Abstract
The areA gene of Aspergillus oryzae was cloned by cross-hybridization with the Aspergillus nidulans areA gene and was found to encode an 866-amino-acid protein that is very similar to other fungal nitrogen regulatory proteins. The A. oryzae areA gene can complement A. nidulans areA loss-of-function mutations. Functional analyses indicated that the N-terminal region of the A. oryzae AreA protein was dispensable for function and revealed a probable acidic activation domain in the protein. C-terminal truncation of the protein resulted in derepression of several nitrogen-controlled activities in A. nidulans, while deletions extending into the conserved GATA type zinc finger region abolished the activator function. The A. oryzae areA gene was inactivated by replacement with the A. oryzae pyrG gene. Strains containing the resulting areA deletion grew as well as the wild-type strain on glutamine but were unable to grow vigorously on other nitrogen sources, including ammonium. While A. oryzae exhibited reduced growth on 10 mM ammonium, the results of growth tests indicated that areA mutants of both A. oryzae and A. nidulans were affected in utilization of low concentrations of ammonium. The levels of the major nitrogen assimilatory enzymes, NADP-linked glutamate dehydrogenase (EC 1.4.1.4) and glutamine synthetase (EC 6.3.1.2), were determined. In both A. oryzae and A. nidulans areA mutants, the NADP-glutamate dehydrogenase levels were reduced, whereas the glutamine synthetase levels were not affected. These results suggest that the AreA protein may play an important role in the regulation of nitrogen assimilation in addition to its previously established regulatory role in nitrogen catabolism.
Nitrogen metabolite repression is a global regulatory control mechanism that governs the expression of a wide range of nitrogen catabolic activities. In Aspergillus nidulans, the areA gene encodes the major nitrogen regulatory protein which activates transcription of many structural genes encoding enzymes for nitrogen source catabolism under nitrogen-limiting conditions. The availability of favored nitrogen sources, such as ammonium and glutamine, prevents expression of enzymes required for the catabolism of less preferred nitrogen sources. Loss-of-function mutants with mutations in the areA gene have been identified, and these mutants have a pleiotropic inability to use a wide range of potential nitrogen sources other than ammonium (2, 20). Rare gain-of-function alleles have also been isolated; these alleles lead to increased utilization of certain nitrogen sources or derepression of areA-controlled activities (8, 19).
The areA gene of A. nidulans has been cloned (6), and corresponding genes have been isolated from other fungal species; these genes include nit-2 of Neurospora crassa (15) and nreA of Penicillium chrysogenum (17). The areA, nit-2, and nreA genes all encode regulatory proteins that contain a single C-X2-C-X17-C-X2-C DNA binding motif which recognizes and binds to DNA sequences containing a core 5′-GATA-3′ sequence (13, 16, 17, 21, 23, 29). The DNA binding domain and extreme C-terminal residues of these proteins are highly conserved.
In view of the central role of the areA gene in the control of nitrogen catabolism, we sought to establish whether the areA gene of Aspergillus oryzae is structurally and functionally homologous to the A. nidulans areA gene. To investigate the phenotype of a loss-of-function mutant, we disrupted the genomic copy of the areA gene in A. oryzae. Our studies revealed that this mutant exhibits reduced ammonium utilization. A comparison with areA mutants of A. nidulans suggested that the AreA protein of each species may play a role in the regulation of ammonium assimilation.
MATERIALS AND METHODS
Aspergillus strains and media.
A. nidulans MH1 (biA1), MH341 (yA1 areA217 riboB2), and MH5699 (yA1 ΔareA::riboB pyroA4 riboB2) were used in this study. The areA217 mutation in MH341 was isolated in an areA102 background, and both mutations are present in this organism (20, 21). The ΔareA::riboB mutation was created in strain MH50 (yA1 areA102 pyroA4 riboB2) with a plasmid containing the riboB gene from pPL3 (24) inserted between the flanking sequences of the areA gene from pAR2-322-6 (11a). A. nidulans strains are available from the Fungal Genetics Stock Center. A. nidulans gene designations have been described previously (7). A. oryzae wild-type strain IFO4177 (Institute for Fermentation, Osaka, Japan) and A. oryzae HowB101 (ΔpyrG) (kindly provided by Howard Brody, Novo Nordisk, Davis, Calif.) were also used. The pyrG deletion was created in wild-type strain IFO4177 with a plasmid containing the flanking regions of the A. oryzae pyrG gene and selection for resistance to 5-fluoroorotic acid. A. oryzae strains are available from T.C. The Aspergillus media used were the media described by Cove (10). Nitrogen sources were used at a final concentration of 10 mM. The mycelia used for enzyme assays were grown in liquid cultures incubated for 16 h at 37°C.
