Abstract
Muconate cycloisomerases play a crucial role in the bacterial degradation of aromatic compounds by converting cis,cis-muconate, the product of catechol ring cleavage, to (4S)-muconolactone. Chloromuconate cycloisomerases catalyze both the corresponding reaction and a dehalogenation reaction in the transformation of chloroaromatic compounds. This study reports the first thorough examination of the substrate specificity of the muconate cycloisomerases from Pseudomonas putida PRS2000 and Acinetobacter “calcoaceticus” ADP1. We show that they transform, in addition to cis,cis-muconate, 3-fluoro-, 2-methyl-, and 3-methyl-cis,cis-muconate with high specificity constants but not 2-fluoro-, 2-chloro-, 3-chloro-, or 2,4-dichloro-cis,cis-muconate. Based on known three-dimensional structures, variants of P. putida muconate cycloisomerase were constructed by site-directed mutagenesis to contain amino acids found in equivalent positions in chloromuconate cycloisomerases. Some of the variants had significantly increased specificity constants for 3-chloro- or 2,4-dichloromuconate (e.g., A271S and I54V showed 27- and 22-fold increases, respectively, for the former substrate). These kinetic improvements were not accompanied by a change from protoanemonin to cis,cis-dienelactone as the product of 3-chloro-cis,cis-muconate conversion. The rate of 2-chloro-cis,cis-muconate turnover was not significantly improved, nor was this compound dehalogenated to any significant extent. However, the direction of 2-chloro-cis,cis-muconate cycloisomerization could be influenced by amino acid exchange. While the wild-type enzyme discriminated only slightly between the two possible cycloisomerization directions, some of the enzyme variants showed a strong preference for either (+)-2-chloro- or (+)-5-chloromuconolactone formation. These results show that the different catalytic characteristics of muconate and chloromuconate cycloisomerases are due to a number of features that can be changed independently of each other.
Despite the persistence of some chloro-substituted aromatic compounds in the environment (15), various bacterial strains show a remarkable capability for the degradation and mineralization of some of these compounds (for recent reviews, see, for example, references 19, 35, and 42). A major route for their aerobic degradation is via ortho cleavage of chlorocatechols, which occur as central intermediates in the degradation pathways of many chloroaromatic substances. A crucial step in the subsequent catabolism is the cycloisomerization of chloro-substituted cis,cis-muconates, resulting in the formation of dienelactones (4-carboxymethylenebut-2-en-4-olides) after the elimination of a chloro-substituent (Fig. 1) (5, 12). This reaction is catalyzed by chloromuconate cycloisomerases (EC 5.5.1.7) (45). Homologous enzymes, muconate cycloisomerases (EC 5.5.1.1), are induced during the degradation of nonchlorinated aromatic compounds via ortho cleavage of catechol. Their function is to convert cis,cis-muconate to (4S)-muconolactone (3, 52), which differs from the dienelactones in that it lacks the additional, exocyclic double bond (Fig. 1).
FIG. 1.
Reactions catalyzed by muconate and chloromuconate cycloisomerases. Letters adjacent to arrows indicate whether the respective reaction is catalyzed by muconate cycloisomerase (MCI) or chloromuconate cycloisomerase (CMCI); the superscript “n” indicates enzymes from gram-negative bacteria (usually β or γ Proteobacteria), and the superscript “p” indicates enzymes from gram-positive bacteria (R. opacus). Numbers adjacent to the arrows indicate whether the reaction is a 1,4- or a 3,6-cycloisomerization. No attempt was made to differentiate between fast turnover and slow turnover.
Muconate cycloisomerase and chloromuconate cycloisomerase were originally identified as being distinct from each other in 3-chlorobenzoate-utilizing Pseudomonas sp. strain B13, in which the former enzyme has considerably lower relative Vmax values and, in general, higher Km values for chloro- and methyl-cis,cis-muconates than the latter enzyme (45). A loss of overall catalytic activity was suggested to have accompanied the acquisition of the broader substrate range by chloromuconate cycloisomerase (32). The substrate specificity of the chloromuconate cycloisomerase carried by pJP4 in 2,4-dichlorophenoxyacetate-degrading Ralstonia eutropha (Alcaligenes eutrophus) JMP134, in contrast, is much more restricted to 2,4-dichloro-, 3-chloro-, and 3-methyl-cis,cis-muconate than that of the strain B13 enzyme, and this fact coincides with high overall turnover rates (28). In addition to the chloromuconate cycloisomerase of pJP4, so far only the muconate and chloromuconate cycloisomerases of chlorophenol-degrading Rhodococcus opacus (Rhodococcus erythropolis) 1CP have been investigated in detail with respect to their substrate specificity (53).
Muconate and chloromuconate cycloisomerases differ from each other not only with respect to substrate specificity but also in many cases with respect to product formation. Muconate cycloisomerases of gram-negative bacteria convert 2-chloro-cis,cis-muconate to mixtures of (+)-2-chloro- and (+)-5-chloromuconolactone by carrying out both 1,4- and 3,6-cycloisomerizations of the substrate (Fig. 1) (57). In contrast, the chloromuconate cycloisomerase of pAC27, which is identical to that of strain B13 (49), as well as the chloromuconate cycloisomerase of pJP4, seems to favor 3,6-cycloisomerization to (+)-5-chloromuconolactone and additionally catalyze dehalogenation of this intermediate to trans-dienelactone (Fig. 1) (28, 45, 59). In gram-positive R. opacus 1CP, both cycloisomerases discriminate between cycloisomerization directions strongly in favor of (+)-5-chloromuconolactone, which is not dehalogenated (53). 3-Chloro-cis,cis-muconate is converted by chloromuconate cycloisomerases to cis-dienelactone (45), but muconate cycloisomerases, in general, form the bacteriotoxic protoanemonin (4).
The cycloisomerases are an excellent system for investigating at the molecular level how enzymes adapt to be able to convert chloro-substituted compounds. The three-dimensional structure of the muconate cycloisomerase from Pseudomonas putida PRS2000 is known at 1.85-Å resolution (18, 21), and that of the chloromuconate cycloisomerase of R. eutropha JMP134(pJP4) is known at 3 Å (23, 25); unfortunately, however, structures with a bound substrate or inhibitor are not yet available. The structural similarities between cycloisomerases and mandelate racemase (31) indicate that, besides Mn2+, Glu327 and Lys167 (P. putida muconate cycloisomerase numbering) are important for the stabilization of a lactonic enol/ enolate as an important intermediate in the catalytic mechanism. A proton is added to this intermediate from Lys169 to yield the final product, (4S)-muconolactone (14). Furthermore, as there is 40 to 44% sequence identity between muconate cycloisomerases from P. putida strains (1, 24) and Acinetobacter “calcoaceticus” ADP1 (50) and chloromuconate cycloisomerases from plasmids pJP4, pAC27, and pP51 (13, 16, 34, 56), the cycloisomerases are more conserved than the dioxygenases and lactone hydrolases (42).
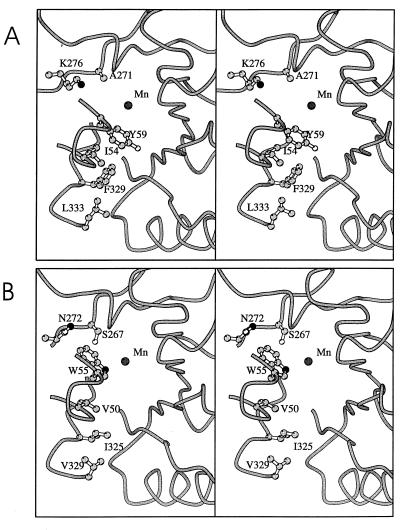
Initial structure comparisons suggested that Val50, Trp55, Ser267, Ile325, and Val329 (chloromuconate cycloisomerase numbering) might be responsible for widening the active site of chloromuconate cycloisomerase of pJP4 in comparison with P. putida muconate cycloisomerase, thereby allowing larger substrates to bind (23) (compare Fig. 2). Multiple sequence alignments were in agreement with this suggestion, since they showed that the amino acids in these positions were essentially conserved among all proteobacterial chloromuconate cycloisomerases, while different amino acids were conserved in the corresponding positions among the proteobacterial muconate cycloisomerases (Ile54, Tyr59, Ala271, Phe329, and Leu333; P. putida muconate cycloisomerase numbering). Lys276 in P. putida muconate cycloisomerase is in contact with one end of the active-site cavity (Fig. 2). Its exchange to asparagine, which is conserved in the corresponding position of chloromuconate cycloisomerases, was likewise assumed to widen the pocket and make it more accessible.
