Abstract
Multipotent mesenchymal stromal cells (MSCs) are ideal candidates for different cellular therapies due to their simple isolation, extensive expansion potential, and low immunogenicity. For various therapeutic approaches, such as bone and cartilage repair, MSCs are expected to contribute by direct differentiation to replace the damaged tissue, while many other applications rely on the secretion of paracrine factors which modulate the immune response and promote angiogenesis. MicroRNAs (miRNAs), which target messenger RNA for cleavage or translational repression, have recently been shown to play critical functions in MSC to regulate differentiation, paracrine activity, and other cellular properties such as proliferation, survival, and migration. The global miRNA expression profile of MSC varies according to the tissue of origin, species, and detection methodology, while also certain miRNAs are consistently found in all types of MSC. The function in MSC of more than 60 different miRNAs has been recently described, which is the subject of this review. A special emphasis is given to miRNAs that have demonstrated a function in MSC in vivo. We also present in detail miRNAs with overlapping effects (i.e., common target genes) and discuss future directions to deepen our understanding of miRNA biology in MSC. These recent discoveries have opened the possibility of modulating miRNAs in MSC, in order to enhance their proregenerative, therapeutic potential.
Keywords: Mesenchymal stem cells, Marrow stromal stem cells, microRNA
Introduction
Multipotent mesenchymal stromal cells (MSCs) are a heterogeneous cell type that can be isolated from a variety of adult tissues. Ex vivo, MSCs are capable of proliferating extensively and their progenies are further capable of differentiating into different cell types such as osteoblasts, adipocytes, and chondrocytes. In addition, MSCs secrete a variety of cytokines and growth factors that have both paracrine and autocrine activities which modulate the immune response and promote angiogenesis, among others [1]. With this therapeutic focus in mind, MSCs are defined as plastic-adherent, fibroblast-like cells that express a panel of mesenchymal, nonhematopoietic markers [2]. It is commonly acknowledged that MSCs can be isolated from virtually any tissue [3], corresponding with vasculature-associated pericytes [4].
MicroRNAs (miRNAs) are 20–22 nucleotide RNA molecules that target messenger RNA (mRNA) for cleavage or translational repression, thus suppressing protein synthesis [5, 6]. Overall, miRNAs are extremely important in specifying cell differentiation and developmental patterning in animals and plants [7]. In human, some miRNAs have been identified as crucial tumor suppressors with high prognostic value and possible therapeutic potential [8]. In human bone marrow-derived MSCs, silencing Dicer or Drosha, two key components in the biogenesis of canonical miRNAs, blocks both osteogenic and adipogenic differentiation [9], establishing that miRNAs are critical regulators of differentiation.
In 2008, Lakshmipathy and Hart published in this journal an early concise review focused on miRNAs in MSCs [10]. Since then, many publications have addressed the role of one or more miRNAs in MSCs and are the body of this new review. The vast majority of these publications describe the effect(s) in MSCs, by either ectopically over-expressing a miRNA or specifically reducing endogenous miRNA levels using antisense oligonucleotides. The majority of the studies have investigated possible target genes for a given miRNA as the underlying mechanism for the observed effect(s). Finally, a few studies (highlighted in this review) have also investigated the effect of miRNAs in MSCs in vivo.
Identifying miRNAs Expressed in MSCs
To determine the global miRNA expression profile of human MSCs, various groups have used microarray-based platforms (Supporting Information Table S1), always comparing two or more different conditions. However, microarray technology gives limited sensitivity in both low and high (saturation) ranges, undermining the determination of relative abundance of miRNAs and detection of low-expressed miRNAs. Deep sequencing technology (RNAseq) quantifies a dynamic range of over 8,000-fold [11]. This method has been applied to identify expression of miRNAs in MSC derived from human ESCs [12] and MSC isolated from adipose tissue [13] bone marrow and umbilical cord [14], which altogether provide information about which miRNAs are common to all MSCs and which may be tissue-specific. In order to establish a basic consensus of what miRNAs are expressed in human MSCs. Table 1 summarizes miRNAs that were detected by at least one of the groups using RNAseq and at least three groups using microarray-based technology. This list of 44 miRNAs shows a high representation of let-7 family members, the miR-23–24–27 clusters (encoded in human chromosomes 9 and 19) and other miRNA families such as miR-10, miR-29, miR-30, and miR-125. Since many miRNAs that share their seed sequence (i.e., family members) show functional redundancy, it is quite feasible that coexpression of different members confers robustness to their function [15].
Table 1.
Consensus microRNAs (miRNAs) expressed in mesenchymal stromal cells (MSCs)
| let-7a | miR-10a | miR-27b | miR-103 | miR-151a | miR-222 |
|---|---|---|---|---|---|
| let-7b | miR-10b | miR-29a | miR-107 | miR-152 | miR-320a |
| let-7c | miR-16 | miR-29b | miR-125a | miR-181a | miR-484 |
| let-7d | miR-21 | miR-30a | miR-125b | miR-191 | miR-1260b |
| let-7e | miR-23a | miR-30d | miR-130a | miR-193a | |
| let-7f | miR-23b | miR-31 | miR-138 | miR-199a | |
| let-7g | miR-24 | miR-99b | miR-143 | miR-214 | |
| let-7i | miR-26a | miR-100 | miR-145 | miR-221 |
Depending on the method for detection and MSC source, distinct miRNA signatures have been described. This table shows miRNAs that were detected among the top 50 in either one of the RNAseq datasets and in at least three different microarray datasets. For complete list of RNAseq and microarrays addressing miRNA expression in MSCs, see Supporting Information Table S1.
miRNAs Involved in Osteogenic Differentiation of MSCs
Most studies addressing the function of miRNAs in MSCs have focused on in vitro differentiation of the cells. MSCs can robustly differentiate into osteoblasts, adipocytes, and chondrocytes, while their potential to differentiate into other cell types such as myoblasts, neurons, or endothelial cells remains controversial.
Multiple miRNAs have been found to enhance osteogenic differentiation of MSCs (Fig. 1 and Table 2). In periodontal ligament tissue-derived MSCs, miR-17 promotes osteogenesis by targeting Smad ubiquitin regulatory factor 1 (SMURF1), a negative regulator of Runt-related transcription factor 2 (RUNX2). Interestingly, MSCs isolated from patients with periodontitis (i.e., under chronic inflammation) express significantly lower mir-17 levels, higher levels of Smurf1, and consequently display lower osteogenic potential [16]. Also miR-20a, which belongs to the same cluster as miR-17 and is a member of the miR-17 subfamily, enhances osteogenesis by directly repressing the translation of the adipogenic transcription factor peroxisome proliferator-activated receptor gamma (PPAR-γ) and the osteogenic inhibitors bone morphogenic protein (BMP) and activin membrane-bound inhibitor and cysteine-rich transmembrane BMP regulator 1 [17]. The other miRNAs of this cluster (miR-18a, miR-19a, miR-19b, and miR-92a) seem not to be expressed in MSCs.