Isolation of the A. oryzae areA gene.
The A. oryzae areA gene was cloned by low-stringency hybridization (40% formamide, 37°C) to the A. nidulans areA gene by using a partial SauIIIA genomic library of A. oryzae IFO4177 DNA in λGEM11 (Promega, Sydney, New South Wales, Australia). Positive lambda clones were isolated, and fragments were subcloned in pBluescript SK+ (Stratagene, La Jolla, Calif.). The subclones were tested for complementation of A. nidulans areA loss-of-function mutations, and 5,643 bp from the complementing region was sequenced manually from double-stranded templates by the dideoxynucleotide termination procedure (31) by using a Sequenase 2.0 DNA sequencing kit (United States Biochemical Corp.).
Partial cDNA clones covering the entire mRNA were isolated from oligo(dT) and randomly primed libraries and were sequenced to confirm the presence of a single intron in the coding region.
Plasmid constructs.
Deletions at the 5′ end of the A. oryzae areA gene were made by subcloning appropriate fragments from pOARESK9 containing a 7-kb SacI fragment into pBluescriptSK+. pToC257 contains a 3.3-kb EagI-XhoI fragment (areA sequences 3′ of nucleotide 879) cloned into a NotI-XhoI-restricted vector, pToC262 contains a 2.9-kb ApaI-XhoI fragment (areA sequence 3′ of nucleotide 1109) cloned into a SalI-ApaI-restricted vector, and pToC255 contains a 2.6-kb SalI-XhoI fragment (areA sequences 3′ of nucleotide 1435) cloned into a SalI-restricted vector.
Plasmids carrying internal deletions of areA sequences also were constructed. pToC181 (deletion of bp 130 to 723 encoding amino acids 44 to 218) was constructed by cloning the SacI (position −204)-PvuII (position 127) and PvuII (position 721)-SalI (position 1434) fragments of pOARESK9 into SacI-SalI-restricted PUC19. The SacI-SalI fragment carrying the deletion was used to replace the wild-type fragment in pOARESK9. pToC206 (deletion of bp 724 to 1044 encoding amino acids 219 to 325) was made by cloning the EcoNI (position 192)-PvuII (position 721) and SmaI (position 1042)-NarI (position 1540) fragments of pOARESK9 into EcoNI-NarI-restricted pOARESK9. pToC180 (deletion of bp 1045 to 2013 encoding amino acids 326 to 648) was made by cutting pOARESK9 with SmaI and religation. Plasmids carrying deletions in the 3′ end of the areA gene were made by reconstructing the gene by using 3′ deletions created by exonuclease III digestion. pToC191 lacked bp 2001 to 2710 and encoded a 644-amino-acid AreA protein followed by a 30-amino-acid peptide (PVPGPPSRSTVSISLISNIFLEPPLIFSHR); pToC183 lacked bp 2318 to 2710 and encoded a 749-amino-acid AreA protein followed by a 29-amino-acid peptide (RGGPPSRSTVSISLISNIFLEPPLIFSHR); and pToC184 lacked bp 2524 to 2710 and encoded an 818-amino-acid AreA protein followed by a 20-amino-acid peptide (EGGPPLEVDGIDKLDIEYLS). pTOC266 was constructed by inserting a 1.8-kb SalI fragment containing the pyrG gene from pJers4 (Howard Brody) into the SalI site of pToC243. pToC243 contained a 2.1-kb HindII-SacI fragment from the 5′ end of the areA gene and a 1.4-kb Asp718-XmaI fragment from the 3′ end of the areA gene. The Asp718 site was not genomic. The two areA fragments were separated by 3.2 kb in the genome and flanked the coding region of the gene.
Transformation.
Protoplasts of A. oryzae and A. nidulans strains were prepared and transformed as described previously (1) by using approximately 3 μg of plasmid DNA. Transformants of A. nidulans and A. oryzae areA mutants were either selected on protoplast regeneration medium containing 10 mM nitrate as the sole nitrogen source or obtained by cotransformation with the riboB plasmid pPL3 (24) on medium containing 10 mM ammonium and lacking riboflavin. Complementing transformants were analyzed by Southern blot analysis, and a range of copy number transformants were identified. The phenotypes of the transformants were found to be independent of copy number. Noncomplementing transformants carrying deleted versions of the A. oryzae areA gene were analyzed by the Southern blotting method to confirm the presence of intact copies of the areA-containing plasmid. In order to inactivate the A. oryzae areA gene, the pyrG mutant HowB101 was transformed with pToC266 linearized by digestion with EcoRI, and PyrG+ transformants were selected on regeneration media containing 5% sodium chlorate and 0.5 mM ammonium sulfate.
