Abstract
Metabolic dysfunction-associated steatohepatitis (MASH), or previously known as non-alcoholic steatohepatitis (NASH), is a severe form of metabolic dysfunction-associated steatotic liver disease (MASLD), or formerly known as non-alcoholic fatty liver disease (NAFLD), which poses a global threat to human health due to its potential to progress to advanced fibrosis, leading to cirrhosis and an increased risk of liver cancer. Recent advances in single-cell methodologies, refined models of disease, and genetic and epigenetic insights have provided a highly nuanced understanding of fibrogenesis in MASH. Although the activation of hepatic stellate cells into myofibroblasts remains a central focus, the substantial heterogeneity of cell types in MASH livers creates opportunities to uncover additional pathways of cell-cell interactions and behavior. Unlike fibrogenesis, mechanisms underlying fibrosis regression in MASH are still inadequately understood. Clarifying the mechanisms behind MASH fibrosis has led to the identification of new therapeutic targets, which may lead to improved clinical outcomes. A refined antifibrotic treatment framework would accelerate progress by improving non-invasive assessment and developing more targeted therapies that preserve hepatocellular function and restore the liver's architectural integrity. These advances in MASH fibrosis epitomize the translational journey from discovery to treatment.
Introduction
The prevalence of obesity is a major health challenge in our rapidly developing world. One serious consequence of the obesity epidemic is the increased incidence of metabolic dysfunction-associated steatotic liver disease (MASLD or previously non-alcoholic fatty liver disease/NAFLD), characterized by excessive fat accumulation (steatosis), and its more severe form, metabolic dysfunction-associated steatohepatitis (MASH or previously non-alcoholic steatohepatitis/NASH), which is associated with inflammation and fibrosis (1-4). Most cases of MASH develop in obese patients but a substantial portion (10-20%) of cases develop in lean individuals for reasons that remain unclear, but which may involve metabolic abnormalities that exist in these lean individuals (5) (6). Up to ~30% of the world’s population has MASLD, and a quarter of them have MASH (7, 8). In the US at least 2 million Americans have ‘high risk’ MASH (defined as those with intermediate stages of disease activity and fibrosis) (9). There are currently no studies that distinguish fibrosis mechanisms or progression between lean and obese MASH.
MASH-related liver disease is predicted to overtake chronic hepatitis C as the leading cause of liver transplantation and is the fastest rising cause of liver cancer (7, 10) and is already the leading indication for liver transplantation among Medicare recipients in the US (11). Despite the scale and urgency of the disease, there are currently no approved drugs for MASH, making it a major unmet need (3, 12). MASH is characterized by steatosis, hepatocyte ballooning, lobular inflammation, and fibrosis. Among these features, the extent of fibrosis is the strongest predictor of patient mortality, and is therefore the focus of drug development efforts (13-16).
The objective of this review is to offer a thorough overview of the molecular mechanisms that drive fibrosis in MASH. Furthermore, we explore the most recent advancements in targeting crucial determinants of MASH fibrosis that may lead to improved outcomes.
The biochemical and cellular landscapes of MASH
Hepatic fibrosis is a pathologic accumulation of extracellular matrix (ECM) molecules comprised of proteins, glycoproteins, and proteoglycans. In normal liver, ECM is sparsely scattered around blood vessels, with basement membrane components present in the subendothelial space of Disse, although the healthy liver lacks an electron-dense basement membrane. Nonetheless, these subendothelial ECM molecules, or the matrisome, support cellular homeostasis of both hepatocytes as well as surrounding endothelial and stellate cells. Progressive injury leads to changes in the quality and quantity of ECM with accumulation of fibrillar collagen (primarily types I and III) initially within the subendothelial space, which coalesce over years to form fibrils that generate bands of collagen. Progressive changes in the composition of the matrisome affect all families of ECM molecules (17, 18), leading to altered biochemical and mechanical features of the liver. Efforts to better understand the biochemistry have laid fallow since the 1980s until recently, when emerging microscopic and proteomic technologies, reagents and single cell analysis have revitalized studies linking ECM to disease. Specifically, accumulation of ECM increases the stiffness of the liver both macro- and microscopically. Microscopically, the enhanced stiffness of the pericellular milieu affects all cells, most notably by activating hepatic stellate cells (HSCs) to increase their fibrogenic activity (19). Concurrently, enhanced organ stiffness disrupts hemodynamics in sinusoids and large vessels. Increasing liver stiffness during fibrosis progression is the basis for diagnostic techniques of transient- and magnetic resonance-elastography to stage MASH fibrosis, as an alternative to liver biopsy.
In addition to increasing liver stiffness, ECM accumulation elicits other changes that can lead to clinical outcomes. In non-cirrhotic stages, soluble signals from the changing ECM composition or fibrogenic cells can impede regeneration, disrupt immune homeostasis, and alter the clearance of systemic and hepatic metabolites. ECM may also generate a physical barrier to trans-sinusoidal transport (‘capillarization’) and concurrently disrupt the function of other resident liver cells besides hepatocytes, in particular sinusoidal endothelial cells. Once fibrosis begins to distort the liver’s architecture, disrupting blood flow and oxygenation of the tissue, the functional and regenerative capacity of liver is impaired to an extent detectable by routine tests including INR, albumin, and platelet count, among others. There likely are progressive functional changes throughout the course of MASH progression, but current diagnostic tests are not sufficiently sensitive to detect them until they are accompanied by marked architectural disruption. Ultimately both fibrosis and the changes it confers throughout the liver contribute to a microenvironment that is permissive for development of hepatocellular carcinoma.
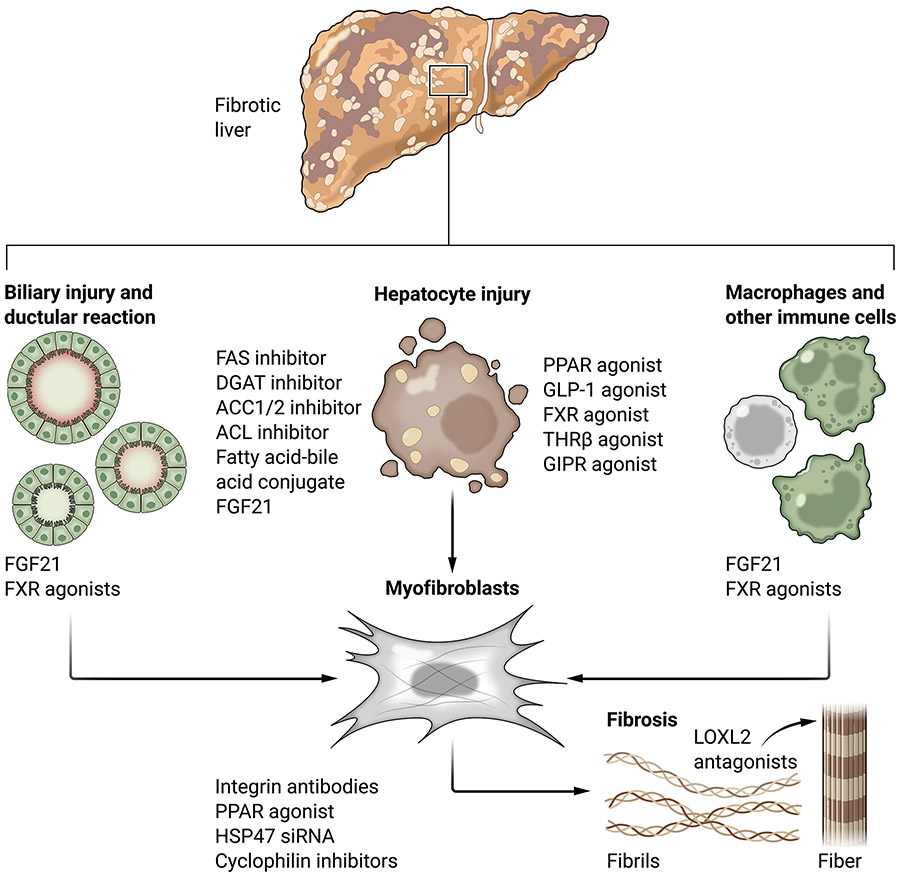
Cellular contributions to hepatic fibrosis in MASH are increasingly understood thanks to rapid advances in imaging, single cell technologies, and publicly available datasets. As a result, a more comprehensive and dynamic understanding of this landscape has emerged (summarized in Figure 1).
Figure 1:
Cellular drivers and potential therapeutics for MASH fibrosis. Potential therapeutics currently undergoing clinical testing to treat MASH are shown in red.
Hepatocyte injury.
Liver fibrosis develops in response to chronic injury to the parenchyme comprised of hepatocytes. In fatty liver disease, increased circulating fatty acid uptake and de novo lipogenesis by hepatocytes promotes lipid accumulation with the generation of reactive oxygen species (ROS) and products of lipid peroxidation. This buildup of toxic byproducts eventually exceeds the liver’s detoxification systems, leading to macromolecular damage, cellular stress, and cell death (4, 20). Hepatocyte death promotes fibrosis indirectly through inflammasome activation and recruitment of inflammatory cells, especially macrophages, that stimulate myofibroblasts and alter extracellular matrix turnover (21, 22). Dying or damaged hepatocytes secrete damage-associated molecular patterns including high mobility group box 1 protein (HMGB1), osteopontin (SPP1), purinoceptor 14 (P2Y14) ligands, apoptotic bodies, microRNA (miRNA), and DNA that can be packaged into microvesicles and directly taken up by HSCs, leading to their activation into collagen-producing myofibroblasts (see “Cellular plasticity”, below) (23-31). Thus far only drugs targeting upstream hepatic lipid accumulation pathways but none of the downstream mediators of cell injury and cell death show signs of efficacy in the clinic. Some of the challenges may be lack of mouse models that recapitulate late-stage disease, as discussed in the disease models section below, and a lack of access to human liver samples for critical validation studies. For a detailed overview of hepatocellular injury and lipotoxicity in MASLD/MASH the readers are referred to two recent reviews (3, 4).
Biliary injury and ductular reaction in MASH.
Injury to cholangiocytes (epithelial cells that line the bile ducts) may lead to cholestasis (bile flow obstruction), provoking the development of a ductular reaction that is histologically defined as a hyperproliferation of biliary ducts, possibly to overcome cholestasis. Ductular reaction in MASH is frequent and strongly correlates with fibrosis severity in patients for unclear reasons (32, 33). Hepatic progenitor cells are typically present within ductular reactions and represent a potential source of transforming growth factor beta (TGFβ and platelet-derived growth factor (PDGF), potent activating stimuli (34-36). Expansion of the hepatic progenitor compartment in disease may increase the release of proliferative and inflammatory stimuli (33). Clarification of progenitor cell biology in liver regeneration and repair is evolving rapidly, but at present there is not yet a clear distinction between bipotential hepatocytes, hepatocyte precursors, and ductular cells, all of which display progenitor features. Better understanding of the progenitor compartment could lead to the development of therapeutic approaches to enhance liver cell regeneration, attenuate fibrosis, and reduce cancer development.
Like hepatocytes, cholangiocytes can undergo lipoapoptosis and senescence in the presence of increased saturated fatty acids, which may provoke release of damage-associated molecular patterns, senescence-associated secretory patterns (such as chemokine ligand 2 (CCL2)), and SPP1, which activate HSCs to generate fibrosis (37-40). It is unclear if these pathways contribute to the correlation between ductular reaction and fibrosis in MASH, however.
Myofibroblasts.
Similar to other tissues, myofibroblasts are the main fibrogenic cells in fibrotic liver. In liver they are derived from resident mesenchymal pericytes, or HSCs. Myofibroblasts are proliferative, migratory, contractile, and secrete extracellular matrix proteins. These functions normally contribute to tissue repair under homeostatic conditions, but sustained activation in chronic liver diseases drives fibrosis, especially in MASH (41) where they are detectable by expression of desmin and alpha smooth muscle actin (αSMA) using immunofluorescence, and by conservation of gene expression based on single-cell RNA-seq (42-46). We recently used single nuclear RNA-seq in murine MASH models and patients with MASH to demonstrate enrichment of pathways in myofibroblasts that regulate extracellular matrix production and cell migration, including matrix metalloproteinase (MMPs), tissue inhibitors of metalloproteinases (TIMPs), and lysyl oxidases (LOXLs) (47-49), similar to earlier studies of mouse fibrosis and human cirrhosis (50, 51). Further informatic subsetting of purified HSCs in the foz/foz mouse model revealed 3 distinct activated cell clusters and 1 inactivated cell cluster, expressing different abundances of classical myofibroblast markers such as alpha smooth muscle actin (Acta2), collagen type 1 alpha 1 (Col1a1), and tissue inhibitors of metalloproteinase 1 (Timp1) at the transcript level. It is not yet established whether these subsets of HSCs with different gene expression profiles represent functionally distinct cell populations with unique biological roles.
The majority of myofibroblasts in chemically-injured mouse models also derive from HSCs, and their depletion reduces liver fibrosis (52-54). Other sources of myofibroblasts include vascular smooth muscle cells, mesothelial cells, and portal fibroblasts, out of which portal myofibroblasts marked by thymocyte differentiation antigen 1 (THY1) may play an outsized role in biliary fibrosis due to their physical localization in the portal region (53, 55-57) (58).
Macrophages and other immune cells.
Macrophages regulate extracellular matrix composition and turnover through the expression of small amounts of collagens (for example COL1A1, collagen type 3 alpha 1 (COL3A1)) and matrix metalloproteases (for example matrix metalloprotease 2 (MMP2), matrix metalloprotease 13 (MMP13) (59, 60). A nuanced appreciation of macrophage function is emerging, with evidence of both pro- and anti-fibrogenic functions based on studies using cell transplantation, chemical manipulation, or depletion (60-63). Macrophages’ primary fibrosis-promoting activity is to stimulate myofibroblasts (59, 64). Macrophages, both the liver resident Kupffer cells and bone marrow monocyte-derived cells recruited in response to injury, secrete cytokines that activate HSCs into myofibroblasts, the most potent of which being TGFβ. Other macrophage-secreted cytokines that may promote HSC activation include PDGF, tumor necrosis factor alpha (TNFα), and interleukin 1 beta ( IL-1β) (65-69). Monocyte-derived macrophages also stimulate HSC migration and recruitment through the secretion of CCL2, chemokine ligand 5 (CCL5), and SPP1 (70-72).
The gradual replacement of liver resident Kupffer cells with monocyte-derived macrophages in injury has two key implications. First, a subset of monocytes recruited into the chronically injured liver differentiate into profibrogenic, triggering receptor expressed on myeloid cells 2 (TREM2)-expressing macrophages (73). Second, monocyte-derived macrophages replace liver resident Kupffer cells that normally clear iron, resulting in iron overload, oxidative stress, and activation of HSCs into myofibroblasts (70, 74-77). In fatty liver, a novel subset of lipid-associated macrophages (LAMs) display a unique ability to metabolize lipids, but their contribution to fibrosis is unknown (70, 76). Our profiling of FAT-MASH macrophages using single nucleus RNA-seq showed no enrichment of ECM-related pathways, only induction of a few select matrix metalloproteinases (MMP2, matrix metalloproteinase 12 (MMP12), matrix metalloproteinase 14 (MMP14)) and decreased collagen type 18 alpha 1 (COL18A1) (48).
Aside from macrophages, other immune cell types may contribute to the inflammatory milieu, including CD4 T-cells, B cells, dendritic cells, innate immune cells, Natural Killer cells, Th17 cells, platelets, and Mast cells but more functional studies are needed (78-89) (90). Natural killer (NK) cells have an intriguing capacity to clear activated HSCs (91-94). NK cytotoxicity may be enhanced by mucosal invariant T (MAIT) cells (95). The HSC-NK cell crosstalk is bidirectional, as activated HSCs can also disrupt cancer dormancy in liver through their secretion of C-X-C motif chemokine 12 (CXCL12) to induce NK cell quiescence that reduces cancer cell killing (94).
Cellular plasticity and heterogeneity.
Fibrogenic cells in the liver exhibit extraordinary plasticity in chronic injury from MASH, especially the activation of HSCs into myofibroblasts. This feature of fibrogenic cells becomes apparent thanks to the remarkable technical advances in single cell analytic methods (see below). HSCs normally exist as quiescent pericyte-like cells that wrap around the vasculature, promote homeostatic hepatocyte turnover through the secretion of growth factors such as hepatocyte growth factor (HGF), and serve as a major storage site for vitamin A. In the presence of signals from the damaged and inflammatory liver, quiescent HSCs “activate” into proliferative, contractile, ECM-remodeling myofibroblasts that also harbor inflammatory features through the secretion of cytokines and chemokines (96) (97, 98). The detection of both quiescent and activated HSCs in mouse liver injury models using single cell RNA-seq reinforces HSC activation as a driver of fibrosis common across etiologies that includes MASH (46-50). Similar evidence is emerging from human cirrhotic patients where multiple HSC subsets can be detected with either quiescent or activated gene expression profiles (47) (99).
Liver macrophages also exhibit considerable plasticity derived from the infiltration of monocytes into the injured liver in response to damage cues and their subsequent differentiation into liver macrophages (100). This process is influenced by the liver microenvironment (101), leading to the identification of several different subsets of monocyte-derived macrophages based on their gene expression. Some monocyte-derived macrophages resemble Kupffer cells, the homeostatic liver resident macrophage, whereas others acquire a classical “M2-like” polarization and promote resolution of inflammation through production of proresolvin mediators such as maresin 1. A maresin 1/ RAR-related orphan receptor alpha (RORα)/12-lipoxygenase autoregulatory circuit prevents inflammation and progression of MASH (102). Within this milieu, some macrophages resemble LAMs as described in the adipose tissue of obese individuals (70, 76). These monocyte-derived macrophages are generally more fibrogenic than Kupffer cells based on their expression of genes related to ECM turnover. LAMs are identified by the expression of TREM2, Tetraspanin CD9 (CD9), and SPP1, which also represent markers of scar-associated macrophages (SAMs) in cirrhotic patients, suggesting a common macrophage lineage in different liver injury models (51). A five-marker signature has recently been described that identified pro-fibrotic macrophages across different tissues, including liver (103). These “Fab 5” cells (CD9+TREM2+ macrophages that express SPP1, transmembrane glycoprotein NMB (GPNMB), fatty acid binding protein 5 (FABP5), and CD63 molecule (CD63)) could represent a candidate macrophage target to reduce fibrogenesis by their inactivation or depletion.
In MASH, liver sinusoidal endothelial cells (LSECs) undergo a morphological transition characterized by the loss of fenestration, lower prevalence of autophagic vacuoles, and the acquisition of an "activated" phenotype. This activation involves the expression of adhesion proteins such as vascular cell adhesion molecule 1 (VCAM-1), which directly recruits pro-inflammatory and pro-fibrogenic immune cells into the liver, while also disrupting the quiescent state of HSCs (104) (105-109). Although the molecular mechanisms through which LSECs contribute to MASH fibrosis are not fully understood, studies in chemically-induced and bile duct ligation (BDL) mouse models of fibrosis suggest that an imbalance between C-X-C motif chemokine receptor 4 (CXCR4) and atypical chemokine receptor 3 (CXCR7) activation in LSECs promotes HSC activation and fibrosis over liver regeneration (110).
Hepatocytes also exhibit plasticity in response to chronic injury in MASH, reactivating several developmental pathways that promote epithelial renewal but also contribute to fibrosis. Notable among these pathways are yes-associated protein 1 (YAP)/tafazzin (TAZ), hedgehog, and, notch, as well as others (111). Increased TAZ abundance in hepatocytes, partly due to cholesterol stabilization, induces Hedgehog expression and secretion that stimulates HSC activation in MASH. Inhibiting hedgehog signaling can attenuate HSC activation and MASH fibrosis (112, 113). Reactivation of notch signaling in hepatocytes in MASH can increase SPP1 expression, which activates HSCs in a paracrine manner. Blocking notch signaling in MASH, both genetically and pharmacologically, can attenuate MASH-associated liver fibrosis (114).
As with all liver cell types, single cell analyses (see next section) of hepatocytes have uncovered critical cellular subsets that have unique disease-promoting or protective features. For example, in experimental MASH and human disease a subset of hepatocytes that express the Ephrin B2 receptor can induce cell-autonomous inflammation that provokes fibrosis, establishing these cells as a potential therapeutic target (49). Conversely, a recent study has identified a subset of hepatocytes that express annexin A2 that preferentially repopulate damaged tissue in acute liver injury (115) – the relevance of this finding to MASH injury is not clear yet.
The impact of “-omic” studies.
In recent years, advancements in genomics and single-cell analyses, largely facilitated by technological breakthroughs in massive parallel sequencing coupled with the development of corresponding informatic pipelines, have provided significant insights into transcriptomic drivers/gene signatures, tissue heterogeneity, and cell-cell communication networks that underlie MASH fibrosis. Through the use of bulk RNA-seq, liver gene signatures were derived from whole liver, isolated hepatocytes, isolated non-parenchymal cells, or plasma that correlated with various disease features and, in some cases, predicted the transition from MASLD to MASH (44, 116-122). Proteomics analysis of patient serum also identified a protein signature that effectively distinguished different stages of fibrosis (123). Further correlation of circulating proteomics with liver transcriptomics has generated a proteo-transcriptomic signature that outperforms current non-invasive methods in predicting patients with progressive MASLD (124).