FIG. 2.
Stereo views of the active-site regions of P. putida muconate cycloisomerase (21) (A) and chloromuconate cycloisomerase of pJP4 (25) (B) drawn with MOLSCRIPT (27). The path of the backbone is shown as a tube, and the side chains in the active site that were mutated in this study are shown as balls and sticks (carbon atoms in light grey, nitrogen atoms in black, and oxygen atoms in white). The position of the active-site manganese ion is shown (dark-grey circle).
So far it is unclear, however, what effects these changes have on the catalytic properties of the enzymes. Also unknown are the substrate specificities of the proteobacterial muconate cycloisomerases for which structure and/or sequence information is available. Furthermore, little is known about how and to what extent various substituents affect binding to and conversion of the substrates by muconate cycloisomerases and whether or not different catalytic features of cycloisomerases are necessarily connected to each other. The present paper aims to fill these gaps in the following ways. First, we characterize fully and for the first time the substrate specificities and catalytic properties of two proteobacterial wild-type muconate cycloisomerases. Second, we similarly characterize seven active-site variants of the P. putida enzyme, chosen because (i) they might widen the active-site cavity and (ii) they are conserved in chloromuconate cycloisomerases (see above). Third, we explore to what extent the different reactivities of chloromuconates can be accounted for merely by their solution chemistry.
(A preliminary report of this work has been presented previously [58]).
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. Plasmid-containing strains were usually grown in Luria-Bertani, 2×YT (38), or TB (55) medium supplemented with 100 μg of ampicillin per ml at 37°C in a rotary shaker. For growth on plates, 1.5% (wt/vol) agar was added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21 (DE3)(pLysS) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) pLysS (Cmr) | 38, 54 |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1 | GIBCO BRL |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15) | Amersham Sculptor-Kit (17, 38) |
| Plasmids | ||
| pPX31 | pUC19 (Apr) with catB from P. putida PRS2000 on a 2.1-kbp PstI fragment | L. N. Ornston (24) |
| pET-11a* | Apr PT7lac derivative of pET-11a containing additional KpnI, XhoI, and SalI cloning sites | J. Altenbuchner (54) |
| pUC BM20 | Apr Plac derivative of pUC18 with larger multiple cloning site | Boehringer Mannheim |
| pBluescript II KS(+) | Apr Plac f1(+) and ColE1 origin | Stratagene (2) |
| pBluescript II SK(+) | Apr Plac f1(+) and ColE1 origin | Stratagene (2) |
| pCATB1 | pET-11a* (Apr) with catB flanked by NdeI and BamHI sites | This study |
| pCATB2 | pUC BM20 (Apr) with catB flanked by NdeI and BamHI sites | This study |
| pCATB3 | pUC BM20 (Apr) with 855-bp HindIII-BamHI fragment of catB | This study |
| pCATB4 | pBluescript II KS(+) (Apr) with 237-bp SalI-HindIII fragment of catB | This study |
| pCATB5 | pBluescript II SK(+) (Apr) with 211-bp SacII-XhoI fragment of catB | This study |
| pCATB6 | pBluescript II SK(+) (Apr) with 319-bp SacII-HincII fragment of catB | This study |
| pCATB51 | pET-11a* (Apr) with NdeI-BamHI fragment carrying I54V-CatB | This study |
| pCATB52 | pET-11a* (Apr) with NdeI-BamHI fragment carrying Y59W-CatB | This study |
| pCATB53 | pET-11a* (Apr) with NdeI-BamHI fragment carrying K276N-CatB | This study |
| pCATB54 | pET-11a* (Apr) with NdeI-BamHI fragment carrying A271S-CatB | This study |
| pCATB55 | pET-11a* (Apr) with NdeI-BamHI fragment carrying F329I-CatB | This study |
| pCATB56 | pET-11a* (Apr) with NdeI-BamHI fragment carrying L333V-CatB | This study |
| pCATB57 | pET-11a* (Apr) with NdeI-BamHI fragment carrying I54V-Y59W-CatB | This study |
| pCATB58 | pET-11a* (Apr) with NdeI-BamHI fragment carrying I54V-F329I-CatB | This study |
| pCATB59 | pET-11a* (Apr) with NdeI-BamHI fragment carrying Y59-F329I-CatB | This study |
| pCATB60 | pET-11a* (Apr) with NdeI-BamHI fragment carrying I54V-Y59W-F329I-CatB | This study |
Apr, resistance to ampicillin; Cmr, resistance to chloramphenicol.
DNA preparation and in vitro manipulation.
Plasmid DNA was isolated either by the method of Lee and Rasheed (29) or by use of Pharmacia FlexiPrep-Kit or Macherey-Nagel Nucleobond AX 100 cartridges. Restriction endonucleases, T4 DNA ligase, and alkaline phosphatase were obtained from GIBCO BRL or New England BioLabs and applied in accordance with the instructions of the manufacturers. Escherichia coli strains were transformed by the method of Chung et al. (8).
Site-directed mutagenesis and sequencing.
To obtain the desired mutations in catB, the muconate cycloisomerase gene, phosphorothioate-based site-directed mutagenesis (40) was carried out with an Amersham Sculptor-Kit. Mutagenesis experiments were performed with single-stranded DNA rescued from E. coli TG1 host strains grown on TYP medium (containing, per liter, 16 g of tryptone, 16 g of yeast extract, 5 g of NaCl, and 2.5 g of K2HPO4) by infection with helper phage R408 (37). The latter was obtained from J. Altenbuchner, Stuttgart, Germany. The host strains contained either pCATB4, pCATB5, or pCATB6, each of which carries short fragments of catB in pBluescript II vectors (Table 1 and Fig. 3). Annealing of the mutagenic oligonucleotides (Table 2) and the mutagenesis procedures were performed as suggested in the Sculptor-Kit manual. Mutations were subsequently verified by the dideoxy sequencing method of Sanger et al. (39) by use of a United States Biochemicals Sequenase 2.0 kit with [α-35S]dATP or by use of the dye terminator thermocycler sequencing protocol (AmpliTaq DNA polymerase; 25 cycles: 98°C, 1 s; 60°C, 15 s or, in the final step, 4 min; model 373 automated sequencer from Applied Biosystems).
FIG. 3.
Plasmids and overall strategy used for the construction and expression of muconate cycloisomerase variants by site-directed mutagenesis. The catB gene of P. putida, its fragments, and mutated derivatives are indicated by boxes; vectors are indicated by lines (black for pUC BM20 and pBluescript II and grey for pET-11a*).
TABLE 2.
Site-directed mutagenesis of catB from P. putida PRS2000 and the respective changes on the DNA and protein levels
| Amino acid sub- stitution | Nucleotide substitution in catBa | Mutagenesis oligonucleotideb |
|---|---|---|
| I54V | ATC→GTC | 5′-CAGGCCACCGACGGTGGTGG-3′ |
| Y59W | TAT→TGG | 5′-CGTAGCCCCAGGCCAGGCCAC-3′ |
| A271S | GCC→TCC | 5′-CGATTTTCAGGGAAAAGATGC-3′ |
| K276N | AAG→AAC | 5′-GCCGCCGTTGTTGGCGATTTTC-3′ |
| F329I | TTC→ATC | 5′-CAGCGGGCCGATCAGCTCGG-3′ |
| L333V | CTG→GTG | 5′-CGGTCAGCACCAGCGGGCCG-3′ |
| I54V-Y59W | ATC→GTC and TAT→TGG | 5′-GCCCCAGGCCAGGCCACCGACG GTG-3′ |
Bases in boldface were exchanged.
