Abstract
Four class IIa bacteriocins (pediocin PA-1, enterocin A, sakacin P, and curvacin A) were purified to homogeneity and tested for activity toward a variety of indicator strains. Pediocin PA-1 and enterocin A inhibited more strains and had generally lower MICs than sakacin P and curvacin A. The antagonistic activity of pediocin-PA1 and enterocin A was much more sensitive to reduction of disulfide bonds than the antagonistic activity of sakacin P and curvacin A, suggesting that an extra disulfide bond that is present in the former two may contribute to their high levels of activity. The food pathogen Listeria monocytogenes was among the most sensitive indicator strains for all four bacteriocins. Enterocin A was most effective in inhibiting Listeria, having MICs in the range of 0.1 to 1 ng/ml. Sakacin P had the interesting property of being very active toward Listeria but not having concomitant high levels of activity toward lactic acid bacteria. Strains producing class IIa bacteriocins displayed various degrees of resistance toward noncognate class IIa bacteriocins; for the sakacin P producer, it was shown that this resistance is correlated with the expression of immunity genes. It is hypothesized that variation in the presence and/or expression of such immunity genes accounts in part for the remarkably large variation in bacteriocin sensitivity displayed by lactic acid bacteria.
Many lactic acid bacteria (LAB), including members of the genera Lactococcus, Lactobacillus, Carnobacterium, Enterococcus, and Pediococcus, are known to secrete small, ribosomally synthesized antimicrobial peptides called bacteriocins (26, 29, 34). Some of these peptides undergo posttranslational modifications (class I bacteriocins), whereas others are not modified (class II bacteriocins) (29, 34). Class II bacteriocins contain between 30 and 60 residues and are usually positively charged at a neutral pH. Studies of a large number of class II bacteriocins have led to subgrouping of these compounds (29, 34). One of the subgroups, class IIa, contains bacteriocins that are characterized by the presence of YGNG and CXXXXCXV sequence motifs in their N-terminal halves as well as by their strong inhibitory effect on Listeria (e.g., 3, 4, 22, 23, 27, 28, 31, 38, 45) (Fig. 1). Because of their effectiveness against the food pathogen Listeria, class IIa bacteriocins have potential as antimicrobial agents in food and feed.
FIG. 1.
Sequence alignment of class IIa bacteriocins. Residue numbering is according to the sequence of pediocin PA-1. Cysteine residues are printed in boldface; the two known class IIa bacteriocins with four cysteine residues are in the upper group. No attempt was made to optimize the alignment in the C-terminal halves of the peptides. Piscicolin 126 is identical to piscicocin V1a (4). Carnobacteriocin BM1 most probably is identical to piscicocin V1b (4). Sakacin P most probably is identical to bavaricin A (30). Curvacin A is identical to sakacin A (2). The consensus sequence includes residues conserved in at least 8 of the 12 sequences shown; 100% conserved residues are underlined.
Class IIa bacteriocins act by permeabilizing the membrane of their target cells (1, 5, 6, 9, 10, 26, 28). The most recent studies on the mode of action of these bacteriocins indicate that antimicrobial activity does not require a specific receptor and is enhanced by (but not fully dependent on) a membrane potential (9, 28). Little is known about bacteriocin structure, and unravelling the relationships between structure and function is one of the great challenges in current bacteriocin research. A logical starting point for structure-function studies is a thorough study of the differences in activity and target cell specificity between naturally occurring homologous bacteriocins. A few such studies have been described, but these suffer from either a very limited number of tested indicator strains or the use of culture supernatants instead of purified bacteriocins (3, 4, 17, 45). The use of purified bacteriocins for comparative analyses is absolutely essential, since it is becoming increasingly evident that bacteriocin producers produce more than one bacteriocin (4, 8, 38, 48; this study).
In the present study, the activities of four pure class IIa bacteriocins (pediocin PA-1, enterocin A, curvacin A, and sakacin P) (Fig. 1) were tested against a large number of LAB as well as several strains of the food pathogen Listeria monocytogenes. The bacteriocins were purified from their respective producer strains by use of an optimized purification protocol yielding highly pure samples. The contribution of disulfide formation was assessed and found to be important for activity. The effects of the purified bacteriocins on (noncognate) class IIa bacteriocin-producing strains are described, and the implications of our findings for immunity and resistance are discussed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Lactobacillus spp., Pediococcus spp., Enterococcus spp., Leuconostoc spp., and Carnobacterium spp. were grown in MRS broth (Oxoid, Unipath Ltd., Basingstoke, Hampshire, England) at 30°C. Lactococci were grown in M17 medium (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% (wt/vol) glucose at 30°C. Listeria spp. were grown on brain heart infusion medium (Difco) at 30°C, and Clostridium spp. were grown on both thioglycolate medium (Oxoid) and reinforced clostridial medium (Oxoid) at 37°C. For cultivating Clostridium spp., the Anaerogen system for anaerobic incubation (Oxoid) was used.
Bacteriocin purification and concentration determination.
