Abstract
The basic/helix–loop–helix (bHLH) family is a major family of transcription factors in plants. Although it has been reported that bHLH plays a defensive role against pathogen infection in plants, there is no comprehensive study on the bHLH-related defence response in rose (Rosa sp.). In this study, a genome-wide analysis of bHLH family genes (RcbHLHs) in rose was carried out, including their phylogenetic relationships, gene structure, chromosome localization and collinearity analysis. Via phylogenetic analysis, a total of 121 RcbHLH genes in the rose genome were divided into 21 sub-groups. These RcbHLHs are unevenly distributed in all 7 chromosomes of rose. The occurrence of gene duplication events indicates that whole-genome duplication and segmental duplication may play a key role in gene duplication. Ratios of non-synonymous to synonymous mutation frequency (Ka/Ks) analysis showed that the replicated RcbHLH genes mainly underwent purification selection, and their functional differentiation was limited. Gene expression analysis showed that 46 RcbHLHs were differentially expressed in rose petals upon B. cinerea infection. It is speculated that these RcbHLHs are candidate genes that regulate the response of rose plants to B. cinerea infection. Virus-induced gene silencing (VIGS) confirmed that RcbHLH112 in rose is a susceptibility factor for infection with B. cinerea. This study provides useful information for further study of the functions of the rose bHLH gene family.
Keywords: bHLH, transcription factor, Botrytis cinerea, phylogenetic analysis, expression pattern
1. Introduction
Transcription factors have been extensively studied in plant growth, development, metabolism and stress response due to their important roles in transcriptional regulation [1]. Transcription factors usually consist of at least DNA-binding domains, transcriptional regulatory domains, oligomerisation sites and nuclear localisation signals [2]. The bHLH gene family is one of the most important transcription factor families in plants. Since the discovery of basic/helix–loop–helix (bHLH) motifs [3] with the ability to bind DNA, members of the bHLH protein superfamily have been found to have more and more functions in the basic physiology and development of animals and plants [4,5,6,7,8]. The bHLH domain consists of about 60 amino acids and has two regions with different functions, i.e., the basic domain and the HLH domain. The basic domain is located at the N-terminus of the bHLH domain and acts as a DNA-binding motif. It consists of about 15 amino acids, usually including 6 basic residues. The HLH region contains two amphiphilic alpha helices and a variable-length linker. Two amphiphilic alpha helices of bHLH proteins can interact to form homodimers or heterodimers [9,10]. Some bHLH proteins have been shown to bind to sequences containing a common core element called the E-box (5′-CANNTG-3′). In addition, nucleotides flanking the core elements may also play a role in binding specificity [11].
bHLH transcription factors are involved in the regulation of various plant processes, including growth, development and response to biotic and abiotic stresses. The function of bHLHs in disease resistance has been characterized in Arabidopsis and many other crops. For example, the wheat bHLH transcription factor gene TabHLH060 increases the susceptibility of transgenic Arabidopsis to Pseudomonas aeruginosa [12]. In tomato, SlybHLH131 increases resistance to yellow leaf curl virus by controlling cell death [13]. Overexpression of jasmonate-responsive OsbHLH034 in rice results in the induction of bacterial blight resistance via an increase in lignin biosynthesis [14]. In addition, bHLHs are also associated with abiotic stress in plants. For example, MdbHLH130 is the drought response bHLH protein in apple that confers drought tolerance in transgenic tobacco [15]. Overexpression of a bHLH gene from Tamarix hispida in Arabidopsis can improve salt and drought tolerance by increasing osmotic potential and reducing the accumulation of reactive oxygen species [16]. In Arabidopsis, bHLH122 is important for drought and osmotic stress resistance and repressing ABA catabolism [17].
Recent research has shown that plant bHLHs can act as a susceptibility gene, negatively regulating plant disease resistance. Zhang and co-authors found that loss of function of the bHLH transcription factor Nrd1 in tomato enhances resistance to Pseudomonas syringae. The mutant plants showed increased immunity due to the suppression of a defence gene, Agp1, by Nrd1. This enhanced immunity is independent of the activation of other immunity-associated genes, indicating that Nrd1 plays a specific role in regulating Agp1 expression and susceptibility to Pseudomonas syringae in tomato [18].
Roses (Rosa sp.) are commercially the most important ornamental plant, generating tens of billions of dollars in value each year [19]. Grey mould disease of roses caused by Botrytis cinerea causes huge losses. There are no reports on the involvement of bHLH transcription factors in rose grey mould resistance. To better understand the involvement of the bHLH genes in rose resistance against B. cinerea, we performed a genome-wide analysis of the bHLH family in rose. We further performed RNA-Seq analysis and showed that a large number of genes encoding bHLH transcription factors were significantly upregulated upon B. cinerea infection, implying that they were involved in the resistance of rose to B. cinerea [20]. Importantly, virus-induced gene silencing (VIGS) further confirmed that RcbHLH112 plays an important role in resistance to B. cinerea as a susceptibility gene.
2. Results
2.1. Identification of RcbHLH Genes in Rose
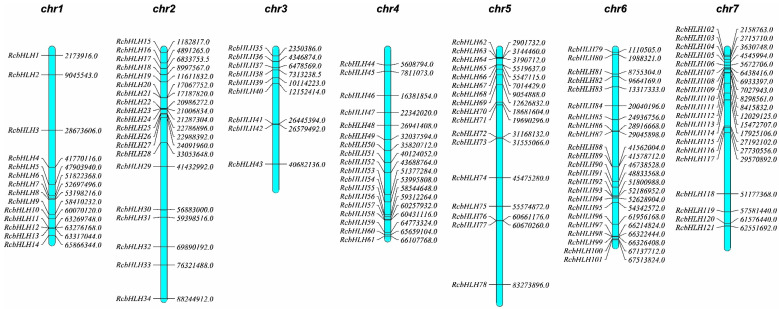
In the process of identifying bHLH family genes in the rose genome, we used the bHLH Hidden Markov Model (HMM) file (PF00010) to perform a Hmmsearch search in the rose genome database, and a total of 136 candidate RcbHLH proteins were obtained. MEME (https://meme-suite.org/meme/) (accessed on 11 July 2022) and Pfam database comparison further confirmed that the extracted protein domain was consistent with the characteristics of the family, and finally 121 RcbHLH gene members were identified in the rose genome, as these 121 protein sequences had a domain profile consistent with a typical bHLH transcription factor. All RcbHLH family genes can be mapped to chromosomes and named RcbHLH1 to RcbHLH121 according to their order on chromosomes (Figure 1).
Figure 1.
Chromosome localization of rose bHLH family members. The physical distribution of each RcbHLH gene is listed on the seven chromosomes of Rose chinensis.
There is a significant difference in the protein size of these RcbHLHs. Among the 121 RcbHLHs, RcbHLH25 has the longest amino acid sequence with 1275 amino acids, while the shortest RcbHLH85 has only 151 amino acids. The average length of RcbHLH protein is 385 aa. The details of all RcbHLH genes are listed in Table 1.
Table 1.
Members of the RcbHLH gene family as predicted in R. chinensis genome sequence.