Figure 1.
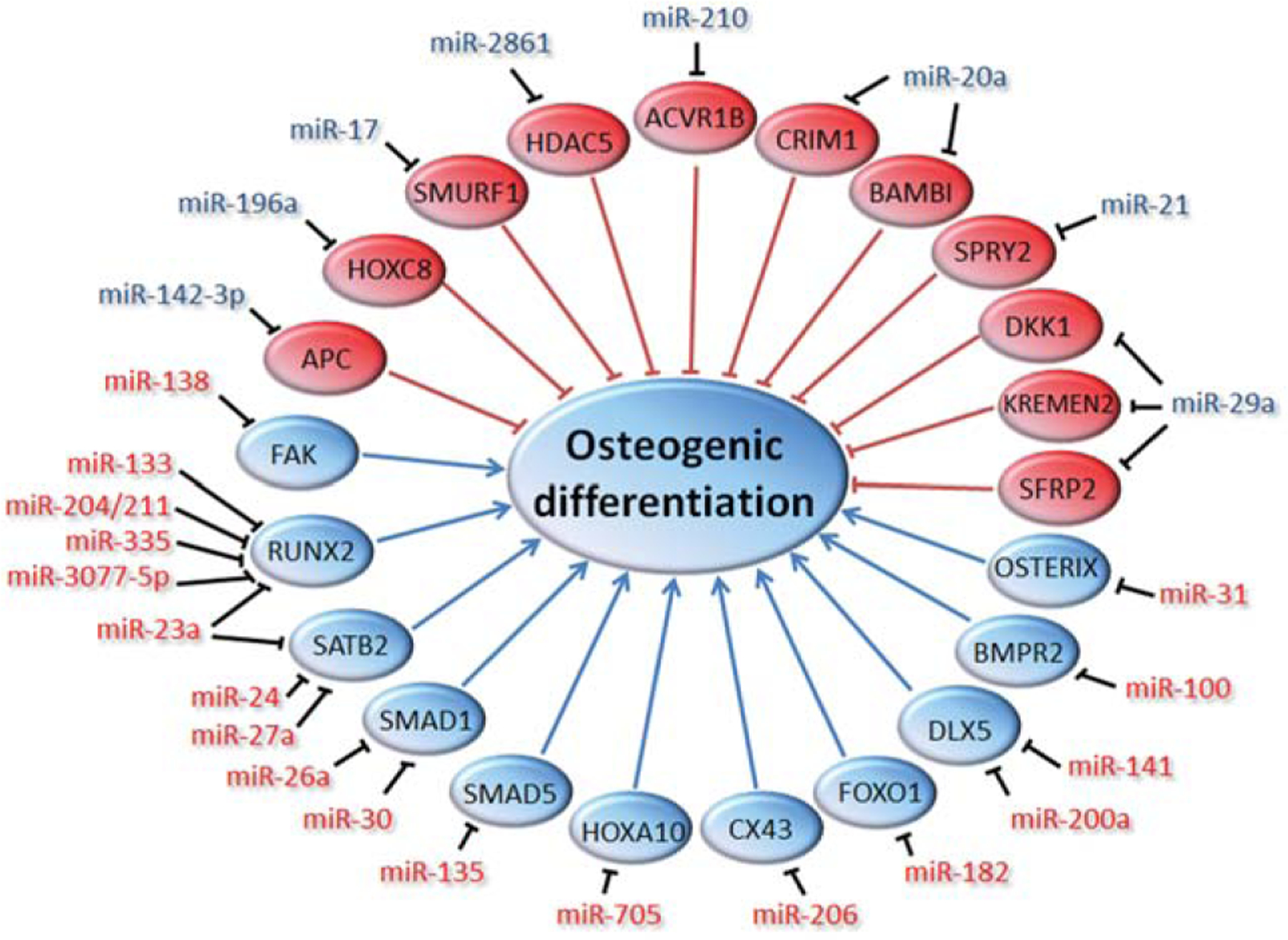
Schematic overview of microRNAs (miRNAs) regulating osteogenic differentiation of mesenchymal stromal cells. Proteins and miRNAs that promote osteogenesis are indicated in blue, while inhibitors are depicted in red.
Table 2.
miRNA with assigned function in MSCs
| miRNA | Effect in MSC | MSC source | Target(s) | Ref. |
|---|---|---|---|---|
| let-7 | Inhibits adipogenesis and migration of cell lines | h-BM | IL6 | [18] |
| miR-10a | Promotes differentiation and inhibits senescence | h-BM | KLF4 | [19] |
| miR-10b | Promotes cell migrationa | m-BM | n.d. | [20] |
| miR-16 | Promotes G1 arrest and myogenesisa | h-BM | n.d. | [21] |
| Inhibits proliferation and angiogenic potential | h-decidua MSC | CCNE1 | [22] | |
| miR-17 | Promotes osteogenesis | h-PO | SMURF1 | [16] |
| miR-20a | Promotes osteogenesis | h-BM | PPARγ, BAMBI, CRIM1 | [17] |
| miR-21 | Promotes survival in hypoxia/serum starvation | r-BM | n.d. | [23] |
| Promotes osteogenesis and adipogenesis | h-AT | SPRY2 | [24] | |
| Promotes osteogenesis | h-BM | SPRY1/2 | [25] | |
| Promotes adipogenesis | h-AT | TGFBR2 | [26] | |
| Inhibits proliferation | h-AT | STAT3 | [27] | |
| miR-23a | Promotes survival in hypoxia/serum starvation | r-BM | n.d. | [23] |
| Inhibits osteogenesis | m-cell line | RUNX2, SATB2 | [28] | |
| miR-23b | Promotes chondrogenesisa | h-BM | PRKACB | [29] |
| miR-24 | Inhibits osteogenesis | m-cell line | SATB2 | [28] |
| Promotes adipogenesis | m-cell line | n.d. | [30] | |
| miR-26a | Inhibits osteogenesis | h-AT | SMAD1 | [31] |
| miR-27a | Inhibits osteogenesis | h-BM, m-cell line | SATB2 | [28, 32] |
| Inhibits adipogenesisa | m-cell line | PPARγ | [33] | |
| miR-27b | Inhibits adipogenesis | h-AT, m-cell line | PPARγ, CEBPα | [34, 35] |
| Promotes immune regulation | r-AT | SDF-1α | [36] | |
| Inhibits cell migrationa | m-BM | SDF-1α | [37] | |
| miR-29a | Promotes osteogenesis | h-BM | DKK1, KREMEN2, SFRP2 | [38] |
| miR-30 | Promotes adipogenesis | h-AT | RUNX2 | [13] |
| Inhibits osteogenesis | m-BM | SMAD1 | [39] | |
| miR-31 | Inhibits adipogenesis | m-cell line | CEBP/A | [30] |
| Inhibits osteogenesis | h-BM | OSTERIX, n.d. | [40, 41] | |
| miR-32 | Reverses high glucose- cell cycle arresta | r-BM | n.d. | [42] |
| miR-100 | Inhibits osteogenesis | h-AT | BMPR2 | [43] |
| miR-107 | Protects against anoxia | r-BM | PDCD10 | [44] |
| miR-124 | Inhibits cardiomyogenesis | r-BM | STAT3 | [45] |
| Inhibits proliferationa | h-BM | n.d. | [46] | |
| miR-125b | Inhibits osteogenesis and proliferation | m-BM | n.d. | [47] |
| Promotes survival | h-BM | P53 | [48] | |
| miR-126 | Promotes angiogenesisa | m-BM | n.d. | [49] |
| Promotes endothelial differentiationa | m-BM | n.d. | [50] | |
| miR-130a | Induces substance P productionb | h-BM | TAC1 | [51] |
| miR-133 | Inhibits osteogenesisa | m-cell line | RUNX2 | [52] |
| Promotes proliferation | r-BM | n.d. | [53] | |
| miR-135 | Inhibits osteogenesis | m-cell line | SMAD5 | [52] |
| miR-138 | Inhibits adipogenesisa | h-AT | EID1 | [54] |
| Inhibits osteogenesis | h-BM | FAK | [55] | |
| miR-140 | Promotes chondrogenesis | h-BM, m-BM, m-cell line | ADAMTS-5, HDAC4 | [56–58] |
| miR-141 | Inhibits osteogenesis | m-cell line | DLX5 | [59] |
| miR-142-3p | Promotes osteogenesis | h-cell line | APC | [60] |
| miR-143 | Promotes adipogenesis | preadipocyte | n.d. | [61] |
| Promotes proliferation | r-BM | ERK5 | [62] | |
| miR-145 | Inhibits chondrogenesis | m-BM | SOX9 | [63, 64] |
| miR-146a-5p | Inhibits immune regulation | m-BM | PTGES2 | [65] |
| Promotes TNF-α-mediated IL-8 | h-BM | n.d. | [66] | |
| Promotes survival against anoxia | r-BM | FAS | [67] | |
| Inhibits migration and promotes proliferation | h-BM and h-UC | SDF-1α, | [14] | |
| miR-148b | Promotes osteogenesis | h-BM | n.d. | [32] |
| miR-155 | Inhibits adipogenesisa | h-cell line | n.d. | [68] |
| Inhibits immune regulation | m-BM | TAB2 | [69] | |
| miR-181 | Inhibits immune suppression | h-BM | TGFBR1, TGFBRAP1 | [70] |