Sequences of mutagenesis oligonucleotides were complementary, in an inverted orientation, to single-stranded templates. Characters in boldface indicate nucleotide changes compared to the wild-type sequence.
Construction of expression plasmids containing complete catB or its variants.
For subcloning of wild-type catB into the NdeI and BamHI sites of expression plasmid pET-11a*, the gene was amplified by PCR with the following primers: 5′-CGTCATATGACAAGCGCGCTGATTGAACG-3′, containing the start codon (bold) in the NdeI site (underlined), and 5′-CGCGGATCCTGCTGATCAGCGACGGGCGAAG-3′, comprising the inverse complement of the 3′ end of catB (bold) and the BamHI site (underlined). The reaction mixture (final volume, 100 μl) initially contained 100 pmol of primer, 20 nmol of each deoxynucleoside triphosphate, 150 pg of pPX31 as the template, and reaction buffer. After heating to 94°C for 4 min, 2.5 U of Taq polymerase (Pharmacia) was added and, under a layer of mineral oil, the mixture was incubated for 30 cycles of the following temperature profile: 94°C, 90 s; 50°C, 90 s; and 72°C, 180 s. After ligation of the NdeI-BamHI-digested PCR product into pET-11a*, the authenticity of the resulting plasmid, pCATB1, was confirmed by sequencing. The sequence of catB in pCATB1, as well as in the starting plasmid pPX31, obtained from L. N. Ornston, New Haven, Conn., was found to differ from the published sequence (24). The following five changes corrected the sequence: position 413, A→T; position 489, T→C; positions 725 and 726, CG→GC; position 786, T→C; and position 798, G→C. The changes at position 413 and positions 725 and 726, respectively, indicate that the muconate cycloisomerase carries a valine instead of a glutamate at position 138 and a serine instead of a threonine at position 242.
Complete catB genes carrying the desired mutations were constructed on the pUC BM20 vector since, after insertion of the NdeI-BamHI fragment with catB (yielding pCATB2), the relevant restriction sites occurred only on the insert (Fig. 3). The inserts from the pCATB4 and pCATB5 derivatives were excised, purified by agarose gel electrophoresis, eluted from the gel by the Gene-clean procedure (Bio 101, Inc., La Jolla, Calif.), and ligated with the larger fragments of pCATB2 isolated after digestion of this plasmid with the same enzymes. The SacII-HincII inserts from pCATB6 derivatives containing the mutation F329I or L333V were isolated and ligated with the large fragment of SacII-HincII-digested pCATB3. The resulting plasmids were subsequently digested with BamHI and HindIII, and an 855-bp fragment was ligated with the large fragment of pCATB2 cut with the same endonucleases. Genes carrying the F329I mutation in addition to the I54V, Y59W, or I54V-Y59W mutations were constructed by digestion of the respective pCATB2 derivatives with BamHI and SacII and ligation of the small fragment (436 bp) carrying the F329I mutation with the respective large fragments (3.1 kbp) carrying the other mutations. The inserts from the pCATB2 derivatives (vector pUC BM20) were checked by DNA sequencing of the complete genes. For expression of the CatB variants, the NdeI-BamHI fragments with altered catB were excised from the pUC BM20 vector and ligated with NdeI-BamHI-digested pET-11a*, yielding plasmids pCATB51 to pCATB60.
Enzyme expression and preparation of cell extracts.
E. coli BL21(DE3)(pLysS) was used as the host strain to express wild-type CatB from pCATB1 and the CatB variants from pCATB51 to pCATB60. One-liter cultures were grown at 37°C in 2×YT medium with ampicillin to an optical density at 546 nm of 0.7. Induction was achieved by adding 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Subsequently, the cultures were incubated at 30°C for 3 to 3.5 h. Cells were harvested by centrifugation for 15 min at 3,000 × g and 4°C, washed with 20 mM Tris-HCl (pH 7.5)–1 mM MnSO4, and stored at −20°C until used. Frozen cells were later resuspended in 20 ml of the same buffer. After the addition of DNase I, the cells were passed twice through a cooled Aminco French pressure cell operated at 115 MPa. Cell debris was removed by centrifugation for 30 min at 130,000 × g and 4°C. The clear supernatant was used for purification on the same day.
Enzyme assays and analysis of kinetic data.
The activities of wild-type muconate cycloisomerase and its derivatives were measured spectrophotometrically at 260 nm and 25°C with a solution which usually contained 30 mM Tris-HCl (pH 8.0), 1 mM MnSO4, and 0.1 mM cis,cis-muconate (33). For the kinetic experiments, the pH was adjusted to 7.5, and at least two independent assays usually at nine different substrate concentrations between 2.5 and 175 μM were performed. When chloro-substituted substrates were used, dienelactone hydrolase partially purified from R. eutropha JMP134 was added in excess. Extinction coefficients usually were taken from Dorn and Knackmuss (9). For the conversion of 2,4-dichloro-cis,cis-muconate, a coefficient of 5,800 M−1 cm−1 (28) was used. As has been observed for other muconate cycloisomerases (4), the conversion of 3-chloro-cis,cis-muconate yielded protoanemonin as the main product (see below); like the substrate, protoanemonin shows strong absorption at 260 nm (51). Nevertheless, an initial linear decrease in the absorption (for less than 1 min) was observed, probably due to the formation of an unstable reaction intermediate. For this initial phase, the difference in the extinction coefficients of the substrate and product(s) was estimated to be ca. 4,000 M−1 cm−1 (the value was difficult to determine precisely because of the short duration of the phase; it might be dependent on the exact conditions). Protein concentrations were determined by the method of Bradford (6) with bovine serum albumin as the standard. Kinetic parameters were calculated by nonlinear regression with the Enzfitter program (Biosoft, Cambridge, United Kingdom). The turnover numbers (kcat values) were calculated by assuming a subunit molecular mass of 40 kDa.
Enzyme purification.
Wild-type muconate cycloisomerase from P. putida and its variants were purified essentially by the same procedures. All chromatographic steps were carried out at room temperature with separation media and a fast protein liquid chromatography system from Pharmacia. Purification was initiated by anion-exchange chromatography by use of a Q Sepharose high-performance HR 16/10 column with 25 mM Tris-HCl (pH 7.5)–1 mM MnSO4 as the buffer and a linear gradient of 0 to 250 mM NaCl over 15 column volumes (300 ml) for elution. The fractions with the highest muconate cycloisomerase activities (eluting at approximately 140 mM NaCl) were combined, and 0.4 M ammonium sulfate was added. After filtration, further purification was achieved by hydrophobic-interaction chromatography on a Phenyl Superose HR 10/10 column equilibrated with 20 mM Tris-HCl (pH 7.5)–1 mM MnSO4–0.4 M (NH4)2SO4. With a decreasing gradient of 0.4 to 0 M ammonium sulfate over 12.5 column volumes (100 ml), the main activity peaks of muconate cycloisomerase and its derivatives occurred at about 30 mM ammonium sulfate. Wild-type muconate cycloisomerase, for example, had a specific activity of 36.1 U/mg in the cell extract (total protein, 316 mg; total activity, 11,400 U). The specific activity increased 4.2-fold in the Q Sepharose high-performance step (yield, 85%). In the final Phenyl Superose step, a specific activity of 160 U/mg was obtained (4.4-fold purification; 51% overall yield). The resulting preparations of wild-type and variant muconate cycloisomerases were at least 95% pure, as estimated with overloaded sodium dodecyl sulfate-polyacrylamide gels (57) stained with Coomassie brilliant blue R-250 (data not shown).
Analyses of product formation.
Product formation during the turnover of 2-chloro- and 3-chloro-cis,cis-muconate was analyzed by reversed-phase high-pressure liquid chromatography (HPLC) with a Grom-SIL 100 C8 column (length, 250 mm; internal diameter, 4.6 mm; Grom, Herrenberg, Germany) under conditions described by Vollmer et al. (57).
