Abstract
Epicidin 280 is a novel type A lantibiotic produced by Staphylococcus epidermidis BN 280. During C18 reverse-phase high-performance liquid chromatography two epicidin 280 peaks were obtained; the two compounds had molecular masses of 3,133 ± 1.5 and 3,136 ± 1.5 Da, comparable antibiotic activities, and identical amino acid compositions. Amino acid sequence analysis revealed that epicidin 280 exhibits 75% similarity to Pep5. The strains that produce epicidin 280 and Pep5 exhibit cross-immunity, indicating that the immunity peptides cross-function in antagonization of both lantibiotics. The complete epicidin 280 gene cluster was cloned and was found to comprise at least five open reading frames (eciI, eciA, eciP, eciB, and eciC, in that order). The proteins encoded by these open reading frames exhibit significant sequence similarity to the biosynthetic proteins of the Pep5 operon of Staphylococcus epidermidis 5. A gene for an ABC transporter, which is present in the Pep5 gene cluster but is necessary only for high yields (G. Bierbaum, M. Reis, C. Szekat, and H.-G. Sahl, Appl. Environ. Microbiol. 60:4332–4338, 1994), was not detected. Instead, upstream of the immunity gene eciI we found an open reading frame, eciO, which could code for a novel lantibiotic modification enzyme involved in reduction of an N-terminally located oxopropionyl residue. Epicidin 280 produced by the heterologous host Staphylococcus carnosus TM 300 after introduction of eciIAPBC (i.e., no eciO was present) behaved homogeneously during reverse-phase chromatography.
Lantibiotics are bacteriocin-like antimicrobial peptides that are characterized by the presence of lanthionine and β-methyllanthionine, which form intramolecular thioether rings and which originate from posttranslational modifications of serine, threonine, and cysteine residues (17, 32, 34). Nisin, which is produced by Lactococcus lactis subsp. lactis, is the most prominent member of this group of peptide antibiotics and is widely used as a natural food preservative (10). Epidermin (1), subtilin (11), and Pep5 (28) are additional well-studied lantibiotics, which act primarily by forming pores in the bacterial membrane (21, 30, 32). In contrast to the activities of the type A lantibiotics mentioned above, the primary activity of the compact, globular, type B lantibiotics, such as duramycin, actagardine, and mersacidin, appears to be directed towards inhibition of specific enzyme functions (6, 32, 48).
All lantibiotics are ribosomally synthesized as prepeptides that consist of an N-terminal leader sequence, which is removed during biosynthesis, and a C-terminal propeptide region. Modification of the propeptide region includes (i) dehydration of specific serine and/or threonine residues, which results in the formation of 2,3-didehydroamino acids, and (ii) addition of thiol groups of cysteine residues to the double bonds of some of these amino acids, which yields the lanthionine and β-methyllanthionine residues (14, 32, 34). These unique biosynthetic reactions are catalyzed by the proteins LanB and LanC, respectively (32). The lanB and lanC genes are normally located close to the structural gene for the prelantibiotic (lanA) and to other genes necessary for lantibiotic production and secretion. The other genes often include genes for a transporter belonging to the ABC transporter family (lanT), a subtilisin-like protease (lanP), a protein or peptide involved in producer self-protection (immunity) (lanI), and proteins similar to members of the family of two-component regulatory proteins (lanR, lanK) (19, 22, 32). Not all of these genes have been detected in all gene clusters, indicating that some of the accessory genes may be located outside the gene cluster or that the gene functions can be provided by host-encoded proteins having similar activities.
Several expression systems for production of lantibiotics have been constructed in order to improve the peptides used in certain applications and to study structure-function relationships (32). While it was possible to produce a large number of mutated peptides, it became obvious that the biosynthetic machinery, including the producer self-protection system, has restrictions concerning which modifications are tolerated (5). Thus, we decided to try to learn more about the interdependence of the structure of lantibiotics, their immunity peptides, and biosynthesis enzymes by analyzing naturally occurring related lantibiotics. Here we describe isolation and characterization of epicidin 280, which is related to the lantibiotic Pep5; the strain which produces epicidin 280, Staphylococcus epidermidis BN 280, exhibits cross-immunity to Pep5. The biosynthetic gene cluster of epicidin 280 includes eciI, eciA, eciP, eciB, and eciC, which are sufficient to produce epicidin 280 in the heterologous host Staphylococcus carnosus TM 300. An additional gene, tentatively designated eciO, codes for an enzyme that is very similar to proteins belonging to the oxidoreductase family; this enzyme could catalyze an N-terminal modification of epicidin 280. As in Pep5, epilancin K7 (39), and lactocin S (37), the first amino acid of epicidin 280 after maturation (i.e., after dehydration of hydroxyl amino acids and proteolytic cleavage) is a didehydro residue. Didehydro residues have been found to be unstable when they are N-terminally exposed after processing (34) and to deaminate to oxoacyl residues. In epilancin K7 the N-terminal oxopropionyl is subsequently reduced to 2-hydroxypropionyl, a modification for which no dedicated biosynthetic gene has been found so far. The newly identified oxidoreductase, EciO, seems to be involved in a similar reduction of an N-terminal oxopropionyl residue in epicidin 280.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and detection of antibacterial activity.
