Abstract
A screening method based on differential staining of the wild type and exopolysaccharide-deficient mutants of Rhizobium (Sinorhizobium) meliloti by the lipophilic dye Sudan Black B is described. Mutants defective in the production of either succinoglycan or EPS II (galactoglucan) were isolated by using this method, which might also prove useful for isolating exopolysaccharide-defective derivatives of other bacteria.
Exopolysaccharide synthesis by Rhizobium (Sinorhizobium) meliloti is critical in order for it to invade the nodules it elicits on alfalfa (Medicago sativa) and establish a productive symbiosis (reviewed in references 16 and 21). Nodulation of leguminous plants by rhizobia is a complex developmental process that requires a series of interactions between the bacterium and its plant host (reviewed in references 10, 11, and 31). Failure of R. meliloti Rm1021 to synthesize either of two exopolysaccharides, succinoglycan and EPS II (galactoglucan), results in a very early block in the invasion process (5a, 9, 12, 20, 33).
Succinoglycan, which is also synthesized by several other bacteria (18), is composed of repeating octasaccharide subunits, each of which consists of a backbone of three glucoses and one galactose, a side chain of four glucoses, and 1-carboxyethylidene (“pyruvyl”), acetyl, and succinyl modifications in a ratio of approximately 1:1:1 (1, 6, 18, 19, 28). EPS II consists of a disaccharide subunit of glucose and galactose carrying acetyl and pyruvyl modifications (17). A specific size class of EPS II oligosaccharides has recently been shown to be required for the symbiotic role of EPS II (15). EPS II is a member of a biologically important class of bacterial exopolysaccharides termed galactoglucans, whose backbones consist of alternating glucose and galactose moieties but which differ with respect to the noncarbohydrate modifications they carry. For example, galactoglucans are produced by isolates of Pseudomonas putida and Pseudomonas fluorescens (27), fluorescent Pseudomonas species (8), Agrobacterium radiobacter and Achromobacter strains (35, 36), and Burkholderia cepacia (4). Galactoglucans have been postulated to function in biofilm formation and to serve as pathogenicity determinants.
The discovery that succinoglycan plays a critical role in R. meliloti-alfalfa symbiosis resulted directly from the observation that succinoglycan-producing colonies of R. meliloti fluoresce under UV light when grown on media containing the laundry whitener Calcofluor (20). This made it very easy to isolate mutants of R. meliloti that were defective in production of succinoglycan, even though there were no other observable changes in the morphology of the colonies. The ability of succinoglycan to bind Calcofluor and fluoresce under UV light greatly facilitated subsequent genetic analyses of succinoglycan’s biosynthesis and its symbiotic roles (9, 13, 14, 20, 23, 24), regulation (5, 7, 26), and degradation (34). However, a combination of DNA sequencing, fine-structure genetic analysis, and biochemical studies (13, 14, 29, 34) later revealed that certain classes of mutants affecting succinoglycan production were not easily detected on the basis of their Calcofluor fluorescence phenotypes. The EPS II exopolysaccharide, which is expressed in a symbiotically active form by the Rm1021 expR101 derivative (12, 15), does not bind Calcofluor, making genetic analysis of its synthesis and regulation more difficult than that of succinoglycan.
In this report, we describe a new screening method for exopolysaccharide-deficient mutants that does not depend on Calcofluor binding. This method can be used to detect mutants of R. meliloti that are defective in the production of either succinoglycan or EPS II and even allows the detection of mutants which are altered in succinoglycan production but cannot be detected on the basis of their Calcofluor fluorescence phenotypes. The idea for this method grew out of our efforts to use the lipophilic dye Sudan Black B (3) to analyze poly-β-hydroxybutyrate (PHB) synthesis by R. meliloti in a manner similar to that used to analyze PHB synthesis by Alcaligenes eutrophus (30). In the course of this work, we found that exopolysaccharide-producing strains of R. meliloti excluded the stain Sudan Black B and that exoB mutants, which synthesize neither succinoglycan nor EPS II (12, 20), readily incorporated this stain. On the basis of this observation, we set out to determine whether this differential uptake of Sudan Black B could be used to screen for mutants defective in succinoglycan production.
