.9) | <0.0001* |
| ALT (IU/L) | 95 | 3,373 (1,830–6,576) | 92 | 3,277 (1,476–5,871) | 0.40 |
| Creatinine (mg/dl) | 95 | 1.4 (0.8–3.1) | 91 | 2.6 (1.1–3.8) | 0.002* |
| INR | 96 | 2.7 (1.9–4.0) | 88 | 3.4 (2.3–4.9) | 0.003* |
| pH | 86 | 7.4 (7.4–7.5) | 80 | 7.4 (7.3–7.5) | 0.23 |
| Lactate (mmol/L) | 70 | 2.8 (1.7–5.4) | 62 | 6.9 (4.2–11.1) | <0.0001* |
| Organ support | |||||
| Vasopressor use | 96 | 8 (9%) | 92 | 39 (42%) | <0.0001* |
| Ventilator use | 96 | 56 (58%) | 92 | 73 (79%) | 0.003* |
| RRT use | 96 | 18 (19%) | 92 | 24 (26%) | 0.29 |
| High-grade HE | 96 | 55 (57%) | 90 | 64 (71%) | 0.07 |
| Late time-point (n = 184) | Survivors (n = 97) | Non-survivors (n = 87) | |||
| Biochemistry | |||||
| Bilirubin (mg/dl) | 90 | 5.6 (3.1–8.3) | 78 | 9.9 (6.7–13.8) | <0.0001* |
| ALT (IU/L) | 92 | 1,142 (611–1991) | 78 | 961 (376–2008) | 0.38 |
| Creatinine (mg/dl) | 92 | 1.2 (0.7–2.6) | 82 | 2.4 (1.3–4.0) | <0.0001* |
| INR | 82 | 1.5 (1.3–1.8) | 74 | 2.3 (1.8–4.3) | <0.0001* |
| pH | 55 | 7.4 (7.4–7.5) | 74 | 7.4 (7.3–7.5) | 0.07 |
| Lactate (mmol/L) | 31 | 1.7 (1.0–2.2) | 40 | 3.7 (2.5–6.7) | <0.0001* |
| Organ support | |||||
| Vasopressor use | 97 | 5 (5%) | 87 | 44 (51%) | <0.0001* |
| Ventilator use | 97 | 49 (51%) | 87 | 74 (85%) | <0.0001* |
| RRT use | 97 | 19 (20%) | 87 | 27 (31%) | 0.09 |
| High-grade HE | 59 | 35 (59%) | 82 | 72 (88%) | 0.0001* |
Non-normally continuous data were analysed using Mann-Whitney U tests and presented as median (IQR). Categorical data were analysed using Fisher’s exact tests and presented as number (%). Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). ALT, alanine aminotransferase; HE, hepatic encephalopathy; INR, international normalised ratio; RRT, renal replacement therapy.
At both time-points, non-survivors had evidence of greater liver injury (significantly elevated bilirubin, INR and lactate values) and multi-organ failure (significantly elevated creatinine values, higher rate of ventilator and vasopressor use). At the late time-point, non-survivors had a greater prevalence of high-grade HE. No difference was observed between groups for patient age, sex or whether patients were listed for transplant.
Comparison of miRNA expression in survivors and non-survivors
Outcomes of quality control and haemolysis experiments are shown in Fig. S1. Minimal haemolysis was observed (5/372 samples) and there was a stable expression of quality control markers for RNA isolation, RT and PCR efficiency; therefore, no samples were excluded from further analysis. Samples were then excluded if they did not express all of the normalising miRNA (miR-23a, −26a and −103). From the early time-point, 2 survivors and 3 non-survivors were excluded (survivors n = 94, non-survivors n = 89). From the late time-point, 1 survivor and 3 non-survivors were excluded (survivors n = 96, non-survivors n = 84). This left 92 survivors and 77 non-survivors with paired samples.
The top 16 miRNA with the highest SD across all samples and time-points underwent supervised 2-way hierarchical clustering and are shown in a heat map in Fig. 1 (SD of all miRNA are shown in Table S3) (Fig. S2 demonstrates unsupervised 2-way hierarchical clustering). Whilst this demonstrates a degree of congruence in miRNA expression within each clinical outcome group and time-point, it also demonstrates that expression of individual miRNA across outcome groups is inconsistent and time-point dependent. Univariate analysis of miRNA expression between outcome groups at both time-points is shown in Tables 2 and 3. After correction for false discovery, whilst significant differences were demonstrated in the expression of certain miRNA at individual time-points (early; miR-150, −16–2 and −29b, late; miR-122, −21, −30a and −503), the performance of these individual miRNAs at discriminating outcome in APAP-ALF was poor (best performing frequently detected miRNA at each time-point; early: miR-150 [AUC 0.64, 95% CI 0.55–0.72], late: miR-122 [AUC 0.63, 95% CI 0.54–0.71]). Converting miRNAs less prevalently expressed into categorical variable identified further associations with outcome at each time-point (early: miR-191, −20a, and −149; late: −149, −17, −191 and −16–2). No significant differences were observed between survivors and non-survivors with paired samples for change in miRNA expression from the early to the late time-point (Table S4).
Fig. 1. Heat map and 2-way hierarchical clustering of miRNA expression in both outcome groups at both time-points.
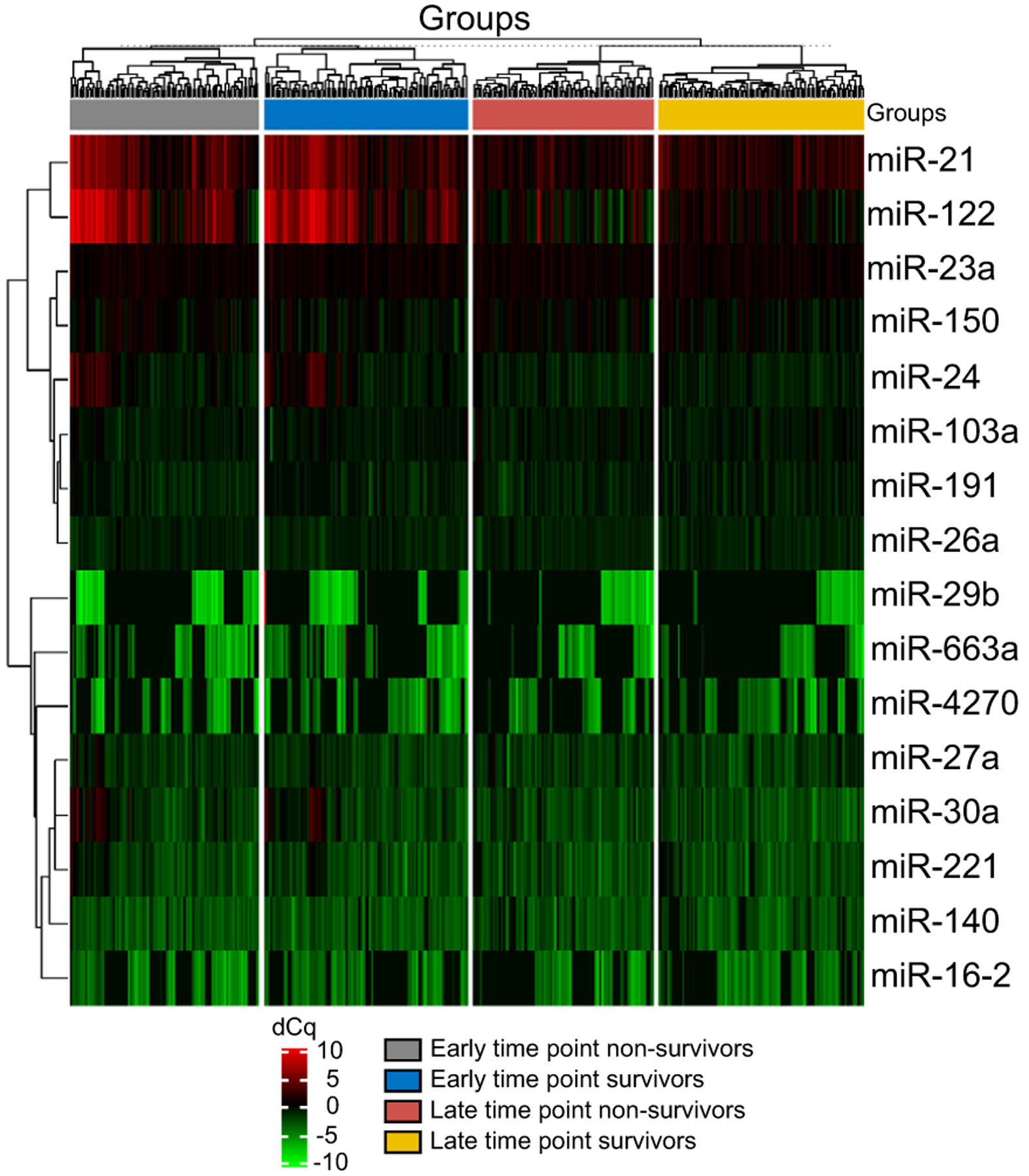
Clustering was performed on all samples and on the top 16 miRNAs with the highest SD using dCq values (miRNA were excluded if not detected in greater than 100 samples). dCq, delta quantification cycle; miR/miRNA, microRNA.