Pediocin PA-1, enterocin A, sakacin P, and curvacin A were purified from the supernatants of early-stationary-phase cultures of Pediococcus acidilactici (35), Enterococcus faecium CTC492 (3), Lactobacillus curvatus LTH1174 (45), and Lactobacillus sake LTH673 (45), respectively. In terms of bacteriocin production, E. faecium CTC492 is identical to E. faecium T136 (8). The bacteriocins were purified to homogeneity essentially as described previously (3, 35, 45). The method consists of ammonium sulfate precipitation followed by three chromatography steps (ion exchange, hydrophobic interaction, and reversed phase). The final, reversed-phase chromatography step (with a fast protein liquid chromatography system supplied by Pharmacia-LKB, Uppsala, Sweden) was repeated two or three times to ensure maximum purity. In contrast to the previously described method, very large volumes of washing buffer (up to 40 times the column volume) were used for the washing steps during ion-exchange and hydrophobic-interaction chromatography. This small change in the protocol contributed considerably to purity.
The purity of the bacteriocins was assessed by 10 steps of Edman degradation with a model 477A sequencer (Applied Biosystems, Foster City, Calif.). Purity was assessed by comparing the yields of the expected amino acids with the yields of other amino acids (that would result from contaminating peptides) in each degradation step. On the basis of these analyses, purity was estimated to be >98% for sakacin P, curvacin A, and pediocin PA-1 and about 95% for enterocin A.
Initially, the concentration of the purified bacteriocins was assessed by two methods: measuring UV absorption at 280 nm and amino acid composition analysis. For the former method, molar extinction coefficients were calculated from the contributions of individual amino acid residues by use of the program PCGENE (IntelliGenetics Inc., Geneva, Switzerland). The differences between the results of the two methods were on the order of 10% for all four bacteriocins (data not shown). The simplest of the two methods (measuring UV absorption at 280 nm) was therefore used for further routine concentration determinations.
The purified bacteriocins were stored at −20°C and used within 1 month of purification. The bacteriocins retained ≥90% of their activity during this period, as indicated by the results of weekly activity assays with a standard set of three indicator strains.
Bacteriocin assay.
Bacteriocin activity was quantified with a microtiter plate assay (21, 24). For the comparative studies, a standardized procedure was used; indicator cells were always derived from a fresh overnight culture and diluted 400 times before use. The subsequent incubation time was 18 to 20 h in all assays. The activities of the four bacteriocins toward a specific strain were always determined in a single assay. The MICs given represent the bacteriocin concentration needed to obtain 50% inhibition of growth. The values are the averages of three independent measurements, which gave standard deviations on the order of 25% of the values. Bacteriocin activity is expressed in units, one bacteriocin unit being the amount of bacteriocin required to reduce the growth of the indicator strain by 50% under the conditions of the assay (200-μl culture volume).
Disulfide bonds.
Disulfide bond formation was assessed by determining the molecular masses of the purified bacteriocins by electrospray ionization mass spectrometry and by determining the number of free thiol groups as described by Ellman (15). In the latter method, the molar extinction coefficient used for 2-nitro-5-thiobenzoate was 14,150 M−1 cm−1. A sample containing a peptide with only one cysteine was included as a positive control.
To test the effect of the reduction of disulfide bonds on bacteriocin activity, bacteriocin assays in which dithiothreitol (DTT; final concentration, 10 mM) was added to the culture medium in the microtiter plate wells were conducted. These experiments were conducted with indicator strains whose growth was not inhibited by the presence of 10 mM DTT alone.
Induction of bacteriocin production.
Under certain conditions, bacteriocin-negative (Bac−) cultures of the sakacin P producer L. sake LTH673 can be obtained, as described previously (7, 13). These cultures have a stable Bac− phenotype, but the bacteriocin-positive (Bac+) phenotype may be restored by adding the pheromone needed for the expression of bacteriocin-related genes (13). In the present study, Bac− and Bac+ cultures of L. sake LTH673 were obtained by making two identical 100-fold dilutions of a Bac− overnight culture. To one of the two dilutions, the appropriate pheromone was added to a final concentration of 50 ng/ml (7, 13). The addition of the pheromone led to the transcription of genes involved in bacteriocin production, as documented previously (7).
Sequence alignments.
Sequence identities and similarities between immunity proteins were determined with the program PALIGN, which is part of the PCGENE software package (33). The genetic code matrix (16) with default gap opening and extension penalties was used. These settings kept the numbers of insertions and deletions equal to or below five for all pairwise sequence alignments. The following groups of amino acids were defined as similar: A, S, and T; D and E; N and Q; R and K; I, L, M, and V; and F, Y, and W.
RESULTS AND DISCUSSION
Class IIa bacteriocin producers produce more than one bacteriocin.
Recent studies of bacteriocin producers suggest that it may be common for LAB to produce more than one bacteriocin. For example, E. faecium CTC492 not only produces the class IIa bacteriocin enterocin A but also produces enterocin B, which does not belong to class IIa. These two bacteriocins were shown to have different inhibitory spectra (8, 14). Another example of the production of multiple bacteriocins by LAB is illustrated by Table 1. The data in Table 1 show that during purification of sakacin P, inhibitory activity toward some strains was lost, whereas inhibitory activity toward other strains was retained (Table 1). The lost activity could be recovered by pooling the supernatant and the pellet obtained in the ammonium sulfate precipitation step at the start of the purification protocol (data not shown). Thus, in addition to sakacin P, L. sake LTH673 produces at least one other bacteriocin. This other bacteriocin(s) has not yet been characterized, but transcription studies have shown that at least one gene encoding a hitherto-uncharacterized bacteriocin-like peptide is expressed in addition to the sakacin P structural gene (7). The data in Table 1 show how erroneous results would have been obtained if the inhibitory spectrum of sakacin P had been assessed with culture supernatants.
