Abstract
Accurate prediction of antigen presentation by human leukocyte antigen (HLA) class II molecules is crucial for rational development of immunotherapies and vaccines targeting CD4+ T cell activation. So far, most prediction methods for HLA class II antigen presentation have focused on HLA-DR because of limited availability of immunopeptidomics data for HLA-DQ and HLA-DP while not taking into account alternative peptide binding modes. We present an update to the NetMHCIIpan prediction method, which closes the performance gap between all three HLA class II loci. We accomplish this by first integrating large immunopeptidomics datasets describing the HLA class II specificity space across all loci using a refined machine learning framework that accommodates inverted peptide binders. Next, we apply targeted immunopeptidomics assays to generate data that covers additional HLA-DP specificities. The final method, NetMHCIIpan-4.3, achieves high accuracy and molecular coverage across all HLA class II allotypeshttps://services.healthtech.dtu.dk/service.php?NetMHCIIpan-4.3.
NetMHCIIpan-4.3 closes the performance gap between HLA-DR, -DP and -DQ and allows for prediction of inverted peptide binders.
INTRODUCTION
Major histocompatibility complex (MHC) class II molecules, also known as human leukocyte antigen (HLA) class II in humans, are expressed on the surface of professional antigen-presenting cells and play a pivotal part in the function of the immune system by presenting antigenic peptides to CD4+ T cells (1, 2). Structurally, these molecules are heterodimers consisting of α and β chains encoded by three different loci (HLA-DR, HLA-DP, and HLA-DQ) that are among the most polymorphic genes in the human genome (3). The majority of these polymorphisms are clustered around the peptide binding domain formed by the α and β chain, giving rise to a broad range of peptide binding specificities (4).
While in HLA-DR, polymorphic variation is primarily defined by the β chain, in HLA-DP and HLA-DQ both α and β chains display polymorphism. Additional diversity can be provided by cis- and trans-dimerization, whereby distinct HLA-DP and HLA-DQ heterodimers are formed with α and β chains either encoded on the same chromosome (referred to as “cis”) or the opposite chromosomes (referred to as “trans”). Although the expression of trans-encoded HLA class II molecules has been confirmed by previous studies (5), evidence suggests that not every α and β chain pairing forms a stable heterodimer (6, 7).
Among all HLA class II molecules, DRB1 molecules have been investigated most extensively because of their established association with different conditions such as autoimmune disorders in particular (2), as well as cancer (8–10) and infectious diseases (11–13). The significance of other class II alleles (HLA-DRB3, 4, and 5; HLA-DQ; and HLA-DP) has been largely overlooked because of their relatively lower expression level and the strong linkage disequilibrium (LD) between these alleles and the corresponding HLA-DRB1, which has overshadowed the role these molecules play in disease susceptibility or protection and their contribution to HLA class II antigenic landscape. However, recent works have demonstrated the importance and function of these molecules in autoimmune diseases (1, 14–16) and transplantation (17–19) at both HLA expression and antigen presentation level that was previously underappreciated.
The HLA class II immunopeptidome of different antigen-presenting cells is inherently a complex mixture of peptides presented by HLA-DRB1, 3, 4, and 5; HLA-DQ; and HLA-DP. Given the distinct roles, these alleles play in the course of disease progression and treatment; using a method that can accurately predict antigen presentation across all HLA class II loci and reliably deconvolute the complete HLA class II immunopeptidome is of utmost importance for resolving the function of each of these molecules. To accomplish this, it is necessary to integrate large-scale, high-quality datasets covering a wide variety of class II molecules and their specificities.
Over the past decade, large immunopeptidome datasets have been acquired by liquid chromatography coupled with mass spectrometry (LC-MS/MS) (20–22). These data, often referred to as eluted ligand (EL) data, contain signals from different steps of HLA class II antigen presentation, such as antigen digestion, HLA loading of ligands, and transport to the cell surface. Hence, they have served as an essential means to enhance our understanding of the rules governing antigen processing and presentation and the development of in silico methods for prediction of HLA class II antigen binding and presentation (4, 23–25). Historically, most immunopeptidomics datasets have been generated by applying HLA-DR–specific antibodies followed by pan–HLA class II antibodies during the affinity purification step before the MS sequencing. In this approach, HLA-DR molecules are purified from the cell or tissue lysate using the HLA-DR–specific antibody while the pan–HLA class II antibody is applied as a means to capture the remaining class II molecules (HLA-DP and HLA-DQ) of the sample (23, 26). However, pan–HLA class II antibodies have demonstrated a rather poor specificity toward both DP and DQ (23, 25) and thereby an overall very low peptide yield for these loci. This has ultimately led to very few datasets describing DQ and DP molecules, resulting in subpar characterization of the role and rules for HLA class II antigen presentation for these molecules. Furthermore, despite the early focus on DR in immunopeptidomics assays, most studies have until recent years ignored the relevance of DRB3, 4, and 5. This meant that only DRB1 alleles were included in HLA typings of most samples used in immunopeptidomics assays. However, Kaabinejadian et al. (24) recently showed that DRB3, 4, and 5 have a substantial role in defining the DR ligandome, which underlines the importance of using full HLA typings to accurately characterize the immunopeptidome across HLA class II molecules (24).
A large variety of methods have been proposed for prediction of HLA class II antigen presentation [earlier methods are reviewed in (27) and recent methods include (25, 28, 29)]. The vast majority of these are trained on MS-immunopeptidome data. A critical challenge associated with the interpretation and analysis of this type of data is the fact that the data most often is multi-allelic (MA), meaning that each peptide in a given dataset can originate from the set of possible HLA molecules expressed in the given sample. This is in contrast to single-allelic (SA) data, e.g. binding affinity data or EL data derived from monoallelic cell lines, in which all peptides originate from only one HLA molecule. Therefore, interpretation and characterization of MA data require sorting the peptides into their most likely HLA restriction, a procedure known as motif deconvolution. Several methods have been proposed for this including MoDec (23) and NNAlign_MA (30). While achieving overall highly comparable results, the two methods differ fundamentally in how the motif deconvolution task is performed. MoDec performs the motif deconvolution in a per-dataset manner, challenging the identification of minority motifs characterized by limited peptide count. In contrast, NNAlign_MA performs the motif deconvolution in a pan-specific manner, leveraging information across all datasets, allowing us to boost the deconvolution accuracy for such minority motifs in situations where they are shared between multiple samples. This has been demonstrated to result in overall superior performance both in terms of the identified number of motifs and annotated peptides during motif deconvolution (30, 31).
Applying the NNAlign_MA framework, we have, in recent papers, demonstrated how the generation of high-quality MS HLA elution datasets combined with powerful and tailored machine learning frameworks can allow us to make profound advances within both the accuracy for prediction and fundamental understanding of the rules defining HLA class II antigen presentation. These advances include a transformed view on the contribution of the HLA-DR3, 4, and 5 molecules in the overall HLA-DR immunopeptidome (24), a confirmation of earlier proposed rules for pairing of HLA-DQA and HLA-DQB chains into functional molecules that greatly limit the diversity of the HLA-DQ functional space (25), and improved predictive accuracy and molecular coverage for both the HLA-DR and HLA-DQ loci.
In terms of the third HLA class II locus, HLA-DP, there have been major advances recently in the characterization of binding motifs. For example, van Balen et al. (32), who generated extensive datasets of eluted HLA-DP ligands, observed a binding motif for HLA-DPB1*05:01, which could be clustered into two separate motifs sharing a mirror symmetry. On the basis of this, they hypothesized that for this molecule, peptides can bind both in a canonical (N- to C-terminal) and inverted (C- to N-terminal) orientation. This inverted binding mode was later confirmed experimentally in independent studies (28, 33). In the context of T cell epitope prediction, Racle et al. (28) showed in a recent publication that by incorporating inverted binding prediction into their MixMHC2pred method, they were able to identify several epitopes bound inverted to DPA1*02:01-DPB1*01:01, which elicited a CD4+ immune response. This illustrates the importance of taking into account the different binding modes of HLA ligands when developing antigen presentation prediction methods.
However, despite these important advances, the accuracy of predictive methods for HLA class II antigen presentation, particularly for HLA-DP, remains low compared to that of HLA class I. Furthermore, it remains unclear to what degree current methods and datasets cover the set of prevalent and relevant HLA class II molecules and if there are still gaps remaining in our characterization and understanding of HLA class II binding specificities.
Here, we seek to address these issues by first closing the performance gap between DR and DQ/DP by integrating large high-quality immunopeptidomics datasets covering all three loci into the NetMHCIIpan machine learning framework and applying an updated version of NNAlign_MA that incorporates prediction of the peptide binding mode (forward versus inverted) into the method training. Using this method, we investigate the predictive performance across HLA-DR, HLA-DQ, and HLA-DP, and how prediction of the inverted binding mode affects the motif deconvolution. Next, we seek to expand the HLA coverage of the developed method by tailored data generation. For this purpose, we apply the developed machine learning model to identify HLA-DP molecules potentially missing from the covered specificity space. Next, we generate high-quality MS datasets for these molecules and illustrate how such tailored data generation improves coverage of the HLA class II specificity space.
RESULTS
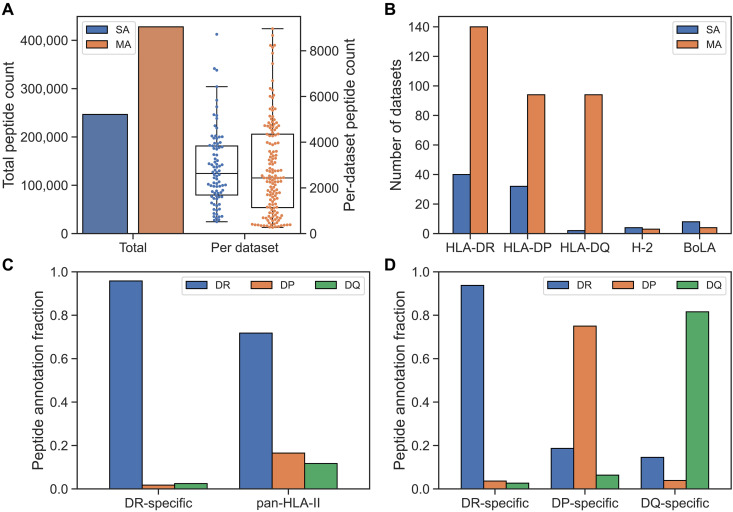
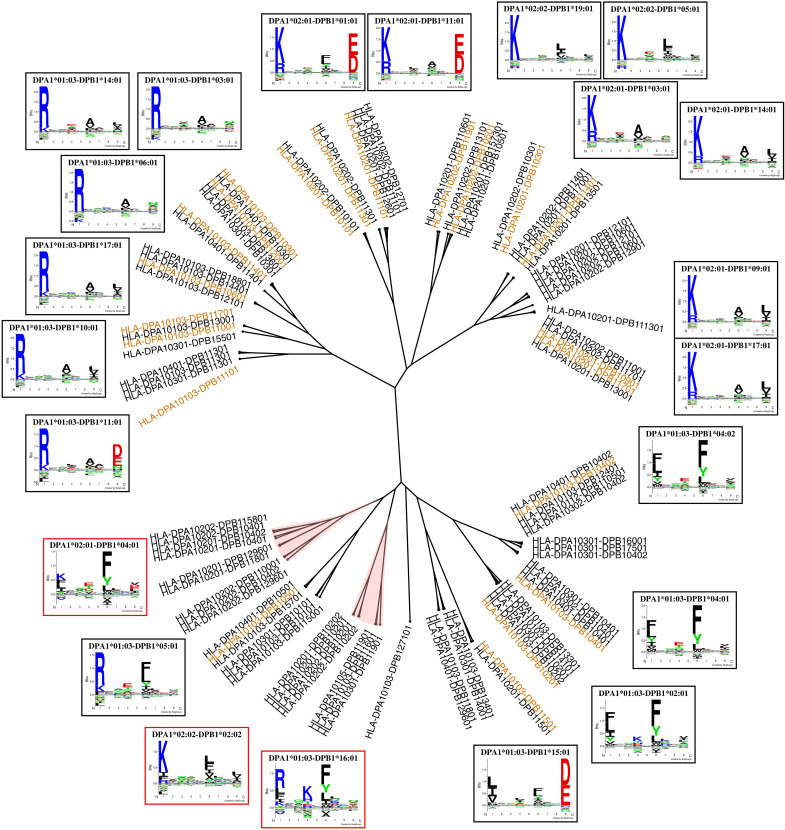
In this study, we set out to complete the journey of characterizing the rules of and developing prediction methods for HLA class II antigen presentation. To achieve this, we first compiled a comprehensive immunopeptidomics dataset, integrating the training data for the NetMHCIIpan-4.2 method including DQ-specific immunopeptidomics data covering 14 HLA-DQ molecules (25) with DP-specific data from van Balen et al. and related studies covering 19 HLA-DP molecules (32–34), along with additional data for HLA-DR (24) and BoLA-DR (see table S1 for more information on all the included datasets) (35). Figure 1A gives an overview of this combined dataset in terms of the SA and MA data categories, illustrating that the majority of the training data are derived from MA datasets. Furthermore, Fig. 1B displays the number of SA and MA datasets per locus, showing that the inclusion of the DP data from van Balen and colleagues (32–34) (termed “Balen_DP” from here on) gives HLA-DP a similar number of SA datasets to HLA-DR. The low number of SA datasets for HLA-DQ should be seen in light of the large number of DQ-specific MA datasets from NetMHCIIpan-4.2.