Table 2.
Early time-point miRNA expression.
| (A) | ||||||
|---|---|---|---|---|---|---|
| miRNA | n | Survivors | n | Non-survivors | p value | AUC (95% CI) |
| miR-103 | 94 | 29.82 (0.35) | 89 | 29.76 (0.29) | 0.25 | |
| miR-122 | 94 | 31.80 (1.17) | 88 | 31.72 (1.40) | 0.67 | |
| miR-140 | 87 | 28.43 (0.47) | 84 | 28.42 (0.43) | 0.86 | |
| miR-149 | 26 | 26.97 (1.21) | 10 | 27.70 (1.05) | 0.10 | |
| miR-150 | 94 | 29.98 (0.48) | 88 | 30.20 (0.43) | 0.002* | 0.64 (0.55–0.72) |
| miR-16-2 | 56 | 27.83 (0.82) | 56 | 27.40 (0.92) | 0.01* | 0.65 (0.55–0.75) |
| miR-17 | 19 | 25.83 (0.88) | 25 | 25.79 (0.91) | 0.91 | |
| miR-191 | 55 | 29.49 (0.34) | 71 | 29.48 (0.32) | 0.97 | |
| miR-200b | 14 | 27.45 (0.81) | 9 | 27.14 (1.01) | 0.42 | |
| miR-20a | 5 | 26.81 (0.81) | 16 | 26.02 (0.97) | 0.12 | |
| miR-21 | 94 | 31.45 (0.77) | 89 | 31.42 (0.84) | 0.81 | |
| miR-221 | 93 | 28.89 (0.68) | 88 | 29.01 (0.64) | 0.24 | |
| miR-23a | 94 | 30.46 (0.21) | 89 | 30.47 (0.18) | 0.77 | |
| miR-24 | 94 | 30.11 (0.65) | 89 | 30.09 (0.68) | 0.86 | |
| miR-26 | 94 | 29.56 (0.28) | 89 | 29.61 (0.26) | 0.22 | |
| miR-27a | 94 | 29.16 (0.47) | 89 | 29.32 (0.50) | 0.03 | 0.60 (0.52–0.68) |
| miR-29b | 33 | 26.28 (1.69) | 36 | 25.32 (0.75) | 0.003* | 0.72 (0.60–0.84) |
| miR-30a | 94 | 29.35 (0.84) | 89 | 29.45 (0.84) | 0.42 | |
| miR-330 | 16 | 26.71 (1.25) | 22 | 26.19 (1.39) | 0.25 | |
| miR-4270 | 50 | 27.24 (1.23) | 38 | 26.94 (1.28) | 0.28 | |
| miR-503 | 14 | 26.47 (1.00) | 13 | 26.23 (1.21) | 0.49 | |
| miR-663 | 53 | 27.23 (1.29) | 47 | 26.91 (1.10) | 0.19 | |
| (B) | ||
|---|---|---|
| miRNA | OR (95% CI) | p value |
| miR-149 | 0.33 (0.15–0.71) | 0.006* |
| miR-16-2 | 1.15 (0.63–2.12) | 0.65 |
| miR-17 | 1.54 (0.80–3.11) | 0.23 |
| miR-191 | 2.80 (1.46–5.49) | 0.002* |
| miR-200b | 0.64 (0.27–1.59) | 0.38 |
| miR-20a | 3.90 (1.47–10.02) | 0.01* |
| miR-29 | 1.26 (0.68–2.32) | 0.54 |
| miR-330 | 1.60 (0.77–3.29) | 0.21 |
| miR-4270 | 0.66 (0.37–1.16) | 0.18 |
| miR-503 | 0.98 (0.43–2.18) | >0.9999 |
| miR-663 | 0.87 (0.49–1.54) | 0.66 |
(A) Results of t tests between survivors and non-survivors. Results given as mean (SD). If p values <0.05, AUC was calculated. (B) For miRNA detected in <85% of samples at the early time-point, reanalysis was performed after converting results to either detected (1) or undetected (0) using Fisher’s exact test – OR for mortality. Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). miR/miRNA, microRNA; OR, odds ratio.
Table 3.
Late time-point miRNA expression.
