Abstract
Plant-pathogenic bacteria produce various extracellular polysaccharides (EPSs) which may function as virulence factors in diseases caused by these bacteria. The EPS levan is synthesized by the extracellular enzyme levansucrase in Pseudomonas syringae, Erwinia amylovora, and other bacterial species. The lsc genes encoding levansucrase from P. syringae pv. glycinea PG4180 and P. syringae pv. phaseolicola NCPPB 1321 were cloned, and their nucleotide sequences were determined. Heterologous expression of the lsc gene in Escherichia coli was found in four and two genomic library clones of strains PG4180 and NCPPB 1321, respectively. A 3.0-kb PstI fragment common to all six clones conferred levan synthesis on E. coli when further subcloned. Nucleotide sequence analysis revealed a 1,248-bp open reading frame (ORF) derived from PG4180 and a 1,296-bp ORF derived from NCPPB 1321, which were both designated lsc. Both ORFs showed high homology to the E. amylovora and Zymomonas mobilis lsc genes at the nucleic acid and deduced amino acid sequence levels. Levansucrase was not secreted into the supernatant but was located in the periplasmic fraction of E. coli harboring the lsc gene. Expression of lsc was found to be dependent on the vector-based Plac promoter, indicating that the native promoter of lsc was not functional in E. coli. Insertion of an antibiotic resistance cassette in the lsc gene abolished levan synthesis in E. coli. A PCR screening with primers derived from lsc of P. syringae pv. glycinea PG4180 allowed the detection of this gene in a number of related bacteria.
The plant pathogens Pseudomonas syringae pv. glycinea PG4180 and P. syringae pv. phaseolicola NCPPB 1321 invade plant leaves, inducing typical leaf spot symptoms on soybeans and bush beans, respectively. Bacterial blight of soybeans, caused by P. syringae pv. glycinea, is characterized by water-soaked leaf spots which soon develop into necrotic lesions surrounded by chlorotic halos, whereas halo blight of beans, caused by P. syringae pv. phaseolicola, is characterized by water-soaked lesions presumably filled with highly hydrated polysaccharides. In general, plant-pathogenic pseudomonads produce various high-molecular-weight polysaccharides, including alginate and levan, which might function as virulence factors (10, 12, 36). Levan formation serves as a taxonomic characteristic of P. syringae independent of pathovar assignments.
Bacterial polysaccharides are found either as a dense layer of more or less regularly arranged polymer structures attached to the bacterial cell walls or as loosely associated exopolysaccharides (EPSs) (4, 47). EPSs are thought to provide a selective advantage for plant-pathogenic bacteria based on their generally hydrophilic and anionic properties. They may improve bacterial fitness by generating a hydrogenated matrix, minimizing direct contact with plant surfaces, preventing recognition by the host, and functioning as detoxifying barriers against plant defense compounds (24, 28, 30). In the case of wilt diseases caused by Ralstonia solanacearum and Clavibacter michiganensis, bacterial EPSs appear to plug the xylem vessels and contribute to plant wilting (1, 10, 29). During epiphytic growth, EPSs protect the bacteria from desiccation, concentrate minerals and nutrients, and may enhance attachment to surfaces (47).
The EPS levan is a β-(2,6) polyfructan with extensive branching through β-(2,1) linkages (Fig. 1) (11). Both linkage types are produced by the extracellular enzyme levansucrase (EC 2.4.1.10) with sucrose as a substrate. This enzyme conducts three reactions: (i) synthesis of levan from sucrose by transfructosylation while releasing glucose, (ii) hydrolysis of levan to monosaccharides of fructose, and (iii) exchange of [14C]glucose in the reaction fructose-2,1-glucose + [14C]glucose to fructose-2,1-[14C]glucose + glucose (17).
FIG. 1.
Structure of the EPS levan as a β-(2,6) polyfructan.
Levansucrases from several bacterial genera, including those from the species Bacillus subtilis, Streptococcus mutans, Zymomonas mobilis, Acetobacter diazotrophicus, Erwinia amylovora, and P. syringae pv. phaseolicola, have been reported and biochemically characterized (2, 9, 16, 20, 32, 39). In contrast to levansucrases from gram-positive bacteria, which differ widely in their biochemical characteristics, those of gram-negative species share some common characteristics, including constitutive expression, molecular mass, and N-terminal amino acid sequences (15, 20, 44).
The P. syringae pv. phaseolicola levansucrase was previously characterized as a 45-kDa extracellular enzyme which is highly stable under an array of tested conditions (20). However, the lsc gene encoding levansucrase from P. syringae has not yet been identified. Knowledge of the genetics of levansucrase is essential for further analysis of the role of levan in plant-microbe interactions. The lack of levan-deficient mutants of P. syringae has deterred previous researchers from gaining definitive evidence regarding the role of levan in host-pathogen interactions. Until now, reporter gene studies or a detailed transcriptional analysis of levansucrase expression have not been available, leaving room for speculation on the constitutive expression of this enzyme in P. syringae, based only on phenotypic observations (40).
Here we report the cloning and nucleotide sequencing of the lsc genes from P. syringae pv. glycinea PG4180 and P. syringae pv. phaseolicola NCPPB 1321. Levansucrase activity was located mostly in the periplasmic fraction and to some extent in the membrane and cytoplasmic fractions of recombinant Escherichia coli, but hardly at all in the extracellular fraction. Using nested deletion analysis, we showed lsc expression in E. coli to be dependent on the vector-based Plac promoter. Insertion of an antibiotic resistance cassette in the recombinant lsc gene abolished levan synthesis in E. coli. The lsc gene could be detected in related P. syringae pathovars by PCR. Our results suggest that lsc genes from gram-negative plant pathogens form a distinct cluster.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. Pseudomonas strains were maintained on mannitol-glutamate (MG) medium (25) at 28°C. Single colonies of P. syringae grown on MG agar for 96 h were resuspended in 5 ml of King’s B medium (27) and incubated overnight on a rotary shaker at 280 rpm and 28°C. Fifty microliters of the overnight culture was then used to inoculate 5 ml of King’s B medium, which was incubated on a rotary shaker at 280 rpm and 18°C for 24 to 48 h. E. coli DH5α (38) was used as a host in cloning and expression studies and was cultured in 5 ml of Luria-Bertani (LB) medium in test tubes at 37°C. Bacterial growth was monitored by measuring the optical density at 600 nm. Cells were lysed by repeated freeze-thaw cycles or by sonication. The protein concentration in cell lysates was determined by the Bradford assay (38). The following antibiotics were added to the media (in micrograms per milliliter): ampicillin, 50; tetracycline, 25; spectinomycin, 25; and streptomycin, 25.