See Table 5 for the conditions used for 2-chloro-cis,cis-muconate conversion. Product formation from 3-chloro-cis,cis-muconate was analyzed by use of 1-ml reaction mixtures containing 30 mM Tris-HCl (pH 7.0 or 7.5), 1 mM MnSO4, and 0.25 or 0.5 mM substrate. Between 0.2 and 2 U of enzyme activity (measured with cis,cis-muconate) was added to ensure that after 10 min (at 25°C), no residual 3-chloro-cis,cis-muconate was detectable by HPLC analyses.
TABLE 5.
Product formation during 2-chloro-cis,cis-muconate turnover by variants of muconate cycloisomerasea
| Variant | Concn of residual substrate (mM) | Concn of the product (mM):
|
5-CML/2-CML ratio | trans-DL/(trans-DL + 5-CML) ratio (%) | ||
|---|---|---|---|---|---|---|
| 2-CML | 5-CML | trans-DL | ||||
| I54V | 0.32 | 0.020 | 0.16 | 0.013 | 8.0 | 7.5 |
| Y59W | 0.38 | 0.088 | 0.0073 | ND | 0.083 | ND |
| A271S | 0.23 | 0.083 | 0.19 | 0.013 | 2.3 | 6.4 |
| K276N | 0.28 | 0.074 | 0.13 | 0.0039 | 1.8 | 2.9 |
| F329I | 0.33 | 0.13 | 0.0023 | <0.001 | 0.018 | ND |
| L333V | 0.18 | 0.085 | 0.23 | 0.024 | 2.7 | 9.4 |
| I54V-Y59W | 0.32 | 0.0081 | 0.17 | 0.017 | 21 | 9.1 |
| I54V-F329I | 0.29 | 0.059 | 0.15 | 0.0078 | 2.5 | 4.9 |
| Y54W-F329I | 0.38 | 0.072 | 0.034 | 0.0025 | 0.47 | 6.8 |
| I54V-Y59W-F329I | 0.29 | 0.042 | 0.15 | 0.0081 | 3.6 | 5.1 |
| None (wild type) | 0.22 | 0.090 | 0.19 | 0.0045 | 2.1 | 2.3 |
The reactions were carried out with 30 mM Tris-HCl (pH 7.0), 1 mM MnSO4, 0.5 mM 2-chloro-cis,cis-muconate, and ca. 0.005 U of enzyme per ml (assayed with 2-chloro-cis,cis-muconate at pH 7.5) at 25°C. Concentrations were determined by HPLC analyses after a reaction time of 70 min. 2-CML, 2-chloromuconolactone; 5-CML, 5-chloromuconolactone; trans-DL, trans-dienelactone. ND, not determined.
Chemicals.
cis,cis-Muconic acid was generously provided by J. E. Adamus (Celgene Corp., Warren, N.J.). 2-Chloro- and 2-fluoro-cis,cis-muconic acid had been prepared previously (53, 57). All other substituted cis,cis-muconates were obtained enzymatically in situ from the respective catechols by use of partially purified chlorocatechol 1,2-dioxygenase from R. eutropha JMP134 (28). These reactions were controlled by repeatedly recording UV absorption spectra (200 to 400 nm) with a double-beam spectrophotometer. 3-Methyl- and 4-methylcatechol were purchased from Aldrich Chemie (Steinheim, Germany), and 4-fluoro-, 4-chloro-, and 3,5-dichlorocatechol were available from previous syntheses (9, 43, 47). cis-Dienelactone was kindly provided by S. R. Kaschabek und W. Reineke (Wuppertal, Germany). Protoanemonin was initially synthesized from trans-acetylacrylic acid as described by Shaw (51). It was later prepared (always freshly on the day of its use as a standard) by converting 3-chloro-cis,cis-muconate with large amounts of P. putida muconate cycloisomerase in the presence of dienelactone hydrolase partially purified from R. eutropha JMP134. The protoanemonin content in standard solutions was estimated from the absorption at 260 nm with 14,000 M−1 cm−1 as the extinction coefficient (51). trans-Dienelactone and (+)-5-chloro- and (+)-2-chloromuconolactone were available from previous syntheses (36, 57).
RESULTS
Kinetics of substrate conversion.
Kinetic analyses of the purified wild-type muconate cycloisomerase from P. putida PRS2000 with different substituted cis,cis-muconates showed, as expected, that cis,cis-muconate was converted with the highest turnover rate and a high affinity (Table 3). In addition, 3-fluoro-, 2-methyl-, and 3-methyl-cis,cis-muconate proved to be good substrates. For 3-fluoro-cis,cis-muconate, a very high kcat value was measured, and for 3-methyl-cis,cis-muconate, the Km value was very low. Analyses of enzyme kinetics with chloromuconate isomers, however, revealed that these compounds were poor substrates for muconate cycloisomerase. This was primarily due to the extremely low kcat values but, for 3-chloro-cis,cis-muconate, low affinity contributed as well. Significant conversion of 2-fluoro-cis,cis-muconate (0.1 mM) could not be detected by photometric measurements (data not shown), but this compound was, like 2,4-dichloro- and 3-chloro-cis,cis-muconate, an inhibitor of cis,cis-muconate conversion. Inhibition studies with 0.025 and 0.05 mM 2-fluoro-cis,cis-muconate, 0.05, 0.075, and 0.10 mM 3-chloro-cis,cis-muconate, and 0.005, 0.0125, and 0.025 mM 2,4-dichloro-cis,cis-muconate, although not definitive, suggested competitive or mixed-type inhibition (data not shown). Assuming the former, by plotting the slopes of the primary Lineweaver-Burk plots against the inhibitor concentrations (48), Ki values were determined to be 3.5 μM for 2,4-dichloro-cis,cis-muconate, 31 μM for 2-fluoro-cis,cis-muconate, and 95 μM for 3-chloro-cis,cis-muconate.
TABLE 3.
Kinetic constants of P. putida muconate cycloisomerase and variants with various substratesa
| Substrate | Kinetic constant of the following enzyme:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
I54V
|
Y59W
|
A271S
|
K276N
|
F329I
|
L333V
|
I54V-F329I
|
|||||||||||||||||
| Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km(min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | |
| cis,cis-Muconate | 40 ± 4 | 12,600 ± 400 | 310 | 96 ± 8 | 2,500 ± 100 | 26 | 47 ± 3 | 1,100 ± 100 | 24 | 35 ± 3 | 7,100 ± 200 | 200 | 74 ± 5 | 2,100 ± 100 | 29 | 47 ± 4 | 2,600 ± 100 | 54 | 52 ± 8 | 68 ± 4 | 1.3 | 140 ± 10 | 540 ± 30 | 3.8 |
| 2-Chloro-cis,cis-muconate | 59 ± 5 | 49 ± 2 | 0.83 | 47 ± 4 | 35 ± 1 | 0.75 | 17 ± 1 | 15 ± 1 | 0.87 | 60 ± 9 | 15 ± 1 | 0.25 | 34 ± 1 | 3.2 ± 0.1 | 0.095 | 37 ± 2 | 48 ± 1 | 1.3 | 36 ± 2 | 0.73 ± 0.02 | 0.020 | 33 ± 2 | 10 ± 0.2 | 0.32 |
| 3-Chloro-cis,cis-muconateb | 320 ± 200 | 64 ± 30 | 0.20 | 45 ± 8 | 200 ± 10 | 4.4 | ND | ND | ND | 180 ± 20 | 960 ± 60 | 5.4 | 550 ± 230 | 380 ± 130 | 0.69 | 690 ± 400 | 460 ± 230 | 0.67 | 76 ± 15 | 52 ± 5 | 0.68 | 210 ± 40 | 110 ± 10 | 0.54 |
| 2,4-Dichloro-cis,cis-muconate | 26 ± 3 | 3.3 ± 0.1 | 0.13 | 93 ± 21 | 7.2 ± 0.8 | 0.078 | ND | ND | ND | 44 ± 8 | 2.9 ± 0.2 | 0.067 | 40 ± 3 | 0.61 ± 0.16 | 0.015 | 78 ± 16 | 32 ± 3 | 0.41 | 39 ± 3 | 0.88 ± 0.02 | 0.022 | 45 ± 6 | 22 ± 1 | 0.49 |
| 3-Fluoro-cis,cis-muconate | 97 ± 21 | 8,900 ± 1,000 | 92 | 74 ± 8 | 1,200 ± 100 | 16 | 150 ± 20 | 3,200 ± 200 | 21 | 100 ± 12 | 6,000 ± 400 | 60 | 230 ± 20 | 5,200 ± 300 | 22 | 150 ± 10 | 8,000 ± 500 | 53 | 63 ± 6 | 4,100 ± 200 | 65 | 100 ± 9 | 4,900 ± 200 | 47 |
| 2-Methyl-cis,cis-muconate | 40 ± 2 | 3,900 ± 100 | 97 | 58 ± 5 | 650 ± 30 | 11 | 69 ± 6 | 3,700 ± 200 | 53 | 59 ± 5 | 6,400 ± 200 | 110 | 180 ± 10 | 1,000 ± 40 | 5.8 | 68 ± 3 | 10,000 ± 200 | 150 | 110 ± 20 | 320 ± 30 | 2.8 | 57 ± 4 | 1,400 ± 100 | 24 |
| 3-Methyl-cis,cis-muconate | 6.0 ± 0.5 | 590 ± 10 | 99 | 7.2 ± 0.7 | 740 ± 20 | 100 | 15 ± 1 | 140 ± 10 | 9.7 | 9.7 ± 0.4 | 680 ± 10 | 71 | 12 ± 1 | 95 ± 2 | 8.2 | 16 ± 1 | 270 ± 10 | 17 | 9.1 ± 1.7 | 54 ± 3 | 6.0 | 6.4 ± 0.6 | 310 ± 10 | 49 |
The Km and kcat values were determined as described in Materials and Methods. Standard deviations were calculated with the Enzfitter program. Km values lower than and kcat or kcat/Km values higher than those for the wild-type enzyme are shown in italic type and underlined if they differ by more than a factor of 2 and in boldface type and double underlined if they differ by a factor of at least 10. ND, not determined.
