Abstract
The plasmid-free Lactococcus lactis subsp. cremoris MG1614 is highly phage sensitive and lacks lactose fermenting ability (Lac) and primary casein degrading ability (Prt). Food grade gene transfer systems were used to sequentially superimpose different phage defense systems on this background, resulting in a gradual increase in resistance to bacteriophage in the derivatives. pLP712, encoding Lac and Prt, was then transferred to one of these hosts, into which plasmids encoding adsorption inhibition, restriction modification, and abortive infection had already been introduced. This resulted in a phage-resistant strain which was successfully used as a single-strain starter for cheddar cheese manufacture under industrial conditions.
Bacteriophage infection is the single most important cause of slow or inhibited acid production by lactococci in commercial dairy fermentations (5, 8, 17). As a result of this constant threat of phage infection, cheese manufacturers nowadays routinely employ highly phage-resistant strains in defined starter systems. In reality, there is a very limited number of such phage-resistant strains which possess traits rendering them suitable for prolonged cheese manufacture (4). In response to this predicament, there has been an intensive worldwide research effort focusing on the molecular analysis of infecting bacteriophage and the phage defense systems which occur naturally in lactococci. These include phage adsorption inhibition (Ads), DNA injection blocking, restriction and modification (R/M), and abortive infection (Abi), and their study has been extensively reviewed in recent years (5, 8, 17). In addition, during the past 10 years, considerable effort has been directed at transferring the different phage resistance mechanisms to industrially used cheese-making lactococci in order to improve their performance in the presence of industrial bacteriophage (2, 13, 16, 22, 24). At present, the conjugal transfer of naturally occurring phage resistance plasmids is the most widely accepted approach for precise genetic improvement of phage resistance in starter cultures destined for consumer products (17). It has also been shown that stacking multiple phage resistance systems in a single strain can significantly enhance the level of resistance mediated over and above that conferred by a single system (3, 7, 15, 16, 19, 25–27). In the majority of cases where stacking has been achieved, recombinant DNA technology was used. However in some instances, the natural food grade approach of bacterial conjugation was employed to stack two Abi systems (3) and to combine R/M with Abi (16, 27). There are limits to the extent to which bacterial conjugation can be used to introduce and artificially stack phage resistance systems within a single strain, principally because of the widespread paucity of easily selectable markers for phage resistance plasmids, leading to difficulty in recognizing genuine phage-resistant transconjugants which may arise (10, 22, 24). In this study, a plasmid-free Lactococcus lactis strain was employed as a prototype strain, and the effect of introducing three distinct potent phage resistance mechanisms, R/M, Abi, and Ads, singly and in combinations of two and three, on the replication of all available phages which were lytic for the plasmid-free recipient was assessed. The phages included four members of the c2/c6A species and two of the 936/P008 species according to the classification system of Jarvis et al. (14). All strains were constructed using nonrecombinant techniques in order to ensure food grade status. Lactose fermenting ability and proteinase activity were then superimposed on a derivative strain harboring all three phage resistance mechanisms, and the resulting culture was assessed for its performance under cheese-making conditions in the presence of high levels of lytic phages.
Construction of phage-resistant derivatives of L. lactis MG1614.
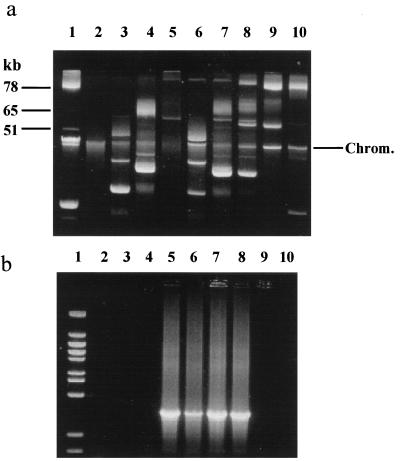
The bacterial conjugation system described by McKay and Baldwin (18) was used for the sequential delivery of plasmids to target strains (Table 1). Construction of the MG1614-derived hosts DPC3343 and DPC3290, containing pAH82 and pAH90, respectively, has been described previously (11). Transfer of the 60.2-kb conjugative plasmid pMRC01, encoding phage resistance and production of or immunity to lacticin 3147, was performed as described previously (2). Putative pMRC01-containing transconjugants were selected on lactose indicator agar plates containing the bacteriocin lacticin 3147 and the antibiotics streptomycin (500 μg/ml) and rifampin (100 μg/ml). Introduction of pLP712 (Lac Prt) from strain NCDO712 (9) was achieved on the basis of selection for acid-producing colonies on lactose indicator agar-lacticin 3147 selective plates. The plasmid isolation procedure of Anderson and McKay (1) was used to confirm the plasmid profiles of transconjugant lactococci, which were analyzed following electrophoresis as described previously (2). Plasmid sizes were confirmed by comparing their mobilities with those of the larger plasmids present in L. lactis subsp. lactis biovar diacetylactis DRC3 (78, 65 and 51 kb). Analysis of the plasmid DNA complements confirmed the presence of pAH82 (20.75 kb), pAH90 (26.75 kb), pMRC01 (60.3 kb), and pLP712 (50 kb) and combinations thereof in MG1614 derivatives (Fig. 1a). Presence of pMRC01 in transconjugants, where it was difficult to visualize, was confirmed by PCR using pMRC01-specific primers 5′-TCTGA TAGGA TCGCC TTAAG-3′ and 5′-TGTTG GTTGA AAAAT TGTTG-3′ (6) (Fig. 1b).