| Gene | Accession Number 1 | Chr. 2 | Position 3 | Intron | Extron | CDS (bp) | Amino Acids | Clade |
|---|---|---|---|---|---|---|---|---|
| RcbHLH1 | RchiOBHm_Chr1g0314891 | 1 | 2,173,916 | 7 | 8 | 1296 | 432 | Ⅸ |
| RcbHLH2 | RchiOBHm_Chr1g0321521 | 1 | 9,045,543 | 1 | 2 | 708 | 236 | Ⅰb |
| RcbHLH3 | RchiOBHm_Chr1g0337011 | 1 | 28,673,606 | 2 | 3 | 1371 | 457 | Ⅱ |
| RcbHLH4 | RchiOBHm_Chr1g0348781 | 1 | 41,770,117 | 5 | 6 | 1404 | 468 | Ⅶ |
| RcbHLH5 | RchiOBHm_Chr1g0355211 | 1 | 47,903,940 | 7 | 8 | 1944 | 648 | Ⅲ(d+e+f) |
| RcbHLH6 | RchiOBHm_Chr1g0360001 | 1 | 51,822,366 | 1 | 2 | 1851 | 617 | Ⅲ(d+e+f) |
| RcbHLH7 | RchiOBHm_Chr1g0360811 | 1 | 52,697,497 | 0 | 1 | 1464 | 488 | Ⅲ(d+e+f) |
| RcbHLH8 | RchiOBHm_Chr1g0361191 | 1 | 53,198,215 | 3 | 4 | 1254 | 418 | Ⅰa |
| RcbHLH9 | RchiOBHm_Chr1g0368211 | 1 | 58,410,232 | 1 | 2 | 708 | 236 | Ⅰb |
| RcbHLH10 | RchiOBHm_Chr1g0370561 | 1 | 60,070,116 | 1 | 2 | 795 | 265 | Ⅴb |
| RcbHLH11 | RchiOBHm_Chr1g0376001 | 1 | 63,269,742 | 1 | 2 | 918 | 306 | Ⅰb |
| RcbHLH12 | RchiOBHm_Chr1g0376011 | 1 | 63,276,165 | 1 | 2 | 570 | 190 | Ⅰb |
| RcbHLH13 | RchiOBHm_Chr1g0376061 | 1 | 63,317,042 | 1 | 2 | 711 | 237 | Ⅰb |
| RcbHLH14 | RchiOBHm_Chr1g0380101 | 1 | 65,866,341 | 2 | 3 | 1098 | 366 | Ⅰa |
| RcbHLH15 | RchiOBHm_Chr2g0085911 | 2 | 1,182,817 | 6 | 7 | 1263 | 421 | Ⅺ |
| RcbHLH16 | RchiOBHm_Chr2g0091241 | 2 | 4,891,265 | 6 | 7 | 1158 | 386 | Ⅹ |
| RcbHLH17 | RchiOBHm_Chr2g0093571 | 2 | 6,833,754 | 6 | 7 | 1743 | 581 | Ⅳd |
| RcbHLH18 | RchiOBHm_Chr2g0096091 | 2 | 8,997,567 | 6 | 7 | 1293 | 431 | Ⅺ |
| RcbHLH19 | RchiOBHm_Chr2g0099391 | 2 | 11,611,832 | 4 | 5 | 954 | 318 | Ⅳb |
| RcbHLH20 | RchiOBHm_Chr2g0105811 | 2 | 17,067,752 | 0 | 1 | 753 | 251 | Ⅷ(a+b+c) |
| RcbHLH21 | RchiOBHm_Chr2g0105931 | 2 | 17,187,821 | 7 | 8 | 1128 | 376 | Ⅶ |
| RcbHLH22 | RchiOBHm_Chr2g0109611 | 2 | 20,986,271 | 3 | 4 | 1056 | 352 | Ⅳa |
| RcbHLH23 | RchiOBHm_Chr2g0109621 | 2 | 21,006,834 | 4 | 5 | 1062 | 354 | Ⅳa |
| RcbHLH24 | RchiOBHm_Chr2g0109941 | 2 | 21,287,305 | 3 | 4 | 1101 | 367 | Ⅲ(a+b+c) |
| RcbHLH25 | RchiOBHm_Chr2g0111201 | 2 | 22,786,895 | 2 | 3 | 3825 | 1275 | Orphan |
| RcbHLH26 | RchiOBHm_Chr2g0111351 | 2 | 22,988,391 | 1 | 2 | 624 | 208 | Ⅴb |
| RcbHLH27 | RchiOBHm_Chr2g0112221 | 2 | 24,091,959 | 2 | 3 | 996 | 332 | Ⅰa |
| RcbHLH28 | RchiOBHm_Chr2g0120331 | 2 | 33,053,648 | 0 | 1 | 849 | 283 | Ⅷ(a+b+c) |
| RcbHLH29 | RchiOBHm_Chr2g0126861 | 2 | 41,432,994 | 2 | 3 | 678 | 226 | Ⅰb |
| RcbHLH30 | RchiOBHm_Chr2g0139261 | 2 | 56,883,003 | 8 | 9 | 1026 | 342 | Ⅹ |
| RcbHLH31 | RchiOBHm_Chr2g0141851 | 2 | 59,398,516 | 0 | 1 | 1308 | 436 | Ⅷ(a+b+c) |
| RcbHLH32 | RchiOBHm_Chr2g0152511 | 2 | 69,890,195 | 8 | 9 | 1617 | 539 | Ⅶ |
| RcbHLH33 | RchiOBHm_Chr2g0160481 | 2 | 76,321,485 | 0 | 1 | 912 | 304 | Ⅷ(a+b+c) |
| RcbHLH34 | RchiOBHm_Chr2g0176421 | 2 | 88,244,910 | 3 | 4 | 1503 | 501 | Ⅲ(a+b+c) |
| RcbHLH35 | RchiOBHm_Chr3g0451111 | 3 | 2,350,386 | 6 | 7 | 999 | 333 | Ⅺ |
| RcbHLH36 | RchiOBHm_Chr3g0454211 | 3 | 4,346,874 | 2 | 3 | 1020 | 340 | Ⅰa |
| RcbHLH37 | RchiOBHm_Chr3g0457291 | 3 | 6,478,569 | 1 | 2 | 591 | 197 | ⅩⅥ |
| RcbHLH38 | RchiOBHm_Chr3g0458701 | 3 | 7,313,238 | 8 | 9 | 2184 | 728 | Ⅶ |
| RcbHLH39 | RchiOBHm_Chr3g0462431 | 3 | 10,114,223 | 0 | 1 | 768 | 256 | Ⅷ(a+b+c) |
| RcbHLH40 | RchiOBHm_Chr3g0465361 | 3 | 12,152,414 | 9 | 10 | 2079 | 693 | ⅩⅢ |
| RcbHLH41 | RchiOBHm_Chr3g0480621 | 3 | 26,445,394 | 5 | 6 | 1032 | 344 | Ⅸ |
| RcbHLH42 | RchiOBHm_Chr3g0480751 | 3 | 26,579,491 | 6 | 7 | 1077 | 359 | Ⅻ |
| RcbHLH43 | RchiOBHm_Chr3g0493491 | 3 | 40,682,138 | 4 | 5 | 891 | 297 | Ⅷ(a+b+c) |
| RcbHLH44 | RchiOBHm_Chr4g0390311 | 4 | 5,608,794 | 2 | 3 | 792 | 264 | Ⅳd |
| RcbHLH45 | RchiOBHm_Chr4g0392401 | 4 | 7,811,073 | 3 | 4 | 678 | 226 | Ⅳa |
| RcbHLH46 | RchiOBHm_Chr4g0399211 | 4 | 16,381,854 | 4 | 5 | 735 | 245 | Ⅲ(a+b+c) |
| RcbHLH47 | RchiOBHm_Chr4g0403251 | 4 | 22,342,021 | 5 | 6 | 465 | 155 | Ⅻ |
| RcbHLH48 | RchiOBHm_Chr4g0405961 | 4 | 26,941,409 | 5 | 6 | 501 | 167 | Ⅹ |
| RcbHLH49 | RchiOBHm_Chr4g0409001 | 4 | 32,037,594 | 5 | 6 | 1302 | 434 | Ⅸ |
| RcbHLH50 | RchiOBHm_Chr4g0412071 | 4 | 35,820,711 | 7 | 8 | 1662 | 554 | Ⅻ |
| RcbHLH51 | RchiOBHm_Chr4g0415421 | 4 | 40,124,051 | 4 | 5 | 1341 | 447 | Ⅰa |
| RcbHLH52 | RchiOBHm_Chr4g0418301 | 4 | 43,688,763 | 2 | 3 | 609 | 203 | Ⅰa |
| RcbHLH53 | RchiOBHm_Chr4g0425781 | 4 | 51,377,284 | 0 | 1 | 681 | 227 | ⅩⅥ |
| RcbHLH54 | RchiOBHm_Chr4g0429161 | 4 | 53,995,807 | 6 | 7 | 558 | 186 | Ⅻ |
| RcbHLH55 | RchiOBHm_Chr4g0434901 | 4 | 58,544,648 | 5 | 6 | 720 | 240 | Ⅻ |
| RcbHLH56 | RchiOBHm_Chr4g0435901 | 4 | 59,312,260 | 3 | 4 | 1062 | 354 | Ⅴb |
| RcbHLH57 | RchiOBHm_Chr4g0437041 | 4 | 60,257,934 | 8 | 9 | 1647 | 549 | Ⅻ |
| RcbHLH58 | RchiOBHm_Chr4g0437281 | 4 | 60,431,122 | 6 | 7 | 1008 | 336 | Ⅴa |
| RcbHLH59 | RchiOBHm_Chr4g0443741 | 4 | 64,773,328 | 7 | 8 | 1464 | 488 | Ⅲ(a+b+c) |
| RcbHLH60 | RchiOBHm_Chr4g0445091 | 4 | 65,659,106 | 5 | 6 | 825 | 275 | Ⅻ |
| RcbHLH61 | RchiOBHm_Chr4g0445691 | 4 | 66,107,770 | 6 | 7 | 1578 | 526 | Ⅹ |