TABLE 1.
Bacterial strains used in this study and results of PCR screening for the lsc gene
| Strain | Relevant characteristics | PCR resulta | Reference or source |
|---|---|---|---|
| E. coli DH5α | Levan− | 38 | |
| P. syringae pv. glycinea | |||
| PG4180 | Levan+ | 1.25 | R. Mitchell |
| GSPB1201, GSPB1548, GSPB1834 | Levan+ | 1.25 | GSPBb |
| Psg 7a/90, Psg 49a/90, Psg 16/83 | Levan+ | 1.25 | B. Völksch |
| P. syringae pv. phaseolicola | |||
| NCPPB1321 | Levan+ | 1.25 | G. E. Jones |
| GSPB796 | Levan+ | 1.25 | GSPB |
| Psph 699, Psph PK2, Psph KZ2W | Levan+ | 1.25 | B. Völksch |
| P. syringae pv. syringae | |||
| Pss 61 | Levan+ | A. Collmer | |
| GSPB1142 | Levan+ | GSPB | |
| Pss 4918, Pss 1391, C72 | Levan+ | 0.9 | B. Völksch |
| FF5 | Levan+ | 0.9 | G. W. Sundin |
| P. syringae pv. tomato | |||
| DSM50315 | Levan+ | 1.0, 1.25, 1.5 | DSMc |
| GSPB487, GSPB479 | Levan+ | 1.4, 1.0 | GSPB |
| DC3000 | Levan+ | 1.25 | D. Cuppels |
| P. syringae pv. pisi | |||
| GSPB1477 | Levan+ | 1.0 | GSPB |
| GSPB104, GSPB1206, GSPB1787 | Levan+ | 1.1 | GSPB |
| P. syringae pv. morsprunorum Pm7 | Levan+ | 1.25 | A. Jones |
| P. syringae pv. lachrymans GSPB82a | Levan+ | 1.25 | GSPB |
| P. syringae pv. atropurpurea MAFF301313 | Levan+ | 0.8 | K. Nishiyama |
| P. syringae pv. maculicola GSPB2145 | Levan+ | GSPB | |
| P. syringae pv. savastanoi GSPB2264 | Levan+ | GSPB | |
| P. putida GSPB1498 | Levan− | GSPB | |
| P. marginalis GSPB92 | Levan+ | GSPB | |
| P. viridiflava GSPB1685 | Levan+ | 0.8 | GSPB |
| P. fluorescens GSPB1714 | Levan+ | GSPB | |
| Acidovorax (Pseudomonas) avenae GSPB1848 | Levan− | GSPB | |
| Ralstonia (Pseudomnas) pickettii GSPB2016 | Levan− | GSPB | |
| P. cichorii GSPB2097 | Levan+ | 0.8 | GSPB |
| P. agarici GSPB2305 | Levan+ | 1.1 | GSPB |
| P. fuscovaginae GSPB2309 | Levan+ | 0.6 | GSPB |
| A. radiobacter K84 | Levan− | DSM | |
| C. michiganensis ATCC 10202 | Levan− | DSM | |
| R. meliloti ATCC 4399 | Levan− | DSM | |
| X. campestris DSM1050 | Levan− | DSM |
Size (in kilobases) of PCR product amplified with primers LSCF and LSCR and genomic DNAs of the respective strains.
GSPB, Göttinger Sammlung phytopathogener Bakterien.
DSM, German Culture Collection for Microorganisms in Braunschweig, Germany.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| pRK7813 | Tcr; cosmid vector used for generation of genomic libraries | 23 |
| pCAM140 | Ampr Smr-Spr; contains Sm-Sp resistance cassette on 2.0-kb EcoRI fragment | 51 |
| pBluescript SK | Ampr; cloning vector | Stratagene, Heidelberg, Germany |
| pCK2, pCK6, pCK9, pCK10 | Tcr; genomic library clones of PG4180; levan+; approx 25-to-35-kb inserts in pRK7813 | This study |
| p4D7, p4C2 | Tcr; genomic library clones of NCPPB 1321; levan+; approx 25-to-35-kb inserts in pRK7813 | This study |
| pSKL3 | Ampr; contains a 3.0-kb PstI fragment from pCK2 in pBluescript; levan+ | This study |
| pSKP3 | Ampr; contains a 3.0-kb PstI fragment from p4D7 in pBluescript; levan+ | This study |
| pSKL3G | Ampr; contains the 3.0-kb PstI fragment from pSKL3 in opposite orientation in pBluescript; levan− | This study |
| pSKL3Sm | Ampr Smr-Spr; carries Smr-Spr cassette inserted in BstEII sites of pSKL3; levan− | This study |
| pDK-A6 | Ampr; nested deletion clone of pSKL3 with 2.8-kb insert; levan+ | This study |
| pDK-B5 | Ampr; nested deletion clone of pSKL3 with 2.8-kb insert; levan+ | This study |
| pDK-C20 | Ampr; nested deletion clone of pSKL3 with 1.8-kb insert; levan+ | This study |
| pDK-C14 | Ampr; nested deletion clone of pSKL3 with 1.6-kb insert; levan− | This study |
| pDK-B4 | Ampr; nested deletion clone of pSKL3 with 2.48-kb insert; levan− | This study |
DNA procedures.
Genomic DNA was isolated from P. syringae by established procedures (45). Agarose gel electrophoresis, restriction digests, purification of DNA fragments from agarose gels, electroporations, PCR, Southern hybridizations, and small-scale plasmid DNA preparations were performed by standard techniques (38). Genomic cosmid libraries of both P. syringae strains were constructed in pRK7813 as described previously (3). Subclones were generated in pBluescript (Stratagene, Heidelberg, Germany). Nested deletion clones were constructed with the Erase-a-Base kit (Promega, Mannheim, Germany). Large-scale preparations of plasmid DNA from E. coli were isolated by alkaline lysis and purified on Qiagen (Hilden, Germany) columns.
DNA sequencing and analysis.