Fig. 1. Overview of immunopeptidomics datasets used in the study.
(A) Total peptide count for SA and MA data (left y axis), along with distribution of peptide count per SA and MA dataset (right y axis). (B) Number of SA and MA datasets per MHC class II locus in the training data. Here, H-2 and BoLA refer to the MHC class II loci in mice and cattle, respectively. (C) Peptide annotation fractions toward DR, DP, and DQ molecules in the motif deconvolution of our final method for the Racle__CD165 dataset. Two samples are shown: one generated with a DR-specific antibody and one with a pan–HLA-II antibody. (D) Peptide annotation fractions toward DR, DP, and DQ molecules in the motif deconvolution of our final method for the IHW09063 dataset. The fractions are shown for samples generated with DR-, DP-, and DQ-specific antibodies.
A key difference between the DQ- and DP-specific datasets and most prior immunopeptidome data available is that the former were generated using DQ- and DP-specific antibodies during the immunoprecipitation step before the MS/MS sequencing step, as described in the introduction. This is in contrast to earlier datasets where pan–HLA class II antibodies were applied in most cases, resulting in low peptide yield for DQ and DP due to the pan–HLA class II antibodies’ poor specificity toward these loci. This scenario changed drastically when anti–HLA-DP and anti–HLA-DQ specific antibodies were applied. In Fig. 1 (C and D), we give examples illustrating this. Here, the fraction of peptides assigned to DR, DP, and DQ in HLA motif deconvolutions is shown for two samples: One sample was handled using the conventional two-step immunoprecipitation pipeline where, first, a pan-DR antibody is applied followed by a pan–class II antibody, and the second was handled using individual DR, DQ, and DP locus-specific antibodies. Figure 1C illustrates the poor DP and DQ specificity of the pan–class II antibody resulting in very low peptide yield for these two loci. In contrast, Fig. 1D demonstrates how this limitation is resolved when applying the three locus-specific antibodies.
Using this expanded data with highly increased peptide yield for DP (and DQ), we trained prediction models for HLA antigen presentation using the NNAlign_MA framework earlier proposed for handling machine learning on MS HLA elution dataset from HLA heterozygous samples, and multilocus HLA expression (30). Before model training, the MS data were processed and enriched with artificial random natural negative peptides (as described in Materials and Methods). To accommodate the inverted binding mode recently observed for some DP molecules (28, 33), we used a modified version of the NNAlign_MA machine learning framework (see fig. S1), which includes an option to simultaneously predict both the binding core offset and the orientation (forward or inverted) of the peptide ligands (for details on the implementation, refer to Materials and Methods). We next trained three initial prediction models to investigate the impact of the Balen_DP data: one without the data and without peptide inversion (wo_Balen_DP), one with the data and without peptide inversion (w_Balen_DP), and one with the data using peptide inversion (w_inversion). These methods were then evaluated using cross-validation on a per-molecule and per-sample basis.
Performance impact of DP data and inverted binding mode
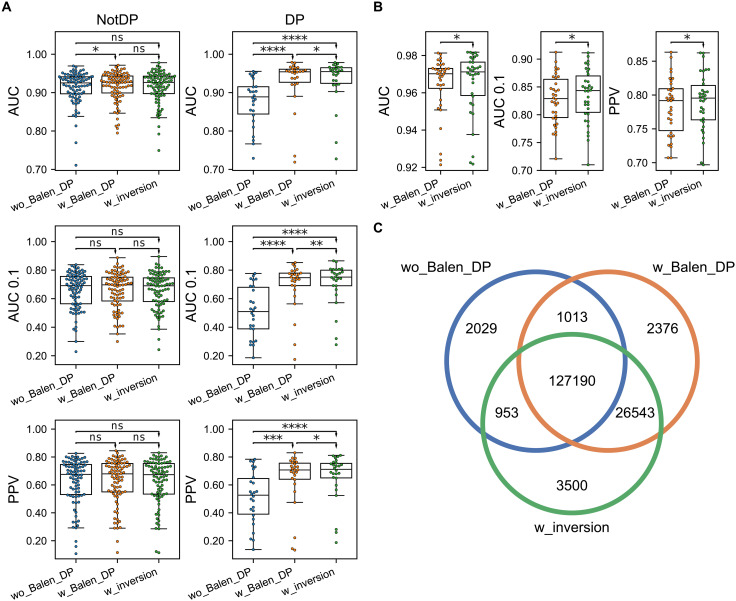
We assessed the predictive performances using three metrics, namely, area under the receiver operating characteristic (ROC) curve (AUC), area under the ROC curve integrated up to a false positive rate of 10% (AUC 0.1), and positive predictive value (PPV) (for more details refer to Materials and Methods). To illustrate how incorporation of the Balen_DP data affected the prediction of HLA class II antigen presentation, we first compared the performances of the two methods trained without peptide inversion (Fig. 2A). For non-DP molecules, a significant increase in AUC was observed in favor of the method with the Balen_DP data (N = 84, P < 0.05, one-tailed binomial test without ties), although both methods had overall similar performance across all metrics. This suggests that the information contained in the Balen_DP data has a limited impact on the method’s learning of other loci’s specificities. On the other hand, a significant performance increase for the method including these data was observed for DP in all metrics (N = 26, P < 0.001 in all metrics, one-tailed binomial tests without ties).
Fig. 2. Impact of HLA-DP data and inverted binding prediction on predictive performance.
Three methods are compared, namely, the method without the Balen_DP data (wo_Balen_DP), the method with the Balen_DP data (w_Balen_DP), and the method including the Balen_DP data trained with peptide inversion (w_inversion). (A) Performance calculated per HLA molecule. The rows correspond to the AUC, AUC 0.1, and PPV performance. The columns correspond to all non-DP molecules (NotDP, N = 84) and all DP molecules (DP, N = 26). (B) Performance per sample in the Balen_DP data for the two methods trained with these data (N = 34). (C) Venn diagram of shared and unique DP ligand annotations in the three methods. Results of one-tailed binomial tests are shown in (A) and (B), with arrowheads indicating the direction of the tests (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant).
When next investigating the model trained including peptide inversion (Fig. 2A), the method achieved similar overall performance on non-DP molecules compared to the other methods, indicating that the updated machine learning framework is able to maintain accuracy across all loci. Furthermore, we found a significant performance increase for DP in all metrics when comparing with the method without inversion including the Balen_DP data (N = 26, P < 0.05 in all metrics, one-tailed binomial tests without ties). Looking at the performance per sample in the Balen_DP data of the methods trained with these data (Fig. 2B), the model with inversion also had significantly improved performance across all three metrics (N = 34, P < 0.02 in all cases, one-tailed binomial tests without ties). This demonstrates that the use of peptide inversion during training has allowed for an improved identification of ligands in the Balen_DP data.
To further quantify this, we next investigated the ligands annotated toward DP across the three methods and visualized their overlaps as a Venn diagram in Fig. 2C. Overall, a total of 163,604 peptides were annotated toward DP with a percentile rank below 20 by at least one of the three methods (the threshold commonly applied to discard HLA irrelevant “contaminants”). Of these, 127,190 (78%) were predicted by all three methods. From the remaining annotations, 26,543 (72%) were predicted only by the two methods including the Balen_DP data, indicating a highly enriched identification of DP ligands in these methods. Investigating the annotations of these 26,543 peptides in the method without the Balen_DP data, the vast majority (~89.5%) were annotated as trash with percentile rank greater than 20, while 6.1% and 4.4% were annotated toward DR and DQ, respectively. In terms of the 3500 uniquely identified DP ligands in the method with inversion, around 61% was annotated as trash in the other two methods. When only considering the ligands that were predicted to bind inverted (1431 of 3500), the percentage of trash annotations in the methods without inversion was increased to 73 and 74% in the models with and without the Balen_DP data, respectively. These results indicate that by considering inversion, our method “rescues” a large proportion of ligands that would otherwise be predicted as nonbinders.
Inverted binding motifs
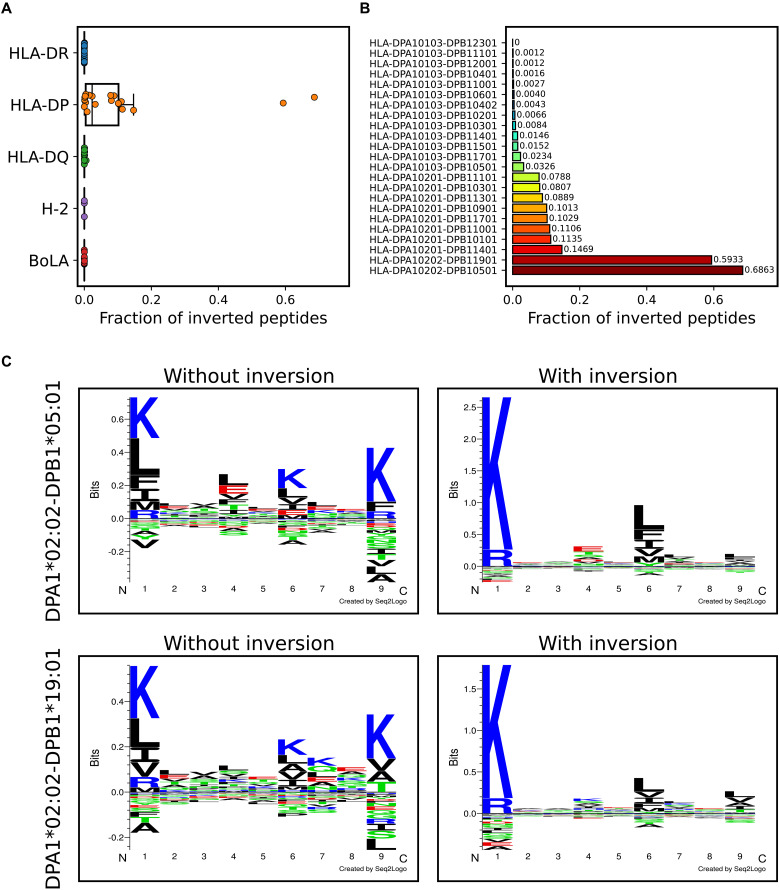
Next, we investigated the presence of inverted peptides across the different HLA class II molecules. Here, the cross-validated predictions from the model trained including inversions were used, and peptides with percentile rank greater than five were discarded to focus the analysis toward highly confident binders. To further reduce the number of noisy annotations for a given molecule, we only included peptides from samples in which at least 5% of the peptides were annotated toward a given locus and where the molecule had at least 5% of that locus’ annotations. Figure 3A gives the result of this analysis and shows the distribution of the percentage of inverted peptides across HLA molecules in the different loci. From this, we find that peptide inversion happens almost exclusively for HLA-DP. When looking at the inversion percentage per DP molecule (Fig. 3B), we see that all the molecules with at least 5% peptide inversion have either DPA1*02:01 or DPA1*02:02 as α chain. Furthermore, the remaining molecules with only a limited proportion of inverted ligands (less than 5%) all share the same DPA1*01:03 α chain. This suggests that the HLA-DP α chain, although bearing a limited specificity-determining role (see later), is the major determinant for the acceptance of the inverted peptide binding mode. These observations are in alignment with recent studies (28, 33), which have shown that widespread inversion of peptide binders is only observed for DP and only for DP molecules with certain α chains (namely, DPA1*02:01 and DPA1*02:02). Although molecules with DPB1*03:01 have not been observed in previous studies to have inverted binders (32), our method predicts a sizable percentage (~8.1%, 83 of 1029 peptides) of inversions for DPA1*02:01-DPB1*03:01.
Fig. 3. Peptide inversion enables accurate DP motif deconvolution.
(A) Proportion of peptide inversion across MHC class II loci. (B) Proportion of inverted peptides per HLA-DP molecule. (C) Identified sequence motifs of HLA-DPA1*02:02-DPB1*05:01 and HLA-DPA1*02:02-DPB1*19:01 for the methods trained without and with peptide inversion.
