Abstract
Biallelic genetic variants in N-acetylneuraminic acid synthase (NANS), a critical enzyme in endogenous sialic acid biosynthesis, are clinically associated with neurodevelopmental disorders. However, the mechanism underlying the neuropathological consequences has remained elusive. Here, we found that NANS mutation resulted in the absence of both sialic acid and protein polysialylation in the cortical organoids and notably reduced the proliferation and expansion of neural progenitors. NANS mutation dysregulated neural migration and differentiation, disturbed synapse formation, and weakened neuronal activity. Single-cell RNA sequencing revealed that NANS loss of function markedly altered transcriptional programs involved in neuronal differentiation and ribosomal biogenesis in various neuronal cell types. Similarly, Nans heterozygous mice exhibited impaired cortical neurogenesis and neurobehavioral deficits. Collectively, our findings reveal a crucial role of NANS-mediated endogenous sialic acid biosynthesis in regulating multiple features of human cortical development, thus linking NANS mutation with its clinically relevant neurodevelopmental disorders.
The human cortical organoid model of sialic acid synthase mutation exhibits aberrant neuronal differentiation and maturation.
INTRODUCTION
Neuronal cell surface glycans control the activity of membrane proteins; modulate cell adhesion, differentiation, and synaptogenesis; and regulate interactions between cells and the extracellular matrix (ECM), which is essential for various neural functions including brain development, learning, and memory (1). In mammalian cells, glycans are primarily terminated with sialic acids on glycoproteins and glycolipids, named sialylation, a biologically important modification that is involved in neurodevelopment and mental disorders. Defects in the glycan biosynthesis or modification pathway lead to inherited metabolic disorders as well as congenital disorders of glycosylation (CDGs) (2). Nearly 80% of individuals with clinically heterogeneous CDGs are associated with neurological impairments, ranging from intellectual developmental disorders (IDDs) and epileptic seizures to cognitive disorders (2). It has been known that sialyltransferase-ST3GAL3 and uridine diphosphate–glucose 6-dehydrogenase mutations result in IDDs and infantile seizures (3, 4), and a variant of glycosylation enzymes is implicated in schizophrenia pathogenesis (5).
Sialic acid belongs to a family of nine-carbon sugar molecules that often attaches to the glycoproteins and gangliosides of neuronal membranes (6). Among all organs in the mammalian, the human central nervous system contains the highest amount of sialic acid, which plays a structural and functional role in brain development (7). Several studies showed that maternal sialic acid supplementation enhances the offspring’s intelligence and cognition performance in rodents and pigs (8, 9). In contrast, defects in sialic acid transfer and transporter due to genetic deficiencies of ST3GAL3, ST3GAL5, and SLC35A1 cause infantile epilepsy, attention deficit, or hyperactivity disorder as well as developmental delays (10, 11). Sialic acid biosynthesis involves a four-step pathway that is regulated by N-acetylmannosamine kinase (GNE), N-acetylneuraminate synthase (NANS), N-acetylneuraminic acid phosphatase, and N-acylneuraminate cytidylyltransferase (CMAS) (12). Loss-of-function mutations in GNE cause a severe progressive muscle-wasting disease but without nervous system abnormalities (13). Mouse models with a deficiency in GNE or CMAS develop early embryonic lethality, indicating a crucial role of sialic acid biosynthesis in embryonic development progresses (14, 15).
NANS (MIM 610442) encodes for an enzyme that converts N-acetylmannosamine (ManNAc)-6-phosphate to ManNAc-9-phosphate, a critical step in endogenous sialic acid biosynthesis (16). A recent study reported that NANS deficiency causes a recessively inherited disorder presenting clinically with severe IDDs, skeletal dysplasia, and dysmorphic features (17, 18), revealing a critical role of sialic acid synthesis in brain development. Knockout of NANS gene impairs cell surface glycan sialylation and sialic acid biosynthesis in human haploid cells and Chinese hamster ovary cells (19). In contrast, the level of free sialic acid in the urine and the sialylation of plasma proteins are not affected in NANS-deficient individuals (7, 18). Nans (NANS in humans) knockdown in zebrafish embryos leads to aberrant skeletal phenotypes and is partially rescued by supplementing exogenous sialic acid (18). In addition, several studies suggested diverse putative functions of sialic acids in migration and differentiation of neural precursors (20), neurite outgrowth (21), and synaptogenesis (22). However, because of the difficulties in accessing human brain tissue from NANS-mutant patients, the role of endogenous sialic acid synthesis in human cortical development has remained largely unclear.
One approach to address this question would be to investigate key features of neocortex development, using appropriate model systems and focusing on the actions of key genes that govern cortical sialic acid synthesis. Recent advances in human induced pluripotent stem cell (iPSC)–derived cerebral organoids and CRISPR-Cas9–mediated genome editing can be combined into a powerful tool for investigating molecular mechanisms involved in neurodevelopmental and neurodegenerative disorders in in vitro systems (23). Human cerebral organoids recapitulate the architectural and functional features of the human brain, containing apical radial glia in the ventricular zone (VZ), human-specific outer radial glia (oRGs) in outer subventricular zone (oSVZ), and deep-layer and upper-layer neurons in cortical plate (CP) (24). A number of research groups have established cerebral organoids to clarify the pathogenesis of autism, Fragile X syndrome, Rett syndrome, Down syndrome, and schizophrenia (25).
Here, we applied NANS homozygous knockout (NANS-KO) iPSC–derived cerebral organoids to investigate genotype-phenotype associations in NANS mutations in human cortical development (26). Nans heterozygous mice were also generated for in vivo neurobehavioral assessment. Our results revealed that NANS mutation caused the absence of sialic acid and protein polysialylation in cortical organoids, leading to reduced neural progenitor proliferation and expansion rate, dysregulated neural migration, impaired synapse formation, and neuronal hypoexcitability. These findings reveal a crucial role of NANS-mediated endogenous biosynthesis of sialic acid in regulating multiple features of human neocortical development.
RESULTS
NANS mutation impairs endogenous sialic acid synthesis and protein polysialylation
Patients with homozygous mutations in the NANS gene are associated with severe developmental delays and loss of gene function (17, 18, 27). To investigate the mode of action of NANS mutation in human cortical development, we have generated a homozygous NANS-KO iPSC line (NCCSEDi001-A-1, https://hpscreg.eu/cell-line/NCCSEDi001-A-1) by inserting the LoxP–green fluorescent protein (GFP)–P2A–Puro–LoxP into exon 1 of the NANS gene (26). In this study, to eliminate GFP expression, we used Cre recombinase to treat the NANS-KO iPSC line and successfully removed the inserted GFP-P2A-Puro cassettes from both alleles (fig. S1, A and B). As anticipated, there was no detection of NANS and GFP protein expression in the NANS-KO iPSC line (fig. S1, C to E). Furthermore, the absence of NANS protein did not affect the iPSC properties such as pluripotent-marker expression, cell growth, self-renewal, and iPSC survival, which was consistent with our previous report (26).
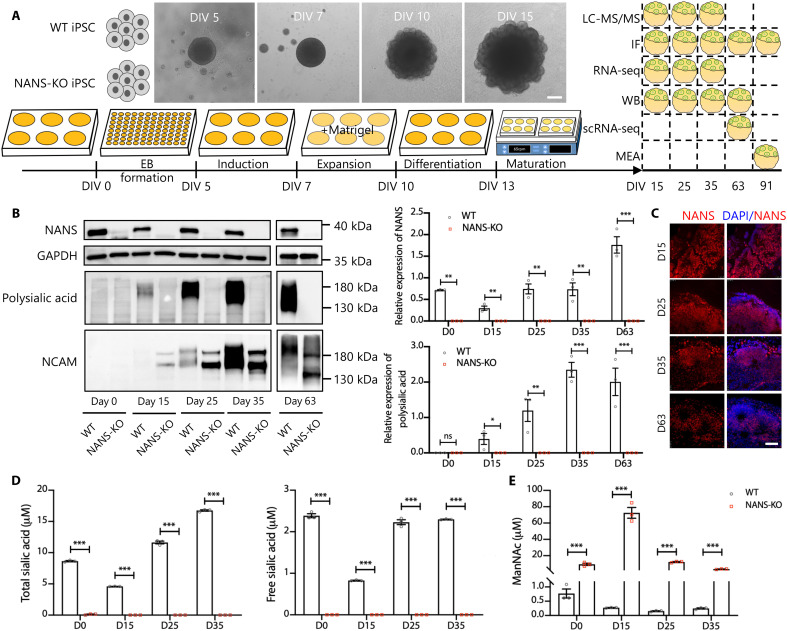
We next generated three-dimensional cerebral organoids (Fig. 1A), which were derived from the wild-type (WT) and NANS-KO iPSCs, to investigate whether NANS mutation affected human cortical development. NANS is an essential enzyme in de novo sialic acid synthesis, and more than 85% of polysialic acid is linked to neural cell adhesion molecule (NCAM) in the brain (28). NCAM polysialylation, which regulates neural cell migration and differentiation during development, plays a crucial role in neurogenesis and synaptic plasticity (29). Consequently, we first examined the expression of NANS protein, and polysialylated proteins in the cerebral organoid were characterized by immunoblotting. The NANS protein was detected at various stages of differentiation in WT cerebral organoids (Fig. 1, B and C). Immunoblotting with the polysialylated-specific monoclonal antibody 735 revealed a typical broad polysialylated protein signal in WT cerebral organoids. The signals were constantly elevated in WT cerebral organoids but completely abolished in NANS-KO cerebral organoids (Fig. 1B). We observed a broad band with an apparent molecular mass ranging from ~120 to 250 kDa, representing the polysialylated form of NCAM in WT cerebral organoid. In contrast, only two distinct protein bands (NCAM isoforms, NCAM-140 and NCAM-180) were detected in NANS-KO cerebral organoids (Fig. 1B), indicating a remarkable reduction in NCAM polysialylation.
Fig. 1. Sialic acid deficit in NANS-KO cerebral organoids.
(A) Scheme illustrating the derivation of cerebral organoids from iPSCs and the various assays used. Scale bar, 200 μm. (B) Representative and quantification of NANS, polysialic acid, and NCAM levels in WT and NANS-KO cerebral organoids through immunoblot analysis at the different cerebral organoid developmental stages. n = 3 biological replicates (five to seven organoids per replicate) from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-way repeated-measure analysis of variance (ANOVA) followed by the Bonferroni’s multiple comparisons test. (C) Confocal imaging of WT cerebral organoids after staining with antibodies against NANS at DIV 15, 25, 35, and 63. Scale bar, 50 μm. (D) Quantification of total sialic acid and free sialic acid in WT and NANS-KO cerebral organoids at DIV 0, 15, 25, and 35, n = 3 biological replicates (one-well iPSCs or five to six organoids per replicate) from three independent experiments. ***P < 0.001, two-way repeated-measure ANOVA followed by the Bonferroni’s multiple comparisons test. (E) Quantification of ManNAc in WT and NANS-KO cerebral organoids at DIV 0, 15, 25, and 35, n = 3 biological replicates (one-well iPSCs or five to six organoids per replicate) from three independent experiments. ***P < 0.001, two-way repeated-measure ANOVA followed by the Bonferroni’s multiple comparisons test. LC-MS/MS, liquid chromatography–tandem mass spectrometry; IF, immunofluorescence; WB, Western blot; scRNA-seq, single-cell RNA sequencing; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; D0, day 0; ns, not significant.