| miR-182 | Inhibits osteogenesis | m-cell line | FOXO1 | [71] |
| miR-193 | Promotes proliferation | r-BM | ING5 | [53] |
| miR-194 | Inhibits chondrogenesis | h-AT | SOX5 | [72] |
| miR-196a | Promotes osteogenesis and inhibits proliferation | h-AT | HOXC8 | [73] |
| miR-199a* | Inhibits chondrogenesis | m-cell line | SMAD1 | [74] |
| miR-200a | Inhibits osteogenesis | m-cell line | DLX5 | [59] |
| miR-204/211 | Inhibits osteogenesis | h-BM | RUNX2 | [75] |
| miR-206 | Induces substance P productionb | h-BM | TAC1 | [51] |
| Inhibits osteogenesis | m-cell line | CX43 | [76] | |
| Promote myogenesis | m-cell line | CX43 | [125] | |
| miR-210 | Promotes osteogenesis | m-cell line | ACVR1B | [77] |
| Promotes survival in hypoxia/serum starvation | r-BM | CASP8AP2 | [23, 44] | |
| Promotes proliferation | h-AT | PTPN2 | [78] | |
| miR-221/222 | Inhibits adipogenesisa | h-cell line | n.d. | [68] |
| miR-302 | Induces substance P productionb | h-BM | TAC1 | [51] |
| miR-335 | Inhibits proliferation, migration and differentiation | h-BM | RUNX2 | [79] |
| miR-369-5p | Inhibits adipogenesis and proliferation | h-BM | FABP4 | [80] |
| miR-371 | Promotes adipogenesis and inhibits proliferation | h-BM | n.d. | [80] |
| miR-449a | Inhibits chondrogenesisa | h-BM | LEF-1 | [81] |
| miR-489 | Inhibits osteogenesis | h-BM | n.d. | [32] |
| miR-499 | Inhibits proliferation | h-BM | n.d. | [80] |
| miR-503 | Promotes survival in hypoxia/serum starvation | r-BM | n.d. | [23] |
| miR-541 | Inhibits osteogenesis | h-BM | n.d. | [82] |
| miR-574-3p | Inhibits chondrogenesis | h-BM | RXRα | [83] |
| miR-705 | Inhibits osteogenesis and promote adipogenesis | m-BM | HOXA10 | [84] |
| miR-886-3p | Inhibits migration | h-BM | SDF-1α | [85] |
| miR-2861 | Promotes osteogenesis | m-BM | HDAC5 | [79] |
| miR-3077-5p | Inhibits osteogenesis and promote adipogenesis | m-BM | RUNX2 | [84] |
Effect was only confirmed by over-expression of the miRNA, not by silencing the endogenous miRNA.
In MSC-derived neuronal cells.
Abbreviations: AT, adipose tissue-derived; BM, bone marrow-derived; h, human; miRNAs, microRNAs; MSCs, mesenchymal stromal cells; m, mouse; n.d., not determined; PO, periodontal ligament; r, rat; UC, umbilical cord.
Experiments using murine MSCs show that miR-2861 promotes osteogenesis by targeting histone deacetylase 5 (HDAC5), which is an enhancer of RUNX2 degradation. Of note, intravenous administration of an inhibitor of miR-2861 in mice leads to reduced RUNX2 levels and loss of bone mass and mutations in miR-2861 are associated with the development of osteoporosis. This strongly suggests that miR-2861 is necessary for normal osteogenesis in vivo [86]. Also miR-210 and miR-196a promote osteogenesis, possibly by targeting activin A receptor type 1B and homeobox protein HOXC8, respectively [73, 77]. These miRNAs shown to promote osteogenesis are likely physiologically relevant, since it has been shown that their levels are regulated either during osteogenic differentiation or in response to an osteogenic stimulus, such as BMP2 or BMP4 [16, 17, 77, 86]. Also miR-148b is upregulated during osteogenesis and promotes osteogenic differentiation, but possible targets in this context have not been identified [32].
Wnt signaling is known to promote osteogenesis and has been shown to increase the expression of miR-29a, which in turn directly binds to the 3′UTR of the Wnt inhibitors Dikkopf-1 (DKK1), the DKK2 cofactor Kremen 2, and secreted frizzled-related protein 2; hence establishing a positive loop for enhanced Wnt-mediated osteogenesis [38]. In addition, miR-142–3p has been found to target the Wnt inhibitor adenomatous polyposis coli, promoting osteogenesis [60]. Another miRNA that promotes osteogenesis is miR-21, possibly by targeting SPROUTY2 [24, 25]. MiR-21 is strongly expressed in MSCs (Table 1) and is highly conserved across species. It was the first mammalian miRNA to be identified [87] and as discussed below plays multiple roles in MSCs, such as to promote adipogenesis [24, 26], inhibit proliferation [27], and enhance cell survival [23].