NADP-GDH and GS assays.
Mycelia were grown overnight on A. nidulans ANM glucose minimal medium (10). Nitrogen sources were used at a final concentration of 10 or 50 mM. NADP-glutamate dehydrogenase (NADP-GDH) and glutamine synthetase (GS) assays were performed essentially as described by Pateman (26). One unit of NADP-GDH activity was defined as 1 nmol of NADP reduced per min per mg of soluble protein, and 1 U of GS activity was defined as 1 nmol of γ-glutamylhydroxymate formed per min per mg of soluble protein. Soluble protein contents were estimated based on the method of Bradford (4) by using Bio-Rad reagents.
Nucleotide sequence accession number.
The nucleotide sequence of the genomic clone determined in this study has been deposited in the GenBank database under accession no. AJ002968.
RESULTS
Cloning and analysis of the A. oryzae areA gene.
The A. oryzae areA gene was isolated from a lambda genomic library by cross-hybridization with the A. nidulans areA gene. A 5,643-bp region was sequenced, and the presence of an intron in the coding region was confirmed by sequencing cDNA clones spanning this region. The gene was predicted to encode an 866-amino-acid protein with high degrees of similarity to the A. nidulans AreA, N. crassa NIT-2, and P. chrysogenum NREA proteins (Fig. 1). A number of regions exhibit extremely high levels of amino acid conservation. The GATA type of DNA binding domain and the residues immediately C terminal to the zinc finger are absolutely conserved in the A. oryzae, A. nidulans, and P. chrysogenum proteins, and the AreA and NIT-2 proteins differ by only two amino acids in this region. Similarly, there is a high level of identity in the C-terminal regions of these proteins; this is particularly evident in the last nine residues, which are identical. Short regions that are identical are also evident in the four proteins.
FIG. 1.
Comparison of nitrogen regulatory proteins: alignment of the predicted A. oryzae AreA sequence (AoAreA) with the AreA sequence of A. nidulans (AnAreA) (accession no. X52491), the NIT-2 sequence of N. crassa (NcNIT2) (accession no. M33956), and the NREA sequence of P. chrysogenum (PcNreA) (accession no. U02612). The sequences were aligned by using the Boxshade program (http://ulrec3.unil.ch/software/Box_form.html). Black backgrounds indicate identical amino acids, and shaded backgrounds indicate similar amino acids as defined by the Boxshade program.
Functional analysis of A. oryzae areA gene.
The A. oryzae gene complemented A. nidulans loss-of-function areA mutants MH341 (areA217) and MH5699 (ΔareA::riboB), restoring the wild-type phenotype with all of the nitrogen sources tested. Complementing transformants were obtained either by direct selection for growth on nitrate or by cotransformation in which the riboB+ gene was used as the selectable marker. Complementation was independent of selection and the site of integration. Transformants were analyzed by the Southern blotting method and were found to contain a range of numbers of copies of the transforming plasmid pOARESK9. No evidence of derepression was seen with multicopy transformants, as shown by their wild-type levels of resistance to 100 mM chlorate, a toxic analog of nitrate, in the presence of ammonium (2). These transformants also did not form milk clearing halos, an indicator of extracellular protease activity, in the presence of ammonium (8). The A. oryzae areA gene, therefore, is functional in A. nidulans and is able to bring about nitrogen regulation in the heterologous species.
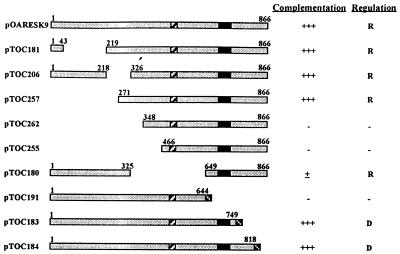
A variety of N-terminal and C-terminal deletion constructs of the A. oryzae areA gene were tested for complementation of A. nidulans areA mutants (Fig. 2). Transformants carrying constructs in which sequences encoding residues N terminal to amino acid 271 were deleted grew as well as wild-type A. nidulans on all of the nitrogen sources tested and exhibited wild-type sensitivity to nitrogen metabolite repression. In contrast, transformation with a construct lacking the internal sequences encoding amino acid residues 326 to 648 of the A. oryzae areA product resulted in transformants with an altered pattern of nitrogen source utilization. Growth on nitrate, nitrite, hypoxanthine, and γ-aminobutyric acid (GABA) as sole nitrogen sources was equivalent to growth of the wild-type strain. However, growth on glutamate and alanine as sole nitrogen sources was poorer than growth of the wild type. Sensitivity to nitrogen metabolite repression was retained in these transformants.