| (A) | ||||||
|---|---|---|---|---|---|---|
| miRNA | n | Survivors | n | Non-survivors | p value | AUC (95% CI) |
| miR-103 | 96 | 29.80 (0.28) | 84 | 29.81 (0.29) | 0.80 | |
| miR-122 | 96 | 30.42 (0.57) | 83 | 30.14 (0.91) | 0.01* | 0.63 (0.54–0.71) |
| miR-140 | 85 | 28.59 (0.59) | 75 | 28.57 (0.49) | 0.75 | |
| miR-149 | 5 | 27.23 (1.65) | 16 | 27.19 (1.11) | 0.96 | |
| miR-150 | 96 | 30.05 (0.42) | 84 | 30.13 (0.43) | 0.19 | |
| miR-16-2 | 61 | 27.78 (0.74) | 37 | 27.42 (0.93) | 0.04 | 0.60 (0.49–0.73) |
| miR-17 | 5 | 26.04 (0.77) | 16 | 26.34 (0.73) | 0.44 | |
| miR-191 | 66 | 29.55 (0.31) | 81 | 29.57 (0.37) | 0.69 | |
| miR-200b | 14 | 26.83 (0.65) | 15 | 26.88 (1.37) | 0.88 | |
| miR-21 | 96 | 30.97 (0.46) | 84 | 30.80 (0.44) | 0.01* | 0.62 (0.54–0.70) |
| miR-221 | 89 | 28.75 (0.55) | 79 | 28.70 (0.60) | 0.55 | |
| miR-23a | 96 | 30.49 (0.18) | 84 | 30.46 (0.20) | 0.42 | |
| miR-24 | 96 | 29.66 (0.32) | 84 | 29.60 (0.34) | 0.27 | |
| miR-26 | 96 | 29.55 (0.27) | 84 | 29.56 (0.26) | 0.73 | |
| miR-27a | 96 | 29.07 (0.53) | 82 | 29.14 (0.53) | 0.40 | |
| miR-29b | 25 | 26.45 (1.17) | 26 | 25.94 (0.98) | 0.09 | |
| miR-30a | 94 | 28.67 (0.58) | 82 | 28.90 (0.55) | 0.008* | 0.61 (0.52–0.69) |
| miR-4270 | 53 | 27.54 (1.06) | 33 | 27.45 (1.13) | 0.71 | |
| miR-503 | 24 | 27.37 (0.73) | 20 | 26.32 (1.43) | 0.003* | 0.74 (0.59–0.90) |
| miR-663 | 32 | 27.25 (1.30) | 29 | 26.83 (1.15) | 0.18 | |
| (B) | ||
|---|---|---|
| miRNA | OR (95% CI) | p value |
| miR-149 | 4.28 (1.61–11.00) | 0.005* |
| miR-16-2 | 0.45 (0.25–0.82) | 0.01* |
| miR-17 | 4.28 (1.61–11.00) | 0.005* |
| miR-191 | 12.27 (3.72–29.27) | <0.0001* |
| miR-200b | 1.27 (0.60–2.75) | 0.69 |
| miR-20a | 4.27 (0.90–20.66) | 0.08 |
| miR-29 | 1.27 (0.66–2.49) | 0.51 |
| miR-330 | 3.62 (0.87–17.90) | 0.15 |
| miR-4270 | 0.53 (0.28–0.95) | 0.07 |
| miR-503 | 0.94 (0.47–1.82) | 0.86 |
| miR-663 | 1.06 (0.58–1.92) | 0.88 |
(A) Results of t tests between survivors and non-survivors. Results given as mean (SD). If p values <0.05, AUC was calculated. (B) For miRNA detected in <85% of samples at the early time-point, reanalysis was performed after converting results to either detected (1) or undetected (0) using Fisher’s exact test – OR for mortality. Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). miR/miRNA, microRNA; OR, odds ratio.
Development of miRNA-based 21-day mortality outcome prediction models
Multiple logistic regression was used to develop miRNA-based 21-day mortality outcome prediction models utilising miRNA with the highest predictive value expressed in greater than 85% of samples if utilised as continuous variables, or greater than 10% of samples if utilised as categorical variables. These cut-offs were used to ensure that these models were applicable to the majority of patients and the miRNA utilised represented true differences between outcome groups.
The early time-point model contained miR-150 and −27a as continuous variables and miR-149, −191 and −20a as categorical variables and included data from 182/183 patients (99.5%) (Fig. 2). All miRNA remained statistically significant within this model (Fig. 2A) and the model discriminated 21-day mortality with an AUC of 0.78 (95% CI 0.71–0.84) (Fig. 2B). The late time-point model contained miR-122 and −30a as continuous variables and miR-149, −191 and −16–2 as categorical variables and included data from 175/180 patients (97.2%) (Fig. 3). These miRNAs also all remained statistically significant within this model (Fig. 3A) and the model discriminated 21-day mortality with an AUC of 0.83 (95% CI 0.76–0.89) (Fig. 3B).
Fig. 2. The early time-point miRNA-based model.
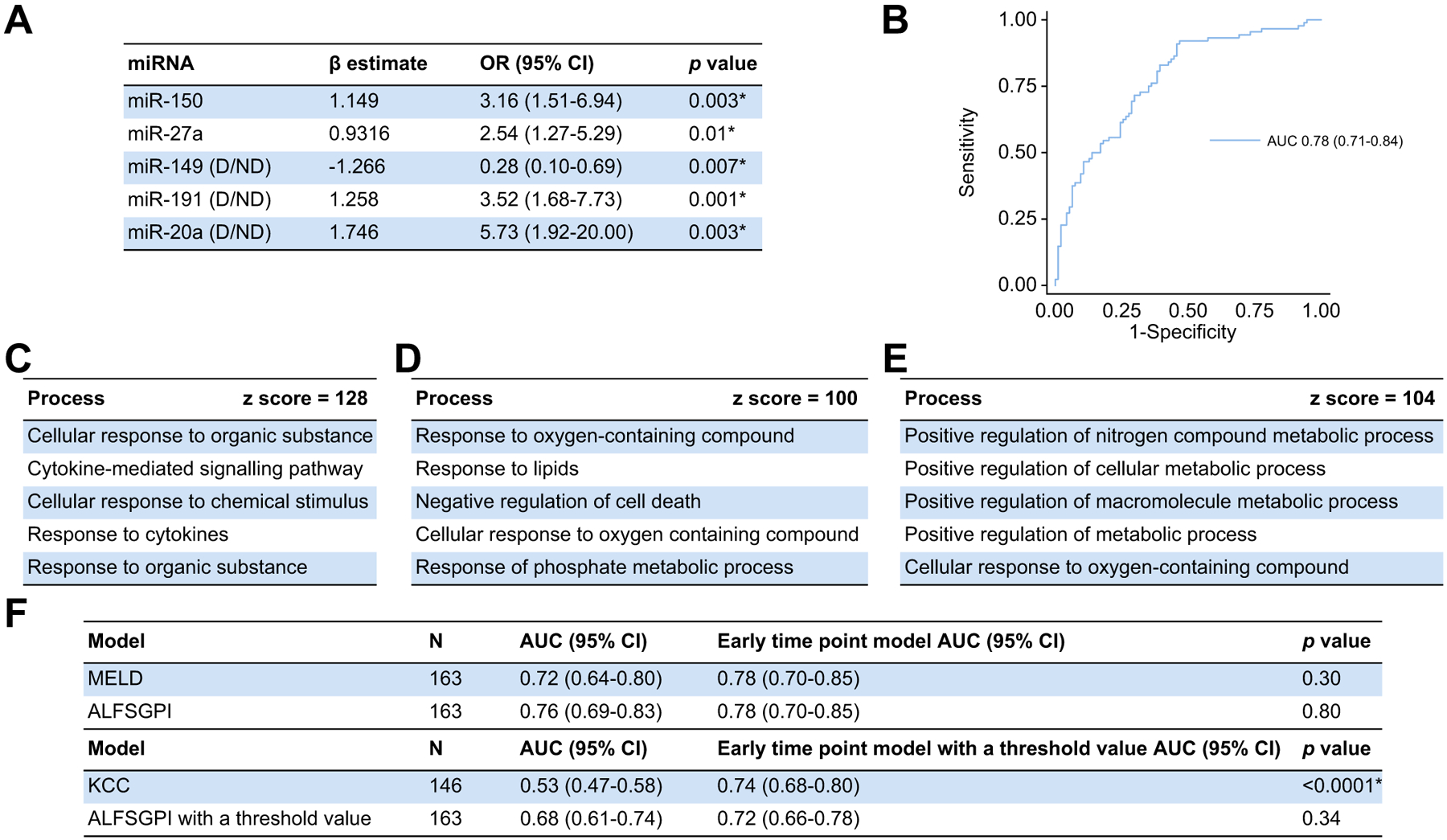
(A) miRNA included within the model. (B) Model performance (n = 182; AUC 0.78 [95% CI 0.71–0.84; p <0.0001*]; pseudo r2 = 0.2213; HL statistic 12.67 [p = 0.12]). (C) MetaCore™ pathway analysis including all miRNA within the model. (D) MetaCore™ pathway analysis including miRNA within both time-point models (miR-149 and −191) (E) MetaCore™ pathway analysis including miRNA only within the early time-point model (miR-20a, −27a, −150). (E) Comparisons with other outcome prediction models with and without threshold values using the DeLong method. Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). ALFSGPI, Acute Liver Failure Study Group prognostic index; HL, Hosmer-Lemeshow; KCC, King’s College criteria; MELD, model for end-stage liver disease; miR/miRNA, microRNA; OR, odds ratio.