Most of the miRNAs that have been described to inhibit osteogenic differentiation in MSCs bind to the mRNA of osteogenic transcription factors, blocking their translation and are therefore potential therapeutic targets for age-related bone diseases. MiR-23a, miR-133, the homologs miR-204 and miR-211, miR-335, and miR-3077–5p all inhibit osteogenesis by directly interacting with the 3′UTR of RUNX2 [28, 52, 75, 79, 84]. Each individual component of the miRNA cluster 23a~27a~24–2 targets special AT-rich sequence-binding protein 2 (SATB2), which synergizes with RUNX2 to promote osteogenesis. Importantly, gain-of-function experiments by restoring SATB2 protein levels reverse the effect of miR-23a, miR-27a, and miR-24, clearly demonstrating the importance of this target gene [28].
The miR-30 family and miR-26a inhibit osteogenesis in MSCs by targeting SMAD1 [31, 39], while miR-31 inhibits osteogenesis [40] and targets Osterix [41]. MiR-135 inhibits osteogenesis by targeting SMAD5 [52], while miR-138 inhibits osteogenesis and targets focal adhesion kinase, an activator of RUNX2 [55]. Most important, miR-138 also controls the osteogenesis of MSCs in vivo, as demonstrated using a murine ectopic bone formation assay [55]. Both miR-141 and miR-200a inhibit osteogenesis and target Distal-less homeobox 5 [59], while miR-100 inhibits osteogenesis possibly by targeting BMP receptor 2 [43]. Of note, injection of miR-182 precursor in zebrafish embryos inhibits skeleton development, by targeting the transcription factor Forkhead box protein O1 [71].
Osteogenesis is reduced upon expression of miR-705, which targets Homeobox protein HOXA10 [84]. Connexin 43 is an important gap junction protein in osteoblasts, which is a target of miR-206. Remarkably, transgenic mice that ectopically express miR-206 in osteoblasts show lower bone density and decreased bone formation rate [76]. Silencing endogenous levels of miR-125b was found to inhibit osteogenesis in a MSC cell line [47], but not in primary culture of MSCs [48], stressing the difference of cell lines to primary cultures of MSCs. Also miR-489 and miR-541 inhibit osteogenesis [32, 82], but their targets in this context are not known.
miRNAs Involved in Adipogenic Differentiation of MSCs
Osteogenesis and adipogenesis are frequently presented as reciprocally regulated events, where a common factor induces commitment into one lineage, while blocking the other [88–90]. Both miR-705 and miR-3077–5p act in this manner, promoting adipogenesis while inhibiting osteogenesis, and are found at higher levels in mice with osteoporosis as compared to healthy mice [84]. Similarly, miR-30 family and miR-24, which we discussed as inhibitors of osteogenesis [28, 39], promote adipogenesis [13, 30]. Also miR-21 promotes adipogenesis possibly by targeting either SPROUTY2 [24] or transforming growth factor beta receptor 2 (TGFBR2) [26]. In contrast, ectopic expression of miR-143 and miR-371 has been shown to promote adipogenesis with no apparent effect on osteogenesis [61, 80], but the mechanism has not been elucidated.
Both miR-27a and miR-27b are reduced during adipogenic differentiation of MSCs. Indeed, both miR-27 family members are strong inhibitors of adipogenesis by targeting the transcription factors PPAR-γ and CCAAT/enhancer-binding protein alpha (C/EBPα) [33–35]. Both miR-27a and miR-27b are increased in epididymal fat tissue from obese ob/ob mice as compared to genetically matched lean animals [35]. It is unclear what exact cell type is responsible for this regulation, but it is presumably a mechanism to control adipose mass, which may have strong implications in the control and assessment of obesity in humans. Since the tumor suppressor miR-31 (discussed above as inhibitor of osteogenesis [40, 41]) also targets C/EBPα, inhibiting adipogenesis [30], it is possible that miR-31 may actually be a general inhibitor of differentiation. Another tumor suppressor, let-7, targets interleukin-6 (IL-6) in MSCs and consequently inhibits adipogenesis [18]. In addition to blocking osteogenesis, miR-138 inhibits adipogenesis, by a mechanism involving direct binding to the nuclear receptor coregulator adenovirus early region 1-A-like inhibitor of differentiation 1 [54]. Similarly, miR-335, which inhibits osteogenesis, also inhibits adipogenesis, [79] although possible targets have not been experimentally tested. Other inhibitors of adipogenesis include miR-24 [30], miR-155, miR-221, miR-222 [68], and miR-369–5p which target fatty acid-binding protein 4 [80].
miRNAs Involved in Chondrogenesis and Other Differentiation Pathways of MSCs
The differentiation of MSCs into chondrocytes is regulated by transcription factors such as SOX9 and growth factors such as the TGF-β superfamily. So far, two miRNAs have been found to promote chondrogenesis in MSCs and both are increased during chondrogenic differentiation. These are miR-23b, which targets the beta catalytic subunit of protein kinase A [29] and miR-140 which acts in part by targeting the 3′ UTR of histone deacetylase 4 (HDAC4) [56] and a disintegrin and metalloproteinase with thrombospondin motifs 5, a key cartilage matrix-degrading protease in osteoarthritis (OA) [57]. The importance of miR-140 during chondrogenesis is also evidenced by the fact that miR-140 is chondrocyte-specific. In addition, levels of miR-140 are significantly lower in cartilage from donors with OA as compared to healthy donors [91] and transgenic mice deficient in miR-140 exhibited age-related OA-like effects like proteoglycan loss and fibrillation of articular cartilage [58]. These observations suggest that miR-140 could be a potential therapeutic target to alleviate OA.
MiR-199a* and miR-145 inhibit chondrogenesis in a MSC line by targeting SMAD1 and SOX9, respectively [63, 64, 74]. Other miRNAs that inhibit chondrogenesis are miR-574–3p and miR-194, which were found to target retinoid X receptor and osteogenic SOX5, respectively [72, 83]. Using a bioinformatic approach, miR-449a was described as a potential regulator of chondrogenesis in part by targeting the 3′UTR of lymphoid enhancer-binding factor-1, a positive regulator of chondrogenesis [81]. However, experimentally, only over-expression of this miRNA decreased chondrogenesis, while silencing the endogenous levels of miR-449a did not affect differentiation.
