Abstract
A novel liquefying α-amylase (LAMY) was found in cultures of an alkaliphilic Bacillus isolate, KSM-1378. The specific activity of purified LAMY was approximately 5,000 U mg of protein−1, a value two- to fivefold greater between pH 5 and 10 than that of an industrial, thermostable Bacillus licheniformis enzyme. The enzyme had a pH optimum of 8.0 to 8.5 and displayed maximum activity at 55°C. The molecular mass deduced from sodium dodecyl sulfate-polyacrylamide gel electrophoresis was approximately 53 kDa, and the apparent isoelectric point was around pH 9. This enzyme efficiently hydrolyzed various carbohydrates to yield maltotriose, maltopentaose, maltohexaose, and maltose as major end products after completion of the reaction. Maltooligosaccharides in the maltose-to-maltopentaose range were unhydrolyzable by the enzyme. The structural gene for LAMY contained a single open reading frame 1,548 bp in length, corresponding to 516 amino acids that included a signal peptide of 31 amino acids. The calculated molecular mass of the extracellular mature enzyme was 55,391 Da. LAMY exhibited relatively low amino acid identity to other liquefying amylases, such as the enzymes from B. licheniformis (68.9%), Bacillus amyloliquefaciens (66.7%), and Bacillus stearothermophilus (68.6%). The four conserved regions, designated I, II, III, and IV, and the putative catalytic triad were found in the deduced amino acid sequence of LAMY. Essentially, the sequence of LAMY was consistent with the tertiary structures of reported amylolytic enzymes, which are composed of domains A, B, and C and which include the well-known (α/β)8 barrel motif in domain A.
α-Amylase (1,4-α-d-glucan glucanohydrolase [EC 3.2.1.1]) and pullulanase (pullulan 6-glucanohydrolase [EC 3.2.1.41]) are amylolytic enzymes of industrial importance, particularly in the food and detergent industries. We have found and characterized some unique debranching enzymes, such as a high-alkaline pullulanase (2), an alkali-resistant neopullulanase (16), and an alkaline isoamylase (3), from cultures of alkaliphilic Bacillus strains, and these enzymes can be used as effective additives in dishwashing and laundry detergents under alkaline conditions, especially when used in combination with α-amylase. We have also found the first known alkaline amylopullulanase from alkaliphilic Bacillus sp. strain KSM-1378 (4), which is very unique in that it efficiently hydrolyzes the α-1,6 linkages of pullulan, as well as the α-1,4 linkages of various carbohydrates at different active sites (1, 13).
Liquefying α-amylases, particularly the Bacillus licheniformis enzyme (BLA) (35), are used widely in technical application fields, such as in bread making, production of glucose and fructose syrup and fuel ethanol from starch materials, and textile treatment. The demand for α-amylase for use in laundry and automatic dishwashing detergents has also been growing for several years (42). However, most of the Bacillus liquefying amylases, such as the enzymes from Bacillus amyloliquefaciens (BAA) and Bacillus stearothermophilus (BSA) (28), including BLA (35), have pH optima of between 5 and 7.5 (44). These neutrophilic enzymes are essentially not good for use in detergents, because the working pH range between 8 and 11 is relevant to washing in detergents (17). Since Horikoshi (15) first reported an alkaline amylase from alkaliphilic Bacillus sp. strain A-40-2, many alkaline amylases have been found in cultures of, for example, Bacillus sp. strain NRRL B-3881 (31), Bacillus sp. strain H-167 (14), Bacillus alcalothermophilus A3-8 (7), and Bacillus sp. strain GM8901 (21). The alkaline amylases from these alkaliphilic Bacillus strains reported to date are all of the saccharifying type, except for the enzymes from Bacillus sp. strain 707 (22, 41) and B. licheniformis TCRDC-B13 (5). However, very limited or no information about enzymatic properties of these two liquefying amylases is available. In this paper, we report the isolation of a novel liquefying α-amylase (LAMY) from cultures of the alkaline amylopullulanase producer Bacillus sp. strain KSM-1378 (13). This enzyme is highly active at alkaline pH compared with those of other liquefying α-amylases reported to date. Furthermore, analysis of the gene for this α-amylase (amyK) indicates that LAMY exhibits low amino acid identity to the reported liquefying α-amylases.
MATERIALS AND METHODS
Organism and culture conditions.
Bacillus sp. strain KSM-1378, a relative of Bacillus firmus, was used (4), which had previously been isolated from a soil sample collected in Tochigi City, Tochigi, Japan. The optimum temperature and pH for growth of this organism were around 30°C and pH 10, respectively. The organism was found to produce an alkaline α-amylase on an alkaline agar plate composed of 1% (wt/vol) soluble starch (Wako Pure Chemical, Osaka, Japan), 0.4% starch azure (Sigma, St. Louis, Mo.), 0.2% tryptone (Difco Laboratories, Detroit, Mich.), 0.1% yeast extract (Difco), 0.2% KH2PO4, 0.1% MgSO4 · 7H2O, 0.1% CaCl2 · 2H2O, 0.001% FeSO4 · 7H2O, 0.0001% MnCl2 · 4H2O, 1.0% Na2CO3 (separately autoclaved), and 1.0% agar (pH 10).
The organism was propagated at 30°C for 2 days in 50-ml aliquots of an alkaline medium placed in 500-ml flasks, with shaking on a reciprocal shaker (125 strokes/min; Iwashiya, Tokyo, Japan). The medium contained 1% (wt/vol) soluble starch (Wako Pure Chemical), 0.2% tryptone, 0.1% yeast extract, 0.2% KH2PO4, 0.1% MgSO4 · 7H2O, 0.1% CaCl2 · 2H2O, 0.001% FeSO4 · 7H2O, 0.0001% MnCl2 · 4H2O, and 1.0% Na2CO3 (separately autoclaved). The final pH of the complete medium was about pH 10. After removal of cells by centrifugation (12,000 × g, 15 min) at 4°C, the supernatant (pH 8.6 to 8.8) was used as the starting material for purification of the enzyme.
Purification of the enzyme.
Enzyme purification was done at a temperature below 4°C. The centrifugal supernatant of the culture broth was treated with ammonium sulfate, and the fraction that precipitated at 60% saturation was collected. The precipitates formed were dissolved in a small volume of 10 mM Tris-HCl (pH 7.5) plus 2 mM CaCl2, and the solution was dialyzed twice over the course of 16 h against 50 vol of the same buffer. The retentate was then applied to a column of DEAE-Toyopearl 650M (10 by 15 cm; Tosoh, Tokyo, Japan) that had been equilibrated with 10 mM Tris-HCl plus 2 mM CaCl2 (pH 7.5). The column was washed with the equilibration buffer, and the nonadsorbed active fractions were combined and concentrated by ultrafiltration (PM-10; 10,000-Mr cutoff; Amicon, Danvers, Mass.). The concentrate was put on a column of CM-Toyopearl 650S (2.5 by 50 cm; Tosoh) that had been equilibrated with 10 mM Tris-HCl (pH 7.5) plus 2 mM CaCl2. The column was initially washed with 300 ml of the equilibration buffer, and proteins were eluted with 2.0-liter linear gradient of 0 to 0.5 M NaCl in the same buffer, at a flow rate of 120 ml h−1. Fractions of 15 ml were collected from the start of the gradient. The active fractions were combined and concentrated by ultrafiltration on a PM-10 membrane. The concentrate was dialyzed overnight against 10 mM Tris-HCl (pH 7.5) plus 2 mM CaCl2. The resulting retentate was used exclusively for further experiments as the final preparation of purified enzyme. For comparison, we also purified a commercially available, thermostable BLA (Termamyl; Novo Nordisk, Bagsvaerd, Denmark) to homogeneity by the method described above.