TABLE 1.
Purification of bacteriocins from L. sake LTH673 and assessment of yields with various indicator strains
| Indicator strain | Total activity, 10−5 bacteriocin units, of:
|
Yield (%)b | |
|---|---|---|---|
| Culture supernatant | Purified sakacin Pa | ||
| Lactobacillus coryneformis NCDO 2740 | 6.6 | 3.4 | 52 |
| L. sake NCDO 2714 | 76 | 4.1 | 5.4 |
| Lactobacillus sp. strain LMG 2804 | 8.5 | 0.09 | 1.1 |
Ten steps of Edman degradation were performed and indicated that the material contained >98% pure sakacin P.
The dramatic indicator strain-dependent variation in yield showed that antimicrobial activity toward some strains had been lost, whereas activity toward other strains had been retained (see the text for details).
Inhibitory spectra.
The activities of the bacteriocins toward various indicator strains are shown in Table 2 and Fig. 2. The results show that pediocin PA-1 and enterocin A are generally more active than sakacin P and curvacin A. Pediocin PA-1 and enterocin A also have a broader inhibitory spectrum (Fig. 2). The magnitude of the difference in activity between pediocin PA-1 and enterocin A on the one hand and sakacin P and curvacin A on the other hand varies widely with the indicator strain used. The latter represents a general phenomenon illustrated by Table 2: when LAB were used as indicator strains, the activity ratio between the various bacteriocins differed from species to species and even from strain to strain. More consistent results were obtained when listeriae were used as indicator strains: enterocin A was 5 to 10 times more active than sakacin P and pediocin PA-1, which were 5 to 10 times more active than curvacin A.
TABLE 2.
Activities of purified class IIa bacteriocins toward various indicator strains
| Indicator strainb | Bacc | Sensitivity (MIC, μg/ml)a to:
|
|||
|---|---|---|---|---|---|
| Pediocin PA-1 | Curvacin A | Sakacin P | Enterocin A | ||
| Lactobacillus curvatus | |||||
| LMG 2353 | >2.0 | >1.0 | >2.0 | >2.0 | |
| LMG 2371 | 0.020 | 0.15 | >0.5 | 0.017 | |
| NCFB 2739 B | >2.0 | >2.0 | >2.0 | >0.5 | |
| L. plantarum | |||||
| NCDO 1869 | 1.5 | >2.0 | >2.0 | 0.40 | |
| LMG 2352 | 0.050 | >2.0 | >2.0 | 0.12 | |
| LMG 2362 | +d | 0.96 | >2.0 | >2.0 | >0.5 |
| LMG 2362 | −d | 0.58 | >2.0 | >2.0 | 0.31 |
| LMG 2375 | 0.017 | >2.0 | >0.5 | 0.40 | |
| DSM 20174 | 0.0094 | >2.0 | >0.5 | 0.015 | |
| NCDO 1193 B | >2.0 | >2.0 | >2.0 | >0.5 | |
| L. sake | |||||
| NCDO 2714 | 0.0004 | 0.0011 | 0.0037 | 0.0001 | |
| LMG 2380 | + | >1.0 | 1.1 | >0.5 | 0.035 |
| LMG 2799 | 0.093 | 0.31 | 0.42 | 0.011 | |
| L. casei | |||||
| NCDO 161 | 0.32 | >2.0 | >2.0 | >2.0 | |
| NCDO 2713 | >2.0 | >2.0 | >2.0 | >0.5 | |
| NCDO 1857 | 0.0084 | 0.98 | >2.0 | 0.0059 | |
| NCDO 2743 | >2.0 | >2.0 | >2.0 | >0.5 | |
| ATCC 334 B | >2.0 | >2.0 | >2.0 | >0.5 | |
| L. coryneformis | |||||
| NCDO 2740 | 0.0005 | 0.0037 | 0.0052 | 0.0003 | |
| NCDO 2741 | 0.011 | 0.050 | 0.0049 | 0.0066 | |
| Lactobacillus sp. strain | |||||
| LMG 2003 | 0.0009 | 0.91 | 0.023 | 0.0026 | |
| NCDO 2503 | 0.016 | 0.27 | >0.5 | 0.36 | |
| LMG 2333 | + | 0.0008 | 0.0040 | 0.0086 | 0.0016 |
| Pediococcus acidilactici | |||||
| NCDO 1859 | 0.0028 | >2.0 | >2.0 | 0.080 | |
| NCDO 521 | 0.29 | >2.0 | >2.0 | >2.0 | |
| NCDO 1851 | 0.0039 | 1.8 | >2.0 | 0.0068 | |
| P. pentosaceus | |||||
| LMG 2001 | (+)e | 0.0030 | >2.0 | >2.0 | 0.0062 |