Rat MSCs in coculture with ventricular myocytes can differentiate into cardiomyocytes in vitro, a process that is inhibited by over-expression of miR-124, where signal transducers and activators of transcription 3 (STAT3) is a putative target [45]. In one of the earliest publications addressing the function of miR-NAs in MSCs, Greco et al. showed that miR-130a, miR-206, and miR-302 target mRNAs encoding the tachykinin peptide hormone family (TAC1), regulating the synthesis of the neurotransmitter substance P in human MSC-derived neuronal cells [51].
miRNAs Involved in Proliferation, Senescence, Migration, and Survival of MSCs
Many miRNAs in MSCs have functions in both differentiation and proliferation. Differentiation is often associated with cell cycle arrest in G0/G1 [92] and accordingly miRNAs often play dual functions in differentiation and cell cycle progression/arrest. This is likely true for miR-21, which promotes osteogenesis and adipogenesis (discussed above) but inhibits proliferation, targeting STAT3 [27]. Similarly, miR-371 promotes adipogenesis and inhibits proliferation, while miR-369–5p inhibits both adipogenesis and proliferation [80]. MiR-210 (which promotes osteogenesis) increases cell proliferation by targeting protein tyrosine phosphatase, nonreceptor type 2 [78]. In contrast, miR-196a, which promotes osteogenesis, inhibits cell proliferation by targeting HOXC8 [73]. In addition, miR-16 has been proposed to promote myogenesis of MSCs, while inducing cell cycle arrest in G1 [21]. Other groups have confirmed miR-16-induced cell cycle arrest in MSCs and have identified cyclin E1 as a target of miR-16 [22]. Overexpression of miR-124 and miR-499 inhibits cell proliferation [46, 80]. Although possible targets in MSCs have not been identified, it is feasible that the mechanism for miR-124 function involves inhibition of STAT3 (see above [45]).
High glucose can induce cell cycle arrest, an effect that can be reversed with miR-32 mimics, which activate AKT signaling while inhibiting the Wnt pathway [42]. MSCs genetically engineered to over-express AKT and Angiopoietin 1 show higher proliferation than control cells, in a manner strongly dependent on expression of miR-143, which targets ERK5 [62], suggesting that miR-143 may also promote proliferation of regular MSC. Both miR-133b and miR-193 promote proliferation of MSCs, where the latter targets inhibitor of growth family member 5, an inhibitor of cyclin-dependent kinase 2 [53]. Similarly, miR-146a-5p promotes proliferation and targets the I-κ-B kinase epsilon suppressor SIKE1 [14]. Recently, miR-10a was found to promote differentiation, while inhibiting senescence of MSCs by targeting KLF4 [19].
In addition to inhibiting osteogenic and adipogenic differentiation, miR-335 inhibits proliferation and migration of MSCs [79]. In contrast, ectopic expression of miR-10b enhances the migration potential of MSC, correlating with a decrease of e-cadherin [20]. In cancer, miR-10b induces cell migration by directly targeting components of the Rho GTPase pathway [93–95], suggesting an additional mechanism for the enhanced migration of MSCs.
miRNAs have also been found to function in apoptotic pathways in MSCs. Blockade of miR-146a (now called miR-146a-5p), which targets FAS ligand (tumor necrosis factor superfamily, member 6), abolishes diazoxide-induced cytoprotective effects against lethal anoxia [67], suggesting a critical role of miR-146a in MSC survival. Also miR-21, miR-23a, miR-210, and miR-503 promote survival of MSC during hypoxia and serum deprivation [23, 44]. MiR-21 may act by preserving mitochondrial function averting the mitochondrial apoptotic pathway [23], while miR-210 may act by targeting the apoptotic protein Caspase 8-associated protein 2 [44]. MiR-210 is in fact the best-known hypoxia-responsive miRNA [96]. Also miR-107, which targets the 3′UTR of programmed cell death-10, is upregulated during anoxia, while loss of function of miR-107 results in increased apoptosis [44]. Maintenance of MSCs in suspension leads to upregulation of miR-125b, which in turn represses p53, protecting cells against anoikis (death by detachment) [48].
miRNAs Involved in Paracrine Effects of MSCs
Recent studies have also focused on the function of miRNAs in the paracrine effects of MSCs. These miRNAs may either modulate expression of proteins secreted by MSCs or be contained in microvesicles and exosomes, to exert their regulatory function in target cells. The most important paracrine effects of MSCs include supporting hematopoietic stem cells (HSCs) in the bone marrow [97–99], promoting angiogenesis and stabilizing blood vessels [4, 100], and to modulating the immune system [101, 102].
Stromal-derived factor 1 (SDF-1α) is a chemokine crucial for both homing and retention of HSCs in the bone marrow [103, 104]. Recently, multiple miRNAs including miR-27b, miR-126, miR-146a-5p, and miR-886–3p have been shown to repress SDF-1α translation by directly binding to the 3′UTR of SDF-1α mRNA [14, 37, 85, 105–109]. Chen et al. found that expression of miR-27b (referred above as inhibitor of adipogenesis) is higher in adipose tissue-derived-MSCs (AT-MSCs) isolated from rats that tolerated orthotopic liver transplants, as compared to recipients that acutely rejected the transplant [36]. Reduction of miR-27b led to enhanced mRNA and protein levels of SDF-1α, while the proliferation of CD4+ T lymphocytes was reduced. This could explain the enhanced tolerance found in AT-MSCs in their model. Expression of miR-126 is restricted to endothelial cells as it locates within intron 7 of the gene EGF-like domain-containing protein 7 [110]. However, MSCs engineered to ectopically express miR-126 induced increased angiogenesis upon transplantation into infarcted heart [49]. The possible mechanisms include increased secretion of angiogenic factors [50], higher retention of the miR-126-expressing MSCs, and even differentiation of MSCs into an endothelial-like phenotype [111]. In a recent publication, Zhang et al. demonstrated that miR-126 is also aberrantly expressed in breast cancer cells [112]. Interestingly, they show that miR-126 and its complementary sequence miR-126* (both originating from the same precursor) target SDF-1α and as a result attraction of MSCs toward the tumor is inhibited, both in vitro and in vivo. Indeed, it had been shown that SDF-1α also acts as a chemokine for MSCs, which express the SDF-1α receptor CXCR4 [113]. Recently, miR-146a-5p that also targets SDF-1α was also shown to affect migration of MSCs [14], further supporting the autocrine and paracrine importance of miRNA-control of SDF-1α. IL-6 induces migration of prostate cancer-derived cells, which can be repressed by let-7 in MSCs. Consequently, repression of endogenous let-7 enhances the ability of MSCs to attract prostate cancer cell lines [18].
The immune-suppressive properties of MSCs both in vitro and in vivo (using a murine model of colitis) are reversed upon ectopic expression of miR-181a, putatively by targeting TGFBR1 and TGF-β receptor-associated protein 1 [70]. One of the best characterized immune suppressive factors released by MSCs is Prostaglandin E2 (PGE2) [101]. It is therefore particularly relevant that miR-146a-5p was found to directly target PGE2 synthase-2 [65]. In addition, miR-146a-5p mediates the increased expression of IL-8 in MSCs in response to TNF-α [66], further highlighting the importance of miR-146a-5p in immune modulatory properties of MSCs. miR-155 inhibits MSC-mediated immune suppression, as evidenced by an increase in the proliferation of T cells in vitro. In this context, miR-155 was found to target the 3′UTR of TAK1-binding protein 2, a regulator of iNOS (NFκB pathway) in MSCs [69].