The reaction with 3-chloro-cis,cis-muconate was experimentally difficult to assay due to protoanemonin formation (see Materials and Methods) (4), so the standard deviations were relatively high.
The wild-type muconate cycloisomerase from A. “calcoaceticus” ADP1, which was investigated for comparison, had significantly higher Km values for all substrates except for 3-fluoro-cis,cis-muconate. Additionally, it was less active with this substrate as well as with cis,cis-muconate, while it had higher activity with 3-methyl-cis,cis-muconate (Table 4). As for the P. putida cycloisomerase, 2-fluoro-cis,cis-muconate and the chloromuconates were poor substrates for the Acinetobacter enzyme. Ki values (determined as described above) were 35 μM for 2,4-dichloro-cis,cis-muconate (with inhibitor concentrations of 0.033, 0.05, and 0.1 mM) and 27 μM for 2-fluoro-cis,cis-muconate (with inhibitor concentrations of 0.033, 0.05, and 0.075 mM).
TABLE 4.
Substrate specificity of the muconate cycloisomerase from A. “calcoaceticus” ADP1a
| Substrate | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) |
|---|---|---|---|
| cis,cis-Muconate | 130 ± 10 | 3,700 ± 200 | 29 |
| 2-Chloro-cis,cis-muconate | 290 ± 90 | 17 ± 4 | 0.06 |
| 3-Chloro-cis,cis-muconate | (<10)b | ||
| 2,4-Dichloro-cis,cis-muconate | (<10)b | ||
| 3-Fluoro-cis,cis-muconate | 100 ± 10 | 2,600 ± 200 | 24 |
| 2-Methyl-cis,cis-muconate | 490 ± 120 | 4,700 ± 1,000 | 9.7 |
| 3-Methyl-cis,cis-muconate | 99 ± 7 | 1,800 ± 100 | 18 |
The Km and kcat values were determined as described in Materials and Methods. Standard deviations were calculated with the Enzfitter program. The measurements were determined with an enzyme preparation described previously (57).
The activity at a substrate concentration of 0.1 mM was below the detection limit for this measurement. Consequently, kcat, Km, and kcat/Km values could not be determined.
With cis,cis-muconate, the purified P. putida muconate cycloisomerase variants showed a more or less pronounced decrease in kcat values and, often, a slight increase in Km values compared to the wild-type enzyme (Table 3). Decreased activities were generally also found with the other good substrates for the wild-type muconate cycloisomerase, such as 3-fluoro-, 2-methyl-, and 3-methyl-cis,cis-muconate (Table 3). However, several of the variants analyzed showed, as we hoped, a significant increase in turnover rates for 3-chloro- and 2,4-dichloro-cis,cis-muconate. Compared to the wild-type enzyme, enzymes with I54V and A271S substitutions showed 22- and 27-fold increases, respectively, in the specificity constant for 3-chloro-cis,cis-muconate (Table 3), due to both higher affinities and higher turnover rates. The F329I and I54V-F329I variants had 3.2- and 3.8-fold-higher specificity constants for 2,4-dichloro-cis,cis-muconate than the wild-type enzyme (Table 3). This effect was caused by 9.7- and 6.7-fold increases in kcat values accompanied by concurrent increases in Km values. An improved specificity constant for 3-chloro- or 2,4-dichloro-cis,cis-muconate was not, however, accompanied by a significant increase in the kcat/Km values for 2-chloro-cis,cis-muconate. The double and triple variants I54V-Y59W, Y59W-F329I, and I54V-Y59W-F329I (not shown in Table 3) were less active than the variants with only one substitution. Like the wild-type muconate cycloisomerase, none of the CatB variants showed significant activity in photometric assays for the conversion of 2-fluoro-cis,cis-muconate (0.1 mM).
Product formation during the conversion of chloro-cis,cis-muconates.
As mentioned above, muconate and chloromuconate cycloisomerases differ not only in their substrate specificities but also with respect to the products formed from chloromuconates (Fig. 1). Thus, it was of considerable interest to investigate whether the muconate cycloisomerase variants would differ from the wild-type enzyme in this respect. HPLC analyses of the reaction mixtures after 3-chloro-cis,cis-muconate turnover revealed that both the P. putida wild-type muconate cycloisomerase and all analyzed mutant derivatives (Y59W was not tested) catalyzed the formation of protoanemonin as the main product. Some cis-dienelactone, the only product formed from 3-chloro-cis,cis-muconate by chloromuconate cycloisomerase, was also detected. For wild-type CatB, the cis-dienelactone concentration was 8% that of protoanemonin. None of the mutations resulted in a higher percentage yield.
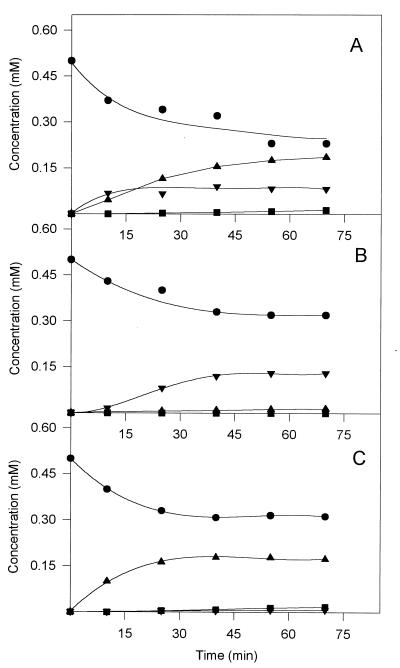
While the P. putida muconate cycloisomerase converts 2-chloro-cis,cis-muconate to a pH-dependent equilibrium mixture of (+)-2-chloromuconolactone, (+)-5-chloromuconolactone, and residual substrate (57), chloromuconate cycloisomerases of gram-negative bacteria catalyze an additional dehalogenation of (+)-5-chloromuconolactone to trans-dienelactone (Fig. 1) (59). As indicated by HPLC analyses, several of the muconate cycloisomerase variants appeared to dehalogenate during 2-chloro-cis,cis-muconate turnover somewhat better than the wild-type enzyme, but none dehalogenated nearly as well as the chloromuconate cycloisomerases (Table 5). Thus, in this respect, the variants still resembled the wild-type muconate cycloisomerase. With some amino acid exchanges, however, discrimination between the cycloisomerization directions of 2-chloro-cis,cis-muconate occurred, in contrast to the situation with the wild-type enzyme, which formed considerable amounts of both (+)-5-chloro- and (+)-2-chloromuconolactone. The I54V and I54V-Y59W variants formed 5-chloromuconolactone almost exclusively (Fig. 4C): with these mutations, the 3,6-cycloisomerization direction was preferred. The F329I and Y59W variants formed 2-chloromuconolactone as the chief product, meaning that the 1,4-cycloisomerization direction was strongly favored (Table 5 and Fig. 4B). The third group of variants, such as A271S, behaved similarly to the wild-type enzyme with respect to the cycloisomerization direction (Fig. 4A).