To determine whether NANS deficiency affected the intracellular concentrations of sialic acid in cerebral organoids, we first measured the concentrations of total and free sialic acid using the 1,2-diamino-4,5-methylenedioxybenzene (DMB)–high-performance liquid chromatography (HPLC) method (see Materials and Methods). Both total sialic acid and free sialic acid were undetectable in NANS-KO cerebral organoids (Fig. 1D). As ManNAc is the precursor for the biosynthesis of sialic acid, the urine level of ManNAc was elevated in NANS-mutant patients (17, 18, 27). In this study, the concentrations of ManNAc were found to increase by approximately 12- to 266-fold from WT to NANS-KO cerebral organoids at different time points (Fig. 1E).
NANS mutant impairs neural progenitor cell proliferation and reduces cerebral organoid size
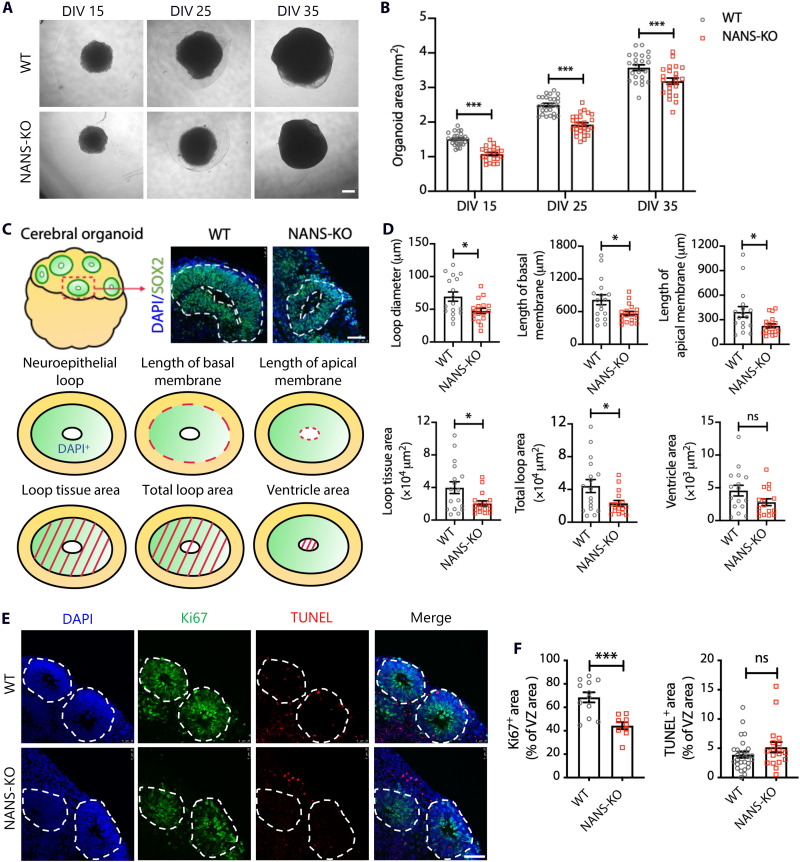
On day 15, WT organoids developed large neuroepithelial loops that persisted from day 25 to day 35, and the overall organoid sizes consistently increased over time (Fig. 2A). The NANS-KO cerebral organoids presented an obviously smaller size and reduced organoid areas as compared to WT organoids (Fig. 2, A and B). To further examine the neuronal structures of NANS-KO cerebral organoids, we assessed the architecture of neuroepithelial loops through the loop diameter, length of the apical and basal membranes, total loop area, loop tissue area and ventricle area in the WT, and NANS-KO cerebral organoids on day 15 in vitro (DIV 15), 25, and 35, respectively (Fig. 2C and fig. S2). Compared to the WT cerebral organoids, morphological parameters of NANS-KO cerebral organoids were significantly reduced except for ventricle area (Fig. 2D and fig. S2).
Fig. 2. NANS regulates cortical neurogenesis in human cerebral organoids.
(A) Bright-field microscopy images of WT and NANS-KO cerebral organoids at DIV 15, 25, and 35. Scale bar, 500 μm. (B) Quantification of cerebral organoid area. n = 21 to 27 organoids from three independent experiments. ***P < 0.001, two-way repeated-measure ANOVA followed by Bonferroni’s multiple comparisons test. (C) A schematic overview of the different parameters of neuroepithelial loops in WT and NANS-KO organoids. Scale bar, 50 μm. (D) Quantification of the parameters in the neuroepithelial loops of WT and NANS-KO cerebral organoids at DIV 35. n = 17 to 18 neuroepithelial loops (four to five neuroepithelial loops per organoid) from two independent experiments, *P < 0.05, Student’s t test. (E) Representative images of WT and NANS-KO cerebral organoids at DIV 35 were stained with Ki67+ and TUNEL+. Scale bar, 50 μm. (F) Quantification of the percentage of Ki67+ and TUNEL+ cells among the total cells in the examined areas. n = 9 to 12 cortical structures (three to five cortical structures per organoid) from two independent experiments, ***P < 0.001, Student’s t test.
Coimmunostaining with the proliferation marker Ki67 and the apoptotic marker terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) was performed to examine the effect of NANS mutation on the proliferation and apoptosis of neural progenitor cells (NPCs) (Fig. 2E and fig. S3). We found a significant reduction in Ki67+ cells at DIV 15, 25, and 35. In contrast, the expression of TUNEL was slightly elevated at DIV 15 and 25 but did not show substantial changes at DIV 35 (Fig. 2F and fig. S3).
To further verify the findings mentioned above, we used a single guide RNA targeting exon 1 of NANS (fig. S4A) and identified a clone with a 31–base pair (bp) deletion in the NANS coding sequence (named H1-NANS-KO; fig. S4B). As shown in fig. S4C, NANS protein was absent in H1-NANS-KO human embryonic stem cells (hESCs). We then generated cerebral organoids from H1-WT and H1-NANS-KO hESC lines, followed by evaluation of aberrant phenotypes in NANS-KO organoids. H1-NANS-KO organoids displayed reduced size, featuring tiny neuroepithelial loops and fewer proliferative NPCs (fig. S5). These results suggested that the decreased proliferation and increased apoptosis of NPCs may be responsible for the smaller size of NANS-KO cerebral organoids at the early stage of corticogenesis.
Transcriptomic analysis reveals an abnormal neuronal migration in NANS-KO organoids
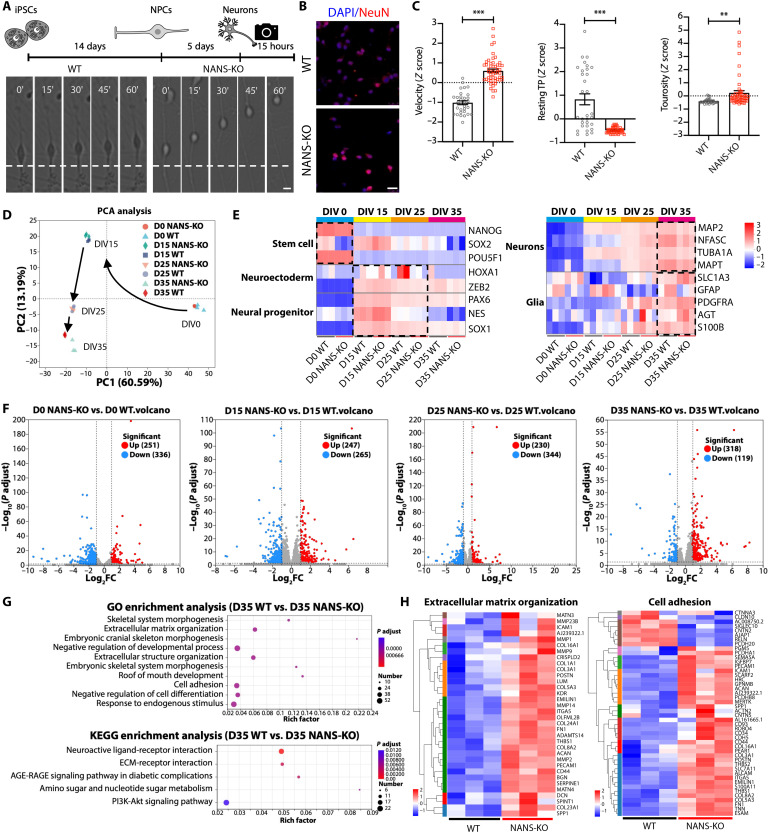
We first performed time-lapse imaging of young migrating neurons to investigate the role of NANS in neuronal migration (Fig. 3, A and B). WT and NANS-KO young neurons were imaged continuously for 15 hours, and dynamic behavior parameters, such as migration speed, resting time points, and tortuosity, were analyzed (Fig. 3C and movies S1 and S2). Notably, compared to the WT young neurons, NANS-KO young neurons displayed significantly faster migration, greater tortuosity, and fewer resting points (Fig. 3C and movies S1 and S2), suggesting that NANS knockout abnormally accelerated radial migration of human cortical neurons.
Fig. 3. Dysregulated gene expression and defective neuronal migration in NANS-KO cerebral organoids.
(A) Scheme depicting the experimental setup of neuronal differentiation and time-lapse imaging between WT and NANS-KO neurons. Scale bar, 10 μm. (B) Confocal imaging of WT and NANS-KO neurons after staining with antibodies against NeuN. Scale bar, 25 μm. (C) Quantification of different parameters of neuronal migration dynamics, tracked using live imaging. Resting TP, resting time points; n = 31 to 53 neurons from two independent experiments, **P < 0.01, ***P < 0.001, Student’s t test. (D) PCA of gene expression over time in WT and NANS-KO cerebral organoids. (E) Heatmap showing distinct developmental stages. DIV 0, stem cell stage; DIV 15, NPC stage; DIV 25, neurogenesis; DIV 35, the early developmental stage of neuronal maturation with concurrent gliogenesis. (F) Volcano plots of DEGs in WT and NANS-KO cerebral organoids at DIV 0, 15, 25, and 35. FC, fold change. (G) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of DEGs in WT and NANS-KO cerebral organoids at DIV 35. (H) Heatmap showing DEGs associated with ECM organization and cell adhesion at DIV 35. AGE-RAGE, advanced glycation end product (AGE) and their receptor (RAGE); PI3K-Akt, phosphoinositide 3-kinase and Akt.
To further understand the molecular basis of these abnormalities in neuronal migration, time-course RNA sequencing (RNA-seq) and principal components analysis (PCA) were performed. The PCA results demonstrated that the main source of variation was related to distinct developmental stages at DIV 0, 15, 25, and 35. This observation is supported by the first principal component (PC1), which explained 60.59% of the variance (Fig. 3D). The findings demonstrate that the transcriptional profiles of both WT and NANS-KO cerebral organoids were more similar at the same differentiation time, indicating that they display similar biological properties during in vitro differentiation (Fig. 3D). At DIV 0, the cell population exhibited a marked expression of pluripotency markers. Subsequently, at DIV 15 and 25, the cerebral organoids were found to be enriched in early neuroectodermal cell and NPC markers and then in neuronal and glia markers at DIV 35 (Fig. 3E). Compared to the WT group, the NANS-KO group showed 251, 247, 230, and 318 up-regulated differentially expressed genes (DEGs) at DIV 0, 15, 25, and 35, respectively; nevertheless, 336, 265, 344, and 119 DEGs were down-regulated (Fig. 3F, figs. S6 to S9, and table S2). Our Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that the neuroactive ligand-receptor interaction pathway was enriched in NANS-KO cerebral organoids at DIV 15, 25, and 35 (figs. S7 to S9). The neuroactive ligand-receptor interaction pathway regulates neuronal impulse reception and transmission. Dysregulation can cause abnormal neuronal responses and is linked to autism spectrum disorder (30). Notably, Gene Ontology (GO) and KEGG analysis of these DEGs revealed substantial enrichment in ECM organization, cell adhesion, and ECM-receptor interaction in NANS-KO cerebral organoids (Fig. 3G). Most of the DEGs in ECM organization and cell adhesion were up-regulated at DIV 35 (Fig. 3H and fig. S10). Then, the short time-series expression miner method (31) was used to identify gene clusters with distinct and representative expression patterns (table S3). In cluster 7, the gene expression–enriched GO pathways mostly include ECM organization and extracellular structure organization (fig. S11). These findings indicate that the altered gene expression profile may contribute to abnormal dynamic behaviors in neuronal migration in NANS-KO cerebral organoids.