Fig. 3. The late time-point miRNA-based model.
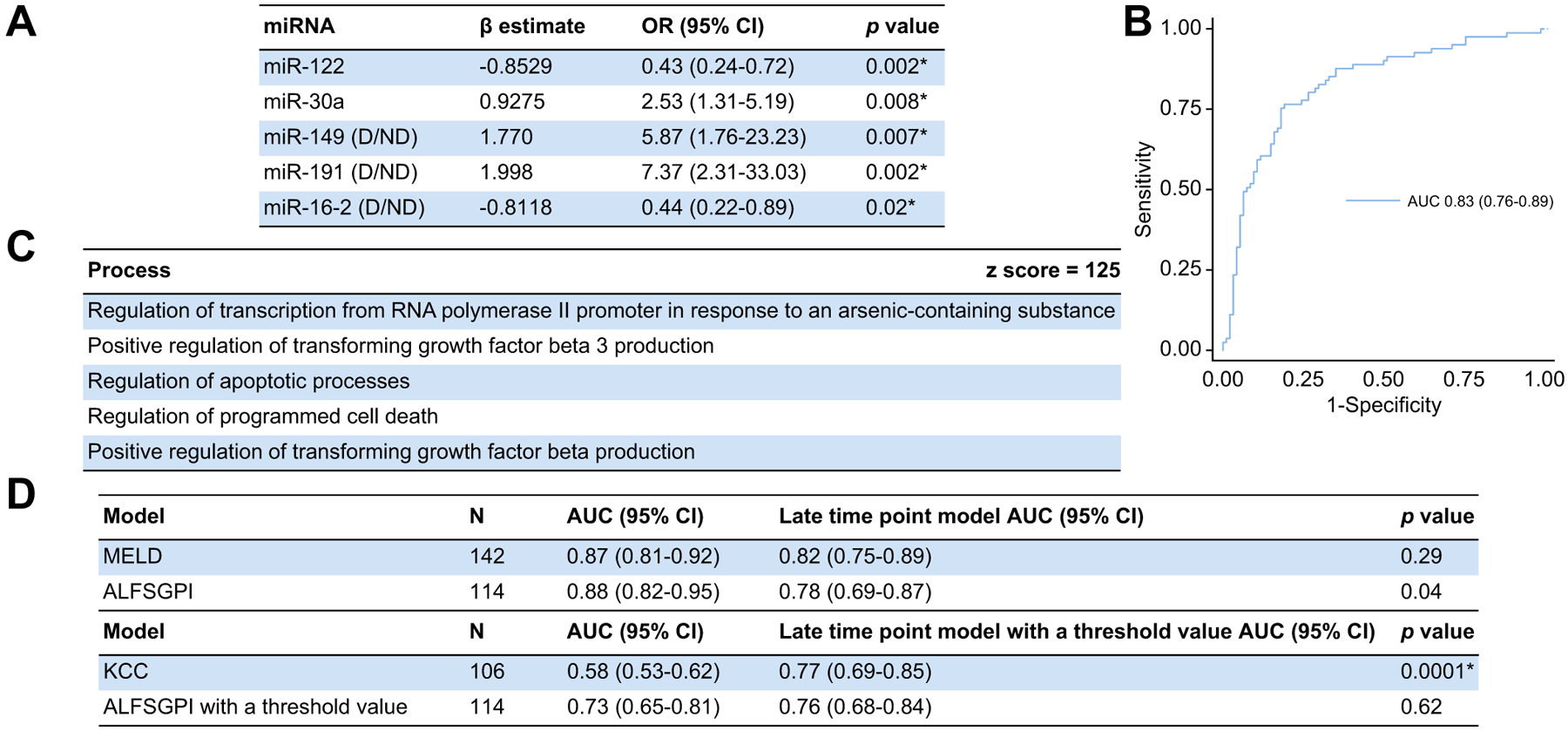
(A) miRNA included within the model. (B) Model performance (n = 175; AUC 0.83 [95% CI 0.76–0.89; p <0.0001*]; pseudo r2 = 0.2767, HL statistic 11.54 [p = 0.17]). (C) MetaCore™ pathway analysis including miRNA within the model. (D) Comparisons with other outcome prediction models with and without threshold values using the DeLong method. Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). ALFSGPI, Acute Liver Failure Study Group prognostic index; HL, Hosmer-Lemeshow; KCC, King’s College criteria; MELD, model for end-stage liver disease; miR/miRNA, microRNA; OR, odds ratio.
MetaCore™ pathway analysis, based on all miRNAs included within the early time-point model, is shown in Fig. 2C. This shows that miRNAs included within this model are implicated in processes associated with a response to cellular injury. Further analysis was performed splitting the miRNAs into those observed in both time-point models (miR-149 and −191) and those not observed in the late time-point model (miR-150, −27a and −20a). The expression of miR-149 and −191 are associated with processes associated with response and mediation of cellular injury (Fig. 2D) and miR-150, −27a and −20a were associated with processes associated with positive regulation of cellular metabolism (Fig. 2E). MetaCore™ pathway analysis was then performed utilising all miRNAs included within the late time-point model (Fig. 3C). This demonstrates that the miRNAs within this model are associated with processes associated with cell-death. All Z scores from these analyses were greater or equal to 100 confirming a strong positive association with the results.
Sensitivity analyses were undertaken to evaluate the performance of the models in patients at the highest risk of non-survival (Fig. S3). At the early time-point, this was performed by sequentially excluding patients with high- and low-grade HE. The early time-point model had comparable performance in patients with high-grade and low-grade HE (Fig. S3A and B). Non-survivors were then excluded by time to death (grouped into days 0–5 and days 6–21) sequentially at both time-points. Whilst the early time-point model performed better at determining patient mortality at days 0–5 (AUC 0.84; 95% CI 0.77–0.91), it was able to discriminate outcome at days 6–21 (AUC 0.72; 95% CI 0.63–0.81) (Fig. S3C and D). The late time-point model performed comparably at days 0–5 and 6–21 (Fig. S3E and F).
To understand the transferability of these models between time-points, the late time-point model was utilised at the early time-point and the early time-point model was utilised at the late time-point in patients with paired samples (Fig. 4A,B). The late time-point model was unable to discriminate 21-day mortality at the early time-point (AUC 0.54; 95% CI 0.45–0.63). However, the early time-point model discriminated 21-day mortality at the late time-point (AUC 0.65; 95% CI 0.56–0.73).