FIG. 4.
Time course of 2-chloro-cis,cis-muconate turnover by muconate cycloisomerase variants A271S (A), F329I (B), and I54V-Y59W (C). The concentrations of 2-chloro-cis,cis-muconate (•), 5-chloromuconolactone (▴), 2-chloromuconolactone (▾), and trans-dienelactone (■) were analyzed by HPLC. Reaction conditions are indicated in Table 5, footnote a.
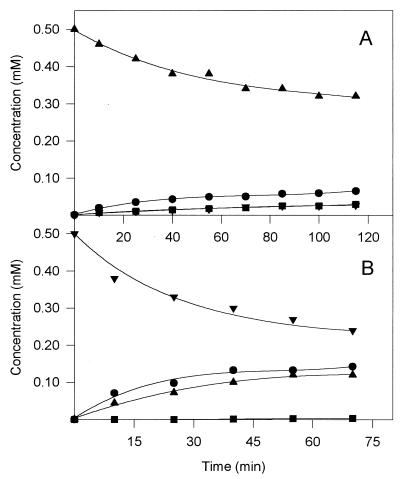
In order to analyze whether the variants that catalyzed the formation of one chloromuconolactone predominantly were able to convert the other possible chlorolactone, turnover experiments with (+)-5-chloromuconolactone and (+)-2-chloromuconolactone and enzyme variants F329I (Fig. 5A) and I54V (Fig. 5B), respectively, were performed. In both cases, they resulted in a mixture of the two chloromuconolactones, indicating that neither mutation abolished the capability to convert the chloromuconolactone of which less was formed during 2-chloro-cis,cis-muconate conversion.
FIG. 5.
Turnover of (+)-5-chloromuconolactone by muconate cycloisomerase variant F329I (A) and of (+)-2-chloromuconolactone by enzyme variant I54V (B). The experiment was performed with 30 mM Tris-HCl (pH 7.0), 1 mM MnSO4, 0.5 mM respective substrate, and ca. 0.005 U of enzyme activity per ml (measured with 2-chloro-cis,cis-muconate) at 25°C. Concentrations of (+)-2-chloromuconolactone (▾), (+)-5-chloromuconolactone (▴), 2-chloro-cis,cis-muconate (•), and trans-dienelactone (■) were analyzed by HPLC.
DISCUSSION
Differential specificities of muconate cycloisomerases.
Since the early studies of Pseudomonas sp. strain B13 (45), it has been known that muconate and chloromuconate cycloisomerases differ in their specificities, especially with respect to the turnover of chlorinated compounds. Later, R. eutropha JMP134 (28, 44), Comamonas (Pseudomonas) acidovorans CA28 (22), and R. opacus 1CP (53) were also reported to contain cycloisomerizing isoenzymes that differ in their kinetic properties and whose induction is dependent on the carbon source. Overall, however, the specificities of cycloisomerases, compared to those of the ring cleavage dioxygenases, have received relatively little attention. It was, for example, unclear whether the results reported for the muconate cycloisomerase of Pseudomonas sp. strain B13 would also be valid for enzymes for which sequence and structural data are available, i.e., the muconate cycloisomerases from A. “calcoaceticus” ADP1 and especially P. putida PRS2000. Kinetic analyses of these enzymes now show that both have the highest specificity constant for cis,cis-muconate (Tables 3 and 4), while 3-fluoro-, 2-methyl-, and 3-methyl-cis,cis-muconate are also good substrates. The specificity constants for the latter three substrates compared to that for cis,cis-muconate were about 30% for the P. putida cycloisomerase and between ca. 30 and 80% for the A. “calcoaceticus” enzyme. The relative specificity constants for the chloromuconates, if they could be determined at all, were below 0.5%. This pattern is very different from that of the R. opacus muconate cycloisomerase, for which, for example, the specificity constant for 2-chloro-cis,cis-muconate is 21% that for cis,cis-muconate (53). This pattern also differs from that reported for the Pseudomonas sp. strain B13 muconate cycloisomerase, which appears to convert 3-chloro-cis,cis-muconate with a relatively high specificity constant, i.e., 2.9% that for cis,cis-muconate (calculated from reference 45), almost 50-fold higher than that calculated from Table 3 for the enzyme from P. putida (0.06%).
Conversion of 2-substituted muconates.
2-Chloro-cis,cis-muconate binds to the P. putida muconate cycloisomerase with a relatively high affinity but with a low kcat (Table 3). This fact is apparently not due to steric effects, since 2-methyl-cis,cis-muconate is converted much faster, while 2-fluoro-cis,cis-muconate, which carries a smaller substituent, is a potent inhibitor but almost not a substrate (rate undetectable in photometric assays).
Why should this be so? Simple chemical substituent effects may be considered to explain the low kcat with 2-chloro- and 2-fluoro-cis,cis-muconate for both possible cycloisomerization directions (Fig. 6). The rate of 1,4-cycloisomerization could be decreased by a negative inductive effect of F or Cl in the α position, which would reduce the charge of the C-l carboxylate and thus slow its attack on the C-4—C-5 double bond (45). Additionally, a positive mesomeric effect of F or Cl could increase the electron density at C-4. The rate of 3,6-cycloisomerization would also be decreased, because the electron-withdrawing effect of the F or Cl would slow down the addition of a proton to the exocyclic carbon of the enol/enolate intermediate. Therefore, there is an electronic incompatibility between fluorine or chlorine at position 2 and the mechanism of cycloisomerization, which should be difficult to overcome. Consistent with this idea, 2-fluoro-cis,cis-muconate is at best converted extremely slowly by other cycloisomerases examined (10, 46, 53). However, the chloromuconate cycloisomerase of Pseudomonas sp. strain B13 and, to a lesser extent, the muconate cycloisomerase of R. opacus 1CP convert 2-chloro-cis,cis-muconate comparatively fast (45, 53). The substituent effects discussed above thus only partly explain why 2-chloro-cis,cis-muconate is a poor substrate for muconate cycloisomerases.
FIG. 6.
Negative inductive effect of the chlorine substituent as a possible reason for the slow turnover of 2-chloro-cis,cis-muconate. Partial negative and positive charges are indicated only for reaction steps which are assumed to be retarded (thin arrows)—for 1,4-cycloisomerization, the formation of the enol/enolate intermediate (postulated by Gerlt and Gassman [14]), and for 3,6-cycloisomerization, the proton addition to the exocyclic carbon.
Although some of our P. putida muconate cycloisomerase variants formed relatively more trans-dienelactone from 2-chloro-cis,cis-muconate than the wild-type enzyme, none were efficient dehalogenators (Table 5); however, several showed dramatic changes in the direction of cycloisomerization. Wild-type CatB formed approximately twofold more (+)-5-chloro- than (+)-2-chloromuconolactone (Table 5) (57). While with some CatB variants (Y59W and F329I) 1,4-cycloisomerization to 2-chloromuconolactone dominated, other variants (I54V and I54V-Y59W) strongly favored 3,6-cycloisomerization to 5-chloromuconolactone (Table 5). The altered product formation must have been due to differences in the productive binding of 2-chloro-cis,cis-muconate. In the structures of muconate and chloromuconate cycloisomerases, I54 or V50 and F329 or I325 occupy very different positions in the active-site cavity (Fig. 2). This fact is consistent with their having different effects on 2-chloro-cis,cis-muconate binding, as has been shown by more detailed structural and modelling studies (41).
Conversion of 3-substituted muconates.
