Abstract
Hypericum perforatum (St. John’s Wort) is a medicinal plant from the Hypericaceae family. Here, we sequenced the whole chloroplast genome of H. perforatum and compared the genome variation among five Hypericum species to discover dynamic changes and elucidate the mechanisms that lead to genome rearrangements in the Hypericum chloroplast genomes. The H. perforatum chloroplast genome is 139,725 bp, exhibiting a circular quadripartite structure with two copies of inverted repeats (IRs) separating a large single-copy region and a small single-copy region. The H. perforatum chloroplast genome encodes 106 unique genes, including 73 protein-coding genes, 29 tRNAs, and 4 rRNAs. Hypericum chloroplast genomes exhibit genome rearrangement and significant variations among species. The genome size variation among the five Hypericum species was remarkably associated with the expansion or contraction of IR regions and gene losses. Three genes—trnK-UUU, infA, and rps16—were lost, and three genes—rps7, rpl23, and rpl32—were pseudogenized in Hypericum. All the Hypericum chloroplast genomes lost the two introns in clpP, the intron in rps12, and the second intron in ycf3. Hypericum chloroplast genomes contain many long repeat sequences, suggesting a role in facilitating rearrangements. Most genes, according to molecular evolution assessments, are under purifying selection.
Keywords: St. John’s wort, phylogeny, codon usage, substitution rate, intron loss, rearrangement
1. Introduction
Hypericum perforatum L., commonly known as St. John’s Wort, a perennial Hypericaceae herb, is widely distributed in temperate areas of Eurasia and northwestern Africa natively and is an indigenous species of China. H. perforatum has been extensively utilized in medicine since the first century AD due to its well-known antidepressant, antibacterial, and anti-inflammatory properties [1]. The species has been naturalized worldwide but has become invasive in Oceania and the American continent. With high levels of genotypic plasticity and variable levels of facultative apomixis, H. perforatum has rapidly spread throughout these continents. The species can be divided into four subspecies based on morphology and geographical distribution [2,3].
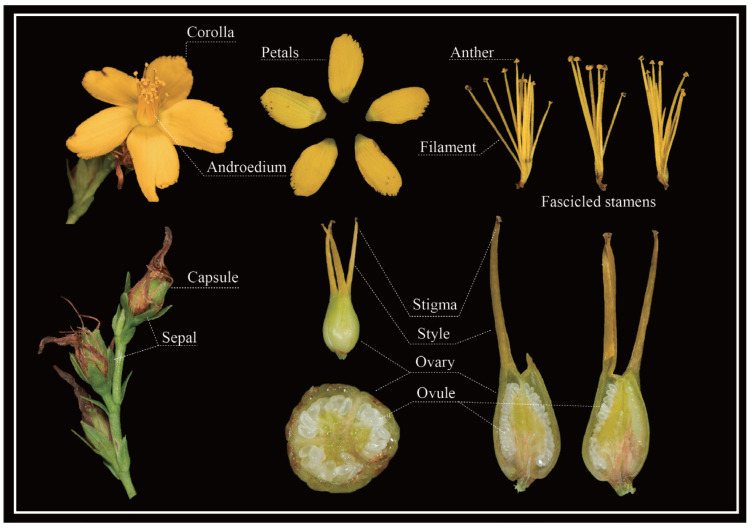
Like other plants in the Hypericum genus, H. perforatum possesses clustered stamens and glands, which are the shared characteristics of the genus. It distinguishes itself from others by its stamens fascicled into three bundles, translucent glands on its stem and leaves, and dark glands on its flowers (Figure 1). H. perforatum displays remarkable variations in its morphology, ploidy, and breeding system, which ranges from sex to apomixis [2,3]. More genetic information is essential to studying this species’ genetics, evolutionary origin, and breeding.
Figure 1.
Floral anatomy of H. perforatum.
Until now, most research on H. perforatum has focused mainly on the properties of its constituent compounds; the genetic background and resources remain scarce. The basic systematical location of this species was revealed by a full Bayesian approach using the internal transcribed spacer and three chloroplast DNA sequence regions [4]. Koch et al. [2] used several chloroplast markers and amplified fragment length polymorphism to present the phylogeographic scenario for the origin of H. perforatum; they also discussed the impact of interspecific gene flow. Using the sequence-related amplified polymorphism (SRAP) markers to access the genetic diversity of H. perforatum in the Qinling Mountains of China, SRAP markers were shown to be closely associated with morphological variability [5]. However, these molecular markers are less variable and this limits the development and utilization of this medicinal plant.
The chloroplast genome has been widely used for reconstructing relationships and species identification at the species level [6,7,8] or as a molecular marker for genetic diversity analyses within a species [9,10,11]. The typical angiosperm chloroplast genome contains a large single-copy region (LSC), a small single-copy region (SSC), and two inverted repeats (IRs) regions. Although highly conserved in their structural organization, chloroplast genomes may exhibit rearrangements, including gene or intron loss. The H. ascyron chloroplast genome, the first completely sequenced one, exhibits dynamic changes in gene and intron content and structure compared to the typical chloroplast genome [12]. However, the mechanisms that lead to rearrangements in Hypericum remain poorly known, indicating a need for further analyses to sequence more Hypericum chloroplast genomes and compare the dynamic changes among species of the genus.
In this study, we sequenced and assembled the whole chloroplast genome of H. perforatum and performed a comparative genome analysis with other Hypericum species. We conducted a comprehensive analysis of H. perforatum, including annotating functional genes, analyzing codon usage, and identifying sequence repeats to examine the pattern of chloroplast genome organization and infer the phylogenetic relationships among Hypericum species.