Fig. 4. Comparing the performances of the early and late time-point models.
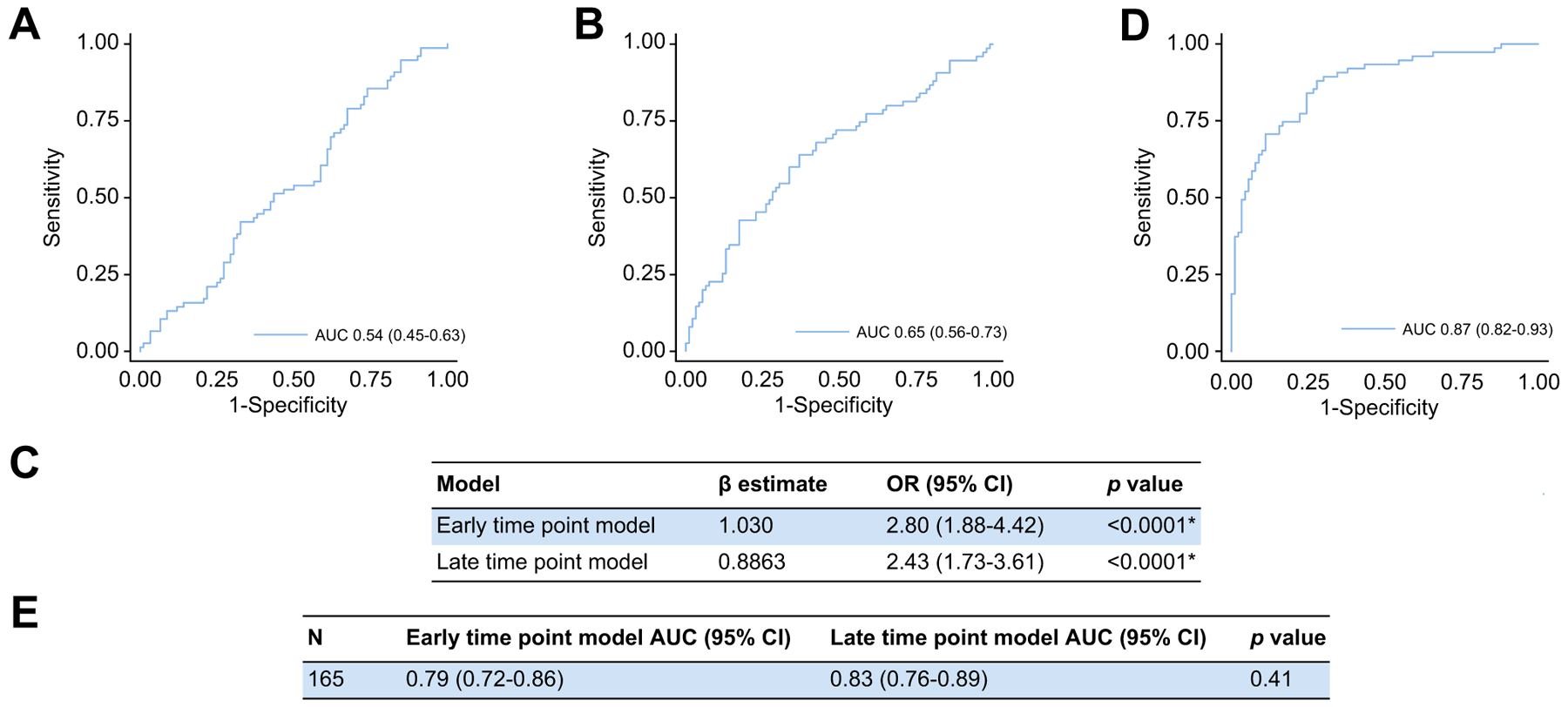
(A) Performance of the late time-point model at the early time-point (n = 167; AUC 0.54; 95% CI 0.45–0.63; p = 0.40). (B) Performance of the early time-point model at the late time-point (n = 165; AUC 0.65; 95% CI 0.56–0.73; p = 0.001*). (C,D) Combined model performance in patients with paired samples (n = 165; AUC 0.87 [95% CI 0.82–0.93; p <0.0001*]; pseudo r2 = 0.4153, HL statistic 14.37 [p = 0.07]). (E) Comparison of both models’ performances in patients with paired samples using the DeLong method. Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). HL, Hosmer-Lemeshow; OR, odds ratio.
Multiple logistic regression combining both models was then used at the appropriate time-point in patients with paired samples (Fig. 4C,D). Each model remained statistically significant within the combined model and an AUC of 0.87 (95% CI 0.82–0.93) was achieved in predicting 21-day mortality. In patients with paired samples, neither model outperformed the other in predicting 21-day mortality when utilised at the appropriate time-point (Fig. 4E).
Incorporating clinical variables
Clinical variables were incorporated as markers of liver injury and critical illness to evaluate whether they would improve prognostication, given their use in classical prognostic models. This also allowed us to adjust the models for clear differences observed in liver injury and critical illness between both cohorts (Table 1). We evaluated the effect of MELD score which utilises laboratory values found to be significantly different between outcome groups at both time-points. In addition, we evaluated vasopressor use which was the organ failure variable associated with the greatest statistically significant difference between survivors and non-survivors at both time-points.
Both models remained statistically significant when adjusted for MELD score and vasopressor use (Fig. 5A,B). Prognostic performance of each model was superior, with the early time-point model achieving an AUC of 0.83 (95% CI 0.78–0.89) and the late time-point model achieving an AUC of 0.91 (95% CI 0.86–0.96) (Fig. 5C,D).
Fig. 5. Combining the models with clinical parameters.