In addition to regulating expression of endogenous proteins in MSCs, miRNA may also work through exosomes. Exosomes are 50–90 nm diameter vesicles released by MSCs and other cell types [114, 115] that contain proteins, mRNAs, and miRNAs, among other molecules and are involved in paracrine signaling between MSCs and nearby cells [116]. Around 60 different miR-NAs have been detected by microarray in MSC-derived exosomes. Commonly, the pre-miRNA form is 200–1,000-fold predominant over the mature species [115]. Exosomes have been shown to be functionally extremely important, as they provide therapeutic benefit against myocardial ischemic injury [117], acute kidney injury [118], stroke [119], and liver fibrosis [120], among others. Nevertheless, the exact mechanisms underlying exosomal-mediated tissue repair remain largely unknown and strongly demands further investigation.
Conclusion: Future Directions
Our knowledge of the function of miRNAs in MSCs has expanded tremendously within the last few years. However, many of these studies present caveats that will be important to address in order to thoroughly understand the underlying molecular mechanisms and find optimal therapeutic applications to this new knowledge: (a) discoveries in immortalized cell lines need to be validated in primary cultures. (b) Silencing endogenous levels of a miRNA is necessary in order to better understand the magnitude of a miRNA-associated effect; notice how miRNA inhibitors commonly exert milder effects as compared to over-expression (to supraphysiological concentrations) of miRNA mimics. (c) To study the function of miRNAs in relevant in vivo models is of highest urgency. (d) Investigators should confirm the specificity of their miRNA detection method, in particular within family members. For example, mature let-7a and let-7c differ in a single nucleotide. Do investigators accurately distinguish between these two let-7 members? (e) Commonly, potential miRNA targets are identified by computational prediction algorithms which typically predict hundreds to thousands of target genes for each miRNA [121]. However, only one or a few of these predicted targets are validated experimentally, typically using a luciferase-reporter system [122]. It is therefore important to perform gain-of-function assays (e.g., by cotransfecting with the target gene without 3′UTR) in order to demonstrate experimentally if the identified miRNA target is biologically relevant. In addition, thorough identification of miRNA targets, as applied to other cell types [123, 124] will be essential.
Progress in the study of the role of miRNAs in MSCs will facilitate the translation of this new field to a therapeutic level, where miRNA can be used as biomarkers or become direct pharmacological targets to treat MSC-associated diseases. A comprehensive understanding of the effects of miR-NAs in MSCs at both molecular and physiological levels is of utmost importance. Having addressed this, MSCs could be genetically modified to alter their miRNA profile, thus enhancing their therapeutic benefit.
Supplementary Material
Acknowledgments
The authors are supported by grant #R01GM099688 (Nolta) from the NIH Common fund transformative research projects program.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have no potential conflicts of interests to declare.
References
- 1.Caplan AI, Correa D. The MSC: An injury drugstore. Cell Stem Cell 2011;9:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horwitz EM, Le Blanc K, Dominici M et al. Clarification of the nomenclature for MSC: The International Society for Cellular Therapy position statement. Cytotherapy 2005;7:393–395. [DOI] [PubMed] [Google Scholar]
- 3.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci 2006;119:2204–2213. [DOI] [PubMed] [Google Scholar]
- 4.Crisan M, Yap S, Casteilla L et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008; 3:301–313. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 6.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science 2005;309: 1519–1524. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, Glasner ME, Yekta S et al. Vertebrate microRNA genes. Science 2003;299: 1540. [DOI] [PubMed] [Google Scholar]
- 8.Kong YW, Ferland-McCollough D, Jackson TJ et al. microRNAs in cancer management. Lancet Oncol 2012;13:e249–258. [DOI] [PubMed] [Google Scholar]
- 9.Oskowitz AZ, Lu J, Penfornis P et al. Human multipotent stromal cells from bone marrow and microRNA: Regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA 2008;105: 18372–18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakshmipathy U, Hart RP. Concise review: MicroRNA expression in multipotent mesenchymal stromal cells. Stem Cells 2008; 26:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh W, Sheng CT, Tan B et al. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics 2010;11(suppl 1):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaragosi LE, Wdziekonski B, Brigand KL et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol 2011;12:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh JY, Huang TS, Cheng SM et al. miR-146a-5p circuitry uncouples cell proliferation and migration, but not differentiation, in human mesenchymal stem cells. Nucleic Acids Res 2013;41:9753–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012;149:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Liu W, Hu C et al. MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis. Stem Cells 2011;29:1804–1816. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JF, Fu WM, He ML et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol 2011;8: 829–838. [DOI] [PubMed] [Google Scholar]
- 18.Sung SY, Liao CH, Wu HP et al. Loss of Let-7 microRNA upregulates IL-6 in bone marrow-derived mesenchymal stem cells triggering a reactive stromal response to prostate cancer. PLoS One 2013;8:e71637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Dong J, Zhang ZH et al. miR-10a restores human mesenchymal stem cell differentiation by repressing KLF4. J Cell Physiol 2013;228:2324–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F, Jing S, Ren T et al. microRNA-10b promotes the migration of mouse bone marrow-derived mesenchymal stem cells and downregulates the expression of E-cadherin. Mol Med Rep 2013;8:1084–1088. [DOI] [PubMed] [Google Scholar]
- 21.Liu JL, Jiang L, Lin QX et al. MicroRNA 16 enhances differentiation of human bone marrow mesenchymal stem cells in a cardiac niche toward myogenic phenotypes in vitro. Life Sci 2012;90:1020–1026. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Fan H, Zhao G et al. miR-16 inhibits the proliferation and angiogenesis-regulating potential of mesenchymal stem cells in severe pre-eclampsia. FEBS J 2012; 279:4510–4524. [DOI] [PubMed] [Google Scholar]
- 23.Nie Y, Han BM, Liu XB et al. Identification of MicroRNAs involved in hypoxia- and serum deprivation-induced apoptosis in mesenchymal stem cells. Int J Biol Sci 2011;7: 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei Y, Bian C, Li J et al. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem 2013;114:1374–1384. [DOI] [PubMed] [Google Scholar]
- 25.Yang N, Wang G, Hu C et al. TNF-alpha suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res 2013;28:559–573. [DOI] [PubMed] [Google Scholar]
- 26.Kim YJ, Hwang SJ, Bae YC et al. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009;27:3093–3102. [DOI] [PubMed] [Google Scholar]
- 27.Kim YJ, Hwang SH, Cho HH et al. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J Cell Physiol 2012;227:183–193. [DOI] [PubMed] [Google Scholar]
- 28.Hassan MQ, Gordon JA, Beloti MM et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24–2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci USA 2010;107:19879–19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ham O, Song BW, Lee SY et al. The role of microRNA-23b in the differentiation of MSC into chondrocyte by targeting protein kinase A signaling. Biomaterials 2012;33: 4500–4507. [DOI] [PubMed] [Google Scholar]