| NCDO 559 | 0.0086 | 0.98 | 0.72 | 0.021 | |
| NCDO 1850 | >2.0 | 1.8 | >2.0 | 0.55 | |
| LMG 2366 | >2.0 | >2.0 | >2.0 | >2.0 | |
| LMG 2722 | 0.0004 | 0.54 | 0.15 | 0.0005 | |
| Enterococcus faecalis NCDO 581 | 0.0007 | 0.0020 | 0.0057 | 0.0013 | |
| E. faecium NCDO 942 | >2.0 | >2.0 | >2.0 | >0.5 | |
| Carnobacterium piscicola | |||||
| LMG 2332 | +f | 0.0002 | 0.0021 | 0.0009 | 0.0006 |
| NCDO 2764 | 0.0027 | 0.017 | 0.0015 | 0.0017 | |
| C. divergens NCDO 2306 | 0.0041 | 0.023 | 0.0033 | 0.00004 | |
| Leuconostoc mesenteroides NCDO 529 | 1.4 | >2.0 | >2.0 | >2.0 | |
| L. cremoris LMG 2724 | 0.025 | >2.0 | >2.0 | >1.0 | |
| Leuconostoc sp. strain NCDO 543 | 0.0009 | >2.0 | 0.70 | >1.0 | |
| Lactococcus lactis | |||||
| Nine different strains | +g | >2.0 | >2.0 | >2.0 | >0.5 |
| LMG 2070 | 0.17 | >2.0 | >0.5 | >0.5 | |
| LMG 2095 | + | >2.0 | 0.91 | >2.0 | >0.5 |
| Clostridium tyrobutyricum (11 different strains) | >2.0 | >2.0 | >2.0 | >0.5 | |
| C. butyricum NCDO 855A | 0.035 | >2.0 | 0.083 | 0.0021 | |
| Listeria monocytogenes | |||||
| LMG 2650 | 0.0047 | 0.049 | 0.0073 | 0.0008 | |
| LMG 2651 | 0.0029 | 0.028 | 0.0036 | 0.0006 | |
| LMG 2652 | 0.0050 | 0.030 | 0.0068 | 0.0004 | |
| LMG 2653 | 0.0050 | 0.049 | 0.0068 | 0.0008 | |
| LMG 2800 | 0.0062 | 0.069 | 0.0083 | 0.0011 | |
| LMG 2801 | 0.0033 | 0.040 | 0.0034 | 0.0005 | |
| LMG 2802 | 0.0013 | 0.028 | 0.0034 | 0.0002 | |
| L. innocua LMG 2654 | 0.0031 | 0.026 | 0.0057 | 0.0003 | |
| L. ivanovii LMG 2803 | 0.0006 | 0.0092 | 0.0002 | <0.0001 | |
Values preceded by a greater-than sign indicate the highest bacteriocin concentration tested.
Strains were from our laboratory collection (LMG) or from well-known culture collections: ATCC, American Type Culture Collection, Rockville, Md.; NCDO and NCFB, National Collection of Food Bacteria, Reading, United Kingdom.
+, the indicator strain is known to produce at least one bacteriocin; −, the indicator strain is capable of producing bacteriocins but has lost the Bac+ phenotype.
This strain produces several bacteriocins that do not belong to class IIa; production can be controlled in a manner similar to that in the sakacin P producer (see the text) (12).
This strain produces pediocin A, a high-molecular-weight “bacteriocin” (80 kDa) (37).
This strain produces the lantibiotic carnobacteriocin UI49 (44).
These strains included strains producing nisin and lactococcin A.
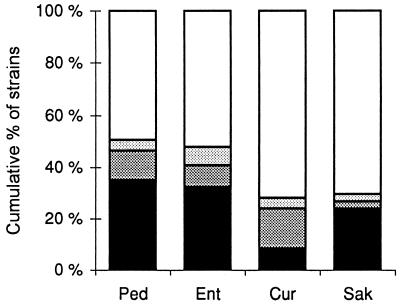
FIG. 2.
Sensitivity of indicator strains. Strains are categorized according to MICs (Table 2): black, <10 ng/ml; dark grey, 10 to 100 ng/ml; light grey, 100 to 500 ng/ml; white, >500 ng/ml. Ped, pediocin PA-1; Ent, enterocin A; Cur, curvacin A; Sak, sakacin P.
Interestingly, sakacin P had modest activity toward LAB but was almost as effective as enterocin A and pediocin PA-1 against listeriae. Because of this combination of high antilisterial activity and a narrow inhibitory spectrum (Fig. 2), sakacin P is perhaps the most promising of the tested bacteriocins for use in LAB fermentations that are prone to Listeria infections.
For analyzing possible synergistic inhibitory effects of class IIa bacteriocins, the activity of every possible one-to-one combination of two of the purified bacteriocins was measured at maximum total concentrations of approximately 1 μg/ml. No synergistic effects were observed (results not shown).
Role of disulfide bonds.