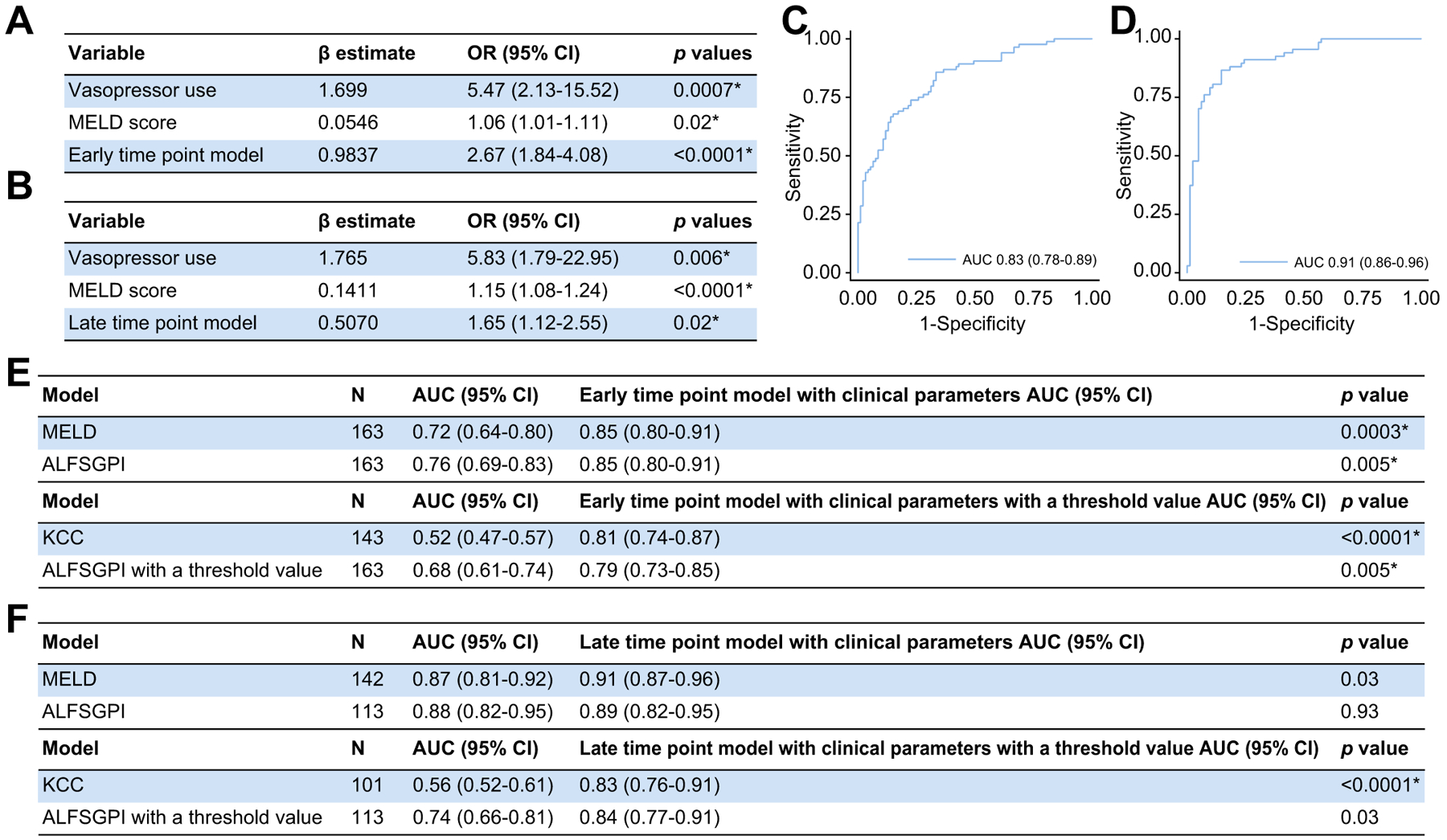
(A) Early time-point model adjusted for MELD and vasopressor use. (B) Late time-point model adjusted for MELD and vasopressor use. (C) Early time-point model performance (n = 177; AUC 0.83 [95% CI 0.78–0.89; p <0.0001*], pseudo r2 = 0.3396, HL statistic 6.83 [p = 0.56]). (D) Late time-point model performance (n = 149; AUC 0.91 [95% CI 0.86–0.96; p <0.0001*), pseudo r2 = 0.5290, HL statistic = 6.74 (p = 0.57)). (E,F) Comparing the early (E) and late (F) time-point models with clinical parameters integrated (with and without threshold values) to other commonly used outcome prediction models using the DeLong method. Statistical significance set as per Benjamini-Hochberg procedure with a false discovery rate of 0.05 (*p <0.026). ALFSGPI, Acute Liver Failure Study Group prognostic index; HL, Hosmer-Lemeshow; KCC, King’s College criteria; MELD, model for end-stage liver disease; OR, odds ratio.
Comparing miRNA-based outcome prediction models with conventional models for 21-day mortality prediction
We selected 3 currently used models for comparison to the miRNA-based outcome prediction models in patients with paired samples at both time-points. The MELD score and ALFSGPI were used as models without threshold values predicting 21-day mortality. The KCC and the ALFSGPI (using the threshold value from the original manuscript5) were used as models with threshold values for predicting 21-day mortality. Performance of these models are shown in Table S5.
We compared the miRNA-based models alone with conventional models and observed no significant differences between either time-point model and the ALFSGPI or MELD score in discriminating 21-day mortality (Fig. 2F, 3D and Table S6). Both models significantly outperformed the KCC.
Given the performance of early and late time-point models incorporating MELD score and vasopressor use, we compared these to the conventional models (Fig. 5E,F and Table S6). The early time-point model incorporating these clinical parameters significantly outperformed the MELD score, ALFSGPI with and without a threshold value and the KCC in predicting 21-day mortality. The late time-point model with clinical variables integrated only outperformed the KCC and the ALFSGPI with a threshold value.
Threshold values for predicting 21-day mortality for all of the miRNA-based models were determined (Table S7). All models significantly outperformed the KCC in predicting 21-day mortality (Figs. 2F, 3D, 5E,F). The late time-point model was significantly outperformed by the ALFSGPI without a threshold value (Table S8B). No models outperformed the MELD score and ALFSGPI without a threshold value. However, the early time-point model with clinical variables incorporated significantly outperformed the ALFSGPI with a threshold value in discriminating 21-day mortality.
Discussion
To our knowledge, we have carried out the largest analysis of serum miRNA expression at multiple time-points in APAP-ALF. Our analysis shows that miRNA-based signatures can discriminate outcome in ALF and, when combined with clinical parameters, our early time-point model outperformed all conventional outcome prediction models in this study.
Whilst our prognostic models have substantially improved performance over present systems, we have demonstrated that individual miRNAs lack utility as biomarkers in isolation. The best performing miRNA which was detected in greater than 85% of samples (miR-150) achieved an AUC of 0.64 (Table 2), comparable to previous reported findings.8 We also demonstrate that serum miRNA expression is dynamic through the clinical course of APAP-ALF and that similar expression may be observed in patients with differing clinical outcomes at different clinical time-points (Fig. 1). This likely reflects that survival from ALF is not determined by a singular biological process but multiple different processes, including regeneration and response to cell-death, which have time-dependent roles in recovery. A clear definition of time of sampling is vital in interpreting the prognostic value of a miRNA. Differing sampling time points may be amongst the reasons why there are conflicting data regarding the expression of certain miRNAs in clinical outcome from APAP toxicity.8,9 Whilst the time course expression of circulating miRNA has been explored in a porcine model of APAP-ALF,11 this has not previously been reported in humans. Further understanding the time-dependent expression of circulating miRNAs may increase the prognostic utility of individual miRNAs as biomarkers.
The early time-point model utilised miRNAs which we have previously observed to be associated with successful regeneration in auxiliary LTx and induce proliferation in an in vitro model.12 Furthermore, MetaCore™ pathway analysis demonstrated that the miRNAs present solely within the early time-point model were associated with positive regulation of increased cellular metabolism; a vital process to facilitate hepatocyte regeneration after injury.23 The late time-point model conversely was comprised of miRNAs that on MetaCore™ pathway analysis were associated with cell-death and have previously been associated with hepatocyte injury and death.9,24–27 It is reasonable therefore to conclude that the early time-point model represents a regeneration-linked miRNA signature and the late time-point model is a cell-death linked miRNA signature.
Conventional models used to predict outcome in ALF have tended to focus on markers of liver injury.4–6 In this study, we not only demonstrate that our regeneration-linked miRNA model predicts 21-day mortality at the early time-point, we show that it can predict 21-day mortality at the late time point (Fig. 4B). When MELD score and vasopressor use were integrated within the early time-point model, this model outperformed all models within this study (Fig. 5E and Table S6c). When a threshold value was applied, the early time-point model significantly outperformed the KCC and the ALFSGPI with a threshold value (Fig. 5E). In comparison, the late time-point model did not outperform the ALFSGPI without a threshold value or MELD score and, when a threshold value was applied, only outperformed the KCC (Fig. 5F and Table S8d). Our findings indicate that biomarkers associated with regeneration have the potential to enhance traditional models of liver injury and organ failure, thereby improving prognostication for patients with ALF.