FIG. 2.
Functional analysis of the A. oryzae areA gene. Constructs encoding various deleted forms of the A. oryzae AreA protein (see text) were transformed into MH5699 (ΔareA::riboB). The abilities of the various constructs to complement for AreA function were assessed by comparing their growth to the growth of the wild-type strain: +++, growth equivalent to wild-type growth on all nitrogen sources tested; ±, reduced growth compared to the wild-type strain when 10 mM glutamate and 10 mM alanine were the sole nitrogen sources; −, no complementation for nitrogen source utilization. The sensitivity to repression by ammonium was assessed relative to that of the wild-type strain on 100 mM sodium chlorate with 10 mM ammonium tartrate and on 10% skim milk with 10 mM ammonium tartrate: R, wild-type levels of chlorate resistance and no milk clearing; D, sensitivity to chlorate toxicity and halos of milk clearing around colonies on the same media. The amino acid coordinates of the predicted A. oryzae AreA proteins are shown. The zinc finger region of protein is indicated by the solid box; the cross-hatched box indicates the putative acidic activation region; and the solid cross-hatched boxes indicate additional non-AreA amino acids encoded in the C-terminal deletion constructs (see text).
Deletion of sequences encoding C-terminal amino acids 645 to 866, which included the DNA binding domain, completely abolished the A. oryzae areA gene function, whereas deletions which truncated the C terminus of the protein but retained the DNA binding domain (amino acids 750 to 866 and 819 to 866) resulted in full activator function. However, these transformants showed increased sensitivity to chlorate and produced extracellular protease in the presence of 10 mM ammonium, indicating that there was a loss of nitrogen metabolite repression.
Creation of A. oryzae areA mutant.
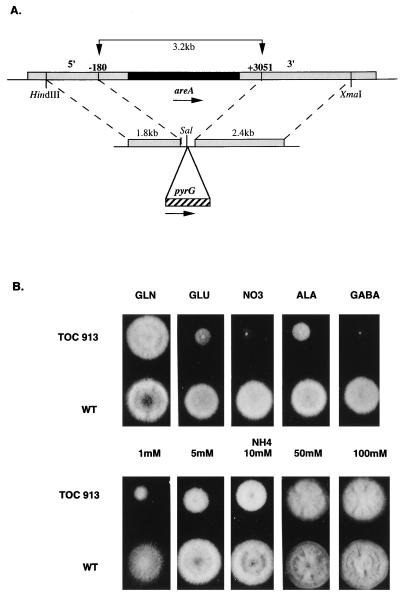
To establish the function of the cloned gene in A. oryzae, the genomic areA sequences were replaced by the A. oryzae pyrG gene (Fig. 3). The deletion construct pToC266 was linearized with EcoRI and transformed into the pyrG mutant HowB101. Initially, PyrG+ transformants were selected on a medium containing 0.5 mM ammonium as the sole nitrogen source along with 5% sodium chlorate, which was added to select against PyrG+ transformants arising from nonhomologous integration events. Wild-type strains are sensitive to chlorate under the conditions used, and A. nidulans areA mutants are not able to use nitrate and hence are chlorate resistant. Only weakly growing PyrG+ transformants were obtained. Genomic Southern blot analysis confirmed that the native areA sequence was replaced with the pyrG gene in these transformants. The growth of three independent isolates (TOC913, TOC919, and TOC920) was tested on a variety of media. These strains grew well on glutamine but exhibited reduced growth on all of the other nitrogen sources tested, including ammonium (Fig. 3). Subsequently, deletion of the areA gene was also achieved with linearized pToC266 by selection for PyrG+ transformants on a medium containing glutamine as the sole nitrogen source. Therefore, in A. oryzae the loss of areA function has a pleiotropic effect on nitrogen source utilization, including utilization of ammonium.
FIG. 3.
Inactivation of the A. oryzae areA gene. (A) Diagram of the construct used to inactivate the genomic A. oryzae areA gene. The shaded regions represent the 5′ and 3′ regions of the areA gene, and the solid region represents the areA coding sequences. In the inactivation construct the selectable marker pyrG (cross-hatched region) was flanked by sequences derived from the 5′ and 3′ ends of the areA gene (see text). This construct was transformed into a ΔpyrG recipient, and PyrG+ transformants were selected. The transformants were screened for the ability to use a variety of nitrogen sources, putative areA inactivation mutants were isolated, and inactivation of the areA gene was confirmed by Southern blot analysis. (B) Growth of the wild-type strain (WT) and ΔareA::pyrG strain TOC913 when glutamine (GLN), sodium glutamate (GLU), sodium nitrate (NO3), alanine (ALA), and GABA (final concentration of each compound, 10 mM) were used as sole nitrogen sources in glucose minimal media. Growth on ammonium tartrate (NH4) was also tested by using a range of concentrations, as indicated. Growth was scored after 2 days of incubation at 37°C.