TABLE 1.
Bacterial strains used in this study
| L. lactis strain | Plasmid content | Relevant genotype | Refer-ence |
|---|---|---|---|
| NCDO712 | Five plasmids including pLP712 | lac prt | 9 |
| MG1614 | None | 9 | |
| DPC5013a | pLP712 | lac prt | This study |
| DPC3343a | pAH82 | r/m | 11 |
| DPC3290a | pAH90 | r/mads | 11 |
| DPC5012a | pMRC01 | abi | 2 |
| DPC5002a | pAH82 pMRC01 | r/mabi | This study |
| DPC5001a | pAH90 pMRC01 | r/mads abi | This study |
| DPC5000a | pAH90 pMRC01 pLP712 | r/mads abi lac prt | This study |
a Derived from strain MG1614. DPC, Dairy Products Research Centre, Teagasc, Moorepark, Fermoy, Co. Cork, Ireland. NCDO, National Collection of Dairy Organisms, Institute of Food Research, Reading, United Kingdom.
FIG. 1.
(a) Plasmid profiles of transconjugant lactococcal strains demonstrating the sequential buildup of phage resistance systems leading to the construction of the strain DPC5000. Lanes: 1, molecular size markers from DRC3 (18); 2, MG1614; 3, DPC3343(pAH82); 4, DPC3290(pAH90); 5, DPC5012(pMRC01); 6, DPC5002(pMRC01, pAH82); 7, DPC5001(pMRC01, pAH90); 8, DPC5000(pMRC01, pAH90, pLP712); 9, DPC5013(pLP712); 10, NCDO712 (contains pLP712 and other native plasmids). The position of the chromosomal band (Chrom.) is indicated. (b) Gel obtained by PCR using pMRC01-specific primers indicating the presence of the pMRC01 plasmid. Lanes are the same as those in panel a except with Lambda BstEII replacing DRC3 as the molecular mass standard (lane 1).
Phage resistance exhibited by MG1614 and derivatives.
Plaque assays were performed by the method of Terzaghi and Sandine (28) as modified by Coakley et al. (2). The efficiency of plaquing (EOP) of a phage on a particular host was determined by dividing the phage titer on the test strain by the titer on the homologous phage-sensitive host. The phage resistance of the various strains (evaluated by means of plaque assays) is presented in Table 2. The plasmid pAH82 harbors an R/M system effective against small isometric headed phages of the 936/P008 group, 712 and sk1. It reduced the EOP of phage 712 by 2 log cycles and that of phage sk1 by 1 log cycle. pAH82 appeared to have no significant effect on any of the four representatives of the c2/c6A group of phages. In contrast, pAH90 encodes an R/M system that gave a higher order of protection against phage 712 but not against sk1, reducing the EOP of the former by 3 log cycles while the additional Ads mechanism encoded by this plasmid gave complete resistance against all the c2/c6A phages (no plaque formation). The molecular nature of these two systems is at present unknown. The Abi system encoded by pMRC01, which is distinct from the previously classified Abi systems on the basis of nucleotide sequence data (6), rendered strains completely resistant to the two 936/P008 phages (712 and sk1), provided a sixfold reduction in c2 plaque size and a fourfold reduction for phage ml3, and prevented plaque formation by phages stl1 and eb1. Superimposing pMRC01 on a strain containing pAH82 provided a double protection against the two 936/P008 phages, although this gave a phenotypic resistance pattern similar to that of the strain possessing pMRC01 alone. Total resistance to all six phages was achieved in the strain containing pMRC01 and pAH90, a background in which all three resistance mechanisms (R/M, Ads, and Abi) are present. This ensures two resistance mechanisms active against each phage species, i.e., R/M and Abi against the 936/P008 group and Abi and Ads against the c2/c6A group. L. lactis DPC5000 contained both of these plasmids in addition to the Lac/Prt plasmid pLP712 and was therefore completely impervious to both phage groups. We are unaware of representatives of other phage groups (14) which are lytic for strain MG1614.