NANS mutation disturbs neuronal differentiation and ribosomal biogenesis pathways
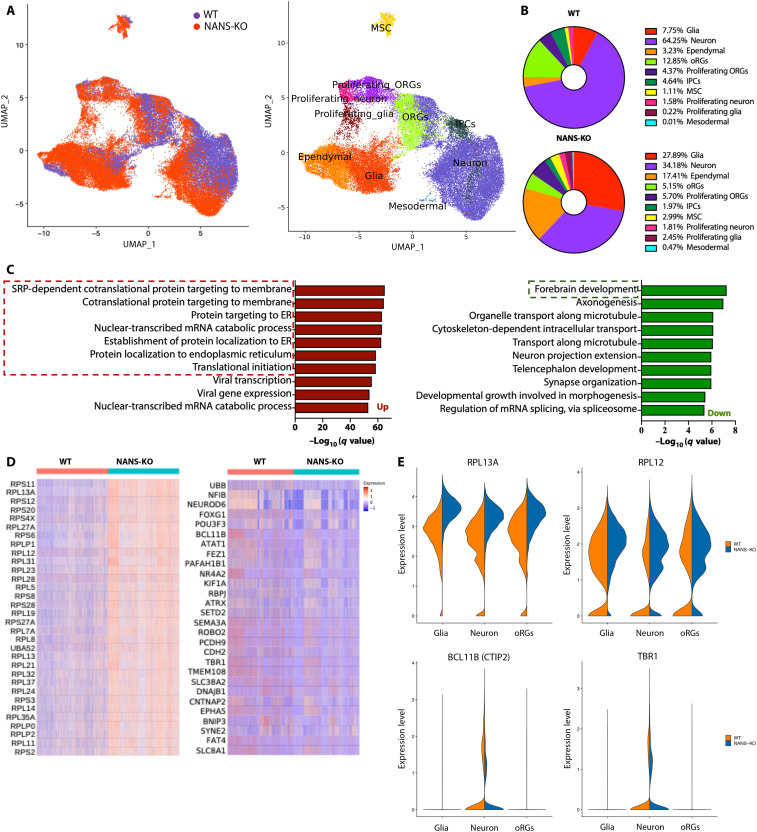
WT and NANS-KO cerebral organoids displayed generally similar cortical layering characteristics (fig. S12). To comprehensively characterize the cell types presented in cerebral organoids and to gain insight into the cell type–specific transcriptomic changes, single-cell RNA-seq was performed to dissect the cellular composition of cerebral organoids. After quality control and filtering, a total of 47,080 single cells were analyzed (Fig. 4A). There was a clear separation between WT and NANS-KO populations in the Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction plot (Fig. 4A). Comparisons of the specific gene expression were then manually annotated on the basis of the expression of known cell type markers (Fig. 4A and table S1). In the WT organoids, 64.25% of the identified single cells expressed neuronal lineage markers, and 7.75% expressed glial cell markers. However, only 34.18% of cells expressed neuronal markers, and 27.89% expressed glial cell markers in NANS-KO cerebral organoids (Fig. 4B). As GO enrichment analysis revealed that the pathways associated with cytosolic ribosome and protein translation were commonly up-regulated in glia, neuron, and oRGs (fig. S13), the DEGs were computed among all cell types with subsequent GO enrichment analysis to identify the mechanistic pathways. The results from GO analysis indicated that the down-regulated genes were enriched in forebrain development, axonogenesis, and synaptic organization in NANS-KO cerebral organoids (Fig. 4, C and D). In contrast, the up-regulated genes were enriched in protein translation and ribosomal function (Fig. 4, C and D). In NANS-KO organoids, marker genes specific for cortical development were expressed at the lower level (Fig. 4E and fig. S14), whereas the expression of ribosomal protein genes was abnormally enhanced in all cell types (Fig. 4E and fig. S14). Collectively, these findings revealed that NANS-KO notably disturbed neuronal differentiation as well as ribosomal biogenesis pathway in cerebral organoids.
Fig. 4. Single-cell RNA-seq reveals abnormal neural differentiation and expression of ribosomal protein genes in NANS-KO cerebral organoids.
(A) UMAP plots reveal differential changes in the population of specific cell types of NANS-KO as compared to WT (left). Ten clusters distinguished by cell type are identified by the expression of specific markers (right). (B) Pie charts comparing cell type composition between WT and NANS-KO cerebral organoids. Numbers indicate the percentage of total cells belonging to the respective cell type. (C) GO enrichment analysis with DEGs is compared between WT and NANS-KO cerebral organoids. The top 10 up and down GO terms are depicted. SRP, signal recognition particle. (D) Heatmap depicting the normalized and scaled gene expression of ribosomal proteins (left) and forebrain development (right). (E) Violin plot represents the expression of PRL13A, RPL12, BCL11B (CTIP2), and TBR1 in glial cells, neurons, and oRGs populations between WT and NANS-KO cerebral organoids.
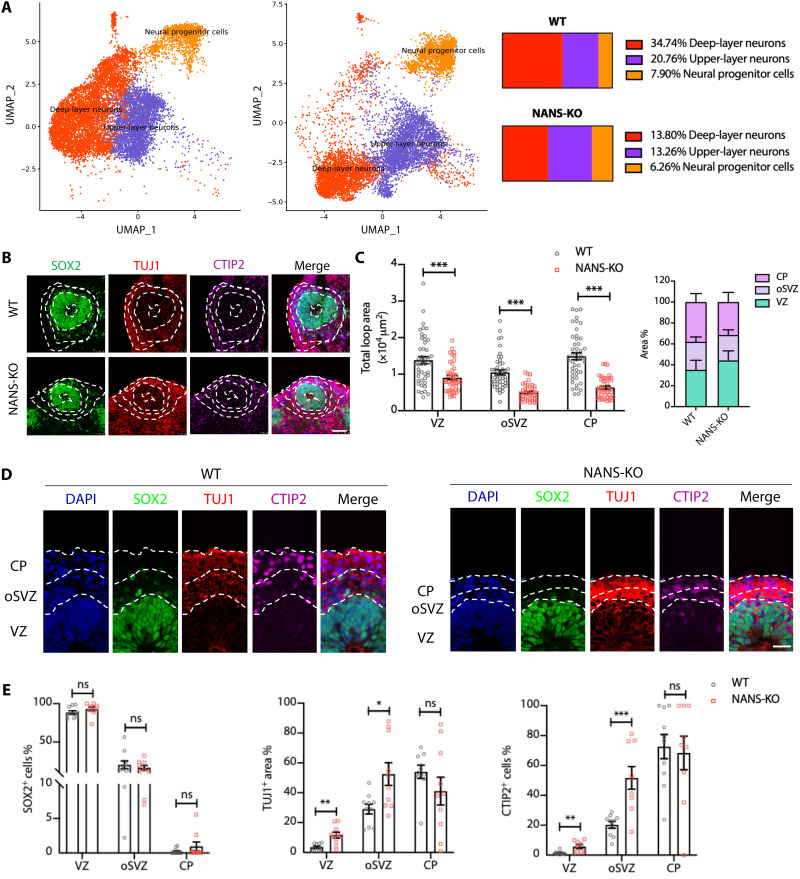
Loss of NANS impairs cortical layer expansion in cerebral organoids
The cortical structures of cerebral organoids display three distinct regions (25). Our single-cell RNA-seq data showed decreases in the percentage of both upper-layer neurons (from 34.74% in WT to 13.8% in NANS-KO cerebral organoid) and deep-layer neurons (from 20.76% in WT to 13.26% in NANS-KO cerebral organoid) (Fig. 5A). To further examine the consequence of altered neuronal differentiation in NANS-KO cerebral organoids, the defined markers were used for labeling cells that resided in VZ, oSVZ, and CP layers of cerebral organoids (Fig. 5, B and D). The VZ was indicated by densely packed SOX2+ NPCs on the apical side, the oSVZ contained mixed populations of SOX2+ NPCs and TUJ1+ immature neurons, and CTIP2 was used to differentiate deep-layer cortical neurons that were mainly expressed in the CP (Fig. 5, B and D). Consistent with the findings depicted in Fig. 2D, the total VZ, oSVZ, and CP loop areas were significantly decreased in NANS-KO cerebral organoids as compared to those in WT cerebral organoids (Fig. 5C). Moreover, the ratio of TUJ1+ and the number of CTIP2+ cells distributed in the VZ and oSVZ areas were significantly elevated in NANS-KO organoids (Fig. 5E). However, the ratio of SOX2+ NPCs in the VZ, oSVZ, and CP showed no substantial differences between NANS-KO and WT organoids (Fig. 5E). Coimmunostaining analysis was performed to determine whether NANS protein was distributed in different cortical structures of WT cerebral organoids. The results showed that more NANS-positive cells were present in the VZ and oSVZ layers than in the CP layer (fig. S15), suggesting that the loss of NANS may disturb neuronal differentiation rather than neuronal maturation.
Fig. 5. NANS mutation disrupts VZ integrity and disturbs corticogenesis in cerebral organoids.
(A) UMAP plots and pie charts comparing NPCs, upper-layer neurons, and deep-layer neurons between WT and NANS-KO cerebral organoids. Numbers indicate the percentage of total cells belonging to the respective cell type. (B) Representative images of the VZ-like area in WT and NANS-KO cerebral organoids. Scale bar, 50 μm. (C) Quantification of the loop area of VZ, oSVZ, and CP. n = 45 to 48, ***P < 0.001, Student’s t test. (D) Representative images of WT and NANS-KO cerebral organoids after staining with DAPI, SOX2, TUJ1, and CTIP2. Scale bar, 10 μm. (E) Statistical analysis of marker distribution across the VZ/oSVZ/CP within organized regions in WT and NANS-KO cerebral organoids. n = 9 to 10, *P < 0.05, **P < 0.01, ***P < 0.001, two-way repeated-measure ANOVA followed by the Bonferroni’s multiple comparisons test.
NANS knockout disrupts synapse formation and synaptic connectivity
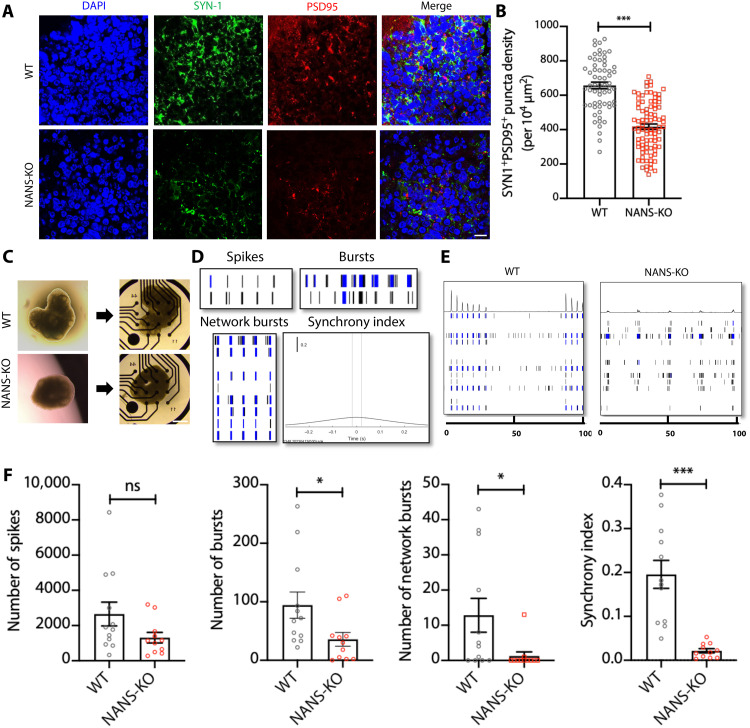
As a reduction in the number of dendritic spines per neuron can weaken synaptic activity and alter synaptic function (32), we then investigated synapse formation and neuronal function in NANS-KO cells. Through immunostaining presynaptic vesicle protein SYNAPSIN 1 (SYN1) and postsynaptic protein PSD95, the density of synaptic puncta was characterized to investigate the effect of NANS loss on synapse formation (Fig. 6A and fig. S16A). When compared to WT cerebral organoids, we observed a significant reduction in the density of SYN1+/PSD95+ synaptic puncta at both DIV 63 and 91 (Fig. 6B and fig. S16B).