To illustrate the improved DP motif deconvolution by considering inverted peptides, sequence logos for DPA1*02:02-DPB1*05:01 and DPA1*02:02-DPB1*19:01 were shown for the models trained with and without inversion in Fig. 3C. Here, the peptides for the method without inversion were filtered as described above. We observe that for the method without inversion, the identified motifs are mirrored around the central position, with the K and R being present at both P1 and P9. In contrast, for the method with inversion, the motifs take into account the dual binding mode, resulting in more clear motifs with the K and R preference being present only at P1.
Correlation between deconvoluted and predicted motifs
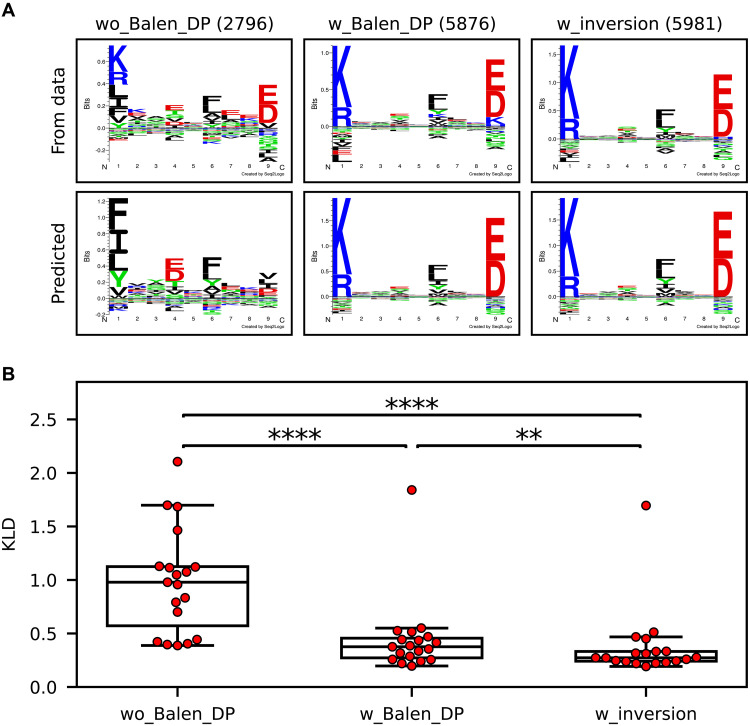
We next investigated the 19 HLA-DP molecules in the Balen_DP data in terms of correlation between the binding motifs obtained from motif deconvolution and the predicted motifs based on random natural peptides to assess to what degree the trained models were capable of learning the individual binding motifs in the MS elution data. Such an analysis is essential because motif deconvolution could appear accurate because of the nature of the task, i.e., placing peptides into a fixed set of buckets, but without the associated prediction model having learned the associated motifs. An example of this is shown in Fig. 4A for the molecule DPA1*02:01-DPB1*01:01. Here, the motifs from the deconvolution of the input data for the different models are all in overall high agreement. However, when looking at the predicted motifs estimated on the basis of the top 1% of 100,000 random natural peptides, the model trained without the Balen_DP data completely fails to learn the correct motif. Moreover, the method with inversion achieves the highest concordance between predicted and observed motifs, as it can position the “K” at P1 instead of P9 for the peptides predicted to bind inverted in the motif deconvolution.
Fig. 4. Correspondence between observed and predicted motifs.
(A) Motif deconvolution and predicted logos for DPA1*02:01-DPB1*01:01. The rows correspond to logos based on either the cross-validation predictions on the Balen_DP data (From data) or the top 1% of predictions on 100,000 random natural peptides (Predicted). The columns correspond to the methods trained without the Balen_DP data (wo_Balen_DP), with the Balen_DP data (w_Balen_DP), and with the Balen_DP data including peptide inversion (w_inversion). For the motif deconvolution logos, predicted contaminant peptides with percentile rank greater than 20 were excluded, and peptide counts are shown in parentheses above each logo. (B) KLD values between PSFMs obtained from motif deconvolution and prediction with random peptides as described above. Each point corresponds to a DP molecule from the Balen_DP data. The stars indicate the results from paired two-sided t tests (N = 19; **P < 0.01 and ****P < 0.0001).
To further quantify this, for each of the three prediction methods described earlier, we constructed position-specific frequency matrices (PSFMs) for each molecule in the Balen_DP data based on the cross-validation predictions across all samples in the Balen_DP data and from the top scoring random natural peptides as described above. Then, for each method, the two-sided Kullback-Leibler divergence (KLD) between the PSFM from the motif deconvolution and the corresponding predicted motif PSFM was then calculated (for details on this metric refer to Materials and Methods). This KLD metric can be interpreted as a “distance” between the binding motifs of two molecules, where lower values indicate more similar motifs. The result of this analysis is shown on Fig. 4B, illustrating that the model trained without the Balen_DP data has significantly higher KLD between the observed and predicted motifs when compared to the other methods (N = 19, P = 5.2 × 10−6 and P = 8.4 × 10−7, paired two-sided t tests). Furthermore, the model including inversion has significantly lower KLD values than the method without inversion (N = 19, P = 0.0077, paired two-sided t tests). An outlier is observed for all three methods, which corresponds to the molecule DPA1*02:01-DPB1*04:01. This molecule is only present in MA samples of the training data and was found to have consistently low peptide counts in all DP-heterozygous datasets, leading to motifs of lower quality compared to the remaining molecules (see more on this later).
Determinants of HLA-DP specificities
In contrast to HLA-DQ, where previous studies have shown that certain α and β chain combinations cannot form stable heterodimers because of structural constraints (25, 36), to our knowledge, no such constraints have been described for HLA-DP. Thus, for HLA-DP, any α and β chain can, in principle, pair to form a stable heterodimer. In light of this, we next investigated the motifs of DP-annotated peptides in DP-heterozygous samples for the model trained with peptide inversion. Here, HLA irrelevant “contaminant” peptides with percentile rank greater than 20 were removed. Figure S2 shows the DP sequence logos obtained from motif deconvolution of these samples. Here, we observe, in most cases, a similar motif for molecule pairs with the same β chain, which suggests that DP specificity is primarily driven by the β chain alone. To quantify this, we calculated the KLDs between the PSFMs of molecules sharing either the same α or β chain within each heterozygous sample and plotted the distribution of KLDs for the two groups (shown in fig. S3). From this analysis, the molecules with the same β chain were found to have significantly lower KLDs than the molecules with shared α chains (N = 27 motif pairs in each group, t = 3.93, P = 0.0003, two-sample unpaired t tests). This indicates that the β chain is the primary specificity defining element for DP molecules, with the α chain having a secondary role in terms of defining the given molecule’s ability to accommodate inverted peptide binders as described earlier.
Molecular coverage of HLA-DP
Given the increased predictive power of HLA class II achieved through integration of DP-specific immunopeptidomics data, we next wanted to investigate the molecular coverage of the models for HLA-DP. For this purpose, we focus only on the method trained without the Balen_DP data and the method trained with inversion including these data. First, we assessed for each method how many DP molecules were properly covered by the training data. Here, for each molecule, the data were filtered as described earlier by only including peptides with percentile rank less than 5 and only considering samples with at least 5% of annotations toward the DP locus and where the molecule received at least 5% of the locus’ peptides. Molecules with at least 50 peptides across all its included samples were then said to have peptide coverage. Here, the method without the Balen_DP data had peptide coverage of 13 DP molecules, while the method with these data had an increased coverage of 24 DP molecules.
Then, a functional coverage was estimated by considering the proportion of a reference set of DP molecules found within a distance of at most 0.05 to the molecules with peptide coverage. Briefly, this reference set was constructed by querying the Allele Frequency Net Database (37) for DP haplotype frequency data, resulting in a set of 167 DP haplotypes. The distance was here defined from the similarity between the HLA pseudo-sequences (see Materials and Methods), and the threshold of 0.05 was estimated on the basis of the distance at which the model trained without the Balen_DP data could reach optimal performance when evaluating on molecules not part of the method’s SA training data (fig. S4).
From this analysis, a significant increase in functional DP coverage was found (P < 0.0004, chi-square test), corresponding to 116 of 167 compared to 82 of 167 covered molecules for the methods with and without the Balen_DP data, respectively. Using the set of functionally covered molecules in each method, we next estimated the DP population coverage by summing their haplotype frequencies. Here, the method without the Balen_DP data had a DP population coverage of ~61%, while the method with the Balen_DP data had a population coverage of ~91%, indicating a highly boosted coverage as a result of including these data.
Next, using the method with inversion, we constructed a DP specificity tree based on the MHCCluster approach (38). Briefly, the list of 167 prevalent DP molecules was reduced to a list of 95 molecules with unique specificities based on the pseudo-sequence (39). Then, distances between molecules were estimated on the basis of correlations between prediction scores of a large set of random natural peptides, resulting in the tree shown in Fig. 5. Investigating the tree, we observe that the model had an overall wide coverage of the different DP specificities, with most branches having at least one molecule with peptide coverage (molecules with at least 50 confident peptide annotations). However, a few branches were found with poor coverage. One such branch includes DPA1*02:01-DPB1*04:01, for which the motif had not been learned properly by the prediction methods as described earlier. This molecule was present in seven samples in the training data, all of which are DP-heterozygous. In all of these samples, this molecule was assigned less than 5% of the DP annotations, resulting in an effective peptide count of 0 (see above). This lack of peptide annotations can either be biological or simply a result of the method not having learned the molecule’s specificity due to lack of high-quality data for this molecule.
Fig. 5. HLA-DP specificity tree.
Orange molecules have peptide coverage corresponding to at least 50 high-confidence ligands. Branches shaded in red correspond to noncovered specificities. Logos in red frames correspond to the identified noncovered molecules that would benefit from additional data.
Another example molecule with poor peptide coverage is DPA1*01:03-DPB1*16:01, which was only part of (homozygous) datasets by Nilsson et al. (25) and Kaabinejadian et al. (24) purified with DQ- and DR-specific antibodies, respectively, resulting in low yield of DP ligands. Therefore, analyzing this cell line with a DP-specific antibody during the purification process could potentially increase the amount of peptides covering this molecule substantially. Furthermore, we looked into the molecules in the tree that were not covered either by at least 50 high-confidence peptides or had a distance greater than 0.05 to the molecules with peptide coverage, yielding a set of 40 noncovered molecules. From these, the molecule with the highest haplotype frequency as identified from the Allele Frequency Net Database was DPA1*02:02-DPB1*02:02. This molecule was found at high frequency among Asians, while it was rare in other populations, resulting in a worldwide population frequency of 2.5%.
On the basis of the above analyses, we next generated MS-immunopeptidomics data using a DP-specific immunoprecipitation method from DP-homozygous cell lines expressing DPA1*01:03-DPB1*16:01, DPA1*02:01-DPB1*04:01, and DPA1*02:02-DPB1*02:02 (for more information on these data refer to Materials and Methods). The total number of peptides eluted from these cell lines, considering all 10- to 25-mer peptides identified at 1% false discovery rate, was 2423, 1797, and 2428 peptides, respectively. After filtering the datasets to remove posttranslational modifications and eliminating redundant peptides, the number of unique 12- to 21-mer peptides in each sample was reduced to 1550, 1259, and 1502, respectively, which were then used to retrain the method with inversion (enriched with random negatives generated as for the other datasets as described in Materials and Methods), to assess their impact on the DP motif deconvolution and molecular coverage.
