Abstract
Endophytic fungi isolated from medicinal ferns serve as significant natural resources for drug precursors or bioactive metabolites. During our survey on the diversity of endophytic fungi from Dicranopteris species (a genus of medicinal ferns) in Guizhou, Apoiospora was observed as a dominant fungal group. In this study, seven Apiospora strains, representing four new species, were obtained from the healthy plant tissues of three Dicranopteris species—D. ampla, D. linearis, and D. pedata. The four new species, namely Apiospora aseptata, A. dematiacea, A. dicranopteridis, and A. globosa, were described in detail with color photographs and subjected to phylogenetic analyses using combined LSU, ITS, TEF1-α, and TUB2 sequence data. This study also documented three new hosts for Apiospora species.
Keywords: seven taxa, asexual morph, dematiaceous conidia, medicinal ferns, phylogeny, taxonomy
1. Introduction
Apiosporaceae, typified by Apiospora Sacc., was established by Hyde et al. [1] to accommodate Arthrinium-like taxa characterized by a basauxic, arthrinium-like conidiogenesis producing apiospores [2,3]. Currently, only three genera, Apiospora, Arthrinium Kunze, and Nigrospora Zimm, are accepted in this family [4,5]. Apiospora Sacc. was introduced by Saccardo, P. A. [6] within the family Apiosporaceae (Amphisphaeriales, Sordariomycetes), with A. montagnei Sacc. as the type species. Due to the morphological similarities between the genera Apiospora and Arthrinium, they were long considered synonymous based on the one fungus one name principle [7,8,9,10,11]. It was not until the study by Pintos, Á. & P. Alvarado [12] that they were clarified as separate genera based on genetic, ecological, and morphological evidence. This delineation was confirmed and supported by subsequent studies [13,14,15,16,17,18,19,20]. Evidence of genome draft was employed within the fungal group Arthrinium/Apiospora for the first time, which also supported them into two separate genera [21]. Currently, 157 epithets under the genus Apiospora are listed in Index Fungorum (September 2023), while 116 epithets are listed in Species Fungorum. Out of these, molecular data have confirmed 91 Apiospora species [12,13,14,15,17,18,19,20,22].
The sexual morph of Apiospora is characterized by immersed to erumpent, multi-loculate, perithecial ascostromata, unitunicate, broadly clavate to cylindric–clavate asci, and hyaline, ellipsoidal, inequilaterally 2-celled ascospores with or without a gelatinous sheath [5,9,10,14,22,23]. The asexual morphs of Apiospora include hyphomycetes and coelomycetes. The hyphomycetous asexual morphs feature septate, subhyaline, or brown conidiophores emerging from basal cells or reduced to conidiogenous cells, basauxic conidiogenous cells, and typically globose to subglobose, aseptate conidia that appear lenticular or obovoid from side view [11,13,16,18,19,24]. The coelomycetous asexual morph of Apiospora is marked by its erumpent, pustulate, coriaceous conidiomata, hyphoid conidiophores, blastic, integrated, determinate, doliiform or cylindrical conidiogenous cells, and oval, brown conidia, which may have a truncate basal scar and a germ slit [2,9,20].
Endophytes are endosymbiotic flora, microorganisms that colonize the internal tissues of healthy plants without causing any direct, noticeable negative effects [25,26]. The taxonomic research of endophytic fungi has become a popular trend not only because of their beneficial effects on plants but also because of compounds including antibiotics and other compounds of therapeutic significance [26,27,28]. That is, endophytic fungi (especially medicinal plants) possess significant potential to discover or synthesize more bioactive compounds and mimic the structure and function of host compounds [25,28,29,30], which shows a new source of potentially useful pharmaceutical compounds [26,28,31].
Dicranopteris Bernh. is an ancient and widespread fern genus belonging to the family Gleicheniaceae (Filicopsida) found in tropical and subtropical ecosystems [32,33]. It belongs to a group of medically important ferns known for their significant pharmacological effects, including removing blood stasis, clearing heat and diuresis, anticancer, antinociception, and anti-inflammation [34,35,36,37,38]. Extracts from Dicranopteris are rich in bioactive compounds and have the potential to yield new structural compounds [34,39].
In this study, we examined endophytic fungi isolated from three Dicranopteris species (D. ampla, D. linearis, and D. pedate) in Guizhou, China, aiming to explore the diversity of fungi with research significance. We isolated nearly a thousand endophytic taxa from various parts of the three Dicranopteris species, belonging to 146 genera, based on NCBI searches of the ITS and LSU sequence data. Among these isolates, Apiospora emerged as a common genus. Within this collection, seven taxa were identified herein as four endophytic Apiospora species new to science, viz. A. aseptata, A. dematiacea, A. dicranopteridis, and A. globosa. To further determine the taxonomic placement of these four Apiospora species, we employed phylogenetic analyses using combined LSU, ITS, TEF1-α, and TUB2 sequence data, complemented by morphological features. A backbone tree of Apiosporaceae is provided in this study.