Fig. 6. NANS mutation impairs synapse formation and neuronal network activity in NANS-KO cerebral organoids.
(A and B) NANS mutation decreases synapse formation in NANS-KO organoids. Representative images (A) and quantification (B) of SYN1+/PSD95+ puncta density in WT and NANS-KO cerebral organoids at DIV 91. n = 63 to 89 cortical layers (five to six cortical layers per organoid) from three independent experiments, ***P < 0.001, Student’s t test. Scale bar, 10 μm. (C) Bright-field images of 300-μm WT and NANS-KO cerebral organoid slice seeded on the MEA plate. Scale bar, 500 μm. (D) Schematic representation of general activity parameters such as spike, burst, network burst, and synchrony index. (E) Representative spike raster plots of WT and NANS-KO cerebral organoids at DIV 91. (F) MEA analysis reveals a reduction in spontaneous neuronal activity in NANS-KO compared with WT cerebral organoids. n = 11 to 12 MEA wells per genotype from one independent experiment, *P < 0.05, ***P < 0.001, Student’s t test.
Neurons are known to form electrical activity and can be seen in the form of network bursts (33). To investigate whether NANS loss of function caused alteration in the neuronal activity, spontaneous electrical activity of iPSC-derived neurons was measured by multielectrode array (MEA). Compared to WT neuronal cells, NANS-KO–derived cortical neurons displayed weakened neuronal excitability, which was evidenced by declines in the mean firing rates, the number of spikes, and bursts (fig. S16, C and D). To further examine the impact of NANS deficiency on neuronal network organization, we seeded the slices of organoids directly onto the MEA plate (Fig. 6C). Our results revealed that mature NANS-KO organoids exhibited a significant reduction in both the number of bursts and network bursts as well as lower synchrony levels compared to WT organoids at DIV 91 (Fig. 6, D to F). Collectively, our results indicated that NANS deficiency resulted in dysregulated formation and function of excitatory synapses in cerebral organoids.
Nans knockout causes cortical neurogenesis impairment and aberrant neurobehaviors in mice
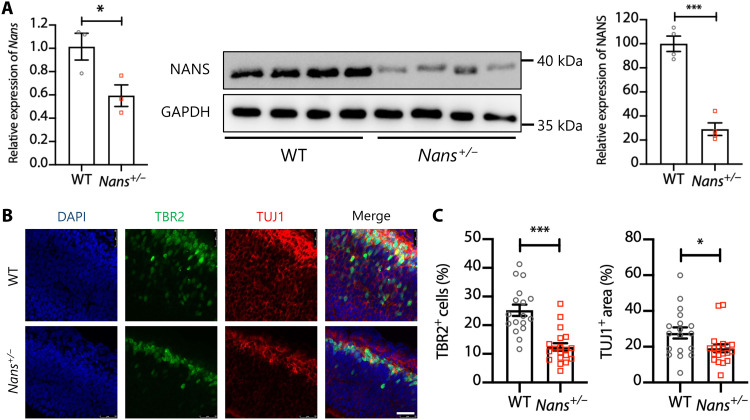
To further characterize the effect of Nans mutations in vivo, we used the CRISPR-Cas9 genome editing system to generate a Nans knockout mouse using guide RNAs (gRNAs) targeted to exons one and six of Nans (fig. S17). However, no homozygous Nans mutants (Nans−/− mice) were identified from heterozygote crosses, and heterozygous offspring appeared normal and fertile. Compared to WT mice, the levels of Nans mRNA and protein were found to be significantly reduced in the cortex of the heterozygous mutants (Nans+/− mice) (Fig. 7A). By labeling intermediate progenitor cell (IPC) marker TBR2 and the early neuronal marker TUJ1, we examined IPCs and granular neurons on coronal cerebral sections obtained from the embryonic day 14.5 brains along the rostral-caudal axis (Fig. 7B). Notably, the TBR2-positive cells and TUJ1-positive areas were significantly reduced in brain VZ/oSVZ regions (Fig. 7C), suggesting cortical neurogenesis impairment in the brain of Nans+/− mice.
Fig. 7. Impaired cortical neurogenesis in Nans+/− mice.
(A) Reverse transcription quantitative polymerase chain reaction (qPCR) (left) and immunoblot analysis (right) of Nans expression in the prefrontal cortex of adult mice (~10 weeks of age) between WT and Nans+/− groups. n = 3 to 4 mice for each genotype (two females and one to two males), *P < 0.05, ***P < 0.001, Student’s t test. (B) Representative images of cerebral cortex sections stained with antibodies against TBR2+ and TUJ1+ in the WT and Nans+/− mice. DAPI stains nuclei. Scale bar, 25 μm. (C) Quantification of the percentage of the TBR2+ cells and the TUJ1+ area. n = 17 to 19 mice for each genotype, *P < 0.05, ***P < 0.001, Student’s t test.
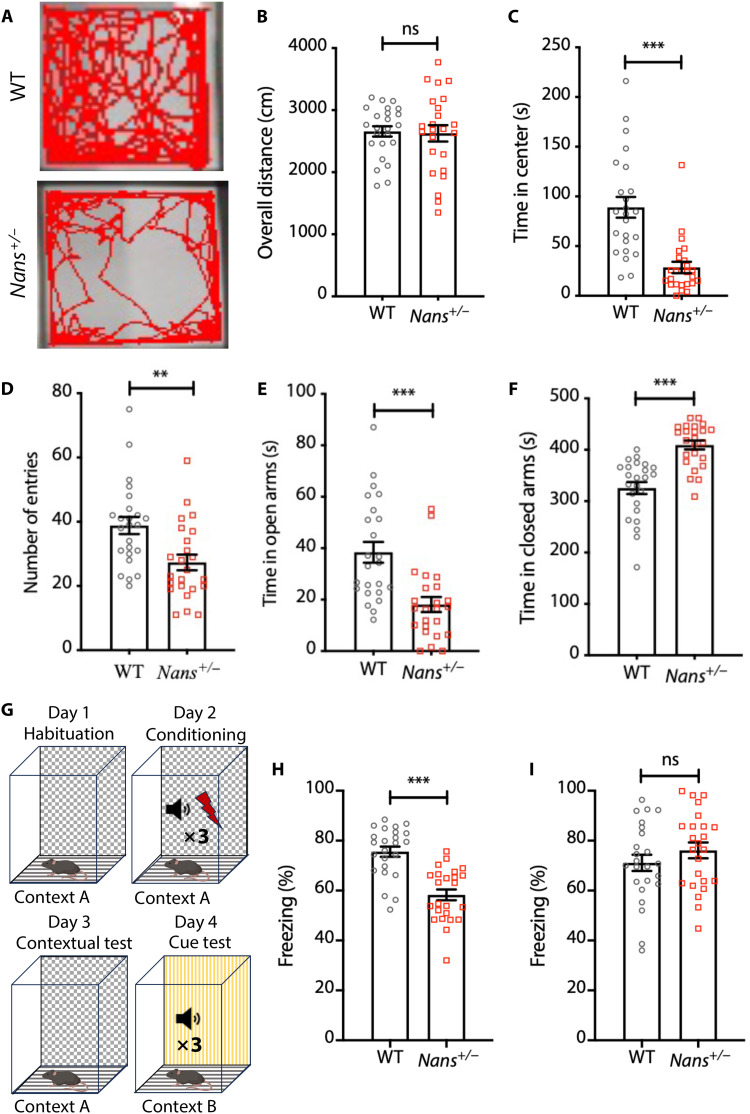
Next, a range of behavioral tests were conducted using the Nans+/− mice, and the WT mice were used as a control. We first used an open-field test to evaluate the overall movement and anxiety. The mice were placed inside a chamber, and their activity was monitored for 10 min (Fig. 8A). No significant differences were observed in the overall distances (Fig. 8B). However, Nans+/− mice spent less time in the center area, indicating elevated anxiety (Fig. 8C). Then, WT and Nans+/− mice were subjected to elevated plus mazes to test anxiety. The number of entries and the time spent in open arms were decreased; in contrast, the time spent in closed arms was significantly increased in Nans+/− mice (Fig. 8, D to F), suggesting anxiety-like behavior in Nans+/− mice. The contextual and cued fear conditioning tests were applied to determine whether Nans mutation caused learning and memory deficits in mice (Fig. 8G). The results show that Nans mutation impaired contextual fear conditioning, but cue fear conditioning was unaffected (Fig. 8, H and I). Results from this series of animal experiments suggested that heterozygous mutation of Nans can cause neurobehavioral abnormalities in mice.
Fig. 8. Behavioral abnormalities in Nans+/− mice.
(A) Representative traces in open-field tests. (B) No difference in the total distance traveled between WT and Nans+/− mice. n = 24 mice for each genotype (12 females and 12 males). (C) Reductions in the time spent in the center in Nans+/− mice. n = 24 mice for each genotype (12 females and 12 males), ***P < 0.001, Student’s t test. (D) Reductions in the number of entries between WT and Nans+/− mice. n = 24 mice for each genotype (12 females and 12 males), **P < 0.01, Student’s t test. (E) Reductions in the time in open arms in Nans+/− mice. n = 24 mice for each genotype (12 females and 12 males), ***P < 0.001, Student’s t test. (F) Increases in the time in closed arms in Nans+/− mice. n = 24 mice for each genotype (12 females and 12 males), ***P < 0.001, Student’s t test. (G) Schematic of the fear conditioning paradigm. (H) Reductions in contextual freezing time in Nans+/− mice after training. n = 24 mice for each genotype (12 females and 12 males), ***P < 0.001, Student’s t test. (I) No difference is observed in the time spent freezing in the cue test, n = 24 mice for each genotype (12 females and 12 males).
DISCUSSION
Because of the difficulties in accessing human brain tissue from NANS-mutant patients, we generated sialic acid–deficient mice and human iPSC–derived cerebral organoids by the genetic disruption of NANS to investigate the role of NANS-dependent endogenous sialic acid synthesis in the cerebral cortex. Combining molecular, cell biological, and animal approaches, our results indicated that NANS mutation resulted in the absence of endogenous sialic acid and protein polysialylation. NANS mutation leads to a global dysregulation in neuronal proliferation, migration, differentiation, as well as synaptic formation and function in cerebral organoids. Furthermore, NANS mutation alters ribosomal functions, which may disturb protein translational processes. These findings provide an insight into the mechanistic understanding of the disrupted cellular and molecular processes in NANS knockout human cortical organoids.
Previous studies showed that genetic inactivation of either one of the key enzymes in the biosynthesis of sialoglycans, Gne or Cmas, causes early embryonic lethality in mice (14, 15). Endothelial cells are hyposialylated, and severe hemorrhages in the neuroepithelium are observed in Gne−/− mice (34). Consistent with Gne or Cmas knockout mice, the Nans−/− mice were characterized by embryonic lethality in this study, and we thus investigated the neurobehaviors of Nans+/− mice. We found that heterozygous Nans mutation caused abnormal cortical neurogenesis and anxiety-like behavior in the mice. To investigate the clinically relevant consequences of NANS mutations, which often occur in biallelic variants (18), we applied human stem cell–derived cerebral organoid models to study structural and functional outcomes relevant to NANS genetic variants.
