Abstract
One of the principal assumptions in symbiosis research is that associated partners have evolved in parallel. We report here experimental evidence for parallel speciation patterns among several partners of the sepiolid squid-luminous bacterial symbioses. Molecular phylogenies for 14 species of host squids were derived from sequences of both the nuclear internal transcribed spacer region and the mitochondrial cytochrome oxidase subunit I; the glyceraldehyde phosphate dehydrogenase locus was sequenced for phylogenetic determinations of 7 strains of bacterial symbionts. Comparisons of trees constructed for each of the three loci revealed a parallel phylogeny between the sepiolids and their respective symbionts. Because both the squids and their bacterial partners can be easily cultured independently in the laboratory, we were able to couple these phylogenetic analyses with experiments to examine the ability of the different symbiont strains to compete with each other during the colonization of one of the host species. Our results not only indicate a pronounced dominance of native symbiont strains over nonnative strains, but also reveal a hierarchy of symbiont competency that reflects the phylogenetic relationships of the partners. For the first time, molecular systematics has been coupled with experimental colonization assays to provide evidence for the existence of parallel speciation among a set of animal-bacterial associations.
Cooperative associations with symbiotic bacteria are a common and ancient theme in the biology of animal and plant life. Thus, it is not surprising that there is often congruency between the evolutionary patterns of closely related host species and their symbiotic partners (1, 6, 11, 13). This congruency, known as parallel cladogenesis, has been revealed by comparing the sequences of genes that have rates of divergence suitable for such studies. However, because most animal-bacterial associations cannot be experimentally initiated, the mechanisms underlying the processes of cospeciation and host-symbiont specificity have not been explored.
The luminescent organ associations between sepiolid squids and luminous bacteria provide an unusually tractable system for the study of the evolution of symbiosis because (i) both the host and symbiont can be cultured and maintained in the laboratory (10, 19); (ii) newly hatched squids are colonized by symbiotic bacteria from the environment, allowing the association to be initiated and monitored experimentally (for review, see reference 26); and (iii) the numerous sepiolid squid species have a wide biogeographic distribution (22). The ease of studying both the squid host and its bacterial symbiont separately or combined under experimental conditions allows one to examine whether a particular squid host can distinguish its own natural symbiont from those of other sepiolid squid species (17) and whether this specificity reflects the evolution of the partnership. While coevolution has been experimentally studied in certain plant-bacterial systems (31, 33), the squid-vibrio symbiosis offers a unique opportunity to do so among animal-bacterial associations.
In the present study, the sequences of one nuclear locus and one mitochondrial locus have been used to generate phylogenetic trees for several species of sepiolids from Indo-West Pacific, Eastern Pacific, Mediterranean, and Atlantic populations. For a subset of the species, we determined the phylogenetic relationships of their symbiotic bacteria. An analysis of these data showed congruency between the derived phylogenetic trees of the hosts and their symbionts. In experiments in which two symbiotic bacterial strains were both present during the colonization of a representative host species, a hierarchy of competitive dominance was derived that mirrored the congruency pattern of these phylogenetic trees. Taken together, these data suggest that cospeciation has occurred during the evolution of the squid-vibrio symbioses and that initial recognition processes are key specificity determinants in these associations. These results provide the first experimentally derived support of cospeciation between animal hosts and their bacterial symbionts.
MATERIALS AND METHODS
Generation of molecular phylogenies.
Squid specimens and their bacterial symbionts were collected alive at nine different geographic locations (Table 1). In addition, formalin- or ethanol-preserved specimens of Euprymna stenodactyla, Sepiola atlantica, Sepiola aurantica, Sepiola ligulata, Sepiola rondoletti, Rossia macrosoma, and Heteroteuthis dispar were obtained from various museum and private collections. Bacterial symbionts were identified as either Vibrio fischeri, Vibrio logei, or Photobacterium leiognathi as previously described (7, 25). Only light organ isolates of sepiolid species that were identified as V. fischeri were compared in this study (7).
TABLE 1.