2. Materials and Methods
2.1. Collection and Isolation
Fresh, healthy plant tissues (leaves, rhizomes, roots, and stems) from three Dicranopteris species were collected along with relevant metadata (date, habitat, and locality). Samples were transported to the laboratory and processed for fungal isolation within 48 h. Healthy tissue pieces were first washed under running tap water. Surface sterilization of plant tissues followed the method described by Nontachaiyapoom et al. [40], with some modifications. To eliminate epiphytic microorganisms, the materials were surface-sterilized on a benchtop by immersing them in 75% (v/v) ethanol for 1–3 min (ca. 1 min for leaves and stems; ca. 3 min for rhizomes and roots). They were then rinsed with sterilized distilled water for 2 min, followed by a soak in 10% (v/v) NaClO for 0.5–2 min (ca. 0.5 min for leaves and stems; ca. 2 min for rhizomes and roots). The tissues were then rinsed with sterile distilled water three times in succession. After drying the sterilized plant tissues on sterilized filter paper, they were cut into approximately 2 mm2 pieces using a sterile blade. These small pieces were placed on fresh potato dextrose agar (PDA) containing antibiotics (50 μg/mL penicillin) and cultivated at 25 °C. Once fungal hyphae growth was observed emerging from the plant segments, the hyphae were picked from the edge of the colonies and transferred to fresh PDA media to obtain the pure cultures.
2.2. Morphological Study and Conservation
Isolates were grown on a PDA for one week, and cultural characteristics such as size, shape, color, and texture were recorded. These characteristics were further examined using a stereomicroscope (SMZ168-BL, Motic, Shanghai, China). Micro-morphological characteristics were described based on cultures that sporulated on either water agar (WA) or PDA [11,16,19,23,41,42]. These were photographed using an ECLIPSE Ni-U compound microscope (Nikon, Tokyo, Japan) equipped with an EOS 90D digital camera (Canon, Tokyo, Japan). Measurements of conidiophores, conidiogenous cells, conidia, and mycelia were conducted using the Tarosoft (R) Image Frame Work (version 0.9.7). Figures and the photoplates were processed with Adobe Illustrator CS6 v. 24.0.1 (Adobe Systems, San Jose, CA, USA). Dried materials were deposited in the Herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (HKAS), Kunming, China, and the Herbarium of Guizhou Academy of Agricultural Sciences (GZAAS), Guiyang, China. Living cultures were deposited at the Kunming Institute of Botany, the Chinese Academy of Sciences (KUNCC), and the Guizhou Culture Collection (GZCC). Faces of Fungi and Index Fungorum numbers were registered in accordance with the guidelines presented in Jayasiri et al. [43] and Index Fungorum (http://www.indexfungorum.org/Names/Names.asp; accessed on 15 September 2023).
2.3. DNA Extraction, PCR Amplification and Sequencing
Genomic DNA was extracted from fresh fungal mycelia using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, Shanghai, China) according to the manufacturer’s instructions. Four primer pairs, namely LR0R and LR5 [44], ITS5 and ITS4 [45], EF2 and EF1-728f [46,47], and T1 and Bt2b [48,49], were employed to amplify the large subunit of the ribosomal DNA (LSU), the internal transcribed spacer (ITS), the elongation factor 1-alpha (TEF1-α), and the β-tubulin (TUB2) gene regions, respectively. The polymerase chain reaction (PCR) was carried out in a 50 μL reaction volume containing 2 μL of DNA template, 2 μL of each forward and reverse primer (10 μM), 25 μL of 2× Taq PCR Master Mix with blue dye (Sangon Biotech, China), and 19 μL of distilled–deionized water. The amplification conditions for LSU, ITS, TEF-1α, and TUB2 were based on the protocol described by Feng et al. [23]. Successful PCR products were sent to Sangon Biotech (Shanghai, China) for purification and sequencing. The sequences generated in this study have been deposited in NCBI GenBank (Table 1).
Table 1.