A previous study reported a normal concentration of free sialic acid in urine as well as normal sialylation of plasma proteins in NANS-deficient patients (18). Whether free sialic acid and sialylation could be down-regulated in the brain with NANS variants is unknown. We found that the sialic acids and polysialic acids were completely absent from NANS-KO cerebral organoids. Nonetheless, our preliminary experiments revealed that exogenous sialic acid treatment was ineffective in ameliorating the neurodevelopmental impairments induced by NANS mutation. This implies that NANS is essential for endogenous sialic acid synthesis in early brain development (35). In the mouse embryo, sialic acids are detected in radial glia of the developing cortex (36), and the mRNA levels of polysialyltransferases peak in the late embryonic and early postnatal mouse brain (37). NANS is also highly expressed in human brains (18). We provided evidence that the NANS protein was highly expressed at different time points during cerebral organoid development and was colocalized with NPCs in the VZ and IPCs in the oSVZ. Furthermore, we also found a notable reduction in cerebral organoid size during differentiation, which was linked to a defect in VZ structures. These observations correlated with the reductions in NPC proliferation after NANS deletion, suggesting that the expansion of NPCs mainly contribute to human cortical neurogenesis (38). Collectively, cortical neurogenesis impairment in NANS-KO organoids may be due to the absence of NANS expression in the VZ and oSVZ, leading to protein hyposialylation in the brain area specific to the developing cortex.
Polysialic acids have been identified in the radial glia of the developing cortex (39), which regulates intercellular space and cell-ECM interaction via their large volume and repulsive field (36). We speculated that NANS loss of function may mechanistically lead to neuronal migration defects. After NANS knockout, GO analysis of the RNA-seq data revealed substantial alterations in the regulatory pathways associated with ECM organization and cell adhesion, including matrix metalloproteinase (MMP) family members (e.g., MMP1, MMP2, MMP9, and MMP14), which were remarkably up-regulated. It has been known that MMPs directly cleave almost all components of the ECM and play a crucial role in ECM remodeling (40). Dysfunctions of these genes are reported in malformations of the developing cortex, such as lissencephaly and periventricular heterotopia (41, 42). In our study, NANS-KO–derived neurons and organoids showed notable changes in different neuronal motility parameters and the absence of both polysialic acid and polysialylated-NCAM. The findings are consistent with a previous study that showed an altered migration of neural precursors and a small cerebral cortex in sialyltransferase double-mutant mice (20). However, NCAM-mutant mice show no cell migration defects in the cortex (32). Further studies are necessary to identify the NANS-dependent sialylated proteins related to neuronal migration in the developing cortex.
By single-cell RNA-seq analysis, our findings revealed a cell type–specific impact of NANS knockout on the development of cerebral organoids. First, the NANS mutations disrupted neuronal differentiation through smaller portions of oRGs and neurons. One possible reason is that polysialic acid may bind to various neurologically active molecules, such as neurotrophins, growth factors, and neurotransmitters (43), thus promoting axonal growth and dendritic morphological change (44). In addition, a small thickness in the oSVZ and CP layers was observed in NANS mutant cerebral organoids. The oRGs are thought to support the developmental and evolutionary expansion of the human neocortex, and the cortex neurons are generated from the radial glial progenitors to guide the cortical columns and neuronal layer formation (45). Our studies identified both structural and functional defects in radial glial progenitors in NANS mutant cerebral organoids, linking endogenous sialic acid synthesis and cortical development. Second, NANS loss markedly affected the pathways associated with ribosomal protein genes, including the cotranslational protein targeting to the membrane and protein localization to endoplasmic reticulum. Furthermore, these alterations were observed in all the major cell types, including neurons, glial cells, and oRGs. Ribosomes play critical roles in the protein synthesis for neurodevelopment, and mutations in ribosomal components result in neurodevelopmental disorders that are present in autism and IDDs (46). For instance, the altered expression of ribosomal proteins is observed in several neurodevelopmental disorders, including Rett and Down syndrome (47, 48). Recent works revealed that ribosomal protein genes are up-regulated in postmortem cortical tissue and iPSC-derived NPCs in patients with autism (49, 50). These reports also support the notion that abnormal ribosomal function is a potential risk factor for neurodevelopmental disorders. Further investigation is required to elucidate the molecular basis underlying these outcomes, including protein-synthesizing ability and nonribosomal functions in NANS-KO cerebral organoids.
Sialylation modification of proteins is necessary for the complex neuronal architecture of the developing cortex, which is thought to be responsible for higher cognitive function (17, 18). The observation of IDDs, neurologic dysfunction, and brain dysplasia in NANS-deficient patients underlines the relationship between sialylation and brain functions (17, 18). Previous literature showed that abnormal formation of neuronal networks and synaptogenesis are associated with neurodevelopment disorders (51). For example, a notable reduction in the amount of spontaneous neuronal activity is present in individuals with autism, leading to a pronounced deficiency in network connectivity (52). Perturbation of sialic acid levels influences a wide range of brain functions, which are involved in structural plasticity and neurogenesis (53). In this study, the delayed synaptogenesis in NANS-KO cerebral organoids resulted in reduced synaptic puncta and dysmaturity of the neuronal networks. NANS-dependent endogenous sialic acid synthesis plays a role in regulating neuronal differentiation, maturation, and function in the early stages of cortical development.
It was reported that genetic variants of the GNE, a gene encoding a bifunctional enzyme critical for sialic acid biosynthesis, can cause reductions in sialic acid supply and muscle-restricted phenotypes but without neurodevelopmental disorders (54). Sialic acid transporter SLC35A1 deficiency causes thrombocytopenia due to megakaryocyte maturation impairment in patients and mice (55). Here, we found that the NANS mutant correlated with endogenous sialic acid synthesis deficiency and neurological disorders during human cerebral organoid development. Further studies using GNE KO and SLC35A1 KO iPSC lines, as well as the generation of mutant iPSC lines from patients with NANS-CDG or the creation of iPSC lines carrying NANS point mutation using CRISPR-Cas9–mediated gene editing, will allow for a more in-depth investigation of the de novo biosynthetic pathways of sialic acids.
Collectively, the defects in the de novo biosynthetic pathways of sialic acids and sialylated glycans are responsible for diverse developmental dysfunctions in human cerebral organoids. In particular, our findings provide an insight into the molecular and neurobehavioral phenotypes of the patient with endogenous sialic acid deficiency or NANS genetic variation.
MATERIALS AND METHODS
Stem cell culture
The NANS-KO iPSC line was generated in our previous study (26) and then treated with Cre recombinase to remove the GFP-P2A-Puro cassette. The hESC line H1 was used to generate H1-NANS-KO line, and the NANS gRNA was from our previous study (26). The iPSC lines (WT and NANS-KO) and hESC lines (H1-WT and H1-NANS-KO) were both cultured in mTeSR1 medium (STEMCELL Technologies) using the Matrigel (Corning)–coated plates and incubated at 5% CO2 at 37°C.
Generation of human cerebral organoids
Organoids were generated using a STEMdiff Cerebral Organoid Kit (Stem Cell Technology) based on our previous protocol with modification (56). On day 0, iPSCs or hESCs at 90% confluence were dissociated into single cells using TrypLE (Gibco). After centrifugation at 500g for 5 min, the iPSCs were resuspended in embryoid body (EB) formation medium with 10 μM Rock inhibitor Y27632 (Stem Cell Technology) and diluted to 5 × 104 cells/ml. Then, 100 μl of the cell suspension was distributed to each well of a low attachment 96-well U-bottom plate (Corning) to develop single EBs, and 100 μl of medium was added to each well every 2 days. On day 5, the EBs were transferred to ultralow attachment six-well plates (Corning) and cultured in induction medium. On day 7, organoids were harvested and embedded in the Matrigel and allowed to grow in the expansion medium in ultralow attachment six-well plates. On day 10, the medium was replaced with differentiation medium containing neurobasal (Gibco), GlutaMax (Gibco, 1:100), nonessential amino acids (Gibco, 1:100), B-27 serum substitute without vitamin A (Gibco), fibroblast growth factor 2 (20 ng/ml, Stem Cell Technology), and epidermal growth factor (20 ng/ml, Stem Cell Technology). On day 13, the medium was replaced with maturation medium containing neurobasal, GlutaMax (1:100), nonessential amino acids (1:100), B-27 serum substitute with vitamin A (Gibco), brain-derived neurotrophic factor (BDNF; 20 ng/ml, PeproTech), and NT3 (20 ng/ml, PeproTech). From day 40 onward, BDNF and NT3 were removed from the maturation medium. The plates were transferred to a shaker for continuous culturing from day 13, and the medium was changed every 3 days.
Immunohistochemistry
Cerebral organoids were fixed in 4% formalin at room temperature for 30 min and then placed in 30% sucrose in phosphate-buffered saline (PBS) overnight. The organoids were embedded in optimal cutting temperature (OCT) solution the next day, followed by dry ice freezing, and then sectioning into 20-μm-thick slices for immunostaining. The sections were washed with PBS to remove the excess OCT and blocked in 10% normal goat serum of PBS containing 0.1% Triton X-100. The sections were incubated with primary antibodies at 4°C overnight and then secondary antibodies for 1 hour at room temperature. The nuclei were counterstained with a 300 nM 4′,6-diamidino-2-phenylindole (DAPI) solution. The sections were mounted on glass slides, and the images were taken using a Leica SP8 confocal microscope. The following primary antibodies were used for immunocytochemistry: SOX2 (rabbit, 1:300; Cell Signaling Technology: 3579), CTIP2 (rat, 1:300; Abcam: AB18465), Ki67 (rabbit, 1:400; Abcam: AB 15580), TUJ1 (mouse, 1:200; Cell Signaling Technology: 4466), NeuN (rabbit, 1:200; Invitrogen: PA5-78499), SATB2 (mouse, 1:500; Abcam: AB51502), glial fibrillary acidic protein (rabbit; 1:1000; Abcam: AB7260), NANS (rabbit, 1:100; Invitrogen: PA5-53960), TBR2 (rabbit,1:200; Abcam: ab183991), PSD95 (mouse, 1:500; Invitrogen: MA1-045), and SYN1 (rabbit, 1:200; Cell Signaling Technology: 5297). The secondary antibodies were Alexa Fluor 488–, 555–, or 647–conjugated donkey anti-rabbit, anti-rat, or anti-mouse immunoglobulin G (Invitrogen, all used at 1:1000 dilutions). The TUNEL assay was performed using a Cell Death Kit (Roche: 12156792910). Images were collected by confocal microscopy (Leica SP8).
Quantitative assessment of organoids
The overall size of the organoids was determined using a calibrated 4× bright-field microscope (Olympus). The organoid area in millimeter square was evaluated by using ImageJ software. Quantification of the different parameters of neuroepithelial loops was performed as previously described (57). In short, the loop diameter and the apical and basal membrane length were calculated in ImageJ. The loop tissue area was the difference between the total loop area and the ventricle area (both assessed using ImageJ software). The numbers of SYN1+/PSD95+ cells were counted using ImageJ software to determine the synaptic puncta density. Cell proliferation and death were measured within neural tube structures by quantifying the number of Ki67+- or TUNEL+-stained nuclei relative to the total number of nuclei stained by DAPI.