Enzyme assay.
α-Amylase activity was routinely measured at 50°C in a 1-ml reaction mixture that contained 0.5 ml of a 1.0% (wt/vol) solution of soluble starch (from potato; Sigma) in 50 mM Tris-HCl buffer (pH 8.5) and 0.1 ml of a suitably diluted solution of enzyme. The reducing sugar formed was measured by the dinitrosalicylic acid procedure (26). One unit of enzymatic activity was defined as the amount of protein that produced 1 μmol of reducing sugar as glucose per min under the conditions of the assay. Maltooligosaccharides in the G3 to G7 range and in the G8 to G15 range were purchased from Hayashibara Biochemical (Kurashiki, Japan) and Funakoshi (Tokyo, Japan), respectively. Other polysaccharides used as substrates were the products of Sigma. Protein was determined with a protein assay kit (Bio-Rad, Richmond, Calif.) with bovine serum albumin as the standard protein. Protein in column effluents was also routinely monitored by measuring the A280.
Electrophoretic analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done essentially as described by Laemmli (23) with slab gels (10% [wt/vol] acrylamide, 70 by 50 mm, 2.0-mm thickness), and samples were stained for protein with Coomassie brilliant blue R-250. Activity staining of amylase in slab gels was done essentially as described previously (13), with agar sheets containing starch azure (Sigma) as replica plates. The slab gel after SDS-PAGE was laid on the replica sheet and left for several hours at room temperature. The bands of protein that were associated with amylase activity were seen as clear zones on a dark blue background on the replica sheet.
Molecular masses were estimated by SDS-PAGE (10% [wt/vol] acrylamide gel) with low-range molecular mass standards (Bio-Rad), which included phosphorylase b (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa).
Isoelectrofocusing of proteins was done with a Multiphore II gel electrofocusing system with a PAG-Plate and a broad pI calibration kit (Pharmacia Fine Chemica AB, Uppsala, Sweden), which included amyloglucosidase (pI 3.50), methyl red (pI 3.75), soybean trypsin inhibitor (pI 4.55), β-lactoglobulin A (pI 5.20), bovine carbonic anhydrase b (pI 5.85), human carbonic anhydrase b (pI 6.55), horse myoglobin-acidic band (pI 6.85), horse myoglobin-basic band (pI 7.35), lentil lectin-acidic band (pI 8.15), lentil lectin-middle band (pI 8.45), lentil lectin-basic band (pI 8.65), and trypsinogen (pI 9.30).
Chromatographic analysis of the products of hydrolysis of carbohydrates.
The hydrolysis products of LAMY were analyzed by thin-layer chromatography (TLC) with a precoated silica gel plate (Kieselgel 60 F254; E. Merck AG, Darmstadt, Germany). After development of the products in a solvent systems of butanol-pyridine-water (6:4:3 or 9:2:2 [vol/vol], the spots were visualized by spraying with diphenylamine-aniline reagent (12) and then baking at 90°C for 30 min. The products were quantified by high-performance liquid chromatography (HPLC). The purified enzyme was incubated at 30°C with substrate in 10 mM potassium phosphate buffer (pH 8.0). Samples were removed at intervals and heated immediately in boiling water for 5 min to terminate the reaction, and the products in them were separated in a carbohydrate column (4.6 by 250 mm; Waters, Milford, Mass.) with acetonitrile-water (70:30 [vol/vol]) as an eluent at a flow rate of 1.4 ml min−1. Each product was quantified by using a data analysis software, 805 Data Station (Waters), with authentic maltooligosaccharides.
NMR spectroscopy.
Spectra were recorded on a JNM A-500 nuclear magnetic resonance (NMR) spectrometer (JEOL, Tokyo, Japan) operated at 20°C and at 500 MHz for protons in deuterated 10 mM sodium phosphate buffer (p2H 7.4). The spectral width, data point, and the number of accumulation were 6,500 Hz, 16K, and 256, respectively. The water resonance was suppressed by selective irradiation. Chemical shifts were measured relative to the calibrated resonance of internal sodium 3-(trimethylsilyl)-1-propane sulfonate (Merck). The substrate used was p-nitrophenyl α-d-maltooctaoside (pNP-G8; Calbiochem, La Jolla, Calif.)
Sequencing of amino-terminal regions of protein.
The enzyme sample was blotted on a polyvinylidene difluoride membrane (Prosorb; Perkin-Elmer, Foster City, Calif.), which had been wetted with methanol. The amino-terminal sequence of the protein was determined directly by a protein sequencer (model 476A; Perkin-Elmer).
Isolation of DNA and transformation.
Genomic DNA from Bacillus sp. strain KSM-1378 was prepared as described by Saito and Miura (34) and plasmid DNA was isolated by the alkaline extraction procedure of Birnboim and Doly (6). Escherichia coli HB101 (F− hsdS20 recA13 ara14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44 leuB6 thi-1) cells were transformed with plasmids by the methods of Hanahan (11). Transformed E. coli cells were grown at 37°C for one day on Luria-Bertani (LB) agar plates supplemented with 0.4% (wt/vol) starch azure and ampicillin (100 μg ml−1).
Southern hybridization.
Genomic DNAs after digestion with restriction enzymes and the subsequent electrophoresis were subjected to Southern hybridization (37). Patterns of hybridization of the digested DNAs, which were labeled with digoxigenin-dUTP with probes, were examined with a digoxigenin DNA labeling detection kit (Boehringer Mannheim, Mannheim, Germany).
Amplification and sequencing of DNA.