Apiospora endophytic isolates were used in this study.
| Taxon | Strain Code | Specimen | Status | Host | Tissues | Substrate (Sproluration) |
|---|---|---|---|---|---|---|
| A. aseptata | KUNCC 23-14169 | HKAS 129875 | H | D. pedata | Leaf | PDA |
| A. dematiacea | KUNCC 23-14202 | HKAS 129910 | H | D. ampla | Stem | PDA |
| A. dicranopteris | GZCC 23-0708 | HKAS 129898 | P | D. ampla | Rhizome | PDA |
| A. dicranopteris | GZCC 23-0712 | HKAS 129895 | P | D. pedata | Leaf | PDA |
| A. dicranopteris | KUNCC23-14177 | GZAAS 23-0780 | P | D. pedata | Root | PDA, WA |
| A. dicranopteris | KUNCC23-14171 | HKAS 129877 | H | D. pedata | Stem | PDA |
| A. globosa | KUNCC 23-14210 | HKAS 129921 | H | D. linearis | Stem | WA |
Note: status: H denotes holotype; P denotes paratype.
2.4. Alignments and Phylogenetic Analyses
The quality of the original sequences was checked using BioEdit v. 7.1.3.0 [50] and assembled with SeqMan v. 7.0.0 (DNASTAR, Madison, WI, USA). Consensus sequences underwent BLASTn analysis in the NCBI GenBank database for preliminary identification of similar sequences. Taxa (Table 2), including type and additional strains of Apiospora species and related genera (Nigrospora and Arthrinium) in Apiosporaceae, were selected for the phylogenetic analyses based on data obtained from Genbank and previous studies [8,11,12,14,19,20,23,41,51]. Sequence alignment was performed using MAFFT v.7.0 (https://mafft.cbrc.jp/alignment/server/; accessed on 20 August 2023) [52] and subsequently manually verified in BioEdit 7.1.3.0 [50]. The phylogenetic relationships, based on a combined LSU–ITS–TEF-1α–TUB2 dataset, were analyzed using both maximum likelihood (ML) and Bayesian inference (BI) criteria.
Table 2.
Taxa used in this study and their GenBank accession numbers.
| Taxa Names | Strains | GenBank Accessions | |||
|---|---|---|---|---|---|
| LSU | ITS | TEF1-α | TUB2 | ||
| Apiospora acutiapicum | KUMCC 20-0210 T | MT946339 | MT946343 | MT947360 | MT947366 |
| Apiospora agari | KUC21333 T | MH498440 | MH498520 | MH544663 | MH498478 |
| Apiospora aquaticum | S 642 T | MK835806 | MK828608 | N/A | N/A |
| Apiospora arctoscopi | KUC21331 T | MH498449 | MH498529 | MN868918 | MH498487 |
| Apiospora arundinis | CBS 449 92 | KF144931 | KF144887 | KF145019 | KF144977 |
| Apiospora arundinis | GZCC 20-0116 T | MW478899 | MW481720 | MW522952 | MW522968 |
| Apiospora aurea | CBS 244.83 T | KF144935 | AB220251 | KF145023 | KF144981 |
| Apiospora balearica | CBS 145129 T | MK014836 | MK014869 | N/A | MK017975 |
| Apiospora bambusae | ALV17304 | MK014841 | MK014874 | MK017951 | MK017980 |
| Apiospora bambusicola | MFLUCC 20-0144 T | MW173087 | MW173030 | MW183262 | N/A |
| Apiospora biserialis | CGMCC 3.20135 T | MW478885 | MW481708 | MW522938 | MW522955 |
| Apiospora camelliae sinensis | LC 5007 T | KY494780 | KY494704 | KY705103 | KY705173 |
| Apiospora chiangraiense | MFLUCC 21-0053 T | MZ542524 | MZ542520 | N/A | MZ546409 |
| Apiospora chromolaenae | MFLUCC 17-1505 T | MT214436 | MT214342 | N/A | N/A |
| Apiospora cordylinae | GUCC 10027 T | N/A | MT040106 | MT040127 | MT040148 |
| Apiospora cyclobalanopsidis | CGMCC 3.20136 T | MW478892 | MW481713 | MW522945 | MW522962 |
| Apiospora descalsii | CBS 145130 T | MK014837 | MK014870 | MK017976 | |
| Apiospora dichotomanthi | LC 4950 T | KY494773 | KY494697 | KY705096 | KY705167 |
| Apiospora dongyingensis | SAUCC 0302 T | OP572424 | OP563375 | OP573264 | OP573270 |