Culture and Sudan Black B staining conditions.
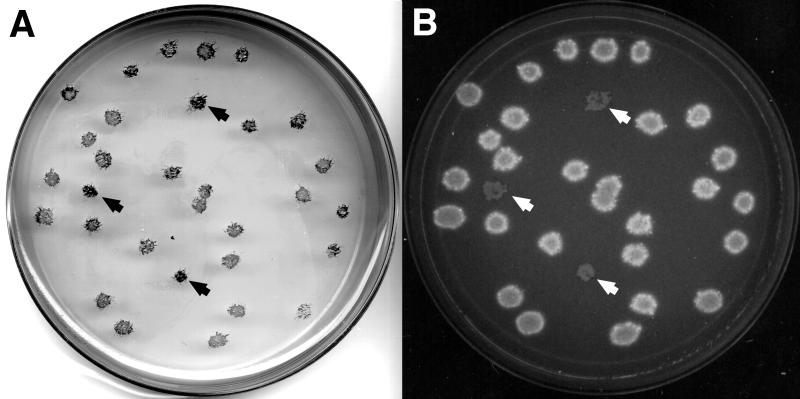
Culture media and growth conditions for R. meliloti were described previously (14, 15). All the R. meliloti strains described in this report are derivatives of Rm1021 (20). Other bacterial strains and plasmids have been described previously (20, 22). To screen for succinoglycan-producing strains, we used MM1 plates (25) with 0.4% fructose. For EPS II strains, we used the minimal medium described by Amemura et al. (2). A 0.02% solution of Sudan Black B (Sigma, St. Louis, Mo.) dissolved in 96% ethanol was used for the staining. All the bacteria were grown as single colonies on plates containing Luria-Bertani (LB) agar or M9 medium. They were then replica plated onto either MM1-fructose plates (succinoglycan-producing strains) or minimal-medium (2) plates (EPS II-producing strains). Approximately 8 ml of the 0.02% Sudan Black B solution was applied to each replica plate and allowed to remain undisturbed for about 10 min. The dye was then decanted, and the plates were gently rinsed by adding 10 ml of 100% ethanol and swirling for a few minutes. Colonies unable to incorporate the Sudan Black B stain appeared white, while colonies able to incorporate the dye appeared bluish black (Fig. 1).
FIG. 1.
Wild-type strain Rm1021 and exoY mutant strain Rm7210 (arrows) were grown on LB agar and then replica plated onto MM1-fructose medium and stained with Sudan Black B (A) or replica plated onto LB-Calcofluor agar and viewed under UV light (B).
Isolation of mutants altered in succinoglycan production by Sudan Black B staining.
We first checked mutants that have transposon insertions in the exoB, exoA, exoF, and exoY genes and are thus completely defective in succinoglycan synthesis (29). The strains were plated as single colonies on LB agar plates and replica plated onto MM1-fructose plates. After 36 h of growth at 30°C, the plates were stained and examined for Sudan Black B staining. As shown in Table 1, all the exo mutants tested stained with Sudan Black B, while the wild-type Rm1021 strain excluded the dye. The staining of the exo mutants appears to be due to Sudan Black B binding to PHB, since an exoY strain with a disruption in the gene encoding PHB synthase (32) was not stained by the dye (data not shown).
TABLE 1.