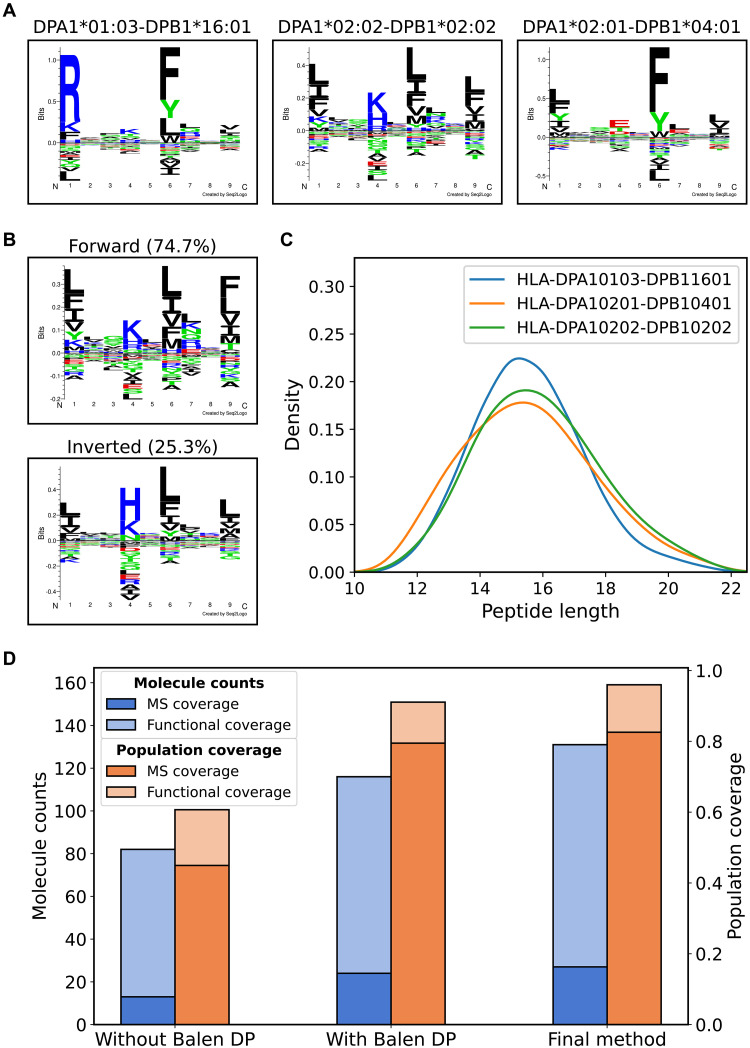
Analyzing the test-set predictions on these samples, the retrained method was able to annotate 678 (53.8%), 715 (47.6%), and 1062 (68.5%) peptides with percentile rank less than 20 toward DPA1*02:01-DPB1*04:01, DPA1*02:02-DPB1*02:02, and DPA1*01:03-DPB1*16:01 from each of the individual datasets, the motifs of which are shown in Fig. 6A. The remaining peptides were coeluted peptides predominantly assigned to HLA-DR and HLA-DQ (fig. S5). A high percentage of inverted binders was predicted for DPA1*02:02-DPB1*02:02 (25.3%), with the inverted peptides having a preference for histidine at P4 (Fig. 6B). Furthermore, the length distributions of the DP-annotated peptides were compared, confirming a normal distribution with a preference for 15-mer peptides, in agreement with the length preference for most HLA class II, including other DP, molecules (Fig. 6C).
Fig. 6. Integration of additional DP data yields improved molecular coverage.
(A) Identified motifs in the DP-specific datasets for DPA1*01:03-DPB1*16:01, DPA1*02:02-DPB1*02:02 (shown as a combined logo), and DPA1*02:01-DPB1*04:01. (B) The motif for DPA1*02:02-DPB1*02:02 shown as two logos corresponding to the forward and inverted binders. (C) Length distributions of the peptides annotated toward the three DP molecules (D) Counts and population coverage of molecules covered either by at least 50 high-confidence ligands (MS coverage) or by pseudo-sequence distance (Functional coverage) for the method trained without the Balen_DP data (Without Balen DP), with the Balen_DP data (With Balen DP), and the final method trained with the additional DP datasets generated in this study (Final method). The left y axis corresponds to the blue bars showing the molecule counts, and the right y axis corresponds to the orange bars showing the population coverage numbers.
Comparing the models trained with and without including these datasets revealed as expected an increased number of DP molecules with peptide coverage (27 compared to 24), resulting in an expanded functional coverage corresponding to 131 of 167 DP molecules (compared to 116 of 167 for the earlier model) and an expanded population coverage of 96% as illustrated in Fig. 6D. Furthermore, fig. S6 displays the DP specificity tree for the final retrained method, showing wide coverage of the specificity space as almost all branches have molecules with either peptide coverage or a distance less than 0.05 to a peptide-covered molecule.
Comparing motif deconvolutions before and after including additional DP data
When investigating the motif deconvolution of the datasets describing DPA1*02:01-DPB1*04:01, we saw a large increase in peptides annotated toward this molecule in the model trained with the additional data as expected (1268 total annotations across all datasets in the retrained method compared to 121 in the previous method). Furthermore, the motifs obtained for this molecule in the heterozygous samples are generally consistent with the motif identified in the additional dataset covering DPA1*02:01-DPB1*04:01 (fig. S7). However, despite this increase in annotations toward DPA1*02:01-DPB1*04:01 and its improved motif consistency, this molecule still had very low contribution in all eight DP-heterozygous datasets containing it (median DP annotation percentage was 4.5%; see fig. S8, A and B). In comparison, molecules with a similar overall peptide annotation count across all datasets in the cross-validation, such as DPA1*02:01-DPB1*03:01 and DPA1*01:03-DPB1*14:01, had much higher contributions.
Another molecule with highly similar behavior is DPA1*02:02-DPB1*04:01, which was found to have very limited DP annotation contribution (4.2%) in the heterozygous dataset Racle__PD42. Comparing this molecule with DPA1*02:01-DPB1*04:01, they were found to have identical pseudo-sequences except for one amino acid variation in the α chain. Given that the DP β chain was observed to be the main determinant of binding specificity as described earlier, we would thus have expected that incorporating additional data for DPA1*02:01-DPB1*04:01 in the training would have aided the deconvolution also toward this molecule. This was however not the case. These findings suggest that both DPA1*02:01-DPB1*04:01 and DPA1*02:02-DPB1*04:01 are poorly functional molecules, which results in these molecules’ limited contribution to the immunopeptidome in their given cell lines.
Impact of context encoding
Earlier work has demonstrated that incorporation of signals of antigen processing identified from residues flanking a given peptide sequence into the training of prediction models for antigen presentation results in substantial boost in performance (40). This form of peptide context has been incorporated into several prediction methods such as NetMHCIIpan and MixMHC2pred [for details on how peptide context is integrated in the NetMHCIIpan method refer to (40)]. However, these works have primarily been focused on HLA-DR, and thus, the impact of context encoding for DP and DQ has until now not been fully elucidated. To investigate the impact of context encoding across all three HLA class II loci, we retrained the method with inversion including peptide context. Here, we observed a significant performance increase for the method with peptide context across all three HLA-II loci and performance metrics (N = 42, N = 28, and N = 32 for DR, DP, and DQ, respectively; P < 2.0 × 10−6 in all cases, one-tailed binomial tests), with HLA-DQ demonstrating the largest improvement (3.5, 9.4, and 5.8 percentage point increase in AUC, AUC 0.1, and PPV, respectively; see fig. S9).
Rediscovering previous findings for DR and DQ
Given the broad coverage of HLA class II specificities in the training data of the final method, we wanted to take a step back and also analyze the method’s predictions for HLA-DR and HLA-DQ. More specifically, for HLA-DR, we wanted to investigate the relative contribution of DRB3, DRB4, and DRB5, and for HLA-DQ, the contribution of cis and trans heterodimers in shaping the DQ ligandome, both of which have been elucidated in recent studies by Kaabinejadian et al. and Nilsson et al., respectively (24, 25). In line with these studies, we first looked into the contribution of DRB3, 4, and 5 relative to DRB1 in samples with DRB1 and at least one secondary DR molecule. We did this by plotting the per-dataset distribution of DR peptide annotation fractions for each pair of molecules (i.e., DRB1 versus DRB3, DRB1 versus DRB4, and DRB1 versus DRB5). The result can be seen in fig. S10A, indicating that in samples with both DRB1 and DRB5, DRB5 had an overall high peptide contribution (median peptide fraction is 0.31). On the other hand, DRB4 had the lowest contribution, while DRB3 had less consistent contribution in agreement with the more polymorphic nature of the DRB3 gene compared to DRB4 and DRB5 (41, 42). These results align well with the findings by Kaabinejadian et al. (24), once again illustrating the importance of including the full HLA-DR typing during motif deconvolution to accurately characterize the DR ligandome.
Furthermore, we analyzed the motif deconvolution of DQ-heterozygous datasets and the role of HLA-DQ ɑ and β chain pairing in shaping the immunopeptidome. A reference list of DQɑ and DQβ chain heterodimers observed as haplotypes (36) was used to define a set of DQ molecules referred to as cis (see Table 1). Any other combination not observed as cis was referred to as “trans-only.” Then, for each DQ molecule in the heterozygous samples, we plotted the average per-dataset peptide annotation fraction, which is shown in fig. S10B. Here, in line with the findings by Nilsson et al. (25), we found that trans-only combinations had consistently low contribution in all DQ-heterozygous datasets, with a significantly higher contribution of cis variants found in DQ-MA datasets compared to trans-only variants (N = 18 and N = 12 for the two groups, t = 3.07, P < 0.005, two-sided unpaired t tests). However, we observed that cis variants present in DQ-SA datasets had an overall higher contribution than cis variants present in DQ-MA datasets, indicating a potential bias toward these molecules.
Table 1. List of HLA-DQ α and β chains that pair to form stable heterodimers.
| α Chain | β Chain |
|---|---|
| DQA1*01 |
DQB1*05 DQB1*06 |
|
DQA1*02 DQA1*03 DQA1*04 DQA1*05 DQA1*06 |
DQB1*02 DQB1*03 DQB1*04 |
While we cannot completely rule out that this bias toward the DQ-SA training data might have an impact on the method’s ability to annotate peptides to trans-only variants, our results are in perfect agreement with rules governing HLA-DQ αβ trans-pairing, which is dictated by the stability of the resulting heterodimer. Specifically, the rules indicate that structural constraints do not favor dimerization of DQA1*01 with DQB1*02, 03, and 04 alleles, all trans-only combinations, resulting in their lack of stability, inefficient assembly, and, therefore, loss of function (7, 36).
HLA class II mega-tree
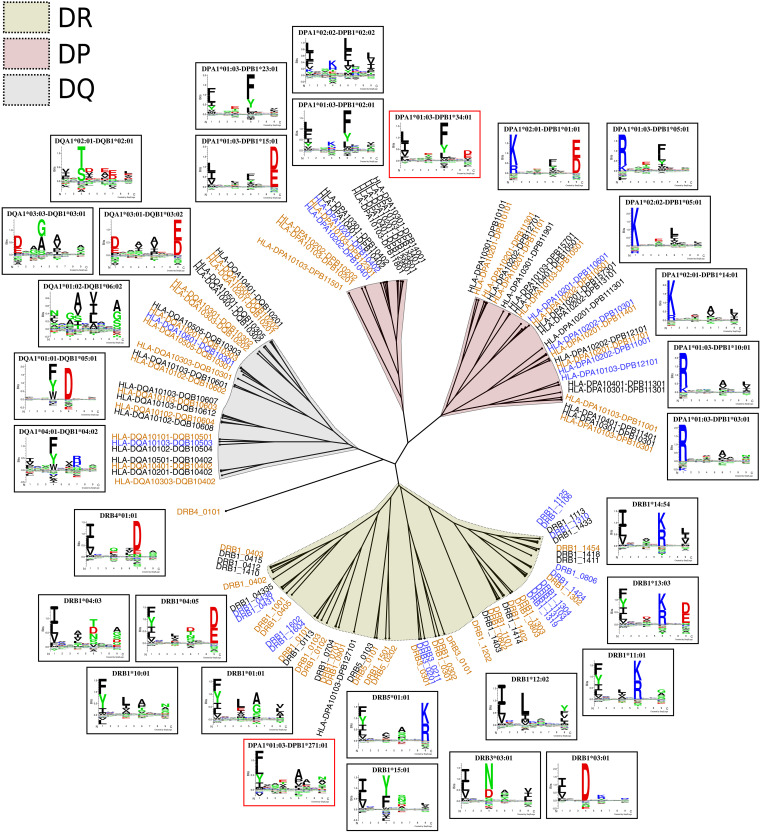
We next estimated the final model’s coverage of HLA-DR and HLA-DQ in a similar way to that of DP. A representative set of 123 DR molecules was retrieved from the IPD-IMGT/HLA database (see Materials and Methods) (43), and for these, we used the Allele Frequency Net Database to estimate their worldwide allelic frequencies. For DQ, we retrieved haplotype frequency data in the same way as for DP, keeping only a subset of 138 molecules known to form stable heterodimers (25, 36). In terms of the number of molecules with peptide coverage, the method covered 24 DQ molecules and 41 DR molecules. From the reference sets of DR and DQ molecules, 105 of 123 DR and 112 of 138 DQ molecules had a distance of at most 0.05 to the molecules with peptide coverage. These molecules corresponded to a population coverage of ~99% for both loci, indicating that the method has nearly full coverage of HLA class II.
To illustrate the overall HLA class II specificity space, we constructed a specificity tree combining HLA-DR, HLA-DP, and HLA-DQ molecules. The lists of molecules per locus used in the population coverage analysis were reduced on the basis of similarity between pseudo-sequences using the Hobohm 1 algorithm (for details refer to Materials and Methods) (44), yielding 53 DR molecules, 40 DP molecules, and 24 DQ molecules with unique specificities. Then, the MHCCluster method was used to construct an overall specificity tree for these molecules. The result of this is shown in Fig. 7. Overall, the molecules in each locus are grouped together in well-defined clusters. A few exceptions can be seen, such as DRB4*01:01, which was positioned alone close to the DQ branch, and DPA1*01:03-DPB1*271:01, which was clustered together with a set of DR molecules. The latter is likely due to this DP molecule being noncovered by our method both in terms of peptide coverage and pseudo-sequence distance.