TABLE 2.
Plaque assays on transconjugants with six lytic phagesa
| L. lactis strain | Plasmid content | Bacterio-phage | No. of plaques | Plaque size (mm) | EOP |
|---|---|---|---|---|---|
| MG1614 | None | 712 | 1.2 × 109 | 0.6 | 1.0 |
| sk1 | 7.5 × 108 | 0.6 | 1.0 | ||
| eb1 | 1.3 × 109 | 0.7 | 1.0 | ||
| c2 | 3.2 × 109 | 3.5 | 1.0 | ||
| ml3 | 1.0 × 1010 | 3.6 | 1.0 | ||
| stl1 | 6.8 × 108 | 2.7 | 1.0 | ||
| DPC5013 | pLP712 | 712 | 1.3 × 109 | 0.6 | 1.1 |
| sk1 | 7.5 × 108 | 0.65 | 1.0 | ||
| eb1 | 1.5 × 109 | 0.65 | 1.2 | ||
| c2 | 3.9 × 109 | 3.4 | 1.2 | ||
| ml3 | 9.6 × 109 | 3.8 | 1.0 | ||
| stl1 | 6.1 × 108 | 2.6 | 0.9 | ||
| DPC3343 | pAH82 | 712 | 4.0 × 107 | 0.55 | 3.3 × 10−2 |
| sk1 | 8.0 × 107 | 0.55 | 1.1 × 10−1 | ||
| eb1 | 1.6 × 109 | 0.7 | 1.2 | ||
| c2 | 2.7 × 109 | 3.2 | 0.8 | ||
| ml3 | 1.2 × 1010 | 3.4 | 1.2 | ||
| stl1 | 7.3 × 108 | 2.4 | 1.1 | ||
| DPC3290 | pAH90 | 712 | 5.0 × 106 | 0.6 | 4.2 × 10−3 |
| sk1 | 8.4 × 107 | 0.65 | 1.1 × 10−1 | ||
| eb1 | — | — | — | ||
| c2 | — | — | — | ||
| ml3 | — | — | — | ||
| stl1 | — | — | — | ||
| DPC5012 | pMRC01 | 712 | — | — | — |
| sk1 | — | — | — | ||
| eb1 | — | — | — | ||
| c2 | 2.8 × 109 | 0.6 | 0.9 | ||
| ml3 | 9.8 × 109 | 0.6 | 1.0 | ||
| stl1 | — | — | — | ||
| DPC5002 | pMRC01, pAH82 | 712 | — | — | — |
| sk1 | — | — | — | ||
| eb1 | — | — | — | ||
| c2 | 3.4 × 109 | 0.65 | 1.1 | ||
| ml3 | 9.6 × 109 | 0.6 | 1.0 | ||
| stl1 | — | — | — | ||
| DPC5001 | pMRC01, pAH90 | 712 | — | — | — |
| sk1 | — | — | — | ||
| eb1 | — | — | — | ||
| c2 | — | — | — | ||
| ml3 | — | — | — | ||
| stl1 | — | — | — | ||
| DPC5000 | pMRC01, pAH90, pLP712 | 712 | — | — | — |
| sk1 | — | — | — | ||
| eb1 | — | — | — | ||
| c2 | — | — | — | ||
| ml3 | — | — | — | ||
| stl1 | — | — | — |
a Plaque data are the averages of three separate experiments. Dashes indicate that no plaques formed. Phages 712 and sk1 belong to the 936/P008 group, and eb1, c2, ml3, and stl1 belong to the c2/c6A group.
Characterization of transconjugant strains.