We have corroborated some of our previous findings evaluating circulating miRNA expression in APAP-ALF with 2 miRNAs from our previous signature incorporated within the early time-point model.13 However, we also observed some differences, which may reflect several differences between this study and our previous work. In our previous study, patients had to fulfil the KCC for inclusion, whilst in this study they required an INR greater than 1.5 and any grade of encephalopathy. It is therefore likely that patients in our previous study had a more severe phenotype of ALF. In our previous studies, we also included patients who underwent LTx which may have impacted on the expression profiles we observed. Our previous study size was relatively small which allowed us to profile the entire miRNome in patients, but impacted on statistical robustness. Additionally, we did not use a threshold value to determine patient mortality with the median time to death in our deceased cohort being 29 days (as opposed to the 21-day mortality used in this study). Finally, samples were taken from patients on admission to critical care as opposed to days 1–2 and 3–5 after enrolment to the study. We have demonstrated in this study that miRNA expression is dynamic over time and that miRNA expression analysis must be undertaken at comparable time-points. These factors likely account for the differences in miRNA expression reported across outcome groups in both studies.
Limitations of this study include the retrospective nested case-control design. However, patients were enrolled and samples with clinical data were collected prospectively and therefore were not subject to recall bias. Investigators were blinded to outcome data when performing miRNA analysis. Given that patients may have been transferred from regional centres and may have required written assent from their next of kin, there is potential for lead time bias. For this reason, our 2 time-points are broad to account for this. Future prospective work should have narrower definitions of time-points. Whilst the methodology utilised in this study allowed us to accurately compare and evaluate the prognostic potential of multiple miRNAs, the practical utility of this methodology in ‘real-world’ time-critical ALF decision-making remains to be determined. However, the field of point-of-care miRNA testing is rapidly evolving28–30 and these technological innovations may improve the clinical utility and applicability of the miRNA signatures we describe. Patients who underwent LTx were excluded from this study as transplant listing decisions for APAP-ALF and organ availability were not consistent between study centres. Therefore, these models may predict patient death that may not have been prevented by LTx. However, the early time-point model demonstrated similar performance in high- and low-grade HE and both models were able to predict mortality after day 5, suggesting that these models may have a role in predicting patients who may benefit from LTx (Fig. S3). There was reduced availability of clinical and laboratory data for patients at the late time-point which reduced the number of patients included in model comparison, particularly for the KCC. Formal mechanistic work is required to investigate the biological processes found on MetaCore™ pathway analysis. Finally, these models require validation in an external cohort. However, this was a large-scale translational study which included patients with APAP-ALF from 16 tertiary LTx centres across the US, the miRNA panel utilised was based on our previous study in APAP-ALF,13 we ensured quality control of each step of miRNA analysis, we adjusted the models for markers of liver injury and critical illness and we compared them to commonly used models for outcome prediction in APAP-ALF. We therefore believe that despite the limitations we highlight, our findings are robust and warrant further evaluation.
In conclusion, we have demonstrated that regeneration-linked and cell-death linked miRNA can be used to predict 21-day mortality in APAP-ALF. A model containing regeneration-linked miRNA, MELD score and vasopressor use significantly outperformed commonly used outcome prediction models. In developing future prognostic models in ALF, biomarkers associated with liver regeneration should be considered.
Supplementary Material
Highlights.
miRNA expression is dynamic across the course of acute liver failure.
At days 1–2, a regeneration-linked miRNA signature discriminates 21-day mortality.
At days 3–5, a cell-death linked miRNA signature discriminates 21-day mortality.
Integrating MELD score and vasopressor use enhances each signatures performance.
The early model with clinical variables outperforms other outcome prediction models.
Acknowledgements
We would like to acknowledge the USALFSG for their support of this study. Members and institutions participating in the Acute Liver Failure Study Group 1998-2020 are as follows: W.M. Lee, M.D. (Principal Investigator); Anne M. Larson, M.D., Iris Liou, M.D., University of Washington, Seattle, WA; Oren Fix, M.D., Swedish Medical Center, Seattle, WA; Michael Schilsky, M.D., Yale University, New Haven, CT; Timothy McCashland, M.D., University of Nebraska, Omaha, NE; J. Eileen Hay, M.B.B.S., Mayo Clinic, Rochester, MN; Natalie Murray, M.D., Baylor University Medical Center, Dallas, TX; A. Obaid S. Shaikh, M.D., University of Pittsburgh, Pittsburgh, PA; Andres Blei, M.D., Northwestern University, Chicago, IL (deceased), Daniel Ganger, M.D., Northwestern University, Chicago, IL; Atif Zaman, M.D., University of Oregon, Portland, OR; Steven H.B. Han, M.D., University of California, Los Angeles, CA; Robert Fontana, M.D., University of Michigan, Ann Arbor, MI; Brendan McGuire, M.D., University of Alabama, Birmingham, AL; Raymond T. Chung, M.D., Massachusetts General Hospital, Boston, MA; Alastair Smith, M.B., Ch.B., Duke University Medical Center, Durham, NC; Robert Brown, M.D., Cornell/Columbia University, New York, NY; Jeffrey Crippin, M.D., Washington University, St Louis, MO; Edwin Harrison, Mayo Clinic, Scottsdale, AZ; Adrian Reuben, M.B.B.S., Medical University of South Carolina, Charleston, SC; Santiago Munoz, M.D., Albert Einstein Medical Center, Philadelphia, PA; Rajender Reddy, M.D., University of Pennsylvania, Philadelphia, PA; R. Todd Stravitz, M.D., Virginia Commonwealth University, Richmond, VA; Lorenzo Rossaro, M.D., University of California Davis, Sacramento, CA; Raj Satyanarayana, M.D., Mayo Clinic, Jacksonville, FL; and Tarek Hassanein, M.D., University of California, San Diego, CA; Constantine J. Karvellas MD, University of Alberta, Edmonton, AB; Jodi Olson MD, University of Kansas, Kansas City, KA; Ram Subramanian MD, Emory, Atlanta, GA; James Hanje MD, Ohio State University, Columbus,OH; Bilal Hameed MD, University of California San Francisco, CA. The University of Texas Southwestern Administrative Group included Grace Samuel, Ezmina Lalani, Carla Pezzia, and Corron Sanders, Ph.D., Nahid Attar, Linda S. Hynan, Ph.D., and the Medical University of South Carolina Data Coordination Unit included Valerie Durkalski, Ph.D., Wenle Zhao, Ph.D., Jaime Speiser, Catherine Dillon, Holly Battenhouse and Michelle Gottfried.
Financial support
This study was supported by the Roche Organ Transplant Research Foundation and the National Institute of Diabetes and Digestive and Kidney Disease (U-01 58369 and R-01 58369 to William M. Lee, MD). The sponsors did not influence any aspect of the study or composition of this manuscript.