| RcbHLH62 | RchiOBHm_Chr5g0004471 | 5 | 2,901,732 | 4 | 5 | 747 | 249 | Ⅳa |
| RcbHLH63 | RchiOBHm_Chr5g0004791 | 5 | 3,144,460 | 3 | 4 | 816 | 272 | Ⅲ(a+b+c) |
| RcbHLH64 | RchiOBHm_Chr5g0004831 | 5 | 3,190,712 | 7 | 8 | 1329 | 443 | Ⅹ |
| RcbHLH65 | RchiOBHm_Chr5g0008581 | 5 | 5,519,637 | 3 | 4 | 573 | 191 | Ⅰb |
| RcbHLH66 | RchiOBHm_Chr5g0008601 | 5 | 5,547,115 | 2 | 3 | 495 | 165 | Ⅰb |
| RcbHLH67 | RchiOBHm_Chr5g0010631 | 5 | 7,014,429 | 1 | 2 | 789 | 263 | Ⅴb |
| RcbHLH68 | RchiOBHm_Chr5g0013411 | 5 | 9,054,888 | 0 | 1 | 741 | 247 | ⅩⅣ |
| RcbHLH69 | RchiOBHm_Chr5g0018101 | 5 | 12,626,832 | 1 | 2 | 861 | 287 | Ⅷ(a+b+c) |
| RcbHLH70 | RchiOBHm_Chr5g0024601 | 5 | 18,681,604 | 1 | 2 | 711 | 237 | Ⅷ(a+b+c) |
| RcbHLH71 | RchiOBHm_Chr5g0025741 | 5 | 19,690,297 | 3 | 4 | 633 | 211 | Ⅲ(a+b+c) |
| RcbHLH72 | RchiOBHm_Chr5g0036871 | 5 | 31,168,132 | 6 | 7 | 1356 | 452 | Ⅹ |
| RcbHLH73 | RchiOBHm_Chr5g0037201 | 5 | 31,555,063 | 4 | 5 | 699 | 233 | Ⅳa |
| RcbHLH74 | RchiOBHm_Chr5g0048491 | 5 | 45,475,282 | 9 | 10 | 2886 | 962 | ⅩⅢ |
| RcbHLH75 | RchiOBHm_Chr5g0053301 | 5 | 55,574,872 | 1 | 2 | 792 | 264 | Ⅴb |
| RcbHLH76 | RchiOBHm_Chr5g0056871 | 5 | 60,661,176 | 3 | 4 | 987 | 329 | Ⅲ(a+b+c) |
| RcbHLH77 | RchiOBHm_Chr5g0056881 | 5 | 60,670,256 | 3 | 4 | 1101 | 367 | Ⅲ(a+b+c) |
| RcbHLH78 | RchiOBHm_Chr5g0077341 | 5 | 83,273,897 | 6 | 7 | 1314 | 438 | Ⅸ |
| RcbHLH79 | RchiOBHm_Chr6g0245181 | 6 | 1,110,505 | 5 | 6 | 1020 | 340 | Ⅳb |
| RcbHLH80 | RchiOBHm_Chr6g0246251 | 6 | 1,988,321 | 2 | 3 | 975 | 325 | Ⅳa |
| RcbHLH81 | RchiOBHm_Chr6g0253641 | 6 | 8,755,304 | 4 | 5 | 1017 | 339 | Ⅷ(a+b+c) |
| RcbHLH82 | RchiOBHm_Chr6g0254731 | 6 | 9,664,169 | 4 | 5 | 855 | 285 | Ⅹ |
| RcbHLH83 | RchiOBHm_Chr6g0257881 | 6 | 13,317,333 | 2 | 3 | 1095 | 365 | ⅩⅣ |
| RcbHLH84 | RchiOBHm_Chr6g0264701 | 6 | 20,040,197 | 7 | 8 | 1272 | 424 | Ⅻ |
| RcbHLH85 | RchiOBHm_Chr6g0268091 | 6 | 24,936,757 | 1 | 2 | 453 | 151 | Ⅲ(d+e+f) |
| RcbHLH86 | RchiOBHm_Chr6g0270891 | 6 | 28,916,670 | 2 | 3 | 732 | 244 | Ⅰb |
| RcbHLH87 | RchiOBHm_Chr6g0271001 | 6 | 29,045,898 | 2 | 3 | 2796 | 932 | Orphan |
| RcbHLH88 | RchiOBHm_Chr6g0278441 | 6 | 41,562,004 | 8 | 9 | 1653 | 551 | Ⅲ(a+b+c) |
| RcbHLH89 | RchiOBHm_Chr6g0278471 | 6 | 41,578,713 | 7 | 8 | 1890 | 630 | Ⅲ(a+b+c) |
| RcbHLH90 | RchiOBHm_Chr6g0283511 | 6 | 46,738,527 | 6 | 7 | 1650 | 550 | Ⅻ |
| RcbHLH91 | RchiOBHm_Chr6g0285491 | 6 | 48,833,566 | 7 | 8 | 1533 | 511 | Ⅶ |
| RcbHLH92 | RchiOBHm_Chr6g0288541 | 6 | 51,800,989 | 10 | 11 | 2190 | 730 | ⅩⅢ |
| RcbHLH93 | RchiOBHm_Chr6g0288981 | 6 | 52,186,951 | 1 | 2 | 1434 | 478 | Ⅲ(d+e+f) |
| RcbHLH94 | RchiOBHm_Chr6g0289601 | 6 | 52,628,904 | 5 | 6 | 882 | 294 | Ⅺ |
| RcbHLH95 | RchiOBHm_Chr6g0291161 | 6 | 54,342,571 | 5 | 6 | 2109 | 703 | Ⅲ(d+e+f) |
| RcbHLH96 | RchiOBHm_Chr6g0301601 | 6 | 61,956,169 | 6 | 7 | 1434 | 478 | Ⅹ |
| RcbHLH97 | RchiOBHm_Chr6g0308101 | 6 | 66,214,825 | 0 | 1 | 750 | 250 | Ⅷ(a+b+c) |
| RcbHLH98 | RchiOBHm_Chr6g0308241 | 6 | 66,322,449 | 1 | 2 | 633 | 211 | ⅩⅣ |
| RcbHLH99 | RchiOBHm_Chr6g0308251 | 6 | 66,326,408 | 5 | 6 | 1002 | 334 | Ⅶ |
| RcbHLH100 | RchiOBHm_Chr6g0309431 | 6 | 67,137,708 | 3 | 4 | 1137 | 379 | Ⅷ(a+b+c) |
| RcbHLH101 | RchiOBHm_Chr6g0310101 | 6 | 67,513,823 | 8 | 9 | 1293 | 431 | Ⅻ |
| RcbHLH102 | RchiOBHm_Chr7g0180121 | 7 | 2,158,763 | 5 | 6 | 816 | 272 | Ⅻ |
| RcbHLH103 | RchiOBHm_Chr7g0181001 | 7 | 2,715,710 | 4 | 5 | 750 | 250 | Ⅳb |
| RcbHLH104 | RchiOBHm_Chr7g0182341 | 7 | 3,630,748 | 4 | 5 | 1092 | 364 | Ⅶ |
| RcbHLH105 | RchiOBHm_Chr7g0183781 | 7 | 4,545,994 | 11 | 12 | 1701 | 567 | Ⅴa |
| RcbHLH106 | RchiOBHm_Chr7g0185551 | 7 | 5,672,706 | 5 | 6 | 855 | 285 | Ⅻ |
| RcbHLH107 | RchiOBHm_Chr7g0186541 | 7 | 6,438,416 | 9 | 10 | 2337 | 779 | ⅩⅢ |
| RcbHLH108 | RchiOBHm_Chr7g0187141 | 7 | 6,933,397 | 1 | 2 | 1407 | 469 | Ⅲ(d+e+f) |
| RcbHLH109 | RchiOBHm_Chr7g0187261 | 7 | 7,027,943 | 1 | 2 | 1350 | 450 | Ⅲ(d+e+f) |
| RcbHLH110 | RchiOBHm_Chr7g0188921 | 7 | 8,298,561 | 3 | 4 | 1593 | 531 | Ⅲ(a+b+c) |
| RcbHLH111 | RchiOBHm_Chr7g0189021 | 7 | 8,415,832 | 7 | 8 | 1041 | 347 | Ⅻ |
| RcbHLH112 | RchiOBHm_Chr7g0193761 | 7 | 12,029,125 | 2 | 3 | 573 | 191 | Ⅰb |
| RcbHLH113 | RchiOBHm_Chr7g0197531 | 7 | 15,472,708 | 7 | 8 | 1920 | 640 | Ⅲ(d+e+f) |
| RcbHLH114 | RchiOBHm_Chr7g0199961 | 7 | 17,925,106 | 1 | 2 | 765 | 255 | Ⅰb |
| RcbHLH115 | RchiOBHm_Chr7g0209751 | 7 | 27,192,104 | 0 | 1 | 2082 | 694 | Ⅲ(d+e+f) |
| RcbHLH116 | RchiOBHm_Chr7g0210101 | 7 | 27,730,558 | 2 | 3 | 963 | 321 | Ⅰa |
| RcbHLH117 | RchiOBHm_Chr7g0212241 | 7 | 29,570,891 | 1 | 2 | 1392 | 464 | Ⅲ(d+e+f) |
| RcbHLH118 | RchiOBHm_Chr7g0227911 | 7 | 51,177,367 | 4 | 5 | 672 | 224 | Ⅸ |
| RcbHLH119 | RchiOBHm_Chr7g0233161 | 7 | 57,581,440 | 3 | 4 | 1068 | 356 | Ⅲ(a+b+c) |
| RcbHLH120 | RchiOBHm_Chr7g0236841 | 7 | 61,576,445 | 1 | 2 | 645 | 215 | Ⅴb |
| RcbHLH121 | RchiOBHm_Chr7g0237511 | 7 | 62,551,696 | 1 | 2 | 1083 | 361 | ⅩⅢ |
1 Available at https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/ (accessed on 5 July 2022 ). 2 Chromosome. 3 Starting position (b).