The assessment of cortical structure–like morphology was conducted using methods similar to those previously described (58). The VZ was characterized by distinct SOX2 immunoreactivity and a cortical structure–like morphology. The oSVZ was identified as the region containing a mixture of SOX2+ and CTIP2+ nuclei, along with TUJ1+ area outside the VZ. The CP was defined as the region extending from the boundary of the oSVZ to the outer surface, containing exclusively CTIP2+ nuclei. Once the boundaries of each layer were determined, the area of each layer was assessed using ImageJ software. CTIP2 and SOX2 were manually quantified in the cortical structures by counting the CTIP2+- and SOX2+-stained nuclei relative to the total number of cells stained by DAPI.
Western blot analysis
Human iPSCs, cerebral organoids, and prefrontal cortex of mice were lysed on ice inside in radioimmunoprecipitation assay buffer supplemented with protease inhibitor cocktail (50 μl/ml; Solarbio) for 30 min. The suspension was centrifuged at 12,000g for 30 min at 4°C, and the supernatant was collected. The proteins were quantified with a bicinchoninic acid (BCA) kit (Beyotime), separated by SDS–polyacrylamide gel electrophoresis (10%), and transferred to a polyvinylidene fluoride membrane (Invitrogen). Immunoblotting analysis was performed using the following antibodies: glyceraldehyde-3-phosphate dehydrogenase (rabbit; 1:1000; Cell Signaling Technology: 2118), NANS (rabbit, 1:1000; Invitrogen: PA5-53960), polysialic acid (rabbit; 1:1000; LifeSpan BioScience Inc.: LS-C777456), and NCAM (mouse, 1:200; Santa Cruz Biotechnology: sc-7326).
Transcriptomic analysis by RNA-seq
The iPSCs and cerebral organoids from WT and NANS-KO groups were collected at DIV 0 (iPSCs), 15, 25, and 35. In the sample preparation process, each sample was composed of either one well of iPSCs or a pool of five to eight organoids derived from a single independent experiment. Three samples were obtained from three separate independent differentiation experiments. Total RNA was isolated using the miRNeasy Mini Kit (QIAGEN, 160040742). The quantity and quality of RNA samples were determined using NanoDrop 2000c (Thermo Fisher Scientific). Total mRNA was prepared with a sample preparation kit from Illumina, followed by cDNA synthesis using a double-stranded cDNA synthesis kit (Invitrogen). Sheared cDNA with lengths of 200 to 300 bp was inserted into a library, quantified using TBS380, and then sequenced with the Illumina HiSeq 4000 (2 × 150 bp read length). The raw paired-end reads were trimmed, using SeqPrep and Sickle using the default parameters. Reads were mapped to GRCh38.p10 using TopHat (version 2.0.0).
The differential expression analysis was performed using the DEseq2 method with Benjamini-Hochberg correction; genes were considered significantly differentially expressed if adjusted P value < 0.05 and |log2 fold change| ≥ 1 between WT and NANS-KO samples. The expression level of each transcript was determined on the basis of the transcripts per million reads method. Scatter, hierarchical clustering heatmap, GO enrichment, and the KEGG enrichment analyses were performed on the Majorbio platform (http://majorbio.com).
Measurement of sialic acid concentration by DMB-HPLC analysis
The cerebral organoids were homogenized in 10 mM tris-HCl (pH 8.0) solution for total sialic acid extraction, and 2 M HCl (final concentration, 0.1 M HCl) was added to the homogenate. Then, it was heated at 80°C for 3 hours to release sialic acid and cooled on ice to terminate the reaction. Cerebral organoids were homogenized in a 10 mM tris-HCl (pH 8.0) solution for free sialic acid extraction. Subsequently, the samples were freeze-thawed with liquid nitrogen three times and clarified by centrifugation. Sialic acid was fluorescently labeled with DMB using a labeling kit (Takara) according to the manufacturer’s protocol. A Shimadzu LC-30 AD HPLC system equipped with a Shimadzu fluorescence detector (set at wavelengths of 373-nm excitation and 448-nm wavelengths) was operated at 0.3 ml/min at a column temperature of 30°C. The column was a Waters Acquity UPLC BEH C8 (1.7 μm, 2.1 mm by 50 mm) column. An isocratic elution method was adopted using solvent A (water) and solvent B [acetonitrile/methanol = 9/7 (v/v)] at a 92:8 (v/v) ratio.
Measurement of ManNAc concentration by LC tandem mass spectrometry analysis
Intracellular concentrations of ManNAc in the cerebral organoids at different time points were detected by using a triple-quadrupole liquid-chromatography tandem mass spectrometry system (LCMS-8060NX, Shimadzu, Japan) coupled with an LC-40D XS system, as previously reported (59). The column was an ACQUITY UPLC BEH Amide column (2.1 mm by 50 mm, 1.7 μm). The mobile phase comprised ultrapure water with 0.5% acetic acid (mobile A) and 0.5% acetonitrile (mobile B). Gradient elution was used at a flow rate of 0.40 ml/min, and the temperature is 40°C. The retention time of ManNAc was 1.5 min, and the analytical process lasted 3 min in total.
Cerebral organoid dissociation and preparation for single-cell RNA-seq
In this study, five to seven cerebral organoids for each group were randomly collected at DIV 63 and subsequently dissociated into a single-cell suspension. The cerebral organoids were kept in the CelLive Tissue Preservation Solution (Singleron) on ice for 30 min. The samples were washed with Hanks’ balanced salt solution three times and then digested with 2 ml of CelLive Tissue Dissociation Solution (Singleron). The reaction was conducted using the Singleron PythoN Automated Tissue Dissociation System (Singleron) at 37°C for 15 min. The solution was then centrifuged at 500g for 5 min and gently resuspended in PBS. Last, the samples were stained with trypan blue (Sigma-Aldrich), and the cellular viability was evaluated microscopically.
Library preparation and single-cell RNA-seq
Single-cell suspensions (1 × 105 cells/ml) in PBS (HyClone) were loaded onto the microfluidic devices through the Singleron Matrix Single Cell Processing System (Singleron). Subsequently, the single-cell RNA-seq libraries were constructed using the GEXSCOPE Single Cell RNA Library Kits (Singleron). Individual libraries were diluted to 4 nM and pooled for sequencing. Last, the pools were sequenced using Illumina HiSeq X with 150-bp paired-end reads.
Single-cell RNA-seq quantifications and statistical analysis
The raw reads were processed with fastQC and fastp to remove low-quality reads. The cell barcode and unique molecular identifiers (UMIs) were extracted after filtering the read one without poly-T tails. Poly-A tails and adaptor were trimmed (fastp V1) before aligning the second read to GRCh38 using the ensemble version 92 gene annotation (fastp 2.5.3a and featureCounts 1.6.2) (60). Reads with the same cell barcode, UMI, and gene were grouped to determine the number of UMIs of genes within each cell. The UMI count tables for each cellular barcode underwent further analysis. The cell type identity of each cluster was performed using the Seurat program (table S1). Seurat program (http://satijalab.org/seurat/, R package, v.3.0.1) was used to analyze RNA-seq data. The UMI count tables were loaded into R with the read.table function. The parameter resolution was set to 0.6 in the FindClusters function for clustering analysis. Genes expressed in more than 10% of the cells in a cluster and with average log (fold change) of greater than 0.25 were selected as DEGs by Seurat v3.1.2 FindMarkers based on Wilcox likelihood-ratio test with default parameters. Heatmaps/dot plots/violin plots displaying the expression of markers used to identify each cell type were generated by Seurat v3.1.2 DoHeatmap/DotPlot/Vlnplot. GO function enrichment analysis was carried out on the gene set using the “clusterProfiler” R package 3.16.1 for the biological functions or pathways significantly associated with the DEGs.
Generation of NPCs and neurons
Neural progenitors were generated with a STEMdiff SMADi Neural Induction Kit (Stem Cell Technology) based on the monolayer culture protocol. On day 0, human iPSCs at 1 × 106 cells/ml concentration were seeded onto the Matrigel-coated six-well plates and cultured in a neural induction medium (STEMdiff Neural Induction Medium supplemented with 10 μM Rock inhibitor Y27632). The cells were passaged three times as single cells through ACCUTASE (Stem Cell Technology), and the medium was changed daily. After the third passage, NPCs were cultured in neural induction medium for 1 day. Then, the medium was replaced with neuron differentiation medium (STEMdiff Forebrain Neuron Differentiation Medium). After 3 days of differentiation, the cells were harvested and cultured in neuron maturation medium (STEMdiff Forebrain Neuron Maturation Medium). The medium was changed every 2 days.
Neuronal migration assays
Images of the neurons were taken for 3 hours using a Leica SP8 confocal microscope at 37°C and 5% CO2. The cells were imaged every 3 min at six chosen positions. Single-cell movement data were derived from time-lapse movies using the Manual Tracking plugin of ImageJ. After the time-lapse imaging, the neuronal identity of the tracked cells was confirmed using NeuN antibody.
MEA recordings
The cortical neurons and cerebral organoids derived from WT and NANS-KO groups were seeded into 24-well MEA plates (Axion Biosystems) coated with poly-l-ornithine (Sigma-Aldrich) and Matrigel (BD Biosciences) (55). For cortical neurons experiment, each well containing 16 low-impedance platinum-integrated microelectrodes was seeded with 5000 NPCs, and neuronal differentiation was induced as described above. For cerebral organoid experiment, one slice (thickness, 300 μm) of each cerebral organoid was placed into an MEA well at DIV 77 and cultured in BrainPhys medium (Stem Cell Technology) for 14 days until the time of measurement. Spontaneous electrical activity was recorded and analyzed using the Maestro Edge MEA platform and Axion’s Integrated Studio (AXIS) software (Axion Biosystems) with a band-pass filter of 10 Hz and 2.5 kHz cutoff frequencies. Spike detection was computed with an adaptive threshold of six times the SD of the estimated noise for each electrode. Bursts were identified using an interspike interval threshold of five spikes with a maximum interspike interval of 100 ms. Network bursts were defined as bursts of more than 10 spikes that occurred in more than 35% of the active electrodes in the well, with a maximum interspike interval of 100 ms. The synchrony index was estimated by the area under the normalized cross-correlogram for a time window of 20 ms and represented a measure of similarity between two spike trains.
Nans knockout mouse generation
Two gRNAs (CGGCGGGTGCGGAAACGCCCGGG and GACTGGCTGCTAAGAACCGTAGG) were designed for targeting exons one and six of mouse Nans (fig. S17). The designed gRNA and Cas9 mRNA were injected into C57BL/6N zygotes. Fifteen pups were obtained from the zygote injection and were genotyped; the Sanger sequencing showed three (one male and two females) mice that were identified with large deletion-carrying mutations in Nans. The Nans+/− mice are maintained on C57BL/6N background. The sequence mapped uniquely to the targeting site through BLAST, reducing the likelihood of off-target mutations. The Nans-F (GTAAGGCTCCGATTGGCTGT) and Nans-R (GCCAACTGTACCGTGGGTTA) were used for Nans genotyping in mice. All animal experiments were approved by the Sichuan University Animal Care and Use Committee and conducted following the Experimental Animal Ethics Committee of Sichuan University.
Real-time quantitative polymerase chain reaction
Total RNA from cortex of Nans+/− mice and their control littermates was extracted using the miRNeasy Mini Kit (QIAGEN) and then processed according to the manufacturer’s instructions. RNA was used to synthesize cDNA [HiScript III All-in-one RT SuperMix perfect for quantitative polymerase chain reaction (qPCR), Vazyme], and real-time qPCR was performed using HiScript Q RT SuperMix for qPCR (Vazyme). The primers used were as follows: mNans-F (AATTCAGCCACGACCAGTACAA) and mNans-R (TTCTTGGCTGTCTTTTCCAGGT). Relative expression levels for Nans were normalized to Gapdh, and data are represented as fold change relative to control levels.