Nucleotide sequencing reactions with nested deletion clones were performed by the dideoxynucleotide method (38) with the Thermo Sequenase fluorescent-labelled primer cycle sequencing kit (Amersham-Buchler, Braunschweig, Germany) and Cy5-labelled T3 and T7 oligonucleotide primers (Pharmacia, Freiburg, Germany). Automated DNA sequencing was accomplished with an ALF Express sequencing apparatus (Pharmacia). Sequence data were aligned and processed with the DNASTAR version 4.1 software package (Lasergene, Madison, Wis.). DNA and protein sequence homology searches of the GenBank, EMBL, PIR, and SWISSPROT databases were performed with the University of Wisconsin Genetics Computer Group (UWGCG) programs BLASTX, FASTEMBL, GAP, BESTFIT, and GROWTREE.
Assays for levansucrase activity.
Qualitative estimation of levansucrase activity in sterile bacterial supernatants, cell lysates, and cell fractions was carried out on water-agar plates containing 5% sucrose. Enzyme activity was visualized by the formation of opalescent slime plugs. Fractionation of P. syringae and E. coli cells was carried out according to the method of Boyd et al. (5). Levansucrase activity was determined by measuring the amount of glucose liberated during incubation with sucrose, using the glucose GOD-Perid assay kit from Boehringer Mannheim (Mannheim, Germany). Samples (10 μl) were mixed with 10 μl of McIllvaine’s buffer (10% sucrose–0.02% sodium azide [pH 6.2]) and 200 μl of test reagent (100 mM potassium phosphate [pH 7] containing 1 U of peroxidase per ml, 10 U of glucose oxidase per ml, 1 mg of 2,2′-azino-di-[3-ethylbenzthiazolinsulfonate-(6)]diammonium salt per ml, and 0.01% sodium azide). The samples were incubated at 20°C, and absorbance was measured at 600 nm at 5 to 10 min intervals for 1 to 2 h. One unit of levansucrase activity represented the amount of enzyme that released 1 μmol of glucose per min.
Anthrone detection assay for levan.
The formation of levan was determined after precipitation with methanol as described previously (18) in equivalents of fructose according to the method of Helbert and Brown (19). Samples were dialyzed, and aliquots of 200 μl were mixed with 80 μl of anthrone (2% in ethyl acetate) and immediately with 1 ml of concentrated sulfuric acid. After 60 min of incubation at 20°C, the extinction was measured at 600 and 492 nm against a blank. Specific detection of ketoses was due to the wavelength used for detection. Lyophilized levan derived from P. syringae pv. phaseolicola cultures served as the standard.
Nucleotide sequence accession number.
The reported nucleotide sequences were deposited with GenBank and EMBL under accession no. AF037443 and AF052289.
RESULTS
Cloning of levansucrase genes of P. syringae pv. glycinea and phaseolicola.
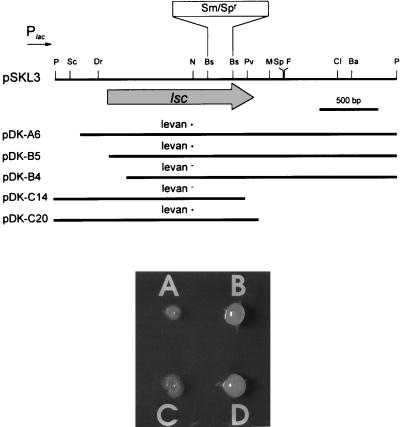
Genomic libraries of P. syringae pv. glycinea PG4180 and P. syringae pv. phaseolicola NCPPB 1321 were prepared by cloning partially Sau3A-digested genomic DNA of both strains into BamHI-digested pRK7813. For both libraries, a total of 960 E. coli recombinants, each containing approximately 25 to 35 kb of insert DNA, were screened for levan synthesis on LB agar plates containing 5% sucrose by overnight incubation at 37°C and subsequent incubation at 18°C. This temperature regime was chosen because optimal levansucrase activity in P. syringae had been reported to occur at 18°C (20). After 10 to 14 days, four genomic library clones of PG4180, designated pCK2, pCK6, pCK9, and pCK10, and two genomic library clones of NCPPB 1321, designated p4D7 and p4C2, exhibited a clearly visible mucoid phenotype which was not observed when these clones were incubated on LB agar plates free of sucrose. Restriction analysis of the inserted DNA revealed a 3.0-kb PstI fragment that was common to all six clones (Fig. 2). Cosmid clones pCK2 and p4D7 were treated with PstI, and restriction fragments were shotgun cloned into pBluescript. Recombinants were screened on LB agar plates containing 5% sucrose. Two pBluescript clones, one derived from pCK2 and designated pSKL3 and the other derived from p4D7 and designated pSKP3, showed the same mucoid phenotype described for the cosmid clones and contained the 3.0-kb PstI fragment as insert DNA (Fig. 3). The biochemical nature of the EPS produced by E. coli harboring pCK2, pSKL3, or pSKP3 was tested by using the anthrone detection assay (18), indicating that the EPS produced was levan (data not shown). A nested deletion analysis from both ends of the insert DNA of pSKL3 was carried out to determine the minimal size of insert DNA required for levan synthesis in E. coli (Fig. 3). Deletion clones were screened for levan synthesis on LB agar plates containing 5% sucrose. Deletion clones pDK-B5 and pDK-C20 represented the smallest inserts still conferring levan synthesis on E. coli. Deletion clones pDK-B4 and pDK-C14 contained the largest inserts which did not confer levansucrase activity on the recombinants (Fig. 3).
FIG. 2.
Restriction analysis of various PstI-digested genomic library clones following agarose gel electrophoresis. The 3.0-kb fragment conferring levan synthesis on E. coli is marked with an arrow. In plasmid pSKL3Sm (lane 6) this fragment is increased in size due to the insertion of an antibiotic resistance cassette. Lanes: 1, pCK2; 2, pCK6; 3, pCK9; 4, pCK10; 5, pSKL3; 6, pSKL3Sm.
FIG. 3.
Restriction map of the 3.0-kb PstI fragment from P. syringae pv. glycinea PG4180 inserted in plasmid pSKL3 and location of nested deletion clones (top) conferring a levan+ or levan− phenotype on E. coli grown on LB agar plates containing 5% sucrose (bottom). The lsc gene (shaded arrow), the orientation of the vector-based Plac promoter (small arrow), and the inserted Sm-Sp antibiotic resistance cassette are indicated. The photograph shows E. coli(pBluescript) (A), E. coli(pSKL3) (B), E. coli(pDK-B4) (C), and E. coli(pDK-A6). The restriction enzymes used are indicated as follows: Ba, BamHI; Bs, BstEII; Cl, ClaI; Dr, DraI; F, FspI; M, MluI; N, NaeI; P, PstI; Pv, PvuI; Sc, ScaI; and Sp, SphI.