Sudan Black B staining of R. meliloti strains
| Strain | Relevant genotype | Stained by Sudan Black B | Reference or source |
|---|---|---|---|
| Rm1021 | Wild type | No | 20 |
| Rm7210 | exoY210::Tn5 | Yes | 20 |
| Rm6024 | exoB94::Tn5-233 | Yes | 12 |
| Rm8049 | exoA::Tn5 | Yes | 20 |
| Rm6089 | exoF::Tn5 | Yes | 20 |
| Rm8545 | exoO::Kan | Yes | 13 |
| Rm8285 | exoW28::Tn5 | Yes | 13 |
| Rm8600 | expR101 | No | 12 |
| Rm9000 | exoY210::Tn5 expR101 | No | 15 |
| Rm8603 | exoF55::Tn5-233 expR101 | No | 12 |
| Rm8601 | exoA32::Tn5-233 expR101 | No | 12 |
| Rm10000 | expA61::Tn5-Tp exoA125::Tn5 expR101 | Yes | 12 |
| Rm8064 | expA125::Tn5 | No | 12 |
| Rm10001 | expA::Tn5 exoA32::Tn5-233 | Yes | J. W. Reed and G. C. Walker |
Utilizing the same system, we screened about 7,000 Tn5-mutagenized Rm1021 bacteria for a loss in their ability to take up Sudan Black B. The mutagenesis was carried out as described previously (23). This screen yielded 30 colonies that stained black. All of these mutants were analyzed in a plasmid complementation assay in which triparental mating was used to introduce cosmids (pEX312, pEX154, or pD56) that carry part or all of the exo region (20, 23). Each mutant was complemented by one or more of these cosmids, indicating that all mutations mapped to the 28-kb exo region. Twenty-six of the mutants were dark under UV light when grown on LB plates containing Calcofluor, and the corresponding mutations were not mapped further since they corresponded to exoB or other well-characterized mutations that cause a Calcofluor-dark phenotype (13, 14, 23).
The remaining four mutants fluoresced under UV light on Calcofluor medium. Two were shown to be exoU mutants, and two were shown to be exoX mutants. exoU mutants are difficult to isolate on the basis of their Calcofluor phenotypes because exoU mutations cause so slight a change in the color of the fluorescing material that it is difficult to detect (13, 14, 23). Encouraged by this observation, we tested exoW and exoO mutants that also form Calcofluor-bright colonies and had not been detected on the basis of their Calcofluor phenotypes (13, 14, 23). Both of these mutants stained with the Sudan Black B dye, indicating that, even though the strains produce truncated forms of succinoglycan able to bind Calcofluor, they do not efficiently exclude the Sudan Black B dye. The exoX gene product is involved in regulating the production of succinoglycan and apparently acts at a posttranslational level (26, 37).
Isolation of mutants altered in EPS II production by Sudan Black B staining.
We then examined whether the Sudan Black B screening procedure could be used to detect R. meliloti mutants defective in the production of EPS II and obtained the results summarized in Table 1. The expR101 mutation permits Rm1021 to synthesize the otherwise cryptic exopolysaccharide EPS II in a symbiotically active form (12, 15). Strains with mutations in exoA or exoY are unable to synthesize succinoglycan and can be stained dark with Sudan Black B. Introduction of expR101 into these strains prevents them from staining with the dye. Adding an expA mutation to these strains abolishes the production of EPS II. This correlates with the ability of the triple mutants to incorporate Sudan Black B. Members of our laboratory subsequently found that additional mutations that interfere with EPS II production and are genetically unlinked to the exp gene cluster can also be detected by this Sudan Black B staining procedure (2a).
Potential applicability to other systems.
The Sudan Black B screening procedure described herein can be used to isolate mutants of R. meliloti that are defective in the production of either succinoglycan or EPS II. Mutants altered in succinoglycan production have previously been isolated on the basis of their Calcofluor fluorescence phenotypes, but until now mutants altered in EPS II production have been isolated only on the basis of changes in their mucoid phenotypes. Although the Sudan Black B method is less convenient than the Calcofluor-based method because of the requirement for replica plating, it may prove useful for analyzing the ability of other bacteria to produce exopolysaccharides which, like EPS II, do not bind Calcofluor. Even in the case of succinoglycan, the Sudan Black B method enabled the isolation of exo mutants that were not identified on the basis of their Calcofluor fluorescence phenotypes.