| Apiospora esporlensis | CBS 145136 T | MK014845 | MK014878 | MK017983 | |
| Apiospora euphorbiae | IMI 285638b | AB220335 | AB220241 | NA | AB220288 |
| Apiospora fermenti | KUC21289 T | MF615213 | MF615226 | MH544667 | MF615231 |
| Apiospora gaoyouensis | CFCC 52301 T | N/A | MH197124 | MH236793 | MH236789 |
| Apiospora garethjonesii | KUMCC 16-0202 T | KY356091 | KY356086 | N/A | N/A |
| Apiospora gelatinosa | KHAS 11962 T | MW478888 | MW481706 | MW522941 | MW522958 |
| Apiospora guiyangensis | HKAS 102403 T | MW240577 | MW240647 | N/A | MW775604 |
| Apiospora guizhouensis | LC 5322 T | KY494785 | KY494709 | KY705108 | KY705178 |
| Apiospora hainanensis | SAUCC 1681 T | OP572422 | OP563373 | OP573262 | OP573268 |
| Apiospora hispanica | IMI 326877 T | AB220336 | AB220242 | N/A | AB220289 |
| Apiospora hydei | CBS 114990 T | KF144936 | KF144890 | KF145024 | KF144982 |
| Apiospora hydei | LC7103 | KY494791 | KY494715 | KY705114 | KY705183 |
| Apiospora hydei | LC7105 | KY494793 | KY494717 | KY705116 | KY705185 |
| Apiospora hydei | SICAUCC 22-0032 | ON185553 | ON183998 | ON221312 | ON221313 |
| Apiospora hyphopodii | MFLUCC 15-0003 T | N/A | KR069110 | N/A | N/A |
| Apiospora hyphopodii | KUMCC 16-0201 | KY356093 | KY356088 | N/A | N/A |
| Apiospora hysterina | ICPM 6889 T | MK014841 | MK014874 | MK017951 | MK017980 |
| Apiospora iberica | CBS 145137 T | MK014846 | MK014879 | N/A | MK017984 |
| Apiospora intestini | CBS 135835 T | KR149063 | KR011352 | KR011351 | KR011350 |
| Apiospora italica | CBS 145138 T | MK014847 | MK014880 | MK017956 | MK017985 |
| Apiospora jatrophae | AMH 9557 T | N/A | JQ246355 | N/A | N/A |
| Apiospora jiangxiensis | LC 4577 T | KY494769 | KY494693 | KY705092 | KY705163 |
| Apiospora kogelbergensis | CBS 113333 K | KF144938 | KF144892 | KF145026 | KF144984 |
| Apiospora koreana | KUC21332 T | MH498444 | MH498524 | MH544664 | MH498482 |
| Apiospora locuta pollinis | LC 11683 T | N/A | MF939595 | MF939616 | MF939622 |
| Apiospora longistroma | MFLUCC 11-0481 T | KU863129 | KU940141 | N/A | N/A |
| Apiospora malaysiana | CBS 102053T | KF144942 | KF144896 | KF145030 | KF144988 |
| Apiospora marianiae | AP18219 T | ON692422 | ON692406 | N/A | ON677186 |
| Apiospora marii | CBS 497.90 T | KF144947 | AB220252 | KF145035 | KF144993 |
| Apiospora marina | KUC21328 T | MH498458 | MH498538 | MH544669 | MH498496 |
| Apiospora mediterranea | IMI 326875 | AB220337 | AB220243 | N/A | AB220290 |
| Apiospora minutispora | 17E-042 T | N/A | LC517882 | LC518889 | LC518888 |
| Apiospora montagnei | AP301120 T | ON692424 | ON692408 | ON677182 | ON677188 |
| Apiospora mori | MFLU 18-2514 T | MW114393 | MW114313 | N/A | N/A |
| Apiospora multiloculata | MFLUCC 21-0023 T | OL873138 | OL873137 | N/A | OL874718 |
| Apiospora mytilomorpha | DAOM 214595 T | N/A | KY494685 | N/A | N/A |
| Apiospora neobambusae | LC 7106 T | KY494794 | KY494718 | KY806204 | KY705186 |
| Apiospora neochinense | CFCC 53036 T | N/A | MK819291 | MK818545 | MK818547 |
| Apiospora neogarethjonesii | HKAS 102408 T | MK070898 | MK070897 | N/A | N/A |
| Apiospora neosubglobosa | JHB 006 | KY356094 | KY356089 | N/A | N/A |
| Apiospora neosubglobosa | KUMCC 16-0203 T | KY356095 | KY356090 | N/A | N/A |
| Apiospora obovata | LC 4940 T | KY494772 | KY494696 | KY705095 | KY705166 |
| Apiospora ovata | CBS 115042 T | KF144950 | KF144903 | KF145037 | KF144995 |
| Apiospora paraphaeosperma | MFLUCC 13-0644 T | KX822124 | KX822128 | N/A | N/A |
| Apiospora phragmitis | CPC 18900 | KF144956 | KF144909 | KF145043 | KF145001 |