2.2. Chromosomal Locations, Whole-Genome Duplication and Microsynteny
The 121 RcbHLH genes identified are unevenly distributed across 7 rose chromosomes (Figure 1). Chromosome 6 has the most RcbHLH genes with 23. There are 20 RcbHLH genes on chromosomes 2 and 7. Chromosome 3 has the fewest RcbHLH genes, only 9. Meanwhile, 12.37% and 13.22% of RcbHLH genes are located in the upper and middle parts of chromosomes 2 and 7, respectively; 9. 92% of the RcbHLH genes are located in the upper and middle parts of chromosome 5; 9.09% and 10.74% of the genes are distributed in the middle and lower parts of chromosomes 1 and 4, respectively; and 19.01% of the RcbHLH genes are distributed on chromosome 6.
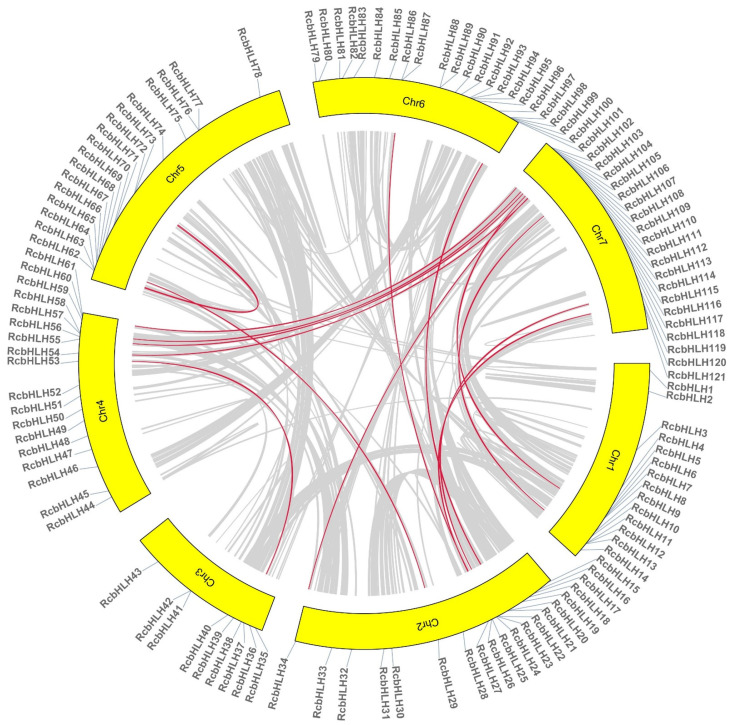
Tandem and segmental duplication play an important role in the expansion of gene families and the generation of new gene functions. On further examination of the repetitive events, we found that there were 16 gene pairs in this family, all of which were whole-genome duplication (WGD) or segmental duplication, while there were gene pairs on different chromosomes, indicating that these genes were paralogous genes. The microsynteny of these RcbHLH genes is shown in Figure 2.
Figure 2.
Microsyntenic analyses of the rose bHLH transcription factors in the Rose chinensis genome. Circular visualization of rose bHLH transcription factors is mapped onto different chromosomes using Circos [21]. The red lines indicate rose bHLH genes with a syntenic relationship. The grey lines represent all syntenic blocks in the genome of Rose chinensis.
To investigate the selective constraints between duplicated RcbHLH genes, the ratios of non-synonymous mutation frequency (Ka) to synonymous mutation frequency (Ks) of 16 gene pairs were calculated (Table 2). In general, Ka/Ks > 1 is consistent with positive selection, whereas Ka/Ks < 1 indicates purifying selection. The Ka/Ks ratio of all 16 repetitive gene pairs is less than 1 (Table 2), indicating limited functionally divergent purifying selection during the evolutionary history of the repetitive RcbHLH genes.
Table 2.
Duplication analysis of the RcbHLH gene family.
| Sequence 1 | Sequence 2 | Ka | Ks | Ka/Ks | Effective Len |
Average S-Sites |
Average N-Sites |
|---|---|---|---|---|---|---|---|
| RcbHLH7 | RcbHLH109 | 0.443081 | NaN | NaN | 1275 | 291.6667 | 983.3333 |
| RcbHLH11 | RcbHLH114 | 0.462303 | NaN | NaN | 597 | 133.8333 | 463.1667 |
| RcbHLH20 | RcbHLH97 | 0.328431 | 2.687639 | 0.1222 | 636 | 140 | 496 |
| RcbHLH21 | RcbHLH99 | 0.48712 | 1.774278 | 0.274545 | 885 | 202.0833 | 682.9167 |
| RcbHLH24 | RcbHLH119 | 0.287838 | 1.620301 | 0.177645 | 1032 | 222.1667 | 809.8333 |
| RcbHLH25 | RcbHLH87 | 0.457511 | 2.840734 | 0.161054 | 2733 | 555.5833 | 2177.417 |
| RcbHLH26 | RcbHLH120 | 0.59898 | 1.798421 | 0.333059 | 555 | 133.8333 | 421.1667 |
| RcbHLH29 | RcbHLH65 | 0.455457 | 4.125944 | 0.110389 | 537 | 122.5 | 414.5 |
| RcbHLH34 | RcbHLH110 | 0.370849 | 2.185043 | 0.169722 | 1443 | 325.9167 | 1117.083 |
| RcbHLH37 | RcbHLH53 | 0.392481 | 1.639823 | 0.239344 | 588 | 153.5833 | 434.4167 |
| RcbHLH54 | RcbHLH111 | 0.438746 | NaN | NaN | 552 | 122.75 | 429.25 |
| RcbHLH55 | RcbHLH106 | 0.435383 | 1.727525 | 0.252027 | 681 | 142.3333 | 538.6667 |
| RcbHLH58 | RcbHLH105 | 0.367022 | 2.17115 | 0.169045 | 981 | 223 | 758 |
| RcbHLH60 | RcbHLH102 | 0.205017 | 1.25652 | 0.163163 | 753 | 174.1667 | 578.8333 |
| RcbHLH62 | RcbHLH73 | 0.230957 | 1.156031 | 0.199784 | 693 | 159.3333 | 533.6667 |
| RcbHLH64 | RcbHLH72 | 0.369998 | 1.720924 | 0.215 | 1137 | 258.75 | 878.25 |
2.3. Phylogenetic and Exon-Intron Structural Analysis of Rose bHLH Genes
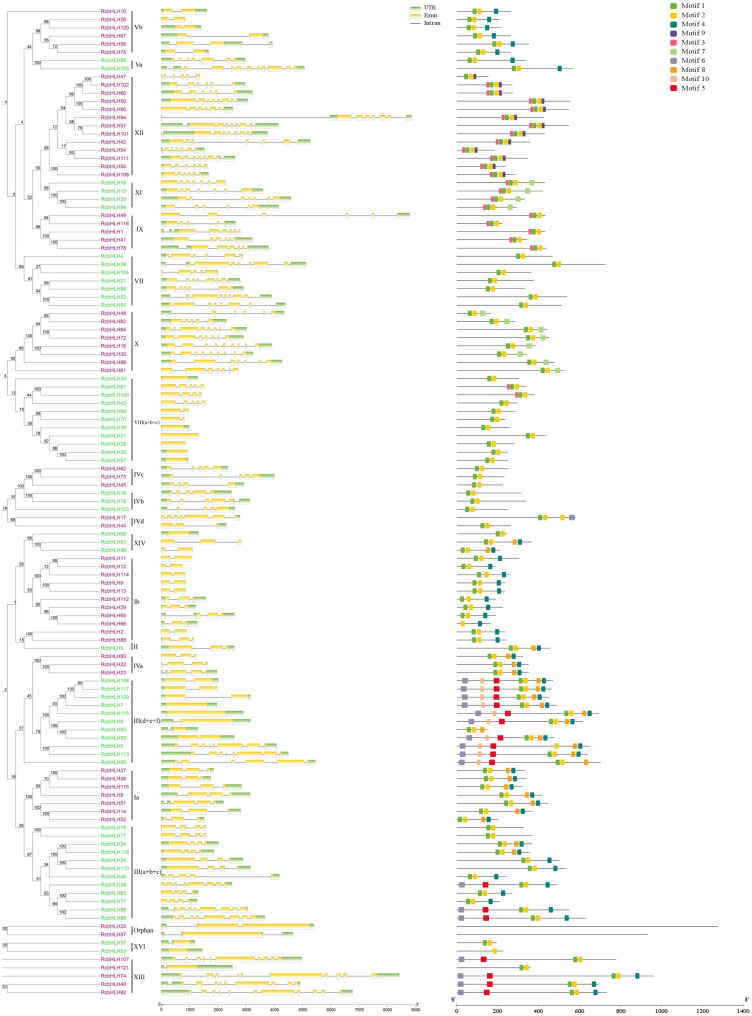