With respect to 3-chloro-cis,cis-muconate, the reasons for the slow turnover and the low affinity are less obvious than for the 2-isomer. 3-Chloro-, 3-fluoro-, and 3-methyl-cis,cis-muconate generally undergo 3,6-cycloisomerization (with bacterial cycloisomerases) to a 4-substituted muconolactone or an end product derived from it (Fig. 1) (4, 7, 12, 20, 26, 30, 43, 45). 3-Methyl-cis,cis-muconate binds to the P. putida muconate cycloisomerase with a very high affinity and is converted with a moderate kcat (Table 3), suggesting that steric problems should be of limited importance. 3-Fluoro-cis,cis-muconate, on the other hand, while bound with a moderate affinity, is transformed at a very high rate, corresponding to the negative inductive effect of the substituent facilitating nucleophilic attack of the C-6 carboxylate on C-3. According to these simple chemical considerations, 3-chloro-cis,cis-muconate might also be expected to be a good substrate for muconate cycloisomerase, which it obviously is not. Therefore, other, more subtle, factors must be assumed to influence its affinity and turnover rate, as they do for 2-chloro-cis,cis-muconate.
A better understanding of the reasons for the inefficient 3-chloro-cis,cis-muconate turnover by the wild-type muconate cycloisomerase may come from the investigation of catalytically improved variants of the P. putida enzyme. Interestingly, 3-chloro-cis,cis-muconate was the substrate for which the most remarkable improvements in the catalytic constants, compared to those for wild-type CatB, were obtained (Table 3). The largest increases in the kcat/Km values were found for A271S (27-fold) and I54V (22-fold), in both cases as a result of increased affinity and higher turnover rate. While some other variants showed improvement with respect to only one of these parameters, none of the modified enzymes avoided the formation of the toxic protoanemonin. Despite the improved 3-chloro-cis,cis-muconate conversion by some of the variants, it must now be questioned whether the increased rates of catalysis were in fact due to a widened active site, as originally planned. Indeed, it appears that the active-site cavity of chloromuconate cycloisomerase is not significantly larger than that of muconate cycloisomerase (41).
Conversion of 2,4-dichloromuconate.
The low specificity constant of the P. putida muconate cycloisomerase for the conversion of 2,4-dichloro-cis,cis-muconate is due solely to a very low kcat with this substrate (Table 3). The affinity of the wild-type enzyme for this substrate is already high and consistently was not increased in any of the variants (Table 3). The kcat for 2,4-dichloro-cis,cis-muconate, in contrast, was 10-fold higher in variant F329I and 7-fold higher in variant I54V-F329I than in wild-type CatB, resulting in both cases in three- to fourfold-higher specificity constants. All variants showing higher 2,4-dichloro-cis,cis-muconate turnover rates also were better catalysts of 3-chloro-cis,cis-muconate conversion, indicating that, to some extent, the same factors are involved in determining the rates for these substrates. The negative inductive effect of the chlorine in position 2 additionally may have contributed to the even lower kcat for 2,4-dichloro-cis,cis-muconate, as discussed above for the 1,4-cycloisomerization of 2-chloro-cis,cis-muconate (Fig. 6).
Evolution of catalytic function.
If, as previously assumed, chloromuconate cycloisomerases were basically low-specificity variants of muconate cycloisomerases (45), a few mutations might be sufficient to bring about a broader substrate range. However, a more rigorous examination of the previously investigated cycloisomerases (4, 57, 59), the inclusion of enzymes from other sources (53), and the construction of variants by site-directed mutagenesis have now shown that the putative evolution from muconate to chloromuconate cycloisomerases was a rather complex process. The following apparently independent features were altered to achieve this change in Proteobacteria: (i) acceleration of 2-chloro-cis,cis-muconate turnover, (ii) discrimination between the two possible cycloisomerization directions of this substrate, (iii) dehalogenation of (+)-5-chloromuconolactone, (iv) acceleration of 3-chloro- and 2,4-dichloro-cis,cis-muconate conversion, and (v) avoidance of protoanemonin formation during 3-chloro-cis,cis-muconate turnover. Simple changes in the binding site cavity, as investigated in this study, may have been responsible for points ii and iv above; other or more complex changes may have been responsible for the remaining effects. We anticipate that further structure-function studies as well as insight from the functionally convergent evolution of the rhodococcal chloromuconate cycloisomerase (11) will help elucidate the nature of these changes.
ACKNOWLEDGMENTS
This work was supported by a grant from the Federal Ministry for Research and Technology (Projekt A10U; Zentrales Schwerpunktprojekt Bioverfahrenstechnik, Stuttgart, Germany) and by grant 1144 from the Academy of Finland to A.G.
We are grateful to H.-J. Knackmuss for providing an excellent environment in which we could do this work. We are indebted to L. N. Ornston and J. E. Houghton for supplying the catB clone and for sharing sequence information prior to its publication. We thank D. H. Pieper and R. Blasco for valuable discussions and for initiating the work resulting in one of the variants. Thanks are due to J. Pleiss for visualizing the active-site structures for us, to J. Altenbuchner for support with the expression system, to R. Schmid for use of the DNA sequencer, to S. Bürger for performing part of the DNA sequencing, and to T. Kajander for Fig. 2.
REFERENCES
- 1.Aldrich T L, Chakrabarty A M. Transcriptional regulation, nucleotide sequence, and localization of the promoter of the catBC operon in Pseudomonas putida. J Bacteriol. 1988;170:1297–1304. doi: 10.1128/jb.170.3.1297-1304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alting-Mees M A, Sorge J A, Short J M. pBluescriptII: multifunctional cloning and mapping vectors. Methods Enzymol. 1992;216:483–495. doi: 10.1016/0076-6879(92)16044-k. [DOI] [PubMed] [Google Scholar]
- 3.Avigad G, Englard S. Stereochemistry of enzymic reactions involved in cis,cis muconic acid utilization. Fed Proc. 1969;28:345. . (Abstract 486.) [Google Scholar]
- 4.Blasco R, Wittich R-M, Mallavarapu M, Timmis K N, Pieper D H. From xenobiotic to antibiotic, formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J Biol Chem. 1995;270:29229–29235. doi: 10.1074/jbc.270.49.29229. [DOI] [PubMed] [Google Scholar]
- 5.Bollag J-M, Briggs G G, Dawson J E, Alexander M. 2,4-d Metabolism: enzymatic degradation of chlorocatechols. J Agric Food Chem. 1968;16:829–833. [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Cain R B, Kirby G W, Rao G V. Stereochemistry of enzymic cyclisation of 3-methyl-cis,cis-muconic acid to form 3- and 4-methylmuconolactone. J Chem Soc D. 1989;1989:1629–1631. [Google Scholar]
- 8.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978;174:85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engesser K H, Auling G, Busse J, Knackmuss H-J. 3-Fluorobenzoate enriched bacterial strain FLB 300 degrades benzoate and all three isomeric monofluorobenzoates. Arch Microbiol. 1990;153:193–199. [Google Scholar]
- 11.Eulberg D, Kourbatova E M, Golovleva L A, Schlömann M. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J Bacteriol. 1998;180:1082–1094. doi: 10.1128/jb.180.5.1082-1094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans W C, Smith B S W, Moss P, Fernley H N. Bacterial metabolism of 4-chlorophenoxyacetate. Biochem J. 1971;122:509–517. doi: 10.1042/bj1220509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlt J A, Gassman P G. Understanding enzyme-catalyzed proton abstraction from carbon acids: details of stepwise mechanisms for β-elimination reactions. J Am Chem Soc. 1992;114:5928–5934. [Google Scholar]
- 15.Ghisalba O. Chemical wastes and their biodegradation—an overview. Experientia. 1983;39:1247–1257. doi: 10.1007/BF01990362. [DOI] [PubMed] [Google Scholar]