2. Results
2.1. Features of the H. perforatum Chloroplast Genome
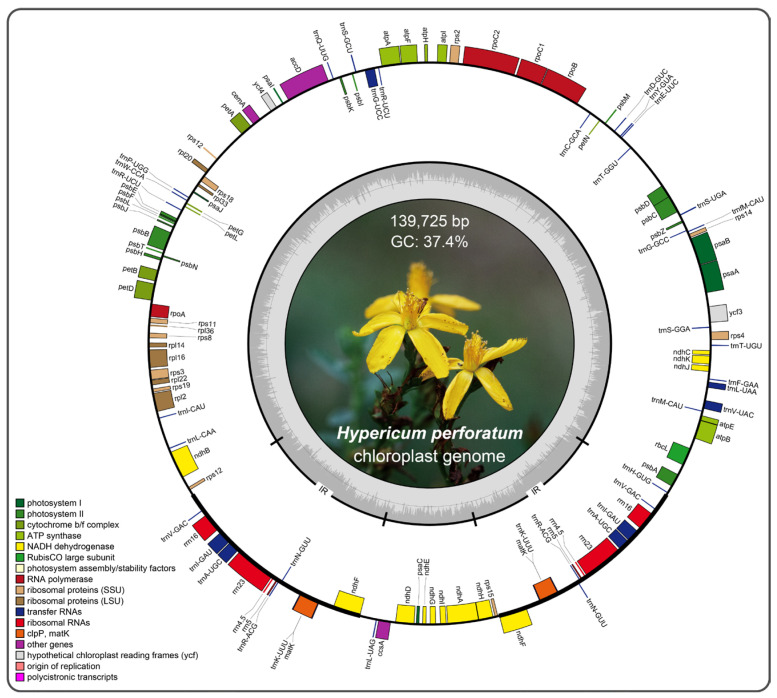
The length of the H. perforatum chloroplast genome was 139,725 bp, exhibiting a circular quadripartite structure with two copies of IRs of 16,766 bp, separating the LSC region (95,109 bp) and SSC region (11,084 bp) (Figure 2, Table 1). We identified 106 unique genes in the H. perforatum chloroplast genome, including 73 protein-coding genes, 29 tRNAs, and four rRNAs (Table S1). Among those genes, 47 were involved in photosynthesis, and 55 were associated with self-replication. Thirteen genes contained an intron; the largest one was found in ndhA (1379 bp). The overall GC content was 37.4%.
Figure 2.
Chloroplast genome map of H. perforatum. Genes inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. Genes within the same functional category are displayed in different colors. The thick lines on the plastid map indicate the inverted repeats (IRs).
Table 1.
Comparison of the chloroplast genome features of Hypericum perforatum and four other Hypericum species from Genbank.
| Species | H. perforatum | H. ascyron | H. hookerianum | H. monogynum | H. uralum |
|---|---|---|---|---|---|
| Genome size (bp) | 139,725 | 162,286 | 138,260 | 138,972 | 136,817 |
| LSC (bp) | 95,109 | 97,542 | 120,848 | 93,123 | 118,902 |
| SSC (bp) | 11,084 | 11,052 | 10,950 | 11,079 | 10,961 |
| IR (bp) | 16,766 | 26,846 | 3231 | 17,385 | 3477 |
| Overall GC content % | 37.4 | 37.4 | 38.1 | 37.8 | 37.9 |
| GC content in LSC % | 35.9 | 36.5 | 38.8 | 36.4 | 38.6 |
| GC content in SSC % | 31.4 | 32.3 | 32.6 | 32.0 | 32.5 |
| GC content in IR % | 43.8 | 40.0 | 34.0 | 43.4 | 34.9 |
| Unique genes | 103 | 107 | 105 | 105 | 105 |
| Unique protein-coding genes | 70 | 74 | 72 | 72 | 72 |
| Unique rRNAs | 4 | 4 | 4 | 4 | 4 |
| Unique tRNAs | 29 | 29 | 29 | 29 | 29 |
| Pseudogenes | 3 | 3 | 3 | 3 | 3 |
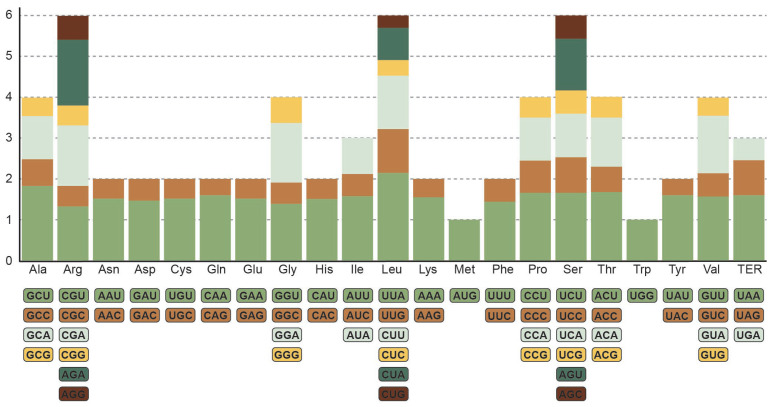
The H. perforatum chloroplast genome encoded 20,096 codons, representing 20 amino acids and a stop codon (Figure 3). The most abundant amino acid was leucine, encoded by six types of codons, with a frequency of 11.13%, and the second most abundant was isoleucine (1710 codons, 8.5% frequency). Cysteine was the least abundant, with a frequency of 1.16%. Additionally, the most frequent codon was the isoleucine-encoding AUU, with 900 occurrences, while the least frequent was the cysteine-encoding UGC, with 56 occurrences. The stop codons were mostly encoded by UAA (39 of 73). Thirty codons had an RSCU value >1, indicating their usage bias in the H. perforatum chloroplast genome, among which 29 ended in A or U (Figure 3). The highest RSCU values were exhibited in leucine-encoding UUA, followed by alanine-encoding GCU and tyrosine-encoding ACU; UGG and AUG exhibited no bias (RSCU = 1).
Figure 3.
RSCU index for each amino acid and stop codon for the H. perforatum chloroplast genome.
2.2. Comparison of Whole Hypericum Chloroplast Genomes
The chloroplast genomes of the studied Hypericum species displayed striking size variations, ranging from 136,817 bp (H. uralum) to 162,286 bp (H. ascyron) (Table 1). The genome size variation was remarkably associated with the expansion or contraction of IR regions, and IR regions ranged from 3231 bp to 26,846 bp. The GC contents displayed significant differences, ranging from 37.1% to 38.1%. Specifically, the GC content in the IR regions was higher than in the LSC and SSC regions, except for the chloroplast genome of H. hookerianum.