In view of the A. oryzae areA mutant phenotype, the growth properties of the mutant were assessed by using a range of ammonium concentrations (Fig. 3). At high ammonium concentrations, the growth of the areA mutant was comparable to wild-type growth. However, as the ammonium concentration in the medium was reduced, the growth of the mutant was progressively poorer. At a concentration of 1 mM, the areA mutant grew poorly, and conidiation was limited compared to wild-type conidiation. Previous studies have suggested that A. nidulans areA mutants grow as well as or not quite as well as the wild type on ammonium (2, 20). Growth tests in which the areA mutants MH341 (areA217) and MH5699 (ΔareA::riboB) were compared to wild-type strain MH1 revealed a similar pattern of concentration-dependent growth responses in the A. nidulans mutants (data not shown). Growth was slightly reduced at an ammonium concentration of 10 mM. Therefore, the loss of AreA function appears to be associated with effects on ammonium utilization in both A. nidulans and A. oryzae. However, the phenotypic effect of loss of AreA function on growth on 10 mM ammonium appeared to be more apparent in A. oryzae than in A. nidulans.
The levels of activity of the major ammonium assimilatory enzymes, NADP-GDH and GS, were determined in wild-type and areA deletion strains of both A. oryzae and A. nidulans (Table 1). The pattern of expression of the two enzymes in wild-type A. oryzae was similar to the pattern of expression observed previously in A. nidulans and N. crassa (26); the levels of NADP-GDH activity were low on glutamate and high on glutamine, whereas the levels of GS activity were low on glutamine and high on glutamate. The levels of NADP-GDH activity were substantially reduced in the areA mutant strains of both species compared to those in the respective wild-type strains on ammonium, whereas the levels of GS activity were not affected. The reduced level of activity in the A. oryzae mutant was found to be a direct consequence of the loss of areA function by retransforming the mutant with the A. oryzae areA gene or the A. nidulans areA gene. Restoration of a functional areA gene simultaneously restored wild-type growth on nitrate or ammonium and wild-type levels of NADP-GDH activity (data not shown). As growth on high levels of ammonium was found to overcome the ammonium utilization phenotype of areA mutants, levels of NADP-GDH activity were determined under these conditions. Growth on 50 mM ammonium did not reverse the reduced levels of NADP-GDH activity observed in areA mutants grown on 10 mM ammonium.
TABLE 1.
NADP-GDH and GS activities in A. nidulans and A. oryzae wild-type and areA mutant strains
| Enzyme | Strain | Relevant genotype | Activity (U) with the following nitrogen sourcesa:
|
|||
|---|---|---|---|---|---|---|
| Glutamate | Ammonium | Ammonium (50 mM) | Glutamine | |||
| NADP-GDH | A. nidulans MH1 | Wild type | 45 (13) | 84 (9) | 48 (8) | 105 (20) |
| A. nidulans MH5699 | ΔareA::riboB | NDb | 28 (4) | 21 (3) | ND | |
| A. oryzae IFO4177 | Wild type | 100 (30) | 149 (26) | 87 (16) | 207 (32) | |
| A. oryzae TOC913 | ΔareA::pyrG | ND | 39 (8) | 18 (4) | ND | |
| GS | A. nidulans MH1 | Wild type | 370 (50) | 121 (26) | ND | 65 (11) |
| A. nidulans MH5699 | ΔareA::riboB | ND | 111 (31) | ND | ND | |
| A. oryzae IFO4177 | Wild type | 278 (8) | 88 (10) | ND | 61 (19) | |
| A. oryzae TOC913 | ΔareA::pyrG | ND | 72 (29) | ND | ND | |
Mycelia were grown for 16 h on glucose minimal media containing nitrogen sources at a concentration of 10 mM unless indicated otherwise. Crude cell extracts were prepared, and NADP-GDH and GS activities were determined as described in the text. Standard errors are shown in parentheses.
ND, not determined.