We used the neighbour-joining method (NJ) method to reconstruct the phylogeny of all RcbHLH genes and constructed a phylogenetic tree. The results of the follow-up analysis of the exon–intron structure are consistent with those of the phylogenetic analysis (Figure 3). Most genes clustered in the same group have similar genetic structures, especially in terms of the number of introns, such as RcbHLH10, RcbHLH26 and RcbHLH120. However, there were some exceptions. For example, RcbHLH58 and RcbHLH105 contain different numbers of introns. In addition, their intron length is very variable, ranging from tens to thousands of nucleotides. These results indicate that there is a highly conservative structure in the RcbHLH subfamily and that there is sequence diversity between different RcbHLH groups.
Figure 3.
Phylogenetic analyses, DNA structures and protein motifs of the bHLH gene family in rose. Complete alignments of all rose bHLH proteins were used to construct a phylogenetic tree using the neighbour-joining method. The left represents gene structures. The green boxes, yellow boxes and grey lines in the exon–intron structure diagram represent UTRs, exons and introns, respectively. The right represents protein motifs in the bHLH members. The colourful boxes delineate different motifs (unit: aa). The scale on the bottom is provided as a reference.
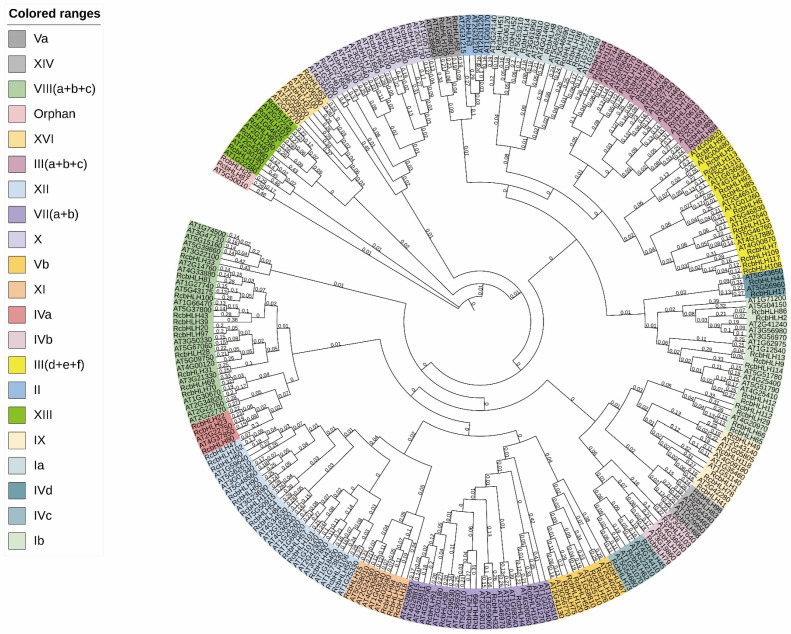
In addition, there is increasing evidence that bHLH transcription factors play a key role in disease resistance in various plant species (Table 3). To assess the evolutionary relationship between RcbHLHs and AtbHLHs genes, we constructed a composite phylogenetic tree (Figure 4). According to The Arabidopsis Information Resources (TAIR) (http://www.arabidopsis.org/) (accessed on 11 July 2022), there are 158 AtbHLH genes in Arabidopsis. These members can be divided into 21 different groups. The results confirmed the previously proposed classification of the bHLH family. The subfamily VIII (a+b+c) contains 31 proteins, and the subfamily IVb contains 3 proteins. The bootstrap values of some branches in the phylogenetic tree are low, which may be due to the short bHLH domain and relatively little information other than highly conserved information.
Table 3.
Plant bHLH family genes involved in disease resistance.
| Gene Name | Gene ID | Species | Pathogens | References |
|---|---|---|---|---|
| SlybHLH131 | Solyc06g051550.2.1 | Solanum lycopersicum | Tomato yellow leaf curl virus | [13] |
| FAMA | AT3G24140 | Arabidopsis thaliana | Botrytis cinerea | [22] |
| AtMYC2 | At1g32640 | Arabidopsis thaliana | Botrytis cinerea | [23] |
| AtbHLH13 | At1g01260 | Arabidopsis thaliana | Botrytis cinerea | [24] |
| OsbHLH6 | Os04g23550 | Oryza sativa | Magnaporthe oryzae | [25] |
| OsbHLH034 | Os02g49480 | Oryza sativa | Xanthomonas oryzae pv. oryzae | [14] |
Figure 4.
Phylogenetic analyses of the rose bHLH transcription factors. Composite phylogenetic tree of rose and Arabidopsis bHLH transcription factors. The bootstrap values are indicated on the nodes of the branches.
2.4. Expression of RcbHLH Genes in Response to B. cinerea Infection
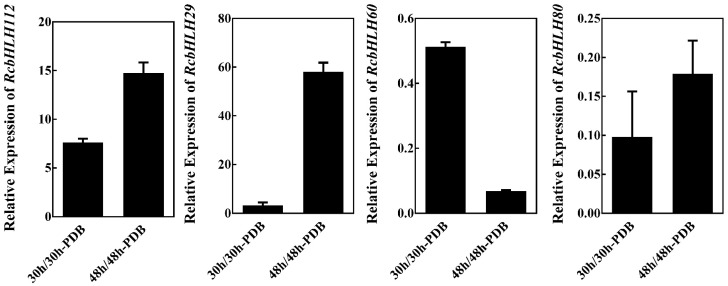
A growing body of evidence from different plant species indicates that plant bHLH transcription factors play an important role in pathogen response. To investigate the role of bHLH genes in B. cinerea resistance in rose, we analysed transcriptome data from rose petals inoculated with the pathogen at 30 and 48 hpi. The 30 hpi time point represents the early response to infection, whereas the 48 hpi time point corresponds to the late response (Table 4). The log2Ratio transformed expression profiles were obtained from the RNA-seq dataset [20]. A total of 21 RcbHLH genes (RcbHLH17, RcbHLH21, RcbHLH29, RcbHLH34, RcbHLH40, RcbHLH44, RcbHLH46, RcbHLH59, RcbHLH62, RcbHLH67, RcbHLH72, RcbHLH75, RcbHLH80, RcbHLH90, RcbHLH99, RcbHLH101, RcbHLH106, RcbHLH108, RcbHLH111, RcbHLH112 and RcbHLH115) were upregulated, suggesting that they may be the key regulators of B. cinerea infection and influence the disease resistance of rose. To further verify the expression profile of RNA-seq, the expression of 4 RcbHLHs was analysed by RT-qPCR. The results of the RT-qPCR analysis were consistent with those of the transcriptome analysis (Figure 5).