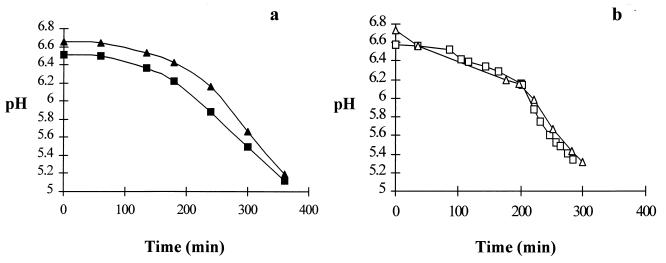
All of the strains in this study (Table 1) were assayed for bacteriocin production by means of the well assay technique as described by Parente and Hill (20) and modified by Ryan et al. (23) by using L. lactis HP as an indicator strain for further confirmation of the transfer of pMRC01. A large zone of inhibition comparable to that produced by the positive control strain DPC5012 was evident around DPC5000, DPC5001, and DPC5002, while no inhibition was associated with the strains MG1614, DPC5013, DPC3343, and DPC3290, as expected. The transconjugant strain DPC5000 was subjected to a starter activity test (Fig. 2a) based on the method of Harrington and Hill (10) in both 10% reconstituted skim milk (RSM) and pasteurized milk, where it performed optimally, reducing the pH to 5.2 after 6 h.
FIG. 2.
pH development in 10% RSM (■) and pasteurized milk (▴) during incubation of DPC5000 over the cheese temperature profile (a) and during an industrial-scale cheddar cheese trial using DPC5000 (□) and a commercial cheddar cheese starter (▵) (b).
Two phage cocktails were used to determine the strength of the phage protection mediated by the three phage resistance systems in strain DPC5000. One was a defined cocktail containing all the available phages which are lytic for strain MG1614, namely phages 712 and sk1 (of the 936/P008 group) and c2, ml3, eb1, and stl1 (of the c2/c6A group). These phages were each present in the cocktail at a minimum titer of 108 PFU/ml. The other phage groups described by Jarvis et al. (14) were not represented, as none has been reported to target MG1614. The other cocktail was undefined, and its source is a number of cheddar cheese plants in Europe and North America and is representative of the current phage situation in a variety of cheddar cheese factories that use defined starter cultures. The phage resistance of L. lactis DPC5000 (Lac+ Prt+ Abi+ R/M+ Ads+) was compared with that of L. lactis DPC5013 (Lac+ Prt+ Abi− R/M− Ads−) by using the method of Heap and Lawrence (12), by which both strains were exposed successively to phage-whey over a period of 10 days (Table 3). Strain DPC5013 failed on day 1 with both phage cocktails, while DPC5000 survived the entire 10-day trial, maintaining maximum activity throughout and reducing the milk pH to 5.08 in the presence of phage on the tenth day. The stability of pMRC01, pAH90, and pLP712 was confirmed by growing strain DPC5000 through 75 generations in GM17 broth at 30°C, after which the culture was plated and plasmid profiles were obtained for 110 isolated colonies. None of the isolates had lost any of the three plasmids.
TABLE 3.
Result of Heap and Lawrence test
| L. lactis strain and phage treatment | Final pH
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | Day 10 | |
| DPC5013 | ||||||||||
| None (phage absent) | 5.19 | 5.24 | 5.14 | 5.18 | 5.20 | 5.16 | 5.21 | 5.15 | 5.16 | 5.18 |
| Defined phage cocktaila | 6.30 | 6.38 | 6.32 | 6.28 | 6.32 | 6.26 | 6.30 | 6.26 | 6.28 | 6.30 |
| Industrial phage cocktailb | 6.31 | 6.41 | 6.30 | 6.31 | 6.36 | 6.18 | 6.18 | 6.36 | 6.28 | 6.22 |
| DPC5000 | ||||||||||
| None (phage absent) | 5.20 | 5.21 | 5.10 | 5.21 | 5.24 | 5.17 | 5.17 | 5.16 | 5.18 | 5.16 |
| Defined phage cocktaila | 5.19 | 5.20 | 5.15 | 5.16 | 5.23 | 5.08 | 5.18 | 5.14 | 5.16 | 5.15 |
| Industrial phage cocktailb | 5.14 | 5.18 | 5.07 | 5.07 | 5.15 | 5.02 | 5.11 | 5.08 | 5.05 | 5.08 |
a The defined phage cocktail was composed of phages c2, ml3, stl1, 712, sk1, and eb1, each with a minimum titer of 1.0 × 108 PFU/ml.
b The industrial phage cocktail was derived from industries in Europe and the United States which employ cheddar cheese starters.
Cheddar cheese manufacture with the single-strain starter L. lactis DPC5000.