Single-cell RNA-seq has enabled researchers to attribute differentially expressed genes to specific cell types. Seminal work from the Henderson group first profiled hepatic cells, including HSCs from fibrotic mouse and patient livers (50, 51), which has since been expanded to MASH by several other groups including ours. Although activated HSCs are the primary fibrogenic cells in MASH, these methods have identified an expanded list of fibrogenic drivers expressed by activated HSCs that could represent antifibrotic drug targets (42, 48, 49, 125). Single-cell studies have also revealed remarkable heterogeneity within different cell types in health and disease, identifying multiple HSC clusters in murine models of MASH fibrosis and human cirrhosis. However, the full functional significance of all of these subclusters is not yet completely understood (42, 99, 126). In models of liver cancer (both cholangiocarcinoma and hepatocellular carcinoma) different subsets of HSC-derived cancer-associated fibroblasts (CAFs) have divergent functions that can either promote or attenuate cancer (97, 98). These important observations help reconcile the many reports that were often at odds in defining the role of CAFs in liver cancer, a serious complication of advanced MASH. The findings are also important therapeutically, as identification of specific markers linked to discrete functions of HSC subsets could facilitate targeting to only subsets of disease-promoting cells, for example using chimeric antigen receptor-T cells (CAR-T) to deplete these subsets only (see below) (127-129).
Applying recent developments in informatic tools and databases (130, 131) has uncovered an altered cell-cell communication network in MASH based on the differential expression of ligands and receptors in different cell types (48, 49). These different communication networks are reflected in the differential expression of ligand-receptor signaling pairs, suggesting that the repertoire of disease drivers – and therefore the underlying signaling events – may change as the disease advances, such that advanced fibrotic stages may respond to different pharmacotherapies than earlier stages (48, 49).
Overall, these advancements in genomics and single-cell genomics provide a promising avenue for understanding the mechanisms of MASH fibrosis and identifying therapeutic targets.
Molecular regulation of fibrogenic cell responses
Genetic determinants.
Large-scale genome-wide association studies (GWAS) have identified several genetic polymorphisms that increase the risk of MASH by affecting the amount of fat in the liver and causing lipotoxicity in hepatocytes. Among these polymorphisms, carriers of the Patatin Like Phospholipase Domain Containing 3 (PNPLA3) allele rs738409[G] (encoding I148M) are most prevalent in Hispanics and display an increased susceptibility to MASLD and MASH. Conversely, carriers of the PNPLA3 allele rs6006460[T] (encoding S453I), enriched in African Americans, are associated with decreased MASLD (132-134). These findings may explain the overrepresentation of Hispanic and underrepresentation of African Americans in MASH clinics. From a mechanistic perspective, the I148M single nucleotide polymorphism (SNP) drives injury and fibrosis by promoting fat accumulation in hepatocytes (135) and enhancing fibrogenic activity of HSCs (136), respectively.
Additionally, after the initial discovery of Membrane Bound O-Acyltransferase Domain Containing 7 (MBOAT7) as a risk gene for alcohol-related cirrhosis, MBOAT7 allele rs641738 was subsequently verified as a risk factor for MASH (137-139). The rs58542926 minor allele of transmembrane 6 superfamily member 2 (TM6SF2) is also reportedly associated with MASLD and sometimes MASH (134, 140-142). Furthermore, the splice variant rs72613567 in the Hydroxysteroid 17-beta Dehydrogenase 13 (HSD17B13) gene has been identified as protective against MASH, with HSD17B13 potentially contributing to MASH fibrosis by promoting pyrimidine catabolism (134, 140-142). A recent study has specifically explored HSD17B13 variant frequencies in Hispanic/Latino patients and those of diverse genetic ancestry (143). Absence of the HSD17B13 protein due to a truncation variant is protective against fibrosis, suggesting that this enzyme generates a potential lipotoxic species, which is absent from those with the variant, however the native substrate and product of this protein have not been established. Furthermore, HSD17B13 is not expressed by HSCs, indicating that the impact of the gene in promoting or protecting from fibrosis is indirect and due to changes in hepatocytes. Most recently, a rare germline mutation in the Cell Death Inducing DFFA Like Effector B (CIDEB) gene, which encodes a structural protein in lipid droplets, has been linked to altered lipid metabolism and reduced risk of MASH (144), but its potential direct impact on HSC fibrogenesis has not been reported.
Although these findings provide valuable insights into the mechanisms underlying MASH, overall, few genetic variants have been mechanistically linked to MASH pathogenesis (145), and each exerts only a modest effect on disease outcome at the population level, suggesting that environmental determinants play a more important role in driving the disease. Nonetheless, as the genetic databases such as the UK Biobank continue to scale up, further variants are likely to emerge with the hope that they may yield informative aggregate disease risk scores when combined with clinical and diagnostic features; such polygenic scores has been shown to improve predictions of disease severity by fibrosis score alone (146).
Signaling pathways.
TGFβ is the major signaling pathway promoting fibrogenesis by myofibroblasts in MASH and other chronic diseases in which there is hepatocyte injury (96). Although studies have long implicated TGFβ1 as the dominant fibrogenic driver, a recent study also implicates TGFβ2 and TGFβ3 as potent fibrogenic stimuli, but which lack the pro-inflammatory effects of TGFβ1 antagonism that have limited the latter’s appeal as a therapeutic target (147). High concentrations of cholesterol and free cholesterol accumulate in HSCs in methionine-choline deficient (MCD) MASH mice, which increases toll-like receptor 4 (TLR4) and sensitizes the cells to TGFβ–induced activation (148). Iron can be shuttled by extracellular vesicles from hepatocytes to HSCs in MASH, stimulating fibrogenesis through iron overload and oxidative stress, an effect that is further exacerbated by TGFβ-mediated suppression of the antioxidant protein cytoglobin (CYGB) (74) (149). Another ligand of the TGFβ-bone morphogenetic protein (BMP) superfamily, bone morphogenetic protein 8B (BMP8B), is upregulated in MASH and signals through mothers against decapentaplegic (SMADs) to drive HSC activation in animal models, reinforcing TGFβ's vital role as a fibrogenic stimulus (150). In addition, TGFβ signaling in hepatocytes drive MASH fibrosis by regulating lipid metabolism and hepatocyte apoptosis (151, 152).
Notch signaling has been identified as an important driver of HSC activation in fibrosis in general (114, 153). However, it may play an even more prominent role in MASH fibrosis, where it is reportedly activated not only in HSCs but also in hepatocytes and endothelial cells. In hepatocytes, activation of TLR4 increases the expression of jagged 1 (JAG1), a well-established notch receptor 1 (NOTCH1) ligand, which leads to SRY-box transcription factor 9 (SOX9)-dependent transcription and secretion of SPP1, stimulating HSC activation (114). Furthermore, HSCs synthesize hyaluronic acid through the expression of hyaluronic acid synthase 2, which promotes fibrogenicity in a NOTCH1-dependent manner by stimulating TLR4 and CD44 expressed on HSCs themselves (154). NOTCH expression has also been detected in endothelial cells of patients with MASH, and its selective depletion or overexpression can reduce or aggravate MASH fibrosis in murine models, respectively, possibly by modulating nitric oxide synthase 3 (eNOS) (155). A nanoparticle-based approach to target γ-secretase inhibitor and block NOTCH activity reduced MASH fibrosis in mice (156).
Ballooning of hepatocytes is a prominent pathologic hallmark of MASH that distinguishes MASH from MASLD and is thought to represent a unique type of cell injury that is associated with lipotoxicity and release of inflammatory and fibrogenic signals. These signals include sonic hedgehog, which activates hedgehog signaling in the surrounding stromal cells (157). On the other hand, genetic depletion of Indian hedgehog from hepatocytes reduces MASH fibrosis and hepatocellular carcinoma in a dietary mouse model, suggesting that different hedgehog ligands may have distinct activities (158). SPP1, a downstream target of hedgehog signaling, is induced in myofibroblasts in MASH, and its antagonism reduces fibrosis in the MCD mouse model (38, 159). SPP1 expression and secretion is also promoted by nuclear factor of activated T cells 4 (NFATc4) and coiled-coil-helix-coiled-coil-helix domain containing 2 (CHCHD2), transcription factors activated in MASH mice and patients (160, 161). The convergence of different fibrogenic pathways in MASH upon SPP1 makes it a potential therapeutic target. Indeed SPP1-deficient mice are protected from MASH fibrosis in the MCD model (162). Other major sources of hedgehog ligands may be reactive-appearing ductular cells that strongly correlate with fibrosis in patients with MASH or in MASH-associated hepatocytes where increased cholesterol intake activate TAZ to drive Indian hedgehog expression and secretion (112, 113, 163).
Additional signaling pathways also directly promote or block HSC activation in MASH fibrosis, including fibroblast growth factor 21 (FGF21) (164), glucagon-like peptide 2/ glucagon-like peptide 2 receptor (165), interleukin 11 (IL11) (46, 166), and chemokine ligand 2 (CCL25)/C-C chemokine receptor type 9 (CCR9) (167).
Epigenetic regulation.
Epigenetic regulation plays a critical role in conveying environmental information that influences gene expression and cellular behavior. Recent studies have shed light on the specific epigenetic changes, including DNA and histone modification, and microRNAs, associated with MASLD.
For instance, miR-331-3p and miR-30c can distinguish between twins, with one developing MASLD and the other not (168). Moreover, certain patterns of DNA methylation at fibrosis-related genes, such as those encoding TGFβ1, COL1A1, PDGFα, peroxisome proliferator-activated receptor alpha (PPARα), and peroxisome proliferator-activated receptor delta (PPARδ), can not only distinguish between patients with mild versus advanced MASLD, but also predict future fibrosis progression (169-172). These findings support the idea that DNA methylation changes play a key role in driving MASLD.
Numerous epigenetic modulators participate in modifying and binding to DNA and histone modifications. Among them, histone deacetylase 2 (HDAC2) and DNA methyltransferase 1 (DNMT1) are believed to induce the endothelial production of insulin like growth factor binding protein 7 (IGFBP7)/ADAM metallopeptidase with thrombospondin type 1 motif 1 (ADAMTS1), which in turn recruits profibrogenic Th17 cells and affects the vascular niche (173). Other epigenetic regulators, such as methyl CpG binding protein 2 (MECP2), enhancer of zeste homolog 1/2 (EZH1/2), bromodomain-containing protein 4 (BRD4), Sirtuin 1 (SIRT1), lysine-specific demethylase 4D (KDM4D), and tet methylcytosine dioxygenase 3 (TET3), can contribute to chemically-induced liver fibrosis (174-178) (179, 180) (181). However, their specific contributions to MASH fibrosis remain to be determined.
Recent studies using single nucleus ATAC-seq to profile the epigenome of hepatocytes from MASH mice have revealed increased NOTCH binding in the Ephrin Type-B Receptor 2 (EPHB2) locus, resulting in the upregulation of EPHB2 as a driver of MASH fibrosis (49). This reinforces the potential therapeutic value of targeting NOTCH activity in MASH.
Metabolic regulation of fibrogenic cells.
The regulation of fibrogenic cells is a complex process that involves various metabolic pathways. In particular, the activation of HSCs in culture is associated with increased glycolysis, lipolysis, and glutaminolysis, which provide the energy needed for this energy-intensive process (as detailed in (182)). However, for MASH, only cholesterol and lipid metabolism have been linked to HSC fibrogenicity in vivo.
Increased energy is needed to support HSC activation. In addition to generation of intermediary metabolites through glycolysis, lipolysis, and glutaminolysis, the cells rely on activation of autophagy to release fatty acids through hydrolysis of retinyl esters (183) linked to endoplasmic reticulum stress (184). Autophagy also provokes the release of miR-29a, which can stimulate fibrogenesis in neighboring cells through paracrine signaling (185).
Studies on different strains of mice have shown that plasma cholesterol strongly correlates with the extent of fibrosis, and mice supplemented with high cholesterol develop increased fibrosis (148, 186). Mechanistically, cholesterol accumulation in cultured HSCs has been found to increase TLR4 abundance by suppressing its endosomal degradation, which sensitizes these cells to TGFβ-induced activation (148). In vivo studies have confirmed that TLR4 knockout MCD mice are protected against the profibrotic effect of cholesterol supplementation (187). Lipoprotein lipase abundance also increases in HSCs in patients with MASLD and MASH, and depletion of lipoprotein lipase selectively in HSCs reduces their activation and liver fibrosis progression in a rodent model of MASLD (188). Mechanistically, lipoprotein lipase promotes free cholesterol accumulation in HSCs, which in turn activates TLR4 signaling and reduces Bambi expression that together sensitizes HSCs to TGFβ-driven activation (188).
Acetyl coenzyme A carboxylase (ACC) is the rate-limiting enzyme in de novo lipogenesis. Mice overexpressing both ACC1 and ACC2 display increased de novo lipogenesis in the liver and spontaneously develop MASH-like symptoms (189). Inhibition of ACC1/2 using small-molecule inhibitors can reduce steatosis in hepatocytes, but it also directly blocks primary HSC activation in culture and reduces liver fibrosis in a choline-deficient, high-fat diet rat model of MASH (190, 191). Similarly, pharmacological or genetic inhibition of ATP citrate lyase, which acts upstream of ACC1/2, reduces steatosis and fibrosis, possibly through direct inhibition of HSC activation (192).
Microbiome impact on fibrosis.
MASH fibrosis is linked to a unique gut microbiome (193-197), and specific microbiome signatures can distinguish human MASH from MASLD (4, 198, 199). These findings implicate the microbiota in driving this critical transition, but fecal transplant studies have yielded mixed results and no single bacterium has emerged as a pathogenic agent across different studies. Transplant of fecal microbiota from lean healthy donors into obese patients improved lipid profiles and reduced liver stiffness in one study (200), whereas in another a synbiotic formation (probiotics + prebiotics) failed to alter fibrosis (201). To establish causality, transplantation of fecal microbiota from patients with MASLD or MASH induced steatosis and fibrosis in antibiotic-treated and germ-free mice (87, 202, 203). Co-housing wildtype mice with mice prone to MASH led to increased disease severity in wildtype mice, presumably due to a transfer of fecal microbiota (204).
There are some data linking specific bacterial strains to fibrosis in MASH including the alcohol-producing Klebsiella pneumoniae and Porphyromonas gingivalis; the latter directly promotes HSC activation in culture (202, 205, 206). On the other hand, the replacement of Ruminococcus faecis, found to be deficient in patients with MASH fibrosis improved fibrosis in the MCD rodent MASH model (198). Bacterial lipopolysaccharides can exacerbate fibrosis in mice fed a high-fat diet, provoking an inflammatory cascade through TLR4 that indirectly activates fibrogenic cells (207). Microbial acetate on the other hand, is protective against MASH fibrosis through activation of the free fatty acid receptor 2 (FFAR2) receptor on hepatocytes and modulation of lipid metabolism (208). MASH dysbiosis also incorporates an altered fungal flora, with some efficacy of antifungals in reducing disease severity and fibrosis in germ-free mice (209). Although still in its early days, microbial research is already demonstrating considerable translational potential.
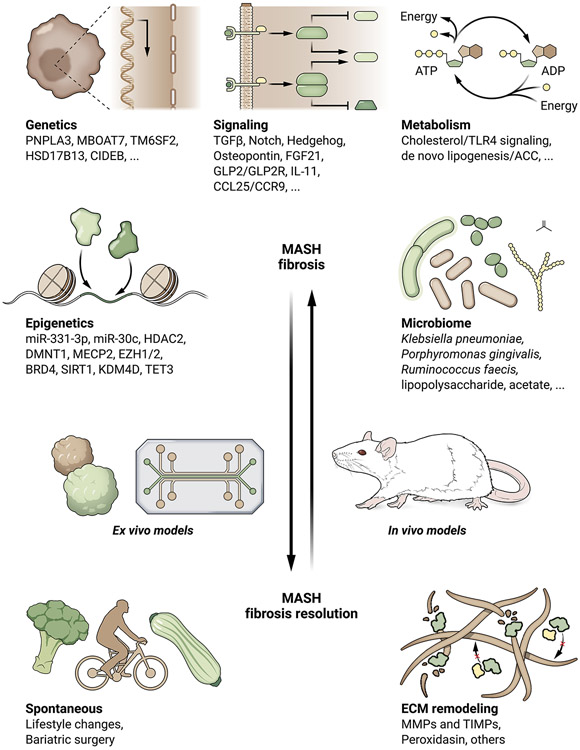
The molecular pathways that drive MASH fibrosis are summarized in Figure 2.
Figure 2:
Molecular regulation of the fibrogenic response. Molecular mechanisms that contribute to MASH fibrosis (left) and MASH fibrosis resolution (right) are depicted.
Models of MASH
Experimental models that recapitulate human disease pathology are central to understanding MASH biology and evaluating candidate drugs. Models are powerful tools to elucidate disease biology owing to the potential to generate gene-altered mice, but cannot predict with absolute certainty the efficacy and safety of therapies in human trials. In vivo, rodent models are the backbone of experimental MASH research. As in humans, the most useful rodent MASH models typically first develop steatosis and then progress to MASH, with increasing fibrosis severity. Deeper interrogation of these models using single cell methodologies will continue to refine the value and limitations of animal models in recapitulating features of human disease.
Rodent models of MASH have been extensively reviewed and comprise 3 broad categories: 1) diet-based; 2) genetic; or 3) mixed, which refers to combined diet, genetic, or chemical perturbations (118, 210-213). One widely used popular mouse model is MCD diet, in which mice rapidly develop severe inflammation and fibrosis in the liver characteristic of human disease pathology, within a period of several weeks. However, these mice do not become obese and therefore fail to develop associated metabolic defects, including insulin resistance. On the other hand, diet-induced MASH models such as the amylin liver NASH (AMLN) and Gubra-amylin NASH (GAN) models follow the natural course of human MASH development in which mice become overweight prior to developing fatty liver, followed by inflammation and fibrosis (214, 215). However, liver disease progression is slow on a Western diet alone in common inbred mouse strains such as C57Bl/6 and thus additional injury may accelerate progression, which can be accomplished by administering low weekly doses of the hepatotoxin carbon tetrachloride (CCl4).
Just as not all obese individuals develop MASLD and MASH, different strains of mice exhibit varying susceptibility to these conditions. For instance, a study involving the expression of human apolipoprotein E*3-Leiden and cholesteryl ester transfer protein in approximately 100 common inbred strains of mice revealed a wide spectrum of fibrosis development. The degree of fibrosis ranged from non-detectable to robust, indicating a major contribution of genetic background to MASH pathology (186).
Among the most often used genetic models of MASLD are mice genetically deficient for leptin (ob/ob) or the leptin receptor (db/db) (216-219). A different adipocyte secreted hormone, adiponectin, has antifibrotic properties but has not yet emerged as a therapeutic, although increased adiponectin is emerging as an indication of therapeutic benefit in clinical trials of metabolic therapy for MASH (220).
Several transgenic animals have been developed to study MASH pathogenesis, including those that overexpress urokinase-type plasminogen activator (MUP-uPA) (221) or sterol regulatory element-binding protein 1 (SREBP1) (222, 223). Additionally, mice deficient in methionine adenosyltransferase 1A (MAT1A) (224), phosphatase and tensin homolog (PTEN) (225), or alternative oxidase (AOX) (226) spontaneously exhibit pathological features of MASH. However, it is worth noting that the etiology of MASH in these models may not precisely parallel human MASH, as these mutations are not observed in patients.
Previous MASLD models that relied only on a single perturbation, for example a Western diet or genetic mutations, typically lack the features or gene expression patterns of human MASH (227). Therefore, several mixed rodent models of MASH combine different stressors. For instance, the diet-induced animal model of non-alcoholic fatty liver disease (DIAMOND) mouse model exposes B6/129 mixed strain mice to a Western diet, leading to the development of hepatocellular carcinoma in most animals between 32-52 weeks (228). The Oz mouse harbors a spontaneous mutation within the Alstrom syndrome 1 (Alms1) gene, similar to patients with Alstrom syndrome, which, when combined with a Western diet on a C57BL/6 background, leads to obesity and diabetes, then MASH, with advanced fibrosis by 12 weeks of age (43, 229, 230). Similarly, another MASH model combines the Agouti yellow mutation driving hyperphagia with a Western diet and fructose water, where mice start to develop fibrosis after 16 weeks (231). Although MASH development in both Oz and Agouti yellow mice follow the natural history of disease progression in humans, both models require mice with specific genetic backgrounds.
Our laboratory uses a simple rodent model of MASH, the “FAT-MASH” mouse, which robustly develops Fibrosis and Tumors, features of advanced disease, after 24 weeks of a Western diet combined with a small weekly dose of the liver toxin CCl4 (45). Dietary cholesterol supplementation in the FAT-MASH mice is necessary to approximate liver cholesterol levels seen in human MASH, since mice are poor cholesterol absorbers compared to humans. Increased cholesterol in turn promotes MASH fibrosis through the TAZ/Indian hedgehog pathway described above (232-234). It is worth noting that the FAT-MASH mice exhibit transcriptomic similarities to human MASH patients (48), and their microbiome changes also resemble human MASH pathology (197).
The prevalence of co-morbid diabetes and MASH is high, with up to 75% of type II diabetes patients also developing MASLD (3). To recapitulate this co-morbidity, a mouse model of diabetes due to beta cell depletion by a low dose of streptozotocin was subjected to subsequently high-fat diet feeding to induce MASH and hepatocellular carcinoma at 20 weeks (235, 236). Although this model recapitulates diabetes and fatty liver, it is unclear if it is generalizable to human MASH because the animals have glucose intolerance due to insulin depletion and not insulin resistance, which is more typical of human MASH.
To enhance the relevance of rodent models to human pathology, animal models of MASH are increasingly incorporating not only pathogenic drivers in liver but also systemic factors, including adipose dysregulation and changes in the microbiome. The microbiome can profoundly influence responsiveness to pharmacotherapies in disease models outside the liver (237, 238), and similar approaches are being explored in MASH. Continued efforts to refine the microbiome's contributions to MASH will help bridge the gap between mouse modeling and human disease (239).
Other shortcomings of current mouse MASH models include the difficulty recapitulating late-stage disease, including cirrhosis and hepatic decompensation, where treatment is most urgently needed. Future models should be designed to model end-stage liver disease and use improved survival as the optimal readout of a successful genetic/pharmacological intervention.