Primer DNAs were designed for the amplification of appropriate regions between specific sites in the genomic DNA. The primer sequences used were as follows: primer A, 5′-TNGAYGCNGTNAARCAYATHAA-3′; primer B, 5′-CGNCANTGNAARCANCTRTTRGTRCT-3′; primer C, 5′-AGCCAATCTCTCGTATAGCTGTA-3′; primer D, 5′-GTACAAAAACACCCTATACATG-3′; primer E, 5′-AATGGWACWATGATGCAKTA-3′; primer F, 5′-CATTTGGCAAATGCCATTCAAA-3′; primer G, 5′-AAAATTGATCCACTTCTGCAG-3′; primer H, 5′-CAGCGCGTGATAATATAAATTTGAAT-3′; and primer I, 5′-AAGCTTCCAATTTATATTGGGTGTAT-3′. They were prepared on a DNA synthesizer (model 392A; Perkin-Elmer) and were purified with a DNA refinement system (model Dnastec-1000; Astec, Fukuoka, Japan). PCR was performed with a DNA thermal cycler (model 480; Perkin-Elmer) with each primer (0.2 μg) plus genomic DNA (1.0 μg) from Bacillus sp. strain KSM-1378 (94°C for 1 min, 55°C for 1 min, and 72°C for 2 min for 30 cycles). The reaction mixture contained 200 μM deoxynucleotide triphosphates, 25 mM KCl, 5 mM (NH4)2SO4, 2.5 U of Pwo DNA polymerase (Boehringer Mannheim) and 10 mM Tris-HCl buffer (pH 8.85) in a reaction volume of 100 μl. Products of PCR were purified with a PCR product purification kit (Boehringer Mannheim), and they were used for sequencing or for subcloning.
Sequencing was performed by the dideoxy chain termination method of Smith et al. (36), by using fluorescent terminators and an automated DNA sequencer (model 373A; Perkin-Elmer). Both strands of the DNA were sequenced, and computer analysis was done with a GENETYX program (SDC Software Development, Tokyo, Japan). Amino acid sequence alignments were done with a GENETYX MAlign program (SDC Software Development).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB008763.
RESULTS AND DISCUSSION
Purification and physicochemical properties of LAMY.
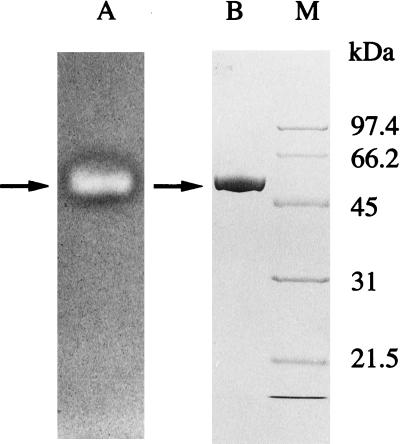
A highly purified preparation of LAMY was obtained by a simple purification procedure with high yield (35%), as summarized in Table 1. Approximately 6.1-fold purification to a specific activity of 5,009 U mg of protein−1 was obtained for the α-amylase activity when measured at 50°C and at pH 8.5 in 50 mM Tris-HCl buffer. The protein was homogeneous, as judged by SDS-PAGE, and the band of protein coincided fairly well with the band that was visualized by activity staining, as shown in Fig. 1. The molecular mass of the purified LAMY was determined to be approximately 53 kDa by SDS-PAGE. The isoelectric point was estimated to be around pH 9. The N-terminal amino acid sequence was HHNGTNGTMMQYFEW. BLA from a commercial product was also purified to homogeneity. The molecular mass and the specific activity of the reference amylase were approximately 53 kDa and 1,600 U/mg of protein, respectively, when measured under our standard assay conditions.
TABLE 1.
Purification of LAMY produced by Bacillus sp. strain KSM-1378
| Purification step | Total amt of protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture filtrate | 353.6 | 292,401 | 827.0 | 100 | 1.0 |
| 60% Ammonium sulfate precipitation | 68.8 | 399,572 | 3,811.9 | 137 | 4.6 |
| DEAE-Toyopearl, unadsorbed | 55.4 | 218,067 | 3,935.3 | 75 | 4.8 |
| CM-Toyopearl chromatography | 20.4 | 102,039 | 5,009.3 | 35 | 6.1 |
FIG. 1.
SDS-PAGE of LAMY purified from cultures of Bacillus sp. strain KSM-1378. The purified enzyme (40 μg) was visualized by activity staining (lane A) and Coomassie brilliant blue staining for protein (lane B). Lane M, molecular mass markers (calibration in kilodaltons). The arrows indicate the positions of the purified enzyme.
Effects of pH and temperature on activity and stability.
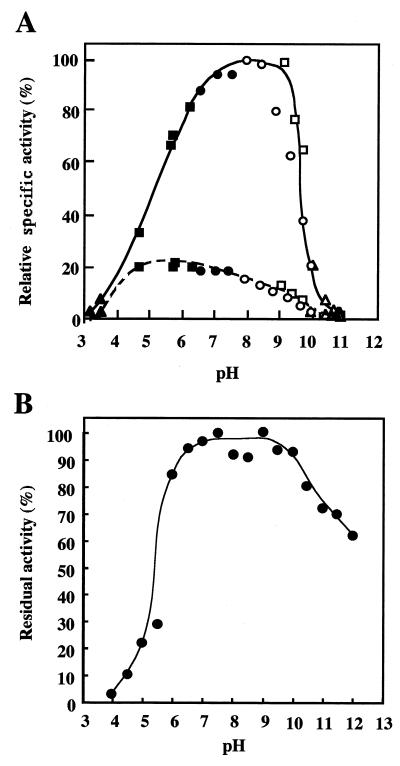
The ranges of pH at which LAMY was active and stable were determined with soluble starch as the substrate. As shown in Fig. 2A, the maximum activity was observed around pH 8.0 to 8.5 when measured in various buffers at 50 mM. More than 50% of the maximum activity was detectable between pH 6 and pH 9. In this pH range, the specific activity of LAMY is two- to fivefold greater than that of BLA. When Britton-Robinson buffers (50 mM phosphoric acid-acetic acid-boric acid; with pH adjusted with NaOH) at different pH values were used, the pH optima were around pH 7.5 to 8.0 for LAMY and pH 5.0 to 6.0 for BLA. To determine the pH stability of LAMY, the enzyme was preincubated at 40°C for 30 min in 10 mM Britton-Robinson buffer and assayed at 50°C in 50 mM Tris-HCl buffer at pH 8.5. LAMY was stable over a range between pH 6 and pH 10 (Fig. 2B).
FIG. 2.
Effect of pH on the activity and stability of LAMY. (A) The pH activity curves of purified LAMY and BLA (each at 0.2 U ml−1) are shown by solid and dotted lines, respectively. The buffers used (50 mM each) were as follows: glycine-HCl, pH 3.0 to 3.5 (▴); acetate, pH 4.0 to 6.0 (■); Tris-HCl, pH 6.5 to 8.5 (•); glycine-NaOH, pH 9.0 to 10.5 (□); glycine-NaCl-NaOH, pH 8.0 to 10.5 (○); and carbonate, pH 10.0 to 11.0 (▵). The values are shown as percentages of the maximum specific activity of LAMY observed at pH 8.0 to 8.5, which is taken as 100%. (B) To assess the pH stability of LAMY, the enzyme (2.0 U ml−1) was preincubated at the indicated pH in 10 mM Britton-Robinson buffer and at 40°C for 30 min, and then samples (0.1 ml) were used for the measurements of the residual activity under the standard conditions of the enzymatic assay. The values are shown as percentages of the original activity, which is taken as 100%.