Overall, the Hypericum chloroplast genomes encoded 103–107 unique genes, including 70–74 protein-coding genes, 29 tRNAs, and four rRNAs (Table 1). Variations were observed when putative pseudogenization or gene losses occurred in some genomes. The ycf1 and ycf2 genes were lost in H. perforatum, H. hookerianum, H. monogynum, and H. uralum. The rps7, rpl23, and rpl32 genes were pseudogenized in all Hypericum species. Additionally, all five genomes seemed to have lost trnK-UUU, infA, and rps16. The two introns in clpP, the intron in rps12, and the second intron in ycf3 were also lost in the five studied genomes.
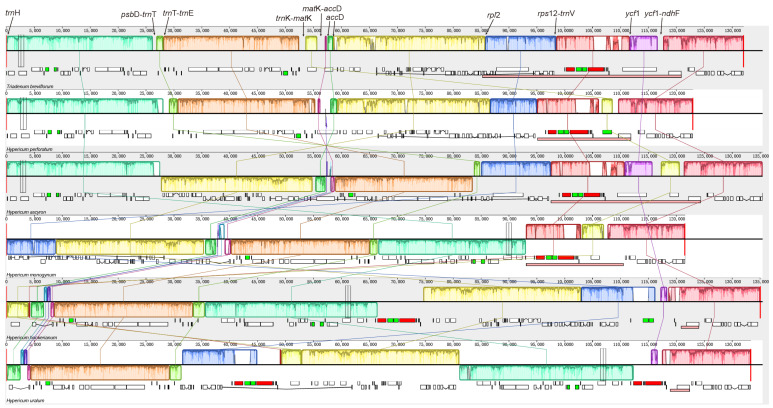
Multiple sequence alignments revealed a highly rearranged structure and large inversions in the five Hypericum chloroplast genomes (Figure 4). Ten synteny blocks were identified among the five chloroplast genomes aligned. H. monogynum and H. hookerianum exhibited a significantly different structure from the other three species. A large inversion (~57 kb) in the LSC region flanking trnE-UUC and rpl2 was found in H. perforatum compared with T. breviflorum. Several small inversions were variably located in the chloroplast genomes, such as the trnT-GGU. The location of matK was very different in the five chloroplast genomes; for example, matK was located in the LSC region of T. breviflorum, H. hookerianum, and H. monogynum, while it was transferred from the LSC region into the IR region in H. perforatum, H. ascyron, and H. uralum.
Figure 4.
Structural alignments of five Hypericum chloroplast genomes using Mauve. The colored blocks represent colinear sequence blocks shared by all chloroplast genomes. Connecting lines indicate the covariate association between blocks. Pink boxes below each plastome block indicate its inverted repeat (IR).
2.3. Analysis of Long Sequence Repeats and SSRs
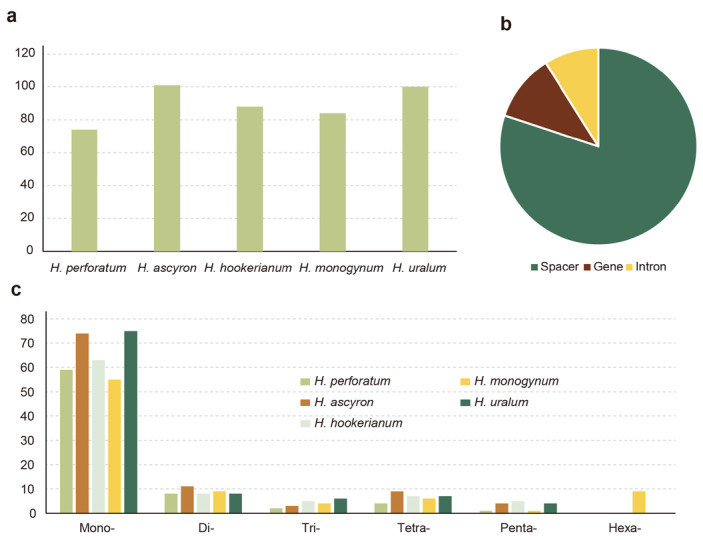
The characteristics of SSRs in the five Hypericum chloroplast genomes were analyzed, and the pattern of SSR distributions is presented in Figure 5 and Table S2. H. perforatum had the lowest number of SSRs (74), while H. ascyron had the highest number (101) among the five species. The most abundant SSRs were A or T nucleotide repeats, which accounted for 70.9% of the total (317 of 447). The most abundant type of dinucleotide SSR was AT. The occurrences of mono-, di-, tri-, tetra-, penta-, and hexanucleotides were 72.9%, 9.8%, 4.5%, 7.4%, 3.4%, and 2.0% of the total, respectively. SSRs were mainly distributed in the intergenic spacer region (359 occurrences, 80%), followed by the coding region (49 occurrences, 11%) and introns (40 occurrences, 9%).
Figure 5.
Number and distribution of SSRs. (a) Total number of SSRs in each chloroplast genome. (b) Distribution of the SSRs in the chloroplast genome. (c) Six types of SSR with their length across the chloroplast genome.