To further improve MASH modeling, in vitro models such as organoids are being developed. Organoids are grown as 3D spheroids in suspension culture and recapitulate in vivo biology more than traditional 2D cultures. MASH organoids can be generated from induced pluripotent stem cells (iPSCs) or embryonic stem cells (ESCs) via stepwise differentiation that parallels liver development, followed by exposure to free fatty acids (240, 241). These MASH organoids derived from iPSCs/ESCs display increased inflammatory and fibrotic responses in addition to increased stiffness, all of which can be reversed with fibroblast growth factor 19 (FGF19) treatment (241). Furthermore, iPSC-derived MASH organoids from different donors have been used to screen for MASH susceptibility loci, leading to the identification of the glucokinase regulator (GCKR) allele rs1260326[T] which increases lipid loading in hepatocytes (242). In a different approach, primary bipotent cells have been isolated from patients with MASH and cultured to differentiate into hepatocyte or cholangiocyte organoids displaying features that align with MASH pathology such as increased fatty acid loading, reduced markers of liver function and proliferative capacity, and sensitivity to apoptosis when compared to similar cultures derived from healthy donors (243). Furthermore, the transcriptomes of these bipotent cell-derived MASH organoid cultures resemble those of human MASH.
Other promising avenues for MASH modeling include the use organ-on-chip systems and precision cut liver slices (PCLSs). These 3D in vitro systems use microfluidics and microphysiological platforms to mimic the liver microenvironment, potentially improving the physiological relevance and reproducibility of in vitro MASH models. These systems consist of layers of primary or immortalized hepatocytes and endothelial cells plated on platforms that allow constant flow of nutrients and oxygen to mimic physiological blood flow. HSCs and/or macrophages are seeded on top of these layers to produce inflammatory and fibrotic mediators, along with the addition of free fatty acids to mimic fatty liver conditions (244).The effectiveness of organ-on-chip systems for MASH modeling has been demonstrated in studies (245-247). As a perhaps simpler alternative to organ-on-chip systems, PCLS cultures are ~200 micron-thick tissue slices cut out of a liver core biopsy that has been shown to maintain cell identity and tissue structure for several days when cultured under specialized conditions (248)(249). However, PCLS requires continued access to human liver core biopsies, which can be challenging to acquire.
Both organoid and organ-on-chip systems hold promise for modeling diseases linked to human germline mutations, as cells derived from patients can be used. Nevertheless, widespread adoption of these systems is hindered by several challenges. These include batch-to-batch variations in primary cells and their proportions, difficulties associated with scaling and maintaining long-term culture, lack of a complete immune component, as well as the complex culture conditions required (245-247). In theory, organ-on-chip systems have the potential to study inter-organ cross-talk, such as those between the liver and intestine; however, the effectiveness of such systems has yet to be proven.
Fibrosis regression in patients and mouse models with MASLD
Natural history of MASLD and MASH fibrosis.
The natural history of MASLD typically starts with fatty liver disease alone followed by inflammation and fibrosis, but we lack a full understanding of disease progression. Additionally patients may have spontaneous improvement without intervention, a feature that could contribute to the surprisingly high rates of response among placebo patients in clinical trials (250-252). Some of these placebo responses could represent inadvertent changes in lifestyle by patients as they participate in clinical trials (253).
Recent studies of patients cured of hepatitis C virus (HCV) have highlighted the liver's impressive ability to resorb advanced fibrosis or cirrhosis once the underlying injury is removed (254). Spontaneous fibrosis regression was also observed in patients with hepatitis B virus (HBV) treated with antivirals to suppress HBV activity (255). Such clinical observations, coupled with the knowledge that fibrosis is the most significant predictor of patient mortality in MASH, focus renewed attention on mechanisms of both spontaneous and pharmacologic-induced fibrosis regression (13-16), as more research is needed to develop clinical interventions that halt progression or reverse the disease.
Clinical interventions such as weight loss resulting from dietary changes, bariatric surgery, pharmacological agents, or the use of farnesoid X receptor (FXR) agonists have been associated with fibrosis regression in patients with MASH (256-258). On the other hand, weight loss did not always correlate with fibrosis regression, as seen in recent clinical trials with pan-PPAR and glucagon like peptide 1 receptor (GLP1R) agonists, suggesting that reduced MASH is probably a more direct and better predictor of fibrosis regression than weight loss. Nevertheless, these interventions suggest that, like HCV and HBV, MASH fibrosis is reversible in non-cirrhotic liver, but unlike in HCV and HBV there is no evidence yet that cirrhosis in MASH is reversible (259).
Earlier stages of MASH fibrosis can regress more easily than advanced stages, a trend observed in other liver diseases (260), which may be explained in part by the marked increase in total collagen and extracellular matrix associated with progressive fibrosis (261). Patterns of fibrosis regression need to be clarified. A recent study using artificial intelligence-based collagen morphometry assessment has indicated that fibrosis regression in MASH starts near the perisinusoidal region (258), but more rigorous validation studies are needed.
Molecular basis of ECM turnover and HSC inactivation.
Animal models reinforce the capacity of the injured liver to regress fibrosis and provide important platform for dissecting underlying mechanisms, although it should be noted that fibrosis in mice regresses incredibly rapidly, which may be in part attributed to the short duration of models (weeks to months in mice vs. years to decades in humans) and initiation of fibrosis at relatively young age (with most studies completed before mice hit “middle age”) (52, 262). In contrast, the rat thioacetamide (TAA) model of fibrosis might represent a possible alternative to mice as TAA-induced fibrosis no longer regresses after 6-8 weeks on the model (263, 264). As noted above, termination of liver injury is the most efficient way to spur fibrosis regression, where activated HSCs are removed via apoptosis or deactivation, reducing the fibrogenic cell population.
Stellate cell activation is regulated through coordinated expression of transcription factors governing fibrogenesis, proliferation, chemotaxis, and contractility, whereas a separate transcriptional program controls cellular deactivation. A growing list of transcription factors promote the quiescent state of HSCs, including transcription factor 21 (TCF21) (265), LIM/homeobox protein 2 (LHX2) (266), GATA binding protein 4 (GATA4) (267), and PPARγ (268). In principle enhancement of their activity could have antifibrotic activity, but this has not been tested yet.
We have only a fragmentary understanding of how the fibrotic scar is degraded (18, 269, 270). Animal models implicate HSCs and immune cells, especially macrophages, as major cellular drivers of extracellular matrix turnover, both through reduced secretion of ECM components and crosslinking enzymes as well as increased secretion of proteases and their inhibitors (51, 52, 64, 262, 270-274).
Although MASH fibrosis regresses spontaneously in experimental models when injury is attenuated (275), few studies have focused on mechanisms of fibrosis regression in MASH. An earlier study implicated a specific subset of macrophages in mice treated with CCl4 that are low-Ly6C-expressing or “Ly6Clo” as a potential source of proteolytic enzymes (276, 277), but neither the human equivalent of these cells nor the specific proteases they secrete have been established. Concurrently, tissue inhibitors of metalloproteases (TIMPs) produced by HSCs are increased in experimental fibrosis, which could block the activity of their cognate enzymes and also prevent apoptosis of activated HSCs (278, 279).
Lysyl oxidases (LOX) enzymes are the major ECM crosslinkers in liver that stabilize collagen or elastin by forming covalent crosslinks. LOX inhibition destabilizes collagen fibers and accelerates fibrosis regression in chemically-induced fibrosis (273), however clinical trials of a neutralizing antibody to lysyl oxidase like 2 (LOXL2) had no effect (280). In MASH fibrosis a different crosslinker, peroxidasin, has been implicated in fibrosis regression, as mice genetically deficient in peroxidasin display accelerated fibrosis regression (281). However, the mechanisms behind peroxidasin-induced fibrosis regression does not require its crosslinking activity towards collagen IV (282); rather, peroxidasin deficiency produces a hypoxic environment that reduces the activation of HSCs and recruits pro-healing macrophages (281).
Diagnostic landscape
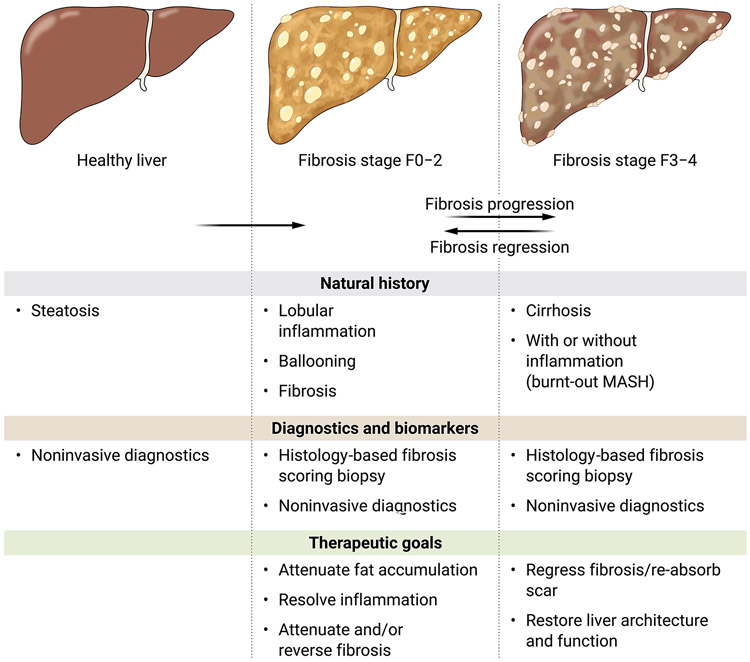
The fervor to clarify the mechanisms of MASH fibrosis has yielded remarkable progress in identifying candidate therapeutic targets (Figure 1). Still, we lack a clear hierarchy of causality that establishes which are the most vulnerable points of therapeutic attack. Indeed, given the remarkable heterogeneity of the disease, including its clinical presentation and extrahepatic co-morbidities, as well as its environmental and genetic factors, MASH may well comprise multiple subtypes, each of which has its own vulnerabilities to therapy. At the same time, although the fundamental diagnosis of MASH and assessment of fibrosis severity have relied on liver biopsy, the field is rapidly moving closer to non-invasive diagnostics that will replace biopsy, thereby accelerating clinical trial enrollment and assessment of therapeutic efficacy for candidate drugs. The following sections lay out in broad strokes the current status of diagnostic and therapeutic development for fibrosis in MASH, emphasizing the most promising directions and unmet needs (summarized in Figure 3). For more nuanced reviews of drug development and clinical trial design, the reader is referred to several recent articles (283-285).
Figure 3:
Diagnostic and therapeutic opportunities in MASH fibrosis.
Among the histologic features of MASH, fibrosis is the only one whose severity correlates with clinical outcomes (286-288). Thus, although the classic histologic features of MASH, including ballooning, lobular inflammation, and steatosis may be linked to necro-inflammatory activity, their improvement does not guarantee therapeutic benefit in terms of reduced complications or mortality. The heightened focus on fibrosis has greatly impacted the design of clinical trials, which now require either complete resolution of MASH or improvement in fibrosis with no worsening of MASH. Still, current clinical trials that rely on biopsy, or future studies that are evaluated using non-invasive biomarkers, will require long term follow-up to ensure that improvements in these parameters reliably correlate with better clinical outcomes. These outcomes include reduced mortality and decreased complications of end stage liver disease, including reduced manifestations of portal hypertension or hepatocellular carcinoma development.
Current clinical trials that rely on fibrosis stage improvement use a discontinuous severity scale from 0-4, based upon an expert pathologist’s assessment. However, these semi-quantitative scoring systems do not quantify the absolute amount of scar or its structural features. Moreover, the systems are prone to substantial sampling variability (289). To improve the quantitative measurement of fibrosis in liver biopsies, several robust platforms have been established that leverage machine or deep learning to precisely quantify not only the amount of scar, but also structural features of the extracellular matrix that may reflect progression or regression (48, 258, 290, 291). Although regulatory agencies do not yet accept these digital methods in lieu of conventional fibrosis staging, their validation and ultimate implementation is inevitable, as they more accurately predict clinical outcomes than conventional fibrosis staging.
As the field seeks non-invasive measures of fibrosis severity, several modalities are being closely scrutinized, alone or in combination. They range from blood tests based on standard lab parameters such as fibrosis-4 (FIB-4) (292, 293), to proprietary tests such as enhanced liver fibrosis test (ELF) (294) and type III collagen propeptide (Pro-C3)/ type V collagen propeptide (Pro-C5) (295). Serum proteomics and metabolomics techniques that measure changes in large numbers of analytes (296, 297) show considerable promise but are not yet widely implemented. Stiffness of the liver using a bedside elastography is increasingly popular for clinical care and in approximating fibrosis severity (298), but is not yet suitable as a stand-alone diagnostic for clinical trials. On the other hand, a more precise assessment of liver stiffness using magnetic resonance elastography (MRE) has been correlated with risk of clinical outcomes (299). Similar data are emerging for another MR imaging approach using corrected T1 weighted imaging (300). Most recently, imaging tests using magnetic resonance imaging (MRI) contrast agents or probes that quantify components of extracellular matrix in liver show promise (301, 302).
Rather than use imaging or assessing circulating molecules from the liver, quantification of functional hepatic reserve is a conceptually attractive approach, because function is likely to correlate more directly with outcomes. Indeed, in fibrotic disorders of other organs functional tests are far more valuable than imaging, for example in lung or kidney. In liver, other emerging technologies assess either clearance of a liver-metabolized substrate (303), or release of proteases that reflect ECM degradation in liver (304).
Despite the continued identification of gene variants that correlate with risk of MASH (305), the value of using genetic information to diagnose and assess risk of progression for clinical management has not been established. Most GWAS studies seeking MASH risk variants have linked their presence to elevated Aspartate Transaminase (AST)/Alanine Transaminase (ALT), which helps identify gene products that modify liver injury, but not necessarily fibrosis. Noninvasive tests for fibrosis (for example using ELF, MRE, FIB-4) could in principle be correlated to GWAS hits. This approach might specifically identify variants directly linked to fibrogenic activity rather than to liver damage. Large patient datasets containing both genetic analyses and data from non-invasive fibrosis markers would be ideal for this purpose, but no such data have been reported yet.
The incorporation of genetics into risk assessment will be most informative when combined with environmental, metabolic, and systemic risk factors to establish more robust diagnostic algorithms. Although SNPs in PNPLA3, for example, predict risk of more advanced disease, this and other SNP genotypes are not yet incorporated into diagnostic algorithms. On the other hand, actionable SNPs such as the protective truncation variant in HSD17B13 (306), may be valuable by designing siRNA therapies that replicate this SNP’s protective action in patients with a disease-promoting variant.
Diagnostics for fibrosis have different contexts of use. They can be applied large scale screening of either general populations or at-risk patients, for example those with type 2 diabetes. Alternatively, they can be used to identify patients with more advanced disease suitable for enrollment in a clinical trial, and finally, they will be necessary to assess response to therapy without requiring liver biopsy. Each of these use-cases imposes different challenges and requirements for validation requested by regulatory agencies (307). Efforts to harmonize data and accelerate progress have been supported by large consortia in North America (NIMBLE)(308) and Europe (LITMUS)(309). Working cooperatively with these consortia has been the Liver Forum, an all-stakeholders working group seeking to improve diagnostics in MASH (310).
Current clinical practice is moving towards the use of multimodality non-invasive diagnostic testing to identify patients at highest risk for advanced fibrosis who may be suitable for either enrollment in a clinical trial or screening for hepatocellular carcinoma (292, 293, 311). Continued progress in diagnostic staging is anticipated, and the need will quicken once drugs are approved for MASH when it will be essential to not only identify suitable candidates for therapy, but also to determine whether patients are responding or instead require other therapies.
Concepts of antifibrotic therapy in MASH
These are still early days in the development of therapies for MASH fibrosis, with no drugs approved yet and continued reliance on liver biopsy to merit U.S. Food and Drug Administration (FDA) and European Medicines Agency (EMA) approvals. Still, a framework of antifibrotic drug development is crystallizing. Because MASLD is part of a systemic disease with multiple co-morbidities, patients are at greater risk of death or illness from extrahepatic diseases, especially heart disease, than from liver disease until hepatic fibrosis is quite advanced (stage F3 or F4). Therefore, the primary goal of treatments for MASH is to attenuate or reverse fibrosis progression before the liver is cirrhotic, and to regress fibrosis if cirrhosis has developed. The therapeutic targets for these two populations are likely to be different, with a focus on attenuating inflammation and lipotoxicity as major goals prior to cirrhosis, whereas in cirrhosis, therapies need to reduce scar content and reverse architectural distortion of the liver.
Currently, mechanisms for those therapies in clinical trials comprise agents that attenuate fat accumulation, inflammatory signaling, and glucose dysregulation. Current liver-directed therapies include inhibitors of lipogenesis whose targets include fatty acid synthase, diacyl glycerol transferase (DGAT), ACC, broad metabolic modulators including thyroid hormone beta (THRβ) agonists, agonists of GLP-1R, PPARs and gastric inhibitory polypeptide receptor (GIPR), as well as a fatty acid-bile acid conjugate (Aramchol) and FGF21, among others (307). Among these, an FDA advisory panel has recommended against approving the FXR agonist obeticholic acid due to safety concerns, despite a significant impact on fibrosis in phase 3 trial. However, the thyroid hormone beta agonist, resmetirom, is awaiting review by an FDA advisory panel. Resmetirom showed a significant benefit in both key endpoints in a phase 3 MASH trial – an improvement in fibrosis without worsening MASH, and a reduction in MASH without worsening fibrosis. Specifically, 30% of patients in the 100 mg resmetirom group (the higher of two doses in the trial) reached the primary endpoint of MASH resolution with more than a 2-point reduction in NAFLD activity score (NAS) and no worsening of fibrosis. Concurrently, 26% of patients receiving 100 mg resmetirom achieved fibrosis improvement of at least one stage with no worsening of NAS compared with 24% of patients who received the 80 mg dose and 14% of patients on placebo. Importantly, the safety profile appears more favorable than obeticholic acid’s. Although these results are very encouraging, it is important to recognize that the improvement in both primary endpoints in this phase 3 resmetirom trial occurred in less than 50% of patients. If the drug is approved, this still raises a key issue of how clinicians will know whether the patient is responding to the drug to justify its continuation. Combination therapies may become necessary to either improve the percentage of responders, as well as improving the magnitude of response.
Therapies intended to reduce hepatic fibrosis or promote its regression are primarily directed to HSCs/myofibroblasts, as these are the primary source of extracellular matrix in MASH. Dampening the inflammatory signals, especially from macrophage subsets that directly stimulate myofibroblasts, is also an appealing strategy. The microbiome is an untapped source of other potential targets whose modulation could attenuate injury or fibrosis (312).
There are several important considerations in the development of antifibrotic therapies for MASH. First, antifibrotic drugs should primarily attack the disease in liver and avoid extrahepatic activity to improve tolerability and safety. This will require careful identification of targets in liver that, where possible, are not shared with similar cells in other organs. Targeting specifically to HSCs is possible either by applying systemic drugs to cell receptors with restricted expression in this cell type (for example, β-PDGFR and others uncovered using single cell technology), or by developing either cell-specific lipid nanoparticles (LNPs) or CAR-T cells. LNPs and CAR-T cells have been combined for antifibrotic therapy in experimental heart fibrosis by administering LNPs that reprogram host lymphocytes into CAR-T cells in order to target cardiac myofibroblasts that express fibroblast activation peptide alpha (FAP) (129). Because FAP is also implicated in hepatic fibrosis (313, 314), this approach merits further evaluation in MASH. An exciting element of this LNP-mediated approach is its capacity to scale administration across many patients and sites by generating them in a central location, which could then distribute widely, similar to the LNP-based coronavirus disease (COVID) vaccines that were administered to billions of individuals.
2) It is unclear whether targeting fibrosis alone without attenuating the upstream drivers elicited by inflammation and injury will be sufficient to block fibrosis accumulation or reverse scarring. Thus, a combination of drugs targeting inflammation and lipotoxicity, as well as directly attenuating fibrogenesis, is a rational approach. One such proof of principle study combining ACC inhibitor/FXR agonist and GLP-1R agonist improved steatosis but awaits validation with a larger follow-up trial using biopsy to measure changes in fibrosis (315). More rigorous systematic efforts to establish rational combination therapies are urgently needed (252).
3) Identification of HSC subpopulations that are the main fibrosis drivers will lead to more targeted, safer therapies (42, 48). This approach was successfully validated by a study that used CAR-T to deplete only HSCs that expressed the senescence marker urokinase plasminogen activator receptor (uPAR), thereby improving inflammation and fibrosis (128). Complete elimination of all HSCs is not a viable strategy, as these cells also carry out important homeostatic functions to preserve hepatocyte mass (316). Conversely, there is a Yin-Yang relationship between fibrosis and regeneration, such that drugs that solely improve fibrosis could still unmask regenerative activity to improve liver function and restore normal architecture, whereas drugs that solely restore hepatocyte health might still unmask an endogenous antifibrotic response. Although still theoretical, this prospect is supported by evidence of fibrosis regression seen in patients cured of viral hepatitis by antivirals, yet these agents have no direct antifibrotic activity.
4) The development of delivery platforms, either viral or LNP-based, that specifically target HSCs will broaden the number of potential antifibrotic targets (317). These technologies could modulate many HSC functions through cell-surface or intracellular delivery of nucleic acids, lipids or proteins at more effective concentrations, with reduced off-target effects than if systemically administered. Indeed, there is already an HSC-targeting liposome currently in a phase 2 trial for patients with hepatic fibrosis that delivers an siRNA to heat shock protein 47 (HSP47), which disrupts normal collagen folding and leads to HSC apoptosis (318).
Points of antifibrotic attack
The majority of therapies in clinical trials for MASH target the upstream pathways that stimulate fibrosis, rather than the fibrogenic cells directly (Figure 1). Nonetheless, the identification of pathways that regulate the activation, deactivation, or apoptosis of HSCs brings the prospect of direct antifibrotic therapy closer to reality (319, 320). At present, the number of therapeutic assets in clinical trials targeting these events is limited (Figure 1), but as delivery methods improve, the prospects will broaden.