L. lactis DPC5000 was employed as a single starter culture for cheddar cheese manufacture. The bulk starter was composed of 10% RSM which had been treated at 90°C for 30 min and cooled to 21°C before inoculation with L. lactis DPC5000. Following overnight growth at 21°C, the bulk starter was added (1.5%) to the cheese vat. A commercially available cheddar cheese starter was prepared and used under identical conditions as a control. Cheese trials were performed as described by Ryan et al. (23). Assessment of pH development in the fermentation (Fig. 2b) indicated that DPC5000 possessed the essential acid-producing capability under manufacturing conditions (4). Cheese compositional analysis 1 week after the fermentation indicated that the DPC5000 cheese had a pH of 5.13, a fat composition of 30.75%, and a salt in moisture ratio of 4.49. This compared very well with the control cheese, which had a pH of 5.16, 31.5% fat, and a salt in moisture ratio of 4.19. This cheese composition data is in agreement with desirable values reported by Pearce and Gilles (21). In addition, there was no difference in terms of yield between the two cheeses.
Discussion.
It has long been recognized that the conjugative properties of many phage resistance plasmids can be exploited to develop cheese starter cultures for industry with improved phage resistance (18, 24). The recognized approach is to take phage-sensitive industrial starter strains and render them less sensitive to phage attack by introducing conjugative phage resistance plasmids. This strategy can be difficult to implement (2, 4) and is critically affected by the choice of recipient strain, the presence or absence of a suitable selectable marker, plasmid incompatibility, and variation in the conjugation methods employed. While the artificial stacking of more than one phage resistance mechanism leading to increased levels of phage insensitivity has been documented (3, 7, 15, 16, 19, 25, 26, 27), in the majority of cases recombinant methodologies were used. Food grade bacterial conjugation was used to stack different Abi and R/M systems by using pTR2030, pTN20, pTRK11, and pTRK68 in different combinations (16, 27). Similar methodologies were used by Coffey et al. (3) to combine two phenotypically similar Abi systems. To date, no stacking approach, whether recombinant or food grade, has included an Ads system. This is significant since Ads is the first host defense encountered by lytic phage. The sequential stacking of the three distinct phage resistance systems Ads, R/M, and Abi by natural means, the superimposition of milk fermenting ability, and the subsequent use of the resulting strain for cheese manufacture therefore represent a significant advance in deliberate strain construction for food use. The initial goal of this study was to compare and contrast the phage resistances encoded by pAH82, pAH90, and pMRC01 singly and in various combinations by using a single plasmid-free L. lactis strain as a background. Having an isogenic host background for all plasmid combinations allows direct comparisons to be made and permits a situation described by Klaenhammer’s group (7, 27) whereby the derivative strains may be rotated in a cheese plant without affecting the cheese manufacturing process or the flavor of the product. The combinations of phage resistance mechanisms which act at three different points of the phage lytic cycle provide excellent protection to strains against phage attack (16). The strain designated DPC5000, harboring the three mechanisms, was derived entirely by food grade approaches and was shown to be highly effective against all the available phages which are lytic for the plasmid-free MG1614. Strain DPC5000 was shown to be stable, possessed acceptable fermentation characteristics, and on the basis of tests described by Heap and Lawrence (12) could be predicted to withstand phage attack in commercial circumstances for extended periods of time.
Acknowledgments
This research was partly funded by grant aid under the Food Sub-Programme of the Operational Programme for Industrial Development, which is administered by the Irish Department of Agriculture, Food and Forestry and supported by national and EU funds. D.O.S. was supported by a Teagasc Walsh fellowship.
We thank Horst Neve for electron microscopic analysis of phage eb1.