Fig. 7. Combined specificity tree for HLA-DR, HLA-DP, and HLA-DQ.
Orange molecules have peptide coverage corresponding to at least 50 high-confidence ligands, and blue molecules have a pseudo-sequence distance of at most 0.05 to an orange molecule. Logos in red frames correspond to noncovered molecules.
NetMHCIIpan-4.3
The final prediction method, titled NetMHCIIpan-4.3, is available as a webserver at https://services.healthtech.dtu.dk/service.php?NetMHCIIpan-4.3. Predictions can be made for all MHC class II molecules of known sequence. Furthermore, the method can also include ligand context encoding. To reduce computational time, the method by default only considers peptide inversion for HLA-DP molecules. However, an option is included to consider inversion for all molecules, which could be useful when e.g., predicting binding toward molecules not characterized before.
CD4+ epitope benchmark
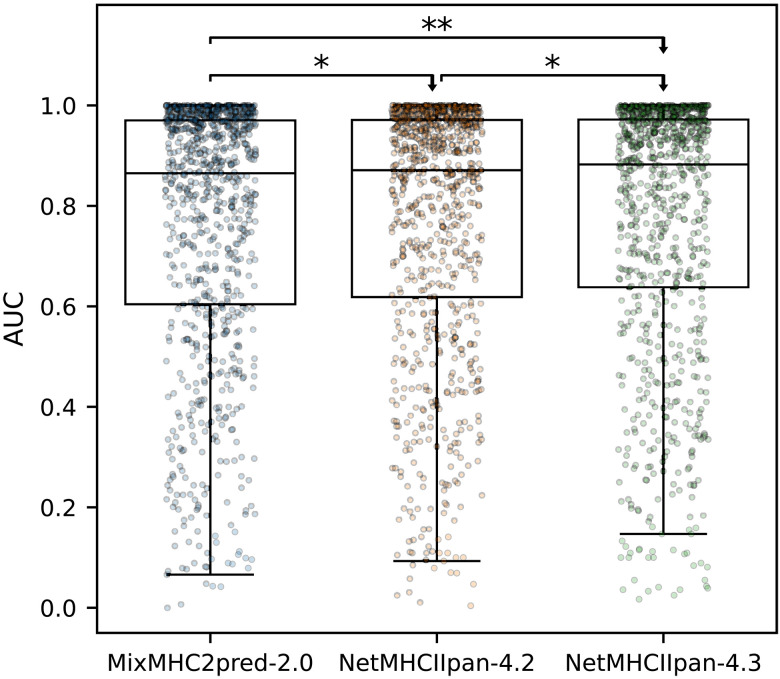
As a final validation of NetMHCIIpan-4.3, we benchmarked its performance in identification of CD4+ epitopes. Here, we compared our method with MixMHC2pred-2.0 (28), a recent update to the MixMHC2pred prediction algorithm, as well as NetMHCIIpan-4.2. In short, we queried the Immune Epitope Database (45) for positive CD4+ T cell epitopes of length 12-21 with known HLA restriction and source protein sequence. Then, for each entry of source protein, epitope, and HLA, we extracted all peptides of the same length as the epitope from the protein sequence, labeling the epitope as positive and the remaining peptides as negatives. To minimize bias, all peptides that were found in the EL training data of NetMHCIIpan-4.3 were removed. For more information on the benchmark data, refer to Materials and Methods. Using each of the included methods, we then predicted binding of each peptide to its given HLA molecule and calculated an AUC per source protein, epitope, and HLA entry. The benchmark result is illustrated in Fig. 8, showing that NetMHCIIpan-4.3 significantly outperforms MixMHC2pred-2.0 and NetMHCIIpan-4.2 (N = 842, P = 0.007 and P = 0.031, one-tailed binomial tests without ties).
Fig. 8. CD4+ epitope benchmark.
Each point is the AUC performance for an epitope, source protein, HLA combination. Results of one-tailed binomial tests without ties are shown, with arrowheads indicating the direction of the test (N = 842; *P < 0.05 and **P < 0.01).
DISCUSSION
Accurate prediction of antigen presentation for HLA class II is crucial for our understanding of the molecular mechanisms underlying the adaptive immune system. In recent years, the generation of large datasets of HLA ligands identified through MS in conjunction with powerful machine learning methods being developed has allowed researchers to make tremendous progress in improving the predictive accuracy for HLA class II. However, until now, most methods have been mainly focused on HLA-DR because of a lack of available high-quality data for especially HLA-DP. Here, we have presented NetMHCIIpan-4.3, which accurately predicts antigen presentation across the entire HLA class II specificity space. This was achieved by integrating high-quality immunopeptidomics datasets for HLA-DP, along with previous datasets describing the specificities of DR and DQ.
Our method was shown to achieve high and comparable performance across all HLA class II loci. Furthermore, the ability to perform accurate motif identification was improved by taking into account the inverted peptide binding mode, which was found to be restricted to a small set of DP molecules primarily defined by HLA-DPA1, in agreement with previous findings. By integrating additional datasets for rationally selected DP molecules, the method’s coverage of DP was extended even further, illustrating the importance of targeted immunopeptidomics assays for generating information-rich high-quality training data. NetMHCIIpan-4.3 was demonstrated to have a population coverage exceeding 96% for all three HLA class II loci based on haplotype frequencies obtained from the Allele Frequency Net Database. In relation to this coverage, one must be aware that the haplotype frequency data used to calculate the population coverage may be affected by a lack of available frequency data for all molecules in a wide range of demographics.
Investigating the pairing of HLA-DPA1 and HLA-DPB1 chains, we found that the β chain was the main determinant of HLA-DP specificities, meaning that most HLA-DP molecules with identical β chain share similar binding motifs, whereas this was not the case for molecules with identical α chain. Furthermore, studying the contribution of individual HLA-DP molecules to the immunopeptidome of heterozygous cell lines revealed that certain DP molecules, such as DPA1*02:01-DPB1*04:01 and DPA1*02:02-DPB1*04:01, share a limited contribution to the immunopeptidome of the given cell lines, suggesting that they might be either poorly functional or have low surface expression. The rs9277534A/G polymorphism at HLA-DPB1 3′ untranslated region (3′UTR) has been associated with transcriptional and cell surface HLA-DPB1 expression in different antigen-presenting cells including B cells. HLA-DPB1 surface expression is substantially higher in cells homozygous for rs9277534-G compared to those homozygous for rs9277534-A. The following DPB1 alleles (02:01, 02:02, 04:01, 04:02, and 17:01) have been reported to have rs9277534A at 3′UTR, which is correlated with reduced surface expression and is potentially one of the sources for the limited contribution of these two DP molecules to the class II immunopeptidome of the cells (46–48).
However, this cannot fully explain our observation, as in this study, we see high peptide counts toward molecules such as HLA-DPA1*02:01-DPB1*17:01 and HLA-DPA1*01:03-DPB1*04:01, underlining that the β chain is not the only factor that determines the level of contribution of an HLA-DP molecule to the immunopeptidome. In the DP heterodimer, positions 85 to 87 of the β chain, as well as the position 31 of the α chain, participate in the formation of the P1 pocket (49) of the peptide-binding region, emphasizing that both α and β chains play a critical role in antigen presentation.
Despite the high number of possible DPA1-DPB1 heterodimers, only a few HLA-DP haplotypes are dominant within most populations, suggesting a potential linkage disequilibrium (LD) between certain DPA1 and DPB1 alleles (50). DPB1 alleles are clustered into two groups on the basis of whether they carry GPM (DPB1*04:01, 02:01, and 04:02) or EAV (DPB1*01:01, 03:01, and 05:01) at positions 85 to 87. On the other hand, a single amino acid polymorphism at position 31 [methionine (M) or glutamine (Q)] divides DPA1 alleles into two groups, each of which can form a heterodimer with a DPB1 allele. The most frequent DPA1 alleles, DPA1*01:03, 02:01, and 02:02, contain M, Q, and Q at position 31, respectively.
When DPA1-DPB1 haplotypes are examined, a near-complete LD is observed between DPA1 alleles with 31Q (DPA1*02:01 and 02:02) and DPB1 alleles with the EAV sequence, while DPA1*01:03 (31M) is nearly always detected on a haplotype with DPB1 alleles carrying a GPM sequence (50). Although these rules are not as strict as the rules defined for DQ α and DQ β dimerization and certain exceptions have been observed among DP haplotypes, they can restrict the possibility of formation of all potential DPA1-DPB1 heterodimers. This may explain the distinct pattern of haplotype frequency observed for this locus, where a small number of DP haplotypes, as few as 15, account for over 80% of cumulative frequency in different populations (51) and is consistent with the idea that particular DP α and β chain combinations may not form a structurally stable heterodimer.
While the HLA-DPA1*02:01-DPB1*17:01 and HLA-DPA1*01:03-DPB1*04:01 molecules both follow this rule, which is in line with their high peptide counts, DPA1*02:01-DPB1*04:01 and DPA1*02:02-DPB1*04:01 molecules are both exceptions, where a DPA1 allele with Q at position 31 has formed a heterodimer with a DPB1 allele bearing GPM sequence at positions 85 to 87. Therefore, less stability of the heterodimer along with possibly low cell surface expression as described earlier might be the reasons why these molecules have a limited role in antigen presentation and contribution to the class II immunopeptidome. Further investigation, for instance, by analysis of immunopeptidome profiles in selected DP heterozygous cell lines is required to assess this and fully define rules associating HLA-DP α and β chain pairing with immunopeptidome contribution.
Last, the tool was benchmarked against a set of earlier developed tools in the context of prediction of known CD4+ epitopes as obtained from the IEDB and was demonstrated to achieve superior performance. It is important to underline that this benchmark is highly biased toward HLA-DR, and hence likely does not fully reflect the performance difference between the different methods that are expected to be most pronounced for DP (and DQ when compared to MixMHC2pred-2.0). Furthermore, we were able to confirm our previous findings regarding the contribution of DRB3, 4, and 5 (24), as well as cis– and trans–HLA-DQ heterodimers in shaping the class II immunopeptidome (25).
In summary, these results highlight the successful integration of high-quality MS EL data generated with loci-specific antibodies. This integration has effectively narrowed the performance gap between HLA-DP (and HLA-DQ) and HLA-DR, leading to enhanced motif characterizations across all three HLA class II loci. As a result, we can now assert that the specificity puzzle of HLA class II molecules has been fully resolved. These findings and the NetMHCIIpan-4.3 tool are expected to serve as a means to broaden our understanding of the molecular role of HLA class II in the initiation of cellular immunity in the context of infectious and autoimmune diseases beyond that of HLA-DR.
MATERIALS AND METHODS
Cell lines and antibody
A group of three homozygous B lymphoblastoid cell lines (BLCL) expressing low-frequency HLA-DP alleles were selected for generation of MS-immunopeptidomics data to further extend the coverage of the HLA-DP specificity tree. IHW09063 (DPA1*01:03-DPB1*16:01) and IHW09066 (DPA1*02:02-DPB1*02:02) were obtained from the International Histocompatibility Working Group Cell and DNA bank housed at the Fred Hutchinson Cancer Research Center, Seattle, WA (www.ihwg.org). IHW09208 (DPA1*02:01-DPB1*04:01) was a gift from J. Gumperz (University of Wisconsin-Madison). The Hybridoma for HLA-DP–specific monoclonal antibody (clone B7/21) was a gift from T. Purcell (Monash University). The anti-human HLA-DP monoclonal antibody was produced in house from the hybridoma cell line and used for affinity purification of total HLA-DP from the BLCLs.
The cells were grown in high-density cultures in roller bottles in complete RPMI medium (Gibco) supplemented with 15% fetal bovine serum (Gibco/Invitrogen Corp) and 1% 100 mM sodium pyruvate (Gibco). Cells were harvested from the suspension, washed with phosphate-buffered saline, and spun down at 4°C for 10 min. The cell pellets were immediately frozen in LN2 and stored at −80°C until downstream processing. The cell lines were subjected to high-resolution HLA typing (HLA-A, HLA-B, HLA-C; DRB1, 3, 4, and 5; DP; and DQ) before large-scale culture and data collection for authentication.