Table 4.
Expression patterns of RcbHLH genes under infection of B. cinerea.
| Gene 2 | Accession Number | Group | log2Ratio 30 hpi |
log2Ratio 48 hpi |
|---|---|---|---|---|
| RcbHLH4 | RchiOBHm_Chr1g0348781 | Ⅶ | −1.02302 | −1.36247 |
| RcbHLH8 | RchiOBHm_Chr1g0361191 | Ⅰa | −1.05954 | −1.91491 |
| RcbHLH16 | RchiOBHm_Chr2g0091241 | Ⅹ | 0 | −1.13271 |
| RcbHLH17 | RchiOBHm_Chr2g0093571 | Ⅳd | 3.07535 | 4.92649 |
| RcbHLH21 | RchiOBHm_Chr2g0105931 | Ⅶ | 0 | 1.03517 |
| RcbHLH29 * | RchiOBHm_Chr2g0126861 | Ⅰb | 0 | 6.04668 |
| RcbHLH32 | RchiOBHm_Chr2g0152511 | Ⅶ | 0 | −16.01 |
| RcbHLH34 | RchiOBHm_Chr2g0176421 | Ⅲ(a+b+c) | 1.07031 | 1.74663 |
| RcbHLH37 | RchiOBHm_Chr3g0457291 | ⅩⅥ | −1.36188 | -- |
| RcbHLH39 | RchiOBHm_Chr3g0462431 | Ⅷ(a+b+c) | −1.48345 | -- |
| RcbHLH40 | RchiOBHm_Chr3g0465361 | ⅩⅢ | 1.40229 | 2.20545 |
| RcbHLH42 | RchiOBHm_Chr3g0480751 | Ⅻ | 0 | −1.48859 |
| RcbHLH44 | RchiOBHm_Chr4g0390311 | Ⅳd | 2.8578 | 4.76511 |
| RcbHLH46 | RchiOBHm_Chr4g0399211 | Ⅲ(a+b+c) | 1.25346 | 3.44205 |
| RcbHLH50 | RchiOBHm_Chr4g0412071 | Ⅻ | −1.46896 | −1.77088 |
| RcbHLH53 | RchiOBHm_Chr4g0425781 | ⅩⅥ | 0 | −1.61078 |
| RcbHLH55 | RchiOBHm_Chr4g0434901 | Ⅻ | 0 | −2.09156 |
| RcbHLH57 | RchiOBHm_Chr4g0437041 | Ⅻ | 1.04454 | -- |
| RcbHLH59 | RchiOBHm_Chr4g0443741 | Ⅲ(a+b+c) | 0 | 1.92286 |
| RcbHLH60 * | RchiOBHm_Chr4g0445091 | Ⅻ | −1.22975 | −4.12905 |
| RcbHLH62 | RchiOBHm_Chr5g0004471 | Ⅳa | 0 | 2.03271 |
| RcbHLH67 | RchiOBHm_Chr5g0010631 | Ⅴb | 1.17478 | 1.30658 |
| RcbHLH72 | RchiOBHm_Chr5g0036871 | Ⅹ | 0 | 2.48493 |
| RcbHLH75 | RchiOBHm_Chr5g0053301 | Ⅴb | −2.3709 | −1.67965 |
| RcbHLH78 | RchiOBHm_Chr5g0077341 | Ⅸ | −1.05564 | −1.12357 |
| RcbHLH80 * | RchiOBHm_Chr6g0246251 | Ⅳa | −3.29702 | −2.05486 |
| RcbHLH84 | RchiOBHm_Chr6g0264701 | Ⅻ | 0 | −1.21958 |
| RcbHLH90 | RchiOBHm_Chr6g0283511 | Ⅻ | 1.64315 | 2.09937 |
| RcbHLH91 | RchiOBHm_Chr6g0285491 | Ⅶ | 0 | −1.17746 |
| RcbHLH92 | RchiOBHm_Chr6g0288541 | ⅩⅢ | 0 | −1.37089 |
| RcbHLH94 | RchiOBHm_Chr6g0289601 | Ⅺ | 0 | −1.06842 |
| RcbHLH99 | RchiOBHm_Chr6g0308251 | Ⅶ | 1.31529 | 2.89317 |
| RcbHLH101 | RchiOBHm_Chr6g0310101 | Ⅻ | 0 | 1.23509 |
| RcbHLH102 | RchiOBHm_Chr7g0180121 | Ⅻ | 0 | −1.80483 |
| RcbHLH104 | RchiOBHm_Chr7g0182341 | Ⅶ | 0 | −1.28473 |
| RcbHLH105 | RchiOBHm_Chr7g0183781 | Ⅴa | −1.38441 | -- |
| RcbHLH106 | RchiOBHm_Chr7g0185551 | Ⅻ | 0 | 1.81513 |
| RcbHLH108 | RchiOBHm_Chr7g0187141 | Ⅲ(d+e+f) | 0 | 2.74536 |
| RcbHLH109 | RchiOBHm_Chr7g0187261 | Ⅲ(d+e+f) | −3.56492 | -- |
| RcbHLH110 | RchiOBHm_Chr7g0188921 | Ⅲ(a+b+c) | 0 | −1.77728 |
| RcbHLH111 | RchiOBHm_Chr7g0189021 | Ⅻ | 1.34134 | 2.0827 |
| RcbHLH112 * | RchiOBHm_Chr7g0193761 | Ⅰb | 1.22723 | 4.82187 |
| RcbHLH115 | RchiOBHm_Chr7g0209751 | Ⅲ(d+e+f) | 0 | 1.34433 |
| RcbHLH116 | RchiOBHm_Chr7g0210101 | Ⅰa | 0 | −3.38838 |
| RcbHLH118 | RchiOBHm_Chr7g0227911 | Ⅸ | 0 | −1.0994 |
| RcbHLH121 | RchiOBHm_Chr7g0237511 | ⅩⅢ | 0 | −1.02865 |
The log2 transformed expression profiles were obtained from the RNA-seq dataset [20]. 2 RcbHLHs upregulated are shown in bold. The genes validated by qPCR were marked with asterisks.
Figure 5.
Validation of RNA-Seq results using qRT-PCR. RhUbi was used as an internal control. Expression profile data of four RcbHLH genes at 30 hpi and 48 hpi after B. cinerea inoculation were obtained using qRT-PCR. Values are the means of three replicates ± SD. The primers used are listed in Table 5.
2.5. RcbHLH112 Is a Susceptibility Gene to B. cinerea in Rose
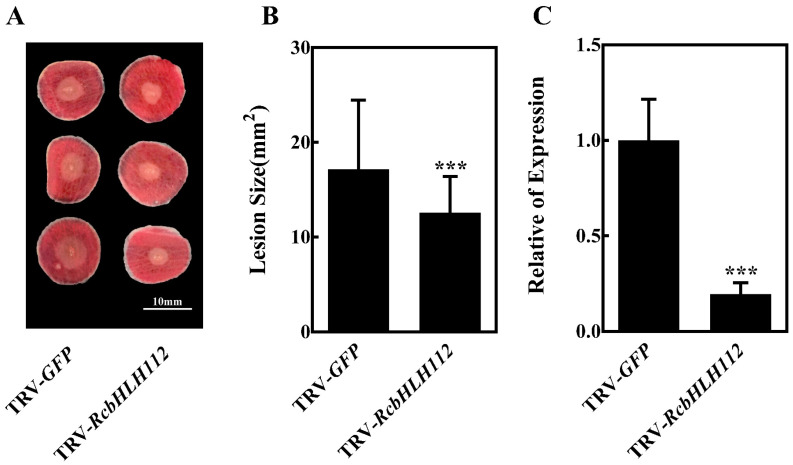
To further investigate the potential role of B. cinerea-induced RcbHLH genes in pathogen resistance, we used VIGS to knock down the expression of RcbHLH112 in rose petals. The reason for selecting RcbHLH112 for this VIGS study was that RcbHLH112 is one of the most upregulated RcbHLHs after B. cinerea infection (Table 4). To silence RcbHLH112 in rose petals, we cloned the 256 bp fragment of RcbHLH112 into the tobacco rattle virus (TRV2) vector to generate TRV-RcbHLH112. Agrobacterium tumefaciens carrying TRV-RcbHLH112 and TRV1 were co-infiltrated into rose petals to produce rose petals with RcbHLH112 silencing. The infiltrated rose petals were then inoculated with B. cinerea. Compared with the control petals (TRV-GFP) inoculated with TRV with a GFP sequence, petals inoculated with TRV-RcbHLH112 showed attenuation of disease symptoms, with a significant reduction in the size of the lesion (Figure 6A,B). In addition, we used RT-qPCR to verify the silencing efficiency of VIGS (Figure 6C). These results show that RcbHLH112 is a susceptibility factor for rose resistance against B. cinerea and that its silencing increases resistance to B. cinerea in rose.