Nucleotide sequence analysis and comparison.
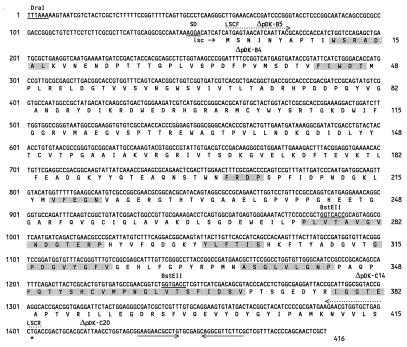
The 3.0-kb insert of pSKL3 and a 1.5-kb DraI subclone of pSKP3 were sequenced on both strands. The insert DNA of pSKL3 derived from P. syringae pv. glycinea PG4180 contained an open reading frame (ORF) of 1,248 bp with a Shine-Dalgarno sequence and a putative stem-loop terminator sequence (Fig. 3 and 4). In the case of the subclone from pSKP3 derived from P. syringae pv. phaseolicola NCPPB 1321, an ORF of 1,296 bp was identified (data not shown). The authenticity of both ORFs was confirmed by a codon preference analysis with the UWGCG program CODONPREFERENCE in its default mode and a P. syringae codon usage table available via the worldwide web (CUTG [Codon Usage Tabulated from GenBank] at http://www.dna.affrc.go.jp). Nucleotide sequence data were compared with database entries by using the FASTEMBL, BLASTX, BESTFIT, and GAP programs. The nucleotide sequence of the 1,248-bp ORF from pSKL3 showed significant similarities over the entire length of the nucleotide sequence to the lsc gene of E. amylovora (69%) and the levU gene of Z. mobilis (60%), both of which encode levansucrase (15, 44). An identity of 64% over approximately 700-bp was observed with the lsdA gene, which encodes levansucrase in A. diazotrophicus (2). All conserved sequence motifs common to levansucrase genes of gram-negative bacteria were identified within this ORF (Fig. 4). The ORF showed weak similarities to levansucrase genes from gram-positive bacteria, such as B. subtilis (46) and Bacillus amyloliquefaciens (48), and to the fructosyl transferase gene of S. mutans (43). The deduced amino acid sequence of the 1,248-bp ORF indicated a protein mass of approximately 45 kDa and showed similarities to the protein sequences of levansucrases from E. amylovora (83%) and Z. mobilis (63%). Similarities to levansucrase sequences from gram-positive species were low. The reported ORF was designated lsc. The insert DNA upstream and downstream of lsc (approximately 500 and 1,250 bp) did not show significant similarities to database entries. The 1,296-bp ORF derived from NCPPB 1321 showed a very high sequence similarity to lsc originated from PG4180 at the nucleotide level (86%) and at the deduced amino acid sequence level (95%). It also was designated lsc and exhibited essentially the same similarities to database entries as the lsc gene from PG4180.
FIG. 4.
Nucleotide sequence and predicted amino acid sequence of the lsc gene from P. syringae pv. glycinea PG4180. Nucleotides and amino acid residues are numbered on the left and right, respectively. The putative ribosome binding site (SD, underlined), the DraI and BstEII restriction sites (underlined) used for insertion of the Smr-Spr cassette, the stop codon (∗), and the putative transcriptional terminator sequences (horizontal arrows) are indicated. Triangles mark the sequence ends of deletion clones used in this study. Primer binding sites used for PCR screening of the lsc gene are indicated by dotted arrows. Shaded boxes mark the conserved regions in levansucrases of gram-negative bacteria (2). SD, Shine-Dalgarno sequence.
Cluster analysis of levansucrases in gram-negative and gram-positive bacteria.
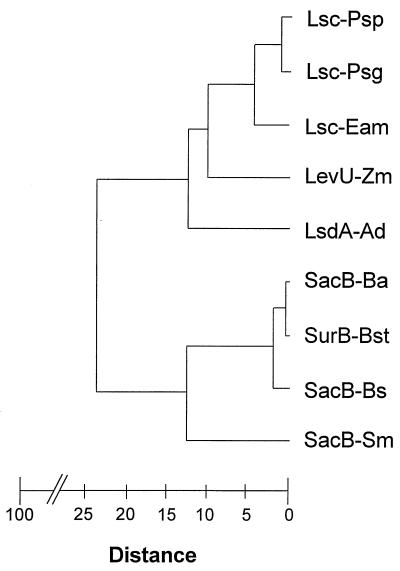
To further compare the sequence relationships of all known bacterial levansucrases, a dendrogram was generated by using the GROWTREE program and the deduced amino acid sequences from five gram-negative and four gram-positive organisms (Fig. 5). Based on the calculated distance matrix and the distance coefficients represented by branch points in this dendrogram, the two sequences from P. syringae form a distinct cluster together with levansucrase from E. amylovora. In contrast, the sequences from the remaining two gram-negative bacteria, Z. mobilis and A. diazotrophicus, branched individually. This was underlined by the Jukes-Cantor algorithm of the program DISTANCES, which estimates the number of substitutions per 100 amino acid residues: this number was 7 for the two P. syringae sequences, 25.5 and 27.8 for the E. amylovora sequence in comparison to either of the P. syringae sequences, and between 64.4 and 84.5 for the levansucrase sequences of the two remaining gram-negative bacteria. As expected, the levansucrase sequences derived from gram-positive bacteria exhibited only a low degree of relatedness to the sequences derived from P. syringae. To confirm the cluster analysis of levansucrase sequences among gram-negative bacteria, the computer program CLUSTAL was also employed (data not shown). These data indicated that the reported levansucrases from plant-pathogenic bacteria are closely related.
FIG. 5.
Dendrogram illustrating the sequence relationship of levansucrases from gram-negative and gram-positive bacteria, including P. syringae pv. glycinea (Lsc-Psg), P. syringae pv. phaseolicola (Lsc-Psp), E. amylovora (Lsc-Eam), Z. mobilis (LevU-Zm), A. diazotrophicus (LsdA-Ad), B. amyloliquifaciens (SacB-Ba), Bacillus stearothermophilus (SurB-Bst), B. subtilis (SacB-Bs), and S. mutans (SacB-Sm). The branch points represent distance coefficients calculated by the GROWTREE program. Distance values range from 0 to 100, with a distance value of 0 representing complete identity between two sequences.