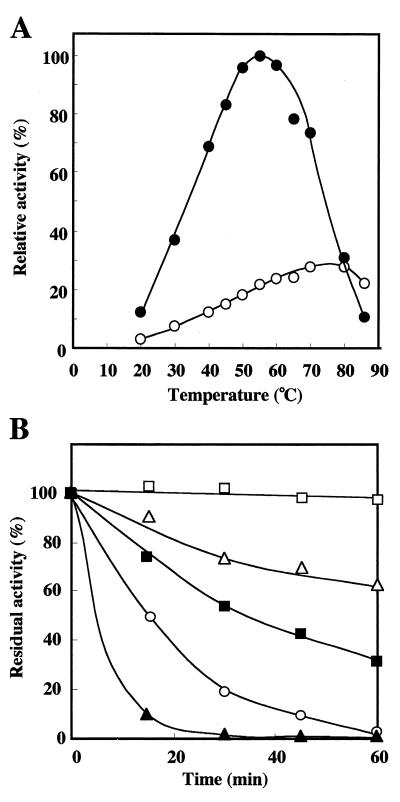
LAMY activity was measured at various temperatures at pH 8.5 in 50 mM Tris-HCl buffer. The optimum temperature for activity of LAMY was around at 55°C (Fig. 3A), while that of BLA was around at 80°C. At 80°C, the specific activities of BLA and LAMY were almost equal, and they were approximately 25% of the maximum activity of LAMY observed at 55°C. The time course of the thermal inactivation of LAMY was monitored at various temperatures and at pH 8.5 in 20 mM Tris-HCl buffer (Fig. 3B). In the absence of CaCl2, the enzyme retained its full activity after 60 min of incubation at 45°C, but only 32 and 3% of the original activity remained after 60 min of incubation at 50 and 60°C, respectively. However, in the presence of 0.1 mM CaCl2, nearly 100 and 65% of the original activity remained at 50 and 60°C, respectively. The enzymatic activity was abolished after being heated at 70°C for 60 min, even in the presence of 0.1 mM Ca2+ ions. In contrast, BLA was quite stable with incubation at 70°C, at least up to 60 min, regardless of whether CaCl2 was present or not.
FIG. 3.
Effect of temperature on the activity and stability of LAMY. (A) The temperature activity curves of purified LAMY (•) and BLA (○) (each at 0.18 U ml−1) are shown. The reactions were done at the indicated temperatures for 10 min and at pH 8.5 in 50 mM Tris-HCl buffer. The values are shown as percentages of the specific activity of LAMY observed at 55°C, which is taken as 100%. (B) For determination of the thermostability of LAMY, the enzyme (1.8 U ml−1) was heated at the indicated temperatures in the presence (at 50°C [□], 60°C [▵], and 70°C [○]) or absence (at 50°C [■] and 60°C [▴]) of 0.1 mM Ca2+ ions in 50 mM Tris-HCl buffer (pH 8.5). Samples (0.1 ml) were used for the determination of the residual activity under the standard conditions of the assay.
Effects of metal ions and chemical reagents.
The purified enzyme was incubated with various cations (1 mM) at 40°C for 30 min, and the residual activities were assayed at 50°C in 50 mM Tris-HCl buffer. Ni2+, Cd2+, Zn2+, and Hg2+ ions strongly inhibited the enzymatic activity by 82, 91, 100, and 100%, respectively. LAMY activity was also inhibited almost completely by iodoacetate (0.5 mM), but not by the thiol inhibitors p-chloromercuribenzoate (0.5 mM) and N-ethylmaleimide (1 mM), as shown in Table 2. The following chemical reagents were without effect on LAMY activity under our assay conditions: citrate (10 mM), diethyl pyrocarbonate (2 mM), and di-isopropyl fluorophosphate (1 mM). When the enzyme was preincubated with 10 mM EDTA or EGTA, the enzyme activity decreased to 10 or 9% of the initial activity, respectively. It has been reported that saccharifying amylases from Bacillus sp. strain A-40-2 (15), Bacillus sp. strain NRRL B-3881 (31), and Bacillus alcalothermophilus A3-8 (7) are all stable in response to EDTA treatment, whereas the liquefying enzymes from Bacillus strains require Ca2+ ions for expression of enzyme activity (44). LAMY activity was completely abolished by a low concentration of N-bromosuccinimide (0.1 mM), which oxidizes tryptophan residues. Tryptophan residues have been suggested to be involved in substrate binding and/or catalysis for an α-amylase (25), a pullulanase (2), and cellulases (20, 45). LAMY was resistant to incubation at 40°C for 1 h with various surfactants, such as SDS, polyoxyethylene alkyl ether, sodium α-sulfonated fatty acid ester, linear-alkylbenzene sulfonate, and alkyl glucoside (each added at 0.1% [wt/vol]) (data not shown).
TABLE 2.
Effects of chemical reagents on the activity of LAMY
| Additive | Reagent concn (mM) | Residual activity (%)a |
|---|---|---|
| None | 100 | |
| N-Bromosuccinimideb | 0.1 | 0 |
| Iodoacetatec | 0.5 | 2 |
| N-Ethylmaleimide | 1 | 107 |
| p-Chloromercuribenzoatec | 0.5 | 80 |
| Citrate | 10 | 98 |
| EDTA | 10 | 10 |
| EGTA | 10 | 9 |
Enzymatic activity was measured after the enzyme had been treated with each inhibitor for 30 min at the indicated temperature and pH in the appropriate buffer. Usually, the enzyme was treated with inhibitor at 30°C and at pH 8.5 in 50 mM Tris-HCl buffer, and an aliquot (0.1 ml) was used for assay of the residual activity in the standard buffer. The values shown are the percentages of the activity without additives, which is taken as 100%.
Preincubated for 15 min at 4°C and at pH 5.0 in 10 mM acetate buffer.
Preincubated for 15 min at 40°C and at pH 5.5 in 10 mM acetate buffer.
Substrate specificity.
The purified LAMY was examined for its ability to hydrolyze various carbohydrates under the standard conditions of the assay. It hydrolyzed soluble starch, amylopectin (from potato), amylose (from potato; degree of polymerization, 17-mers), glycogen (from oyster), and dextrin (from corn) at a relative rate of 100:114:37:83:7. No reaction was observed on dextran, pullulan, and α-, β-, and γ-cyclodextrins. When soluble starch was used as the substrate, the hydrolysis ratio (amount of reducing sugar formed as glucose [initial amount of total reducing sugars as glucose]−1 × 100) after completion of the reaction (a 20-h digestion) was approximately 48%.