Geographical collection sites and symbiont characteristics for specimens of squids from the families Sepiolidae and Loliginidae
| Species | Collection site | Representative luminescent organ symbiont strains | Luminescence on seawater nutrient agar medium |
|---|---|---|---|
| Euprymna morsei | Seto Sea, Japan | EM17a and EM24a | Bright |
| Euprymna scolopes | Kane’ohe Bay, Hawai’i | ES114a and ES12a | Nonvisible |
| Euprymna stenodactyla | Solomon Islands | NAb | |
| Euprymna tasmanica | Crib Point, Australia | ET101a and ET104a | Dim |
| Heteroteuthis dispar | Atlantic Ocean | NA | |
| Loliolus noctiluca | Crib Point, Australia | LN101c | Bright |
| Sepietta oweniana | Banyuls-sur-mer, France | No organ | |
| Sepiola affinis | Banyuls-sur-mer, France | SA1a and SA18a | Moderately bright |
| Sepiola atlantica | Vigo, Spain | NA | |
| Sepiola aurantica | Adriatic Sea | NA | |
| Sepiola intermedia | Banyuls-sur-mer, France | SI6a and SI8a | Moderately bright |
| Sepiola ligulata | Banyuls-sur-mer, France | NA | |
| Sepiola robusta | Banyuls-sur-mer, France | SR5a | Moderately bright |
| Sepiola rondoleti | Adriatic Sea | NA | |
| Rossia macrosoma | Atlantic Ocean | No organ | |
| Rossia pacifica | San Diego, Calif. | No organ |
V. fischeri.
NA, symbiont isolates not available.
P. leiognathi.
Squid DNA was extracted from fresh tissue (1 to 10 g) by homogenization in 100 mM Tris-HCl (pH 8), containing 1.4 M NaCl, 20 mM EDTA, 2% (wt/vol) hexadecyltrimethylammonium bromide, 0.2% (vol/vol) β-mercaptoethanol, 0.1% (wt/vol) polyvinylpyrrolidone, and 0.1% (wt/vol) sodium dodecyl sulfate (SDS). The homogenate was incubated at 60°C for 60 min, an equal volume of a chloroform-isoamyl alcohol mixture (24:1) was then added, and the suspension was centrifuged at 10,000 × g for 10 min. A two-thirds volume of cold isopropanol was added to the resulting aqueous phase, and this solution was placed at −20°C overnight to precipitate the DNA. The DNA was then pelleted, resuspended in 2.5 ml of TE buffer (10 mM Tris-HCl and 1 mM EDTA [pH 8.0]) containing 2.6 g of CsCl2, and centrifuged at 270,000 × g for 8 h at 20°C. The isolated genomic DNA fraction was then reprecipitated with 95% ethanol, washed with 70% ethanol, dried, and resuspended in TE buffer.
To isolate DNA from fixed animal tissues, between 1 and 10 mg of tissue was extracted at 65°C in 500 μl of a buffer containing 1 mM NaCl, 1 mM Tris-HCl, 0.1 mM EDTA (pH 7.5), and 0.2% (vol/vol) SDS, to which was added 250 μl of 7 M ammonium acetate (12). The homogenate was centrifuged, and the DNA was precipitated, washed, and resuspended in TE buffer as described above.
DNA templates for sequence analyses were obtained by PCR amplification. Each 100-μl PCR mixture contained 1 μl of template DNA solution, 0.01 μM (each) PCR primer, and 200 μM total deoxynucleoside triphosphates. DNA from sepiolid species was amplified with PCR primers specific for either their internal transcribed spacer (ITS) region between the 18S and 25S rRNA genes (1,425 to 1,450 bp in length, including ITS region 1, the 5.8S gene, and ITS region 2) or their cytochrome oxidase subunit I (COI) sequence (700 bp in length). For the ITS region, the PCR primer sequences were those reported by Goff et al. (9). The amplification conditions used were 25 cycles of 94°C for 75 s, 55°C for 2 min, and 72°C for 4 min. The ITS regions for several species could not be amplified because of the poor condition of the template DNA, particularly from formalin-fixed specimens. Because the ITS exists in several genomic copies, two or three different ITS fragments from an individual of each species were cloned and sequenced to estimate the extent of both interclonal and within-individual variation. For the COI region, universal primers (8) were used under the following amplification conditions: 25 cycles of 94°C for 50 s, 50°C for 75 s, and 72°C for 90 s. PCR fragments were directly sequenced in both directions by using an automated sequencer. Sequencing of the entire ITS region required an additional two internal primers: 5′-TCGTCGATCGGAGACGCGGC-3′ (forward) and 5′-CCTCCACAGTGTTTCTTCAC-3′ (reverse).