Expression of levansucrase from P. syringae pv. glycinea PG4180 in E. coli.
For comparison, cell pellets of selected E. coli constructs were lysed and lysates combining cytoplasmic, membrane, and periplasmic proteins were screened for levansucrase activity (Table 3). The spatial distance and orientation of the lsc start codon to the vector-based Plac promoter influenced recombinant levan synthesis. Deletion clone pDK-A6, which was characterized by the shortest distance (220 bases) between the translational start codon of lsc and the Plac promoter, showed the highest levansucrase activity. Moreover, levansucrase activity was not detected in cell lysates of E. coli harboring pSKL3G, which contained the same insert DNA as pSKL3 but carried the lsc gene in the opposite orientation to Plac (Table 3). These data suggested that lsc transcription in E. coli was not controlled by its native promoter. Similar results were obtained for the lsc gene from P. syringae pv. phaseolicola NCPPB 1321 (data not shown). The relative distribution of levansucrase activities was assayed in the extracellular, periplasmic, cytoplasmic, and membrane fractions of selected E. coli recombinants (Table 3). Fractions were generated by the method of Boyd et al. (5). The enzyme was not significantly secreted into the culture supernatants. It was detectable in the cytoplasmic and membrane fractions in low abundance. Approximately 79 to 93% of the total levansucrase activity was found in the periplasmic fractions regardless of the size of the recombinant’s insert, indicating that the enzyme was mostly associated with this compartment in E. coli cultures. In contrast, P. syringae secreted levansucrase into the supernatant as expected (Table 3). Deletion clone pDK-C14 lacked the C-terminal 36 codons of levansucrase and failed to exhibit levansucrase activity. Deletion clone pDK-B5 exhibited levan synthesis although the N-terminal 14 bases of lsc were deleted in this construct (Fig. 3 and 4), suggesting a translational fusion between the nonfunctional lacI gene of pBluescript and the lsc gene. Furthermore, this indicated that the deleted region of levansucrase was not required for enzymatic activity. In E. coli (pDK-B5) approximately 11% of the total levansucrase activity was found in the cytoplasm (Table 3), suggesting that transport across the inner membrane might have been affected.
TABLE 3.
Levansucrase activity in different cell compartments of P. syringae and E. colia
| Bacterial strain | Enzyme sp act (U/μg of protein) | % Total enzyme activity in:
|
|||
|---|---|---|---|---|---|
| Cytoplasm | Membrane | Periplasm | Extracellular fraction | ||
| P. syringae PG4180 | 16.1 ± 0.6 | 0.2 ± 0.1 | 0.5 ± 0.2 | 46.4 ± 0.4 | 52.9 ± 1.4 |
| E. coli(pSKL3) | 4.5 ± 2.5 | 0.6 ± 0.2 | 6.1 ± 1.6 | 93.3 ± 1.7 | 0 |
| E. coli(pDK-A6) | 15.2 ± 2.2 | 0.5 ± 0.2 | 8.1 ± 0.6 | 90.9 ± 0.9 | 0.5 ± 0.1 |
| E. coli(pDK-B5) | 2.0 ± 0.8 | 10.8 ± 1.6 | 8.2 ± 0.9 | 79.2 ± 2.3 | 1.8 ± 0.1 |
| E. coli(pDK-C20) | 3.7 ± 1.5 | 4.4 ± 0.6 | 6.8 ± 0.2 | 88.8 ± 0.6 | 0 |
| E. coli(pDK-B4) | 0 | ||||
| E. coli(pDK-C14) | 0 | ||||
| E. coli(pDK-C20) | 0 | ||||
| E. coli(pSKL3G) | 0 | ||||
| E. coli(pSKL3Sm) | 0 | ||||
| E. coli(pBluescript) | 0 | ||||
Data represent the averages (± standard deviations) of three to six replicates.
Mutagenesis of levansucrase in E. coli.
A 2.0-kb streptomycin-spectinomycin resistance (Smr-Spr) cassette was excised from pCAM140 (51) as an EcoRI fragment, blunt end filled, and ligated to pSKL3 which had been linearized with the restriction enzyme BstEII and blunt end filled by using Klenow enzyme. The new construct was designated pSKL3Sm (Fig. 2 and 3). E. coli harboring pSKL3Sm did not show levansucrase activity (Table 3), indicating that the antibiotic cassette insertion had disrupted the lsc gene and confirming that the insert DNA of pSKL3 is responsible for the expression of levansucrase in E. coli.
Screening for the lsc gene in related pseudomonads by PCR and Southern blot analysis.
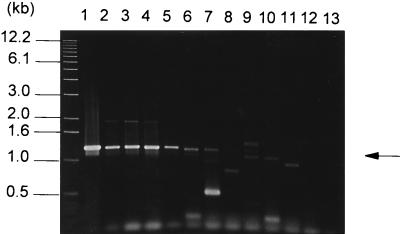
Primers LSCF (5′ATGAGTAACATCAATTAC3′) and LSCR (5′TCAGCTCAGCACCACGTTCT3′) (Fig. 4), delineating a 1,248-bp DNA fragment, were used to amplify DNA sequences from 44 bacterial strains representing 10 pathovars of P. syringae, 9 other Pseudomonas species, and Xanthomonas campestris, Rhizobium meliloti, C. michiganensis, and Agrobacterium radiobacter. The results are given in Table 1, and representative data are shown in Fig. 6. As expected, no PCR amplification was observed in levan-negative strains, indicating the specificity of the oligonucleotide primers that we used. A 1.25-kb product was amplified in all tested P. syringae isolates belonging to the pathovars glycinea, phaseolicola, morsprunorum, and lachrymans as well as in P. syringae pv. tomato strains DC3000 and DSM50315. A slightly smaller product was amplified from genomic DNA of Pseudomonas agarici and three of four P. syringae pv. pisi isolates. When PCR products were digested with the restriction enzyme BstEII, no restriction fragment length polymorphism was observed among samples from P. syringae pv. glycinea, phaseolicola, morsprunorum, and lachrymans (data not shown). Among the other strains tested there was variability in the amplification product sizes ranging from 0.9 to 1.4 kb, suggesting nonspecific annealing of the primers or variable lsc gene sizes (Fig. 6). To test this, DNA-DNA hybridization analysis was carried out with the lsc gene of P. syringae pv. glycinea PG4180 as the DNA probe and PCR products from other P. syringae pathovars. Experiments were carried out under high-stringency conditions as described earlier (49). The 1.25-kb PCR products from the genomic DNAs of all P. syringae pv. glycinea, phaseolicola, morsprunorum, and lachrymans strains hybridized to the lsc gene from PG4180 (data not shown). For the genomic DNAs of P. syringae pv. tomato strains, hybridization of a 1.25-kb PCR product was observed in strain DSM50315 but not in strain DC 3000. PCR products from P. syringae pv. tomato strains GSPB487 and GSPB479 did not hybridize with the probe used in this study. Likewise, PCR products derived from the genomic DNAs of all remaining levan-producing strains did not react with the lsc probe of PG4180, indicating that the observed PCR products from these strains were due to nonspecific annealing. These data suggest that the primer pair cannot be used for the detection of the lsc gene in general but is suitable to identify closely related lsc genes. In order to screen for lsc homologs in those levan-producing P. syringae strains for which no hybridizing PCR products were observed, Southern hybridization analyses under different stringency conditions were carried out with the lsc gene as the probe and restriction digests of genomic DNAs of various strains (Fig. 7). At the lowest-stringency conditions, with a hybridization temperature of 55°C and two 7-min washes with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 55°C, restriction fragments of genomic DNA from PCR-positive strains, as well as from strains producing levan but not reacting in the PCR, hybridized to the probe. As expected, genomic DNA from levan-negative isolates did not react with the probe. Under higher-stringency conditions (a hybridization temperature of 60 to 68°C) the signals for PCR-negative strains gradually disappeared (data not shown).