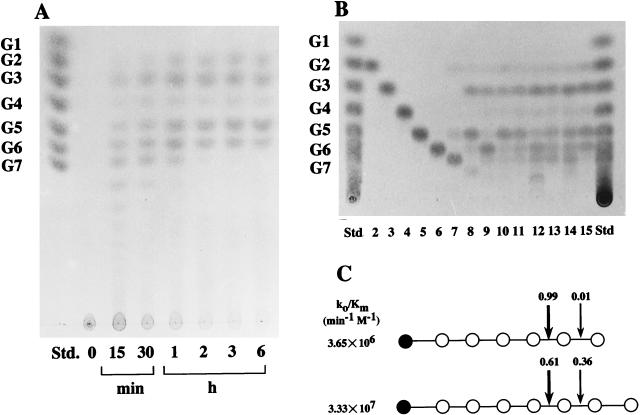
Purified LAMY is unique in that at the early stage of hydrolysis of soluble starch, the enzyme produced G5 to G7 and larger sizes in quantities much greater than G2 and G3, as analyzed by TLC (Fig. 4A). G1 and G4 were formed at this stage in trace amounts. On further incubation, G3 and G5 increased, while G6 to G8 decreased. When analyzed quantitatively by high-performance liquid chromatography (HPLC), the typical molar ratios of products at equilibrium (after 20 h) were as follows: G7, 0.38 mM; G6, 2.02 mM; G5, 2.16 mM; G4, 0.51 mM; G3, 3.05 mM; G2, 2.05 mM; and G1, 0.59 mM. The action patterns of the enzyme on amylose, amylopectin, and glycogen were very similar to that on soluble starch. Therefore, LAMY is classified as liquefying-type amylase, because large maltooligosaccharides in the G4 to G6 range were generated from starch (18, 40, 44). The main products of starch hydrolysis by saccharifying-type enzymes are G1, G2, and G3 (44).
FIG. 4.
Patterns of hydrolysis of soluble starch and maltooligosaccharides by LAMY. (A) Products from soluble starch (0.2% [wt/vol]). The reaction (0.2 U ml−1) was done at 30°C and at pH 8.5 in 50 mM Tris-HCl buffer. Samples were taken at the indicated intervals and boiled for 5 min to terminate the reaction. The products formed were analyzed by TLC with butanol-pyridine-water (6:3:4 [vol/vol]) as the solvent system. Std denotes authentic maltooligosaccharides. (B) Maltooligosaccharides in the G2 to G15 range (0.2% [wt/vol]), shown in lanes 2 through 15, respectively were hydrolyzed for 15 min by LAMY (0.2 U ml−1) under the standard conditions of the assay. The products formed were boiled for 5 min, and then they were analyzed by TLC with butanol-pyridine-water (9:2:2 [vol/vol]) as the solvent system. (C) Cleaved-bond distribution in the hydrolysis of maltooligosaccharides G7 and G8 by LAMY. All reactions were performed at 40°C and at pH 8.5 in 50 mM Tris-HCl buffer. The products formed were boiled for 5 min, and then they were quantified by HPLC, as described in Materials and Methods. Solid symbols (•) represent nonreducing ends of the substrates. The numerals indicate the cleavage frequency of the bond. Catalytic efficiency for each substrate is expressed as k0/Km.
Under the standard conditions of the reaction, LAMY could not act on maltooligosaccharides in the G2 to G6 range, but could hydrolyze the substrates in the G7 to G15 (or larger) range to generate G3, G5, and G6 as the major end products, as shown in Fig. 4B. When excess LAMY was used, only G6, but not G1 through G5, was cleaved slightly to yield G1 and G5. The Km and Vmax values were 6.37 mM and 16.6 V min−1 for G7, 2.18 mM and 58.5 V min−1 for G8, and 0.89 mM and 136.1 V min−1 for G17, respectively. The frequency distribution of bond cleavage of maltooligosaccharides was analyzed by HPLC with p-nitrophenyl (PNP)-labeled and nonlabeled maltooligosaccharides as substrates. LAMY hydrolyzed pNP-G8 to generate G5, G6, PNP-G3, and PNP-G2 as the major end products, indicating that it cleaves the α-glucosidic linkages at the 5th and 6th positions from the nonreducing end of the substrate. As shown in Fig. 4C, G7 was hydrolyzed to yield G2 almost exclusively (99% of the total products), and G8 was hydrolyzed to yield both G3 (61%) and G2 (36%) as the end products. The catalytic efficiencies were 3.33 × 107 for G8 and 3.65 × 106 min−1 M−1 for G7. These results indicate that larger maltooligosaccharides are better substrates for LAMY.
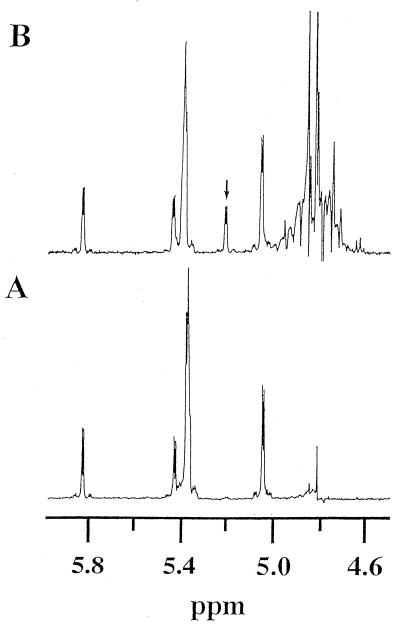
The anomeric form of the products was examined by 1H-NMR spectrometry and polarimetry. Figure 5A shows a 1H-NMR spectrum of the anomeric region of pNP-G8 in deuterated 10 mM phosphate buffer (p2H 7.4). In a spectrum recorded 40 min after addition of LAMY (72 μg/ml), an α-anomeric proton signal of the hydrolyzed product appeared at 5.21 ppm (doublet, J = 3.5 Hz), as shown in Fig. 5B. In addition, optical rotation of the reaction solution containing 1 mM amylose (degree of polymerization, 17-mers) was decreased sharply after completion of the reaction (data not shown). These results indicate that the product(s) from the substrate has an α-configuration, and, hence, LAMY is classified as an α-amylase.
FIG. 5.
1H-NMR spectra of the anomeric proton regions of pNP-G8 and its products of hydrolysis by LAMY. (A) Before addition of enzyme. (B) Forty minutes after addition of enzyme (72 μg ml−1). The enzymatic reaction was performed at 20°C and at p2H 7.4. The arrow in the upper panel indicates a newly appearing doublet signal. Signals around at 5.4 ppm and at 5.82 ppm (doublet) are due to protons at C-2 to C-8 and C-1, respectively, of the G8 moiety of the substrate. The detailed experimental procedure is described in Materials and Methods.
Cloning of the gene for LAMY.