S. epidermidis BN 280, which produces epicidin 280, was maintained as a glycerol stock culture at −70°C and was grown on tryptone soy agar (Difco, Augsburg, Germany) supplemented with 20 μg of tetracycline per ml or in tryptone soy broth (TSB) at 37°C. Recombinant bacteria were plated onto tryptone soy agar containing 40 μg of ampicillin per ml or 20 μg of chloramphenicol per ml; pUC18+ (Pharmacia, Uppsala, Sweden) and pCU1 (2) were used as cloning vectors. The level of antibacterial activity was determined by the deferred antagonism test (28), and the MICs of the purified peptides were determined as previously described (4). The following strains were used as indicators: S. epidermidis 5 (the wild-type producer of Pep5), S. epidermidis 5 Pep5− (a variant of S. epidermidis 5 that has been cured of the plasmid which carries the Pep5 gene cluster; 8), S. epidermidis 280 (the wild-type producer of epicidin 280), Micrococcus luteus ATCC 4698, S. carnosus TM 300 (33), and Staphylococcus simulans 22 (28).
Purification of epicidin 280.
Epicidin 280 was purified by using the method described previously for Pep5 purification (28). Two-liter TSB cultures were inoculated with 200 μl of a S. epidermidis BN 280 overnight culture and incubated at 37°C for 18 to 22 h. The antibacterial polypeptide was purified from the supernatant by hydrophobic interaction chromatography (Serdolit AD-2; Serva, Heidelberg, Germany), cation-exchange chromatography (CM Sephadex C-25; Pharmacia), and reverse-phase high-pressure liquid chromatography (HPLC) as described previously (23). Active fractions were rechromatographed and lyophilized before they were used for mass spectrometry, amino acid analysis, and MIC determinations.
Mass spectrometry and amino acid analysis.
The lyophilized fractions were analyzed with a model API III triple quadrupole mass spectrometer equipped with an IonSpray source (Sciex, Thornhill, Canada) (44). The amino acid composition was analyzed by precolumn derivatization with o-phthaldialdehyde (29). For proteolytic cleavage with chymotrypsin, 1 mg of purified peptide was incubated at 26°C in the presence of 15 μg of chymotrypsin (Sigma, Munich, Germany) in 0.2 M ethylmorpholine buffer (pH 8)–2 mM dithioerythreitol for 15 min.
Peptide sequencing.
Prior to analysis with a model 476A protein sequencer (Applied Biosystems, Weiterstadt, Germany), purified epicidin was modified by using a three step procedure that included thiol addition, peroxidation with trifluoroperacetic acid, and a second thiol addition (24).
Molecular cloning and DNA preparation.
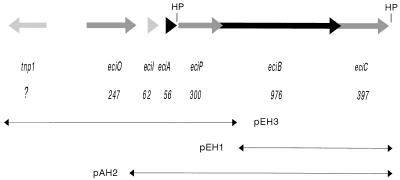
DNA was isolated by previously described procedures by using different DNA purification kits (Qiagen, Hilden, Germany). Restriction digestion and DNA ligation were performed by following the recommendations of the supplier (Boehringer, Mannheim, Germany). Cells of Escherichia coli JM 109 or TB1 (15, 46) were transformed by electroporation with a Bio-Rad Gene Pulser (Bio-Rad, Mississauga, Canada) as described previously (23). A 1.9-kb DNA fragment which harbored eciIAP and the beginning of eciB was excised from pEH3 (Fig. 1) by digestion with RcaI (made blunt with the Klenow fragment) and XbaI. This fragment was cloned into pEH1 (Fig. 1), which contained the downstream part of the epicidin gene cluster, after digestion with SalI (made blunt with the Klenow fragment) and XbaI. The resulting insert harbored eciIAPBC and was ligated into the shuttle vector pCU1 after EcoRI-PstI digestion. S. carnosus TM 300 was transformed with pAH2 (Fig. 1) by the protoplast transformation method (4).
FIG. 1.
Organization of the epicidin 280 gene cluster. The number of amino acids encoded by each gene is indicated below each gene locus, and the arrows indicate the relative directions of transcription. Subcloned fragments are shown below the gene cluster. The positions of putative hairpins (HP) are indicated. The nucleotide and amino acid sequences have been deposited in the EMBL data bank under accession no. Y14023.
The oligonucleotides used for PCR and sequencing were obtained from Eurogentec (Seraing, Belgium) or MWG Biotech (Ebersberg, Germany) or were prepared with a model 391 PCR Mate (Applied Biosystems). Southern blotting of S. epidermidis BN 280 DNA was performed with digoxigenin-labeled (Boehringer) oligonucleotides designed to detect different overlapping parts of the epicidin 280 gene cluster.
DNA sequence determination and analysis.
Double-stranded DNA was sequenced either by Sequiserve (Vaterstetten, Germany) or with a model A.L.F. DNA sequencer (Pharmacia Biotech, Uppsala, Sweden). The BLITZ@EMBL-Heidelberg service and ClustalW (1.4) software were used to perform a sequence analysis.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the EMBL data library under accession no. Y14023.
RESULTS
Purification of epicidin 280.
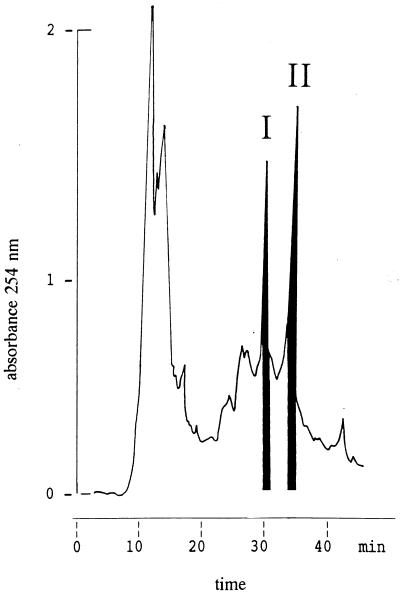
Epicidin 280 was purified from supernatants of 2-liter TSB cultures of S. epidermidis BN 280 by using a method originally developed for isolation of Pep5 (28). The final purification step on a C18 reverse-phase HPLC column yielded two distinct active peaks eluting at 32 and 34% acetonitrile (Fig. 2, peaks I and II, respectively).The average molecular masses of the peak I and II compounds were 3,136 ± 1.5 and 3,133 ± 1.5 Da, respectively, as determined by mass spectrometry. Typically, we obtained 500 μg of pure peptide per liter of culture supernatant.