DNA was isolated from strains of symbiotic bacteria (Table 1) that were cultured from the luminescent organs of freshly collected squid hosts as previously described (3). The bacterial DNA recovered from the luminescent organs of fixed host specimens was not suitable for sequencing. To extract DNA, individual colonies were homogenized in 200 μl of a buffer composed of 20 mM Tris-HCl and 0.05 mM EDTA (pH 7.4), and containing 5% (wt/vol) Chelex-100 resin (Bio-Rad Laboratories, Richmond, Calif.). The homogenate was incubated at 80°C for 25 min and then boiled for 10 min to denature proteins and lyse the cells. The cell debris was pelleted by centrifugation, and the supernatant fluid was used as the bacterial DNA template for PCR. DNA was amplified with primers specific for the glyceraldehyde phosphate dehydrogenase (gapA) gene (15) as previously described (16). To derive phylogenetic trees for both the hosts and the symbionts, DNA sequences were analyzed by parsimony analysis with PAUP 3.1.1 (3), and the maximum likelihood calculations were made with PHYLIP, version 3.5 (24), and fastDNAml (24), with no assumption of a molecular clock.
Colonization experiments.
For determinations of either the extent of colonization or the degree of competitiveness between different strains of symbiotic bacteria, we used the standard colonization assay as previously described (17, 27). Briefly, newly hatched Euprymna scolopes squid were placed in vials containing 5 ml of seawater, to which approximately 104 CFU of either one or, for competition experiments, two strains of symbiotically competent bacteria had been added. After 12 h of incubation, the juvenile squid were transferred to vials containing 5 ml of seawater without symbiotic bacteria. The progress of light organ colonization was periodically monitored by measuring the luminescence of each squid with a sensitive photometer (model 3000; Biospherical Instruments, San Diego, Calif.). Because bacterial cells from light organ homogenates have essentially a 100% plating efficiency (27), the actual extent of colonization could be calculated from the number of CFU arising from aliquots of light organ homogenates that were plated on seawater nutrient agar medium (3). Different clutches of E. scolopes eggs were used in at least three replicate competition experiments to ensure that the results were not affected by any interclutch variation in colonization characteristics. Forty-eight hours after inoculation, the relative degree of colonization by the two competing bacterial strains from different host species was quantified by plating homogenates of light organs of E. scolopes juveniles exposed to mixtures of the strains (17). As previously described (23), the relative abundance of each strain in the light organ was determined by the visually distinct and genetically stable differences in the luminescence intensities of colonies of these different strains.
Nucleotide sequence accession number.
The sequences reported in this paper have been deposited in the GenBank database under the following accession numbers: ITS sequences, AF031881 to AF031885, AF034558 to AF034565, and AF034844; COI sequences, AF035701 to AF035715 and AF036912; and gapA sequences, AF034845 to AF034851.
RESULTS
Molecular phylogenies.
The ITS and COI sequences of squid specimens were compared for within-species variation and between-species divergence. No differences were found in the >1,400-bp ITS locus between separate clones derived from an individual animal, and less than 0.1% sequence variation occurred between any two or three individuals of the same squid species at the ITS locus. Sequence divergence rates ranged from 2% (between species) to 12% (between species in the same genus and outgroups). Similarly, there was no variation observed between individuals of the same species at the COI locus, and the sequence divergence rates ranged from 3.5% to 22%.