Isolation and purification of HLA-DP–bound peptides
HLA-DP molecules were purified from the cells by affinity chromatography using the anti-human HLA-DP–specific antibody (clone B7/21). Immunoaffinity columns were generated by coupling 1.5 mg of the purified antibody to 1 ml of matrix (CNBr-activated Sepharose 4 Fast Flow, Amersham Pharmacia Biotech, Orsay, France). Frozen cell pellets were pulverized using Retsch Mixer Mill MM400; resuspended in lysis buffer composed of tris (pH 8.0; 50 mM), IGEPAL 0.5%, NaCl (150 mM), and cOmplete protease inhibitor cocktail (Roche, Mannheim, Germany); and incubated at 4°C for 1 hour on a rotary shaker. Lysates were centrifuged in an Optima XPN-80 ultracentrifuge (Beckman Coulter, IN, USA) at 4°C for 90 min (200,000g). Cleared supernatants were filtered using a 0.45-μm filter and loaded on immunoaffinity columns overnight at 4°C. Columns were washed sequentially with 10 column volumes of wash buffers at pH:8.0 and were eluted with 0.2 M acetic acid. The HLA was denatured, and the peptides were isolated by adding glacial acetic acid (up to 10%) and heat (76°C for 10 min). The mixture of peptides and HLA-DP was subjected to reverse-phase high-performance liquid chromatography (RP-HPLC).
Fractionation of the HLA/peptide mixture by RP-HPLC
RP-HPLC was used to reduce the complexity of the peptide mixture eluted from the affinity column. First, the eluate was dried under vacuum using a CentriVap concentrator (Labconco, Kansas City, MO, USA). The solid residue was dissolved in 10% acetic acid and fractionated over a 150-mm-long Gemini C18 column, with pore size of 110 Å and particle size of 5 μm (Phenomenex, Torrance, CA, USA), using a Shimadzu Nexera instrument (Shimadzu Scientific Instruments, Pittsburg, PA, USA). An acetonitrile (ACN) gradient was run at pH 2 using a two-solvent system. Solvent A contained 2% ACN in water, and solvent B contained 5% water in ACN. Both solvent A and solvent B contained 0.1% trifluoroacetic acid. The column was preequilibrated at 2% solvent B. The sample was loaded on the column in a period of 18 min using a solvent system composed of 2% solvent B. Then, a two-segment gradient was run at a flow rate of 160 μl/min: four to 40% solvent B for 40 min, followed by 40 to 80% solvent B for 8 min (24). Fractions were collected in 2-min intervals using a Gilson FC 203B fraction collector (Gilson, Middleton, Wi, USA), and the ultraviolet absorption profile of the eluate was recorded at 215-nm wavelength.
Nano–LC-MS/MS analysis
Peptide-containing HPLC fractions were dried and resuspended in a solvent composed of 10% acetic acid, 2% ACN, and iRT peptides (Biognosys, Schlieren, Switzerland) as internal standards. Fractions were applied individually to an Eksigent nanoLC 415 nanoscale RP-HPLC (AB Sciex, Framingham, MA, USA), including a 5-mm-long, 350–μm–internal diameter ChromXP C18 trap column with 3-μm particles and 120-Å pores and a 15-cm-long ChromXP C18 separation column (internal diameter, 75 μm) packed with the same medium (AB Sciex, Framingham, MA, USA). An ACN gradient was run at pH 2.5 using a two-solvent system. Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in 95% ACN in water. The column was preequilibrated at 2% solvent B. Samples were loaded at a flow rate of 5 μl/min onto the trap column and run through the separation column at 300 nl/min with two linear gradients: ten to 40% B for 70 min, followed by 40 to 80% B for 7 min.
The column effluent was ionized using the NanoSpray III ion source of an AB Sciex TripleTOF 5600 quadruple time-of-flight mass spectrometer (AB Sciex, Framingham, MA, USA) with the source voltage set to 2400 V. Information-dependent analysis of peptide ions was acquired on the basis of a survey scan in the TOF-MS positive-ion mode over a range of 300 to 1250 mass/charge ratio (m/z) for 0.25 s. Following each survey scan, up to 22 ions with a charge state of 2 to 5 and intensity of at least 200 counts per second were subjected to collision-induced dissociation for MS/MS over a maximum period of 3.3 s. Selection of a particular ion m/z was excluded for 30 s after three initial MS/MS experiments. Dynamic collision energy was used to automatically adjust the collision voltage based on ion size and charge. PeakView Software version 1.2.0.3 (AB Sciex, Framingham, MA, USA) was used for data visualization.
Peptide data analysis
Peptide sequences were identified using PEAKS Studio 11 software (Bioinformatics Solutions, Waterloo, Canada) at a precursor mass error tolerance of 30 ppm and a fragment mass error tolerance of 0.02 Da. A database composed of Swiss-Prot Homo sapiens (taxon identifier 9606) and iRT peptide sequences was used as the reference for database search. Variable posttranslational modifications including acetylation, deamidation, pyroglutamate formation, oxidation, sodium adducts, phosphorylation, and cysteinylation were included in database search. Identified peptides were further filtered at a peptide false discovery rate of 1% using PEAKS decoy-fusion algorithm.
Training data
We started out by collecting binding affinity and EL datasets from previous publications, including the training data for NetMHCIIpan-4.2 (25), NetBoLAIIpan-1.0 (35), and additional HLA-DR peptide ligands from Kaabinejadian et al. (24), as well as a small set of additional unpublished BoLA EL data (provided by S. Wilkowsky; sample id: HFX231_IPP_RP_BBOVIS). These data were combined with DP EL data from van Balen et al. and related studies (32–34) consisting of 34 samples covering a total of 19 DP molecules. Of these 34 datasets, 30 are SA datasets and 4 are MA datasets.
In addition, we included immunopeptidomics data made specifically for this study, which was generated from three different cell lines (IHW09063, IHW09066, and IHW09208). For the IHW09063 cell line, we also included sets of DR and DQ affinity-purified peptides identified with PEAKS Studio 11 from the same samples used by Nilsson et al. (25) and Kaabinejadian et al. (24). These peptide sets were used instead of the previous peptide sets identified in the aforementioned publications.
Aside from the datasets from the works of Nilsson et al. and Fisch et al. (25, 35), which were already preprocessed, all datasets were filtered as described earlier to exclude possible contaminant peptides and MHC class I binders, resulting in peptides of length 12-21 (24). These peptides were then mapped against the human (or cattle in the case of the BoLA data) proteome to define source protein context. Here, around 2.3% of peptides with no reference match were discarded. The EL data were then enriched with random natural peptides assigned as negatives. This enrichment was done in a per–sample id manner by uniformly sampling 12-21 mer peptides, such that the amount of negatives was equal to five times the number of peptides for the most prevalent peptide length in the given sample.
The final EL dataset includes a total of 675,364 positive and 6,886,973 negative peptides from a total of 237 EL samples, covering a total of 142 MHC class II molecules. Furthermore, the binding affinity data consists of 129,110 data points covering 80 class II molecules. An overview of all the datasets used in the study in terms of peptide counts, HLA types, dataset type (BA and EL) and processing method (preprocessed or filtered) are provided in table S1. The complete dataset was partitioned into five subsets for use in cross-validation using the common-motif approach, such that peptides with a subsequence overlap of nine or more amino acids were placed in the same partition (52).
Training of prediction models using NNAlign_MA
To accommodate the DP data from van Balen et al. (32–34) containing inverted binders, we developed an extension of the NNAlign_MA method that includes an option to consider peptide inversion during training and prediction. With this option, inverted binding can be predicted by reversing the peptide sequence and its encoding in the network input layer. Furthermore, the peptide binding mode is encoded in the input layer with either 0 or 1 for forward and inverted binding, respectively.
When training with inversion, we apply an initial burn-in period of two epochs in which no peptides are inverted, after which inversion is considered in the remaining epochs. At the beginning of each epoch with inversion allowed, the optimal binding mode (forward versus inverted) is assigned to each peptide by selecting the mode that yields the highest prediction score from the network. Afterward, the annotated binding modes are used in the backpropagation. When making predictions with the trained network ensembles, each peptide’s inversion state (inverted or noninverted) is reported as the majority vote between the networks. In case of ties, the noninverted mode is reported.
Each trained method is an ensemble consisting of 100 models corresponding to two different architectures with either 100 or 120 neurons in the single hidden layer, five different cross-validation folds, and 10 random initializations. All models were trained with stochastic gradient descent using backpropagation. Furthermore, each training was performed for 300 epochs without early stopping using a learning rate of 0.05. The training included a burn-in period of 20 epochs in which only SA data were used to update the model parameters. The remaining epochs included both SA and MA data. Furthermore, a P1 burn-in period was used, in which only peptides with one of the following amino acids in the first position of their binding core are considered: ILVMFYWRK. This P1 alphabet was extended with R and K compared to that in the original NNAlign_MA method to accommodate the P1 anchor amino acid preference in some DP binding motifs. For the models trained without peptide inversion, a standard P1 burn-in of two epochs was used. On the other hand, in the models trained with inversion, the P1 burn-in was extended to four epochs for the method to learn the P1 amino acid preferences for both forward and inverted peptide binders.
Initially, we trained three prediction methods to investigate the impact of the DP data from van Balen and colleagues (32–34) on predictive performance. Here, one method was trained without these data and without inversion, and two models were trained including the data, either without or with using peptide inversion. An additional method, entitled NetMHCIIpan-4.3, was trained with inversion including the DP data generated for this study in the training. Furthermore, this final model was also trained with peptide context. Here, peptide context is defined as three residues flanking the peptide’s N and C termini within the source protein, as well as the first three residues from the peptide’s N and C termini, respectively, all concatenated into a 12-mer amino acid sequence (40).
Performance evaluation
Performance was evaluated using cross-validation by concatenating the five EL cross-validation prediction folds for each method and then calculating the performance on a per–HLA molecule or per–sample id basis. For the per-molecule evaluation, only HLA molecules with at least 25 positive peptides in all methods were included, this to ensure a level of certainty in the calculated metrics. The cross-validation performance was then evaluated in terms of AUC, AUC 0.1, and PPV. Here, PPV is defined as the number of true positives in the top N predictions for a given sample, where N is the total number of positives for the given sample.
Correspondence between motifs
To assess the similarity between sequence motifs, we used PSFMs to represent a given set of peptide binding cores. Then, each PSFM was represented as a single vector by concatenating each of the nine positions’ vectors with 20 values. A symmetric KLD between two vectors a and b was then calculated using the following formula
| (1) |
Here, only positions where each vector’s value was greater than 0 were included to avoid division by 0.
Pseudo-sequence distance metric
Distances between HLA class II molecules were estimated using the following relation
| (2) |
where s(X,Y) is the summed BLOSUM50 similarity between molecules X and Y in terms of their pseudo-sequences (53). Here, the pseudo-sequence refers to a set of 34 polymorphic residues in the HLA sequence (15 from the α chain and 19 from the β chain) concatenated into a single sequence (39).
Allelic and haplotype frequencies
For DR, a reference set of 123 molecules was defined by considering DRB molecules with full-length sequence data as obtained from the IPD-IMGT/HLA database (retrieved April 2022) (43), filtered to only keep molecules with unique HLA pseudo-sequences (39). For pseudo-sequences mapping to multiple molecules, the molecule with the lowest second-field number in the allele name (e.g., DRB1*09:01) was chosen. On the basis of this list, worldwide allelic frequencies were estimated by querying the Allele Frequency Net Database (37). In short, frequencies were obtained for each HLA-DR allele from an average over worldwide populations of size 100 and above, weighted by population size capped at the maximum values of 10,000.
For each of the DP and DQ loci, we retrieved high-resolution haplotype frequency data from the Allele Frequency Net Database, in populations of size 100 and above. For HLA-DQ, only haplotypes corresponding to known stable DQ heterodimers (listed in Table 1) were included (36). Next, capping the maximum population size at 1000, we calculated the weighted average haplotype frequencies on the basis of the population sizes. This resulted in lists of 167 DP and 138 DQ haplotypes.
Specificity trees
Each specificity tree was based on predictions for a set of 100,000 random 13-17 mer peptides, which were done for each included molecule. For the given set of molecules to include in the tree, the top 1% of random peptides in terms of prediction score was retrieved for each molecule. Then, the union of these top 1% peptide sets was used for the specificity tree calculation using the MHCCluster method (38). The method builds a set of 100 distance matrices using bootstrapping, each calculated by pairwise correlations between the prediction scores for each pair of molecules, and summarizes these matrices into a consensus tree. All trees were drawn using the Iroki tree viewer (54).
For the HLA-DP specificity trees, the set of DP haplotype molecules was reduced to a list of molecules with unique pseudo-sequences. Then, each pseudo-sequence was mapped to a molecule name matching that sequence. By default, any DP molecule in the DP data from the work of van Balen and colleagues (32–34) was used to represent a given pseudo-sequence; otherwise, the name of the molecule with the highest haplotype frequency among the possible candidates was chosen. This resulted in a set of 96 DP molecules, which were included in the DP specificity trees.