Antagonism of stellate cell/myofibroblast activation.
TGFβ1 was the first, and remains the most potent fibrogenic stimulus towards HSCs and other tissue myofibroblasts, but as noted above, TGFβ2 and TGFβ3 merit consideration as safer therapeutic targets (147). Because its pleiotropic effects include growth inhibition that attenuates cancer, however, systemic neutralization of this cytokine is not safe. Instead, targeting of latent TGFβ by antagonizing the cytokine’s local activation at the HSC surface may avoid its systemic liabilities. Molecules to block HSC-specific integrins that activate latent TGFβ are being developed for this purpose, and show promise both in preclinical studies and early clinical trials. An alternative strategy to attenuate TGFβ activation, albeit less potent, is to block angiotensin receptors.
Other approaches are evolving. Antagonism of the cell surface receptor galectin 3 may be efficacious in patients with advanced fibrosis, with current trial results pending (NCT02462967, NCT04607655). Pirfenidone, an approved antifibrotic drug in pulmonary fibrosis, is being explored in hepatic fibrosis (NCT04099407). Cyclophilin inhibition has pleiotropic activity, including inhibition of HSC activation (NCT05402371). Elegant studies have implicated acid ceramidase as an activating stimulus through YAP/TAZ activation, and therefore inhibition of acid ceramidase is being pursued therapeutically (321). Indeed, Yap has been identified as a rheostat in HSCs that can broadly regulate cellular activation (322). Interesting recent work has demonstrated a potent antifibrotic, pro-regenerative effect of inhibiting the claudin-1 receptor through its effects not only on hepatocytes, but also in HSCs (323).
Stellate cell deactivation.
Several studies have defined a unique transcriptional program that regulates deactivation of HSCs, however none of these targets have progressed to clinical testing yet. Nonetheless, as noted above, several transcription factors whose activity downregulates HSC activation include TCF21, PPARγ, LHX2, GATA4, and GATA Binding Protein 6 (GATA6) (324).
Promotion of stellate cell clearance.
As described above, methods are available in animal models to selectively deplete fibrogenic cells in injured tissue using lipid nanoparticles, with or without CAR-T cells. Although not yet advanced to clinical studies, these approaches bring the prospect of targeted gene delivery closer to reality by delivering signals that clear profibrotic subsets of HSCs (such as senescent HSCs) or to deliver mediators that will provoke selective apoptosis (for example, siRNA to HSP47).
Concluding remarks
Awareness of MASLD and MASH among liver specialists and scientists has only surfaced in the past ~15 years. At the same time, many in the scientific community and most of the public remain unaware of these diseases’ impact, despite hundreds of millions of patients being affected worldwide. The strides made in defining basic mechanisms of disease are heartening, but only the beginning. With the recognition that fibrosis is the main determinant of liver outcomes, efforts are intensifying to target fibrogenic cells and pathways driving the scarring response. While there remain significant unmet needs both in the clinical and experimental settings, a growing repertoire of therapeutic targets and diagnostics, combined with major technical advances in cell analysis, cell targeting, and disease modeling augur well for discovering effective antifibrotic therapies, alone or in combination with other agents. This success is urgently needed.
Funding:
1R01 DK136016-01, American Gastroenterological Association Research Scholar AGA2020-13-03 to SW. 5R01DK128289-03, 5R01 DK121154-04, 5P30CA196521-08 to SLF.
Footnotes
Competing interests: Consulting (SLF): 89 Bio, Amgen, Axcella Health, Blade Therapeutics, Bristol Myers Squibb, Cargene, Cellarity, ChemomAb, Fate Therapeutics, Forbion, Galmed, Gordian Biotechnology, Glycotest, Glympse Bio, Hepgene, Idorsia, In sitro, Korro Bio, Ochre Bio, Merck, Metrea, Morphic Therapeutics, North Sea Therapeutics, Novartis, Pfizer Pharmaceuticals, Pliant, Prosciento, Resolution Therapeutics, Sagimet, Satellite Bio, Scholar Rock, Surrozen, Takeda Pharmaceuticals. Stock options (SLF): Blade Therapeutics, Escient, Galectin Galmed, Genfit, Glympse, Hepgene, Lifemax, Metacrine, Morphic Therapeutics, Nimbus, North Sea, Therapeutics, Scholar Rock. Research activities with commercial entities (SLF): Morphic Therapeutics; Novo Nordisk; Abalone Bio (SBIR Grant); Espervita, Galmed, Pionyr. United States provisional patent application 63/345,236 claiming "the use of neurotrophic tyrosine receptor kinase (Trk) inhibitors for liver disease and other diseases" (SW & SLF).
References
- 1.Hossain P, Kawar B, El Nahas M, Obesity and Diabetes in the Developing World — A Growing Challenge. New England Journal of Medicine 356, 213–215 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Diehl AM, Day C, Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. New England Journal of Medicine 377, 2063–2072 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ, Mechanisms of NAFLD development and therapeutic strategies. Nature Medicine 24, 908–922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loomba R, Friedman SL, Shulman GI, Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Leung JC-F, Loong TC-W, Wei JL, Wong GL-H, Chan AW-H, Choi PC-L, Shu SS-T, Chim AM-L, Chan HL-Y, Wong VW-S, Histological severity and clinical outcomes of nonalcoholic fatty liver disease in nonobese patients. Hepatology 65, 54–64 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Younes R, Bugianesi E, NASH in Lean Individuals. Semin Liver Dis 39, 86–95 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ, Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 67, 123–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riazi K, Azhari H, Charette JH, Underwood FE, King JA, Afshar EE, Swain MG, Congly SE, Kaplan GG, Shaheen AA, The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 7, 851–861 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Vilar-Gomez E, Vuppalanchi R, Mladenovic A, Samala N, Gawrieh S, Newsome PN, Chalasani N, Prevalence of High-risk Nonalcoholic Steatohepatitis (NASH) in the United States: Results From NHANES 2017-2018. Clin Gastroenterol Hepatol 21, 115–124 e117 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Younossi ZM, Henry L, Epidemiology of non-alcoholic fatty liver disease and hepatocellular carcinoma. JHEP Rep 3, 100305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stepanova M, Kabbara K, Mohess D, Verma M, Roche-Green A, AlQahtani S, Ong J, Burra P, Younossi ZM, Nonalcoholic steatohepatitis is the most common indication for liver transplantation among the elderly: Data from the United States Scientific Registry of Transplant Recipients. Hepatol Commun 6, 1506–1515 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowe IA, Wong VW, Loomba R, Treatment Candidacy for Pharmacologic Therapies for NASH. Clin Gastroenterol Hepatol 20, 1209–1217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekstedt M, Hagström H, Nasr P, Fredrikson M, Stål P, Kechagias S, Hultcrantz R, Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 61, 1547–1554 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Hagström H, Nasr P, Ekstedt M, Hammar U, Stål P, Hultcrantz R, Kechagias S, Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. Journal of Hepatology 67, 1265–1273 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Angulo P, Kleiner DE, Dam-Larsen S, Adams LA, Bjornsson ES, Charatcharoenwitthaya P, Mills PR, Keach JC, Lafferty HD, Stahler A, Haflidadottir S, Bendtsen F, Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 149, 389–397.e310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, Sebastiani G, Ekstedt M, Hagstrom H, Nasr P, Stal P, Wong VW-S, Kechagias S, Hultcrantz R, Loomba R, Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology 65, 1557–1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolin CE, Arteel GE, The Matrisome, Inflammation, and Liver Disease. Semin Liver Dis 40, 180–188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massey VL, Dolin CE, Poole LG, Hudson SV, Siow DL, Brock GN, Merchant ML, Wilkey DW, Arteel GE, The hepatic “matrisome” responds dynamically to injury: Characterization of transitional changes to the extracellular matrix in mice. Hepatology 65, 969–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG, Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol 293, G1147–1154 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kakisaka K, Cazanave SC, Fingas CD, Guicciardi ME, Bronk SF, Werneburg NW, Mott JL, Gores GJ, Mechanisms of lysophosphatidylcholine-induced hepatocyte lipoapoptosis. Am J Physiol-Gastr L 302, G77–G84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G, Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54, 133–144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wree A, Eguchi A, McGeough MD, Pena CA, Johnson CD, Canbay A, Hoffman HM, Feldstein AE, NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology 59, 898–910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan S-S, Jiang JX, Wu J, Halsted C, Friedman SL, Zern MA, Torok NJ, Phagocytosis of apoptotic bodies by hepatic stellate cells induces NADPH oxidase and is associated with liver fibrosis in vivo. Hepatology 43, 435–443 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Canbay A, Taimr P, Torok N, Higuchi H, Friedman S, Gores GJ, Apoptotic Body Engulfment by a Human Stellate Cell Line Is Profibrogenic. Laboratory Investigation 83, 655–663 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Watanabe A, Hashmi A, Gomes DA, Town T, Badou A, Flavell RA, Mehal WZ, Apoptotic hepatocyte DNA inhibits hepatic stellate cell chemotaxis via toll-like receptor 9. Hepatology 46, 1509–1518 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Arriazu E, Ge X, Leung T-M, Magdaleno F, Lopategi A, Lu Y, Kitamura N, Urtasun R, Theise N, Antoine DJ, Nieto N, Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut 66, 1123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Povero D, Panera N, Eguchi A, Johnson CD, Papouchado BG, de Araujo Horcel L, Pinatel EM, Alisi A, Nobili V, Feldstein AE, Lipid-Induced Hepatocyte-Derived Extracellular Vesicles Regulate Hepatic Stellate Cells via MicroRNA Targeting Peroxisome Proliferator-Activated Receptor-γ. Cellular and Molecular Gastroenterology and Hepatology 1, 646–663.e644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y-S, Kim SY, Ko E, Lee J-H, Yi H-S, Yoo YJ, Je J, Suh SJ, Jung YK, Kim JH, Seo YS, Yim HJ, Jeong W-I, Yeon JE, Um SH, Byun KS, Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci Rep-Uk 7, 3710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernández A, Reyes D, Geng Y, Arab JP, Cabrera D, Sepulveda R, Solis N, Buist-Homan M, Arrese M, Moshage H, Extracellular vesicles derived from fat-laden hepatocytes undergoing chemical hypoxia promote a pro-fibrotic phenotype in hepatic stellate cells. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1866, 165857 (2020). [DOI] [PubMed] [Google Scholar]
- 30.An P, Wei L-L, Zhao S, Sverdlov DY, Vaid KA, Miyamoto M, Kuramitsu K, Lai M, Popov YV, Hepatocyte mitochondria-derived danger signals directly activate hepatic stellate cells and drive progression of liver fibrosis. Nature communications 11, 2362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mederacke I, Filliol A, Affo S, Nair A, Hernandez C, Sun Q, Hamberger F, Brundu F, Chen Y, Ravichandra A, Huebener P, Anke H, Shi H, Martínez García de la Torre RA, Smith JR, Henderson NC, Vondran FWR, Rothlin CV, Baehre H, Tabas I, Sancho-Bru P, Schwabe RF, The purinergic P2Y14 receptor links hepatocyte death to hepatic stellate cell activation and fibrogenesis in the liver. Science Translational Medicine 14, eabe5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson MM, Jonsson JR, Powell EE, Brunt EM, Neuschwander–Tetri BA, Bhathal PS, Dixon JB, Weltman MD, Tilg H, Moschen AR, Purdie DM, Demetris AJ, Clouston AD, Progressive Fibrosis in Nonalcoholic Steatohepatitis: Association With Altered Regeneration and a Ductular Reaction. Gastroenterology 133, 80–90 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Zhao L, Westerhoff M, Pai RK, Choi W-T, Gao Z-H, Hart J, Centrilobular ductular reaction correlates with fibrosis stage and fibrosis progression in non-alcoholic steatohepatitis. Modern Pathology 31, 150–159 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Ramm GA, Nair VG, Bridle KR, Shepherd RW, Crawford DHG, Contribution of Hepatic Parenchymal and Nonparenchymal Cells to Hepatic Fibrogenesis in Biliary Atresia. The American Journal of Pathology 153, 527–535 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grappone C, Pinzani M, Parola M, Pellegrini G, Caligiuri A, DeFranco R, Marra F, Herbst H, Alpini G, Milani S, Expression of platelet-derived growth factor in newly formed cholangiocytes during experimental biliary fibrosis in rats. Journal of Hepatology 31, 100–109 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Malizia G, Brunt EM, Peters MG, Rizzo A, Broekelmann TJ, McDonald JA, Growth factor and procollagen type I gene expression in human liver disease. Gastroenterology 108, 145–156 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Tabibian JH, O'Hara SP, Splinter PL, Trussoni CE, LaRusso NF, Cholangiocyte senescence by way of N-ras activation is a characteristic of primary sclerosing cholangitis. Hepatology 59, 2263–2275 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syn W-K, Choi SS, Liaskou E, Karaca GF, Agboola KM, Oo YH, Mi Z, Pereira TA, Zdanowicz M, Malladi P, Chen Y, Moylan C, Jung Y, Bhattacharya SD, Teaberry V, Omenetti A, Abdelmalek MF, Guy CD, Adams DH, Kuo PC, Michelotti GA, Whitington PF, Diehl AM, Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology 53, 106–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan SK, Ingham SA, Mohr AM, Wehrkamp CJ, Ray A, Roy S, Cazanave SC, Phillippi MA, Mott JL, Saturated free fatty acids induce cholangiocyte lipoapoptosis. Hepatology 60, 1942–1956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiba M, Sasaki M, Kitamura S, Ikeda H, Sato Y, Nakanuma Y, Participation of bile ductular cells in the pathological progression of non-alcoholic fatty liver disease. Journal of Clinical Pathology 64, 564 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Schuster R, Younesi F, Ezzo M, Hinz B, The Role of Myofibroblasts in Physiological and Pathological Tissue Repair. Cold Spring Harbor Perspectives in Biology, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal SB, Liu X, Ganguly S, Dhar D, Pasillas MP, Ricciardelli E, Li RZ, Troutman TD, Kisseleva T, Glass CK, Brenner DA, Heterogeneity of HSCs in a Mouse Model of NASH. Hepatology 74, 667–685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ganguly S, Muench GA, Shang L, Rosenthal SB, Rahman G, Wang R, Wang Y, Kwon HC, Diomino AM, Kisseleva T, Soorosh P, Hosseini M, Knight R, Schnabl B, Brenner DA, Dhar D, Nonalcoholic Steatohepatitis and HCC in a Hyperphagic Mouse Accelerated by Western Diet. Cellular and Molecular Gastroenterology and Hepatology 12, 891–920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcher A-B, Bendixen SM, Terkelsen MK, Hohmann SS, Hansen MH, Larsen BD, Mandrup S, Dimke H, Detlefsen S, Ravnskjaer K, Transcriptional regulation of Hepatic Stellate Cell activation in NASH. Sci Rep-Uk 9, 2324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuchida T, Lee YA, Fujiwara N, Ybanez M, Allen B, Martins S, Fiel MI, Goossens N, Chou H-I, Hoshida Y, Friedman SL, A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. Journal of Hepatology 69, 385–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao X-Y, Ji Y, Li C, Guo L, Zhou L, Chen Z, Leon-Mimila P, Chung MT, Kurabayashi K, Opp J, Campos-Pérez F, Villamil-Ramírez H, Canizales-Quinteros S, Lyons R, Lumeng CN, Zhou B, Qi L, Huertas-Vazquez A, Lusis AJ, Xu XZS, Li S, Yu Y, Li JZ, Lin JD, Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Molecular Cell 75, 644–660.e645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z-Y, Keogh A, Waldt A, Cuttat R, Neri M, Zhu S, Schuierer S, Ruchti A, Crochemore C, Knehr J, Bastien J, Ksiazek I, Sánchez-Taltavull D, Ge H, Wu J, Roma G, Helliwell SB, Stroka D, Nigsch F, Single-cell and bulk transcriptomics of the liver reveals potential targets of NASH with fibrosis. Sci Rep-Uk 11, 19396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Li K, Pickholz E, Dobie R, Matchett KP, Henderson NC, Carrico C, Driver I, Borch Jensen M, Chen L, Petitjean M, Bhattacharya D, Fiel MI, Liu X, Kisseleva T, Alon U, Adler M, Medzhitov R, Friedman SL, An autocrine signaling circuit in hepatic stellate cells underlies advanced fibrosis in nonalcoholic steatohepatitis. Science Translational Medicine 15, eadd3949 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao Y, Batmanov K, Hu W, Zhu K, Tom AY, Guan D, Jiang C, Cheng L, McCright SJ, Yang EC, Lanza MR, Liu Y, Hill DA, Lazar MA, Hepatocytes demarcated by EphB2 contribute to the progression of nonalcoholic steatohepatitis. Science Translational Medicine 15, eadc9653 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, Wallenborg K, Picelli S, Zagorska A, Pendem SV, Hudson TE, Wu MM, Budas GR, Breckenridge DG, Harrison EM, Mole DJ, Wigmore SJ, Ramachandran P, Ponting CP, Teichmann SA, Marioni JC, Henderson NC, Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Reports 29, 1832–1847.e1838 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, Taylor RS, Efremova M, Vento-Tormo R, Carragher NO, Kendall TJ, Fallowfield JA, Harrison EM, Mole DJ, Wigmore SJ, Newsome PN, Weston CJ, Iredale JP, Tacke F, Pollard JW, Ponting CP, Marioni JC, Teichmann SA, Henderson NC, Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Troeger JS, Mederacke I, Gwak GY, Dapito DH, Mu X, Hsu CC, Pradere JP, Friedman RA, Schwabe RF, Deactivation of Hepatic Stellate Cells During Liver Fibrosis Resolution in Mice. Gastroenterology 143, 1073–1083.e1022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere J-P, Schwabe RF, Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nature communications 4, 2823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puche JE, Lee YA, Jiao J, Aloman C, Fiel MI, Muñoz U, Kraus T, Lee T, Yee HF Jr, Friedman SL, A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology 57, 339–350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Troeger JS, Mederacke I, Gwak GY, Dapito DH, Mu X, Hsu CC, Pradere JP, Friedman RA, Schwabe RF, Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 143, 1073–1083 e1022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W, He H, Wang T, Su N, Zhang F, Jiang K, Zhu J, Zhang C, Niu K, Wang L, Yuan X, Liu N, Li L, Wei W, Hu J, Single-Cell Transcriptomic Analysis Reveals a Hepatic Stellate Cell–Activation Roadmap and Myofibroblast Origin During Liver Fibrosis in Mice. Hepatology 74, 2774–2790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei L, Bruneau A, El Mourabit H, Guegan J, Folseraas T, Lemoinne S, Karlsen TH, Hoareau B, Morichon R, Gonzalez-Sanchez E, Goumard C, Ratziu V, Charbord P, Gautheron J, Tacke F, Jaffredo T, Cadoret A, Housset C, Portal fibroblasts with mesenchymal stem cell features form a reservoir of proliferative myofibroblasts in liver fibrosis. Hepatology 76, 1360–1375 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Katsumata LW, Miyajima A, Itoh T, Portal fibroblasts marked by the surface antigen Thy1 contribute to fibrosis in mouse models of cholestatic liver injury. Hepatol Commun 1, 198–214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramachandran P, Matchett KP, Dobie R, Wilson-Kanamori JR, Henderson NC, Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nature Reviews Gastroenterology & Hepatology 17, 457–472 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP, Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. The Journal of Clinical Investigation 115, 56–65 