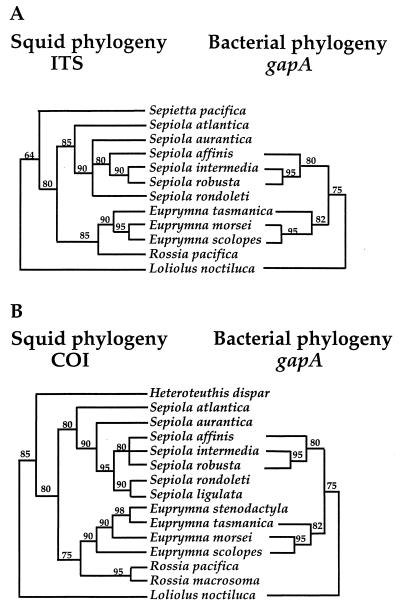
The branching patterns of phylogenetic trees derived from sequences of each of these two loci, by either the parsimony or maximum-likelihood method of analysis, were similar. However, while in all cases Euprymna species clustered together, as did species of Sepiola (Fig. 1), the ITS and COI data sets revealed some minor differences in the species relationships within these clades. Specifically, the relative position of Euprymna morsei varied slightly between the trees. In addition, whereas the ITS data resolved the relationship between the species Sepiola affinis, Sepiola intermedia, and Sepiola robusta, the COI data did not. In both trees, species of the Pacific genera, Euprymna, which bear light organs, and Rossia, which do not, clustered more closely together than Euprymna and Sepiola, which both bear light organs. Similarly, species of the Mediterranean genera, Sepiola, which bear light organs, and Sepietta, which do not, clustered more closely together. The ITS locus of Heteroteuthis dispar, a mesopelagic squid from the Atlantic, could not be completely recovered for sequencing. However, the analyses of COI sequences of this species supported it as an outgroup to all species examined other than Loliolus noctiluca, a loliginid squid with a bacterial light organ, that formed the family-level outgroup for both the ITS and COI loci.
FIG. 1.
Phylogenetic trees of squid host species and their symbiotic bacterial isolates. Each tree was inferred by comparison of the unambiguously aligned positions of either the ITS, the COI, or the gapA sequence. Identical branching patterns were obtained for each locus when analyzed by either the parsimony (30) or maximum-likelihood (24) method. The values given at each node are the percentages of 100 bootstrap resamplings and subsequent heuristic searches that support the relationships between the clades.
The 700-bp gapA sequences obtained from either of the two bacterial isolates that were obtained from the same squid species (Table 1) were identical. Bacterial symbionts isolated from squid species in either the Euprymna clade (strains EM17, ES114, and ET101) or the Sepiola clade (strains SA1, SI6, and SR5) showed a 94 to 98% gapA sequence identity within each clade, but exhibited only an 81 to 88% sequence identity between the two clades (Table 2).
TABLE 2.
Percent gapA sequence identity between different strains of bacterial symbionts
| Symbiont strain | % Identity to symbiont strain (squid host)
|
||||||
|---|---|---|---|---|---|---|---|
| SR5 (S. robusta) | SI6 (S. intermedia) | SA1 (S. affinis) | ES114 (E. scolopes) | EM17 (E. morsei) | ET101 (E. tasmanica) | LN101 (L. noctiluca) | |
| SR5 | 100 | 98 | 95 | 88 | 85 | 82 | 75 |
| SI6 | 100 | 94 | 86 | 83 | 84 | 75 | |
| SA1 | 100 | 85 | 84 | 81 | 77 | ||
| ES114 | 100 | 98 | 96 | 76 | |||
| EM17 | 100 | 95 | 75 | ||||
| ET101 | 100 | 74 | |||||
| LN101 | 100 | ||||||
Phylogenetic analysis of the gapA locus of the symbiotic bacteria revealed a branching pattern that aligned exactly with that of the host ITS tree (Fig. 1A), whereas the symbiont tree aligned similarly, but not exactly, with the host COI tree (Fig. 1B).
Competitive dominance.
When presented individually, all bacterial strains isolated from Euprymna species were capable of initiating and maintaining a typical level of symbiotic colonization of juvenile E. scolopes; i.e., at 24 and 48 h postinoculation, there were between 3 × 105 and 10 × 105 CFU per light organ (data not shown and reference 28). In contrast, SA1, the Sepiola light organ symbiont strain, while able to colonize E. scolopes juveniles, was less effective, reaching a population level of no more than 3 × 105 CFU per light organ. Strain LN101, the light organ symbiont of L. noctiluca, belongs to the distinct luminous species P. leiognathi (Table 1) and has no ability to colonize the E. scolopes light organ.