| Apiospora phyllostachydis | MFLUCC 18-1101 T | MH368077 | MK351842 | MK340918 | MK291949 |
| Apiospora piptatheri | CBS 145149 T | MK014860 | MK014893 | N/A | N/A |
| Apiospora pseudomarii | GUCC 10228 T | N/A | MT040124 | MT040145 | MT040166 |
| Apiospora pseudoparenchymatica | LC7234 T | KY494819 | KY494743 | KY705139 | KY705211 |
| Apiospora pseudorasikravindrae | KUMCC 20-0208 T | N/A | MT946344 | MT947361 | MT947367 |
| Apiospora pseudosinensis | CPC 21546 T | KF144957 | KF144910 | KF145044 | N/A |
| Apiospora pseudospegazzinii | CBS 102052 T | KF144958 | KF144911 | KF145045 | KF145002 |
| Apiospora pterosperma | CPC 20193 T | KF144960 | KF144913 | KF145046 | KF145004 |
| Apiospora pusillisperma | KUC21321 T | MH498453 | MH498533 | MN868930 | MH498491 |
| Apiospora qinlingensis | CFCC 52303 T | N/A | MH197120 | MH236795 | MH236791 |
| Apiospora rasikravindrae | NFCCI 2144 | N/A | JF326454 | N/A | N/A |
| Apiospora rasikravindrae | LC5449 | KY494789 | KY494713 | KY705112 | KY705182 |
| Apiospora sacchari | CBS 372.67 | KF144964 | KF144918 | KF145049 | KF145007 |
| Apiospora saccharicola | CBS 831.71 | KF144969 | KF144922 | KF145054 | KF145012 |
| Apiospora sargassi | KUC21228 T | KT207696 | KT207746 | MH544677 | KT207644 |
| Apiospora sasae | CBS 146808 T | MW883797 | MW883402 | N/A | MW890120 |
| Apiospora septata | CGMCC 3.20134 T | MW478890 | MW481711 | MW522943 | MW522960 |
| Apiospora serenensis | IMI 326869 T | AB220344 | AB220250 | N/A | AB220297 |
| Apiospora setariae | MT492005 | N/A | MT492005 | MW118457 | MT497467 |
| Apiospora setostroma | KUMCC 19-0217 | MN528011 | MN528012 | MN527357 | N/A |
| Apiospora sichuanensis | HKAS 107008 T | MW240578 | MW240648 | N/A | MW775605 |
| Apiospora sorghi | URM 93000 T | N/A | MK371706 | N/A | MK348526 |
| Apiospora sp. | SAUCC 1429 | OQ615287 | OQ592558 | N/A | N/A |
| Apiospora sp. | SAUCC 1430 | OQ615286 | OQ592557 | N/A | N/A |
| Apiospora sphaerosperma | CBS114314 | KF144951 | KF144904 | KF145038 | KF144996 |
| Apiospora stipae | CBS 146804 T | MW883798 | MW883403 | MW890082 | MW890121 |
| Apiospora subglobosa | MFLUCC 11-0397 T | KR069113 | KR069112 | N/A | N/A |
| Apiospora subrosea | LC 7292 T | KY494828 | KY494752 | KY705148 | KY705220 |
| Apiospora taeanensis | KUC21322T | N/A | MH498515 | MH544662 | MH498473 |
| Apiospora thailandica | MFLUCC 15-0202 T | KU863133 | KU940145 | N/A | N/A |
| Apiospora tropica | MFLUCC 21 0056 T | OK491653 | OK491657 | N/A | N/A |
| Apiospora vietnamensis | IMI 99670 T | KX986111 | KX986096 | N/A | KY019466 |
| Apiospora xenocordella | CBS 478 86 T | KF144970 | KF144925 | KF145055 | KF145013 |
| Apiospora yunnana | MFLUCC 15 1002 T | KU863135 | KU940147 | N/A | N/A |
| Arthrinium austriacum | GZU 345006 | MW208860 | MW208929 | N/A | N/A |
| Arthrinium caricicola | CBS 145127 | MK014838 | MK014871 | N/A | MK017977 |
| Arthrinium cf. sporophleoides | GZU 345102 | MW208866 | MW208944 | N/A | N/A |
| Arthrinium crenatum | AG 19066 T | MW208861 | MW208931 | N/A | N/A |
| Arthrinium japonicum | IFO 31098 | AB220358 | AB220264 | N/A | AB220311 |
| Arthrinium luzulae | AP7619 3 T | MW208863 | MW208937 | N/A | N/A |
| Arthrinium curvatum var. minus | CBS 145131 | MK014839 | MK014872 | N/A | MK017978 |
| Arthrinium morthieri | GZU 345043 | MW208864 | MW208938 | MW221920 | MW221926 |
| Arthrinium puccinioides | CBS 145150 | MK014861 | MK014894 | N/A | MK017998 |
| Arthrinium sphaerospermum | AP25619 | MW208865 | MW208943 | N/A | N/A |