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivera CA, Bradford BU, Hunt KJ, Adachi Y, Schrum LW, Koop DR, Burchardt ER, Rippe RA, Thurman RG, Attenuation of CCl4-induced hepatic fibrosis by GdCl3 treatment or dietary glycine. Am J Physiol-Gastr L 281, G200–G207 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Karlmark KR, Weiskirchen R, Zimmermann HW, Gassler N, Ginhoux F, Weber C, Merad M, Luedde T, Trautwein C, Tacke F, Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50, 261–274 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Thomas JA, Pope C, Wojtacha D, Robson AJ, Gordon-Walker TT, Hartland S, Ramachandran P, Van Deemter M, Hume DA, Iredale JP, Forbes SJ, Macrophage therapy for murine liver fibrosis recruits host effector cells improving fibrosis, regeneration, and function. Hepatology 53, 2003–2015 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Fallowfield JA, Mizuno M, Kendall TJ, Constandinou CM, Benyon RC, Duffield JS, Iredale JP, Scar-Associated Macrophages Are a Major Source of Hepatic Matrix Metalloproteinase-13 and Facilitate the Resolution of Murine Hepatic Fibrosis. The Journal of Immunology 178, 5288 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Pradere J-P, Kluwe J, De Minicis S, Jiao J-J, Gwak G-Y, Dapito DH, Jang M-K, Guenther ND, Mederacke I, Friedman R, Dragomir A-C, Aloman C, Schwabe RF, Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 58, 1461–1473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrasek J, Bala S, Csak T, Lippai D, Kodys K, Menashy V, Barrieau M, Min S-Y, Kurt-Jones EA, Szabo G, IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. The Journal of Clinical Investigation 122, 3476–3489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong L, Yamasaki G, Johnson RJ, Friedman SL, Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. The Journal of Clinical Investigation 94, 1563–1569 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellerbrand C, Stefanovic B, Giordano F, Burchardt ER, Brenner DA, The role of TGFβ1 in initiating hepatic stellate cell activation in vivo. Journal of Hepatology 30, 77–87 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Cai B, Dongiovanni P, Corey KE, Wang X, Shmarakov IO, Zheng Z, Kasikara C, Davra V, Meroni M, Chung RT, Rothlin CV, Schwabe RF, Blaner WS, Birge RB, Valenti L, Tabas I, Macrophage MerTK Promotes Liver Fibrosis in Nonalcoholic Steatohepatitis. Cell Metabolism 31, 406–421.e407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Remmerie A, Martens L, Thoné T, Castoldi A, Seurinck R, Pavie B, Roels J, Vanneste B, De Prijck S, Vanhockerhout M, Binte Abdul Latib M, Devisscher L, Hoorens A, Bonnardel J, Vandamme N, Kremer A, Borghgraef P, Van Vlierberghe H, Lippens S, Pearce E, Saeys Y, Scott CL, Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver. Immunity 53, 641–657.e614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miura K, Yang L, van Rooijen N, Ohnishi H, Seki E, Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physiol-Gastr L 302, G1310–G1321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seki E, De Minicis S, Gwak G-Y, Kluwe J, Inokuchi S, Bursill CA, Llovet JM, Brenner DA, Schwabe RF, CCR1 and CCR5 promote hepatic fibrosis in mice. The Journal of Clinical Investigation 119, 1858–1870 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen Y, Lambrecht J, Ju C, Tacke F, Hepatic macrophages in liver homeostasis and diseases-diversity, plasticity and therapeutic opportunities. Cellular & Molecular Immunology 18, 45–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao H, Jin Z, Bandyopadhyay G, Wang G, Zhang D, Rocha K. C. e., Liu X, Zhao H, Kisseleva T, Brenner DA, Karin M, Ying W, Aberrant iron distribution via hepatocyte-stellate cell axis drives liver lipogenesis and fibrosis. Cell Metabolism 34, 1201–1213.e1205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran S, Baba I, Poupel L, Dussaud S, Moreau M, Gélineau A, Marcelin G, Magréau-Davy E, Ouhachi M, Lesnik P, Boissonnas A, Le Goff W, Clausen BE, Yvan-Charvet L, Sennlaub F, Huby T, Gautier EL, Impaired Kupffer Cell Self-Renewal Alters the Liver Response to Lipid Overload during Non-alcoholic Steatohepatitis. Immunity 53, 627–640.e625 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, Keren-Shaul H, David E, Zmora N, Eldar SM, Lubezky N, Shibolet O, Hill DA, Lazar MA, Colonna M, Ginhoux F, Shapiro H, Elinav E, Amit I, Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 178, 686–698.e614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, Bruni CM, Ouyang Z, Li RZ, Sun X, Vu BT, Pasillas MP, Ego KM, Gosselin D, Link VM, Chong L-W, Evans RM, Thompson BM, McDonald JG, Hosseini M, Witztum JL, Germain RN, Glass CK, Niche-Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity 52, 1057–1074.e1057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rai RP, Liu Y, Iyer SS, Liu S, Gupta B, Desai C, Kumar P, Smith T, Singhi AD, Nusrat A, Parkos CA, Monga SP, Czaja MJ, Anania FA, Raeman R, Blocking integrin α4β7-mediated CD4 T cell recruitment to the intestine and liver protects mice from western diet-induced non-alcoholic steatohepatitis. Journal of Hepatology 73, 1013–1022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kennedy L, Meadows V, Sybenga A, Demieville J, Chen L, Hargrove L, Ekser B, Dar W, Ceci L, Kundu D, Kyritsi K, Pham L, Zhou T, Glaser S, Meng F, Alpini G, Francis H, Mast Cells Promote Nonalcoholic Fatty Liver Disease Phenotypes and Microvesicular Steatosis in Mice Fed a Western Diet. Hepatology 74, 164–182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng S, Yang W, Yao D, Tang S, Hou J, Chang X, A comparative study on roles of natural killer T cells in two diet-induced non-alcoholic steatohepatitis-related fibrosis in mice. Annals of Medicine 54, 2233–2245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Syn W-K, Htun Oo Y, Pereira TA, Karaca GF, Jung Y, Omenetti A, Witek RP, Choi SS, Guy CD, Fearing CM, Teaberry V, Pereira FEL, Adams DH, Diehl AM, Accumulation of natural killer T cells in progressive nonalcoholic fatty liver disease. Hepatology 51, 1998–2007 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grohmann M, Wiede F, Dodd GT, Gurzov EN, Ooi GJ, Butt T, Rasmiena AA, Kaur S, Gulati T, Goh PK, Treloar AE, Archer S, Brown WA, Muller M, Watt MJ, Ohara O, McLean CA, Tiganis T, Obesity Drives STAT-1-Dependent NASH and STAT-3-Dependent HCC. Cell 175, 1289–1306 e1220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malehmir M, Pfister D, Gallage S, Szydlowska M, Inverso D, Kotsiliti E, Leone V, Peiseler M, Surewaard BGJ, Rath D, Ali A, Wolf MJ, Drescher H, Healy ME, Dauch D, Kroy D, Krenkel O, Kohlhepp M, Engleitner T, Olkus A, Sijmonsma T, Volz J, Deppermann C, Stegner D, Helbling P, Nombela-Arrieta C, Rafiei A, Hinterleitner M, Rall M, Baku F, Borst O, Wilson CL, Leslie J, O’Connor T, Weston CJ, Chauhan A, Adams DH, Sheriff L, Teijeiro A, Prinz M, Bogeska R, Anstee N, Bongers MN, Notohamiprodjo M, Geisler T, Withers DJ, Ware J, Mann DA, Augustin HG, Vegiopoulos A, Milsom MD, Rose AJ, Lalor PF, Llovet JM, Pinyol R, Tacke F, Rad R, Matter M, Djouder N, Kubes P, Knolle PA, Unger K, Zender L, Nieswandt B, Gawaz M, Weber A, Heikenwalder M, Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nature Medicine 25, 641–655 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Maricic I, Marrero I, Eguchi A, Nakamura R, Johnson CD, Dasgupta S, Hernandez CD, Nguyen PS, Swafford AD, Knight R, Feldstein AE, Loomba R, Kumar V, Differential Activation of Hepatic Invariant NKT Cell Subsets Plays a Key Role in Progression of Nonalcoholic Steatohepatitis. J Immunol 201, 3017–3035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Herck MA, Vonghia L, Kwanten WJ, Julé Y, Vanwolleghem T, Ebo DG, Michielsen PP, De Man JG, Gama L, De Winter BY, Francque SM, Diet Reversal and Immune Modulation Show Key Role for Liver and Adipose Tissue T Cells in Murine Nonalcoholic Steatohepatitis. Cellular and Molecular Gastroenterology and Hepatology 10, 467–490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karl M, Hasselwander S, Zhou Y, Reifenberg G, Kim YO, Park K-S, Ridder DA, Wang X, Seidel E, Hövelmeyer N, Straub BK, Li H, Schuppan D, Xia N, Dual roles of B lymphocytes in mouse models of diet-induced nonalcoholic fatty liver disease. Hepatology 76, 1135–1149 (2022). [DOI] [PubMed] [Google Scholar]
- 87.Barrow F, Khan S, Fredrickson G, Wang H, Dietsche K, Parthiban P, Robert S, Kaiser T, Winer S, Herman A, Adeyi O, Mouzaki M, Khoruts A, Hogquist KA, Staley C, Winer DA, Revelo XS, Microbiota-Driven Activation of Intrahepatic B Cells Aggravates NASH Through Innate and Adaptive Signaling. Hepatology 74, 704–722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carter JK, Friedman SL, Hepatic Stellate Cell-Immune Interactions in NASH. Frontiers in Endocrinology 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gomes AL, Teijeiro A, Buren S, Tummala KS, Yilmaz M, Waisman A, Theurillat JP, Perna C, Djouder N, Metabolic Inflammation-Associated IL-17A Causes Non-alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell 30, 161–175 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Peiseler M, Schwabe R, Hampe J, Kubes P, Heikenwalder M, Tacke F, Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease - novel insights into cellular communication circuits. J Hepatol 77, 1136–1160 (2022). [DOI] [PubMed] [Google Scholar]
- 91.Muhanna N, Abu Tair L, Doron S, Amer J, Azzeh M, Mahamid M, Friedman S, Safadi R, Amelioration of hepatic fibrosis by NK cell activation. Gut 60, 90–98 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Abu-Tair L, Axelrod JH, Doron S, Ovadya Y, Krizhanovsky V, Galun E, Amer J, Safadi R, Natural Killer Cell-Dependent Anti-Fibrotic Pathway in Liver Injury via Toll-Like Receptor-9. PLoS One 8, e82571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, Wu Y, Shen W, Wang B, Yuan X, Crosstalk between NK cells and hepatic stellate cells in liver fibrosis (Review). Mol Med Rep 25, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Correia AL, Guimaraes JC, Auf der Maur P, De Silva D, Trefny MP, Okamoto R, Bruno S, Schmidt A, Mertz K, Volkmann K, Terracciano L, Zippelius A, Vetter M, Kurzeder C, Weber WP, Bentires-Alj M, Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature 594, 566–571 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Jiang X, Peng Y, Liu L, Wang Y, Li M, Li W, Huang F, Zheng C, Xu F, Hu Q, Wei W, Dong S, Zhao Q, MAIT cells ameliorate liver fibrosis by enhancing the cytotoxicity of NK cells in cholestatic murine models. Liver international : official journal of the International Association for the Study of the Liver 42, 2743–2758 (2022). [DOI] [PubMed] [Google Scholar]
- 96.Tsuchida T, Friedman SL, Mechanisms of hepatic stellate cell activation. Nature Reviews Gastroenterology & Hepatology 14, 397–411 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Filliol A, Saito Y, Nair A, Dapito DH, Yu LX, Ravichandra A, Bhattacharjee S, Affo S, Fujiwara N, Su H, Sun Q, Savage TM, Wilson-Kanamori JR, Caviglia JM, Chin L, Chen D, Wang X, Caruso S, Kang JK, Amin AD, Wallace S, Dobie R, Yin D, Rodriguez-Fiallos OM, Yin C, Mehal A, Izar B, Friedman RA, Wells RG, Pajvani UB, Hoshida Y, Remotti HE, Arpaia N, Zucman-Rossi J, Karin M, Henderson NC, Tabas I, Schwabe RF, Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis. Nature 610, 356–365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Affo S, Nair A, Brundu F, Ravichandra A, Bhattacharjee S, Matsuda M, Chin L, Filliol A, Wen W, Song X, Decker A, Worley J, Caviglia JM, Yu L, Yin D, Saito Y, Savage T, Wells RG, Mack M, Zender L, Arpaia N, Remotti HE, Rabadan R, Sims P, Leblond AL, Weber A, Riener MO, Stockwell BR, Gaublomme J, Llovet JM, Kalluri R, Michalopoulos GK, Seki E, Sia D, Chen X, Califano A, Schwabe RF, Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell 39, 866–882 e811 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Payen VL, Lavergne A, Alevra Sarika N, Colonval M, Karim L, Deckers M, Najimi M, Coppieters W, Charloteaux B, Sokal EM, El Taghdouini A, Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep 3, 100278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guilliams M, Scott CL, Liver macrophages in health and disease. Immunity 55, 1515–1529 (2022). [DOI] [PubMed] [Google Scholar]
- 101.Lavin Y, Mortha A, Rahman A, Merad M, Regulation of macrophage development and function in peripheral tissues. Nature Reviews Immunology 15, 731–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han Y-H, Shin K-O, Kim J-Y, Khadka DB, Kim H-J, Lee Y-M, Cho W-J, Cha J-Y, Lee B-J, Lee M-O, A maresin 1/RORα/12-lipoxygenase autoregulatory circuit prevents inflammation and progression of nonalcoholic steatohepatitis. The Journal of Clinical Investigation 129, 1684–1698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fabre T, Barron AMS, Christensen SM, Asano S, Bound K, Lech MP, Wadsworth MH 2nd, Chen X, Wang C, Wang J, McMahon J, Schlerman F, White A, Kravarik KM, Fisher AJ, Borthwick LA, Hart KM, Henderson NC, Wynn TA, Dower K, Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci Immunol 8, eadd8945 (2023). [DOI] [PubMed] [Google Scholar]
- 104.Hammoutene A, Rautou P-E, Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. Journal of Hepatology 70, 1278–1291 (2019). [DOI] [PubMed] [Google Scholar]
- 105.DeLeve LD, Wang X, Guo Y, Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescence. Hepatology 48, 920–930 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie G, Wang X, Wang L, Wang L, Atkinson RD, Kanel GC, Gaarde WA, DeLeve LD, Role of Differentiation of Liver Sinusoidal Endothelial Cells in Progression and Regression of Hepatic Fibrosis in Rats. Gastroenterology 142, 918–927.e916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen L, Gu T, Li B, Li F, Ma Z, Zhang Q, Cai X, Lu L, Delta-like ligand 4/DLL4 regulates the capillarization of liver sinusoidal endothelial cell and liver fibrogenesis. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1866, 1663–1675 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Furuta K, Guo Q, Pavelko KD, Lee J-H, Robertson KD, Nakao Y, Melek J, Shah VH, Hirsova P, Ibrahim SH, Lipid-induced endothelial vascular cell adhesion molecule 1 promotes nonalcoholic steatohepatitis pathogenesis. The Journal of Clinical Investigation 131, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hammoutene A, Biquard L, Lasselin J, Kheloufi M, Tanguy M, Vion A-C, Mérian J, Colnot N, Loyer X, Tedgui A, Codogno P, Lotersztajn S, Paradis V, Boulanger CM, Rautou P-E, A defect in endothelial autophagy occurs in patients with non-alcoholic steatohepatitis and promotes inflammation and fibrosis. Journal of Hepatology 72, 528–538 (2020). [DOI] [PubMed] [Google Scholar]
- 110.Ding B-S, Cao Z, Lis R, Nolan DJ, Guo P, Simons M, Penfold ME, Shido K, Rabbany SY, Rafii S, Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 505, 97–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Loft A, Alfaro AJ, Schmidt SF, Pedersen FB, Terkelsen MK, Puglia M, Chow KK, Feuchtinger A, Troullinaki M, Maida A, Wolff G, Sakurai M, Berutti R, Ekim Üstünel B, Nawroth P, Ravnskjaer K, Diaz MB, Blagoev B, Herzig S, Liver-fibrosis-activated transcriptional networks govern hepatocyte reprogramming and intra-hepatic communication. Cell Metabolism 33, 1685–1700.e1689 (2021). [DOI] [PubMed] [Google Scholar]
- 112.Wang X, Zheng Z, Caviglia JM, Corey KE, Herfel TM, Cai B, Masia R, Chung RT, Lefkowitch JH, Schwabe RF, Tabas I, Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metabolism 24, 848–862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang X, Cai B, Yang X, Sonubi OO, Zheng Z, Ramakrishnan R, Shi H, Valenti L, Pajvani UB, Sandhu J, Infante RE, Radhakrishnan A, Covey DF, Guan K-L, Buck J, Levin LR, Tontonoz P, Schwabe RF, Tabas I, Cholesterol Stabilizes TAZ in Hepatocytes to Promote Experimental Non-alcoholic Steatohepatitis. Cell Metabolism 31, 969–986.e967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu C, Kim K, Wang X, Bartolome A, Salomao M, Dongiovanni P, Meroni M, Graham MJ, Yates KP, Diehl AM, Schwabe RF, Tabas I, Valenti L, Lavine JE, Pajvani UB, Hepatocyte Notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Science Translational Medicine 10, eaat0344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matchett KP, Wilson-Kanamori JR, Portman JR, Kapourani CA, Fercoq F, May SB, Mackey JBG, Brice M, Zajdel E, Beltran MS, Sutherland EF, Wilson JC, Wallace SJ, Kitto L, Younger NT, Dobie R, Oniscu GC, Wigmore SJ, Ramachandiran P, Vallejos CA, Carragher NO, Simpson KJ, Kendall TJ, Rule JA, Lee WM, Hoare M, Weston CJ, Marioni JC, Teichmann ST, Bird TG, Carlin LM, Henderson NC, Multimodal decoding of human liver regeneration. bioRxiv, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Govaere O, Cockell S, Tiniakos D, Queen R, Younes R, Vacca M, Alexander L, Ravaioli F, Palmer J, Petta S, Boursier J, Rosso C, Johnson K, Wonders K, Day CP, Ekstedt M, Orešič M, Darlay R, Cordell HJ, Marra F, Vidal-Puig A, Bedossa P, Schattenberg JM, Clément K, Allison M, Bugianesi E, Ratziu V, Daly AK, Anstee QM, Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Science Translational Medicine 12, eaba4448 (2020). [DOI] [PubMed] [Google Scholar]
- 117.Vandel J, Dubois-Chevalier J, Gheeraert C, Derudas B, Raverdy V, Thuillier D, Gaal L, Francque S, Pattou F, Staels B, Eeckhoute J, Lefebvre P, Hepatic Molecular Signatures Highlight the Sexual Dimorphism of Nonalcoholic Steatohepatitis (NASH). Hepatology 73, 920–936 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Santhekadur PK, Kumar DP, Sanyal AJ, Preclinical models of non-alcoholic fatty liver disease. Journal of Hepatology 68, 230–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cazanave S, Podtelezhnikov A, Jensen K, Seneshaw M, Kumar DP, Min H-K, Santhekadur PK, Banini B, Mauro AG, Oseini AM, Vincent R, Tanis KQ, Webber AL, Wang L, Bedossa P, Mirshahi F, Sanyal AJ, The Transcriptomic Signature Of Disease Development And Progression Of Nonalcoholic Fatty Liver Disease. Sci Rep-Uk 7, 17193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, Cherrington NJ, Analysis of Global and Absorption, Distribution, Metabolism, and Elimination Gene Expression in the Progressive Stages of Human Nonalcoholic Fatty Liver Disease. Drug Metabolism and Disposition 39, 1954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van Koppen A, Verschuren L, van den Hoek AM, Verheij J, Morrison MC, Li K, Nagabukuro H, Costessi A, Caspers MPM, van den Broek TJ, Sagartz J, Kluft C, Beysen C, Emson C, van Gool AJ, Goldschmeding R, Stoop R, Bobeldijk-Pastorova I, Turner SM, Hanauer G, Hanemaaijer R, Uncovering a Predictive Molecular Signature for the Onset of NASH-Related Fibrosis in a Translational NASH Mouse Model. Cellular and Molecular Gastroenterology and Hepatology 5, 83–98.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lefebvre P, Lalloyer F, Baugé E, Pawlak M, Gheeraert C, Dehondt H, Vanhoutte J, Woitrain E, Hennuyer N, Mazuy C, Bobowski-Gérard M, Zummo FP, Derudas B, Driessen A, Hubens G, Vonghia L, Kwanten WJ, Michielsen P, Vanwolleghem T, Eeckhoute J, Verrijken A, Van Gaal L, Francque S, Staels B, Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARα-regulated dermatopontin. JCI Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Corey KE, Pitts R, Lai M, Loureiro J, Masia R, Osganian SA, Gustafson JL, Hutter MM, Gee DW, Meireles OR, Witkowski ER, Richards SM, Jacob J, Finkel N, Ngo D, Wang TJ, Gerszten RE, Ukomadu C, Jennings LL, ADAMTSL2 protein and a soluble biomarker signature identify at-risk non-alcoholic steatohepatitis and fibrosis in adults with NAFLD. Journal of Hepatology 76, 25–33 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Govaere O, Hasoon M, Alexander L, Cockell S, Tiniakos D, Ekstedt M, Schattenberg JM, Boursier J, Bugianesi E, Ratziu V, Investigators L, Daly AK, Anstee QM, A proteo-transcriptomic map of non-alcoholic fatty liver disease signatures. Nat Metab 5, 572–578 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fred RG, Steen Pedersen J, Thompson JJ, Lee J, Timshel PN, Stender S, Opseth Rygg M, Gluud LL, Bjerregaard Kristiansen V, Bendtsen F, Hansen T, Pers TH, Single-cell transcriptome and cell type-specific molecular pathways of human non-alcoholic steatohepatitis. Sci Rep-Uk 12, 13484 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carter J, Wang S, Friedman SL, Ten Thousand Points of Light: Heterogeneity Among the Stars of NASH Fibrosis. Hepatology 74, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aghajanian H, Kimura T, Rurik JG, Hancock AS, Leibowitz MS, Li L, Scholler J, Monslow J, Lo A, Han W, Wang T, Bedi K, Morley MP, Linares Saldana RA, Bolar NA, McDaid K, Assenmacher CA, Smith CL, Wirth D, June CH, Margulies KB, Jain R, Pure E, Albelda SM, Epstein JA, Targeting cardiac fibrosis with engineered T cells. Nature 573, 430–433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Amor C, Feucht J, Leibold J, Ho YJ, Zhu C, Alonso-Curbelo D, Mansilla-Soto J, Boyer JA, Li X, Giavridis T, Kulick A, Houlihan S, Peerschke E, Friedman SL, Ponomarev V, Piersigilli A, Sadelain M, Lowe SW, Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rurik JG, Tombacz I, Yadegari A, Mendez Fernandez PO, Shewale SV, Li L, Kimura T, Soliman OY, Papp TE, Tam YK, Mui BL, Albelda SM, Pure E, June CH, Aghajanian H, Weissman D, Parhiz H, Epstein JA, CAR T cells produced in vivo to treat cardiac injury. Science 375, 91–96 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R, CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nature Protocols 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 131.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, Myung P, Plikus MV, Nie Q, Inference and analysis of cell-cell communication using CellChat. Nature communications 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH, Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics 40, 1461–1465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E, Bugianesi E, Maggioni M, Fracanzani AL, Fargion S, Day CP, Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 51, 1209–1217 (2010). [DOI] [PubMed] [Google Scholar]