Mouse behavioral testing
Eight- to 12-week-old age- and sex-matched littermate mice were used for behavioral tests. To minimize variations stemming from age, sex, and genetic background, sex-matched pairs of WT and Nans+/− siblings were derived from the same litters.
In the open-field test, each mouse was placed in a 45 cm by 45 cm by 40 cm open-field box and left undisturbed for 10 min after an acclimatization period of 2 hours in the behavioral testing room (Fig. 8A). In the fear conditioning experiment, mice were allowed to explore freely for 6 min on day 1. On the training day, after 2 min in the conditioning chamber, mice received three pairings of a tone (3 kHz, 90 dB, 20 s) with a coterminating foot shock (0.7 mA, 2 s) at 120, 180, and 240 s, respectively. They then remained in the chamber for an additional minute before being returned to their home cages. Contextual memory was assessed in the original training chamber 24 hours following the training phase, while cued memory was evaluated in the different context 48 hours following the training phase. Levels of freezing (no movement for 0.75 s) were automatically measured by the ANY-Maze video tracking system (Stoelting) (Fig. 8G). In the elevated plus maze, the mice were placed in the center of the maze, facing one of the enclosed arms, and being undisturbed for 5 min.
Quantification and statistical analysis
All biological experiments were performed with multiple independent batches, which were considered independent experiments. The exact sample sizes and independent experiments are indicated in the figure legends. Statistical analyses were performed using GraphPad Prism (version 8) and Stata/IC (version 15.0). The data are represented as the mean ± SEM. Statistical comparisons between WT and NANS-KO were performed using the unpaired Student’s t test. In case of three and more groups, one-way or two-way analysis of variance (ANOVA) with appropriate post hoc testing was performed. Statistical test details can be found in the respective figure legends. The statistical significance was determined by *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant nos. 82071494 to X.C., 81871043 to X.C., and 81272459 to Yinglan Zhao); 1·3·5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (grant no. ZYGD23011 to X.C.); the Open Research Fund of State Key Laboratory of Southwestern Chinese Medicine Resources (SKLTCM2022024 to Q.B.); the Chengdu Science and Technology Project (2022-YF05-01496-SN to Q.B.); and the Project of Medical Science and Technology of Health Commission of Sichuan Province (21PJ064 to Q.B.).
Author contributions: Conceived and designed the experiments, interpretation of data, financial support, and wrote the manuscript: Q.B. Performed the experiments, acquisition of data, and analysis and interpretation of data: Methodology: Y.D., H.Z., M.L., Haxiaoyu Liu, and Y.H. Data acquisition and statistical analysis: A.Z., F.Q., L.J., L.W., Y.C., Hongchun Li, X.W., Yue Zhao, and M.Q. Interpretation of findings and revised the manuscript: N.Z., W.K., and Ying Zhao. Conceived and supervised this research, financial support, and revised the manuscript: Yinglan Zhao and X.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq data are accessible through GEO Series accession number GSE233460 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE233460).
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Legends for tables S1 to S3
Legends for movies S1 and S2
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
Movies S1 and S2
REFERENCES AND NOTES
- 1.L. R. Conroy, T. R. Hawkinson, L. E. A. Young, M. S. Gentry, R. C. Sun, Emerging roles of N-linked glycosylation in brain physiology and disorders. Trends Endocrinol. Metab. 32, 980–993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.L. A. Wolfe, D. Krasnewich, Congenital disorders of glycosylation and intellectual disability. Dev. Disabil. Res. Rev. 17, 211–225 (2013). [DOI] [PubMed] [Google Scholar]
- 3.L. van Diepen, F. F. R. Buettner, D. Hoffmann, C. T. Thiesler, O. V. B. U. Halbach, V. V. B. U. Halbach, L. R. Jensen, D. Steinemann, S. Edvardson, O. Elpeleg, A. Schambach, R. Gerardy-Schahn, A. W. Kuss, A patient-specific induced pluripotent stem cell model for west syndrome caused by ST3GAL3 deficiency. Eur. J. Hum. Genet. 26, 1773–1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.H. Hengel, C. Bosso-Lefèvre, G. Grady, E. Szenker-Ravi, H. Li, S. Pierce, É. Lebigot, T. T. Tan, M. Y. Eio, G. Narayanan, K. H. Utami, M. Yau, N. Handal, W. Deigendesch, R. Keimer, H. M. Marzouqa, M. Gunay-Aygun, M. J. Muriello, H. Verhelst, S. Weckhuysen, S. Mahida, S. Naidu, T. G. Thomas, J. Y. Lim, E. S. Tan, D. Haye, M. A. A. P. Willemsen, R. Oegema, W. G. Mitchell, T. M. Pierson, M. V. Andrews, M. C. Willing, L. H. Rodan, T. S. Barakat, M. van Slegtenhorst, R. H. Gavrilova, D. Martinelli, T. Gilboa, A. M. Tamim, M. O. Hashem, M. D. AlSayed, M. M. Abdulrahim, M. Al-Owain, A. Awaji, A. A. H. Mahmoud, E. A. Faqeih, A. Al Asmari, S. M. Algain, L. A. Jad, H. M. Aldhalaan, I. Helbig, D. A. Koolen, A. Riess, I. Kraegeloh-Mann, P. Bauer, S. Gulsuner, H. Stamberger, A. Y. J. Ng, S. Tang, S. Tohari, B. Keren, L. E. Schultz-Rogers, E. W. Klee, S. Barresi, M. Tartaglia, H. Mor-Shaked, S. Maddirevula, A. Begtrup, A. Telegrafi, R. Pfundt, R. Schüle, B. Ciruna, C. Bonnard, M. A. Pouladi, J. C. Stewart, A. Claridge-Chang, D. J. Lefeber, F. S. Alkuraya, A. S. Mathuru, B. Venkatesh, J. J. Barycki, M. A. Simpson, S. S. Jamuar, L. Schöls, B. Reversade, Loss-of-function mutations in UDP-glucose 6-dehydrogenase cause recessive developmental epileptic encephalopathy. Nat. Commun. 11, 595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. E. Williams, R. G. Mealer, E. M. Scolnick, J. W. Smoller, R. D. Cummings, Aberrant glycosylation in schizophrenia: A review of 25 years of post-mortem brain studies. Mol. Psychiatry 25, 3198–3207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. L. Schnaar, R. Gerardy-Schahn, H. Hildebrandt, Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 94, 461–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.B. Wang, Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29, 177–222 (2009). [DOI] [PubMed] [Google Scholar]
- 8.B. Wang, B. Yu, M. Karim, H. Hu, Y. Sun, P. McGreevy, P. Petocz, S. Held, J. Brand-Miller, Dietary sialic acid supplementation improves learning and memory in piglets. Am. J. Clin. Nutr. 85, 561–569 (2007). [DOI] [PubMed] [Google Scholar]
- 9.E. Oliveros, E. Vázquez, A. Barranco, M. Ramírez, A. Gruart, J. M. Delgado-García, R. Buck, R. Rueda, M. J. Martín, Sialic acid and sialylated oligosaccharide supplementation during lactation improves learning and memory in rats. Nutrients 10, 1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O. Rivero, J. Alhama-Riba, H. P. Ku, M. Fischer, G. Ortega, P. Álmos, D. Diouf, D. van den Hove, K. P. Lesch, Haploinsufficiency of the attention-deficit/hyperactivity disorder risk gene St3gal3 in mice causes alterations in cognition and expression of genes involved in myelination and sialylation. Front. Genet. 12, 688488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.T. Hennet, Diseases of glycosylation beyond classical congenital disorders of glycosylation. Biochim. Biophys. Acta 1820, 1306–1317 (2012). [DOI] [PubMed] [Google Scholar]
- 12.S. J. Moons, G. J. Adema, M. T. Derks, T. J. Boltje, C. Büll, Sialic acid glycoengineering using N-acetylmannosamine and sialic acid analogs. Glycobiology 29, 433–445 (2019). [DOI] [PubMed] [Google Scholar]
- 13.I. Eisenberg, N. Avidan, T. Potikha, H. Hochner, M. Chen, T. Olender, M. Barash, M. Shemesh, M. Sadeh, G. Grabov-Nardini, I. Shmilevich, A. Friedmann, G. Karpati, W. G. Bradley, L. Baumbach, D. Lancet, E. Ben Asher, J. S. Beckmann, Z. Argov, S. Mitrani-Rosenbaum, The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat. Genet. 29, 83–87 (2001). [DOI] [PubMed] [Google Scholar]
- 14.M. Schwarzkopf, K. P. Knobeloch, E. Rohde, S. Hinderlich, N. Wiechens, L. Lucka, I. Horak, W. Reutter, R. Horstkorte, Sialylation is essential for early development in mice. Proc. Natl. Acad. Sci. U.S.A. 99, 5267–5270 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Abeln, I. Albers, U. Peters-Bernard, K. Flächsig-Schulz, E. Kats, A. Kispert, S. Tomlinson, R. Gerardy-Schahn, A. Münster-Kühnel, B. Weinhold, Sialic acid is a critical fetal defense against maternal complement attack. J. Clin. Invest. 129, 422–436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.G. P. Bhide, K. J. Colley, Sialylation of N-glycans: Mechanism, cellular compartmentalization and function. Histochem. Cell Biol. 147, 149–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. den Hollander, A. Rasing, M. A. Post, W. M. Klein, M. M. Oud, M. M. Brands, L. de Boer, U. F. H. Engelke, P. van Essen, S. A. Fuchs, C. A. Haaxma, B. O. Jensson, L. A. J. Kluijtmans, A. Lengyel, K. D. Lichtenbelt, E. Østergaard, G. Peters, R. Salvarinova, M. E. H. Simon, K. Stefansson, Ó. Thorarensen, U. Ulmen, K. L. M. Coene, M. A. Willemsen, D. J. Lefeber, C. D. M. van Karnebeek, NANS-CDG: Delineation of the genetic, biochemical, and clinical spectrum. Front. Neurol. 12, 668640 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. D. M. Van Karnebeek, L. Bonafé, X. Y. Wen, M. Tarailo-Graovac, S. Balzano, B. Royer-Bertrand, A. Ashikov, L. Garavelli, I. Mammi, L. Turolla, C. Breen, D. Donnai, V. Cormier, D. Heron, G. Nishimura, S. Uchikawa, B. Campos-Xavier, A. Rossi, T. Hennet, K. Brand-Arzamendi, J. Rozmus, K. Harshman, B. J. Stevenson, E. Girardi, G. Superti-Furga, T. Dewan, A. Collingridge, J. Halparin, C. J. Ross, M. I. Van Allen, A. Rossi, U. F. Engelke, L. A. J. Kluijtmans, E. Van Der Heeft, H. Renkema, A. De Brouwer, K. Huijben, F. Zijlstra, T. Heisse, T. Boltje, W. W. Wasserman, C. Rivolta, S. Unger, D. J. Lefeber, R. A. Wevers, A. Superti-Furga, NANS-mediated synthesis of sialic acid is required for brain and skeletal development. Nat. Genet. 48, 777–784 (2016). [DOI] [PubMed] [Google Scholar]
- 19.A. P. Willems, L. Sun, M. A. Schulz, W. Tian, A. Ashikov, M. van Scherpenzeel, E. Hermans, H. Clausen, Z. Yang, D. J. Lefeber, Activity of N-acylneuraminate-9-phosphatase (NANP) is not essential for de novo sialic acid biosynthesis. Biochim. Biophys. Acta Gen. Subj. 1863, 1471–1479 (2019). [DOI] [PubMed] [Google Scholar]