Figure 6.
Functional analysis of rose bHLH transcription factor gene RcbHLH112. (A) Compromised B. cinerea resistance upon silencing of RcbHLH112 at 60 hpi post inoculation. A recombinant tobacco rattle virus (TRV) targeting RcbHLH112 (TRV-RcbHLH112) was used for the gene silencing, and a TRV with GFP sequence (TRV-GFP) was used as the control. (B) Quantification of B. cinerea disease lesions on TRV-RcbHLH112- and TRV-GFP-inoculated rose petal discs. The graph shows the lesion size from three biological replicates (n = 48) with the standard deviation. (C) Expression of RcbHLH112 relative to that in the control at 6 days post silencing, before the infection with B. cinerea (0 hpi). All statistical analyses were performed using Student’s t-test; *** p < 0.001.
3. Discussion
The bHLH genes play important roles in plant growth, development and defence. In this study, we comprehensively analysed the RcbHLH family, including phylogeny, gene structure, chromosome localization, gene duplication events, sequence characteristics and expression profile analysis. We demonstrated that RcbHLH112 is involved in the regulation of resistance to B. cinerea in rose.
It was found that the number of RcbHLH genes in rose (121) was lower than that in Arabidopsis (158), rice (167), potato (124) and maize (208) [26,27,28], indicating that the bHLH gene has expanded to different degrees in different plants. Gene replication plays a very important role in gene family expansion. In this study, 16 replication events were identified in 56 RcbHLHs, all of which involved segmental duplication. The Ka/Ks ratio of the 16 RcbHLH repeats indicates that the RcbHLH gene family is under purifying selection, suggesting a highly conserved evolution. The phylogenetic relationship of bHLH between rose and Arabidopsis showed that most evolutionary branches contained different numbers of AtbHLH and RcbHLH proteins, indicating that the two species showed conservative evolution. These results suggest that the species-specific bHLH gene was lost in rose or gained in the Arabidopsis phylogeny after divergence from the most recent common ancestor.
The role of RcbHLH in B. cinerea resistance is still unclear. In this study, we constructed a phylogenetic tree of known resistance-related bHLHs and found that the bHLHs involved in disease resistance were distributed in groups Ia, Ib, IVb, IVc and Ⅲ(d+e+f). According to the expression in response to B. cinerea infection, we identified 21 RcbHLHs that could be involved in B. cinerea resistance in rose petals. Interestingly, most of the RcbHLH genes induced by B. cinerea are associated with segmental duplication events. The RcbHLH112 belonging to Ib was on the same evolutionary branch as the B. cinerea resistance-related bHLH found in many different species and was significantly induced by B. cinerea at 30 hpi and 48 hpi. Therefore, RcbHLH112 should be considered as an important candidate gene involved in the regulation of disease resistance in rose. The results of VIGS in rose petals showed that silencing of RcbHLH112 improved resistance to B. cinerea, indicating that it is a susceptibility factor of rose in B. cinerea infection process.
4. Materials and Methods
4.1. Identification and Characteristics of the bHLH Genes in Rose Genome
The complete genome data were downloaded from the Rosa chinensis ‘Old Blush’ genome website https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/ (accessed on 5 July 2022) for local alignment and analysis. To identify the non-redundant bHLH genes in the rose genome, first, the common protein sequence of the bHLH Hidden Markov Model (HMM) (PF00010) was downloaded from the Pfam website (http://pfam.xfam.org) (accessed on 11 July 2022). Then, using the HMM profile as a query, the rose genome was searched using the hmmblast function and all sequences were identified as containing bHLH domains with an E-value of <1 × 10−3 in rose. Finally, the protein and DNA sequences of the above rose bHLH members were extracted using the TBTools tool, and all candidate RcbHLHs were verified using the functional structure identified by MEME (https://meme-suite.org/meme/) (accessed on 11 July 2022) and the Pfam database to determine the final family members.
4.2. Mapping bHLH Genes on Rose Chromosomes
The physical locations of 121 genes were extracted from the genomic gff3 annotation file of rose. Mapchart 2.2 software was used to visualise the distribution of bHLH genes on 7 rose chromosomes [29].
4.3. Phylogenetic Analyses and Structure Analysis
A total of 158 Arabidopsis bHLH protein sequences were collected from TAIR (http://www.arabidopsis.org/) (accessed on 11 July 2022). The bHLH protein sequences of Arabidopsis thaliana and rose were compared using ClustalW. The bHLH sequence alignments were used for phylogenetic analysis. The phylogenetic tree was constructed using MEGA6 software, calculating the advance distance via p-distance, estimating the amino acid substitution at each site, performing 1000 bootstrap sampling steps and constructing the phylogenetic tree via the NJ method [30]. The gene structure map and functional structure map of RcbHLH were completed using TBtools [31].
4.4. Collinearity Analyses and Calculation of Ratios of Non-Synonymous (Ka) to Synonymous (Ks) Nucleotide Substitution
We used TBtools to analyse the collinearity of bHLH members and calculate the ratio of Ka/Ks [32].
4.5. Expression of RcbHLHs in Response to B. cinerea
The RNA-Seq data of rose petals infected with B. cinerea can be obtained from the National Center for Biotechnology Information (NCBI) database, accession number PRJNA414570. Clean sequencing reads were mapped to the rose reference genome. Reads per kb per million reads (RPKM) were used to obtain gene expression level. The gene expression level of RcbHLH was calculated as reads per kb per million reads. Differential expression analysis based on Log2 fold change was analysed using DEseq2. To verify the results of RNA-Seq, quantitative PCR (qPCR) was used to analyse the expression of 4 RcbHLH genes. Therefore, total RNA was extracted from rose petals 30 and 48 h after inoculation using the hot borate method [33]. First-strand cDNA was synthesised using HiScript II Q Select RT SuperMix (Vazyme) in 20 μL reaction volume, and 1 μg DNase-treated RNA was used. SYBR Green Master Mix (Takara, Dalian, China) was used for the qPCR reaction, and detection was performed on a StepOnePlus real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). RcUBI2 was used as an internal control. Expression was analysed via the delta–delta–CT method. All primers used for qPCR are listed in Table 5.
Table 5.
List of primers used in this study.
| Gene Name | Accession Number | Primer Sequence (5′-3′) | Amplicon Length | Ta | Tm | Amplification Efficiency |
|---|---|---|---|---|---|---|
| RcbHLH29 | RchiOBHm_Chr2g0126861 | F: GGTTCCACCCTAGAGGTTGTT | 110 bp | 60 °C | 81.69 | 2.005 |
| R: CTGCACGGACTAGGTGAAGT | ||||||
| RcbHLH60 | RchiOBHm_Chr4g0445091 | F: CGATGAGTTTGGACCACCGA | 116 bp | 60 °C | 84.1 | 1.972 |
| R: CCTCAGCTTTGGCCTCAAGA | ||||||
| RcbHLH80 | RchiOBHm_Chr6g0246251 | F: ACACAAACCAAGTGGGGGTT | 102 bp | 60 °C | 85.27 | 1.968 |
| R: GTTCCCTGACTGGCCTTCAA | ||||||
| RcbHLH112 | RchiOBHm_Chr7g0193761 | F: CGATCTTGCAGCCTCCTACA | 120 bp | 60 °C | 82.43 | 2.024 |
| R: CAACCTTGATCCGACCACCA | ||||||
| RcUBI2 | RchiOBHm_Chr1g0359561 | F: GCCCTGGTGCGTTCCCAACTG | 82 bp | 60 °C | 82.43 | 2.024 |
| R: CCTGCGTGTCTGTCCGCATTG |
Ta: amplification temperature; Tm: melting temperature.