- 16.Ghosal D, You I-S. Operon structure and nucleotide homology of the chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Gene. 1989;83:225–232. doi: 10.1016/0378-1119(89)90108-x. [DOI] [PubMed] [Google Scholar]
- 17.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, England: Cambridge University; 1984. [Google Scholar]
- 18.Goldman A, Ollis D L, Steitz T A. Crystal structure of muconate lactonizing enzyme at 3 Å resolution. J Mol Biol. 1987;194:143–153. doi: 10.1016/0022-2836(87)90723-6. [DOI] [PubMed] [Google Scholar]
- 19.Häggblom M M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992;103:29–72. doi: 10.1111/j.1574-6968.1992.tb05823.x. [DOI] [PubMed] [Google Scholar]
- 20.Harper D B, Blakley E R. The metabolism of p-fluorobenzoic acid by a Pseudomonas sp. Can J Microbiol. 1971;17:1015–1023. doi: 10.1139/m71-162. [DOI] [PubMed] [Google Scholar]
- 21.Helin S, Kahn P C, Guha B L, Mallows D G, Goldman A. The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 Å resolution. J Mol Biol. 1995;254:918–941. doi: 10.1006/jmbi.1995.0666. [DOI] [PubMed] [Google Scholar]
- 22.Hinteregger C, Ferschl A, Loidl M, Streichsbier F. Metabolism of aniline and 3-chloroaniline in Pseudomonas acidovorans CA28: evidence of isofunctional muconate cycloisomerases. J Basic Microbiol. 1993;33:301–309. [Google Scholar]
- 23.Hoier H, Schlömann M, Hammer A, Glusker J P, Carrell H L, Goldman A, Stezowski J J, Heinemann U. Crystal structure of chloromuconate cycloisomerase from Alcaligenes eutrophus JMP134 (pJP4) at 3 Å resolution. Acta Crystallogr D. 1994;50:75–84. doi: 10.1107/S090744499300900X. [DOI] [PubMed] [Google Scholar]
- 24.Houghton J E, Brown T M, Appel A J, Hughes E J, Ornston L N. Discontinuities in the evolution of Pseudomonas putida cat genes. J Bacteriol. 1995;177:401–412. doi: 10.1128/jb.177.2.401-412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleywegt G J, Hoier H, Jones T A. A re-evaluation of the crystal structure of chloromuconate cycloisomerase. Acta Crystallogr D. 1996;52:858–863. doi: 10.1107/S0907444995008936. [DOI] [PubMed] [Google Scholar]
- 26.Knackmuss H-J, Hellwig M, Lackner H, Otting W. Cometabolism of 3-methylbenzoate and methylcatechols by a 3-chlorobenzoate utilizing Pseudomonas: accumulation of (+)-2,5-dihydro-4-methyl- and (+)-2,5-dihydro-2-methyl-5-oxo-furan-2-acetic acid. Eur J Appl Microbiol. 1976;2:267–276. [Google Scholar]
- 27.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 28.Kuhm A E, Schlömann M, Knackmuss H-J, Pieper D H. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP 134. Biochem J. 1990;266:877–883. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S-Y, Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques. 1990;9:676–679. [PubMed] [Google Scholar]
- 30.Mazur P, Pieken W A, Budihas S R, Williams S E, Wong S, Kozarich J W. cis,cis-Muconate lactonizing enzyme from Trichosporon cutaneum: evidence for a novel class of cycloisomerases in eucaryotes. Biochemistry. 1994;33:1961–1970. doi: 10.1021/bi00173a045. [DOI] [PubMed] [Google Scholar]
- 31.Neidhardt D J, Kenyon G L, Gerlt J A, Petsko G A. Mandelate racemase and muconate lactonizing enzyme are mechanistically distinct and structurally homologous. Nature (London) 1990;347:692–694. doi: 10.1038/347692a0. [DOI] [PubMed] [Google Scholar]
- 32.Ngai K-L, Ornston L N. Abundant expression of Pseudomonas genes for chlorocatechol metabolism. J Bacteriol. 1988;170:2412–2413. doi: 10.1128/jb.170.5.2412-2413.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ornston L N. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. III. Enzymes of the catechol pathway. J Biol Chem. 1966;241:3795–3799. [PubMed] [Google Scholar]
- 34.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reineke W. Degradation of chlorinated aromatic compounds by bacteria: strain development. In: Chaudhry G R, editor. Biological degradation and bioremediation of toxic chemicals. Portland, Oreg: Dioscorides Press; 1994. pp. 416–454. [Google Scholar]
- 36.Reineke W, Knackmuss H-J. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol. 1984;47:395–402. doi: 10.1128/aem.47.2.395-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russel M, Kidd S, Kelley M R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45:333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sayers J R, Krekel C, Eckstein F. Rapid high-efficiency site-directed mutagenesis by the phosphorothioate approach. BioTechniques. 1992;13:592–596. [PubMed] [Google Scholar]
- 41.Schell, U., S. Helin, T. Kajander, M. Schlömann, and A. Goldman. Structural basis for the activity of two muconate cycloisomerase variants towards substituted muconates. Unpublished data. [PubMed]
- 42.Schlömann M. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 43.Schlömann M, Fischer P, Schmidt E, Knackmuss H-J. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J Bacteriol. 1990;172:5119–5129. doi: 10.1128/jb.172.9.5119-5129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlömann M, Schmidt E, Knackmuss H-J. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J Bacteriol. 1990;172:5112–5118. doi: 10.1128/jb.172.9.5112-5118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem J. 1980;192:339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt E, Knackmuss H-J. Production of cis,cis-muconate from benzoate and 2-fluoro-cis,cis-muconate from 3-fluorobenzoate by 3-chlorobenzoate degrading bacteria. Appl Microbiol Biotechnol. 1984;20:351–355. [Google Scholar]
- 47.Schmidt E, Remberg G, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Halogenated muconic acids as intermediates. Biochem J. 1980;192:331–337. doi: 10.1042/bj1920331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segel I H. Enzyme kinetics. New York, N.Y: John Wiley & Sons, Inc.; 1975. [Google Scholar]
- 49.Seibert, V., S. Lakner, and M. Schlömann. Unpublished results.
- 50.Shanley M S, Harrison A, Parales R E, Kowalchuk G, Mitchell D J, Ornston L N. Unusual G+C content and codon usage in catIJF, a segment of the ben-cat supra-operonic cluster in the Acinetobacter calcoaceticus chromosome. Gene. 1994;138:59–65. doi: 10.1016/0378-1119(94)90783-8. [DOI] [PubMed] [Google Scholar]
- 51.Shaw E. A synthesis of protoanemonin. The tautomerism of acetylacrylic acid and of penicillic acid. J Am Chem Soc. 1946;68:2510–2513. doi: 10.1021/ja01216a024. [DOI] [PubMed] [Google Scholar]
- 52.Sistrom W R, Stanier R Y. The mechanism of formation of β-ketoadipic acid by bacteria. J Biol Chem. 1954;210:821–836. [PubMed] [Google Scholar]
- 53.Solyanikova I P, Maltseva O V, Vollmer M D, Golovleva L A, Schlömann M. Characterization of muconate and chloromuconate cycloisomerase from Rhodococcus erythropolis 1CP: indications for functionally convergent evolution among bacterial cycloisomerases. J Bacteriol. 1995;177:2821–2826. doi: 10.1128/jb.177.10.2821-2826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 55.Tartof K D, Hobbs C A. Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus. 1987;9:12. [Google Scholar]
- 56.van der Meer J R, Eggen R I L, Zehnder A J B, de Vos W M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991;173:2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vollmer M D, Fischer P, Knackmuss H-J, Schlömann M. Inability of muconate cycloisomerases to cause dehalogenation during conversion of 2-chloro-cis,cis-muconate. J Bacteriol. 1994;176:4366–4375. doi: 10.1128/jb.176.14.4366-4375.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vollmer M D, Hoier H, Schell U, Gröning J, Pleiss J, Goldman A, Schlömann M. Differences in the catalytic properties of muconate and chloromuconate cycloisomerases analyzed by site-directed mutagenesis. Biospektrum special issue. 1995. p. 24. . (Abstract KV002.) [Google Scholar]
- 59.Vollmer M D, Schlömann M. Conversion of 2-chloro-cis,cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerases of pJP4 and pAC27. J Bacteriol. 1995;177:2938–2941. doi: 10.1128/jb.177.10.2938-2941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]