- 30.Sun F, Wang J, Pan Q et al. Characterization of function and regulation of miR-24–1 and miR-31. Biochem Biophys Res Commun 2009;380:660–665. [DOI] [PubMed] [Google Scholar]
- 31.Luzi E, Marini F, Sala SC et al. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res 2008;23:287–295. [DOI] [PubMed] [Google Scholar]
- 32.Schoolmeesters A, Eklund T, Leake D et al. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One 2009;4:e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SY, Kim AY, Lee HW et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun 2010;392: 323–328. [DOI] [PubMed] [Google Scholar]
- 34.Karbiener M, Fischer C, Nowitsch S et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPAR-gamma. Biochem Biophys Res Commun 2009;390:247–251. [DOI] [PubMed] [Google Scholar]
- 35.Lin Q, Gao Z, Alarcon RM et al. A role of miR-27 in the regulation of adipogenesis. FEBS J 2009;276:2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen KD, Goto S, Hsu LW et al. Identification of miR-27b as a novel signature from the mRNA profiles of adipose-derived mesenchymal stem cells involved in the tolerogenic response. PLoS One 2013;8:e60492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu MH, Li CZ, Hu CJ et al. microRNA-27b suppresses mouse MSC migration to the liver by targeting SDF-1alphain vitro. Biochem Biophys Res Commun 2012;421:389–395. [DOI] [PubMed] [Google Scholar]
- 38.Kapinas K, Kessler C, Ricks T et al. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 2010;285:25221–25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu T, Zhou H, Hong Y et al. miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem 2012;287:7503–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J, Yang T, Han J et al. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem 2011;112:1844–1856. [DOI] [PubMed] [Google Scholar]
- 41.Baglio SR, Devescovi V, Granchi D et al. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 2013;527:321–331. [DOI] [PubMed] [Google Scholar]
- 42.Zhu G, Chai J, Ma L et al. Downregulated microRNA-32 expression induced by high glucose inhibits cell cycle progression via PTEN upregulation and Akt inactivation in bone marrow-derived mesenchymal stem cells. Biochem Biophys Res Commun 2013; 433:526–531. [DOI] [PubMed] [Google Scholar]
- 43.Zeng Y, Qu X, Li H et al. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett 2012;586:2375–2381. [DOI] [PubMed] [Google Scholar]
- 44.Kim HW, Mallick F, Durrani S et al. Concomitant activation of miR-107/PDCD10 and Hypoxamir-210/Casp8ap2 and their role in cytoprotection during ischemic preconditioning of stem cells. Antioxid Redox Signal 2012; 17:1053–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai B, Li J, Wang J et al. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem Cells 2012;30:1746–1755. [DOI] [PubMed] [Google Scholar]
- 46.Laine SK, Alm JJ, Virtanen SP et al. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem 2012;113:2687–2695. [DOI] [PubMed] [Google Scholar]
- 47.Mizuno Y, Yagi K, Tokuzawa Y et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun 2008;368:267–272. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Cohen DM, Chen CS. miR-125b is an adhesion-regulated microRNA that protects mesenchymal stem cells from anoikis. Stem Cells 2012;30:956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JJ, Zhou SH. Mesenchymal stem cells overexpressing MiR-126 enhance ischemic angiogenesis via the AKT/ERK-related pathway. Cardiol J 2011;18:675–681. [DOI] [PubMed] [Google Scholar]
- 50.Huang F, Zhu X, Hu XQ et al. Mesenchymal stem cells modified with miR-126 release angiogenic factors and activate Notch ligand Delta-like-4, enhancing ischemic angiogenesis and cell survival. Int J Mol Med 2013;31: 484–492. [DOI] [PubMed] [Google Scholar]
- 51.Greco SJ, Rameshwar P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc Natl Acad Sci USA 2007;104:15484–15489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z, Hassan MQ, Volinia S et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA 2008;105:13906–13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Huang W, Wu Y et al. MicroRNA-193 pro-proliferation effects for bone mesenchymal stem cells after low-level laser irradiation treatment through inhibitor of growth family, member 5. Stem Cells Dev 2012;21:2508–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Bian C, Zhou H et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev 2011;20:259–267. [DOI] [PubMed] [Google Scholar]
- 55.Eskildsen T, Taipaleenmaki H, Stenvang J et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA 2011;108:6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuddenham L, Wheeler G, Ntounia-Fousara S et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 2006;580:4214–4217. [DOI] [PubMed] [Google Scholar]
- 57.Miyaki S, Nakasa T, Otsuki S et al. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum 2009;60:2723–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyaki S, Sato T, Inoue A et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev 2010;24:1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and-200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem 2009;284:19272–19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu W, Ye Y, Zhang W et al. miR1423p promotes osteoblast differentiation by modulating Wnt signaling. Mol Med Rep 2013;7: 689–693. [DOI] [PubMed] [Google Scholar]
- 61.Esau C, Kang X, Peralta E et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 2004;279:52361–52365. [DOI] [PubMed] [Google Scholar]
- 62.Lai VK, Ashraf M, Jiang S et al. MicroRNA-143 is a critical regulator of cell cycle activity in stem cells with co-overexpression of Akt and angiopoietin-1 via transcriptional regulation of Erk5/cyclin D1 signaling. Cell Cycle 2012;11:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J Biol Chem 2012;287:916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang B, Guo H, Zhang Y et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One 2011;6:e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matysiak M, Fortak-Michalska M, Szymanska B et al. MicroRNA-146a negatively regulates the immunoregulatory activity of bone marrow stem cells by targeting prostaglandin E2 synthase-2. J Immunol 2013;190: 5102–5109. [DOI] [PubMed] [Google Scholar]
- 66.Perng DW, Yang DM, Hsiao YH et al. miRNA-146a expression positively regulates tumor necrosis factor-alpha-induced interleukin-8 production in mesenchymal stem cells and differentiated lung epithelial-like cells. Tissue Eng Part A 2012;18:2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki Y, Kim HW, Ashraf M et al. Diazoxide potentiates mesenchymal stem cell survival via NF-kappaB-dependent miR-146a expression by targeting Fas. Am J Physiol Heart Circ Physiol 2010;299:H1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skarn M, Namlos HM, Noordhuis P et al. Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev 2012;21:873–883. [DOI] [PubMed] [Google Scholar]
- 69.Xu C, Ren G, Cao G et al. miR-155 regulates immune modulatory properties of mesenchymal stem cells by targeting TAK1-binding protein 2. J Biol Chem 2013;288: 11074–11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu L, Wang Y, Fan H et al. MicroRNA-181a regulates local immune balance by inhibiting proliferation and immunosuppressive properties of mesenchymal stem cells. Stem Cells 2012;30:1756–1770. [DOI] [PubMed] [Google Scholar]
- 71.Kim KM, Park SJ, Jung SH et al. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 2012;27:1669–1679. [DOI] [PubMed] [Google Scholar]
- 72.Xu J, Kang Y, Liao WM et al. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One 2012;7:e31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim YJ, Bae SW, Yu SS et al. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res 2009;24:816–825. [DOI] [PubMed] [Google Scholar]