FIG. 2.
Elution profile of epicidin 280 as determined by C18 reverse-phase HPLC. The lantibiotic was purified from 2 liters of an S. epidermidis BN 280 culture supernatant by using Serdolit AD-2 (Serva) and cation-exchange chromatography. An aliquot containing desalted and freeze-dried peptides was injected. Peaks I and II were the active fractions.
Amino acid composition and sequence.
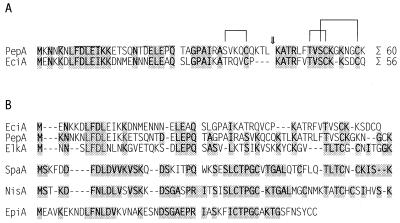
The amino acid compositions of the intact peak I peptide and its chymotrypsin fragments are shown in Table 1; the data obtained for peak II were identical. The chymotrypsin fragmentation data were useful for elucidation of the structure of Pep5, when fragmentation produced two fragments having similar sizes (18); a chymotrypsin fragmentation analysis was performed for epicidin 280 to localize the unmodified serine residue found in the peptide hydrolysate and to obtain information concerning whether there is a thioether bridge connecting the two fragments or whether a Pep5 type of bridging pattern is present. As has been observed with several other lantibiotics, N-terminal sequencing failed. However, after the peptide was subjected to a series of modification reactions designed to overcome lantibiotic sequence blocks (24), the following sequence was obtained: XLGPAIKAXRQVXPKAXRFVXVXXKKXDXQ, where X is either serine, threonine, or cysteine (these amino acids usually undergo modifications in lantibiotics). The results obtained agree well with the nucleotide sequence of the structural gene (see below) and show that epicidin 280 contains three lanthionine-methyllanthionine residues. Also, a Pep5 type of bridging pattern is probably present (Fig. 3A), as the positions of the modified residues are comparable and two Pep5-like chymotrypsin fragments were obtained (18). This precludes the possibility of a head-to-tail bridge and makes involvement of threonine 17 in bridge formation unlikely; previous studies indicated that cleavage of Pep5 by the endopeptidase ArgC does not occur when a bridge is in close proximity (5).
TABLE 1.
Amino acid composition of epicidin 280
| Amino acid | No. of residues in:
|
|||
|---|---|---|---|---|
| Active epicidin 280a | Peak I peptideb | Fragment 1c | Fragment 2c | |
| Ala | 3 | 3.5 | 2.5 | 1 |
| Asp | 1 | 1 | 1 | |
| Arg | 2 | 2 | 1 | 1 |
| Cys | 3 | |||
| Gln | 2 | 2 | 1 | 1 |
| Gly | 1 | 1 | 1 | |
| Ile | 1 | 1 | 1 | |
| Leu | 1 | 1 | 1 | |
| Lys | 4 | 3.5 | 1.5 | 2 |
| Phe | 1 | 1 | 1 | |
| Pro | 2 | NDd | ND | ND |
| Ser | 3 | 1 | 1 | |
| Thr | 4 | |||
| Val | 3 | 3 | 1 | 2 |
| Lan/MeLane | 3 | 1 | 2 | |
Amino acid composition based on translation of the eciA nucleotide sequence.
The intact peptide was analyzed twice by using different purified batches. The peak II peptide was analyzed in parallel and gave identical results.
Fragments 1 and 2 resulted from chymotrypsin digestion of epicidin 280 peak I. The numbers of amino acid residues were determined from the molar ratios relative to arginine and isoleucine.
ND, not detectable after o-phthaldialdehyde derivatization.
Lan/MeLan, lanthionine–β-methyl-lanthionine.
FIG. 3.
Amino acid sequences of the prepeptides of Pep5 and epicidin 280 (A) and comparison of these sequences with the sequences of selected type A lantibiotics (B). (A) Identical residues are indicated by shading. The thioether bridges of mature Pep5 are indicated by brackets. The chymotrypsin cleavage site is indicated by an arrow. (B) The proteolytic cleavage site is indicated by a gap. Conserved residues are indicated by shading.
MICs of epicidin 280 compared to those of Pep5 and K18P Pep5.
Epicidin 280 exhibited the same spectrum of activity as Pep5 but was less active against the indicator strains by factors ranging from 8 to 64 (Table 2). With both peptides, S. carnosus TM 300 was the most sensitive strain tested; the MICs were 1.0 and 175 ng/ml for Pep5 and epicidin 280, respectively.
TABLE 2.
MICs of epicidin 280, Pep5, and K18P Pep5
| Indicator strain | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|
| Pep5 | Epicidin 280 peak I | Epicidin 280 peak II | K18P Pep5a | |
| S. simulans 22 | 0.0047 | 0.304 | 0.605 | 2.0 |
| S. epidermidis 5 Pep5− | 0.605 | 3.85 | 9.7 | 2.0 |
| M. luteus ATCC 4698 | 1.2 | 9.7 | 19.3 | NDb |
| S. carnosus TM 300 | 0.001 | 0.175 | 0.304 | 1.0 |
| S. epidermidis 5 | 19 | 70 | 70 | ND |
| S. epidermidis BN 280 | 1.0 | ND | ND | 125.0 |
Data from reference 4.
ND, not determined.