| Arthrinium sporophleum | CBS 145154 | MK014865 | MK014898 | N/A | MK018001 |
| Nigrospora aurantiaca | CGMCC 3.18130 T | KX986098 | KX986064 | KY019295 | KY019465 |
| Nigrospora bambusae | CGMCC 3.18327 T | NG 069455 | KY385307 | KY385313 | KY385319 |
| Nigrospora camelliae sinensis | CGMCC 3.18125 T | KX986103 | KX985986 | KY019293 | KY019460 |
| Nigrospora chinensis | CGMCC 3.18127 T | KX986107 | KX986023 | KY019422 | KY019462 |
| Nigrospora gorlenkoana | CBS 480.73 T | KX986109 | KX986048 | KY019420 | KY019456 |
| Nigrospora guilinensis | CGMCC 3.18124 T | KX986113 | KX985983 | KY019292 | KY019459 |
| Nigrospora hainanensis | CGMCC 3.18129 T | KX986112 | KX986091 | KY019415 | KY019464 |
| Nigrospora lacticolonia | CGMCC 3.18123 T | KX986105 | KX985978 | KY019291 | KY019458 |
| Nigrospora musae | CBS 319.34 T | KX986110 | MH855545 | KY019419 | KY019455 |
| Nigrospora oryzae | LC2693 | KX986101 | KX985944 | KY019299 | KY019471 |
| Nigrospora osmanthi | CGMCC 3.18126 T | KX986106 | KX986010 | KY019421 | KY019461 |
| Nigrospora pyriformis | CGMCC 3.18122 T | KX986100 | KX985940 | KY019290 | KY019457 |
| Nigrospora rubi | LC2698 T | KX986102 | KX985948 | KY019302 | KY019475 |
| Nigrospora saccharicola | CGMCC 3.19362 T | N/A | MN215788 | MN264027 | MN329951 |
| Nigrospora sphaerica | LC7298 | KX986097 | KX985937 | KY019401 | KY019606 |
| Nigrospora vesicularis | CGMCC 3.18128 T | KX986099 | KX986088 | KY019294 | KY019463 |
| Nigrospora zimmermanii | CBS 290.62 T | KY806276 | KY385309 | KY385311 | KY385317 |
| Seiridium phylicae | CPC 19962 T | NG 042759 | LT853092 | LT853189 | LT853239 |
| Seiridium phylicae | CPC 19965 | KC005809 | LT853093 | LT853190 | LT853240 |
Note: T denotes type strains; “N/A” indicates no data are available in GenBank.
Maximum likelihood (ML) analysis was conducted on the CIPRES web portal (https://www.phylo.org/portal2/home.action; accessed on 20 August 2023) using the RAxML-HPC Blackbox (8.2.10) tool with rapid bootstrap analysis and 1000 bootstrap replicates [53,54]. The final tree was selected from the suboptimal trees of each run by comparing likelihood scores under the GTRGAMMA substitution model.
Posterior probabilities (PP) [55] were calculated using the Bayesian Markov Chain Monte Carlo (BMCMC) sampling method in MrBayes 3.2.7a via CIPRES [53]. The appropriate substitution model best fitting the DNA evolution model for the combined dataset was determined using MrModeltest v.2.3 [56]. For the LSU, ITS, and TUB2 datasets, GTR+I+G was selected, whereas HKY+I+G was selected for TEF1-α. Four simultaneous Markov chains run for 1 million generations, with trees sampled every 100 generations, yielding 10,000 trees. The first 2000 trees, representing the burn-in phase, were discarded, and the remaining 8000 trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree [57].
Phylogenetic trees were visualized using FigTree v. 1.4.4 [58] and adjusted using Adobe Illustrator CS6 (Adobe Systems, San Jose, CA, USA).
3. Results
3.1. Phylogenetic Analysis
Seven endophytic taxa with asexual morphs, isolated from three Dicranopteris species, were identified as A. aseptata, A. dematiacea, A. dicranopteridis, and A. globosa spp. nov. within the genus Apiospora (Apiosporaceae, Amphisphaeriales) (Table 1). The combined LSU (840 bp), ITS (623 bp), TEF1-α (534 bp), and TUB2 (207 bp) sequence alignment comprised 135 taxa, with Seiridium phylicae (CPC 19962 and CPC 19965) serving as the outgroup taxa.