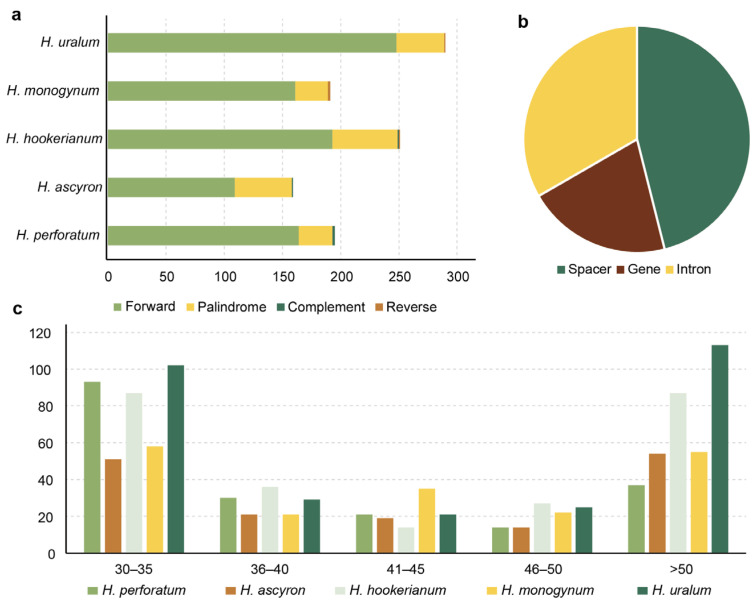
The REPuter data were screened and provided information about the four repeat sequences in the five studied chloroplast genomes (Figure 6 and Table S3). In total, 1086 pairs of long repeats were identified in the five genomes, including 875 forward, 203 palindromic, four reverse, and four complementary repeats (Figure 6a). H. uralum had the highest numbers, with 248 forward, 41 palindromic, one reverse, and no complementary repeats. H. ascyron displayed the lowest numbers with 109 forward, 49 palindromic, one reverse, and no complementary repeats. Most long repeats were distributed in the non-coding regions and a few in coding regions, such as accD, clpP, and rps18 (Figure 6b and Table S3). Overall, long repeats varied from 30 bp to 10,138 bp, but most were 30–35 bp and >50 bp (Figure 6c). The longest repeat occurred in the rps7-trnV and trnH-trnV genes of H. hookerianum.
Figure 6.
Dispersed repeats in five Hypericum chloroplast genomes. (a) Types and numbers of repeats. (b) Distribution of the repeats. (c) Repeat lengths in the five Hypericum chloroplast genomes.
2.4. Molecular Evolution of the Hypericum Chloroplast Genome
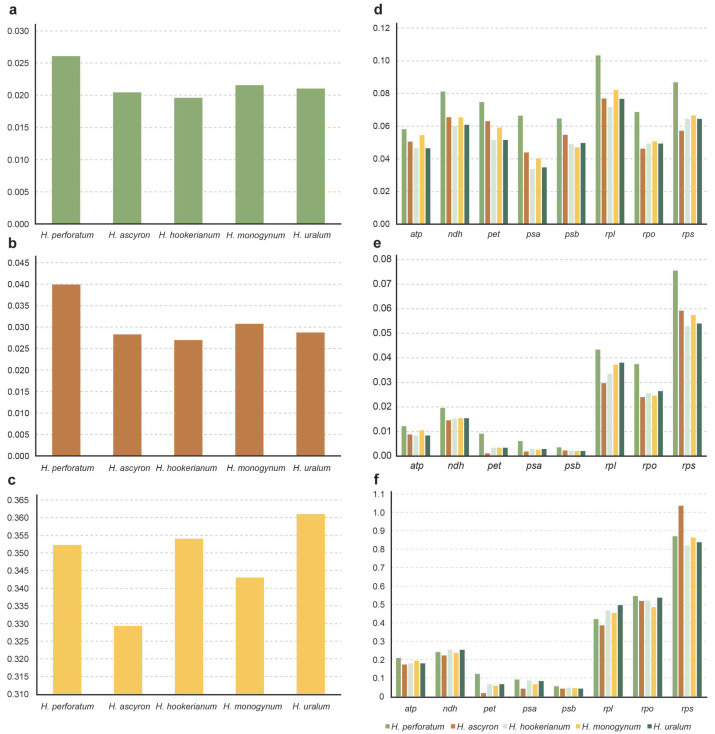
To decipher the situation of evolutionary rates among Hypericum species, we calculated the dN, dS, and ω values for 63 single protein genes, gene groups, and combinations of all 63 protein-coding genes for each Hypericum species, using T. breviflorum as a reference (Figure 7 and Table S4). The t-test for dN, dS, and ω values indicated significant differences for each of the considered Hypericum species, signifying variable molecular evolution rates among genes.
Figure 7.
Evolutionary values of dN, dS, and ω in five Hypericum species. (a–c) dS, dN, and ω values of the five species. (d–f) dS, dN, and ω of different gene groups in the five species.
The mean dN value was 0.022, and the highest dN value was associated with rps18 (0.127). Five genes (petG, psbI, psbL, psbM, and psbT) had non-synonymous substitution sites and displayed a dN = 0. The mean dS value was 0.065, and the highest dS value was associated with rpl22 (0.154), followed by ccsA (0.129) and ndhH (0.128). The psbF gene had no synonymous substitution sites and displayed a dS = 0. Most ω values were <1, indicating that purifying selection acted primarily on most genes. Positive selection occurred in nine genes (cemA, rpl14, rpl2, rpl33, rps14, rps18, rps19, rps2, and rps3), where at least one species pair had a ω > 1. The cemA gene had the highest average ω value (2.542).
Among the different gene groups, the rps group had the highest dN value (0.068), followed by rpl (0.039) and rpo (0.028), and the photosynthetic genes (psa and psb) had the lowest dN values. The rpl gene exhibited the highest value (0.082), and psa had the lowest (0.044). The ω values of gene groups were <0.5, except rpo and rps groups (Figure 7). The ω value was >1 in the rps group in H. ascyron, indicating the presence of negative selection in this group.
At the species level, H. perforatum displayed the highest dN value (0.026), indicating a higher evolution rate for this species, while H. hookerianum had the lowest dN value (0.019). The dS values displayed no significant difference among the five species. All ω values were <0.5 (between 0.329–0.361), indicating that the frequency of non-synonymous substitutions was much lower than that of synonymous substitutions and were under purifying selection.