FIG. 6.
PCR detection of genes homologous to lsc from P. syringae pv. glycinea PG4180 with the primers LSCF and LSCR derived from the N- and C-terminal regions of lsc. Genomic DNAs from various bacterial strains were used as templates. Lanes: 1, E. coli(pSKL3); 2, P. syringae pv. glycinea PG4180; 3, P. syringae pv. phaseolicola NCPPB 1321; 4, P. syringae pv. morsprunorum Pm7; 5, P. syringae pv. lachrymans GSPB82a; 6, P. syringae pv. pisi GSPB104; 7, P. agarici GSPB2305; 8, P. syringae pv. atropurpurea MAFF301313; 9, P. syringae pv. tomato DSM50315; 10, P. syringae pv. pisi GSPB1477; 11, P. syringae pv. syringae FF5; 12, Pseudomonas putida GSPB1498; 13, X. campestris DSM1050. The arrow indicates the 1.25-kb PCR product typical of the lsc gene.
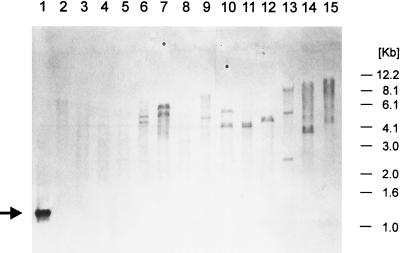
FIG. 7.
Southern hybridization analysis of total SalI-digested genomic DNA from various pseudomonads hybridized with a digoxigenin-labeled probe specific to the PCR-amplified lsc gene of P. syringae pv. glycinea PG4180. The PCR product used as a DNA probe is marked by a black arrow. Molecular size markers are outlined to the right. Lanes: 1, PCR product of the lsc gene from PG4180; 2, P. viridiflava GSPB1686; 3, P. cichorii GSPB2097; 4, P. fuscovaginae GSPB2309; 5, P. marginalis GSPB92; 6, P. syringae pv. maculicola GSPB2145; 7, P. syringae pv. savastanoi GSPB2264; 8, P. fluorescens GSPB1714; 9, P. syringae pv. syringae Pss 61; 10, P. syringae pv. atropurpurea MAFF301313; 11, P. syringae pv. pisi GSPB104; 12, P. syringae pv. glycinea PG4180; 13, P. syringae pv. phaseolicola GSPB796; 14, P. syringae pv. lachrymans GSPB82a; 15, P. syringae pv. tomato DSM50312.
DISCUSSION
To understand the role of the P. syringae EPS levan in plant-microbe interactions, genetic tools are required for monitoring levansucrase expression in response to environmental and plant-borne factors. Hettwer et al. (20) had previously reported the enzymatic characteristics of purified levansucrase from P. syringae pv. phaseolicola. Despite the long-term use of levan formation for the taxonomic identification of P. syringae (6, 41), the gene encoding levansucrase had not been characterized in P. syringae. According to the cluster analysis carried out in this study, the lsc genes of P. syringae pv. glycinea and phaseolicola resemble lsc genes of other gram-negative bacteria, especially that of E. amylovora, and showed weak sequence similarities with levansucrase genes from gram-positive bacteria. These data confirmed reports on the conservation of lsc genes in plant-associated gram-negative bacteria (2, 15, 20, 44) and demonstrated that levansucrases from plant pathogens had the highest degree of similarity with each other. Since epiphytically growing E. amylovora and P. syringae may simultaneously inhabit the surface of plants, one probable scenario could have involved horizontal gene transfer between these organisms in the phylloplane.
The predicted sizes of the two lsc gene products (45 and 47.5 kDa, respectively) matched the reported size of the extracellular levansucrase in P. syringae pv. phaseolicola (20). As described for E. amylovora and Z. mobilis, the predicted sequence motif WSRADAL in the N-terminal region of levansucrase from PG4180 was very close to the translational start codon, confirming previous results of the N-terminal sequencing of levansucrase in P. syringae pv. phaseolicola (20). This indicated that levansucrase is not cleaved during the transport process and is subject to a signal peptide-independent export mechanism. Signal peptide-mediated transport processes were reported or suggested for the levansucrases of B. subtilis and A. diazotrophicus, respectively (2, 7, 35).
Levansucrase activity in E. coli harboring lsc was detected mostly in the periplasmic and to some extent in the membrane and cytoplasmic fractions, but not in the extracellular fraction. Similar results were reported for levansucrase secretion in E. amylovora (15). In contrast to the well-understood process of levansucrase export in B. subtilis (7, 35), no information is currently available on how P. syringae or E. amylovora export this enzyme into the periplasm and extracellular space. For E. amylovora, Geier and Geider (15) had reported that transport of levansucrase across the inner membrane was abolished when the C-terminal part of the lsc gene was mutated. Whether or not the C terminus of levansucrase of P. syringae plays a role in transmembrane processes remains to be elucidated in the native organism. Furthermore, E. coli cells obviously lack the additional factor(s) guiding levansucrase across the outer membrane. In gram-negative pathogens at least three pathways to translocate exoproteins across the inner and outer membranes have been characterized (37), and it remains to be determined by which of these mechanisms levansucrase is exported in P. syringae. Charkowski et al. (8) recently dissected the hrp (for hypersensitive response and pathogenicity) gene-mediated type III secretion pathway in P. syringae pv. syringae and found evidence for two genetically distinguishable stages for the transport of virulence factors across the inner and outer membranes.