- 134.Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, Burt AD, Bedossa P, Palmer J, Liu Y-L, Aithal GP, Allison M, Yki-Järvinen H, Vacca M, Dufour J-F, Invernizzi P, Prati D, Ekstedt M, Kechagias S, Francque S, Petta S, Bugianesi E, Clement K, Ratziu V, Schattenberg JM, Valenti L, Day CP, Cordell HJ, Daly AK, Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort☆. Journal of Hepatology 73, 505–515 (2020). [DOI] [PubMed] [Google Scholar]
- 135.BasuRay S, Smagris E, Cohen JC, Hobbs HH, The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 66, 1111–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bruschi FV, Claudel T, Tardelli M, Caligiuri A, Stulnig TM, Marra F, Trauner M, The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 65, 1875–1890 (2017). [DOI] [PubMed] [Google Scholar]
- 137.Buch S, Stickel F, Trépo E, Way M, Herrmann A, Nischalke HD, Brosch M, Rosendahl J, Berg T, Ridinger M, Rietschel M, McQuillin A, Frank J, Kiefer F, Schreiber S, Lieb W, Soyka M, Semmo N, Aigner E, Datz C, Schmelz R, Brückner S, Zeissig S, Stephan A-M, Wodarz N, Devière J, Clumeck N, Sarrazin C, Lammert F, Gustot T, Deltenre P, Völzke H, Lerch MM, Mayerle J, Eyer F, Schafmayer C, Cichon S, Nöthen MM, Nothnagel M, Ellinghaus D, Huse K, Franke A, Zopf S, Hellerbrand C, Moreno C, Franchimont D, Morgan MY, Hampe J, A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nature Genetics 47, 1443–1448 (2015). [DOI] [PubMed] [Google Scholar]
- 138.Thangapandi VR, Knittelfelder O, Brosch M, Patsenker E, Vvedenskaya O, Buch S, Hinz S, Hendricks A, Nati M, Herrmann A, Rekhade DR, Berg T, Matz-Soja M, Huse K, Klipp E, Pauling JK, Wodke JA, Miranda Ackerman J, Bonin M. v., Aigner E, Datz C, von Schönfels W, Nehring S, Zeissig S, Röcken C, Dahl A, Chavakis T, Stickel F, Shevchenko A, Schafmayer C, Hampe J, Subramanian P, Loss of hepatic Mboat7 leads to liver fibrosis. Gut 70, 940–950 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mancina RM, Dongiovanni P, Petta S, Pingitore P, Meroni M, Rametta R, Boren J, Montalcini T, Pujia A, Wiklund O, Hindy G, Spagnuolo R, Motta BM, Pipitone RM, Craxi A, Fargion S, Nobili V, Kakela P, Karja V, Mannisto V, Pihlajamaki J, Reilly DF, Castro-Perez J, Kozlitina J, Valenti L, Romeo S, The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 150, 1219–1230 e1216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu Y-L, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JBS, Allison MED, Alexander GJ, Piguet A-C, Anty R, Donaldson P, Aithal GP, Francque S, Van Gaal L, Clement K, Ratziu V, Dufour J-F, Day CP, Daly AK, Anstee QM, TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nature communications 5, 4309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, Motta BM, Kaminska D, Rametta R, Grimaudo S, Pelusi S, Montalcini T, Alisi A, Maggioni M, Kärjä V, Borén J, Käkelä P, Di Marco V, Xing C, Nobili V, Dallapiccola B, Craxi A, Pihlajamäki J, Fargion S, Sjöström L, Carlsson LM, Romeo S, Valenti L, Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 61, 506–514 (2015). [DOI] [PubMed] [Google Scholar]
- 142.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC, Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics 46, 352–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rutledge SM, Soper ER, Ma N, Pejaver V, Friedman SL, Branch AD, Kenny EE, Belbin GM, Abul-Husn NS, Association of HSD17B13 and PNPLA3 With Liver Enzymes and Fibrosis in Hispanic/Latino Individuals of Diverse Genetic Ancestries. Clin Gastroenterol Hepatol, (2023). [DOI] [PubMed] [Google Scholar]
- 144.Verweij N, Haas ME, Nielsen JB, Sosina OA, Kim M, Akbari P, De T, Hindy G, Bovijn J, Persaud T, Miloscio L, Germino M, Panagis L, Watanabe K, Mbatchou J, Jones M, LeBlanc M, Balasubramanian S, Lammert C, Enhorning S, Melander O, Carey DJ, Still CD, Mirshahi T, Rader DJ, Parasoglou P, Walls JR, Overton JD, Reid JG, Economides A, Cantor MN, Zambrowicz B, Murphy AJ, Abecasis GR, Ferreira MAR, Smagris E, Gusarova V, Sleeman M, Yancopoulos GD, Marchini J, Kang HM, Karalis K, Shuldiner AR, Della Gatta G, Locke AE, Baras A, Lotta LA, Germline Mutations in CIDEB and Protection against Liver Disease. N Engl J Med 387, 332–344 (2022). [DOI] [PubMed] [Google Scholar]
- 145.Vujkovic M, Ramdas S, Lorenz KM, Guo X, Darlay R, Cordell HJ, He J, Gindin Y, Chung C, Myers RP, Schneider CV, Park J, Lee KM, Serper M, Carr RM, Kaplan DE, Haas ME, MacLean MT, Witschey WR, Zhu X, Tcheandjieu C, Kember RL, Kranzler HR, Verma A, Giri A, Klarin DM, Sun YV, Huang J, Huffman JE, Creasy KT, Hand NJ, Liu CT, Long MT, Yao J, Budoff M, Tan J, Li X, Lin HJ, Chen YI, Taylor KD, Chang RK, Krauss RM, Vilarinho S, Brancale J, Nielsen JB, Locke AE, Jones MB, Verweij N, Baras A, Reddy KR, Neuschwander-Tetri BA, Schwimmer JB, Sanyal AJ, Chalasani N, Ryan KA, Mitchell BD, Gill D, Wells AD, Manduchi E, Saiman Y, Mahmud N, Miller DR, Reaven PD, Phillips LS, Muralidhar S, DuVall SL, Lee JS, Assimes TL, Pyarajan S, Cho K, Edwards TL, Damrauer SM, Wilson PW, Gaziano JM, O'Donnell CJ, Khera AV, Grant SFA, Brown CD, Tsao PS, Saleheen D, Lotta LA, Bastarache L, Anstee QM, Daly AK, Meigs JB, Rotter JI, Lynch JA, Regeneron Genetics C, Geisinger-Regeneron Discov EHRC, Consortium EP, Program VAMV, Rader DJ, Voight BF, Chang KM, A multiancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for nonalcoholic fatty liver disease with histological and radiological validation. Nat Genet 54, 761–771 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.De Vincentis A, Tavaglione F, Jamialahmadi O, Picardi A, Antonelli Incalzi R, Valenti L, Romeo S, Vespasiani-Gentilucci U, A Polygenic Risk Score to Refine Risk Stratification and Prediction for Severe Liver Disease by Clinical Fibrosis Scores. Clin Gastroenterol Hepatol 20, 658–673 (2022). [DOI] [PubMed] [Google Scholar]
- 147.Sun T, Huang Z, Liang WC, Yin J, Lin WY, Wu J, Vernes JM, Lutman J, Caplazi P, Jeet S, Wong T, Wong M, DePianto DJ, Morshead KB, Sun KH, Modrusan Z, Vander Heiden JA, Abbas AR, Zhang H, Xu M, N'Diaye EN, Roose-Girma M, Wolters PJ, Yadav R, Sukumaran S, Ghilardi N, Corpuz R, Emson C, Meng YG, Ramalingam TR, Lupardus P, Brightbill HD, Seshasayee D, Wu Y, Arron JR, TGFbeta2 and TGFbeta3 isoforms drive fibrotic disease pathogenesis. Sci Transl Med 13, (2021). [DOI] [PubMed] [Google Scholar]
- 148.Tomita K, Teratani T, Suzuki T, Shimizu M, Sato H, Narimatsu K, Okada Y, Kurihara C, Irie R, Yokoyama H, Shimamura K, Usui S, Ebinuma H, Saito H, Watanabe C, Komoto S, Kawaguchi A, Nagao S, Sugiyama K, Hokari R, Kanai T, Miura S, Hibi T, Free cholesterol accumulation in hepatic stellate cells: Mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 59, 154–169 (2014). [DOI] [PubMed] [Google Scholar]
- 149.Okina Y, Sato-Matsubara M, Matsubara T, Daikoku A, Longato L, Rombouts K, Thanh Thuy LT, Ichikawa H, Minamiyama Y, Kadota M, Fujii H, Enomoto M, Ikeda K, Yoshizato K, Pinzani M, Kawada N, TGF-β1-driven reduction of cytoglobin leads to oxidative DNA damage in stellate cells during non-alcoholic steatohepatitis. Journal of Hepatology 73, 882–895 (2020). [DOI] [PubMed] [Google Scholar]
- 150.Vacca M, Leslie J, Virtue S, Lam BYH, Govaere O, Tiniakos D, Snow S, Davies S, Petkevicius K, Tong Z, Peirce V, Nielsen MJ, Ament Z, Li W, Kostrzewski T, Leeming DJ, Ratziu V, Allison MED, Anstee QM, Griffin JL, Oakley F, Vidal-Puig A, Bone morphogenetic protein 8B promotes the progression of non-alcoholic steatohepatitis. Nature Metabolism 2, 514–531 (2020). [DOI] [PubMed] [Google Scholar]
- 151.Yang L, Roh YS, Song J, Zhang B, Liu C, Loomba R, Seki E, Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology 59, 483–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu D, Wang K, Li K, Xu R, Chang X, Zhu Y, Sun P, Han X, Ets-1 deficiency alleviates nonalcoholic steatohepatitis via weakening TGF-β1 signaling-mediated hepatocyte apoptosis. Cell Death Dis 10, 458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bansal R, van Baarlen J, Storm G, Prakash J, The interplay of the Notch signaling in hepatic stellate cells and macrophages determines the fate of liver fibrogenesis. Sci Rep 5, 18272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang YM, Noureddin M, Liu C, Ohashi K, Kim SY, Ramnath D, Powell EE, Sweet MJ, Roh YS, Hsin IF, Deng N, Liu Z, Liang J, Mena E, Shouhed D, Schwabe RF, Jiang D, Lu SC, Noble PW, Seki E, Hyaluronan synthase 2-mediated hyaluronan production mediates Notch1 activation and liver fibrosis. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fang Z-Q, Ruan B, Liu J-J, Duan J-L, Yue Z-S, Song P, Xu H, Ding J, Xu C, Dou G-R, Wang L, Notch-triggered maladaptation of liver sinusoidal endothelium aggravates nonalcoholic steatohepatitis through endothelial nitric oxide synthase. Hepatology 76, 742–758 (2022). [DOI] [PubMed] [Google Scholar]
- 156.Richter LR, Wan Q, Wen D, Zhang Y, Yu J, Kang J. k., Zhu C, McKinnon EL, Gu Z, Qiang L, Pajvani UB, Targeted Delivery of Notch Inhibitor Attenuates Obesity-Induced Glucose Intolerance and Liver Fibrosis. ACS Nano 14, 6878–6886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rangwala F, Guy CD, Lu J, Suzuki A, Burchette JL, Abdelmalek MF, Chen W, Diehl AM, Increased production of sonic hedgehog by ballooned hepatocytes. The Journal of Pathology 224, 401–410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chong YC, Lim TE, Fu Y, Shin EM, Tergaonkar V, Han W, Indian Hedgehog links obesity to development of hepatocellular carcinoma. Oncogene 38, 2206–2222 (2019). [DOI] [PubMed] [Google Scholar]
- 159.Sahai A, Malladi P, Melin-Aldana H, Green RM, Whitington PF, Upregulation of osteopontin expression is involved in the development of nonalcoholic steatohepatitis in a dietary murine model. Am J Physiol-Gastr L 287, G264–G273 (2004). [DOI] [PubMed] [Google Scholar]
- 160.Du M, Wang X, Yuan L, Liu B, Mao X, Huang D, Yang L, Huang K, Zhang F, Wang Y, Luo X, Wang C, Peng J, Liang M, Huang D, Huang K, Targeting NFATc4 attenuates non-alcoholic steatohepatitis in mice. Journal of Hepatology 73, 1333–1346 (2020). [DOI] [PubMed] [Google Scholar]
- 161.Li Y, Xiu W, Xu J, Chen X, Wang G, Duan J, Sun L, Liu B, Xie W, Pu G, Wang Q, Wang C, Increased CHCHD2 expression promotes liver fibrosis in nonalcoholic steatohepatitis via Notch/osteopontin signaling. JCI Insight 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schulien I, Hockenjos B, Schmitt-Graeff A, Perdekamp MG, Follo M, Thimme R, Hasselblatt P, The transcription factor c-Jun/AP-1 promotes liver fibrosis during non-alcoholic steatohepatitis by regulating Osteopontin expression. Cell Death & Differentiation 26, 1688–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Machado MV, Michelotti GA, Pereira TA, Xie G, Premont R, Cortez-Pinto H, Diehl AM, Accumulation of duct cells with activated YAP parallels fibrosis progression in non-alcoholic fatty liver disease. Journal of Hepatology 63, 962–970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu P, Zhang Y, Liu Y, Yuan Q, Song L, Liu M, Liu Z, Yang Y, Li J, Li D, Ren G, Fibroblast growth factor 21 attenuates hepatic fibrogenesis through TGF-β/smad2/3 and NF-κB signaling pathways. Toxicol Appl Pharm 290, 43–53 (2016). [DOI] [PubMed] [Google Scholar]
- 165.Fuchs S, Yusta B, Baggio LL, Varin EM, Matthews D, Drucker DJ, Loss of Glp2r signaling activates hepatic stellate cells and exacerbates diet-induced steatohepatitis in mice. JCI Insight 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Widjaja AA, Viswanathan S, Wei Ting JG, Tan J, Shekeran SG, Carling D, Lim W-W, Cook SA, IL11 stimulates ERK/P90RSK to inhibit LKB1/AMPK and activate mTOR initiating a mesenchymal program in stromal, epithelial, and cancer cells. iScience 25, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Morikawa R, Nakamoto N, Amiya T, Chu P.-s., Koda Y, Teratani T, Suzuki T, Kurebayashi Y, Ueno A, Taniki N, Miyamoto K, Yamaguchi A, Shiba S, Katayama T, Yoshida K, Takada Y, Ishihara R, Ebinuma H, Sakamoto M, Kanai T, Role of CC chemokine receptor 9 in the progression of murine and human non-alcoholic steatohepatitis. Journal of Hepatology 74, 511–521 (2021). [DOI] [PubMed] [Google Scholar]
- 168.Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, Loomba R, Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut 65, 1546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley–Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM, Relationship Between Methylome and Transcriptome in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 145, 1076–1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hardy T, Zeybel M, Day CP, Dipper C, Masson S, McPherson S, Henderson E, Tiniakos D, White S, French J, Mann DA, Anstee QM, Mann J, Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 66, 1321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zeybel M, Hardy T, Robinson SM, Fox C, Anstee QM, Ness T, Masson S, Mathers JC, French J, White S, Mann J, Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clinical Epigenetics 7, 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Johnson ND, Wu X, Still CD, Chu X, Petrick AT, Gerhard GS, Conneely KN, DiStefano JK, Differential DNA methylation and changing cell-type proportions as fibrotic stage progresses in NAFLD. Clinical Epigenetics 13, 152 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang H, Ma Y, Cheng X, Wu D, Huang X, Chen B, Ren Y, Jiang W, Tang X, Bai T, Chen Y, Zhao Y, Zhang C, Xiao X, Liu J, Deng Y, Ye T, Chen L, Liu H-M, Friedman SL, Chen L, Ding B-S, Cao Z, Targeting epigenetically maladapted vascular niche alleviates liver fibrosis in nonalcoholic steatohepatitis. Science Translational Medicine 13, eabd1206 (2021). [DOI] [PubMed] [Google Scholar]
- 174.Ding N, Hah N, Yu RT, Sherman MH, Benner C, Leblanc M, He M, Liddle C, Downes M, Evans RM, BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci U S A 112, 15713–15718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Barcena-Varela M, Caruso S, Llerena S, Alvarez-Sola G, Uriarte I, Latasa MU, Urtasun R, Rebouissou S, Alvarez L, Jimenez M, Santamaria E, Rodriguez-Ortigosa C, Mazza G, Rombouts K, Jose-Eneriz E. San, Rabal O, Agirre X, Iraburu M, Santos-Laso A, Banales JM, Zucman-Rossi J, Prosper F, Oyarzabal J, Berasain C, Avila MA, Fernandez-Barrena MG, Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma. Hepatology 69, 587–603 (2019). [DOI] [PubMed] [Google Scholar]
- 176.Martin-Mateos R, De Assuncao TM, Arab JP, Jalan-Sakrikar N, Yaqoob U, Greuter T, Verma VK, Mathison AJ, Cao S, Lomberk G, Mathurin P, Urrutia R, Huebert RC, Shah VH, Enhancer of Zeste Homologue 2 Inhibition Attenuates TGF-beta Dependent Hepatic Stellate Cell Activation and Liver Fibrosis. Cell Mol Gastroenterol Hepatol 7, 197–209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Grindheim JM, Nicetto D, Donahue G, Zaret KS, Polycomb Repressive Complex 2 Proteins EZH1 and EZH2 Regulate Timing of Postnatal Hepatocyte Maturation and Fibrosis by Repressing Genes With Euchromatic Promoters in Mice. Gastroenterology 156, 1834–1848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dong F, Jiang S, Li J, Wang Y, Zhu L, Huang Y, Jiang X, Hu X, Zhou Q, Zhang Z, Bao Z, The histone demethylase KDM4D promotes hepatic fibrogenesis by modulating Toll-like receptor 4 signaling pathway. EBioMedicine 39, 472–483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA, MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 138, 705–714, 714 e701-704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hardy T, Zeybel M, Day CP, Dipper C, Masson S, McPherson S, Henderson E, Tiniakos D, White S, French J, Mann DA, Anstee QM, Mann J, Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 66, 1321–1328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu Y, Sun X, Zhang R, Cao T, Cai S-Y, Boyer JL, Zhang X, Li D, Huang Y, A Positive Feedback Loop of TET3 and TGF-β1 Promotes Liver Fibrosis. Cell reports 30, 1310–1318.e1315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Trivedi P, Wang S, Friedman SL, The Power of Plasticity—Metabolic Regulation of Hepatic Stellate Cells. Cell Metabolism 33, 242–257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL, Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 142, 938–946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hernandez-Gea V, Hilscher M, Rozenfeld R, Lim MP, Nieto N, Werner S, Devi LA, Friedman SL, Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol 59, 98–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yu X, Elfimova N, Muller M, Bachurski D, Koitzsch U, Drebber U, Mahabir E, Hansen HP, Friedman SL, Klein S, Dienes HP, Hosel M, Buettner R, Trebicka J, Kondylis V, Mannaerts I, Odenthal M, Autophagy-Related Activation of Hepatic Stellate Cells Reduces Cellular miR-29a by Promoting Its Vesicular Secretion. Cell Mol Gastroenterol Hepatol 13, 1701–1716 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hui ST, Kurt Z, Tuominen I, Norheim F,Davis RC, Pan C, Dirks DL, Magyar CE, French SW, Chella Krishnan K, Sabir S, Campos-Pérez F, Méndez-Sánchez N, Macías-Kauffer L, León-Mimila P, Canizales-Quinteros S, Yang X, Beaven SW, Huertas-Vazquez A, Lusis AJ, The Genetic Architecture of Diet-Induced Hepatic Fibrosis in Mice. Hepatology 68, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tomita K, Teratani T, Suzuki T, Shimizu M, Sato H, Narimatsu K, Okada Y, Kurihara C, Irie R, Yokoyama H, Shimamura K, Usui S, Ebinuma H, Saito H, Watanabe C, Komoto S, Kawaguchi A, Nagao S, Sugiyama K, Hokari R, Kanai T, Miura S, Hibi T, Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 59, 154–169 (2014). [DOI] [PubMed] [Google Scholar]