2.5. Phylogenetic Inferences in Clusioids Clade of Malpighiales
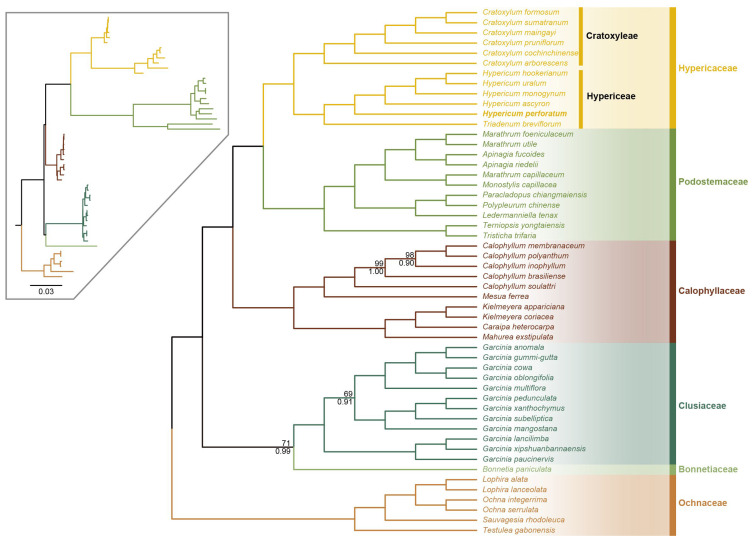
To infer the phylogenetic relationships among Hypericum species and other genera in clusioids clade of Malpighiales, ML and BI trees were constructed based on 63 coding genes (Figure 8). The topological structures produced by both methods had consistent phylogenies and were highly resolved. The Hypericaceae and Podostemaceae families formed a clade and were related to the Calophyllaceae family with strong support (BS = 100% and PP = 1.00). The Hypericaceae family was split into two clades—Cratoxyleae and Hypericeae—with strong support (BS = 100 and PP = 1.0). The Cratoxyleae tribe included six Cratoxylum species, and C. arborescens was the first diverged species with high supported values. The Hypericeae tribe included the Hypericum and Triadenum genera. The five Hypericum species formed a clade, and H. perforatum was the first diverged species.
Figure 8.
Phylogenetic reconstruction of the Hypericaceae family based on the 63 protein-coding genes. ML bootstrap support value and Bayesian posterior probability are presented at each node. The cladogram on the upper left corner displays the branch length of the phylogenetic tree. Species from the same family are showing in the same color.
3. Discussion
3.1. Structural Variations in Hypericum Chloroplast Genomes
The study of chloroplast structure, content, and arrangement has increased significantly over the past few decades, particularly with the advent of high-throughput sequencing [6,8,13], which has completely changed many facets of evolutionary biology. For instance, despite numerous findings supporting the overall conserved structure of the chloroplast genome in land plants, a growing body of evidence points to variations among different plant lineages, casting doubt on the idea that plant lineages share an overall conserved structure. Several plant groups, such as Cyperaceae [14], Opuntieae [15], Lobeliaceae [16], and Medicago [17], exhibited a high number of structural rearrangements. In this study, we sequenced the chloroplast genome of H. perforatum and analyzed the evolution history of the Hypericum genus, revealing structural variations (Figure 4). The Hypericum chloroplast genome displayed dynamic changes, including inversions, rearrangement, and IR boundary shifts (Figure 4).
Several mechanisms can explain the rise and maintenance of structural variations in the chloroplast genomes. One cause includes inversions and deletions that can result from intra- or intermolecular homologous recombination, leading to structural rearrangements of the chloroplast genome [18,19].
Repeats, especially long ones, which occur very frequently at rearrangement endpoints, were highly linked with rearrangements in the chloroplast genome. Such associations are found in several plant lineages, such as Jasminum, Menodora [20], Trifolium [21], and Carex [22]. We identified many long repeats in the Hypericum chloroplast genome (Figure 6), suggesting that the repeats might facilitate rearrangements [12].
The integrity of chloroplastic DNA is thought to deteriorate during plant growth due to the degradation of broken but unrepaired molecules, making chloroplast DNA recombination, replication, and repair processes essential for its maintenance [23]. Due to their harsh environment, cacti are exposed to many factors that can cause DNA damage [15,24]. Like cacti, Hypericum species usually occupy harsh environments, such as rocky terrain, calcareous areas, dry-to-moist grasslands, acidic fens, or shallow swamps [25], which could explain the significant variation observed in the Hypericum chloroplast genomes.
About 20% of angiosperm species have biparental chloroplast inheritance [26,27], and the chloroplast genome rearrangement events are related to this inheritance pattern [7]. Hypericum is also a group with biparental chloroplast inheritance [26]. This biparental inheritance might influence the Hypericum chloroplast genome rearrangements.
3.2. Genome Size Variations and Gene Losses
Typical angiosperm chloroplast genomes range from 130 kb to 170 kb; in related species, the genome size is very similar. However, the chloroplast genome size displayed striking variations among the five Hypericum species. The genome size of T. breviflorum and Cratoxylum cochinchinense, the sister groups of Hypericum, were 167,693 bp and 157,103 bp, respectively [28,29]. These results indicate that the smaller genome size of Hypericum was contracted during the evolutionary process, according to the phylogenetic relationships of Hypericaceae. IR boundary shifts and gene losses affected the chloroplast genome size. The IR regions of H. hookerianum and H. uralum were only 3231 bp and 3477 bp, respectively, which could lead to a smaller chloroplast genome. The ycf2 and ycf1 genes were the first and second longest genes in the chloroplast genome, and the total length of the two genes amounted to about 12 kb. The two genes were lost in the H. perforatum chloroplast genome, which may explain the smaller genome size of this species. In addition, the chloroplast genome size of H. monogynum was similar to that of H. perforatum. However, the IR length of the H. monogynum chloroplast genome was 11 kb, suggesting that the genome size variations in Hypericum involve multiple evolutionary mechanisms.