In all of the competition experiments using symbionts from sepiolid squids, it was observed that 12 h following inoculation, the two strains were present in the light organ at approximately a 1:1 ratio, indicating that these strains were equally competent in initiating a symbiotic colonization (data not shown). However, after 48 h, the native strain, ES114, had achieved a greater than 20-fold advantage over any of the nonnative strains tested (Table 3). When strains from the other two Euprymna species were presented to juveniles of E. scolopes, the strain from E. morsei (EM17), exhibited a fourfold competitive dominance over the Euprymna tasmanica strain (ET101); however, the E. tasmanica strain outcompeted the symbiont from S. affinis (SA1). Thus, the hierarchy among symbiont strains isolated from sepiolid species mirrors the relatedness between the squid-symbiont pairs as derived from the molecular phylogenetic analyses (Fig. 1).
TABLE 3.
Infection of juvenile E. scolopes with mixed inocula of squid light organ symbionts
| Strains used in competition | No. of animals testeda | Mean proportion found in light organs at 48 hb |
|---|---|---|
| ES114 and EM17 | 37 | 98:2 |
| ES114 and ET101 | 35 | 95:5 |
| EM17 and ET101 | 17 | 75:25 |
| ET101 and SA1 | 30 | 90:10 |
| ES114 and LN101 | 21 | 100:0 |
Total number of animals tested in three or four experiments for each pair of strains. All of the animals became colonized by at least one strain in these experiments.
Mean averages of individual ratios of bacterial strains were arc sine transformed (31) to determine the significance of each treatment. All mean values were significantly different to within P ≤ 0.05.
DISCUSSION
In this paper, we present (i) the derived molecular phylogenies of an array of sepiolid squids and their symbiont strains, (ii) an analysis of these phylogenies that revealed parallel cladogenesis between these partners, and (iii) an experimental demonstration that a hierarchy exists in the ability of bacterial symbionts from different species of squids to colonize the Hawaiian squid, E. scolopes.
The systematic relationships among the Sepiolidae have not been adequately revealed by morphological data alone (2), and the fossil record of this group is limited. Thus, molecular data present a particularly valuable approach to resolving the relationships within this family. The phylogenies derived from ITS and COI sequences separated the Sepiola species and the Euprymna species into monophyletic genera (Fig. 1). In addition, the arrangement of squid species on these trees suggests either that luminescent organs arose independently in the evolution of these two genera or that they were lost within members of the genera Sepietta and Rossia.
Although the ITS and COI data resolved the Sepiola species and Euprymna species into two independent groupings, some variability existed between the branching patterns at the species level. Specifically, in the phylogeny derived by using the COI data, E. morsei was more closely related to E. tasmanica and E. stenodactyla than it was to E. scolopes, whereas the ITS data suggested that E. morsei is the sister taxon to E. scolopes (Fig. 1). In addition, the COI tree did not resolve the relationship of the Sepiola species; however, although the COI locus was less useful in certain aspects of these analyses, consideration of the ITS and the COI loci together provided a stronger basis of support for possible coordinate processes occurring in the evolution of these symbioses. The value of data derived from nuclear (ITS), relative to mitochondrial (COI), gene sequences in revealing phylogenies is a controversial issue (14, 20, 21). Thus, analyses based on these two separate gene trees still leave the species tree unresolved. Even with additional molecular phylogenetic data and more knowledge of the life histories of the animals, this issue may not be resolved. However, other results in this paper may be viewed as providing additional support for the ITS tree, i.e., the gapA phylogeny for the bacterial symbionts (Fig. 1B) and the competitive hierarchy from the colonization experiments (Table 3).
The phylogenies derived from the analyses of the squid ITS and the symbiont gapA loci support congruent evolution of host-symbiont pairs. Similar studies with other symbiotic relationships using molecular and/or morphological data have provided evidence that many associations (e.g., chemoautotrophic symbioses, ant-fungal mutualisms, and aphid-Buchnera relationships) are phylogenetically congruent (1, 4, 5, 13). While molecular phylogenetic analysis has been a powerful method to approach these questions, under some circumstances, it has not been able to resolve patterns of parallel cladogenesis (11). Such problems with phylogenetic analyses would benefit from independent measures of congruent evolution.