In contrast to its homolog in E. amylovora (15), the lsc gene of strain PG4180 was not transcribed from its native promoter in E. coli. We assume that initial screening for levan synthesis within the genomic libraries of PG4180 and NCPPB 1321 had selected transcriptional fusions between the vector-based Plac and the lsc gene. Alternatively, other promoters present on the insert DNA of the cosmid clones but absent from that of the pBluescript subclones could have been responsible for the transcription of lsc in E. coli.
Moderate or low levels of expression, the immediate transport into the periplasm, or intrinsic characteristics of the P. syringae levansucrase might account for the absence of lethal effects of this enzyme on E. coli cells. Cell lysis and cell death in E. coli and various other gram-negative bacteria growing on sucrose had been reported when the B. subtilis levansucrase gene was expressed in those organisms (13), a phenomenon that has been successfully used to counterselect for insertions within the gene.
Disruption of the lsc gene abolished levan formation in E. coli, underlining the fact that the cloned gene was functional. The generation of levansucrase-deficient mutants in P. syringae by homologous recombination and their assessment in planta is currently under way in our laboratories. Levan-deficient mutants of E. amylovora spread less rapidly in host tissue, although levan did not appear to be required for pathogenicity in E. amylovora, demonstrating the role of levan formation as a virulence factor (14).
Despite quantitative differences in levan formation (22), lsc genes seemed to be highly conserved among P. syringae pv. glycinea, phaseolicola, morsprunorum, and lachrymans, as shown by PCR screening and Southern blotting. Genetic fingerprinting analyses with plant-pathogenic bacteria had previously indicated an intimate relatedness among these pathovars in comparison with other P. syringae pathovars (31, 33, 50).
Levan and alginate are the two main EPSs produced by P. syringae (12, 17). In contrast to the biosynthesis of the highly complex EPS in Ralstonia solanacearum, where a complex regulatory network has been uncovered (21, 42), at present little information on the regulation of EPS synthesis in P. syringae is available. Alginate and levan production might not be coordinately controlled at all. Instead, the polysaccharides might have distinct functions during different steps of the infection process. Recently, characterization of the alginate biosynthetic gene cluster in P. syringae pv. syringae has shown that the algD promoter is dependent on temperature and osmolarity (34). Moreover, effects of the copper ion concentration on alginate biosynthesis in the same organism have been reported (26).
Since levan formation seems to be multifunctional during the epiphytic and pathogenic life of P. syringae, we speculate that gene expression and/or secretion mechanisms for levansucrase undergo regulation by different abiotic and biotic environmental factors. Molecular tools to study these factors are now available, and appropriate investigations are currently in progress.
ACKNOWLEDGMENTS
We thank all the researchers listed in Table 1 for providing bacterial strains and K. Geider, C. L. Bender, and G. W. Sundin for stimulating discussions.
This work was financed by the Max Planck Society and by the Deutsche Forschungsgemeinschaft.
Ursula Hettwer and Frank R. Jaeckel contributed equally to this work.
REFERENCES
- 1.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta J, Hernandez L, Coego A, Suarez V, Balmori E, Menendez C, Petit-Glatron M F, Chambert R, Selman-Housein G. Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiology. 1996;142:1077–1085. doi: 10.1099/13500872-142-5-1077. [DOI] [PubMed] [Google Scholar]
- 3.Barta T M, Kinscherf T G, Willis D K. Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. J Bacteriol. 1992;174:3021–3029. doi: 10.1128/jb.174.9.3021-3029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beveridge T J, Graham L L. Surface layers of bacteria. Microbiol Rev. 1991;55:684–705. doi: 10.1128/mr.55.4.684-705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradbury J F. Identification of cultivable bacteria from plants and plant tissue cultures by use of simple classical methods. Acta Hortic. 1986;225:27–37. [Google Scholar]
- 7.Chambert R, Petit-Glatron M F. Secretion mechanism of Bacillus subtilis levansucrase: characterization of the second step. J Gen Microbiol. 1988;134:1205–1214. doi: 10.1099/00221287-134-5-1205. [DOI] [PubMed] [Google Scholar]
- 8.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedonder R. Levansucrase from Bacillus subtilis. Methods Enzymol. 1966;8:500–506. [Google Scholar]
- 10.Denny T P. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 11.El-Banoby F, Rudolph K. Structure and function of the extracellular polysaccharide of Pseudomonas syringae pv. syringae synthesized in sucrose containing media. In: Rudolph K, Burr T J, Mansfield J W, Vivian A, von Kitzell J, editors. Developments in plant pathology. 9. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 271–276. [Google Scholar]
- 12.Fett W F, Osman S F, Dunn M F. Characterization of exopolysaccharides produced by plant-associated fluorescent pseudomonads. Appl Environ Microbiol. 1989;55:579–583. doi: 10.1128/aem.55.3.579-583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geider K, Bellemann P, Bernhard F, Chang J C, Geier G. Exopolysaccharides in the interaction of the fire-blight pathogen Erwinia amylovora with its host cells. In: Henecke H, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Press; 1991. pp. 212–217. [Google Scholar]
- 15.Geier G, Geider K. Characterization and influence on virulence of the levansucrase gene from the fireblight pathogen Erwinia amylovora. Physiol Mol Plant Pathol. 1993;42:387–404. [Google Scholar]
- 16.Gross M, Geier G, Rudolph K, Geider K. Levan and levansucrase synthesized by the fireblight pathogen Erwinia amylovora. Physiol Mol Plant Pathol. 1992;40:371–381. [Google Scholar]
- 17.Gross M, Rudolph K. Studies on the extracellular polysaccharides (EPS) produced in vitro by Pseudomonas syringae pv. phaseolicola. II. Characterization of levan, alginate, and LPS. J Phytopathol. 1987;119:206–215. [Google Scholar]
- 18.Gross M, Rudolph K. Studies on the extracellular polysaccharide (EPS) produced in vitro by Pseudomonas syringae pv. phaseolicola. III. Kinetics of levan and alginate formation in batch culture and demonstration of levansucrase activity in crude EPS. J Phytopathol. 1987;119:289–297. [Google Scholar]
- 19.Helbert J R, Brown K D. Factors influencing quantitative determination of methyl pentoses and ketohexoses with anthrone. Anal Chem. 1955;27:1791–1796. [Google Scholar]
- 20.Hettwer U, Gross M, Rudolph K. Purification and characterization of an extracellular levansucrase from Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1995;177:2834–2839. doi: 10.1128/jb.177.10.2834-2839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Carney B F, Denny T P, Weissinger A K, Schell M A. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J Bacteriol. 1995;177:1259–1267. doi: 10.1128/jb.177.5.1259-1267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeckel, F. R., U. Hettwer, K. Rudolph, and M. S. Ullrich. Unpublished data.