ACKNOWLEDGMENTS
We thank Brett Pellock for his careful reading of the manuscript.
This work was supported by U.S. Public Health Service grant GM31030 to G.C.W. J.E.G. was supported by a Post-Doctoral Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. M.L. carried out his research as part of the Undergraduate Research Opportunities Program at the Massachusetts Institute of Technology. L.B.W. was supported in part by a National Science Foundation predoctoral fellowship.
REFERENCES
- 1.Aman P, McNeil M, Franzen L-E, Darvill A G, Albersheim P. Structural elucidation, using HPLC-MS and GLC-MS, of the acidic exopolysaccharide secreted by Rhizobium meliloti strain Rm1021. Carbohydr Res. 1981;95:263–282. [Google Scholar]
- 2.Amemura A, Moori K, Harada T. Purification and properties of a specific, inducible β-glucanase, succinoglycan depolymerase, from Flavobacterium. Biochim Biophys Acta. 1974;334:398–409. [Google Scholar]
- 2a.Anderson, T., J. González, B. Pellock, and G. C. Walker. Unpublished results.
- 3.Burdon K L. Fatty material in bacteria and fungi revealed by staining dried, fixed slide preparations. J Bacteriol. 1946;52:665–678. doi: 10.1128/jb.52.6.665-678.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cérantola S, Marty N, Montrozier H. Structural studies of the acidic exopolysaccharide produced by a strain of Burkholderia cepacia isolated from cystic fibrosis. Carbohydr Res. 1996;285:59–67. doi: 10.1016/s0008-6215(96)90170-6. [DOI] [PubMed] [Google Scholar]
- 5.Cheng H-P, Walker G C. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J Bacteriol. 1998;180:20–26. doi: 10.1128/jb.180.1.20-26.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cheng H-P, Walker G C. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chouly C, Colquhuon I J, Jodelet A, York G, Walker G C. NMR studies of succinoglycan repeating-unit octasaccharides from Rhizobium meliloti and Agrobacterium radiobacter. Int J Biol Macromol. 1995;17:357–363. doi: 10.1016/0141-8130(96)81846-0. [DOI] [PubMed] [Google Scholar]
- 7.Doherty D, Leigh J A, Glazebrook J, Walker G C. Rhizobium meliloti mutants that overproduce the R. meliloti acidic Calcofluor-binding exopolysaccharide. J Bacteriol. 1988;170:4249–4256. doi: 10.1128/jb.170.9.4249-4256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fett W F, Wells J M, Cescutti P, Wijey C. Identification of exopolysaccharides produced by fluorescent pseudomonads associated with commercial mushroom (Agaricus bisporus) production. Appl Environ Microbiol. 1995;61:513–517. doi: 10.1128/aem.61.2.513-517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finan T M, Hirsch A M, Leigh J A, Johansen E, Kuldau G A, Deegan S, Walker G C, Signer E R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985;40:869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- 10.Fisher R F, Long S R. Rhizobium-plant signal exchange. Nature. 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 11.Geurts R, Franssen H. Signal transduction in Rhizobium-induced nodule formation. Plant Physiol (Rockville) 1996;112:447–453. doi: 10.1104/pp.112.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the Calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 13.Glucksmann M A, Reuber T L, Walker G C. Family of glycosyl transferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glucksmann M A, Reuber T L, Walker G C. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González J E, Reuhs B L, Walker G C. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González J E, York G M, Walker G C. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene. 1996;179:141–146. doi: 10.1016/s0378-1119(96)00322-8. [DOI] [PubMed] [Google Scholar]
- 17.Her G-R, Glazebrook J, Walker G C, Reinhold V N. Structural studies of a novel exopolysaccharide produced by a mutant of Rhizobium meliloti strain Rm1021. Carbohydr Res. 1990;198:305–312. doi: 10.1016/0008-6215(90)84300-j. [DOI] [PubMed] [Google Scholar]