To clone and sequence amyK, we first designed primers A and B from two common regions, DAVKHIK and DVTFVDNHD, respectively, of amino acid sequences of typical liquefying α-amylases, such as BLA (46), BAA (39), and BSA (29). The PCR was conducted to amplify a 0.3-kb fragment (fragment A) with the Bacillus sp. strain KSM-1378 DNA as the template and primers A and B. To assist in the choice of restriction enzymes for inverse PCR, we performed Southern hybridization analysis. The genomic DNA from Bacillus sp. strain KSM-1378 was digested with some endonucleases, and the resultant digests were subjected to agarose gel electrophoresis, and then the DNA bands were transferred onto a nylon membrane and allowed to hybridize with digoxigenin-labeled primer C, which was a 5′-terminal complementary sequence of fragment A. A 1.0-kb XbaI fragment hybridized with the probe, and the XbaI site was located 1.0 kb upstream or downstream of the XbaI site in fragment A. The Bacillus sp. strain KSM-1378 genomic DNA (0.5 μg) was digested with XbaI and ligated under conditions that favored the generation of monomeric circles by T4 DNA ligase. The first inverse PCR was conducted to amplify a 0.7-kb DNA fragment (fragment B), with the self-circulated molecules as the template and primers C and D. The primers had been synthesized on the basis of the results of sequencing of fragment A. The sequence of fragment B included a sequence that encoded the deduced amino acid sequence, which was identical to the C-terminal amino acid sequence of typical liquefying α-amylases, and also included a putative stop codon, TAA.
We designed and synthesized primer E, which was deduced from the N-terminal amino acid sequence of the purified LAMY from culture broth of Bacillus sp. strain KSM-1378. The PCR experiment was performed to amplify a 0.7-kb fragment (fragment C), with the genomic DNA from Bacillus sp. strain KSM-1378 as the template and primers C and E. Southern hybridization analysis showed that a HindIII site was found 0.5 kb upstream of the N-terminal sequence in the amplified fragment. Therefore, inverse PCR experiments were conducted to amplify a 0.8-kb DNA fragment (fragment D) by using HindIII self-circularized DNA molecules as the template and suitably synthesized primers F and G. The resulting fragment, D, contained a putative regulatory region and a sequence that corresponded to the amino acid sequence HHNGTNGTMMQYFEW, which was identical to the N-terminal sequence of LAMY produced by Bacillus sp. strain KSM-1378. Using the genomic DNA from Bacillus sp. strain KSM-1378 as the template and primers H and I, we amplified a 1.8-kb fragment (fragment E) by PCR, which would contain the entire amyK gene. The 1.8-kb fragment E was inserted into the SmaI site of pUC19. The resulting plasmid was introduced into E. coli HB101. When the recombinant E. coli strain was grown on the LB agar plate containing starch azure and ampicillin at 37°C for 1 day, clear halos were found to form around the colonies. This indicates that an amylase gene on fragment E was actually expressed in E. coli (data not shown).
The entire nucleotide sequence of amyK.
The 1.8-kb fragment E containing the amylase gene was sequenced. The fragment contains a single open reading frame (ORF), which begins with an ATG codon at nucleotide 1 and ends with a TAA codon at nucleotide 1545 in the 1,786-bp nucleotide sequence determined. Upstream from this ORF, the putative ribosome-binding sequence AGGAGA is found, separated 11 bp from the initiation codon ATG. The sequence at nucleotides between −20 and −50 resembles the consensus sequence of the sigma A (sigA, or ςA)-type promoter of Bacillus subtilis (27). This sequence consists of TTGACT as the potential −35 region and TAAATT as the potential −10 region, separated by 19 bp (data not shown).
Amino acid sequence analysis.
The ORF in the nucleotide sequence encoded 516 amino acid residues, as shown in Fig. 6. The amino acid sequence deduced from amyK contains a short, relatively basic hydrophilic region, from amino acid −31 to amino acid −26, followed by a hydrophobic region that extends from amino acid −27 to amino acid −1. This hydrophilic-hydrophobic sequence resembles signal peptides of Bacillus (32). A deduced sequence that was identical to the N-terminal 15 amino acid residues of LAMY secreted by Bacillus sp. strain KSM-1378, HHNGTNGTMMQYFEW, was found at amino acids 1 to 15. The residues AQA in the hydrophobic region might be the recognition site of a signal peptidase. If this putative signal peptide were cleaved on the C-terminal side of Ala−1, the molecular mass of the extracellular mature enzyme (from 1 to 485) would be 55,391 Da, a value close to the 53 kDa determined by SDS-PAGE of LAMY that was purified from the culture broth of Bacillus sp. strain KSM-1378.
FIG. 6.
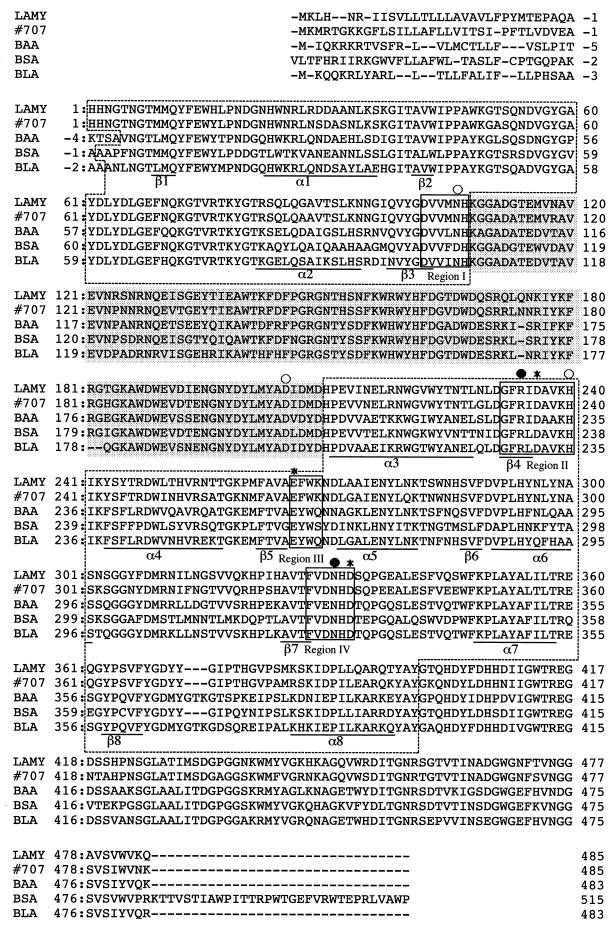
Amino acid sequence alignment of LAMY, strain 707 enzyme, BAA, BSA, and BLA. Each numbering starts after the respective signal peptide. The four conserved regions, I, II, III, and IV (30), are boxed. Domains A and B are indicated by dotted boxes and shading, respectively. The remaining C-terminal sequence corresponds to domain C. Underlined sequences correspond to the secondary structure elements α1 to α8 and β1 to β8 of the BLA (α/β)8 barrel (domain A) according to Machius et al. (24). BLA residues involved in the catalytic site (★), calcium binding site (○), and chloride binding site (•) are indicated above the sequences. Sequence accession numbers (Swiss-Prot database) were as follows: strain 707 enzyme, P19571; BAA, P00692; BSA, P06279; and BLA, P06278.