4.6. VIGS and B. cinerea Inoculation Assays
To generate the TRV-RcbHLH112 constructor, the 256 bp fragment of RcbHLH112 was amplified using a pair of primers, RcbHLH112-F(5’-GGGGGACAAGTTTGTACAAAAAAGCAGGCTTCTGAGGAAGAAGGAGCCGAAG-3’) and RcbHLH112-R(5’-GGGGGACCACTTTGTACAAGAAAGCTGGGTCCTCAGCTTAGCCTTGTGGAGT-3’).
The VIGS process involved taking individual petals from the outermost whorls of the rose at the second stage of flowering. A 15 mm disc was then cut from the centre of each petal. Agrobacterium tumefaciens cultures containing constructs expressing TRV1 and TRV2 were mixed 1:1 and infiltrated into the petal disc under vacuum [34]. On day 6 after infection, the petal disc was inoculated with B. cinerea. A minimum of 48 petal discs were used for genes, and VIGS was repeated at least three times. After inoculation with B. cinerea, Student’s t-test was performed to determine the significance of lesion size.
5. Conclusions
In this study, a genome-wide analysis of the RcbHLH family genes was performed, including phylogenetic relationship, collinearity and expression analysis. A total of 121 non-redundant bHLH family members were identified. These RcbHLH family genes were classified into 21 groups based on phylogeny and conserved domains. Expression analysis showed that the transcriptional regulation of some RcbHLH family genes was induced by B. cinerea infection in rose petals. Furthermore, plant bHLHs involved in disease resistance tended to cluster on the same branch of the phylogenetic tree. Based on these analyses, we used VIGS to further demonstrate that RcbHLH112 is a susceptibility factor of rose in B. cinerea infection process. The information provided by these results can promote further functional analysis of the RcbHLH gene in rose.
Abbreviations
| hpi | hours post inoculation; |
| bHLH | basic/helix–loop–helix; |
| Ka/Ks | Ratios of non-synonymous to synonymous mutation frequencies; |
| ABA | abscisic acid; |
| VIGS | virus-induced gene silencing; |
| HMM | Hidden Markov Model; |
| WGD | whole-genome duplication; |
| Ka | non-synonymous mutation frequency; |
| Ks | synonymous mutation frequency; |
| NJ | neighbour-joining method; |
| TAIR | The Arabidopsis Information Resources; |
| TRV2 | tobacco rattle virus; |
| NCBI | National Center for Biotechnology Information; |
| RPKM | reads per kb per million reads. |
Author Contributions
C.D., X.H. and Z.Z. conceived and designed the experiments. C.D., J.G., S.Z., D.S. and Z.Z. analysed the data and wrote the paper. X.H., S.Z. and N.J. performed the experiments. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable. Our research did not involve any human or animal subjects, material or data. The plant materials used in this study were provided by the China Agricultural University and are freely available for research purposes, following institutional, national and international guidelines.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analysed during the current study has been included within supplemental data. The plant materials are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding Statement
This study was supported by the Special Project for Science and Technology Cooperation and Exchange of Shanxi Province (Grant No. 202204041101017) to Chao Ding and Zhao Zhang. The funders played no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Dou L., Zhang X., Pang C., Song M., Wei H., Fan S., Yu S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genom. 2014;289:1103–1121. doi: 10.1007/s00438-014-0872-y. [DOI] [PubMed] [Google Scholar]
- 2.Cao Y., Han Y., Li D., Lin Y., Cai Y. MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development. Front. Plant Sci. 2016;7:577. doi: 10.3389/fpls.2016.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murre C., McCaw P.S., Vaessin H., Caudy M., Jan L., Jan Y., Cabrera C.V., Buskin J.N., Hauschka S.D., Lassar A.B., et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 4.Ledent V., Vervoort M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toledo-Ortiz G., Huq E., Quail P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skinner M.K., Rawls A., Wilson-Rawls J., Roalson E.H. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation. 2010;80:1–8. doi: 10.1016/j.diff.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateo J.L., Berg D.L.v.D., Haeussler M., Drechsel D., Gaber Z.B., Castro D.S., Robson P., Lu Q.R., Crawford G.E., Flicek P., et al. Characterization of the neural stem cell gene regulatory network identifies OLIG2 as a multifunctional regulator of self-renewal. Genome Res. 2015;25:41–56. doi: 10.1101/gr.173435.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imayoshi I., Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Ellenberger T., Fass D., Arnaud M., Harrison S.C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 10.Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massari M.E., Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F., Lin R., Feng J., Qiu D., Chen W., Xu S. Wheat bHLH transcription factor gene, TabHLH060, enhances susceptibility of transgenic Arabidopsis thaliana to Pseudomonas syringae. Physiol. Mol. Plant Pathol. 2015;90:123–130. doi: 10.1016/j.pmpp.2015.04.007. [DOI] [Google Scholar]
- 13.Wang J., Hu Z., Zhao T., Yang Y., Chen T., Yang M., Yu W., Zhang B. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum) BMC Genom. 2015;16:39. doi: 10.1186/s12864-015-1249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onohata T., Gomi K. Overexpression of jasmonate-responsive OsbHLH034 in rice results in the induction of bacterial blight resistance via an increase in lignin biosynthesis. Plant Cell Rep. 2020;39:1175–1184. doi: 10.1007/s00299-020-02555-7. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Q., Fan Z., Qiu L., Che Q., Wang T., Li Y., Wang Y. MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Front. Plant Sci. 2020;11:543696. doi: 10.3389/fpls.2020.543696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji X., Nie X., Liu Y., Zheng L., Zhao H., Zhang B., Huo L., Wang Y. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 2016;36:193–207. doi: 10.1093/treephys/tpv139. [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Tai H., Li S., Gao W., Zhao M., Xie C., Li W. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014;201:1192–1204. doi: 10.1111/nph.12607. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N., Hecht C., Sun X., Fei Z., Martin G.B. Loss of function of the bHLH transcription factor Nrd1 in tomato enhances resistance to Pseudomonas syringae. Plant Physiol. 2022;190:1334–1348. doi: 10.1093/plphys/kiac312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liorzou M., Pernet A., Li S., Chastellier A., Thouroude T., Michel G., Malécot V., Gaillard S., Briée C., Foucher F., et al. Nineteenth century French rose (Rosa sp.) germplasm shows a shift over time from a European to an Asian genetic background. J. Exp. Bot. 2016;67:4711–4725. doi: 10.1093/jxb/erw269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Cao X., Shi S., Zhao N., Li D., Fang P., Chen X., Qi W., Zhang Z. Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet. 2018;19:62. doi: 10.1186/s12863-018-0668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Yang R., Chen H. The Arabidopsis thaliana Mediator subunit MED8 regulates plant immunity to Botrytis Cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS ONE. 2018;13:e0193458. doi: 10.1371/journal.pone.0193458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzo O., Chico J.M., Saénchez-Serrano J.J., Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H., Gao H., Liu B., Fan M., Wang J., Wang C., Tian H., Wang L., Xie C., Wu D., et al. bHLH13 Regulates Jasmonate-Mediated Defense Responses and Growth. Evol. Bioinform. 2018;14:1176934318790265. doi: 10.1177/1176934318790265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Q., Lu H., Guo H., Wang Y., Zhao P., Li Y., Wang F., Xu J., Mo X., Mao C. OsbHLH6 interacts with OsSPX4 and regulates the phosphate starvation response in rice. Plant J. 2021;105:649–667. doi: 10.1111/tpj.15061. [DOI] [PubMed] [Google Scholar]
- 26.Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J., et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R., Zhao P., Kong N., Lu R., Pei Y., Huang C., Ma H., Chen Q. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes. 2018;9:54. doi: 10.3390/genes9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T., Lv W., Zhang H., Ma L., Li P., Ge L., Li G. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 2018;18:235. doi: 10.1186/s12870-018-1441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voorrips R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C., Xia R., Chen H., He Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018;1:289660 [Google Scholar]
- 32.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.-H., Jin H., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L., Ma N., Jia Y., Zhang Y., Feng M., Jiang C.-Z., Ma C., Gao J. An Ethylene-Induced Regulatory Module Delays Flower Senescence by Regulating Cytokinin Content. Plant Physiol. 2017;173:853–862. doi: 10.1104/pp.16.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Schiff M., Dinesh-Kumar S.P. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study has been included within supplemental data. The plant materials are available from the corresponding author on reasonable request.