S. epidermidis BN 280 and S. epidermidis 5, the strain which produces Pep5, exhibited cross-immunity in the deferred antagonism test; i.e., there were no inhibition zones when both strains were cross-tested as producer and indicator strains. The MICs obtained with purified peptides were 1 μg of Pep5 per ml for S. epidermidis BN 280, 70 μg of epicidin 280 per ml for S. epidermidis 5, and 9.7 μg of epicidin 280 per ml for the Pep5-sensitive strain S. epidermidis 5 Pep−, a variant of S. epidermidis 5 which has lost the biosynthetic gene cluster. These data support the cross-immunity test results and show that the immunity peptide PepI is to some extent effective against epicidin 280. The MICs for epicidin 280 peak I and peak II differed by a factor of two for each test strain, indicating that the peak I peptide has a slightly higher specific activity.
Identification and cloning of the epicidin 280 gene cluster.
Attempts to construct a gene probe from the amino acid sequence information failed; in retrospect, this can be explained by codon usage in the segment of eciA that is unusual for staphylococci. The epicidin 280 gene cluster was then identified by PCR by using the following primers which hybridize with conserved sequence motifs of lanB and lanC (36), respectively: 5′-GAAA(CT)(AT)TATAGATATGGTGG-3′ for the LanB consensus sequence ETYRYGG and 5′-CCATA(AG)CACCAAGCATCTCT-3′ for the LanC consensus sequence RDAWCYG. The reaction yielded a 1.2-kb PCR product when plasmid DNA of S. epidermidis BN 280 was used as the template, demonstrating that the epicidin 280 gene cluster is plasmid encoded. Determination of the nucleotide sequence of this PCR product allowed synthesis of an additional oligonucleotide (5′-TTATCTGACATTTCTCAC-3′), which hybridized with a 3.9-kb EcoRI-XbaI fragment of plasmid pCH01. S. epidermidis BN 280 was found to contain five plasmids, the largest of which is pCH01 (length, >40 kb). Additional fragments were cloned by using the same approach.
A total of 7.4 kb of DNA comprising the complete nucleotide sequence of the epicidin 280 gene cluster was sequenced (Fig. 1). This sequence contained five open reading frames organized in the same order as the open reading frames in the Pep5 gene cluster; these open reading frames code for putative proteins with high levels of amino acid sequence similarity to the lantibiotic biosynthetic proteins PepI, PepA, PepP, PepB, and PepC. The first of these genes, eciI, codes for a 62-amino-acid (7-kDa) peptide which is the only homolog of PepI (26) identified so far. The similarity of this peptide to PepI (level of similarity, 74.2%) (Fig. 4A) and the cross-immunity, as determined by the deferred antagonism test, suggest that EciI acts as an immunity peptide against epicidin 280 and that the producer self-protection mediated by EciI should be based on the same molecular mechanism as the producer self-protection mediated by PepI. In addition, EciI exhibits a significant level of similarity (42.1%) to the product of ORF57 (Fig. 4A), an open reading frame in the gene cluster of the lantibiotic lactocin S (38), and a significant level of similarity (42.9%) to the immunity peptide of the nonlantibiotic bacteriocin divergicin (45) of Carnobacterium divergens (Fig. 4B). Together with PepI, these peptides form a new group of immunity peptides which is obviously not restricted to lantibiotics. However, it is interesting that the lantibiotic immunity peptides belonging to this new group are found solely in strains having intracellular leader peptidases, whereas strains with extracellular processing obviously need another type of producer self-protection.
FIG. 4.
Comparison of the sequences of the immunity peptides EciI, PepI, and ORF57 (A) and comparison of the sequences of EciI and DviA (B). Residues conserved in two peptides are indicated by asterisks, and conservatively exchanged residues are indicated by dots; solid squares and colons indicate residues conserved in three peptides.
The start codon of eciA is located 64 bp downstream of eciI. A total of 58.9% (33 of 56) of the amino acids of EciA are identical to PepA amino acids, and the overall level of similarity is 75.0%. Both leader peptides consist of 26 amino acids, whereas the propeptide region of epicidin 280 is four amino acids shorter than the propeptide region of PepA (Fig. 3A). This deletion considerably shortens the flexible central part of epicidin 280 compared to Pep5. Moreover, a proline is located in this part of the peptide. These differences may be responsible for the reduced antibiotic activity of epicidin 280 compared to Pep5. The primary gene product, EpiA, differs in another remarkable way from the nisin type A lantibiotic prepeptides; position 2 of the leader peptide is occupied by an alanine residue instead of a proline residue, a residue which has previously been observed to be strictly conserved in this group of lantibiotics (Fig. 3B) (7). However, site-directed mutagenesis in the nisin leader peptide and molecular modelling of the interaction between the prepeptide and the leader peptidase have shown that the conserved proline is not essential for cleavage of the leader peptide of nisin (42).
Following eciA, eciP is 87 bp downstream of the stop codon of eciA and overlaps (by 11 bp) the open reading frame of eciB, which in turn extends 13 bp beyond a potential start codon for eciC. A gene locus for an ABC transporter, which has been found in all lantibiotic gene clusters identified previously, is missing in the epicidin 280 gene cluster. EciB (976 amino acids, 117 kDa), EciC (397 amino acids, 45 kDa), and EciP (300 amino acids, 34 kDa) are very similar to the respective Pep5 biosynthetic enzymes (levels of sequence similarity, 68 to 75%). Therefore, analogous to the Pep5 biosynthetic enzymes, EciB and EciC are probably the dehydrating and thioether-forming enzymes, respectively, and EciP should be a cytoplasmic, subtilisin-like protease which removes the leader peptide (23). In Fig. 5 the amino acid sequences of EciB, EciC, and EciP are compared with the sequences of the respective proteins produced by the Pep5 gene cluster and with consensus sequences deduced from all LanB, LanC, and LanP proteins identified so far (36). All residues which are thought to be involved in catalytic activity are conserved in the three epicidin biosynthesis proteins; a few motifs have single amino acid substitutions, but the overall level of similarity is high. Some characteristic amino acid differences found only in the Pep5 biosynthetic proteins are conserved in the respective epicidin 280 biosynthetic proteins (e.g., Tyr in the fifth conserved PepB and EciB motif and Glu in the second conserved PepC and EciC motif).