DISCUSSION
The areA gene of A. oryzae has both structural and functional similarity to the areA gene of A. nidulans. The DNA binding domains of members of the GATA class of proteins are highly conserved, and the DNA binding domain of the A. oryzae protein is identical to the DNA binding domain of A. nidulans AreA. The predicted 866-amino-acid protein is similar in size to the 876-amino-acid A. nidulans areA product and the 835-amino-acid P. chrysogenum nreA product. These proteins are smaller than the predicted 1,036-amino-acid N. crassa Nit-2 protein. Much of the heterogeneity in length and amino acid sequence in these fungal regulatory proteins is in the N-terminal region. Interestingly, where N-terminal deletions have been examined, there is evidence that at least the first 100 residues of the protein are not essential for gene function (17, 22). Furthermore, there appear to be cryptic promoters and translation initiation points within the A. nidulans areA and P. chrysogenum nreA coding sequences that allow expression in the absence of the native 5′ sequences (3, 17). The data presented here indicate that the same flexibility is present for expression of the A. oryzae areA gene. Within the sensitivity of the complementation assay, the dispensable region in the A. oryzae gene product (271 amino acids) encompasses nearly one-third of the entire protein. It is interesting that the deletions remove a number of small regions of N-terminal amino acids that are conserved in all species. Data for A. nidulans indicate that a loss of these sequences results in only subtle alterations in AreA function (22).
An internal deletion in the A. oryzae areA coding sequence leading to formation of an AreA protein lacking amino acids 326 to 648 may have resulted in a protein with reduced activator capacity. The A. nidulans transformants with this deletion were able to grow well on nitrogen sources such as nitrate and GABA but exhibited reduced growth on alanine and glutamate. The differences in nitrogen source utilization may reflect differences in the requirement for AreA activation of structural gene expression. The catabolism of nitrogen sources such as alanine may be more dependent on AreA function than the catabolism of nitrate and GABA, in which strong activation by the pathway-specific regulators NirA and AmdR, respectively (5, 25), may compensate for a weak AreA contribution. Analysis of the A. oryzae AreA protein revealed a possible acidic activation domain (amino acid residues 509 to 516) located in this region and in a position similar to the positions of acidic regions identified in the genes equivalent to the AreA gene in other fungal species (17).
As in the A. nidulans homolog, C-terminal deletions in A. oryzae areA result in derepression. There is strong evidence, obtained from A. nidulans and N. crassa, that the extreme C terminus of the protein is critical for sensitivity to repression; most likely the C terminus interacts with the nmr-1 gene product or its equivalent (30, 32). The extreme conservation of the C-terminal residues, coupled with the in vivo evidence that there is derepression in the absence of these residues, suggests strongly that A. oryzae AreA has a similar structure. In addition, studies performed with A. nidulans indicate that the 3′ untranslated region (3′ UTR) of the areA gene is also involved in nitrogen control (30). Comparisons of the 3′ UTR of the two genes have suggested that sequences in this region may also be conserved. Sequences designated B and B* by Platt et al. (30) are conserved in the A. oryzae 3′ UTR sequence. In addition, the A. nidulans 3′ UTR sequence contains a direct repeat (A and A*). The repeat is absent in the P. chrysogenum 3′ UTR; only the first element is present. In the A. oryzae gene, the first element is also highly conserved, whereas the second element is divergent, which supports the A. nidulans data that suggest that only one of the repeats is required for regulation of mRNA stability.
Cloning of the A. oryzae areA gene has allowed disruption of the areA gene. A loss of areA function results in a pleiotropic inability to use a wide variety of nitrogen sources. Surprisingly, the A. oryzae mutant also exhibited clearly reduced growth on 10 mM ammonium as the sole nitrogen source compared with the wild type. This phenotype was a direct consequence of the loss of areA function as complementation with either the A. oryzae gene or the A. nidulans gene restored growth on all nitrogen sources, including ammonium. It is likely that the poor growth of the A. oryzae mutant on ammonium is due to effects on ammonium uptake. The poor-growth phenotype was most striking at low concentrations, whereas growth on high ammonium concentrations was comparable to wild-type growth. While the phenotype of the A. oryzae mutant was apparent when 10 mM ammonium was used, tests performed with A. nidulans areA mutants also revealed that utilization of ammonium was affected when low ammonium concentrations were used. Previous studies performed with A. nidulans have shown that synthesis of the ammonium transport system(s) is controlled by internal concentrations of ammonium or glutamine (9, 27). It has been suggested that areA may regulate expression of an ammonium transport system other than the system affected in the methylammonium-resistant meaA mutant as the growth of double mutants on ammonium is poorer than the growth of single mutants (2). Similarly, double mutants of N. crassa carrying both a nit-2 mutation and the methylamine-resistant mea-1 mutation are not able to grow on ammonium (12).