Mass spectrometry analysis showed that the molecular masses of purified pediocin PA-1, enterocin A, curvacin A, and sakacin P were 4.5, 4.7, 2.2, and 3.2 Da, respectively, lower than those expected when cysteine residues were assumed to be in a reduced state. Considering the standard deviation in the mass determinations (∼1 Da), these data are in accordance with the notion that all cysteine residues are oxidized and are involved in disulfide bonds. In accordance with this observation, the Ellman assay did not reveal any free cysteine residues in the bacteriocins, whereas it did reveal free cysteine residues in a one-cysteine-containing control peptide (results not shown). Thus, pediocin PA-1 and enterocin A indeed contain two disulfide bonds, whereas sakacin P and curvacin A contain one. It was previously shown that the two disulfide bonds in pediocin PA-1 are formed between Cys9 and Cys14 and between Cys24 and Cys44 (23).
Interestingly, the two most active bacteriocins in this study were the ones with two disulfide bonds, suggesting a correlation between these two properties. To investigate the contribution of disulfide bond formation to bacteriocin activity, three indicator strains whose growth was not inhibited by DTT were selected for testing bacteriocin activity under reducing conditions. As shown in Table 3, DTT dramatically reduced the activities of pediocin PA-1 and enterocin A, whereas the activities of sakacin P and curvacin A were only moderately reduced. Thus, in the presence of DTT, the two-disulfide-bond-containing bacteriocins pediocin PA-1 and enterocin A were no longer generally more potent than the one-disulfide-bond-containing bacteriocins sakacin P and curvacin A.
TABLE 3.
Effect of reduction on antimicrobial activitya
| Strain | Sensitivity to:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Pediocin PA-1
|
Enterocin A
|
Curvacin A
|
Sakacin P
|
|||||
| Standard | + DTT | Standard | + DTT | Standard | + DTT | Standard | + DTT | |
| L. sake NCDO 2714 | 0.4 | 45 (0.9) | 0.1 | 19 (0.5) | 1.1 | 4.5 (24) | 3.7 | 29 (13) |
| Lactobacillus sp. strain LMG 2333 | 0.8 | 8.2 × 102 (0.1) | 1.6 | 1.9 × 102 (0.9) | 4.0 | 48 (8.3) | 8.6 | 54 (16) |
| P. pentosaceus LMG 2722 | 0.4 | 3.2 × 102 (0.1) | 0.5 | 84 (0.6) | 5.4 × 102 | >20 × 102 (<27) | 1.5 × 102 | 5.4 × 102 (28) |
The standard microtiter plate assay was used, with or without the addition of 10 mM DTT (final concentration) to each well. No growth inhibition occurred in control cultures without bacteriocin but with 10 mM DTT. Sensitivity is reported as MIC in nanograms per milliliter. Values in parentheses indicate the percentages of antimicrobial activity under reducing conditions relative to that under standard (nonreducing) conditions.
These results suggest that the high levels of activity of pediocin PA-1 and enterocin A may be due at least in part to the extra disulfide bond present in the C-terminal region. The effect of DTT on the activity of sakacin P and curvacin A suggests that the disulfide bond between fully conserved Cys9 and Cys14 is important but not crucial for activity. This conclusion is in accordance with previous studies on carnobacteriocin B2 (in which the bond between Cys9 and Cys14 is not formed) (38) and leucocin A (which retains considerable activity after reduction and modification of the cysteines) (22). Remarkably, quite opposite results have also been reported. In one study, pediocin PA-1 was found to lose all of its activity upon reduction (10). Furthermore, the activity of mesentericin Y105 was shown to be reduced at least 2,000-fold upon modification or mutation of Cys9 and Cys14 (18). At present, we have no explanation for the apparent inconsistencies among these results.
Several authors have discussed possible structural models for a class IIa bacteriocin in a membrane environment (4, 9, 17, 20, 28). The bacteriocins are unstructured in watery solutions, whereas they adopt a partly helical structure in more hydrophobic environments (14, 18, 20, 42). It has been suggested that the C-terminal half of class IIa bacteriocins forms a hydrophobic or amphiphilic transmembrane α helix, permitting the formation of a so-called “barrel-stave” (36) poration complex (4, 17, 29). Obviously, this simple structural model is incompatible with the presence of a disulfide bond formed by cysteine residues located at the beginning and end of this putative transmembrane helix (as would be the case for pediocin PA-1 and enterocin A). Recent studies of the three-dimensional structure of leucocin A (leucocin A has only the Cys9-Cys14 disulfide bond) (Fig. 1) show that the most C-terminal portion of this bacteriocin has a rather extended (nonhelical) structure, folding back onto a preceding helical region that comprises residues 17 to 31 (20). Extrapolating these observations to enterocin A and pediocin PA-1, one may speculate that the extra disulfide bond in these two bacteriocins stabilizes their structures by covalently coupling the C-terminal residue to a cysteine in the helical region.
Immunity.
LAB producing class IIa bacteriocins generally were not particularly sensitive to noncognate class IIa bacteriocins, but there was a tendency for the producers of the less potent sakacin P and curvacin A to be sensitive to the more potent pediocin PA-1 and enterocin A (Table 4). Remarkably, L. sake Lb706 and L. curvatus LTH1174, which are known to produce identical bacteriocins, displayed different behaviors in terms of bacteriocin sensitivity.