FIG. 5.
Consensus sequences of the biosynthetic proteins LanB, LanC, and LanP (36) compared with the sequences of the corresponding regions of the Pep5 (23) and epicidin 280 related proteins. The arrows indicate amino acids that may have catalytic relevance (36). The numbers of amino acids between the conserved motifs are indicated.
Upstream of eciI there is an open reading frame (designated eciO) which ends 75 bp upstream of the eciI start codon. eciO encodes a putative 247-amino-acid (27-kDa) protein that exhibits 48% sequence similarity to and 35% identity with a hypothetical Bacillus subtilis oxidoreductase (47) (EMBL data bank accession no. P42317). Sequence alignment revealed that EciO belongs to the short-chain dehydrogenase-reductase protein family (Fig. 6) (16). Most of the proteins in this family catalyze the first reduction step in the fatty acid biosynthesis pathway. In this reaction a 3-oxoacyl acyl carrier protein is reduced to a (3R)-3-hydroxyacyl acyl carrier protein, and NADPH acts as a coenzyme (3). The proposed N-terminal NADP binding site (GlyXXXGlyXGly motif) and the active site (Tyr-151, Lys-155) of these enzymes were identified in EciO by sequence alignment.
FIG. 6.
Comparison of the sequences of EciO and the putative oxidoreductase (YXJF) of B. subtilis (47). The asterisks indicate conserved residues, and conservatively exchanged residues are indicated by dots; amino acids that may have catalytic relevance are indicated by shading.
Approximately 850 bp upstream of eciO on the complementary strand we located a gene which encodes a putative transposase with a high level of sequence similarity to the 224-amino-acid transposase for insertion-like element IS431MEC and for IS257 in transposon Tn4003 of Staphylococcus aureus (9, 27). This gene is preceded by an inverted repeat characteristic of IS257. Such insertion sequences are common in staphylococcal plasmids and are often associated with genes which confer resistance to several substances, such as trimethoprim or tetracycline (9).
We predicted that there are two potential hairpins immediately downstream of eciA (free energy value, −55.9 kJ/mol) and downstream of eciC (free energy value, −71.6 kJ/mol) (Fig. 1), which is again in good agreement with the Pep5 gene cluster data (23). The latter hairpin could serve as a transcriptional terminator, whereas the former may allow partial readthrough.
Heterologous expression of epicidin 280.
In order to show that the genes identified are sufficient for epicidin 280 production and to obtain information concerning whether eciO is involved in modification of the lantibiotic, S. carnosus TM 300 was transformed with a recombinant plasmid containing eciIAPBC. The resulting clones produced epicidin 280, and the peptide yields were comparable to those of the wild-type producer. In contrast to S. epidermidis 280, heterologous expression gave rise to only one antimicrobial peptide, which eluted at 34% acetonitrile during reverse-phase HPLC. This peak apparently corresponds to the second peak identified during separation of the wild-type peptides; its molecular mass was determined to be 3.135 ± 1.5 Da. Thus, EciO is obviously not essential for production of active epicidin 280 in general but is essential for formation of one of the two active forms of this lantibiotic.
DISCUSSION
Epicidin 280 is a novel 30-amino-acid lantibiotic with 75% sequence similarity to the type A lantibiotic Pep5 (23), a tricyclic, 34-amino-acid peptide with a molecular mass of 3,488 Da (Fig. 3A). The epicidin 280 biosynthesis gene cluster, eciIAPBC, is organized in the same order on plasmid pCH01 of S. epidermidis BN 280 as the pepIAPBC genes of the Pep5 gene cluster found on plasmid pED503 of S. epidermidis 5 (23). An ABC transporter, which typically is involved in export of lantibiotics, is missing in the epicidin gene cluster. Although it is possible that a gene for a dedicated epicidin 280 exporter is not located in the vicinity of the epicidin 280 gene cluster, we presume that a nondedicated host transporter takes over this function (4, 12, 13). This assumption is based on previous deletion studies performed with the Pep5 gene cluster. It was observed that in the absence of PepT, the Pep5 production rate was not zero but decreased to about 10 to 30% of the wild-type rate (4). Under the same growth conditions, the epicidin 280 yields obtained in this study (0.5 mg/liter) are more comparable to the PepT deletion yields than the Pep5 wild-type producer yields (60 to 100 mg/liter). In addition, the epidermin gene cluster from the wild-type producer S. epidermidis Tü3298 contains the gene epiT′T", which is inactivated by two internal deletions and a frameshift mutation. Complementation with gdmT, an intact transporter gene of the homologous gallidermin gene cluster, enhances epidermin production 2-fold in the wild-type strain and up to 10-fold when the epidermin gene cluster is expressed in the heterologous host S. carnosus TM 300 (25).