The ability to manipulate experimentally the sepiolid-bacterial associations offers a series of methods that provide a powerful complementation to phylogenetic analyses. In the present study, we tested whether parallel cladogenesis was reflected in the dynamics of the initiation of this symbiosis. The hierarchy that was observed among the symbionts of the seven squid species tested directly reflected the relative ITS-gapA phylogenies derived for the host animals and their specific symbionts and provided additional support for the evolution of strain specificity and partner fidelity in squid light organ associations. The hierarchical patterns that we observed in the colonization experiments may also provide insight into the processes that occur during divergence of coevolving species. The data showed two patterns: (i) Euprymna symbionts were capable of infecting E. scolopes juveniles fully, but demonstrated a competitive dominance hierarchy; and (ii) Sepiola symbionts were less competitive than the Euprymna strains. These data suggest either that the Euprymna and the Sepiola associations evolved independently or that their divergence included a two-step process, i.e., a change in symbiotic characters that influenced competitiveness, followed by a loss of traits that allowed full colonization.
Because these symbioses can be experimentally initiated, study of the sepiolid associations promises to advance our understanding of host-symbiont evolution. Future studies will pursue two directions: (i) to continue to use the variety of sepiolid species and their symbionts to study parallel evolutionary relationships and (ii) to study the mechanisms that drive the molecular basis for specificity and speciation. For example, examination of the several sympatric species of Sepiola in the Mediterranean (18) will reveal whether each of these squid species is coevolving with a specific lineage of symbionts, or whether instead they all share a common pool of symbionts. In addition, the discovery that symbionts from divergent Euprymna species express different degrees of colonization dominance provides a model for identifying the biochemistry underlying the evolution of specificity and recognition in a host-bacterial interaction. Finally, as has been the case in the plant root nodule symbioses, molecular genetic manipulation of bacterial light organ symbionts has begun to reveal the genes involved in symbiotic competency (26, 32) and should lead ultimately to an understanding of the biochemical mechanisms underlying species specificity in this cooperative partnership.
ACKNOWLEDGMENTS
We thank P. Baumann, N. Davies, G. Roderick, and members of the M.M.N. and E.G.R. laboratories for their comments and suggestions. Squid specimens were caught or donated by S. von Boletzky, A. Guerra, F. G. Hochberg, M. Norman, T. Okutani, R. Villanueva, and R. Young. The ITS external primers were obtained from L. Goff. Automated sequencing was performed with the assistance of G. Bernardi. PAUP analysis was performed under the auspices of D. Eernisse.
This research was supported by National Science Foundation grants OCE-9321645 (Marine Biotechnology Fellowship) to M.K.N. and IBN-96-01155 (to M.M.N. and E.G.R.), as well as by Office of Naval Research awards N00014-91-J-1357 and N00014-93-I-0846 to M.M.N. and E.G.R., respectively, and NIH R01 RR10926 (to E.G.R. and M.M.N.).
REFERENCES
- 1.Baumann P, Moran N A, Baumann L. The evolution and genetics of aphid endosymbionts. BioScience. 1997;47:12–20. [Google Scholar]
- 2.Bello G. A key for the identification of the Mediterranean sepiolids (Mollusca: Cephalopoda) Bull Inst Oceanogr (Monaco) 1995;16:41–56. [Google Scholar]
- 3.Boettcher K J, Ruby E G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cary S C, Warren W, Anderson E, Giovannoni S J. Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol Mar Biol Biotechnol. 1993;2:51–62. [PubMed] [Google Scholar]
- 5.Distel D L, Felbeck H, Cavanaugh C M. Evidence for phylogenetic congruence among sulfur-oxidizing chemoautotrophic bacterial endosymbionts and their bivalve hosts. J Mol Evol. 1994;38:533–542. [Google Scholar]
- 6.Doyle J J. Phylogeny of the legume family: an approach to understanding the origins of nodulation. Annu Rev Ecol Syst. 1994;25:325–349. [Google Scholar]
- 7.Fidopiastis P, von Boletzky S, Ruby E G. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J Bacteriol. 1998;180:59–64. doi: 10.1128/jb.180.1.59-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 9.Goff L J, Moon D A, Nyvall P, Stache B, Mangin K, Zuccarello G. The evolution of parasitism in the red algae: molecular comparisons of adelphoparasites and their hosts. J Phycol. 1996;32:297–312. [Google Scholar]
- 10.Hanlon R T, Claes M F, Ashcraft S E, Dunlap P A. Laboratory culture of the sepiolid squid Euprymna scolopes: a model system for bacterial-animal symbiosis. Biol Bull. 1997;192:364–374. doi: 10.2307/1542746. [DOI] [PubMed] [Google Scholar]
- 11.Haygood M G, Distel D L. Polymerase chain reaction and 16s rRNA gene sequences from the luminous bacterial symbionts of two deep sea anglerfishes. Nature. 1993;363:154–156. doi: 10.1038/363154a0. [DOI] [PubMed] [Google Scholar]
- 12.Herke, S. Personal communication.