When suitably aligned, the deduced amino sequence of LAMY exhibits only 66.7, 68.6, and 68.9% identity to those of BAA, BSA, and BLA, respectively (Fig. 6). The deduced amino acid sequence of G6-producing α-amylase from alkaliphilic Bacillus sp. strain 707 (41) has the highest identity to that of LAMY (83.5%), although the physicochemical and enzymatic properties of the strain 707 enzyme have not yet been described. The sequence of LAMY shows almost no homology to those of the saccharifying α-amylases reported to date (data not shown).
The deduced amino acid sequence of LAMY was compared with that of BLA that contains three domains, A, B, and C, as first reported by Buisson et al. (8) for porcine pancreatic α-amylase. Essentially, LAMY has the core of the (α/β)8 barrel domain; the structure elements are underlined and labeled by α1 to α8 for α-helices and β1 to β8 for β-sheets in Fig. 6. The amino acid sequences of domains A, B, and C in LAMY and BLA showed 68.0, 72.5, and 67.8% identity, respectively. Four conserved regions (I through IV) which are necessary for the catalytic activity of α-amylase (30, 33) are well conserved in LAMY as Asp102 to His107, Gly232 to His240, Glu266 to Lys269, and Phe328 to Asp333 for regions I, II, III, and IV, respectively. They are believed to form the active center, the substrate binding site, and the calcium binding site. The three BLA residues (Asn104, Asp200, and His235) involved in calcium binding are conserved in LAMY as Asn106, Asp205, and His240. The BLA residues for chloride binding, Arg229 and Asn326, correspond to Arg234 and Asn331 in LAMY. Like other liquefying α-amylases, LAMY has a distinct internal sequence, a B domain loop (Arg171 to Tyr200), which is believed to play an important role in liquefaction of starch (30). Thus, the overall structure of LAMY may be basically similar to that of BLA, but their catalytic properties are significantly different. For instance, BLA is a rather acidic enzyme, whereas LAMY is highly active at the semialkaline pH side (Fig. 2A). Furthermore, BLA is stable and active at high temperature and stable at least after being heated at 70°C for 60 min, and the optimum temperature for its activity is observed at around 80°C (Fig. 3A). In contrast, LAMY is rapidly inactivated at 60°C (Fig. 3B).
Declerck et al. (9, 10) and Joyet et al. (19) have independently reported hyperthermostable mutants of BLA; two substitutions in the amino acid sequence, His133Ile (or His133Tyr) and Ala209Val (or Ala209Ile), can together increase in the half-life of BLA at 90°C up to 10-fold. It is very interesting that the original amino acid sequence of LAMY conserves the corresponding amino acid residues at amino acids 135 (Tyr) and 214 (Ile), respectively. Suzuki et al. (38) compared the deduced amino acid sequences of BAA and BLA and demonstrated that the thermostability of BAA was drastically improved by deletion of Arg176 and Gly177. The thermostability of LAMY can undoubtedly be improved by site-directed mutagenesis, since the amino acid sequences of LAMY, strain 707 enzyme, BAA, and BSA all conserve the corresponding Arg and Gly residues at the respective amino acid positions (Fig. 6). Recently, the substitution of both Ser187Asp and His133Tyr in the amino acid sequence of BLA was found to increase the specific activity of the enzyme threefold (43). The high specific activity of LAMY, compared with that of BLA, may be related to its original amino acid sequence that conserves Tyr and Asp at amino acids 135 and 192, respectively.
We have already improved the thermostability of LAMY by site-directed mutagenesis and are now analyzing the acquired thermostability of mutant proteins by computer-aided structure modeling. The results will shortly be published elsewhere.
REFERENCES
- 1.Ara K, Igarashi K, Hagihara H, Sawada K, Kobayashi T, Ito S. Separation of functional domains for the α-1,4 and α-1,6 hydrolytic activities of a Bacillus amylopullulanase by limited proteolysis with papain. Biosci Biotechnol Biochem. 1996;60:634–639. doi: 10.1271/bbb.60.634. [DOI] [PubMed] [Google Scholar]
- 2.Ara K, Igarashi K, Saeki K, Kawai S, Ito S. Purification and some properties of an alkaline pullulanase from alkalophilic Bacillus sp. KSM-1876. Biosci Biotechnol Biochem. 1992;56:62–65. [Google Scholar]
- 3.Ara K, Saeki K, Ito S. Purification and characterization of an alkaline isoamylase from an alkalophilic strain of Bacillus. J Gen Microbiol. 1993;139:781–786. [Google Scholar]
- 4.Ara K, Saeki K, Igarashi K, Takaiwa M, Uemura T, Hagihara H, Kawai S, Ito S. Purification and characterization of an alkaline amylopullulanase with both α-1,4 and α-1,6 hydrolytic activity from alkalophilic Bacillus sp. KSM-1378. Biochim Biophys Acta. 1995;1243:315–324. doi: 10.1016/0304-4165(94)00148-q. [DOI] [PubMed] [Google Scholar]
- 5.Bajpai P, Bajpai P K. High-temperature alkaline α-amylase from Bacillus licheniformis TCRDC-B13. Biotechnol Bioeng. 1989;33:72–78. doi: 10.1002/bit.260330110. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer E W, Ingle M B. Extracellular alkaline amylase from a Bacillus species. J Bacteriol. 1972;110:992–1000. doi: 10.1128/jb.110.3.992-1000.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buisson G, Duée E, Haser R, Payan F. Three dimensional structure of porcine pancreatic α-amylase at 2.9 Å resolution. Role of calcium in structure and activity. EMBO J. 1987;6:3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Declerck N, Joyet P, Trosset J Y, Garnier J, Gaillardin C. Hyperthermostable mutants of Bacillus licheniformis α-amylase: multiple amino acid replacements and molecular modelling. Protein Eng. 1995;8:1029–1037. doi: 10.1093/protein/8.10.1029. [DOI] [PubMed] [Google Scholar]
- 10.Declerck N, Machius M, Chambert R, Wiegand G, Huber R, Gaillardin C. Hyperthermostable mutants of Bacillus licheniformis α-amylase: thermodynamic studies and structural interpretation. Protein Eng. 1997;10:541–549. doi: 10.1093/protein/10.5.541. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hansen S A. The thin-layer chromatographic method for the identification of mono-, di-, and trisaccharides. J Chromatogr. 1975;107:224–226. [Google Scholar]