For the final HLA-II specificity tree, the sets of DR, DP, and DQ molecules were first sorted individually by their haplotype (or allelic in the case of DR) frequencies in descending order. Then, the Hobohm 1 algorithm (44) was used to reduce each reference list to a shorter list of molecules. The algorithm goes through the list of sequences and keeps track of a list of “unique” sequences, and only adds a new sequence to this list if it is not similar to any of the current sequences in the list. Here, a pseudo-sequence similarity threshold of 0.95 was used, meaning that any pseudo-sequence that had a pseudo-sequence distance of less than 0.05 to any sequence in the unique list was discarded. By sorting the initial lists on the basis of frequencies, the most frequent molecules are placed at the top and are thus more likely to appear in the final reduced lists. This resulted in sets of 53 DR, 40 DP, and 26 DQ molecules, which were included in the overall HLA-II specificity tree.
CD4+ epitope benchmark
We queried the Immune Epitope Database (45) for positive CD4+ T cell epitopes of length 12-21 without posttranslational modifications and with full four-digit HLA typing. Here, only epitope, HLA pairs with at least three positive assays were included. Furthermore, only epitopes with known source protein ID and which were not found in a negative assay were considered. The source proteins from which negative peptides were generated were downloaded from the UniProt database (55). For each {epitope, allele, protein} combination in which the epitope could be mapped to the protein sequence, all overlapping peptides with the same length as the epitope were extracted from the protein sequence, and all peptides beside the epitope were labeled as negatives. One combination was discarded, as its allele (DRB1*07:03) was not supported by MixMHC2pred. Furthermore, all peptides that were found in the EL training data of NetMHCIIpan-4.3 were not included in the evaluation. The final benchmark dataset consisted of 842 {epitope, allele, protein} combinations covering 40 HLA-DR, 13 HLA-DQ, and 4 HLA-DP molecules. As the positive epitopes are usually tested for T cell response individually, the peptide context information is not relevant in this benchmark, and, therefore, all methods were run without inclusion of peptide context encoding.
Data visualization
Data visualizations in the manuscript figures were created in Python 3.10.0 using the Matplotlib (version 3.7.2) and seaborn (version 0.12.2) libraries. Sequence logos were generated with Seq2Logo-2.0 (56).
Statistical analysis
All statistical tests were made in Python 3.10.0 using the SciPy library (version 1.11.1), applying a standard significance level of 0.05 in each test. In the cross-validation and benchmark performance evaluation, one-tailed binomial tests were applied. In these tests, the alternative hypothesis is that one method is more likely to have better performance on a given dataset/molecule than the other method.
Acknowledgments
We would like to thank S. Wilkowsky for providing one of the BoLA samples used in the training data of the prediction methods. We thank R. Buchli (Pure Protein LLC) for providing the B7/21 affinity columns for this study. We also thank S. Cate (University of Oklahoma Health Sciences Center) and S. Osborn (Pure MHC LLC) for HLA typing of the BLCLs and helpful discussions.
Funding: Research reported in this publication was supported in part by the National Cancer Institute (NCI), under award number U24CA248138, and the National Institute of Allergy and Infectious Diseases (NIAID), under award number 75N93019C00001.
Author contributions: Conceptualization: M.N. Methodology: M.N., S.K., H.Y., and J.B.N. Investigation: M.N., J.B.N., S.K., and H.Y. Visualization: J.B.N. Supervision: M.N., M.G.D.K., P.B., and W.H.H. Writing (original draft): J.B.N., M.N., and S.K.. Writing (review and editing): All authors.
Competing interests: S.K. is an employee at Pure MHC LLC. The remaining authors declare no competing interests.
Data and materials availability: The MS proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE (57) partner repository with the dataset identifier PXD044810. Furthermore, the training and evaluation data for NetMHCIIpan-4.3 is available on the NetMHCIIpan-4.3 webserver at https://services.healthtech.dtu.dk/service.php?NetMHCIIpan-4.3. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Legend for table S1
Other Supplementary Material for this manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.S. Tsai, P. Santamaria, MHC class II polymorphisms, autoreactive T-cells, and autoimmunity. Front. Immunol. 4, 321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. T. Arango, C. Perricone, S. Kivity, E. Cipriano, F. Ceccarelli, G. Valesini, Y. Shoenfeld, HLA-DRB1 the notorious gene in the mosaic of autoimmunity. Immunol. Res. 65, 82–98 (2017). [DOI] [PubMed] [Google Scholar]
- 3.M. Van Lith, R. M. McEwen-Smith, A. M. Benham, HLA-DP, HLA-DQ, and HLA-DR have different requirements for invariant chain and HLA-DM. J. Biol. Chem. 285, 40800–40808 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.B. Reynisson, C. Barra, S. Kaabinejadian, W. H. Hildebrand, B. Peters, M. Nielsen, Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J. Proteome Res. 19, 2304–2315 (2020). [DOI] [PubMed] [Google Scholar]
- 5.S. Tollefsen, K. Hotta, X. Chen, B. Simonsen, K. Swaminathan, I. I. Mathews, L. M. Sollid, C. Y. Kim, Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule. J. Biol. Chem. 287, 13611–13619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.W. W. Kwok, G. T. Nepom, Structural and functional constraints on HLA class II dimers implicated in susceptibility to insulin dependent diabetes mellitus. Baillieres Clin. Endocrinol. Metab. 5, 375–393 (1991). [DOI] [PubMed] [Google Scholar]
- 7.W. W. Kwok, S. Kovats, P. Thurtle, G. T. Nepom, HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J. Immunol. 150, 2263–2272 (1993). [PubMed] [Google Scholar]
- 8.M. Kilian, R. Sheinin, C. L. Tan, M. Friedrich, C. Krämer, A. Kaminitz, K. Sanghvi, K. Lindner, Y. C. Chih, F. Cichon, B. Richter, S. Jung, K. Jähne, M. Ratliff, R. M. Prins, N. Etminan, A. von Deimling, W. Wick, A. Madi, L. Bunse, M. Platten, MHC class II-restricted antigen presentation is required to prevent dysfunction of cytotoxic T cells by blood-borne myeloids in brain tumors. Cancer Cell 41, 235–251 (2023). [DOI] [PubMed] [Google Scholar]
- 9.I. A. Park, S. H. Hwang, I. H. Song, S. H. Heo, Y. A. Kim, W. S. Bang, H. S. Park, M. Lee, G. Gong, H. J. Lee, Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLOS ONE 12, e0182786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D. B. Johnson, M. V. Estrada, R. Salgado, V. Sanchez, D. B. Doxie, S. R. Opalenik, A. E. Vilgelm, E. Feld, A. S. Johnson, A. R. Greenplate, M. E. Sanders, C. M. Lovly, D. T. Frederick, M. C. Kelley, A. Richmond, J. M. Irish, Y. Shyr, R. J. Sullivan, I. Puzanov, J. A. Sosman, J. M. Balko, Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 7, 10582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.N. T. P. Lan, M. Kikuchi, V. T. Q. Huong, D. Q. Ha, T. T. Thuy, V. D. Tham, H. M. Tuan, V. Van Tuong, C. T. P. Nga, T. Van Dat, T. Oyama, K. Morita, M. Yasunami, K. Hirayama, Protective and enhancing HLA alleles, HLA-DRB1*0901 and HLA-A*24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS Negl. Trop. Dis. 2, e304 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. J. Dunstan, N. T. Hue, B. Han, Z. Li, T. T. B. Tram, K. S. Sim, C. M. Parry, N. T. Chinh, H. Vinh, N. P. H. Lan, N. T. V. Thieu, P. V. Vinh, S. Koirala, S. Dongol, A. Arjyal, A. Karkey, O. Shilpakar, C. Dolecek, J. N. Foo, L. T. Phuong, M. N. Lanh, T. Do, T. Aung, D. N. Hon, Y. Y. Teo, M. L. Hibberd, K. L. Anders, Y. Okada, S. Raychaudhuri, C. P. Simmons, S. Baker, P. I. W. De Bakker, B. Basnyat, T. T. Hien, J. J. Farrar, C. C. Khor, Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat. Genet. 46, 1333–1336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.V. Grimaldi, L. Sommese, A. Picascia, A. Casamassimi, F. Cacciatore, A. Renda, P. De Rosa, M. L. Montesano, C. Sabia, C. Fiorito, G. De Iorio, C. Napoli, Association between human leukocyte antigen class I and II alleles and hepatitis C virus infection in high-risk hemodialysis patients awaiting kidney transplantation. Hum. Immunol. 74, 1629–1632 (2013). [DOI] [PubMed] [Google Scholar]
- 14.D. Stepniak, M. Wiesner, A. H. De Ru, A. K. Moustakas, J. W. Drijfhout, G. K. Papadopoulos, P. A. Van Veelen, F. Koning, Large-scale characterization of natural ligands explains the unique gluten-binding properties of HLA-DQ2. J. Immunol. 180, 3268–3278 (2008). [DOI] [PubMed] [Google Scholar]
- 15.M. Tafti, H. Hor, Y. Dauvilliers, G. J. Lammers, S. Overeem, G. Mayer, S. Javidi, A. Iranzo, J. Santamaria, R. Peraita-Adrados, J. L. Vicario, I. Arnulf, G. Plazzi, S. Bayard, F. Poli, F. Pizza, P. Geisler, A. Wierzbicka, C. L. Bassetti, J. Mathis, M. Lecendreux, C. E. H. M. Donjacour, A. Van Der Heide, R. Heinzer, J. Haba-Rubio, E. Feketeova, B. Högl, B. Frauscher, A. Benetó, R. Khatami, F. Cañellas, C. Pfister, S. Scholz, M. Billiard, C. R. Baumann, G. Ercilla, W. Verduijn, F. H. J. Claas, V. Dubois, J. Nowak, H. P. Eberhard, S. Pradervand, C. N. Hor, M. Testi, J. M. Tiercy, Z. Kutalik, DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep 37, 19–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.X. Hu, A. J. Deutsch, T. L. Lenz, S. Onengut-Gumuscu, B. Han, W. M. Chen, J. M. M. Howson, J. A. Todd, P. I. W. De Bakker, S. S. Rich, S. Raychaudhuri, Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, 898–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. A. Fernández-Viña, J. P. Klein, M. Haagenson, S. R. Spellman, C. Anasetti, H. Noreen, L. A. Baxter-Lowe, P. Cano, N. Flomenberg, D. L. Confer, M. M. Horowitz, M. Oudshoorn, E. W. Petersdorf, M. Setterholm, R. Champlin, S. J. Lee, M. De Lima, Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood 121, 4603–4610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.P. van Balen, S. A. P. Van Luxemburg-Heijs, M. Van De Meent, C. A. M. Van Bergen, C. J. M. Halkes, I. Jedema, J. H. F. Falkenburg, Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Transplantation 101, 2850–2854 (2017).28252558 [Google Scholar]
- 19.T. Meurer, P. Crivello, M. Metzing, M. Kester, D. A. Megger, W. Chen, P. A. van Veelen, P. van Balen, A. M. Westendorf, G. Homa, S. E. Layer, A. T. Turki, M. Griffioen, P. A. Horn, B. Sitek, D. W. Beelen, J. H. F. Falkenburg, E. Arrieta-Bolaños, K. Fleischhauer, Permissive HLA-DPB1 mismatches in HCT depend on immunopeptidome divergence and editing by HLA-DM. Blood 137, 923–928 (2021). [DOI] [PubMed] [Google Scholar]
- 20.A. W. Purcell, S. H. Ramarathinam, N. Ternette, Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 14, 1687–1707 (2019). [DOI] [PubMed] [Google Scholar]
- 21.A. Nelde, D. J. Kowalewski, S. Stevanović, Purification and identification of naturally presented MHC class I and II ligands. Methods Mol. Biol. 1988, 123–136 (2019). [DOI] [PubMed] [Google Scholar]
- 22.F. Marino, C. Chong, J. Michaux, M. Bassani-Sternberg, High-throughput, fast, and sensitive immunopeptidomics sample processing for mass spectrometry. Methods Mol. Biol 1913, 67–79 (2019). [DOI] [PubMed] [Google Scholar]