- 188.Teratani T, Tomita K, Furuhashi H, Sugihara N, Higashiyama M, Nishikawa M, Irie R, Takajo T, Wada A, Horiuchi K, Inaba K, Hanawa Y, Shibuya N, Okada Y, Kurihara C, Nishii S, Mizoguchi A, Hozumi H, Watanabe C, Komoto S, Nagao S, Yamamoto J, Miura S, Hokari R, Kanai T, Lipoprotein Lipase Up-regulation in Hepatic Stellate Cells Exacerbates Liver Fibrosis in Nonalcoholic Steatohepatitis in Mice. Hepatology Communications 3, 1098–1112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen Z-P, O'Neill HM, Ford RJ, Palanivel R, O'Brien M, Hardie DG, Macaulay SL, Schertzer JD, Dyck JRB, van Denderen BJ, Kemp BE, Steinberg GR, Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nature Medicine 19, 1649–1654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bates J, Vijayakumar A, Ghoshal S, Marchand B, Yi S, Kornyeyev D, Zagorska A, Hollenback D, Walker K, Liu K, Pendem S, Newstrom D, Brockett R, Mikaelian I, Kusam S, Ramirez R, Lopez D, Li L, Fuchs BC, Breckenridge DG, Acetyl-CoA carboxylase inhibition disrupts metabolic reprogramming during hepatic stellate cell activation. Journal of Hepatology 73, 896–905 (2020). [DOI] [PubMed] [Google Scholar]
- 191.Ross TT, Crowley C, Kelly KL, Rinaldi A, Beebe DA, Lech MP, Martinez RV, Carvajal-Gonzalez S, Boucher M, Hirenallur-Shanthappa D, Morin J, Opsahl AC, Vargas SR, Bence KK, Pfefferkorn JA, Esler WP, Acetyl-CoA Carboxylase Inhibition Improves Multiple Dimensions of NASH Pathogenesis in Model Systems. Cellular and Molecular Gastroenterology and Hepatology 10, 829–851 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morrow MR, Batchuluun B, Wu J, Ahmadi E, Leroux JM, Mohammadi-Shemirani P, Desjardins EM, Wang Z, Tsakiridis EE, Lavoie DCT, Reihani A, Smith BK, Kwiecien JM, Lally JSV, Nero TL, Parker MW, Ask K, Scott JW, Jiang L, Paré G, Pinkosky SL, Steinberg GR, Inhibition of ATP-citrate lyase improves NASH, liver fibrosis, and dyslipidemia. Cell Metabolism 34, 919–936.e918 (2022). [DOI] [PubMed] [Google Scholar]
- 193.Leung H, Long X, Ni Y, Qian L, Nychas E, Siliceo SL, Pohl D, Hanhineva K, Liu Y, Xu A, Nielsen HB, Belda E, Clément K, Loomba R, Li H, Jia W, Panagiotou G, Risk assessment with gut microbiome and metabolite markers in NAFLD development. Science Translational Medicine 14, eabk0855 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lelouvier B, Servant F, Paisse S, Brunet AC, Benyahya S, Serino M, Valle C, Ortiz MR, Puig J, Courtney M, Federici M, Fernandez-Real JM, Burcelin R, Amar J, Changes in blood microbiota profiles associated with liver fibrosis in obese patients: A pilot analysis. Hepatology 64, 2015–2027 (2016). [DOI] [PubMed] [Google Scholar]
- 195.Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, Bross C, Durelle J, Goyal NP, Hamilton G, Holtz ML, Lavine JE, Mitreva M, Newton KP, Pan A, Simpson PM, Sirlin CB, Sodergren E, Tyagi R, Yates KP, Weinstock GM, Salzman NH, Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease. Gastroenterology 157, 1109–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE, Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 25, 1054–1062 e1055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Carter JK, Bhattacharya D, Borgerding JN, Fiel MI, Faith JJ, Friedman SL, Modeling dysbiosis of human NASH in mice: Loss of gut microbiome diversity and overgrowth of Erysipelotrichales. PLoS One 16, e0244763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lee G, You HJ, Bajaj JS, Joo SK, Yu J, Park S, Kang H, Park JH, Kim JH, Lee DH, Lee S, Kim W, Ko G, Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nature communications 11, 4982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Caussy C, Hsu C, Lo MT, Liu A, Bettencourt R, Ajmera VH, Bassirian S, Hooker J, Sy E, Richards L, Schork N, Schnabl B, Brenner DA, Sirlin CB, Chen CH, Loomba R, N. i. T. C. Genetics of, Link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology 68, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ng SC, Xu Z, Mak JWY, Yang K, Liu Q, Zuo T, Tang W, Lau L, Lui RN, Wong SH, Tse YK, Li AYL, Cheung K, Ching JYL, Wong VWS, Kong APS, Ma RCW, Chow EYK, Wong SKH, Ho ICH, Chan PKS, Chan FKL, Microbiota engraftment after faecal microbiota transplantation in obese subjects with type 2 diabetes: a 24-week, double-blind, randomised controlled trial. Gut 71, 716 (2022). [DOI] [PubMed] [Google Scholar]
- 201.Scorletti E, Afolabi PR, Miles EA, Smith DE, Almehmadi A, Alshathry A, Childs CE, Del Fabbro S, Bilson J, Moyses HE, Clough GF, Sethi JK, Patel J, Wright M, Breen DJ, Peebles C, Darekar A, Aspinall R, Fowell AJ, Dowman JK, Nobili V, Targher G, Delzenne NM, Bindels LB, Calder PC, Byrne CD, Synbiotics Alter Fecal Microbiomes, But Not Liver Fat or Fibrosis, in a Randomized Trial of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 158, 1597–1610.e1597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, Zhao X, Li N, Li S, Xue G, Cheng W, Li B, Li H, Lin W, Tian C, Zhao J, Han J, An D, Zhang Q, Wei H, Zheng M, Ma X, Li W, Chen X, Zhang Z, Zeng H, Ying S, Wu J, Yang R, Liu D, Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metabolism 30, 675–688.e677 (2019). [DOI] [PubMed] [Google Scholar]
- 203.Chiu C-C, Ching Y-H, Li Y-P, Liu J-Y, Huang Y-T, Huang Y-W, Yang S-S, Huang W-C, Chuang H-L, Nonalcoholic Fatty Liver Disease Is Exacerbated in High-Fat Diet-Fed Gnotobiotic Mice by Colonization with the Gut Microbiota from Patients with Nonalcoholic Steatohepatitis. Nutrients 9, 1220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez J-P, Shulman GI, Gordon JI, Hoffman HM, Flavell RA, Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nagasaki A, Sakamoto S, Chea C, Ishida E, Furusho H, Fujii M, Takata T, Miyauchi M, Odontogenic infection by Porphyromonas gingivalis exacerbates fibrosis in NASH via hepatic stellate cell activation. Sci Rep-Uk 10, 4134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Furusho H, Miyauchi M, Hyogo H, Inubushi T, Ao M, Ouhara K, Hisatune J, Kurihara H, Sugai M, Hayes CN, Nakahara T, Aikata H, Takahashi S, Chayama K, Takata T, Dental infection of Porphyromonas gingivalis exacerbates high fat diet-induced steatohepatitis in mice. Journal of Gastroenterology 48, 1259–1270 (2013). [DOI] [PubMed] [Google Scholar]
- 207.Imajo K, Fujita K, Yoneda M, Nozaki Y, Ogawa Y, Shinohara Y, Kato S, Mawatari H, Shibata W, Kitani H, Ikejima K, Kirikoshi H, Nakajima N, Saito S, Maeyama S, Watanabe S, Wada K, Nakajima A, Hyperresponsivity to Low-Dose Endotoxin during Progression to Nonalcoholic Steatohepatitis Is Regulated by Leptin-Mediated Signaling. Cell Metabolism 16, 44–54 (2012). [DOI] [PubMed] [Google Scholar]
- 208.Aoki R, Onuki M, Hattori K, Ito M, Yamada T, Kamikado K, Kim Y-G, Nakamoto N, Kimura I, Clarke JM, Kanai T, Hase K, Commensal microbe-derived acetate suppresses NAFLD/NASH development via hepatic FFAR2 signalling in mice. Microbiome 9, 188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Demir M, Lang S, Hartmann P, Duan Y, Martin A, Miyamoto Y, Bondareva M, Zhang X, Wang Y, Kasper P, Bang C, Roderburg C, Tacke F, Steffen H-M, Goeser T, Kruglov A, Eckmann L, Stärkel P, Fouts DE, Schnabl B, The fecal mycobiome in non-alcoholic fatty liver disease☆. Journal of Hepatology 76, 788–799 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hunter H, de Gracia Hahn D, Duret A, Im YR, Cheah Q, Dong J, Fairey M, Hjalmarsson C, Li A, Lim HK, McKeown L, Mitrofan C-G, Rao R, Utukuri M, Rowe IA, Mann JP, Weight loss, insulin resistance, and study design confound results in a meta-analysis of animal models of fatty liver. eLife 9, e56573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ibrahim SH, Hirsova P, Malhi H, Gores GJ, Animal Models of Nonalcoholic Steatohepatitis: Eat, Delete, and Inflame. Digest Dis Sci 61, 1325–1336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Brenner DA, Of Mice and Men and Nonalcoholic Steatohepatitis. Hepatology 68, 2059–2061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Im YR, Hunter H, de Gracia Hahn D, Duret A, Cheah Q, Dong J, Fairey M, Hjalmarsson C, Li A, Lim HK, McKeown L, Mitrofan C-G, Rao R, Utukuri M, Rowe IA, Mann JP, A Systematic Review of Animal Models of NAFLD Finds High-Fat, High-Fructose Diets Most Closely Resemble Human NAFLD. Hepatology 74, 1884–1901 (2021). [DOI] [PubMed] [Google Scholar]
- 214.Boland ML, Oro D, Tolbol KS, Thrane ST, Nielsen JC, Cohen TS, Tabor DE, Fernandes F, Tovchigrechko A, Veidal SS, Warrener P, Sellman BR, Jelsing J, Feigh M, Vrang N, Trevaskis JL, Hansen HH, Towards a standard diet-induced and biopsy-confirmed mouse model of non-alcoholic steatohepatitis: Impact of dietary fat source. World J Gastroenterol 25, 4904–4920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Clapper JR, Hendricks MD, Gu G, Wittmer C, Dolman CS, Herich J, Athanacio J, Villescaz C, Ghosh SS, Heilig JS, Lowe C, Roth JD, Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am J Physiol Gastrointest Liver Physiol 305, G483–495 (2013). [DOI] [PubMed] [Google Scholar]
- 216.Polyzos SA, Kountouras J, Mantzoros CS, Leptin in nonalcoholic fatty liver disease: A narrative review. Metabolism 64, 60–78 (2015). [DOI] [PubMed] [Google Scholar]
- 217.Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, Lallone RL, Burley SK, Friedman JM, Weight-Reducing Effects of the Plasma Protein Encoded by the obese Gene. Science 269, 543–546 (1995). [DOI] [PubMed] [Google Scholar]
- 218.Hummel KP, Dickie MM, Coleman DL, Diabetes, a New Mutation in the Mouse. Science 153, 1127–1128 (1966). [DOI] [PubMed] [Google Scholar]
- 219.Sahai A, Malladi P, Pan X, Paul R, Melin-Aldana H, Green RM, Whitington PF, Obese and diabetic db/db mice develop marked liver fibrosis in a model of nonalcoholic steatohepatitis: role of short-form leptin receptors and osteopontin. Am J Physiol-Gastr L 287, G1035–G1043 (2004). [DOI] [PubMed] [Google Scholar]
- 220.Marques V, Afonso MB, Bierig N, Duarte-Ramos F, Santos-Laso A, Jimenez-Aguero R, Eizaguirre E, Bujanda L, Pareja MJ, Luis R, Costa A, Machado MV, Alonso C, Arretxe E, Alustiza JM, Krawczyk M, Lammert F, Tiniakos DG, Flehmig B, Cortez-Pinto H, Banales JM, Castro RE, Normann A, Rodrigues CMP, Adiponectin, Leptin, and IGF-1 Are Useful Diagnostic and Stratification Biomarkers of NAFLD. Front Med (Lausanne) 8, 683250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nakagawa H, Umemura A, Taniguchi K, Font-Burgada J, Dhar D, Ogata H, Zhong Z, Valasek Mark A., Seki E, Hidalgo J, Koike K, Kaufman Randal J., Karin M, ER Stress Cooperates with Hypernutrition to Trigger TNF-Dependent Spontaneous HCC Development. Cancer Cell 26, 331–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS, Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes & Development 12, 3182–3194 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nakayama H, Otabe S, Ueno T, Hirota N, Yuan X, Fukutani T, Hashinaga T, Wada N, Yamada K, Transgenic mice expressing nuclear sterol regulatory element–binding protein 1c in adipose tissue exhibit liver histology similar to nonalcoholic steatohepatitis. Metabolism 56, 470–475 (2007). [DOI] [PubMed] [Google Scholar]
- 224.Lu SC, Alvarez L, Huang Z-Z, Chen L, An W, Corrales FJ, Avila MA, Kanel G, Mato JM, Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proceedings of the National Academy of Sciences 98, 5560–5565 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim Y-J, Sherwin R, Devaskar S, Lesche R, Magnuson MA, Wu H, Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity. Proceedings of the National Academy of Sciences 101, 2082–2087 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Fan C-Y, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK, Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty Acyl-CoA oxidase: implications for peroxisome proliferator-activated receptor α natural ligand metabolism. Journal of Biological Chemistry 273, 15639–15645 (1998). [DOI] [PubMed] [Google Scholar]
- 227.Teufel A, Itzel T, Erhart W, Brosch M, Wang XY, Kim YO, von Schönfels W, Herrmann A, Brückner S, Stickel F, Dufour J-F, Chavakis T, Hellerbrand C, Spang R, Maass T, Becker T, Schreiber S, Schafmayer C, Schuppan D, Hampe J, Comparison of Gene Expression Patterns Between Mouse Models of Nonalcoholic Fatty Liver Disease and Liver Tissues From Patients. Gastroenterology 151, 513–525.e510 (2016). [DOI] [PubMed] [Google Scholar]
- 228.Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, Kumar DP, Daita K, Min H-K, Mirshahi F, Bedossa P, Sun X, Hoshida Y, Koduru SV, Contaifer D, Warncke UO, Wijesinghe DS, Sanyal AJ, A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. Journal of Hepatology 65, 579–588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Farrell GC, Mridha AR, Yeh MM, Arsov T, Van Rooyen DM, Brooling J, Nguyen T, Heydet D, Delghingaro-Augusto V, Nolan CJ, Shackel NA, McLennan SV, Teoh NC, Larter CZ, Strain dependence of diet-induced NASH and liver fibrosis in obese mice is linked to diabetes and inflammatory phenotype. Liver International 34, 1084–1093 (2014). [DOI] [PubMed] [Google Scholar]
- 230.Arsov T, Silva DG, O’Bryan MK, Sainsbury A, Lee NJ, Kennedy C, Manji SSM, Nelms K, Liu C, Vinuesa CG, de Kretser DM, Goodnow CC, Petrovsky N, Fat Aussie—A New Alström Syndrome Mouse Showing a Critical Role for ALMS1 in Obesity, Diabetes, and Spermatogenesis. Molecular Endocrinology 20, 1610–1622 (2006). [DOI] [PubMed] [Google Scholar]
- 231.St. Rose K, Yan J, Xu F, Williams J, Dweck V, Saxena D, Schwabe RF, Caviglia JM, Mouse model of NASH that replicates key features of the human disease and progresses to fibrosis stage 3. Hepatology Communications 6, 2676–2688 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Teratani T, Tomita K, Suzuki T, Oshikawa T, Yokoyama H, Shimamura K, Tominaga S, Hiroi S, Irie R, Okada Y, Kurihara C, Ebinuma H, Saito H, Hokari R, Sugiyama K, Kanai T, Miura S, Hibi T, A high-cholesterol diet exacerbates liver fibrosis in mice via accumulation of free cholesterol in hepatic stellate cells. Gastroenterology 142, 152–164 e110 (2012). [DOI] [PubMed] [Google Scholar]
- 233.Ioannou GN, The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol Metab 27, 84–95 (2016). [DOI] [PubMed] [Google Scholar]
- 234.Wang X, Cai B, Yang X, Sonubi OO, Zheng Z, Ramakrishnan R, Shi H, Valenti L, Pajvani UB, Sandhu J, Infante RE, Radhakrishnan A, Covey DF, Guan KL, Buck J, Levin LR, Tontonoz P, Schwabe RF, Tabas I, Cholesterol Stabilizes TAZ in Hepatocytes to Promote Experimental Non-alcoholic Steatohepatitis. Cell Metab 31, 969–986 e967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Fujii M, Shibazaki Y, Wakamatsu K, Honda Y, Kawauchi Y, Suzuki K, Arumugam S, Watanabe K, Ichida T, Asakura H, Yoneyama H, A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Medical Molecular Morphology 46, 141–152 (2013). [DOI] [PubMed] [Google Scholar]
- 236.Sadi G, Baloğlu MC, Pektaş MB, Differential Gene Expression in Liver Tissues of Streptozotocin-Induced Diabetic Rats in Response to Resveratrol Treatment. PLOS ONE 10, e0124968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B, Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028.e1013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA, Anastasakis DG, Sarshad AA, Leonardi I, Collins N, Blatter JA, Han S-J, Tamoutounour S, Potapova S, Foster St. Claire MB, Yuan W, Sen SK, Dreier MS, Hild B, Hafner M, Wang D, Iliev ID, Belkaid Y, Trinchieri G, Rehermann B, Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, eaaw4361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ratziu V, Friedman SL, Why do so many NASH trials fail? Gastroenterology, (2020). [DOI] [PubMed] [Google Scholar]
- 240.Ramli MNB, Lim YS, Koe CT, Demircioglu D, Tng W, Gonzales KAU, Tan CP, Szczerbinska I, Liang H, Soe EL, Lu Z, Ariyachet C, Yu KM, Koh SH, Yaw LP, Jumat NHB, Lim JSY, Wright G, Shabbir A, Dan YY, Ng H-H, Chan Y-S, Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 159, 1471–1486.e1412 (2020). [DOI] [PubMed] [Google Scholar]
- 241.Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, Thompson W, Karns RA, Mayhew CN, McGrath PS, McCauley HA, Zhang R-R, Lewis K, Hakozaki S, Ferguson A, Saiki N, Yoneyama Y, Takeuchi I, Mabuchi Y, Akazawa C, Yoshikawa HY, Wells JM, Takebe T, Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metabolism 30, 374–384.e376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kimura M, Iguchi T, Iwasawa K, Dunn A, Thompson WL, Yoneyama Y, Chaturvedi P, Zorn AM, Wintzinger M, Quattrocelli M, Watanabe-Chailland M, Zhu G, Fujimoto M, Kumbaji M, Kodaka A, Gindin Y, Chung C, Myers RP, Subramanian GM, Hwa V, Takebe T, En masse organoid phenotyping informs metabolic-associated genetic susceptibility to NASH. Cell 185, 4216–4232.e4216 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.McCarron S, Bathon B, Conlon DM, Abbey D, Rader DJ, Gawronski K, Brown CD, Olthoff KM, Shaked A, Raabe TD, Functional Characterization of Organoids Derived From Irreversibly Damaged Liver of Patients With NASH. Hepatology 74, 1825–1844 (2021). [DOI] [PubMed] [Google Scholar]
- 244.Feaver RE, Cole BK, Lawson MJ, Hoang SA, Marukian S, Blackman BR, Figler RA, Sanyal AJ, Wamhoff BR, Dash A, Development of an in vitro human liver system for interrogating nonalcoholic steatohepatitis. JCI Insight 1, e90954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Feaver RE, Cole BK, Lawson MJ, Hoang SA, Marukian S, Blackman BR, Figler RA, Sanyal AJ, Wamhoff BR, Dash A, Development of an in vitro human liver system for interrogating nonalcoholic steatohepatitis. JCI Insight 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kostrzewski T, Maraver P, Ouro-Gnao L, Levi A, Snow S, Miedzik A, Rombouts K, Hughes D, A Microphysiological System for Studying Nonalcoholic Steatohepatitis. Hepatology Communications 4, 77–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Freag MS, Namgung B, Reyna Fernandez ME, Gherardi E, Sengupta S, Jang HL, Human Nonalcoholic Steatohepatitis on a Chip. Hepatology Communications 5, 217–233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.van de Bovenkamp M, Groothuis GM, Meijer DK, Olinga P, Precision-cut fibrotic rat liver slices as a new model to test the effects of anti-fibrotic drugs in vitro. J Hepatol 45, 696–703 (2006). [DOI] [PubMed] [Google Scholar]
- 249.Paish HL, Reed LH, Brown H, Bryan MC, Govaere O, Leslie J, Barksby BS, Garcia Macia M, Watson A, Xu X, Zaki MYW, Greaves L, Whitehall J, French J, White SA, Manas DM, Robinson SM, Spoletini G, Griffiths C, Mann DA, Borthwick LA, Drinnan MJ, Mann J, Oakley F, A Bioreactor Technology for Modeling Fibrosis in Human and Rodent Precision-Cut Liver Slices. Hepatology, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shamseddeen H, Vilar-Gomez E, Chalasani N, Myers RP, Subramanian GM, Shlevin HH, Allgood AE, Orman ES, Spontaneous Fluctuations in Liver Biochemistries in Patients with Compensated NASH Cirrhosis: Implications for Drug Hepatotoxicity Monitoring. Drug Saf 43, 281–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Albhaisi S, Sanyal AJ, Applying Non-Invasive Fibrosis Measurements in NAFLD/NASH: Progress to Date. Pharmaceut Med 33, 451–463 (2019). [DOI] [PubMed] [Google Scholar]
- 252.Ratziu V, Charlton M, Rational combination therapy for NASH: Insights from clinical trials and error. J Hepatol, (2023). [DOI] [PubMed] [Google Scholar]
- 253.Ng CH, Xiao J, Lim WH, Chin YH, Yong JN, Tan DJH, Tay P, Syn N, Foo R, Chan M, Chew N, Tan EXX, Huang DQ, Dan YY, Tamaki N, Siddiqui MS, Sanyal AJ, Loomba R, Noureddin M, Muthiah MD, Placebo effect on progression and regression in NASH: Evidence from a meta-analysis. Hepatology 75, 1647–1661 (2022). [DOI] [PubMed] [Google Scholar]
- 254.D'Ambrosio R, Aghemo A, Rumi MG, Ronchi G, Donato MF, Paradis V, Colombo M, Bedossa P, A morphometric and immunohistochemical study to assess the benefit of a sustained virological response in hepatitis C virus patients with cirrhosis. Hepatology 56, 532–543 (2012). [DOI] [PubMed] [Google Scholar]
- 255.Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Aguilar Schall R, Bornstein JD, Kitrinos KM, Subramanian GM, McHutchison JG, Heathcote EJ, Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet 381, 468–475 (2013). [DOI] [PubMed] [Google Scholar]
- 256.Vilar-Gomez E, Martinez-Perez Y, Calzadilla-Bertot L, Torres-Gonzalez A, Gra-Oramas B, Gonzalez-Fabian L, Friedman SL, Diago M, Romero-Gomez M, Weight Loss Through Lifestyle Modification Significantly Reduces Features of Nonalcoholic Steatohepatitis. Gastroenterology 149, 367–378.e365 (2015). [DOI] [PubMed] [Google Scholar]