The dataset contained 2204 characters after alignment. The matrix presented 1181 distinct alignment patterns, with 25.29% being completely undetermined characters or gaps. Base frequencies and rates were A = 0.235259, C = 0.247975, G = 0.262058, and T = 0.254709; substitution rates were AC = 1.153887, AG = 2.614757, AT = 1.016680, CG = 0.924945, CT = 4.569867, and GT = 1.000000, with a tree length of 4.582782. The distribution shape parameter α equaled 0.227323. The tree topologies generated from both RAxML and Bayesian analyses were similar, showing no significant conflicts. The best-scoring RAxML tree is shown in Figure 1, with a final likelihood value of −25,698.717456. This phylogenetic tree revealed that the new species Apiospora aseptata (KUNCC 23-14169) clusters with two unidentified Apiospora taxa (SAUCC 1429 and SAUCC 1430), albeit with weak support. Apiospora dematiacea is closely related to the species, which includes four taxa of Apiospora hydei (LC7105, LC7103, CBS 114990, and SICAUCC 22-0032). Four isolates representing the new species, Apiospor dicranopteris (GZCC 23-0708, GZCC 23-0712, KUNCC 23-14177, and KUNCC 23-14171), form a distinct clade, which is basal to A. koreana (KUC21332) and A. qinlingensis (CFCC 52303). Lastly, Apiospor globosa forms its own clade, being a sister to A. neosubglobosa (KUMCC 16-0203 and JHB 006).
Figure 1.
The phylogenetic tree generated from ML analysis is based on a concatenated LSU–ITS–TEF1α–TUB2 dataset for the family Apiosporaceae. Bootstrap support values for ML greater than 60% and Bayesian posterior probabilities (PPs) greater than 0.95 were indicated above or below the nodes as ML/PP. Seiridium phylicae (CPC 19962 and CPC 19965) were selected as the outgroup taxa. The newly generated sequences are shown in red.
3.2. Taxonomy
Apiospora aseptata, J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 2)
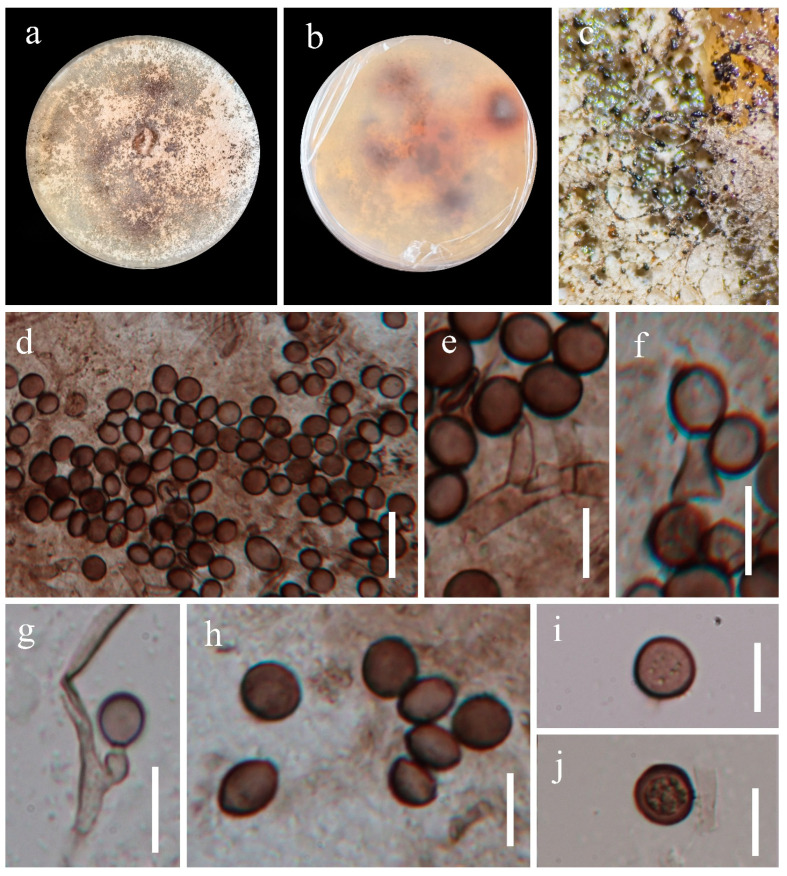
Figure 2.
Apiospora aseptata (HKAS 129875, holotype). (a,b) Cultures on PDA from above and below; (c) colonies on PDA; (d–g) conidiogenous cells with conidia; (h–j) conidia. Scale bars: (d) 20 μm; (e–j) 10 μm.
Index Fungorum number: IF901115; Facesoffungi number: FoF14873.
Etymology: referring to the aseptate conidia.