- 23.J. Racle, J. Michaux, G. A. Rockinger, M. Arnaud, S. Bobisse, C. Chong, P. Guillaume, G. Coukos, A. Harari, C. Jandus, M. Bassani-Sternberg, D. Gfeller, Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 37, 1283–1286 (2019). [DOI] [PubMed] [Google Scholar]
- 24.S. Kaabinejadian, C. Barra, B. Alvarez, H. Yari, W. H. Hildebrand, M. Nielsen, Accurate MHC motif deconvolution of immunopeptidomics data reveals a significant contribution of DRB3, 4 and 5 to the Total DR Immunopeptidome. Front. Immunol. 13, 835454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. B. Nilsson, S. Kaabinejadian, H. Yari, B. Peters, C. Barra, L. Gragert, W. Hildebrand, M. Nielsen, Machine learning reveals limited contribution of trans-only encoded variants to the HLA-DQ immunopeptidome. Commun Biol. 6, 442 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Nielsen, N. Ternette, C. Barra, The interdependence of machine learning and LC-MS approaches for an unbiased understanding of the cellular immunopeptidome. Expert Rev. Proteomics 19, 77–88 (2022). [DOI] [PubMed] [Google Scholar]
- 27.M. Nielsen, M. Andreatta, B. Peters, S. Buus, Immunoinformatics: Predicting Peptide–MHC binding. Annu. Rev. Biomed. Data Sci. 3, 191–215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. Racle, P. Guillaume, J. Schmidt, J. Michaux, A. Larabi, K. Lau, M. A. S. Perez, G. Croce, R. Genolet, G. Coukos, V. Zoete, F. Pojer, M. Bassani-Sternberg, A. Harari, D. Gfeller, Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes. Immunity 56, 1359–1375 (2023). [DOI] [PubMed] [Google Scholar]
- 29.J. G. Abelin, D. Harjanto, M. Malloy, P. Suri, T. Colson, S. P. Goulding, A. L. Creech, L. R. Serrano, G. Nasir, Y. Nasrullah, C. D. McGann, D. Velez, Y. S. Ting, A. Poran, D. A. Rothenberg, S. Chhangawala, A. Rubinsteyn, J. Hammerbacher, R. B. Gaynor, E. F. Fritsch, J. Greshock, R. C. Oslund, D. Barthelme, T. A. Addona, C. M. Arieta, M. S. Rooney, Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity 51, 766–779.e17 (2019). [DOI] [PubMed] [Google Scholar]
- 30.B. Alvarez, B. Reynisson, C. Barra, S. Buus, N. Ternette, T. Connelley, M. Andreatta, M. Nielsen, NNAlign_MA; MHC peptidome deconvolution for accurate MHC binding motif characterization and improved T-cell epitope predictions. Mol. Cell. Proteomics 18, 2459–2477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. T. Refsgaard, C. Barra, X. Peng, N. Ternette, M. Nielsen, NetMHCphosPan—Pan-specific prediction of MHC class I antigen presentation of phosphorylated ligands. ImmunoInformatics. 1–2, 100005 (2021). [Google Scholar]
- 32.P. van Balen, M. G. D. Kester, W. De Klerk, P. Crivello, E. Arrieta-Bolaños, A. H. De Ru, I. Jedema, Y. Mohammed, M. H. M. Heemskerk, K. Fleischhauer, P. A. Van Veelen, J. H. F. Falkenburg, Immunopeptidome analysis of HLA-DPB1 allelic variants reveals new functional hierarchies. J. Immunol. 204, 3273–3282 (2020). [DOI] [PubMed] [Google Scholar]
- 33.S. Klobuch, J. J. Lim, P. van Balen, M. G. D. Kester, W. de Klerk, A. H. de Ru, C. R. Pothast, I. Jedema, J. W. Drijfhout, J. Rossjohn, H. H. Reid, P. A. van Veelen, J. H. F. Falkenburg, M. H. M. Heemskerk, Human T cells recognize HLA-DP–bound peptides in two orientations. Proc. Natl. Acad. Sci. U.S.A. 119, e2214331119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. Laghmouchi, M. G. D. Kester, C. Hoogstraten, L. Hageman, W. de Klerk, W. Huisman, E. A. S. Koster, A. H. de Ru, P. van Balen, S. Klobuch, P. A. van Veelen, J. H. F. Falkenburg, I. Jedema, Promiscuity of peptides presented in HLA-DP molecules from different immunogenicity groups is associated with T-cell cross-reactivity. Front. Immunol. 13, 831822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Fisch, B. Reynisson, L. Benedictus, A. Nicastri, D. Vasoya, I. Morrison, S. Buus, B. R. Ferreira, I. K. F. De Miranda Santos, N. Ternette, T. Connelley, M. Nielsen, Integral use of immunopeptidomics and immunoinformatics for the characterization of antigen presentation and rational identification of BoLA-DR-presented peptides and epitopes. J. Immunol. 206, 2489–2497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.E. W. Petersdorf, M. Bengtsson, M. Horowitz, C. M. Kallor, S. R. Spellman, E. Spierings, T. A. Gooley, P. Stevenson; International Histocompatibility Working Group in Hematopoietic Cell Transplantation , HLA-DQ heterodimers in hematopoietic cell transplantation. Blood 139, 3009–3017 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.F. F. Gonzalez-Galarza, A. McCabe, E. J. M. dos Santos, L. Takeshita, G. Ghattaoraya, A. R. Jones, D. Middleton, Allele frequency net database. Methods Mol. Biol. 1802, 49–62 (2018). [DOI] [PubMed] [Google Scholar]
- 38.M. C. F. Thomsen, C. Lundegaard, S. Buus, O. Lund, M. Nielsen, MHCcluster, a method for functional clustering of MHC molecules. Immunogenetics 65, 655–665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.E. Karosiene, M. Rasmussen, T. Blicher, O. Lund, S. Buus, M. Nielsen, NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 65, 711–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. Barra, B. Alvarez, S. Paul, A. Sette, B. Peters, M. Andreatta, S. Buus, M. Nielsen, Footprints of antigen processing boost MHC class II natural ligand predictions. Genome Med. 10, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Robinson, M. J. Waller, P. Parham, J. G. Bodmer, S. G. E. Marsh, IMGT/HLA Database—A sequence database for the human major histocompatibility complex. Nucleic Acids Res. 29, 210–213 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. Faner, E. James, L. Huston, R. Pujol-Borrel, W. W. Kwok, M. Juan, Reassessing the role of HLA-DRB3 T-cell responses: Evidence for significant expression and complementary antigen presentation. Eur. J. Immunol. 40, 91–102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. Robinson, D. J. Barker, X. Georgiou, M. A. Cooper, P. Flicek, S. G. E. Marsh, IPD-IMGT/HLA Database. Nucleic Acids Res. 48, D948–D955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U. Hobohm, M. Scharf, R. Schneider, C. Sander, Selection of representative protein data sets. Protein Sci. 1, 409–417 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. Martini, M. Nielsen, B. Peters, A. Sette, The immune epitope database and analysis resource program 2003–2018: reflections and outlook. Immunogenetics 72, 57–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.T. Meurer, E. Arrieta-Bolaños, M. Metzing, M. M. Langer, P. van Balen, J. H. Frederik Falkenburg, D. W. Beelen, P. A. Horn, K. Fleischhauer, P. Crivello, Dissecting genetic control of HLA-DPB1 expression and its relation to structural mismatch models in hematopoietic stem cell transplantation. Front. Immunol. 9, 2236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.E. W. Petersdorf, M. Malkki, C. O’hUigin, M. Carrington, T. Gooley, M. D. Haagenson, M. M. Horowitz, S. R. Spellman, T. Wang, P. Stevenson, High HLA-DP expression and graft-versus-host disease. N. Eng. J. Med. 373, 599–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.B. Schöne, S. Bergmann, K. Lang, I. Wagner, A. H. Schmidt, E. W. Petersdorf, V. Lange, Predicting an HLA-DPB1 expression marker based on standard DPB1 genotyping: Linkage analysis of over 32,000 samples. Hum. Immunol. 79, 20–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.G. Díaz, M. Amicosante, D. Jaraquemada, R. H. Butler, M. V. Guillén, M. Sánchez, C. Nombela, J. Arroyo, Functional analysis of HLA-DP polymorphism: A crucial role for DPβ residues 9, 11, 35, 55, 56, 69 and 84-87 in T cell allorecognition and peptide binding. Int. Immunol. 15, 565–576 (2003). [DOI] [PubMed] [Google Scholar]
- 50.J. A. Hollenbach, A. Madbouly, L. Gragert, C. Vierra-Green, S. Flesch, S. Spellman, A. Begovich, H. Noreen, E. Trachtenberg, T. Williams, N. Yu, B. Shaw, K. Fleischhauer, M. Fernandez-Vina, M. Maiers, A combined DPA1∼DPB1 amino acid epitope is the primary unit of selection on the HLA-DP heterodimer. Immunogenetics 64, 559–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.L. E. Creary, N. Sacchi, M. Mazzocco, G. P. Morris, G. Montero-Martin, W. Chong, C. J. Brown, A. Dinou, C. Stavropoulos-Giokas, C. Gorodezky, S. Narayan, S. Periathiruvadi, R. Thomas, D. De Santis, J. Pepperall, G. E. ElGhazali, Z. Al Yafei, M. Askar, S. Tyagi, U. Kanga, S. R. Marino, D. Planelles, C.-J. Chang, M. A. Fernández-Viña, High-resolution HLA allele and haplotype frequencies in several unrelated populations determined by next generation sequencing: 17th International HLA and immunogenetics Workshop joint report. Hum. Immunol. 82, 505–522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. Nielsen, C. Lundegaard, O. Lund, Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 8, 238 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.I. Hoof, B. Peters, J. Sidney, L. E. Pedersen, A. Sette, O. Lund, S. Buus, M. Nielsen, NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R. M. Moore, A. O. Harrison, S. M. McAllister, S. W. Polson, K. Eric Wommack, Iroki: Automatic customization and visualization of phylogenetic trees. PeerJ. 8, e8584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. Bateman, M. J. Martin, S. Orchard, M. Magrane, R. Agivetova, S. Ahmad, E. Alpi, E. H. Bowler-Barnett, R. Britto, B. Bursteinas, H. Bye-A-Jee, R. Coetzee, A. Cukura, A. da Silva, P. Denny, T. Dogan, T. G. Ebenezer, J. Fan, L. G. Castro, P. Garmiri, G. Georghiou, L. Gonzales, E. Hatton-Ellis, A. Hussein, A. Ignatchenko, G. Insana, R. Ishtiaq, P. Jokinen, V. Joshi, D. Jyothi, A. Lock, R. Lopez, A. Luciani, J. Luo, Y. Lussi, A. MacDougall, F. Madeira, M. Mahmoudy, M. Menchi, A. Mishra, K. Moulang, A. Nightingale, C. S. Oliveira, S. Pundir, G. Qi, S. Raj, D. Rice, M. R. Lopez, R. Saidi, J. Sampson, T. Sawford, E. Speretta, E. Turner, N. Tyagi, P. Vasudev, V. Volynkin, K. Warner, X. Watkins, R. Zaru, H. Zellner, A. Bridge, S. Poux, N. Redaschi, L. Aimo, G. Argoud-Puy, A. Auchincloss, K. Axelsen, P. Bansal, D. Baratin, M. C. Blatter, J. Bolleman, E. Boutet, L. Breuza, C. Casals-Casas, E. de Castro, K. C. Echioukh, E. Coudert, B. Cuche, M. Doche, D. Dornevil, A. Estreicher, M. L. Famiglietti, M. Feuermann, E. Gasteiger, S. Gehant, V. Gerritsen, A. Gos, N. Gruaz-Gumowski, U. Hinz, C. Hulo, N. Hyka-Nouspikel, F. Jungo, G. Keller, A. Kerhornou, V. Lara, P. Le Mercier, D. Lieberherr, T. Lombardot, X. Martin, P. Masson, A. Morgat, T. B. Neto, S. Paesano, I. Pedruzzi, S. Pilbout, L. Pourcel, M. Pozzato, M. Pruess, C. Rivoire, C. Sigrist, K. Sonesson, A. Stutz, S. Sundaram, M. Tognolli, L. Verbregue, C. H. Wu, C. N. Arighi, L. Arminski, C. Chen, Y. Chen, J. S. Garavelli, H. Huang, K. Laiho, P. McGarvey, D. A. Natale, K. Ross, C. R. Vinayaka, Q. Wang, Y. Wang, L. S. Yeh, J. Zhang, P. Ruch, D. Teodoro, UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. C. F. Thomsen, M. Nielsen, Seq2Logo: A method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Y. Perez-Riverol, A. Csordas, J. Bai, M. Bernal-Llinares, S. Hewapathirana, D. J. Kundu, A. Inuganti, J. Griss, G. Mayer, M. Eisenacher, E. Pérez, J. Uszkoreit, J. Pfeuffer, T. Sachsenberg, Ş. Yilmaz, S. Tiwary, J. Cox, E. Audain, M. Walzer, A. F. Jarnuczak, T. Ternent, A. Brazma, J. A. Vizcaíno, The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, –D442, D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Legend for table S1
Table S1