- 257.Dixon JB, Bhathal PS, Hughes NR, O'Brien PE, Nonalcoholic fatty liver disease: Improvement in liver histological analysis with weight loss. Hepatology 39, 1647–1654 (2004). [DOI] [PubMed] [Google Scholar]
- 258.Naoumov NV, Brees D, Loeffler J, Chng E, Ren Y, Lopez P, Tai D, Lamle S, Sanyal AJ, Digital pathology with artificial intelligence analyses provides greater insights into treatment-induced fibrosis regression in NASH. Journal of Hepatology 77, 1399–1409 (2022). [DOI] [PubMed] [Google Scholar]
- 259.Lassailly G, Caiazzo R, Ntandja-Wandji LC, Gnemmi V, Baud G, Verkindt H, Ningarhari M, Louvet A, Leteurtre E, Raverdy V, Dharancy S, Pattou F, Mathurin P, Bariatric Surgery Provides Long-term Resolution of Nonalcoholic Steatohepatitis and Regression of Fibrosis. Gastroenterology 159, 1290–1301 e1295 (2020). [DOI] [PubMed] [Google Scholar]
- 260.Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R, Fibrosis Progression in Nonalcoholic Fatty Liver vs Nonalcoholic Steatohepatitis: A Systematic Review and Meta-analysis of Paired-Biopsy Studies. Clinical Gastroenterology and Hepatology 13, 643–654.e649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rosselli M, MacNaughtan J, Jalan R, Pinzani M, Beyond scoring: a modern interpretation of disease progression in chronic liver disease. Gut 62, 1234–1241 (2013). [DOI] [PubMed] [Google Scholar]
- 262.Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, Evans SM, Dillmann W, Glass CK, Brenner DA, Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proceedings of the National Academy of Sciences 109, 9448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Farrell G, Schattenberg JM, Leclercq I, Yeh MM, Goldin R, Teoh N, Schuppan D, Mouse Models of Nonalcoholic Steatohepatitis: Toward Optimization of Their Relevance to Human Nonalcoholic Steatohepatitis. Hepatology 69, 2241–2257 (2019). [DOI] [PubMed] [Google Scholar]
- 264.Muller A, Machnik F, Zimmermann T, Schubert H, Thioacetamide-induced cirrhosis-like liver lesions in rats--usefulness and reliability of this animal model. Exp Pathol 34, 229–236 (1988). [DOI] [PubMed] [Google Scholar]
- 265.Nakano Y, Kamiya A, Sumiyoshi H, Tsuruya K, Kagawa T, Inagaki Y, A Deactivation Factor of Fibrogenic Hepatic Stellate Cells Induces Regression of Liver Fibrosis in Mice. Hepatology 71, 1437–1452 (2020). [DOI] [PubMed] [Google Scholar]
- 266.Wandzioch E, Kolterud A, Jacobsson M, Friedman SL, Carlsson L, Lhx2−/− mice develop liver fibrosis. Proc Natl Acad Sci U S A 101, 16549–16554 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Arroyo N, Villamayor L, Diaz I, Carmona R, Ramos-Rodriguez M, Munoz-Chapuli R, Pasquali L, Toscano MG, Martin F, Cano DA, Rojas A, GATA4 induces liver fibrosis regression by deactivating hepatic stellate cells. JCI Insight 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hazra S, Miyahara T, Rippe RA, Tsukamoto H, PPAR Gamma and Hepatic Stellate Cells. Comp Hepatol 3 Suppl 1, S7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kisseleva T, Brenner D, Molecular and cellular mechanisms of liver fibrosis and its regression. Nature Reviews Gastroenterology & Hepatology 18, 151–166 (2021). [DOI] [PubMed] [Google Scholar]
- 270.Nakano Y, Kamiya A, Sumiyoshi H, Tsuruya K, Kagawa T, Inagaki Y, A novel deactivation factor of fibrogenic hepatic stellate cells induces regression of liver fibrosis in mice. Hepatology n/a, (2019). [DOI] [PubMed] [Google Scholar]
- 271.Calvente CJ, Tameda M, Johnson CD, del Pilar H, Lin YC, Adronikou N, De Mollerat Du Jeu X, Llorente C, Boyer J, Feldstein AE, Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. The Journal of Clinical Investigation 129, 4091–4109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Jiao J, Sastre D, Fiel MI, Lee UE, Ghiassi-Nejad Z, Ginhoux F, Vivier E, Friedman SL, Merad M, Aloman C, Dendritic cell regulation of carbon tetrachloride–induced murine liver fibrosis regression. Hepatology 55, 244–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Liu SB, Ikenaga N, Peng Z-W, Sverdlov DY, Greenstein A, Smith V, Schuppan D, Popov Y, Lysyl oxidase activity contributes to collagen stabilization during liver fibrosis progression and limits spontaneous fibrosis reversal in mice. The FASEB Journal 30, 1599–1609 (2016). [DOI] [PubMed] [Google Scholar]
- 274.Sato T, Head KZ, Li J, Dolin CE, Wilkey D, Skirtich N, Smith K, McCreary DD, Liu S, Beier JI, Singhi AD, McEnaney RM, Merchant ML, Arteel GE, Fibrosis resolution in the mouse liver: Role of Mmp12 and potential role of calpain 1/2. Matrix Biology Plus 17, 100127 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Farooq M, Hameed H, Dimanche-Boitrel M-T, Piquet-Pellorce C, Samson M, Le Seyec J, Switching to Regular Diet Partially Resolves Liver Fibrosis Induced by High-Fat, High-Cholesterol Diet in Mice. Nutrients 14, 10.3390/nu14020386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ramachandran P, Iredale JP, Fallowfield JA, Resolution of liver fibrosis: basic mechanisms and clinical relevance. Semin Liver Dis 35, 119–131 (2015). [DOI] [PubMed] [Google Scholar]
- 277.Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, Williams MJ, Dunbar DR, Manning JR, van Rooijen N, Fallowfield JA, Forbes SJ, Iredale JP, Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci U S A 109, E3186–3195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Murphy FR, Issa R, Zhou X, Ratnarajah S, Nagase H, Arthur MJ, Benyon C, Iredale JP, Inhibition of apoptosis of activated hepatic stellate cells by tissue inhibitor of metalloproteinase-1 is mediated via effects on matrix metalloproteinase inhibition: implications for reversibility of liver fibrosis. J Biol Chem 277, 11069–11076 (2002). [DOI] [PubMed] [Google Scholar]
- 279.Iredale JP, Tissue inhibitors of metalloproteinases in liver fibrosis. Int J Biochem Cell Biol 29, 43–54 (1997). [DOI] [PubMed] [Google Scholar]
- 280.Barry-Hamilton V, Spangler R, Marshall D, McCauley S, Rodriguez HM, Oyasu M, Mikels A, Vaysberg M, Ghermazien H, Wai C, Garcia CA, Velayo AC, Jorgensen B, Biermann D, Tsai D, Green J, Zaffryar-Eilot S, Holzer A, Ogg S, Thai D, Neufeld G, Van Vlasselaer P, Smith V, Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat Med 16, 1009–1017 (2010). [DOI] [PubMed] [Google Scholar]
- 281.Sojoodi M, Erstad DJ, Barrett SC, Salloum S, Zhu S, Qian T, Colon S, Gale EM, Jordan VC, Wang Y, Li S, Ataeinia B, Jalilifiroozinezhad S, Lanuti M, Zukerberg L, Caravan P, Hoshida Y, Chung RT, Bhave G, Lauer GM, Fuchs BC, Tanabe KK, Peroxidasin Deficiency Re-programs Macrophages Toward Pro-fibrolysis Function and Promotes Collagen Resolution in Liver. Cellular and Molecular Gastroenterology and Hepatology 13, 1483–1509 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Soudi M, Zamocky M, Jakopitsch C, Furtmüller PG, Obinger C, Molecular Evolution, Structure, and Function of Peroxidasins. Chemistry & Biodiversity 9, 1776–1793 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Harrison SA, Allen AM, Dubourg J, Noureddin M, Alkhouri N, Challenges and opportunities in NASH drug development. Nat Med 29, 562–573 (2023). [DOI] [PubMed] [Google Scholar]
- 284.Bellucci E, Chiereghin F, Pacifici F, Donadel G, De Stefano A, Malatesta G, Valente MG, Guadagni F, Infante M, Rovella V, Noce A, Tesauro M, Di Daniele N, Della Morte D, Pastore D, Novel therapeutic approaches based on the pathological role of gut dysbiosis on the link between nonalcoholic fatty liver disease and insulin resistance. Eur Rev Med Pharmacol Sci 27, 1921–1944 (2023). [DOI] [PubMed] [Google Scholar]
- 285.Brennan PN, Dillon JF, McCrimmon R, Advances and Emerging Therapies in the Treatment of Non-alcoholic Steatohepatitis. touchREV Endocrinol 18, 148–155 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ratziu V, A critical review of endpoints for non-cirrhotic NASH therapeutic trials. J Hepatol 68, 353–361 (2018). [DOI] [PubMed] [Google Scholar]
- 287.Hagstrom H, Nasr P, Ekstedt M, Hammar U, Stal P, Hultcrantz R, Kechagias S, Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol 67, 1265–1273 (2017). [DOI] [PubMed] [Google Scholar]
- 288.Angulo P, Kleiner DE, Dam-Larsen S, Adams LA, Bjornsson ES, Charatcharoenwitthaya P, Mills PR, Keach JC, Lafferty HD, Stahler A, Haflidadottir S, Bendtsen F, Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 149, 389–397 e310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ratziu V, Charlotte F, Heurtier A, Gombert S, Giral P, Bruckert E, Grimaldi A, Capron F, Poynard T, Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology 128, 1898–1906 (2005). [DOI] [PubMed] [Google Scholar]
- 290.Wang Y, Wong GL, He FP, Sun J, Chan AW, Yang J, Shu SS, Liang X, Tse YK, Fan XT, Hou J, Chan HL, Wong VW, Quantifying and monitoring fibrosis in non-alcoholic fatty liver disease using dual-photon microscopy. Gut 69, 1116–1126 (2020). [DOI] [PubMed] [Google Scholar]
- 291.De Rudder M, Bouzin C, Nachit M, Louvegny H, Vande Velde G, Jule Y, Leclercq IA, Automated computerized image analysis for the user-independent evaluation of disease severity in preclinical models of NAFLD/NASH. Lab Invest 100, 147–160 (2020). [DOI] [PubMed] [Google Scholar]
- 292.Udompap P, Therneau TM, Canning RE, Benson JT, Allen AM, Performance of American Gastroenterological Association Clinical Care Pathway for the risk stratification of patients with nonalcoholic fatty liver disease in the US population. Hepatology 77, 931–941 (2023). [DOI] [PubMed] [Google Scholar]
- 293.Canivet CM, Costentin C, Irvine KM, Delamarre A, Lannes A, Sturm N, Oberti F, Patel PJ, Decaens T, Irles-Depe M, Fouchard I, Hermabessiere P, Roux M, Barthelon J, Cales P, Powell EE, de Ledinghen V, Boursier J, Validation of the new 2021 EASL algorithm for the noninvasive diagnosis of advanced fibrosis in NAFLD. Hepatology 77, 920–930 (2023). [DOI] [PubMed] [Google Scholar]
- 294.Wong VW, Tak WY, Goh GBB, Cheng PN, Lawitz EJ, Younossi ZM, Vuppalanchi R, Younes Z, Alkhouri N, Wang L, Liu J, Kersey K, Myers RP, Harrison SA, Goodman Z, Trauner M, Romero-Gomez M, Anstee QM, Nguyen MH, Okanoue T, Performance of Noninvasive Tests of Fibrosis Among Asians, Hispanic, and non-Hispanic Whites in the STELLAR Trials. Clin Gastroenterol Hepatol 21, 90–102 e106 (2023). [DOI] [PubMed] [Google Scholar]
- 295.Karsdal MA, Daniels SJ, Holm Nielsen S, Bager C, Rasmussen DGK, Loomba R, Surabattula R, Villesen IF, Luo Y, Shevell D, Gudmann NS, Nielsen MJ, George J, Christian R, Leeming DJ, Schuppan D, Collagen biology and non-invasive biomarkers of liver fibrosis. Liver international : official journal of the International Association for the Study of the Liver 40, 736–750 (2020). [DOI] [PubMed] [Google Scholar]
- 296.Luo Y, Wadhawan S, Greenfield A, Decato BE, Oseini AM, Collen R, Shevell DE, Thompson J, Jarai G, Charles ED, Sanyal AJ, SOMAscan Proteomics Identifies Serum Biomarkers Associated With Liver Fibrosis in Patients With NASH. Hepatol Commun 5, 760–773 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Corey KE, Pitts R, Lai M, Loureiro J, Masia R, Osganian SA, Gustafson JL, Hutter MM, Gee DW, Meireles OR, Witkowski ER, Richards SM, Jacob J, Finkel N, Ngo D, Wang TJ, Gerszten RE, Ukomadu C, Jennings LL, ADAMTSL2 protein and a soluble biomarker signature identify at-risk non-alcoholic steatohepatitis and fibrosis in adults with NAFLD. J Hepatol 76, 25–33 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Loomba R, Huang DQ, Sanyal AJ, Anstee QM, Trauner M, Lawitz EJ, Ding D, Ma L, Jia C, Billin A, Huss RS, Chung C, Goodman Z, Wong VW, Okanoue T, Romero-Gomez M, Abdelmalek MF, Muir A, Afdhal N, Bosch J, Harrison S, Younossi ZM, Myers RP, Liver stiffness thresholds to predict disease progression and clinical outcomes in bridging fibrosis and cirrhosis. Gut 72, 581–589 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Gidener T, Dierkhising RA, Mara KC, Therneau TM, Venkatesh SK, Ehman RL, Yin M, Allen AM, Change in serial liver stiffness measurement by magnetic resonance elastography and outcomes in NAFLD. Hepatology 77, 268–274 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Bachtiar V, Kelly MD, Wilman HR, Jacobs J, Newbould R, Kelly CJ, Gyngell ML, Groves KE, McKay A, Herlihy AH, Fernandes CC, Halberstadt M, Maguire M, Jayaratne N, Linden S, Neubauer S, Banerjee R, Repeatability and reproducibility of multiparametric magnetic resonance imaging of the liver. PLoS One 14, e0214921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ning Y, Zhou IY, Roberts JD Jr., Rotile NJ, Akam E, Barrett SC, Sojoodi M, Barr MN, Punshon T, Pantazopoulos P, Drescher HK, Jackson BP, Tanabe KK, Caravan P, Molecular MRI quantification of extracellular aldehyde pairs for early detection of liver fibrogenesis and response to treatment. Sci Transl Med 14, eabq6297 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhou IY, Tanabe KK, Fuchs BC, Caravan P, Collagen-targeted molecular imaging in diffuse liver diseases. Abdom Radiol (NY) 45, 3545–3556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Wieland A, Etzion O, Ali RO, Levy E, Kleiner DE, Helmke SM, Heller T, Everson GT, HepQuant SHUNT Detects Portal Hypertension in Early Stages of Clinically Compensated Chronic Liver Disease. Clin Gastroenterol Hepatol, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Cazanave SC, Warren AD, Pacula M, Touti F, Zagorska A, Gural N, Huang EK, Sherman S, Cheema M, Ibarra S, Bates J, Billin AN, Liles JT, Budas GR, Breckenridge DG, Tiniakos D, Ratziu V, Daly AK, Govaere O, Anstee QM, Gelrud L, Luther J, Chung RT, Corey KE, Winckler W, Bhatia S, Kwong GA, Peptide-based urinary monitoring of fibrotic nonalcoholic steatohepatitis by mass-barcoded activity-based sensors. Sci Transl Med 13, eabe8939 (2021). [DOI] [PubMed] [Google Scholar]
- 305.Trepo E, Valenti L, Update on NAFLD genetics: From new variants to the clinic. J Hepatol 72, 1196–1209 (2020). [DOI] [PubMed] [Google Scholar]
- 306.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, Liu Y, Kozlitina J, Stender S, Wood GC, Stepanchick AN, Still MD, McCarthy S, O'Dushlaine C, Packer JS, Balasubramanian S, Gosalia N, Esopi D, Kim SY, Mukherjee S, Lopez AE, Fuller ED, Penn J, Chu X, Luo JZ, Mirshahi UL, Carey DJ, Still CD, Feldman MD, Small A, Damrauer SM, Rader DJ, Zambrowicz B, Olson W, Murphy AJ, Borecki IB, Shuldiner AR, Reid JG, Overton JD, Yancopoulos GD, Hobbs HH, Cohen JC, Gottesman O, Teslovich TM, Baras A, Mirshahi T, Gromada J, Dewey FE, A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med 378, 1096–1106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Harrison SA, Allen AM, Dubourg J, Noureddin M, Alkhouri N, Challenges and opportunities in NASH drug development. Nat Med, (2023). [DOI] [PubMed] [Google Scholar]
- 308.Sanyal AJ, Shankar SS, Calle RA, Samir AE, Sirlin CB, Sherlock SP, Loomba R, Fowler KJ, Dehn CA, Heymann H, Kamphaus TN, Non-Invasive Biomarkers of Nonalcoholic Steatohepatitis: the FNIH NIMBLE project. Nat Med 28, 430–432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rasmussen DGK, Anstee QM, Torstenson R, Golding B, Patterson SD, Brass C, Thakker P, Harrison S, Billin AN, Schuppan D, Dufour JF, Andersson A, Wigley I, Shumbayawonda E, Dennis A, Schoelch C, Ratziu V, Yunis C, Bossuyt P, Karsdal MA, NAFLD and NASH biomarker qualification in the LITMUS consortium - Lessons learned. J Hepatol 78, 852–865 (2023). [DOI] [PubMed] [Google Scholar]
- 310.Noureddin M, Jones C, Alkhouri N, Gomez EV, Dieterich DT, Rinella ME, Nashnet, Screening for Nonalcoholic Fatty Liver Disease in Persons with Type 2 Diabetes in the United States Is Cost-effective: A Comprehensive Cost-Utility Analysis. Gastroenterology 159, 1985–1987 e1984 (2020). [DOI] [PubMed] [Google Scholar]
- 311.Long MT, Noureddin M, Lim JK, AGA Clinical Practice Update: Diagnosis and Management of Nonalcoholic Fatty Liver Disease in Lean Individuals: Expert Review. Gastroenterology 163, 764–774 e761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Sharpton SR, Schnabl B, Knight R, Loomba R, Current Concepts, Opportunities, and Challenges of Gut Microbiome-Based Personalized Medicine in Nonalcoholic Fatty Liver Disease. Cell Metab 33, 21–32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Lee J, Byun J, Shim G, Oh YK, Fibroblast activation protein activated antifibrotic peptide delivery attenuates fibrosis in mouse models of liver fibrosis. Nature communications 13, 1516 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Yang AT, Kim YO, Yan XZ, Abe H, Aslam M, Park KS, Zhao XY, Jia JD, Klein T, You H, Schuppan D, Fibroblast Activation Protein Activates Macrophages and Promotes Parenchymal Liver Inflammation and Fibrosis. Cell Mol Gastroenterol Hepatol 15, 841–867 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Alkhouri N, Herring R, Kabler H, Kayali Z, Hassanein T, Kohli A, Huss RS, Zhu Y, Billin AN, Damgaard LH, Buchholtz K, Kjaer MS, Balendran C, Myers RP, Loomba R, Noureddin M, Safety and efficacy of combination therapy with semaglutide, cilofexor and firsocostat in patients with non-alcoholic steatohepatitis: A randomised, open-label phase II trial. J Hepatol 77, 607–618 (2022). [DOI] [PubMed] [Google Scholar]
- 316.Trinh V. C.-h., Lee TF, Lemmoine S, Ray KC, Ybanez MD, Tsuchida T, Carter JK, Agudo J, Brown BD, Akat KM, Friedman SL, Lee YA, Hepatic stellate cells maintain liver homeostasis through paracrine neurotrophin-3 signaling. Science signaling, in press (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Vyas K, Patel MM, Insights on drug and gene delivery systems in liver fibrosis. Asian J Pharm Sci 18, 100779 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Lawitz EJ, Shevell DE, Tirucherai GS, Du S, Chen W, Kavita U, Coste A, Poordad F, Karsdal M, Nielsen M, Goodman Z, Charles ED, BMS-986263 in Patients with Advanced Hepatic Fibrosis: 36-Week Results from a Randomized, Placebo-Controlled Phase 2 Trial. Hepatology, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wiering L, Subramanian P, Hammerich L, Hepatic Stellate Cells: Dictating Outcome in Nonalcoholic Fatty Liver Disease. Cell Mol Gastroenterol Hepatol, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tacke F, Puengel T, Loomba R, Friedman SL, An integrated view of anti-inflammatory and antifibrotic targets for the treatment of NASH. Journal of Hepatology, in press (2023). [DOI] [PubMed] [Google Scholar]
- 321.Alsamman S, Christenson SA, Yu A, Ayad NME, Mooring MS, Segal JM, Hu JK, Schaub JR, Ho SS, Rao V, Marlow MM, Turner SM, Sedki M, Pantano L, Ghoshal S, Ferreira DDS, Ma HY, Duwaerts CC, Espanol-Suner R, Wei L, Newcomb B, Mileva I, Canals D, Hannun YA, Chung RT, Mattis AN, Fuchs BC, Tager AM, Yimlamai D, Weaver VM, Mullen AC, Sheppard D, Chen JY, Targeting acid ceramidase inhibits YAP/TAZ signaling to reduce fibrosis in mice. Sci Transl Med 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Du K, Maeso-Diaz R, Oh SH, Wang E, Chen T, Pan C, Xiang K, Dutta RK, Wang XF, Chi JT, Diehl AM, Targeting YAP-mediated hepatic stellate cell death susceptibility and senescence for treatment of liver fibrosis. Hepatology, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Roehlen N, Saviano A, El Saghire H, Crouchet E, Nehme Z, Del Zompo F, Juhling F, Oudot MA, Durand SC, Duong FHT, Cherradi S, Gonzalez Motos V, Almeida N, Ponsolles C, Heydmann L, Ostyn T, Lallement A, Pessaux P, Felli E, Cavalli A, Sgrignani J, Thumann C, Koutsopoulos O, Fuchs BC, Hoshida Y, Hofmann M, Vyberg M, Viuff BM, Galsgaard ED, Elson G, Toso A, Meyer M, Iacone R, Schweighoffer T, Teixeira G, Moll S, De Vito C, Roskams T, Davidson I, Heide D, Heikenwalder M, Zeisel MB, Lupberger J, Mailly L, Schuster C, Baumert TF, A monoclonal antibody targeting nonjunctional claudin-1 inhibits fibrosis in patient-derived models by modulating cell plasticity. Sci Transl Med 14, eabj4221 (2022). [DOI] [PubMed] [Google Scholar]
- 324.Wang S, Friedman SL, Hepatic fibrosis: A convergent response to liver injury that is reversible. J Hepatol 73, 210–211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]