TABLE 4.
Bacteriocin sensitivity of LAB producing class IIa bacteriocins
| Strain | Reference | Bacteriocin produced | Sensitivity (MIC, μg/ml) to:
|
|||
|---|---|---|---|---|---|---|
| Pediocin PA-1 | Curvacin A | Sakacin P | Enterocin A | |||
| L. sake LTH673 | 7, 45 | Sakacin P | >1.0 | 0.63 | >0.7 | 0.021 |
| L. sake LTH673 | 7, 45 | Bac− phenotypea | 0.020 | 0.037 | 0.057 | 0.0056 |
| P. acidilactici | 35 | Pediocin PA-1 | >1.0 | >2.0 | >0.7 | >0.6 |
| L. curvatus LTH1174 | 45 | Curvacin Ab | 0.12 | >2.0 | 0.39 | 0.061 |
| L. sake Lb706 | 2 | Sakacin Ab | >1.0 | >2.0 | >0.7 | >0.6 |
| E. faecium CTC492 | 3 | Enterocin A | >1.0 | >2.0 | >0.7 | >0.6 |
These results do not permit discrimination between insensitivity (the cause of which is not known) and immunity provided by a specific immunity protein. To obtain this discrimination, we exploited the fact that the expression of genes involved in the production of sakacin P can be controlled (7, 13). Cultures of L. sake LTH673 which have a stable Bac− phenotype and in which the transcription of bacteriocin-related genes (including the spiA gene, encoding immunity for sakacin P) is switched off can be obtained. As shown in Table 4, L. sake LTH673 Bac− cells were sensitive to all four bacteriocins. L. sake LTH673 Bac+ cells, which were obtained after induction of a Bac− culture with an appropriate pheromone and which are known to express the spiA gene (7), were much less sensitive. Within the time frame of the experiments, spontaneous development of insensitivity to class IIa bacteriocins was not observed. Thus, the low sensitivity or immunity of Bac+ cells of L. sake LTH673 to various (noncognate) class IIa bacteriocins is correlated with the expression of the sakacin P gene cluster (including spiA).
Table 5 shows that sequence similarities between proteins that (putatively) provide immunity to class IIa bacteriocins are generally low but that most of the proteins do clearly resemble several others. A certain similarity between the proteins was also suggested by secondary structure predictions (40, 41), which indicated that all 12 proteins shown in Table 5 were largely α helical but were devoid of transmembrane helices (14). Studies with L. sake LTH673 indicated that, despite the low degree of sequence homology, the immunity proteins shown in Table 5 provide (partial) immunity to various class IIa bacteriocins.
TABLE 5.
Sequence similarities between proteins that (putatively) provide immunity to class IIa bacteriocinsa
| Immunity protein | % Identity or similarity to:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CarBM1 | SakA | CarB2 | Orf-β3 | SakP | LeuA | MesY105 | EntA | OrfY | PedPA-1 | EntP | Bac31 | |
| CarBM1 | 49 | 19 | 17 | 18 | 20 | 20 | 19 | 24 | 25 | 44 | 50 | |
| SakA (2) | 65 | 19 | 19 | 17 | 21 | 24 | 21 | 22 | 18 | 47 | 42 | |
| CarB2 (39) | 31 | 33 | 26 | 23 | 11 | 12 | 15 | 18 | 5 | 19 | 24 | |
| Orf-β3 | 31 | 31 | 37 | 46 | 28 | 22 | 27 | 22 | 21 | 20 | 22 | |
| SakP (25) | 25 | 22 | 34 | 62 | 18 | 18 | 12 | 17 | 17 | 23 | 17 | |
| LeuA (49) | 38 | 32 | 21 | 38 | 32 | 73 | 38 | 40 | 11 | 22 | 21 | |
| MesY105 | 35 | 32 | 20 | 41 | 29 | 81 | 40 | 36 | 9 | 20 | 22 | |
| EntA | 31 | 31 | 19 | 40 | 19 | 50 | 47 | 27 | 20 | 14 | 17 | |
| OrfY | 35 | 29 | 27 | 36 | 26 | 51 | 45 | 39 | 14 | 17 | 16 | |
| PedPA-1 (50) | 35 | 39 | 19 | 27 | 28 | 20 | 18 | 31 | 23 | 19 | 22 | |
| EntP | 59 | 58 | 33 | 31 | 31 | 35 | 32 | 25 | 28 | 30 | 52 | |
| Bac31 | 66 | 54 | 31 | 32 | 30 | 30 | 32 | 29 | 21 | 34 | 67 | |
Sequence identity (upper-right triangle) and similarity (lower-left triangle) between proteins that (putatively) provide immunity to class IIa bacteriocins are shown. References are given for proteins for which the immunity function has been demonstrated through functional studies. The proteins displayed are the (putative) immunity proteins for carnobacteriocin BM1 (CarBM1) (38), carnobacteriocin B2 (CarB2) (38, 39), sakacin A (SakA) (2), sakacin P (SakP) (25), leucocin A (LeuA) (22, 49), enterocin A (EntA) (3), mesentericin Y105 (MesY105) (19), pediocin PA-1 (PedPA-1) (32, 50), enterocin P (EntP) (11), and bacteriocin 31 (Bac31) (47). Other putative immunity proteins shown are the products of orfY (see the text) and of orf-β3 (an open reading frame that follows the genes encoding carnobacteriocin B2 and the CarB2 immunity protein) (38).