We identified in the epicidin 280 gene cluster a putative oxidoreductase gene (eciO) just upstream of eciI at the site where the transporter gene pepT is located in the Pep5 gene cluster; in contrast to pepT, eciO is located on the same strand as the rest of the gene cluster. A sequence comparison clearly identified EciO as an oxidoreductase which could reduce an N-terminal oxopropionyl residue in epicidin 280. Such residues occur in lantibiotics when dehydrated serine or threonine residues occupy position +1 of the prepeptide. Proteolytic removal of the leader peptide leads to N-terminal dehydroalanine or dehydrobutyrine residues, which are not stable and spontaneously deaminate to oxopropionyl or oxobutyryl residues like those found in Pep5 (18), epilancin K7 (39), and lactocin S (37). In epilancin K7, the oxopropionyl residue was found to be reduced to the respective hydroxypropionyl group, and this led to speculation concerning whether there is a dedicated oxidoreductase present and necessary for this reaction (39). However, a gene for such an enzyme was not found in the direct vicinity of the epilancin K7 structural gene, elkA, although the sequence information for the epilancin K7 gene cluster is only fragmentary so far. Therefore, it was suggested that the N-terminal oxopropionyl residue may be the coincidental substrate of a nondedicated oxidoreductase (39). The presence of eciO strongly suggests that such an enzyme has a dedicated function. Moreover, the presence of eciO could explain why we obtained two different epicidin 280 molecules which differ in mass by approximately 3 Da. Even given the statistical error of the mass determination (±1.5 Da), it is conceivable that we obtained the oxopropionyl and 2-hydroxypropionyl forms of epicidin 280. Structural variations that could explain the occurrence of two peptide peaks, such as the formation of different bridging patterns, could not account for the observed difference in mass; besides, such variations should certainly result in significant changes in biological activities, whereas the activities of the two peptides differ by a factor of only two for all of the indicators tested.
Introduction of eciIAPBC into the heterologous host S. carnosus TM 300 revealed that these genes are sufficient to direct epicidin 280 production in this host. Apparently, epicidin 280 production does not depend on additional factors, such as two-component regulatory systems and dedicated ABC transporters; alternatively, the functions of these factors are provided by the S. carnosus host strain. Heterologous expression in the absence of eciO yielded only one epicidin 280 peptide peak. This may indicate that in the wild-type strain, epicidin 280 is produced as a mixture of peptides that have oxopropionyl and 2-hydroxypropionyl groups at their N termini; such peptides would have calculated monoisotopic masses of about 3,135 and 3,137 Da, which were approximately the masses observed for the two wild-type peptides. However, it is obvious that the statistical error of the mass determinations does not permit further interpretation. We are currently expressing and purifying EciO to determine its precise role in lantibiotic modification and are attempting to characterize the N-terminal modification(s) of the peptide by nuclear magnetic resonance.
The sequence similarity values for all of the gene products of the epicidin 280 and Pep5 gene clusters are in a narrow range, 68 to 75%, suggesting that the entire gene cluster is been mobilized horizontally rather than individual genes. The levels of similarity are unusual; the previously determined values were found to be higher for natural variants, such as epidermin and gallidermin or nisin A and nisin Z, and much lower for more distantly related lantibiotics (1, 32). For example, the LanB proteins EpiB, NisB, SpaB, and PepB have only 16 to 29% identical residues, whereas EciB and PepB exhibit 54.9% amino acid sequence identity. According to the previously published definition (32), epicidin 280 and Pep5 certainly are not sufficiently related to be considered natural variants. However, cross-immunity between two bacteriocin producers clearly indicates that the immunity peptides are functionally related, a feature which seems to be hardly possible in the absence of significant structural and conformational similarities of the lantibiotics. This in turn supports the hypothesis that Pep5 and epicidin 280 have identical bridging patterns. In this context it is interesting to compare Pep5 and epicidin 280 with nisin and subtilin. The latter compounds also have identical bridging patterns, and about 60% of the amino acids in their prepeptides, nisA and spaA, are identical. In contrast, the levels of similarity for the biosynthetic enzymes B, C, and T range from 20 to 30%. Nisin-producing lactococcal strains and the subtilin producer B. subtilis ATCC 6639 have not been reported to exhibit cross-immunity, although this may be difficult to assess because the B. subtilis strain produces additional antibiotic substances which interfere with a simple deferred antagonism test. Moreover, the immunity mechanism is more complicated than the immunity mechanism observed with Pep5 and epicidin 280; apparently, an additional ABC transporter supports the immunity proteins NisI and SpaI (20, 35). The latter proteins exhibit no sequence similarity but share a lipoprotein signal sequence (32).
MIC determinations revealed that epicidin 280 has reduced antibacterial activities compared to Pep5 for the indicator organisms tested. The elongated type A lantibiotics, such as Pep5, nisin, and epidermin, are able to form potential-dependent pores in bacterial cytoplasmic membranes (31). Recently, it has been suggested that the peptides remain surface bound during the pore formation process and enter the membrane in a bent conformation, thereby fusing the outer and inner leaflets of the membrane (40, 41). Conformational studies demonstrated that all of the pore-forming type A lantibiotics contain a central flexible hinge region, and subsequent structure-function studies performed with nisin, epidermin, and Pep5 proved that flexibility is essential for activity (32). This flexible central region in Pep5 extends from Glu-14 to Phe-23. Epicidin 280 lacks four amino acids in this region and in addition has a proline at position 16. Because of these differences, epicidin 280 may be assumed to be more rigid, which could explain its lower level of bactericidal activity; also, the genetically engineered peptide K18P Pep5 (Table 2) is less active by a factor of 500 than wild-type Pep5 (4). In addition, there is a clear difference in charge distribution between the C-terminal parts of epicidin 280 and Pep5, which is mainly caused by Asp-28 in epicidin 280. While Pep5 has a net charge of +7 with the charges distributed randomly throughout the peptide, epicidin 280 has a charge of +4 and has a net neutral C-terminal region. Analysis of nisin Z mutants showed that negative charges in the C-terminal region of the lantibiotic are detrimental for antimicrobial activity (43). The reduced antimicrobial activity of epicidin 280 may also explain why, in contrast to Pep5 (4), heterologous expression in S. carnosus is possible. This species and S. simulans are more sensitive to Pep5 than any other known bacteria are, and the immunity levels achieved in S. carnosus may not be sufficient to prevent producing cells from being killed by the heterologous expressed product when it has a particularly high specific activity.