Two coding genes, infA and rps16, and the tRNA gene trnK-UUU have been lost during the evolution of the Hypericum chloroplast genome. In angiosperms, the trnK-UUU intron includes the matK gene and has been found in Cuscuta [30], Isoetes [31], and Helicanthes [32]. The matK gene was retained as a free-standing gene and was transferred to the IR region in the Hypericum chloroplast genome. The loss of trnK-UUU might be linked to this mutational event. The phylogenetic results showed that the infA gene was lost in Malpighiales [12]. Numerous losses of rps16 were recorded in seed plants [33,34]. Furthermore, three genes (rps7, rpl23, and rpl32) appeared to be pseudogenes due to frameshift indels or internal stop codons in all five Hypericum chloroplast genomes. The rps7, rps16, rpl23, and rpl32 genes encode ribosomal proteins, and ribosomes are necessary for the chloroplast translational apparatus [35]. Many studies have demonstrated that gene losses or pseudogenes in the chloroplast genome are often associated with functional gene transfer to the nuclear genome, leading to substitution by a nuclear-encoded protein [35,36]. For example, the functional transfer of rpl22 has been observed in Fagaceae and Passiflora [37,38,39]; infA undergoes multiple independent transfers to the nucleus [40], and rps16 was discovered in Populus and Medicago [17,41]. All five genes—infA, rps7, rps16, rpl23, and rpl32—were found to have functional replacements in the nuclear genome of H. ascyron [12]. Further studies are required to evaluate whether these lost genes transfer to the nuclear genome in other Hypericum species.
4. Materials and Methods
4.1. Plant Materials, DNA Extraction, and Sequencing
The material of H. perforatum was collected from Yining, Xinjiang, China, and voucher specimens were deposited in the Museum of Beijing Forestry University (BJFC). Identification was conducted by Zhixiang Zhang. The chloroplast genomes of four Hypericum species of H. ascyron (MZ424306), H. hookerianum (MZ714015), H. monogynum (ON080517) and H. uralum (OP580940) were downloaded from GenBank and added to this study for analysis.
Total DNA was extracted using a modified cetyltrimethylammonium bromide method [42]. The DNA quality was detected by 0.8% agarose gel electrophoresis. The total DNA was fragmented using an ultrasonic homogenizer and used to construct the 350-bp insert library. The library was sequenced using the DNBSEQ-T7 PE150 platform at Novogene Biotechnology Co., Ltd. (Tianjin, China), and approximately 5 GB of data were obtained.
4.2. Chloroplast Genome Assembly and Annotation
A quality control of raw data was performed by Trimmomatic 0.36 [43] using the default parameters. The chloroplast genome of H. perforatum was assembled using the GetOrganelle toolkit [44] with k-mer lengths of 95, and the genome annotation was performed using the Plastid Genome Annotator (PGA) software (https://github.com/quxiaojian/PGA) [45]. Furthermore, Geneious Prime was used to manually check and revise the annotation errors and missing genes. The annotated chloroplast genome was submitted to GenBank with the accession number OR449303, and the genome map was drawn and visualized using OGDRAW [46]. We calculated codon usage and the relative synonymous codon usage (RSCU) using the MEGA-X program [47] to display the codon usage bias of H. perforatum chloroplast genome.
4.3. Analysis of Simple Sequence Repeats and Dispersed Repeats
Dispersed repeats and simple sequence repeats (SSRs) were identified in the five Hypericum chloroplast genomes using the REPuter online program [48] and Perl script MISA [49], respectively. The REPuter program was employed to identify four types of dispersed repeats, including forward, reverse, palindromic, and complement, in the five Hypericum chloroplast genomes. The minimal repeat size was set to 30 bp, and the hamming distance was set to 3 after removing one IR copy from each chloroplast genome. MISA was used to identify the SSRs, with minimal iterations of 10 repeat motifs for mononucleotides, five for dinucleotides, four for trinucleotides, and three for the rest.
4.4. Molecular Evolution in Hypericum Chloroplast Genomes
We estimated the non-synonymous substitution (dN), synonymous substitution (dS), and dN/dS ratio (ω) to address the evolutionary features among the Hypericum chloroplast genomes. This estimation allowed us to elucidate the evolutionary rates and the role of natural selection in molecular evolution. Triadenum breviflorum was used as the reference species for these analyses. All the protein-coding genes were extracted according to the genome annotations and aligned using MAFFT. The dN, dS, and ω values were estimated by YN100 in the PAMLX software [50]. We calculated these values for gene groups with the same function and the species level combined with all coding genes for each species.
4.5. Comparison of Hypericum Chloroplast Genomes
We determined synteny and identified possible rearrangements among the five Hypericum chloroplast genomes using Mauve v2.4.1 [51] in Geneious Prime with default parameters. The chloroplast genome of T. breviflorum was used as a reference. This analysis utilized mauveAligner as the alignment algorithm.
4.6. Phylogenetic Analysis
To pinpoint systematic positions of Hypericaceae and reconstruct phylogenetic relationships among the Hypericaceae species, we selected the representative chloroplast genomes of the clusioids clade in the Malpighiales order and all the published chloroplast genome of Hypericaceae from GenBank. Sixty-three protein-coding genes were extracted according to the chloroplast genome annotation. All the genes were aligned using MAFFT with default parameters, and trimAl [52] was used to trim the unreliable alignment regions.
Maximum likelihood (ML) and Bayesian inference (BI) were used to infer the phylogenetic relationships. The ML tree was run using RAxML v8.1.24 [53], and 1000 bootstrap repeats were set to assess node support. The BI tree was inferred using MrBayes 3.2 [54], with a Markov chain Monte Carlo algorithm for 5,000,000 generations, using four simultaneous chains with a random starting tree and default priors. The first 25% of generations were discarded as burn-in, and the remaining trees were determined from the 50% majority-rule consensus to estimate posterior probabilities.
5. Conclusions
In this study, the chloroplast genome of H. perforatum was sequenced and compared with four other published Hypericum species including H. ascyron, H. hookerianum, H. monogynum, and H. uralum. Our analysis revealed the Hypericum chloroplast genomes exhibit genome rearrangement and significant variations among species. Variation of Hypericum chloroplast genomes is remarkable in genome size related to contractions, expansions of IR regions, and the loss of several genes. Three genes were lost and three genes were pseudogenized in Hypericum. All the Hypericum chloroplast genomes contained a rich number of long repeat sequences, suggesting a role in facilitating genome rearrangements.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms242216130/s1.