- 13.Hinkle G, Wetterer J K, Schultz T R, Sogin M L. Phylogeny of the attine ant fungi based on analysis of small subunit rRNA gene sequences. Science. 1994;266:1695–1697. doi: 10.1126/science.7992052. [DOI] [PubMed] [Google Scholar]
- 14.Hoelzer G A. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees revisited. Evolution. 1997;51:622–626. doi: 10.1111/j.1558-5646.1997.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence J G, Ochman H, Hartl D L. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137:1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- 16.Lee K-H. Ecology of Vibrio fischeri, the light organ symbiont of the Hawaiian sepiolid squid Euprymna scolopes. Ph.D. thesis. Los Angeles: University of Southern California; 1994. [Google Scholar]
- 17.Lee K-H, Ruby E G. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J Bacteriol. 1994;176:1985–1991. doi: 10.1128/jb.176.7.1985-1991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangold K, von Boletzky S. Mediterranean cephalopod fauna. In: Clarke M R, Trueman E R, editors. The Mollusca. Vol. 12. San Diego, Calif: Academic Press; 1988. pp. 315–330. [Google Scholar]
- 19.McFall-Ngai M J, Ruby E G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 20.Moore W S. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution. 1995;49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. [DOI] [PubMed] [Google Scholar]
- 21.Moore W S. Mitochondrial-gene trees versus nuclear-gene trees, a reply to Hoelzer. Evolution. 1997;51:627–629. doi: 10.1111/j.1558-5646.1997.tb02452.x. [DOI] [PubMed] [Google Scholar]
- 22.Nesis K N. Cephalopods of the world. Neptune City, N.J: T.F.H. Publications; 1982. pp. 119–136. [Google Scholar]
- 23.Nishiguchi M K, Ruby E G, McFall-Ngai M J. Phenotypic bioluminescence as an indicator of competitive dominance in the Euprymna-Vibrio symbiosis. In: Hastings J W, Kricka L J, Stanley P E, editors. Bioluminescence and chemiluminescence: molecular reporting with photons. J. New York, N.Y: Wiley & Sons; 1997. pp. 123–126. [Google Scholar]
- 24.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 25.Reichelt J L, Baumann P. Taxonomy of the marine, luminous bacteria. Arch Mikrobiol. 1973;94:283–330. doi: 10.1007/BF00424970. [DOI] [PubMed] [Google Scholar]
- 26.Ruby E G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 27.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 28.Ruby E G, Lee K-H. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokal R R, Rohlf F J. Biometry. W. H. New York, N.Y: Freeman & Co.; 1981. [Google Scholar]
- 30.Swofford D L. PAUP-phylogenetic analysis using parsimony, version 3.1.1. Champaign, Ill: Illinois Natural History Survey; 1990. [Google Scholar]
- 31.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visick K L, Ruby E G. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson H H, Spoerke J M, Parker M A. Divergence in symbiotic compatibility in a legume-Bradyrhizobium mutualism. Evolution. 1996;50:470–477. doi: 10.1111/j.1558-5646.1996.tb03920.x. [DOI] [PubMed] [Google Scholar]