A surprising result of this study was the clear indication that AreA is involved in expression of NADP-GDH in both A. nidulans and A. oryzae. In vitro assays have shown that the areA mutants of both A. nidulans and A. oryzae synthesize lower levels of NADP-GDH than the wild-type strains synthesize. As this enzyme is the major route for ammonium assimilation when ammonium levels are high, reduced enzyme levels could contribute to reduced growth on ammonium if the rate of assimilation into glutamate becomes a limiting factor. However, this is unlikely as the growth defect was reversed by high ammonium concentrations even though the NADP-GDH levels were low. Therefore, it appears that the uptake of ammonium rather than the subsequent incorporation of ammonium into glutamate is the limiting factor. The conversion of glutamate into glutamine by GS does not appear to be influenced by a mutation in the areA gene in either species. It is interesting that nit-2 mutants of N. crassa grow as well as the wild type on solid medium but produce less mycelial mass on ammonium in liquid medium (12, 28). Furthermore, Dantzig et al. (11) and Dunn-Coleman et al. (12) found that nit-2 mutants produce only basal levels of NADP-GDH. Therefore, effects on ammonium utilization and NADP-GDH levels may be common features of the loss of the major nitrogen regulatory protein in other fungal species. This may have implications for biotechnology applications. While inactivation of the major nitrogen control activator offers the potential for reduced protease synthesis and hence increased yields of expressed protein, the possible pleiotropic effects of the mutation may also be important in determining appropriate culture conditions.
This study provided evidence that AreA is involved, directly or indirectly, in expression of a key enzyme of nitrogen assimilation. The possibility that AreA is directly involved is supported by the fact that several potential AreA–NIT-2 binding sites (5′-GATAA-3′) are present in the 5′ regions of gdhA and am, the structural genes for NADP-GDH in A. nidulans and N. crassa, respectively (14, 18). It is significant that the influence of AreA on NADP-GDH levels is apparent when the ammonium-grown conditions are examined. This suggests that the protein plays an active role under nitrogen-sufficient conditions in addition to its established role as a transcriptional activator in response to nitrogen limitation.
ACKNOWLEDGMENTS
M.J.H. and M.A.D. acknowledge the support of the Australian Research Council.
T.C. acknowledges Kirsten L. Petersen for excellent technical assistance and Howard Brody, Novo Nordisk Biotech Inc., Davis, Calif., for providing the pyrG deletion strain HowB101 and pJer4 containing the A. oryzae pyrG gene. M.J.H. and M.A.D. thank Jane Copsey, Helene Martin, and Julie Sharp for expert technical assistance and Alex Andrianopoulos for assistance with the computer analysis.
REFERENCES
- 1.Andrianopoulos A, Hynes M J. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Cell Biol. 1988;8:3532–3541. doi: 10.1128/mcb.8.8.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arst H N, Jr, Cove D J. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- 3.Arst H N, Jr, Sheerin A. Translational initiation competence, ’leaky scanning’ and translational reinitiation in areA mRNA of Aspergillus nidulans. Mol Microbiol. 1996;19:1019–1024. doi: 10.1046/j.1365-2958.1996.470976.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Burger G, Strauss J, Scazzochio C, Lang B F. nirA, the pathway specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol Cell Biol. 1991;11:5746–5755. doi: 10.1128/mcb.11.11.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caddick M X, Arst H N, Taylor L H, Johnson R I, Brownlee A G. Cloning of the regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. EMBO J. 1986;5:1087–1090. doi: 10.1002/j.1460-2075.1986.tb04326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clutterbuck A J. Aspergillus nidulans genetics. In: King R C, editor. Handbook of genetics. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 447–510. [Google Scholar]
- 8.Cohen B L. Ammonium repression of extracellular protease in Aspergillus nidulans. J Gen Microbiol. 1972;71:293–299. [Google Scholar]
- 9.Cook R J, Anthony C. Regulation by glutamine of ammonium transport in Aspergillus nidulans. J Gen Microbiol. 1978;109:275–286. [Google Scholar]
- 10.Cove D J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;133:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 11.Dantzig A H, Weigmann F L, Jr, Nason A. Regulation of glutamate dehydrogenase in nit-2 and am mutants of Neurospora crassa. J Bacteriol. 1979;137:1333–1339. doi: 10.1128/jb.137.3.1333-1339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Davis, M. A., and M. J. Hynes. Unpublished data.