Interestingly, the sakacin P producer L. sake LTH673 contains at least one more putative immunity gene (7) (orfY in Table 5), which is transcribed in Bac+ cells only (14). This immunity gene is not coupled to a cognate bacteriocin gene (7). In addition to L. sake LTH673, several other LAB seem to contain immunity genes that are not directly associated with a gene encoding a cognate bacteriocin. Copies of the sakacin P immunity gene have been detected in the chromosome of the sakacin A producer L. sake Lb706 and, most importantly, in the chromosome of an L. sake strain that does not produce any bacteriocin at all (25). The producer of carnobacteriocins BM1 and B2 contains at least three (putative) immunity genes (38, 39). In addition to putative immunity genes for the two class IIa bacteriocins that are produced (designated CarBM1 and CarB2 in Table 5), a third putative immunity gene is present downstream of the structural gene for carnobacteriocin B2 (the protein is designated Orf-β3 in Table 5). Studies of transcription have indicated that the orf-β3 gene is cotranscribed with the genes encoding carnobacteriocin B2 and its cognate immunity protein (43).
Extrapolating the above-mentioned observations, one may speculate that LAB from genera containing class IIa bacteriocin producers generally possess one or more immunity genes for class IIa bacteriocins. These genes may show various degrees of homology and may be expressed to various extents. These suggestions could provide an explanation for the remarkable variation in bacteriocin sensitivity that is observed for closely related LAB strains.
ACKNOWLEDGMENTS
We are grateful to Stephen Bayne, Novo Nordisk, Gentofte, Denmark, for carrying out mass spectrometry analyses and to Grethe Kobro and May-Britt Selvåg Hovet for technical assistance. We thank Tone Katla at Matforsk, The Norwegian Food Research Institute, for assistance in work with pathogenic strains.
This work was supported in part by a grant from the Nordic Industry Fund. M.B.B. was supported by a grant from the Norwegian Research Council.
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of Gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson L, Holck A. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Bacteriol. 1995;177:2125–2137. doi: 10.1128/jb.177.8.2125-2137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aymerich T, Holo H, Håvarstein L S, Hugas M, Garriga M, Nes I F. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol. 1996;62:1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhugaloo-Vial P, Dousset X, Metivier A, Sorokine O, Anglade P, Boyaval P, Maron D. Purification and amino acid sequences of piscicocins V1a and V1b, two class IIa bacteriocins secreted by Carnobacterium piscicola V1 that display significantly different levels of specific inhibitory activity. Appl Environ Microbiol. 1996;62:4410–4416. doi: 10.1128/aem.62.12.4410-4416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhunia A K, Johnson M C, Ray B, Kalchayanand N. Mode of action of pediocin AcH from Pediococcus acidilactici H on sensitive bacterial strains. J Appl Bacteriol. 1991;70:25–30. [Google Scholar]
- 6.Bruno M E C, Montville T J. Common mechanistic action of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1993;59:3003–3010. doi: 10.1128/aem.59.9.3003-3010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brurberg M B, Nes I F, Eijsink V G H. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 8.Casaus P, Nilsen T, Cintas L M, Nes I F, Hernández P E, Holo H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology. 1997;143:2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Shapira R, Eisenstein M, Montville T J. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl Environ Microbiol. 1997;63:524–531. doi: 10.1128/aem.63.2.524-531.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chikindas M L, García-Garcera M J, Driessen A J M, Ledeboer A M, Nissen-Meyer J, Nes I F, Abee T, Konings W N, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cintas L M, Casaus P, Håvarstein L S, Hernández P E, Nes I F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol. 1997;63:4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep D B, Håvarstein L S, Nes I F. Characterization of the locus responsible for bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eijsink V G H, Brurberg M B, Middelhoven P J, Nes I F. Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J Bacteriol. 1996;178:2232–2237. doi: 10.1128/jb.178.8.2232-2237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eijsink, V. G. H., M. Skeie, M. B. Brurberg, and I. F. Nes. Unpublished observations.