- 13.Hatada Y, Igarashi K, Ozaki K, Ara K, Hitomi J, Kobayashi T, Kawai S, Watabe T, Ito S. Amino acid sequence and molecular structure of an alkaline amylopullulanase from Bacillus that hydrolyzes α-1,4 and α-1,6 linkages in polysaccharides at different active sites. J Biol Chem. 1996;271:24075–24083. doi: 10.1074/jbc.271.39.24075. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Akiba T, Horikoshi K. Production and purification of new maltohexaose-forming amylases from alkalophilic Bacillus sp. H-167. Agric Biol Chem. 1988;52:443–448. [Google Scholar]
- 15.Horikoshi K. Production of alkaline amylases by alkalophilic microorganisms. II. Alkaline amylase produced by Bacillus no. A-40-2. Agric Biol Chem. 1971;35:1783–1791. [Google Scholar]
- 16.Igarashi K, Ara K, Saeki K, Ozaki K, Kawai S, Ito S. Nucleotide sequence of the gene that encodes a neopullulanase from an alkalophilic Bacillus. Biosci Biotechnol Biochem. 1992;56:514–516. doi: 10.1271/bbb.56.514. [DOI] [PubMed] [Google Scholar]
- 17.Ito S. Alkaline cellulases from alkaliphilic Bacillus: enzymatic properties, genetics, and application to detergents. Extremophiles. 1997;1:61–66. doi: 10.1007/s007920050015. [DOI] [PubMed] [Google Scholar]
- 18.Iwasa S, Aoshima H, Hiromi K, Hatano H. Subsite affinity of bacterial liquefying α-amylase evaluated from the rate parameter of linear substrates. J Biochem. 1974;75:969–978. doi: 10.1093/oxfordjournals.jbchem.a130499. [DOI] [PubMed] [Google Scholar]
- 19.Joyet P, Declerck N, Gaillardin C. Hyperthermostable variants of highly thermostable alpha-amylase. Biotechnology. 1992;10:1579–1583. doi: 10.1038/nbt1292-1579. [DOI] [PubMed] [Google Scholar]
- 20.Kawaminami S, Ozaki K, Sumitomo N, Hayashi Y, Ito S, Shimada I, Arata Y. A stable isotope-aided NMR study of the active site of an endoglucanase from a strain of Bacillus. J Biol Chem. 1994;269:28752–28756. [PubMed] [Google Scholar]
- 21.Kim T U, Gu B G, Jeong J Y, Byun S M, Shin Y C. Purification and characterization of maltotetraose-forming alkaline α-amylase from an alkalophilic Bacillus strain, GM8901. Appl Environ Microbiol. 1995;61:3105–3112. doi: 10.1128/aem.61.8.3105-3112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura K, Tsukamoto A, Ishii Y, Takano T, Yamane K. Cloning of a gene for maltohexaose producing amylase of an alkalophilic Bacillus and hyper-production of the enzyme in Bacillus subtilis cells. Appl Microbiol Biotechnol. 1988;27:372–377. [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Machius M, Wiegand G, Huber R. Crystal structure of calcium-depleted Bacillus licheniformis α-amylase at 2.2 Å resolution. J Mol Biol. 1995;246:545–549. doi: 10.1006/jmbi.1994.0106. [DOI] [PubMed] [Google Scholar]
- 25.Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure of possible catalytic residues of Taka-amylase A. J Biochem. 1984;95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 26.Miller G L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 27.Moran C P, Jr, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Lusick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 28.Manning G B, Campbell L L. Thermostable α-amylase of Bacillus stearothermophilus. J Biol Chem. 1961;236:2952–2957. [PubMed] [Google Scholar]
- 29.Nakajima R, Imanaka T, Aiba S. Nucleotide sequence of the Bacillus stearothermophilus α-amylase gene. J Bacteriol. 1985;163:401–406. doi: 10.1128/jb.163.1.401-406.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylase. Appl Microbiol Biotechnol. 1986;23:355–360. [Google Scholar]
- 31.Ozaki, A., and A. Tanaka. February 1990. Heat-stable alkaline amylase from Bacillus. Japanese kokai koho patent 9,049,584.
- 32.Perlman D, Halvorson H O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- 33.Qian M, Haser R, Buisson G, Duée E, Payan F. The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2 Å. Biochemistry. 1994;33:6284–6294. doi: 10.1021/bi00186a031. [DOI] [PubMed] [Google Scholar]
- 34.Saito H, Miura K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 35.Saito N. A thermophilic extracellular α-amylase from Bacillus licheniformis. Arch Biochem Biophys. 1973;155:290–298. doi: 10.1016/0003-9861(73)90117-3. [DOI] [PubMed] [Google Scholar]
- 36.Smith L M, Sanders J Z, Kaiser R J, Hughes P, Dodd C, Connell C R, Heiner C, Kent S B H, Hood L E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;21:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 37.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki Y, Ito N, Yuuki T, Yamagata H, Udaka S. Amino acid residues stabilizing a Bacillus α-amylase against irreversible thermoinactivation. J Biol Chem. 1989;264:18933–18938. [PubMed] [Google Scholar]
- 39.Takkinen K, Pettersson R F, Kalkkinen N, Palva I, Söderlund H, Kääriäinen L. Amino acid sequence of α-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983;258:1007–1013. [PubMed] [Google Scholar]
- 40.Thoma J A, Rao G V K, Brothers C, Spradlin J. Subsite mapping of enzymes. J Biol Chem. 1971;246:5621–5635. [PubMed] [Google Scholar]
- 41.Tsukamoto A, Kimura K, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the maltohexaose-producing amylase gene from an alkalophilic Bacillus sp. #707 and structural similarity to liquefying type α-amylases. Biochem Biophys Res Commun. 1988;151:25–31. doi: 10.1016/0006-291x(88)90554-2. [DOI] [PubMed] [Google Scholar]
- 42.UpaDek H, Kottwitz B. Application of amylases in detergents. In: van Ee J H, Misset O, Baas E J, editors. Enzymes in detergency. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 203–212. [Google Scholar]
- 43.van der Laan, J. M. May 1995. Novel amylolytic enzymes derived from the B. licheniformis α-amylase, having improved characteristics. International patent WO 95/35382.
- 44.Yamamoto T The Amylase Research Society of Japan, editors. Handbook of amylases and related enzymes. Oxford, England: Pergamon Press; 1988. Bacterial α-amylase (liquefying- and saccharifying types) of Bacillus subtilis and related bacteria; pp. 40–45. [Google Scholar]
- 45.Yoshimatsu T, Ozaki K, Shikata S, Ohta Y, Koike K, Kawai S, Ito S. Purification and characterization of alkaline endo-1,4-β-glucanases from alkalophilic Bacillus sp. KSM-635. J Gen Microbiol. 1990;136:1973–1979. [Google Scholar]
- 46.Yuuki T, Nomura T, Tezuka H, Tsuboi A, Yamagata H, Tsukagoshi N, Udaka S. Complete nucleotide sequence of a gene for heat- and pH-stable α-amylase of Bacillus licheniformis: comparison of the amino acid sequences of three bacterial liquefying α-amylases deduced from the DNA sequences. J Biochem. 1985;98:1147–1156. doi: 10.1093/oxfordjournals.jbchem.a135381. [DOI] [PubMed] [Google Scholar]