- 20.K. Angata, V. Huckaby, B. Ranscht, A. Terskikh, J. D. Marth, M. Fukuda, Polysialic acid-directed migration and differentiation of neural precursors are essential for mouse brain development. Mol. Cell. Biol. 27, 6659–6668 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. Schiff, I. Röckle, H. Burkhardt, B. Weinhold, H. Hildebrandt, Thalamocortical pathfinding defects precede degeneration of the reticular thalamic nucleus in polysialic acid-deficient mice. J. Neurosci. 31, 1302–1312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.G. Di Cristo, B. Chattopadhyaya, S. J. Kuhlman, Y. Fu, M. C. Bélanger, C. Z. Wu, U. Rutishauser, L. Maffei, Z. J. Huang, Activity-dependent PSA expression regulates inhibitory maturation and onset of critical period plasticity. Nat. Neurosci. 10, 1569–1577 (2007). [DOI] [PubMed] [Google Scholar]
- 23.M. Kampmann, CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 16, 465–480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.I. Kelava, M. A. Lancaster, Stem cell models of human brain development. Cell Stem Cell 18, 736–748 (2016). [DOI] [PubMed] [Google Scholar]
- 25.K. W. Kelley, S. P. Pașca, Human brain organogenesis: Toward a cellular understanding of development and disease. Cell 185, 42–61 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Q. Bu, H. Zhang, Q. Liu, Y. Dai, Q. Wei, A. Xue, Y. Huang, K. Zhong, Y. Huang, H. Gao, X. Cen, Generation of an NANS homozygous knockout human induced pluripotent stem cell line by the insertion of GFP-P2A-Puro via CRISPR/Cas9 editing. Stem Cell Res. 49, 102052 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Y. Masunaga, G. Nishimura, K. Takahashi, T. Hishiyama, M. Imamura, K. Kashimada, M. Kadoya, Y. Wada, N. Okamoto, D. Oba, H. Ohashi, M. Ikeno, Y. Sakamoto, M. Fukami, H. Saitsu, T. Ogata, Clinical and molecular findings in three Japanese patients with N-acetylneuraminic acid synthetase-congenital disorder of glycosylation (NANS-CDG). Sci. Rep. 12, 17079 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.H. Cremer, R. Lange, A. Christoph, M. Plomann, G. Vopper, J. Roes, R. Brown, S. Baldwin, P. Kraemer, S. Scheff, D. Barthels, K. Rajewsky, W. Wille, Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature 367, 455–459 (1994). [DOI] [PubMed] [Google Scholar]
- 29.H. Hildebrandt, M. Mühlenhoff, R. Gerardy-Schahn, Polysialylation of NCAM. Adv. Exp. Med. Biol. 663, 95–109 (2010). [DOI] [PubMed] [Google Scholar]
- 30.X. Shao, Z. Hu, C. Hu, Q. Bu, G. Yan, P. Deng, L. Lv, D. Wu, Y. Deng, J. Zhao, R. Zhu, Y. Li, H. Li, Y. Xu, H. Yang, Y. Zhao, X. Cen, Taurine protects methamphetamine-induced developmental angiogenesis defect through antioxidant mechanism. Toxicol. Appl. Pharmacol. 260, 260–270 (2012). [DOI] [PubMed] [Google Scholar]
- 31.J. Ernst, Z. Bar-Joseph, STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. B. Chidambaram, A. G. Rathipriya, S. R. Bolla, A. Bhat, B. Ray, A. M. Mahalakshmi, T. Manivasagam, A. J. Thenmozhi, M. M. Essa, G. J. Guillemin, R. Chandra, M. K. Sakharkar, Dendritic spines: Revisiting the physiological role. Prog. Neuropsychopharmacol. Biol. Psychiatry 92, 161–193 (2019). [DOI] [PubMed] [Google Scholar]
- 33.T. J. Heikkilä, L. Ylä-Outinen, J. M. A. Tanskanen, R. S. Lappalainen, H. Skottman, R. Suuronen, J. E. Mikkonen, J. A. K. Hyttinen, S. Narkilahti, Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp. Neurol. 218, 109–116 (2009). [DOI] [PubMed] [Google Scholar]
- 34.H. Wedekind, E. Kats, A. C. Weiss, H. Thiesler, C. Klaus, A. Kispert, R. Horstkorte, H. Neumann, B. Weinhold, A. Münster-Kühnel, M. Abeln, Uridine diphosphate-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase deletion in mice leads to lethal intracerebral hemorrhage during embryonic development. Glycobiology 31, 1478–1489 (2021). [DOI] [PubMed] [Google Scholar]
- 35.B. Wang, Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv. Nutr. 3, 465S–472S (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.K. Ono, H. Tomasiewicz, T. Magnuson, U. Rutishauser, N-CAM mutation inhibits tangential neuronal migration and is phenocopied by enzymatic removal of polysialic acid. Neuron 13, 595–609 (1994). [DOI] [PubMed] [Google Scholar]
- 37.I. Oltmann-Norden, S. P. Galuska, H. Hildebrandt, R. Geyer, R. Gerardy-Schahn, H. Geyer, M. Mühlenhoff, Impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid synthesis during postnatal mouse brain development. J. Biol. Chem. 283, 1463–1471 (2008). [DOI] [PubMed] [Google Scholar]
- 38.D. V. Hansen, J. H. Lui, P. R. L. Parker, A. R. Kriegstein, Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). [DOI] [PubMed] [Google Scholar]
- 39.H. Li, J. Babiarz, J. Woodbury, N. Kane-Goldsmith, M. Grumet, Spatiotemporal heterogeneity of CNS radial glial cells and their transition to restricted precursors. Dev. Biol. 271, 225–238 (2004). [DOI] [PubMed] [Google Scholar]
- 40.B. Yue, Biology of the extracellular matrix: An overview. J. Glaucoma 23, S20–S23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.I. Y. Buchsbaum, P. Kielkowski, G. Giorgio, A. C. O’Neill, R. Di Giaimo, C. Kyrousi, S. Khattak, S. A. Sieber, S. P. Robertson, S. Cappello, ECE 2 regulates neurogenesis and neuronal migration during human cortical development. EMBO Rep. 21, e48204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. Bershteyn, T. J. Nowakowski, A. A. Pollen, E. Di Lullo, A. Nene, A. Wynshaw-Boris, A. R. Kriegstein, Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20, 435–449.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.C. Sato, K. Kitajima, Polysialylation and disease. Mol. Aspects Med. 79, 100892 (2021). [DOI] [PubMed] [Google Scholar]
- 44.M. Hane, S. Matsuoka, S. Ono, S. Miyata, K. Kitajima, C. Sato, Protective effects of polysialic acid on proteolytic cleavage of FGF2 and proBDNF/BDNF. Glycobiology 25, 1112–1124 (2015). [DOI] [PubMed] [Google Scholar]
- 45.I. Evsyukova, C. Plestant, E. S. Anton, Integrative mechanisms of oriented neuronal migration in the developing brain. Annu. Rev. Cell Dev. Biol. 29, 299–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Hetman, L. P. Slomnicki, Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J. Neurochem. 148, 325–347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D. C. Rodrigues, M. Mufteev, R. J. Weatheritt, U. Djuric, K. C. H. Ha, P. J. Ross, W. Wei, A. Piekna, M. A. Sartori, L. Byres, R. S. F. Mok, K. Zaslavsky, P. Pasceri, P. Diamandis, Q. Morris, B. J. Blencowe, J. Ellis, Shifts in ribosome engagement impact key gene sets in neurodevelopment and ubiquitination in Rett syndrome. Cell Rep. 30, 4179–4196.e11 (2020). [DOI] [PubMed] [Google Scholar]
- 48.H. Naora, I. Takai, M. Adachi, H. Naora, Altered cellular responses by varying expression of a ribosomal protein gene: Sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J. Cell Biol. 141, 741–753 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. V. Lombardo, Ribosomal protein genes in post-mortem cortical tissue and iPSC-derived neural progenitor cells are commonly upregulated in expression in autism. Mol. Psychiatry 26, 1432–1435 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.K. Griesi-Oliveira, M. S. Fogo, B. G. G. Pinto, A. Y. Alves, A. M. Suzuki, A. G. Morales, S. Ezquina, O. J. Sosa, G. J. Sutton, D. Y. Sunaga-Franze, A. P. Bueno, G. Seabra, L. Sardinha, S. S. Costa, C. Rosenberg, E. C. Zachi, A. L. Sertie, D. Martins-de-Souza, E. M. Reis, I. Voineagu, M. R. Passos-Bueno, Transcriptome of iPSC-derived neuronal cells reveals a module of co-expressed genes consistently associated with autism spectrum disorder. Mol. Psychiatry 26, 1589–1605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.P. Lanillos, D. Oliva, A. Philippsen, Y. Yamashita, Y. Nagai, G. Cheng, A review on neural network models of schizophrenia and autism spectrum disorder. Neural Netw. 122, 338–363 (2020). [DOI] [PubMed] [Google Scholar]
- 52.M. C. Marchetto, H. Belinson, Y. Tian, B. C. Freitas, C. Fu, K. C. Vadodaria, P. C. Beltrao-Braga, C. A. Trujillo, A. P. D. Mendes, K. Padmanabhan, Y. Nunez, J. Ou, H. Ghosh, R. Wright, K. J. Brennand, K. Pierce, L. Eichenfield, T. Pramparo, L. T. Eyler, C. C. Barnes, E. Courchesne, D. H. Geschwind, F. H. Gage, A. Wynshaw-Boris, A. R. Muotri, Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 22, 820–835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. Turbpaiboon, W. Siripan, P. Nimnoi, G. P. Sreekanth, W. Wiriyarat, B. Tassaneetrithep, S. Chompoopong, Neural cell adhesion molecule (NCAM) and polysialic acid-NCAM expression in developing ICR mice. Asian Biomed. 12, 179–187 (2019). [Google Scholar]
- 54.N. Carrillo, M. C. Malicdan, M. Huizing, GNE Myopathy: Etiology, diagnosis, and therapeutic challenges. Neurotherapeutics 15, 900–914 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.X. Ma, Y. Li, Y. Kondo, H. Shi, J. Han, Y. Jiang, X. Bai, S. A. Archer-Hartmann, P. Azadi, C. Ruan, J. Fu, L. Xia, Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. Haematologica 106, 759–769 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Huang, Y. Dai, M. Li, L. Guo, C. Cao, Y. Huang, R. Ma, S. Qiu, X. Su, K. Zhong, Y. Huang, H. Gao, Q. Bu, Exposure to cadmium induces neuroinflammation and impairs ciliogenesis in hESC-derived 3D cerebral organoids. Sci. Total Environ. 797, 149043 (2021). [DOI] [PubMed] [Google Scholar]
- 57.X.-Y. Tang, D. Wang, X.-Y. Zhang, M. Xu, Y. Liu, Optimized protocol for analysis of neural stem proliferation in human-pluripotent-stem-cell-derived cerebral organoids. STAR Protoc. 4, 102169 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.X. Qian, Y. Su, C. D. Adam, A. U. Deutschmann, S. R. Pather, E. M. Goldberg, K. Su, S. Li, L. Lu, F. Jacob, P. T. T. Nguyen, S. Huh, A. Hoke, S. E. Swinford-Jackson, Z. Wen, X. Gu, R. C. Pierce, H. Wu, L. A. Briand, H. I. Chen, J. A. Wolf, H. Song, G.-L. Ming, Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell 26, 766–781.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Fang, X. Xu, M. Zhang, Y. Shi, G. Gu, W. Liu, B. Class, C. Ciccone, W. A. Gahl, M. Huizing, N. Carrillo, A. Q. Wang, Quantitation of cytidine-5′-monophospho-N-acetylneuraminic acid in human leukocytes using LC–MS/MS: Method development and validation. Biomed. Chromatogr. 34, e4735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Y. Liao, G. K. Smyth, W. Shi, FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Legends for tables S1 to S3
Legends for movies S1 and S2
Tables S1 to S3
Movies S1 and S2