- 12.Dunn-Coleman N S, Nassiff M D, Garrett R H. Isolation and characterisation of a methylammonium resistant mutant of Neurospora crassa. Curr Genet. 1984;8:423–427. doi: 10.1007/BF00433908. [DOI] [PubMed] [Google Scholar]
- 13.Feng B, Xiao X, Marzluf G A. Recognition of specific nucleotide bases and cooperative binding by the trans-acting nitrogen regulatory protein NIT2 of Neurospora crassa. Nucleic Acids Res. 1993;21:3989–3996. doi: 10.1093/nar/21.17.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frederick G D, Kinsey J A. Nucleotide sequence and nuclear protein binding of the two regulatory sequences upstream of the am (GDH) gene in Neurospora. Mol Gen Genet. 1990;221:148–154. doi: 10.1007/BF00261714. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y-H, Marzluf G A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1990;10:1056–1065. doi: 10.1128/mcb.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y-H, Marzluf G A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci USA. 1990;87:5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas H, Bauer B, Redl B, Stoffler G, Marzluf G A. Molecular cloning and analysis of nre, the major nitrogen regulatory gene of Penicillium chrysogenum. Curr Genet. 1995;27:150–158. doi: 10.1007/BF00313429. [DOI] [PubMed] [Google Scholar]
- 18.Hawkins A R, Gurr S J, Montague P, Kinghorn J R. Nucleotide sequence and regulation of expression of the Aspergillus nidulans gdhA gene encoding NADP-dependent glutamate dehydrogenase. Mol Gen Genet. 1989;218:105–111. doi: 10.1007/BF00330572. [DOI] [PubMed] [Google Scholar]
- 19.Hynes M J, Pateman J A. The genetic analysis of regulation of amidase synthesis in Aspergillus nidulans. I. Mutants able to utilize acrylamide. Mol Gen Genet. 1970;108:97–106. doi: 10.1007/BF02430516. [DOI] [PubMed] [Google Scholar]
- 20.Hynes M J. Studies on the role of the areA gene in the regulation of nitrogen catabolism in Aspergillus nidulans. Aust J Biol Sci. 1975;28:301–313. doi: 10.1071/bi9750301. [DOI] [PubMed] [Google Scholar]
- 21.Kudla B, Caddick M X, Langdon T, Martinez-Rossi N, Bennett C F, Sibley S, Davies R W, Arst H N. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langdon T, Sheerins A, Ravagnani A, Gielkens M, Caddick M X, Arst H N., Jr Mutational analysis reveals dispensability of the N-terminal region of the Aspergillus transcription factor mediating nitrogen metabolite repression. Mol Microbiol. 1995;17:877–888. doi: 10.1111/j.1365-2958.1995.mmi_17050877.x. [DOI] [PubMed] [Google Scholar]
- 23.Merika M, Orkin S H. DNA binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakley C E, Weil C F, Ktrez P L, Oakley B R. Cloning of the riboB locus of Aspergillus nidulans. Gene. 1987;53:293–298. doi: 10.1016/0378-1119(87)90019-9. [DOI] [PubMed] [Google Scholar]
- 25.Parsons L M, Davis M A, Hynes M J. Identification of functional regions of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Microbiol. 1992;6:2999–3007. doi: 10.1111/j.1365-2958.1992.tb01758.x. [DOI] [PubMed] [Google Scholar]
- 26.Pateman J A. Regulation of synthesis of glutamate dehydrogenase and glutamine synthetase in micro-organisms. Biochem J. 1969;115:769–775. doi: 10.1042/bj1150769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pateman J A, Dunn E, Kinghorn J A, Forbes E C. The transport of ammonium and methylammonium in wild-type and mutant cells of Aspergillus nidulans. Mol Gen Genet. 1974;133:225–236. doi: 10.1007/BF00267672. [DOI] [PubMed] [Google Scholar]
- 28.Perrine K G, Marzluf G A. Amber nonsense mutations in regulatory and structural genes of the nitrogen control circuit of Neurospora crassa. Curr Genet. 1986;10:677–684. doi: 10.1007/BF00410916. [DOI] [PubMed] [Google Scholar]
- 29.Peters D G, Caddick M X. Direct analysis of native and chimeric GATA specific DNA binding proteins from Aspergillus nidulans. Nucleic Acids Res. 1994;22:5164–5172. doi: 10.1093/nar/22.24.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platt A, Langdon T, Arst H N, Jr, Kirk D, Tollervey D, Mates Sanchez J M, Caddick M X. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 1996;15:2791–2801. [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao X, Fu Y-H, Marzluf G A. The negative-acting NMR regulatory protein of Neurospora crassa binds to and inhibits the DNA-binding activity of the positive-acting nitrogen regulatory protein NIT2. Biochemistry. 1995;34:8861–8868. doi: 10.1021/bi00027a038. [DOI] [PubMed] [Google Scholar]