- 15.Ellman G L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 16.Feng D F, Johnson M S, Doolittle R F. Aligning amino acid sequencing: comparison of commonly used methods. J Mol Evol. 1984;21:112–125. doi: 10.1007/BF02100085. [DOI] [PubMed] [Google Scholar]
- 17.Fimland G, Blingsmo O R, Sletten K, Jung G, Nes I F, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleury Y, Dayem M A, Montagne J J, Chaboisseau E, Le Caer J P, Nicolas P, Delfour A. Covalent structure, synthesis and structure-function studies of mesentericin Y 10537, a defensive peptide from Gram-positive bacteria Leuconostoc mesenteroides. J Biol Chem. 1996;271:14421–14429. doi: 10.1074/jbc.271.24.14421. [DOI] [PubMed] [Google Scholar]
- 19.Fremaux C, Héchard Y, Cenatiempo Y. Mesentericin Y105 gene clusters in Leuconostoc mesenteroides Y105. Microbiology. 1995;141:1637–1645. doi: 10.1099/13500872-141-7-1637. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher N L, Sailer M, Niemczura W P, Nakashima T T, Stiles M E, Vederas J C. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry. 1997;36:15062–15072. doi: 10.1021/bi971263h. [DOI] [PubMed] [Google Scholar]
- 21.Geis A, Singh J, Teuber M. Potential of lactic streptococci to produce bacteriocin. Appl Environ Microbiol. 1983;45:205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson J T, Chopko A L, Van Wassenaar P D. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 24.Holo H, Nilssen Ø, Nes I F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hühne K, Holck A, Axelsson L, Kroeckel L. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology. 1996;142:1437–1448. doi: 10.1099/13500872-142-6-1437. [DOI] [PubMed] [Google Scholar]
- 26.Jack R W, Tagg J R, Ray B. Bacteriocins of Gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack R W, Wan J, Gordon J, Harmark K, Davidson B E, Hillier A J, Wettenhall R E H, Hickey M W, Coventry M J. Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl Environ Microbiol. 1996;62:2897–2903. doi: 10.1128/aem.62.8.2897-2903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser A L, Montville T J. Purification of the bacteriocin bavaricin MN and characterization of its mode of action against Listeria monocytogenes Scott A cells and lipid vesicles. Appl Environ Microbiol. 1996;62:4529–4535. doi: 10.1128/aem.62.12.4529-4535.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 30.Larsen A G, Vogensen F K, Josephsen J. Antimicrobial activity of lactic acid bacteria isolated from sour doughs; purification and characterization of bavaricin A, a bacteriocin produced by Lactobacillus bavaricus MI401. J Appl Bacteriol. 1993;75:113–122. doi: 10.1111/j.1365-2672.1993.tb02755.x. [DOI] [PubMed] [Google Scholar]
- 31.Larsen A G, Norrung B. Inhibition of Listeria monocytogenes by bavaricin A, a bacteriocin produced by Lactobacillus bavaricus MI401. Lett Appl Microbiol. 1993;17:132–134. [Google Scholar]
- 32.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers E W, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 34.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V G H, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 35.Nieto Lozano J C, Nissen Meyer J, Sletten K, Pelaz C, Nes I F. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol. 1992;138:1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- 36.Ojcius D M, Young J D-E. Cytolytic pore-forming proteins and peptides: is there a common structural motif? Trends Biochem Sci. 1991;16:225–229. doi: 10.1016/0968-0004(91)90090-i. [DOI] [PubMed] [Google Scholar]
- 37.Piva A, Headon D R. Pediocin A, a bacteriocin produced by Pediococcus pentosaceus FBB61. Microbiology. 1994;140:697–702. doi: 10.1099/00221287-140-4-697. [DOI] [PubMed] [Google Scholar]
- 38.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 39.Quadri L E N, Sailer M, Terebiznik M R, Roy K L, Vederas J C, Stiles M E. Characterization of the protein conferring immunity to the antimicrobial peptide carnobacteriocin B2 and expression of carnobacteriocins B2 and BM1. J Bacteriol. 1995;177:1144–1151. doi: 10.1128/jb.177.5.1144-1151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sailer M, Helms G L, Henkel T, Niemczura W P, Stiles M E, Vederas J C. 15N- and 13C-labeled media from Anabaena sp. for universal isotopic labeling of bacteriocins: NMR resonance assignments of leucocin A from Leuconostoc gelidum and nisin A from Lactococcus lactis. Biochemistry. 1993;32:310–318. doi: 10.1021/bi00052a039. [DOI] [PubMed] [Google Scholar]
- 43.Saucier L, Paradkar A S, Frost L S, Jensen S E, Stiles M E. Transcriptional analysis and regulation of carnobacteriocin production in Carnobacterium piscicola LV17. Gene. 1997;188:271–277. doi: 10.1016/s0378-1119(96)00822-0. [DOI] [PubMed] [Google Scholar]
- 44.Stoffels G, Nissen-Meyer J, Gudmundsdottir A, Sletten K, Holo H, Nes I F. Purification and characterization of a new bacteriocin isolated from a Carnobacterium sp. Appl Environ Microbiol. 1992;58:1417–1422. doi: 10.1128/aem.58.5.1417-1422.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tichaczek P S, Nissen-Meyer J, Nes I F, Vogel R F, Hammes W P. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from L. sake LTH673. Syst Appl Microbiol. 1992;15:460–468. [Google Scholar]
- 46.Tichaczek P S, Vogel R F, Hammes W P. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch Microbiol. 1993;160:279–283. doi: 10.1007/BF00292077. [DOI] [PubMed] [Google Scholar]
- 47.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol. 1996;178:3583–3593. doi: 10.1128/jb.178.12.3585-3593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Belkum M J, Hayema B J, Jeeninga R E, Kok J, Venema G. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991;57:492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Belkum M J, Stiles M E. Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl Environ Microbiol. 1995;61:3573–3579. doi: 10.1128/aem.61.10.3573-3579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venema K, Kok J, Marugg J D, Toonen M Y, Ledeboer A M, Venema G, Chikindas M L. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]