- 74.Lin EA, Kong L, Bai XH et al. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem 2009; 284:11326–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J, Zhao L, Xing L et al. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010;28:357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inose H, Ochi H, Kimura A et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA 2009;106:20794–20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizuno Y, Tokuzawa Y, Ninomiya Y et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett 2009;583:2263–2268. [DOI] [PubMed] [Google Scholar]
- 78.Kim JH, Park SG, Song SY et al. Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell Death Dis 2013;4:e588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tome M, Lopez-Romero P, Albo C et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ 2011; 18:985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bork S, Horn P, Castoldi M et al. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369–5p and up-regulated by microRNA-371. J Cell Physiol 2011;226:2226–2234. [DOI] [PubMed] [Google Scholar]
- 81.Paik S, Jung HS, Lee S et al. miR-449a regulates the chondrogenesis of human mesenchymal stem cells through direct targeting of lymphoid enhancer-binding factor-1. Stem Cells Dev 2012;21:3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eguchi T, Watanabe K, Hara ES et al. OstemiR: A novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One 2013;8:e58796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guerit D, Philipot D, Chuchana P et al. Sox9-regulated miRNA-574–3p inhibits chondrogenic differentiation of mesenchymal stem cells. PLoS One 2013;8:e62582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao L, Yang X, Su X et al. Redundant miR-3077–5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis 2013;4:e600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pillai MM, Yang X, Balakrishnan I et al. MiR-886–3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS One 2010;5:e14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Xie H, Liu W et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest 2009;119:3666–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lagos-Quintana M, Rauhut R, Lendeckel W et al. Identification of novel genes coding for small expressed RNAs. Science 2001;294: 853–858. [DOI] [PubMed] [Google Scholar]
- 88.Hong JH, Hwang ES, McManus MT et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 2005;309:1074–1078. [DOI] [PubMed] [Google Scholar]
- 89.Jeon MJ, Kim JA, Kwon SH et al. Activation of peroxisome proliferator-activated receptor-gamma inhibits the Runx2-mediated transcription of osteocalcin in osteoblasts. J Biol Chem 2003;278:23270–23277. [DOI] [PubMed] [Google Scholar]
- 90.Chen X, Hausman BS, Luo G et al. Protein kinase inhibitor gamma reciprocally regulates osteoblast and adipocyte differentiation by downregulating leukemia inhibitory factor. Stem Cells 2013;31:2789–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol 2012;8:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet 2008;9: 115–128. [DOI] [PubMed] [Google Scholar]
- 93.Liu Z, Zhu J, Cao H et al. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol 2012;40:1553–1560. [DOI] [PubMed] [Google Scholar]
- 94.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007; 449:682–688. [DOI] [PubMed] [Google Scholar]
- 95.Sasayama T, Nishihara M, Kondoh T et al. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 2009;125:1407–1413. [DOI] [PubMed] [Google Scholar]
- 96.Huang X, Le QT, Giaccia AJ. MiR-210—Micromanager of the hypoxia pathway. Trends Mol Med 2010;16:230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendez-Ferrer S, Michurina TV, Ferraro F et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010;466:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ding L, Saunders TL, Enikolopov G et al. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012;481: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greenbaum A, Hsu YM, Day RB et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 2013;495:227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sacchetti B, Funari A, Michienzi S et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007;131:324–336. [DOI] [PubMed] [Google Scholar]
- 101.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol 2008;8:726–736. [DOI] [PubMed] [Google Scholar]
- 102.Le Blanc K, Mougiakakos D. Multipotent mesenchymal stromal cells and the innate immune system. Nat Rev Immunol 2012;12:383–396. [DOI] [PubMed] [Google Scholar]
- 103.Peled A, Petit I, Kollet O et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science 1999;283:845–848. [DOI] [PubMed] [Google Scholar]
- 104.Nagasawa T, Hirota S, Tachibana K et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 1996;382:635–638. [DOI] [PubMed] [Google Scholar]
- 105.Leone V, D’Angelo D, Rubio I et al. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab 2011;96:E1388–1398. [DOI] [PubMed] [Google Scholar]
- 106.van Solingen C, de Boer HC, Bijkerk R et al. MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1(1)/Lin(−) progenitor cells in ischaemia. Cardiovasc Res 2011;92:449–455. [DOI] [PubMed] [Google Scholar]
- 107.Zhang Y, Yang P, Sun T et al. miR-126 and miR-126(*) repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol 2013;15:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nicolas FE, Pais H, Schwach F et al. Experimental identification of microRNA-140 targets by silencing and overexpressing miR-140. RNA 2008;14:2513–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet 2011;43:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kuhnert F, Mancuso MR, Hampton J et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 2008;135:3989–3993. [DOI] [PubMed] [Google Scholar]
- 111.Huang F, Fang ZF, Hu XQ et al. Overexpression of miR-126 promotes the differentiation of mesenchymal stem cells toward endothelial cells via activation of PI3K/Akt and MAPK/ERK pathways and release of paracrine factors. Biol Chem 2013;394:1223–1233. [DOI] [PubMed] [Google Scholar]
- 112.Zhang Y, Yang P, Sun T et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol 2013;15:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ponte AL, Marais E, Gallay N et al. The in vitro migration capacity of human bone marrow mesenchymal stem cells: Comparison of chemokine and growth factor chemotactic activities. Stem Cells 2007;25:1737–1745. [DOI] [PubMed] [Google Scholar]
- 114.Fevrier B, Raposo G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol 2004;16: 415–421. [DOI] [PubMed] [Google Scholar]
- 115.Chen TS, Lai RC, Lee MM et al. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res 2010;38:215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Valadi H, Ekstrom K, Bossios A et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9: 654–659. [DOI] [PubMed] [Google Scholar]
- 117.Lai RC, Arslan F, Lee MM et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res 2010;4:214–222. [DOI] [PubMed] [Google Scholar]
- 118.Bruno S, Grange C, Deregibus MC et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol 2009;20:1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xin H, Li Y, Liu Z et al. Mir-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells 2013;31:2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li T, Yan Y, Wang B et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev 2013;22:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Friedman RC, Farh KK, Burge CB et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA 2003;9:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beitzinger M, Peters L, Zhu JY et al. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol 2007;4:76–84. [DOI] [PubMed] [Google Scholar]
- 124.Tan LP, Seinen E, Duns G et al. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res 2009;37:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anderson C, Catoe H, Werner R. Mir-206 regulates connexin-43 expression during skeletal muscle development. Nucleic Acids Res 2006;34:5863–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