Author Contributions
Conceptualization, project administration W.D. and Z.Z.; Data curation, investigation, conceptualization, and resources, X.L., Y.B., Y.W. and Y.C.; Funding acquisition, W.D. and Z.Z.; Writing-review and editing, visualization, and supervision, X.L. and W.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The chloroplast genome of H. monogynum is deposited in the GenBank database under the following accession number: OR449303.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was financially supported by the Survey of Herbaceous Germplasm Resources in Shandong Province (Grant No. Lu Financial [2021]1), Science and Technology Basic Resources Investigation Program of China (Grant No. 2021FY100200), and CACMS Innovation Fund (Grant No.CI2021A03909).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zhou W., Wang Y., Li B., Petijová L., Hu S., Zhang Q., Niu J., Wang D., Wang S., Dong Y., et al. Whole-genome sequence data of Hypericum perforatum and functional characterization of melatonin biosynthesis by N-acetylserotonin O-methyltransferase. J. Pineal Res. 2021;70:e12709. doi: 10.1111/jpi.12709. [DOI] [PubMed] [Google Scholar]
- 2.Koch M.A., Scheriau C., Betzin A., Hohmann N., Sharbel T.F. Evolution of cryptic gene pools in Hypericum perforatum: The influence of reproductive system and gene flow. Ann. Bot. 2013;111:1083–1094. doi: 10.1093/aob/mct065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molins M.P., Corral J.M., Aliyu O.M., Koch M.A., Betzin A., Maron J.L., Sharbel T.F. Biogeographic variation in genetic variability, apomixis expression and ploidy of St. John’s wort (Hypericum perforatum) across its native and introduced range. Ann. Bot. 2014;113:417–427. doi: 10.1093/aob/mct268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nürk N.M., Madriñán S., Carine M.A., Chase M.W., Blattner F.R. Molecular phylogenetics and morphological evolution of St. John’s wort (Hypericum; Hypericaceae) Mol. Phylogenet. Evol. 2013;66:1–16. doi: 10.1016/j.ympev.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 5.He M., Wang Z. Genetic diversity of Hypericum perforatum collected from the Qinling Mountains of China. Biochem. Syst. Ecol. 2013;50:232–239. doi: 10.1016/j.bse.2013.03.051. [DOI] [Google Scholar]
- 6.Li E., Liu K., Deng R., Gao Y., Liu X., Dong W., Zhang Z. Insights into the phylogeny and chloroplast genome evolution of Eriocaulon (Eriocaulaceae) BMC Plant Biol. 2023;23:32. doi: 10.1186/s12870-023-04034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong W., Li E., Liu Y., Xu C., Wang Y., Liu K., Cui X., Sun J., Suo Z., Zhang Z., et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 2022;20:92. doi: 10.1186/s12915-022-01297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong W.-P., Sun J.-H., Liu Y.-L., Xu C., Wang Y.-H., Suo Z.-L., Zhou S.-L., Zhang Z.-X., Wen J. Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae) J. Syst. Evol. 2022;60:1263–1280. doi: 10.1111/jse.12802. [DOI] [Google Scholar]
- 9.Guo C., Liu K., Li E., Chen Y., He J., Li W., Dong W., Suo Z. Maternal Donor and Genetic Variation of Lagerstroemia indica Cultivars. Int. J. Mol. Sci. 2023;24:3606. doi: 10.3390/ijms24043606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J., Wang Y., Qiao P., Zhang L., Li E., Dong W., Zhao Y., Huang L. Pueraria montana Population Structure and Genetic Diversity Based on Chloroplast Genome Data. Plants. 2023;12:2231. doi: 10.3390/plants12122231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Wang J., Garran T.A., Liu H., Lin H., Luo J., Yuan Q., Sun J., Dong W., Guo L. Genetic diversity and population divergence of Leonurus japonicus and its distribution dynamic changes from the last interglacial to the present in China. BMC Plant Biol. 2023;23:276. doi: 10.1186/s12870-023-04284-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claude S.-J., Park S., Park S. Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae) BMC Plant Biol. 2022;22:135. doi: 10.1186/s12870-022-03515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin Q., Li J., Zeng S., Xu Y., Han F., Yu U. The complete plastomes of red fleshed pitaya (Selenicereus monacanthus) and three related Selenicereus species: Insights into gene losses, inverted repeat expansions and phylogenomic implications. Physiol. Mol. Biol. Plants. 2022;28:123–137. doi: 10.1007/s12298-021-01121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C., Ruhlman T.A., Jansen R.K. Rate accelerations in plastid and mitochondrial genomes of Cyperaceae occur in the same clades. Mol. Phylogenet. Evol. 2023;182:107760. doi: 10.1016/j.ympev.2023.107760. [DOI] [PubMed] [Google Scholar]
- 15.Köhler M., Reginato M., Jin J.-J., Majure L.C. More than a spiny morphology: Plastome variation in the prickly pear cacti (Opuntieae) Ann. Bot. 2023:mcad098. doi: 10.1093/aob/mcad098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knox E.B. DNA Sequence Analysis of an Inversion Hot Spot in Lobeliaceae Plastomes. Plants. 2022;11:2863. doi: 10.3390/plants11212863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao Y.X., He X.F., Song R., Wang X.M., Zhang H., Aili R., Chao Y.H., Shen Y.H., Yu L.X., Zhang T.J., et al. Recent structural variations in the Medicago chloroplast genomes and their horizontal transfer into nuclear chromosomes. J. Syst. Evol. 2023;61:627–642. doi: 10.1111/jse.12900. [DOI] [Google Scholar]
- 18.Wang W., Lanfear R. Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants. Genome Biol. Evol. 2019;11:3372–3381. doi: 10.1093/gbe/evz256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang J.-S., Zhang H.-R., Wang Y.-R., Liang S.-Q., Mao Z.-Y., Zhang X.-C., Xiang Q.-P. Distinctive evolutionary pattern of organelle genomes linked to the nuclear genome in Selaginellaceae. Plant J. 2020;104:1657–1672. doi: 10.1111/tpj.15028. [DOI] [PubMed] [Google Scholar]