- 18.Hisamatsu M, Abe J, Amemura A, Harada T. Structural elucidation of succinoglycan and related polysaccharides from Agrobacterium and Rhizobium by fragmentation with two special β-d-glycanases and methylation analysis. Agric Biol Chem. 1980;44:1049–1055. [Google Scholar]
- 19.Jansson P-E, Kenne L, Lindberg B, Ljunggren H, Ruden U, Svensson S. Demonstration of an octasaccharide repeating unit in the extracellular polysaccharide of R. meliloti by sequential degradation. J Am Chem Soc. 1977;99:3812–3815. doi: 10.1021/ja00453a049. [DOI] [PubMed] [Google Scholar]
- 20.Leigh J A, Signer E R, Walker G C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 22.Long S, McCune S, Walker G C. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J Bacteriol. 1988;170:4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long S, Reed J W, Himawan J, Walker G C. Genetic analysis of a cluster of genes required for synthesis of the Calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol. 1988;170:4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller P, Hynes M, Kapp D, Niehaus K, Pühler A. Two classes of Rhizobium meliloti infection mutants differ in exopolysaccharide production and in coinoculation properties with nodulation mutants. Mol Gen Genet. 1988;211:17–26. [Google Scholar]
- 25.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16: identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 26.Reed J W, Capage M, Walker G C. Rhizobium meliloti exoG and exoJ mutations affect the ExoX-ExoY system for modulation of exopolysaccharide production. J Bacteriol. 1991;173:3776–3788. doi: 10.1128/jb.173.12.3776-3788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed R R, Costerton J W. Purification and characterization of adhesive exopolysaccharides from Pseudomonas putida and Pseudomonas fluorescens. Can J Microbiol. 1987;33:1080–1090. doi: 10.1139/m87-189. [DOI] [PubMed] [Google Scholar]
- 28.Reinhold B B, Chan S Y, Reuber T L, Marra A, Walker G C, Reinhold V N. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J Bacteriol. 1994;176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuber T L, Walker G C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 30.Steinbüchel A, Schlegel H G. Physiology and molecular genetics of poly(β-hydroxyalkanoic acid) synthesis in Alcaligenes eutrophus. Mol Microbiol. 1991;5:535–542. doi: 10.1111/j.1365-2958.1991.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 31.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willis L B, Walker G C. The phbC (poly-β-hydroxybutyrate synthase) gene of Rhizobium (Sinorhizobium) meliloti and characterization of phbC mutants. Can J Microbiol. 1998;44:554–564. doi: 10.1139/w98-033. [DOI] [PubMed] [Google Scholar]
- 33.Yang C, Signer E R, Hirsch A M. Nodules initiated by Rhizobium meliloti exopolysaccharide mutants lack a discrete, persistent nodule meristem. Plant Physiol (Rockville) 1992;98:143–151. doi: 10.1104/pp.98.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.York G M, Walker G C. The Rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low-molecular-weight succinoglycan. Mol Microbiol. 1997;25:117–134. doi: 10.1046/j.1365-2958.1997.4481804.x. [DOI] [PubMed] [Google Scholar]
- 35.Zevenhuizen L P T M. Exocellular pyruvate containing galactoglucan of Acromobacter spp. Arch Microbiol. 1974;96:75–82. [PubMed] [Google Scholar]
- 36.Zevenhuizen L P T M. Diversity of polysaccharide synthesis within a restricted group of bacteria. In: Crescenzi V, Dea I C M, Paoletti S, Stivala S S, Sutherland I, editors. Biomedical and biotechnological advances in industrial polysaccharides. New York, N.Y: Gordon and Breach Science Publishers; 1989. pp. 301–311. [Google Scholar]
- 37.Zhan H, Leigh J A. Two genes that regulate exopolysaccharide production in Rhizobium meliloti. J Bacteriol. 1990;172:5254–5259. doi: 10.1128/jb.172.9.5254-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]