ACKNOWLEDGMENTS
This work was supported by the Commission of the European Communities (grant BIOT-CT91-0265 within the BRIDGE framework), by the Deutsche Forschungsgemeinschaft (grants Sa 292 6-3 and SFB 323 to J.M and G.J.), and by the BONFOR Program of the Medical Faculty, University of Bonn.
We gratefully acknowledge M. Eschbach-Bludau and C. Szekat for expert technical assistance and A. Hoffmann for excellent contributions to the heterologous expression experiments.
REFERENCES
- 1.Allgaier H, Jung G, Werner R G, Schneider U, Zähner H. Epidermin: sequencing of a heterodet tetracyclic 21-peptide amide antibiotic. Eur J Biochem. 1986;160:9–22. doi: 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- 2.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, Entian K-D, Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 3.Barberis-Maino L, Ryffel C, Kayser F H, Berger-Bächi B. Complete nucleotide sequence of IS431mec in Staphylococcus aureus. Nucleic Acids Res. 1990;18:5548. doi: 10.1093/nar/18.18.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierbaum G, Reis M, Szekat C, Sahl H-G. Construction of an expression system for mutagenesis of the lantibiotic Pep5. Appl Environ Microbiol. 1994;60:2876–2883. doi: 10.1128/aem.60.12.4332-4338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierbaum G, Szekat C, Josten M, Heidrich C, Kempter C, Jung G, Sahl H-G. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl Environ Microbiol. 1996;62:385–392. doi: 10.1128/aem.62.2.385-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brötz H, Bierbaum G, Markus A, Molitor E, Sahl H-G. Mode of action of the lantibiotic mersacidin: inhibition of peptidoglycan biosynthesis via a novel mechanism? Antimicrob Agents Chemother. 1995;39:714–719. doi: 10.1128/AAC.39.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vos W M, Kuipers O P, Van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 8.Ersfeld-Dreßen H, Sahl H-G, Brandis H. Plasmid involvement in production and immunity to the staphylococcin-like peptide Pep5. J Gen Microbiol. 1984;130:3029–3035. doi: 10.1099/00221287-130-11-3029. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie M T, Lyon B R, Loo L S L, Matthews P R, Stewart P R, Skurray R A. Homologous direct repeat sequences associated with mercury, methicillin, tetracycline and trimethoprim resistance determinants in Staphylococcus aureus. FEMS Lett. 1987;43:165–171. [Google Scholar]
- 10.Gross E, Morell J L. The structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 11.Gross E, Kiltz H H, Nebelin E. Subtilin, VI. Die Struktur des Subtilins. Hoppe-Seyler’s Z Physiol Chem. 1973;354:810–812. [PubMed] [Google Scholar]
- 12.Higgins C F, Hiles I D, Salmond G P C, Gill D R, Downie J A, Evans I J, Holland I B, Gray L, Buckel S D, Bell A W, Hermodson M A. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986;323:448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- 13.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 14.Ingram L C. Synthesis of the antibiotic nisin: formation of lanthionine and methyl-lanthionine. Biochim Biophys Acta. 1969;184:216–219. doi: 10.1016/0304-4165(69)90121-4. [DOI] [PubMed] [Google Scholar]
- 15.Johnston T C, Thompson R B, Baldwin T O. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the beta subunit of bacterial luciferase. J Biol Chem. 1986;261:4805–4811. [PubMed] [Google Scholar]
- 16.Jornvall H, Persson B, Krook M, Atrian S, Gonzales-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 17.Jung G. Lantibiotics—ribosomally synthesised biologically active polypeptides containing sulphide rings and α,β-didehydroamino acids. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom; 1991. pp. 2–34. [Google Scholar]
- 18.Kellner R, Jung G, Josten M, Kaletta C, Entian K-D, Sahl H-G. Pep5: structure elucidation of a large lantibiotic. Angew Chem Int Ed Engl. 1989;28:616–619. [Google Scholar]
- 19.Klein C, Kaletta C, Schnell N, Entian K-D. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1992;58:132–142. doi: 10.1128/aem.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein C, Entian K-D. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis. Appl Environ Microbiol. 1994;60:2793–2801. doi: 10.1128/aem.60.8.2793-2801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kordel M, Benz R, Sahl H-G. Mode of action of the staphylococcin-like peptide Pep5: voltage-dependent depolarization of bacterial and artificial membranes. J Bacteriol. 1988;170:84–88. doi: 10.1128/jb.170.1.84-88.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuipers O P, Beerthuyzen M M, Siezen R J, De Vos W M. Characterisation of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of the expression of nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–292. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 23.Meyer C, Bierbaum G, Heidrich C, Reis M, Süling J, Iglesias-Wind M I, Kempter C, Molitor E, Sahl H-G. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of Pep5 and PepC; evidence for a role of PepC in the thioether formation. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- 24.Meyer H E, Heber M, Eisermann B, Korte H, Metzger J W, Jung G. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal Biochem. 1994;223:185–190. doi: 10.1006/abio.1994.1571. [DOI] [PubMed] [Google Scholar]