- 23.Jones J D G, Gutterson N. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene. 1987;61:299–306. doi: 10.1016/0378-1119(87)90193-4. [DOI] [PubMed] [Google Scholar]
- 24.Kasapis S, Morris E R, Gross M, Rudolph K. Solution properties of levan polysaccharide from Pseudomonas syringae pv. phaseolicola, and its possible role as a blocker of recognition during pathogenesis. Carbohydr Polym. 1994;23:55–64. [Google Scholar]
- 25.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 26.Kidambi S P, Sundin G W, Palmer D A, Chakrabarty A M, Bender C L. Copper as a signal for alginate synthesis in Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1995;61:2172–2179. doi: 10.1128/aem.61.6.2172-2179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 28.Kiraly Z, El-Zahaby H M, Klement Z. Role of extracellular polysaccharide (EPS) slime of plant pathogenic bacteria in protecting cells to reactive oxygen species. J Phytopathol. 1997;145:59–68. [Google Scholar]
- 29.Leigh J A, Coplin D L. Exopolysaccharides in plant-bacterial interactions. Annu Rev Microbiol. 1992;46:307–346. doi: 10.1146/annurev.mi.46.100192.001515. [DOI] [PubMed] [Google Scholar]
- 30.Lindow S E. Determinants of epiphytic fitness in bacteria. In: Andres J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 295–314. [Google Scholar]
- 31.Louws F J, Fulbright D W, Taylor Stephens C, de Bruijn F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyness E W, Doelle H W. Levansucrase from Zymomonas mobilis. Biotechnol Lett. 1983;5:345–350. [Google Scholar]
- 33.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peñaloza-Vázquez A, Kidambi S P, Chakrabarty A M, Bender C L. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J Bacteriol. 1997;179:4464–4472. doi: 10.1128/jb.179.14.4464-4472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petit-Glatron M F, Benyahia F, Chambert R. Secretion of Bacillus subtilis levansucrase: a possible two-step mechanism. Eur J Biochem. 1987;163:379–387. doi: 10.1111/j.1432-1033.1987.tb10810.x. [DOI] [PubMed] [Google Scholar]
- 36.Rudolf K, Gross M, Ebrahim-Nesbat F, Noellenburg M, Zomorodian A. The role of extracellular polysaccharides as virulence factors for phytopathogenic pseudomonads and xanthomonads. In: Kado C I, Corsa J H, editors. Molecular mechanisms of bacterial virulence. Dordrecht, The Netherlands: Kluwer Academic Press; 1994. pp. 357–378. [Google Scholar]
- 37.Salmond G P C, Reeves P J. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Sato S, Koga T, Inoue M. Isolation and some properties of extracellular d-glucosyltransferases and d-fructosyltransferases from Streptococcus mutans serotypes c, e, and f. Carbohydr Res. 1984;134:293–304. [Google Scholar]
- 40.Sauerstein J, Reuter G. Nachweis und Charakterisierung einer konstitutiven Levansucrase und einer epigenetisch regulierten Saccharase in Pseudomonas syringae pv. phaseolicola. J Basic Microbiol. 1988;28:667–672. doi: 10.1002/jobm.3620280927. [DOI] [PubMed] [Google Scholar]
- 41.Schaad N W. Laboratory guide for identification of plant pathogenic bacteria. St. Paul, Minn: Bacterial Committee of the American Phytopathological Society; 1988. [Google Scholar]
- 42.Schell M A. To be or not to be: how Pseudomonas solanacearum decides whether or not to express virulence genes. Eur J Plant Pathol. 1996;102:459–569. [Google Scholar]
- 43.Shiroza T, Kuramitsu H K. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J Bacteriol. 1988;170:810–816. doi: 10.1128/jb.170.2.810-816.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song K B, Joo H K, Rhee S K. Nucleotide sequence of levansucrase gene (levU) of Zymomonas mobilis ZM1 (ATCC 10988) Biochim Biophys Acta. 1993;1173:320–324. doi: 10.1016/0167-4781(93)90130-6. [DOI] [PubMed] [Google Scholar]
- 45.Staskawicz B J, Dahlbeck D, Keen N T. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA. 1984;81:6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinmetz M, Le Coq D, Aymerich S, Gonzy-Treboul G, Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985;200:220–228. doi: 10.1007/BF00425427. [DOI] [PubMed] [Google Scholar]
- 47.Sutherland I W. Biosynthesis and composition of gram-negative bacterial extracellular and wall polysaccharides. Annu Rev Microbiol. 1985;39:243–270. doi: 10.1146/annurev.mi.39.100185.001331. [DOI] [PubMed] [Google Scholar]
- 48.Tang L B, Lenstra R, Borchert T V, Nagarajan V. Isolation and characterization of levansucrase-encoding gene from Bacillus amyloliquefaciens. Gene. 1990;96:89–93. doi: 10.1016/0378-1119(90)90345-r. [DOI] [PubMed] [Google Scholar]
- 49.Ullrich M, Bereswill S, Völksch B, Fritsche W, Geider K. Molecular characterization of field isolates of Pseudomonas syringae pv. glycinea differing in coronatine production. J Gen Microbiol. 1993;139:1927–1937. doi: 10.1099/00221287-139-8-1927. [DOI] [PubMed] [Google Scholar]
- 50.Weingart H, Völksch B. Genetic fingerprinting of Pseudomonas syringae using ERIC-, REP-, and IS50-PCR. J Phytopathol. 1997;145:339–345. [Google Scholar]
- 51.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]