Abbreviations
- APAP
acetaminophen
- ALF
acute liver failure
- ALFSG
Acute Liver Failure Study Group
- ALFSGPI
Acute Liver Failure Study Group prognostic index
- cDNA
complementary DNA
- HE
hepatic encephalopathy
- INR
international normalised ratio
- KCC
King’s College criteria
- LTx
liver transplantation
- miRNA
microRNA
- MELD
model for end-stage liver disease
- OR
odds ratio
- Cq
quantification cycle
- qPCR
quantitative PCR
- RT
reverse transcription
Footnotes
Conflict of interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2021.03.013
Data availability statement
Data is available upon request to the corresponding author.
References
Author names in bold designate shared co-first authorship
- [1].O’Grady JG, Schalm SW, Williams R. Acute liver failure: redefining the syndromes. Lancet 1993;342:273–275. [DOI] [PubMed] [Google Scholar]
- [2].Reuben A, Tillman H, Fontana RJ, Davern T, McGuire B, Stravitz RT, et al. Outcomes in adults with acute liver failure between 1998 and 2013: an observational cohort study. Ann Intern Med 2016;164:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bernal W, Hyyrylainen A, Gera A, Audimoolan VK, McPhail MJW, Auzinger G, et al. Lessons from look-back in acute liver failure? A single centre experience of 3300 patients. J Hepatol 2013;59:74–80. [DOI] [PubMed] [Google Scholar]
- [4].O’Grady JG, Alexander GJ, Hayllar KM, Williams R. Early indicators of prognosis in fulminant hepatic failure. Gastroenterology 1989;97:439–445. [DOI] [PubMed] [Google Scholar]
- [5].Koch DG, Tillman H, Durkalski V, Lee WM, Reuben A. Development of a model to predict transplant-free survival of patients with acute liver failure. Clin Gastroenterol Hepatol 2016;14:1199–1206.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mochida S, Nakayama N, Matsui A, Nagoshi S, Fujiwara K. Re-evaluation of the Guideline published by the Acute Liver Failure Study Group of Japan in 1996 to determine the indications of liver transplantation in patients with fulminant hepatitis. Hepatol Res 2008;38:970–979. [DOI] [PubMed] [Google Scholar]
- [7].McPhail MJW, Farne H, Senvar N, Wendon JA, Bernal W. Ability of King’s College criteria and model for end-stage liver disease scores to predict mortality of patients with acute liver failure: a meta-analysis. Clin Gastroenterol Hepatol 2016;14. 516–525.e5. [DOI] [PubMed] [Google Scholar]
- [8].John K, Hadem J, Krech T, Wahl K, Manns MP, Dooley S, et al. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology 2014;60:1346–1355. [DOI] [PubMed] [Google Scholar]
- [9].Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DGN, Antoine DJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011;54:1767–1776. [DOI] [PubMed] [Google Scholar]
- [10].Hayes CN, Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int J Mol Sci 2016;17:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Baker LA, Lee KCL, Palacios Jimenez C, Alibhai H, Chang YM, Leckie PJ, et al. Circulating microRNAs reveal time course of organ injury in a porcine model of acetaminophen-induced acute liver failure. PloS One 2015;10: e0128076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Salehi S, Brereton HC, Arno MJ, Darling D, Quaglia A, O’Grady J, et al. Human liver regeneration is characterized by the coordinated expression of distinct microRNA governing cell cycle fate. Am J Transpl 2013;13:1282–1295. [DOI] [PubMed] [Google Scholar]
- [13].Salehi S, Tavabie OD, Verma S, McPhail MJW, Farzaneh F, Bernal W, et al. Serum MicroRNA signatures in recovery from acute and chronic liver injury and selection for liver transplantation. Liver Transpl 2020;26:811–822. [DOI] [PubMed] [Google Scholar]
- [14].von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ 2007;335:806–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Conn HO, Leevy CM, Vlahcevic ZR, Rodgers JB, Maddrey WC, Seeff L, et al. Comparison of lactulose and neomycin in the treatment of chronic portal-systemic encephalopathy. A double blind controlled trial. Gastroenterology 1977;72:573–583. [PubMed] [Google Scholar]
- [16].Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM, Kosberg CL, et al. A model to predict survival in patients with end-stage liver disease. Hepatology 2001;33:464–470. [DOI] [PubMed] [Google Scholar]
- [17].QIAGEN. QIAGEN kit handbooks. QIAGEN; 2020. Accessed online 22/07/2020, https://www.qiagen.com/gb/service-and-support/learning-hub/search-resources/#filters=%7BF321478C-FDDA-437F-BE0B-87001D9936D3%7D. [Google Scholar]
- [18].Russo MW, Steuerwald N, Norton HJ, Anderson WE, Foureau D, Chalasani N, et al. Profiles of miRNAs in serum in severe acute drug induced liver injury and their prognostic significance. Liver Int 2017;37:757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Xiaomin M, et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Elhendawy M, Abdul-Baki EA, Abd-Elsalam S, Hagras MM, Zidan AA, Abdel-Naby AY, et al. MicroRNA signature in hepatocellular carcinoma patients: identification of potential markers. Mol Biol Rep 2020;47:4945–4953. [DOI] [PubMed] [Google Scholar]
- [21].DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–845. [PubMed] [Google Scholar]
- [22].Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- [23].Kumar S, Zou Y, Bao Q, Wang M, Dai G. Proteomic analysis of immediate-early response plasma proteins after 70% and 90% partial hepatectomy. Hepatol Res 2013;43:876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yuan FH, Chen YL, Zhao Y, Liu ZM, Nan CC, Zheng BL, et al. microRNA-30a inhibits the liver cell proliferation and promotes cell apoptosis through the JAK/STAT signaling pathway by targeting SOCS-1 in rats with sepsis. J Cell Physiol 2019;234:17839–17853. [DOI] [PubMed] [Google Scholar]
- [25].Zhang B, Wang X, Deng J, Zheng H, Liu W, Chen S, et al. p53-dependent upregulation of miR-16–2 by sanguinarine induces cell cycle arrest and apoptosis in hepatocellular carcinoma. Canc Lett 2019;459:50–58. [DOI] [PubMed] [Google Scholar]
- [26].Feng Q, Zhang H, Nie X, Li Y, Chen WD, Wang YD. miR-149* suppresses liver cancer progression by down-regulating tumor necrosis factor receptor 1-associated death domain protein expression. Am J Pathol 2020;190:469–483. [DOI] [PubMed] [Google Scholar]
- [27].Pan W, Wang L, Zhang XF, Zhang H, Zhang J, Wang G, et al. Hypoxia-induced microRNA-191 contributes to hepatic ischemia/reperfusion injury through the ZONAB/Cyclin D1 axis. Cell Death Differ 2019;26:291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vliegenthart ADB, Berends C, Potter CMJ, Kersaudy-Kerhoas M, Dear JW. MicroRNA-122 can be measured in capillary blood which facilitates point-of-care testing for drug-induced liver injury. Br J Clin Pharmacol 2017;83:2027–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Williams MR, Stedtfeld RD, Stedtfeld TM, Tiedje, Hashsham SA. Quantification of microRNAs directly from body fluids using a base-stacking isothermal amplification method in a point-of-care device. Biomed Microdev 2017;19:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Behrmann O, Hugle M, Bronsert P, Herde B, Heni J, Schramm M, et al. A lab-on-a-chip for rapid miRNA extraction. PloS One 2019;14: e0226571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available upon request to the corresponding author.