- 20.Lee H.-L., Jansen R.K., Chumley T.W., Kim K.-J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- 21.Sabir J., Schwarz E., Ellison N., Zhang J., Baeshen N.A., Mutwakil M., Jansen R., Ruhlman T. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 2014;12:743–754. doi: 10.1111/pbi.12179. [DOI] [PubMed] [Google Scholar]
- 22.Xu S., Teng K., Zhang H., Gao K., Wu J., Duan L., Yue Y., Fan X. Chloroplast genomes of four Carex species: Long repetitive sequences trigger dramatic changes in chloroplast genome structure. Front. Plant Sci. 2023;14:1100876. doi: 10.3389/fpls.2023.1100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oldenburg D.J., Bendich A.J. DNA maintenance in plastids and mitochondria of plants. Front. Plant Sci. 2015;6:883. doi: 10.3389/fpls.2015.00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J., Li J., Zuo Y., Qin Q., Zeng S., Rennenberg H., Deng H. Plastome variations reveal the distinct evolutionary scenarios of plastomes in the subfamily Cereoideae (Cactaceae) BMC Plant Biol. 2023;23:132. doi: 10.1186/s12870-023-04148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst E. Hypericum: The Genus Hypericum. CRC Press; Boca Raton, FL, USA: 2003. [Google Scholar]
- 26.Corriveau J.L., Coleman A.W. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988;75:1443–1458. doi: 10.1002/j.1537-2197.1988.tb11219.x. [DOI] [Google Scholar]
- 27.Zhang Q., Liu Y., Sodmergen Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- 28.Huang J., Wang Y., Xu S., He J., Zhang Z. The complete chloroplast genome of Cratoxylum cochinchinense (Hypericaceae) Mitochondrial DNA Part B. 2019;4:3452–3453. doi: 10.1080/23802359.2019.1674216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudmoon R., Kaewdaungdee S., Tanee T., Siripiyasing P., Ameamsri U., Syazwan S.A., Lee S.Y., Chaveerach A. Characterization of the plastid genome of Cratoxylum species (Hypericaceae) and new insights into phylogenetic relationships. Sci. Rep. 2022;12:18810. doi: 10.1038/s41598-022-23639-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeal J.R., Kuehl J.V., Boore J.L., Leebens-Mack J., dePamphilis C.W. Parallel Loss of Plastid Introns and Their Maturase in the Genus Cuscuta. PLoS ONE. 2009;4:e5982. doi: 10.1371/journal.pone.0005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira J.B.S., Giulietti A.M., Pires E.S., Laux M., Watanabe M.T.C., Oliveira R.R.M., Vasconcelos S., Oliveira G. Chloroplast genomes of key species shed light on the evolution of the ancient genus Isoetes. J. Syst. Evol. 2021;59:429–441. doi: 10.1111/jse.12693. [DOI] [Google Scholar]
- 32.Darshetkar A.M., Pable A.A., Nadaf A.B., Barvkar V.T. Understanding parasitism in Loranthaceae: Insights from plastome and mitogenome of Helicanthes elastica. Gene. 2023;861:147238. doi: 10.1016/j.gene.2023.147238. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz E.N., Ruhlman T.A., Sabir J.S.M., Hajrah N.H., Alharbi N.S., Al-Malki A.L., Bailey C.D., Jansen R.K. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 2015;53:458–468. doi: 10.1111/jse.12179. [DOI] [Google Scholar]
- 34.Johansson J.T. There large inversions in the chloroplast genomes and one loss of the chloroplast generps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae) Plant Syst. Evol. 1999;218:133–143. doi: 10.1007/BF01087041. [DOI] [Google Scholar]
- 35.Wicke S., Schneeweiss G.M., Depamphilis C.W., Muller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevill P.G., Howell K.A., Cross A.T., Williams A.V., Zhong X., Tonti-Filippini J., Boykin L.M., Dixon K.W., Small I. Plastome-wide rearrangements and gene losses in Carnivorous Droseraceae. Genome Biol. Evol. 2019;11:472–485. doi: 10.1093/gbe/evz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabah S.O., Shrestha B., Hajrah N.H., Sabir M.J., Alharby H.F., Sabir M.J., Alhebshi A.M., Sabir J.S.M., Gilbert L.E., Ruhlman T.A., et al. Passiflora plastome sequencing reveals widespread genomic rearrangements. J. Syst. Evol. 2019;57:1–14. doi: 10.1111/jse.12425. [DOI] [Google Scholar]
- 38.Cauz-Santos L.A., da Costa Z.P., Callot C., Cauet S., Zucchi M.I., Bergès H., van den Berg C., Vieira M.L.C. A Repertory of Rearrangements and the Loss of an Inverted Repeat Region in Passiflora Chloroplast Genomes. Genome Biol. Evol. 2020;12:1841–1857. doi: 10.1093/gbe/evaa155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jansen R.K., Saski C., Lee S.B., Hansen A.K., Daniell H. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011;28:835–847. doi: 10.1093/molbev/msq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millen R.S., Olmstead R.G., Adams K.L., Palmer J.D., Lao N.T., Heggie L., Kavanagh T.A., Hibberd J.M., Gray J.C., Morden C.W., et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueda M., Nishikawa T., Fujimoto M., Takanashi H., Arimura S., Tsutsumi N., Kadowaki K. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25:1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- 42.Li J., Wang S., Jing Y., Wang L., Zhou S. A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 2013;48:72–78. [Google Scholar]
- 43.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin J.-J., Yu W.-B., Yang J.-B., Song Y., dePamphilis C.W., Yi T.-S., Li D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu X.-J., Moore M.J., Li D.-Z., Yi T.-S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:50. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beier S., Thiel T., Munch T., Scholz U., Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu B., Yang Z. PAMLX: A graphical user interface for PAML. Mol. Biol. Evol. 2013;30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- 51.Darling A.E., Mau B., Perna N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandros S. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chloroplast genome of H. monogynum is deposited in the GenBank database under the following accession number: OR449303.