- 25.Peschel A, Schnell N, Hille M, Entian K-D, Götz F. Secretion of the lantibiotics epidermin and gallidermin: sequence, influence on epidermin production, and regulation of gdmT and gdmH by EpiQ. Mol Gen Genet. 1997;254:312–318. doi: 10.1007/s004380050421. [DOI] [PubMed] [Google Scholar]
- 26.Reis M, Eschbach-Bludau M, Iglesias-Wind M I, Kupke T, Sahl H-G. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl Environ Microbiol. 1994;60:2876–2883. doi: 10.1128/aem.60.8.2876-2883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouch D A, Messerotti L J, Loo L S, Jackson C A, Skurray R A. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol. 1989;3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 28.Sahl H-G, Brandis H. Production, purification and chemical properties of an antistaphylococcal agent produced by Staphylococcus epidermidis. J Gen Microbiol. 1981;127:377–384. doi: 10.1099/00221287-127-2-377. [DOI] [PubMed] [Google Scholar]
- 29.Sahl H-G, Grossgarten M, Widger W R, Cramer W A, Brandis H. Structural similarities of the staphylococcin-like peptide Pep5 to the peptide antibiotic nisin. Antimicrob Agents Chemother. 1985;27:836–840. doi: 10.1128/aac.27.5.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahl H-G, Kordel M, Benz R. Voltage-dependent depolarization of bacterial membranes and artificial lipid bilayers by the peptide antibiotic nisin. Arch Microbiol. 1987;149:120–124. doi: 10.1007/BF00425076. [DOI] [PubMed] [Google Scholar]
- 31.Sahl H-G. Pore formation in bacterial membranes by cationic lantibiotics. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom; 1991. pp. 347–358. [Google Scholar]
- 32.Sahl H-G, Jack R W, Bierbaum G. Lantibiotics: biosynthesis and biological activities of peptides with unique posttranslational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 33.Schleifer K H, Fischer U. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int J Syst Bacteriol. 1982;32:153–156. [Google Scholar]
- 34.Schnell N, Entian K-D, Schneider U, Götz F, Zähner H, Kellner R, Jung G. Prepeptide sequence of epidermin, a ribosomally synthesised antibiotic with four sulfide rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 35.Siegers K, Entian K-D. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microbiol. 1995;61:1082–1089. doi: 10.1128/aem.61.3.1082-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siezen R J, Kuipers O P, De Vos W M. Comparison of the lantibiotic gene clusters and encoded proteins. Antonie Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 37.Skaugen M, Nissen-Meyer J, Jung G, Stevanovic S, Sletten K, Abildgaard C I M, Nes I F. In vivo conversion of l-serine to d-alanine in a ribosomally synthesized polypeptide. J Biol Chem. 1994;269:27183–27186. [PubMed] [Google Scholar]
- 38.Skaugen M, Abildgaard C I M, Nes I F. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol Gen Genet. 1997;253:674–686. doi: 10.1007/s004380050371. [DOI] [PubMed] [Google Scholar]
- 39.Van de Kamp M, Van den Hooven H W, Konings R N H, Bierbaum G, Sahl H-G, Kuipers O P, Siezen R J, De Vos W M, Hilbers C W, Van de Ven F J M. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Cloning and characterization of the epilancin K7 encoding gene and NMR analysis of mature epilancin K7. Eur J Biochem. 1995;230:587–600. doi: 10.1111/j.1432-1033.1995.tb20600.x. [DOI] [PubMed] [Google Scholar]
- 40.Van den Hooven H W, Doeland C A E, Van de Kamp M, Konings R N H, Hilbers C W, Van de Ven F J M. Three-dimensional structure of the lantibiotic nisin in the presence of membrane-mimetic micelles of dodecylphosphocholine and of sodium dodecylsulphate. Eur J Biochem. 1996;235:382–393. doi: 10.1111/j.1432-1033.1996.00382.x. [DOI] [PubMed] [Google Scholar]
- 41.Van den Hooven H W, Spronk C A E M, Van de Kamp M, Konings R N H, Hilbers C W, Van de Ven F J M. Surface location and orientation of the lantibiotic nisin bound to membrane-mimicking micelles of dodecylphosphocholine and of sodium dodecylsulphate. Eur J Biochem. 1996;235:394–403. doi: 10.1111/j.1432-1033.1996.00394.x. [DOI] [PubMed] [Google Scholar]
- 42.Van der Meer J R, Rollema H S, Siezen R J, Beerthuyzen M M, Kuipers O P, De Vos W M. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J Biol Chem. 1994;269:3555–3562. [PubMed] [Google Scholar]
- 43.Van Kraaij C, Breukink E, Rollema H S, Siezen R J, Demel R A, De Kruijff B, Kuipers O P. Influence of charge differences in the C-terminal part of nisin on antimicrobial activity and signaling capacity. Eur J Biochem. 1997;247:114–120. doi: 10.1111/j.1432-1033.1997.00114.x. [DOI] [PubMed] [Google Scholar]
- 44.Weil H-P, Beck-Sickinger A G, Metzger J, Stevanovic S, Jung G, Josten M, Sahl H-G. Biosynthesis of the lantibiotic Pep5. Isolation and characterization of a prepeptide containing dehydroamino acids. Eur J Biochem. 1990;194:217–223. doi: 10.1111/j.1432-1033.1990.tb19446.x. [DOI] [PubMed] [Google Scholar]
- 45.Worobo R W, Van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, K.-I., H. Sano, S. Seki, M. Oda, M. Fujimura, and Y. Fujita. 1995. Unpublished data. [DOI] [PubMed]
- 48.Zimmermann N, Metzger J W, Jung G. The tetracyclic lantibiotic actagardine: 1H-NMR and 13C-NMR assignments and revised primary structure. Eur J Biochem. 1995;228:786–797. [PubMed] [Google Scholar]