REFERENCES
- 1.Anderson D G, McKay L L. A simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coakley M, Fitzgerald G F, Ross R P. Application and evaluation of the phage resistance- and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl Environ Microbiol. 1997;63:1434–1440. doi: 10.1128/aem.63.4.1434-1440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffey A G, Fitzgerald G F, Daly C. Identification and characterisation of a plasmid encoding abortive infection from Lactococcus lactis subsp. lactis UC811. Neth Milk Dairy J. 1989;43:229–244. [Google Scholar]
- 4.Coffey A G, Coakley M, McGarry A, Fitzgerald G F, Ross R P. Increasing phage resistance of cheese starters: a case study using Lactococcus lactis DPC4268. Lett Appl Microbiol. 1997;26:51–55. [Google Scholar]
- 5.Daly C, Fitzgerald G F, Davis R. Biotechnology of lactic acid bacteria with special reference to bacteriophage resistance. Antonie Leeuwenhoek. 1996;70:99–111. doi: 10.1007/BF00395928. [DOI] [PubMed] [Google Scholar]
- 6.Dougherty B A, Hill C, Weldman J F, Richardson D R, Venter J C, Ross R P. Sequence and analysis of the 60 kb conjugative bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol. 1998;29:1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x. [DOI] [PubMed] [Google Scholar]
- 7.Durmaz E, Klaenhammer T R. A starter culture rotation strategy incorporating paired restriction/modification and abortive infection bacteriophage defenses in a single Lactococcus lactis strain. Appl Environ Microbiol. 1995;61:1266–1273. doi: 10.1128/aem.61.4.1266-1273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garvey P, van Sinderen D, Twomey D P, Hill C, Fitzgerald G F. Molecular genetics of bacteriophage and natural phage defence systems in the genus Lactococcus. Int Dairy J. 1995;5:905–947. [Google Scholar]
- 9.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrington A, Hill C. Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis 425A by using the conjugal plasmid pNP40. Appl Environ Microbiol. 1991;57:3405–3409. doi: 10.1128/aem.57.12.3405-3409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington A, Hill C. Plasmid involvement in the formation of a spontaneous bacteriophage insensitive mutant of Lactococcus lactis. FEMS Microbiol Lett. 1992;96:135–142. doi: 10.1016/0378-1097(92)90393-3. [DOI] [PubMed] [Google Scholar]
- 12.Heap H A, Lawrence R C. The selection of starter strains for cheesemaking. N Z J Dairy Sci Technol. 1976;2:16–20. [Google Scholar]
- 13.Jarvis A W, Heap H H, Limsowtin G K Y. Resistance against industrial bacteriophages conferred by plasmid pAJ1106 and related plasmids. Appl Environ Microbiol. 1989;55:1537–1543. doi: 10.1128/aem.55.6.1537-1543.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis A W, Fitzgerald G F, Mata M, Mercenier A, Neve H, Powell I B, Ronda C, Saxelin M, Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32:2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- 15.Josephsen J, Klaenhammer T R. Stacking of three different restriction and modification systems in Lactococcus lactis by cotransformation. Plasmid. 1990;23:71–75. doi: 10.1016/0147-619x(90)90046-f. [DOI] [PubMed] [Google Scholar]
- 16.Klaenhammer T R. Genetic characterisation of multiple mechanisms of phage defence from a prototype phage-insensitive strain Lactococcus lactis ME2. J Dairy Sci. 1989;72:3429–3442. [Google Scholar]
- 17.Klaenhammer T R, Fitzgerald G F. Bacteriophage and bacteriophage resistance. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Chapman and Hall; 1994. pp. 106–168. [Google Scholar]
- 18.McKay L L, Baldwin K A. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl Environ Microbiol. 1984;46:68–74. doi: 10.1128/aem.47.1.68-74.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLandsborough L A, Sechaud L, McKay L L. Synergistic effects of abiE or abiF from pNP40 when cloned in combination with abiD from pBF61. J Dairy Sci. 1988;81:362–368. [Google Scholar]
- 20.Parente E, Hill C. A comparison of factors affecting the production of two bacteriocins from lactic acid bacteria. J Appl Bacteriol. 1992;73:290–298. [Google Scholar]
- 21.Pearce K N, Gilles J. Comparison and grade of cheddar cheese manufactured over three seasons. N Z J Dairy Sci Technol. 1979;14:63–71. [Google Scholar]
- 22.Powell I B, Romano G M, Hillier A J, Davidson B E. Genetic enhancement of phage resistance in a commercial cheese starter. Aust J Dairy Technol. 1994;49:30–33. [Google Scholar]
- 23.Ryan M P, Rea M C, Hill C, Ross R P. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol. 1996;62:612–619. doi: 10.1128/aem.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders M E, Leonhard P J, Sing W D, Klaenhammer T R. Conjugal strategy for construction of fast acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986;52:1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schouler C, Gautier M, Ehrlich S D, Chopin M C. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol Microbiol. 1998;28:169–178. doi: 10.1046/j.1365-2958.1998.00787.x. [DOI] [PubMed] [Google Scholar]
- 26.Sing W D, Klaenhammer T R. Characterisation of restriction-modification plasmids from Lactococcus lactis ssp. cremoris and their effects when combined with pTR2030. J Dairy Sci. 1991;74:1133–1144. [Google Scholar]
- 27.Sing W D, Klaenhammer T R. A strategy for rotation of different bacteriophage defenses in a lactococcal single-strain starter culture system. Appl Environ Microbiol. 1993;59:365–372. doi: 10.1128/aem.59.2.365-372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]