Culture characteristics: Colonies on PDA are medium circular, spread, flat with an entire edge, with thin aerial hyphae, reaching ca. 50 mm diam after 10 d at 25 °C, grey-brown from above, yellow–brown in reverse. Mycelium consists of septate, branched, hyaline to brown hyphae.
Description: Endophytic in the healthy roots of Dicranopteris pedata. Sexual morph: Undetermined. Asexual morph: Conidiophores are cylindrical, septate, branched, rough-walled, flexuous, and often reduced to conidiogenous cells. Conidiogenous cells ca. 3.5 µm wide, aggregated in clusters on hyphae, solitary, mono-polyblastic, cylindrical to subglobose, hyaline to brown. Conidia amerospores, aseptate, globose or sub globose, 7–9.5 (–13) µm diam. ( = 8 µm, n = 40) in surface view, subglobose; 6–8.5 × 5–7 µm ( = 7 × 6 µm, n = 20) from side view, lenticular with a pale longitudinal germ slit, smooth to finely roughened, occasionally micro guttules, pale brown to brown.
Material examined: China, Guizhou Province, Qianxinan Buyi, and Miao Autonomous Prefecture, Chengheng County, isolated from the healthy leaf of Dicranopteris pedata near the roadside, 16 March 2022, J.Y. Zhang, 138-3 (HKAS 129875, holotype); ex-type living cultures, KUNCC 23-14169.
GenBank accession numbers: (LSU) OR590335, (ITS) OR590341, (TEF1-α) OR634949, and (TUB) OR634943.
Notes: Apiospora aseptata aligns well with the characteristics of the genus Apiospora and is most similar to A. pseudoparenchymaticum in the shape of its conidiogenous cells and conidia. However, they differ in conidial size [11]. Apiospora aseptata has noticeably smaller conidia than A. pseudoparenchymaticum (7–9.5 (–13) µm diam. vs. 13.5–27 × 12–23.5 µm). In phylogenetic analysis, Apiospora aseptata clusters with Apiospora sp. strains (SAUCC 1429 and SAUCC 1430) and forms a sister relationship with the clade that includes A. arctoscopi (KUC21331), A. jiangxiensis (LC 4577), and A. obovata (LC 4940). Unfortunately, we could not compare the morphological characteristics of the Apiospora sp. strains (SAUCC 1429 and SAUCC 1430), as the morphology of these strains has not been reported. Based on both phylogeny and morphology, we introduce Apiospora aseptate as a new species.
Apiospora dematiacea J. Y. Zhang and Y. Z. Lu, sp. Nov. (Figure 3)
Figure 3.
Apiospora dematiacea (HKAS 129910, Holotype). (a,b) Cultures on PDA from above and below; (c) colonies on PDA; (d–i) conidiogenous cells with conidia; (j–p) conidia. Scale bars: (d) 50 μm; (e–l) 20 μm; (m–p) 10 μm.
Index Fungorum number: IF901116; Facesoffungi number: FoF14874.
Holotype: HKAS 129910
Etymology: referring to its dematiaceous spore.
Culture characteristics: Colonies on PDA medium circular, cottony, edge entire, flat, spreading, with abundant aerial mycelia, zonate with one concentric circle, reaching 47 mm diam after 10 d at 25 °C, white from above, yellowish white to grey to light yellow from center to edge in reverse. Vegetative hypha septate, branched, hyaline to light brown.
Description: Endophytic in the stems of Dicranopteris ampla. Sexual morph: Undetermined. Asexual morph: Conidiophores reduced to conidiogenous cells, cylindrical, septate, hyaline. Conidiogenous cells are cylindrical to subglobose, aggregated in clusters on hyphae, smooth, hyphae-like, and hyaline to brown. Conidia aseptate, globose to ellipsoid in surface view, 14.5–18(–20) µm diam. ( = 16.5 µm, n = 30), lenticular to lageniform from side view, 18.5–23(–25) × 10–13 µm diam. ( = 21.5 × 11.5 µm, n = 30), with longitudinal, pale germ slit. Sterile cells up to 32 µm long, 9–12(–16) µm ( = 10.5 µm, n = 20) wide, elongated, mixed among conidia brown, rarely truncate, and have a darkened scar at the base.
Material examined: CHINA, Guizhou Province, Qianxinan Buyi and Miao Autonomous Prefecture, Ceheng County (24°59′44″ N 105°50′16″ E), isolated from the healthy stem of Dicranopteris ampla near the roadside, 16 March 2022, J.Y. Zhang, 307-1 (HKAS 129910, Holotype), ex-living cultures, KUNCC 23-14202.
GenBank accession numbers: (LSU) OR590339, (ITS) OR590346, (TEF1-α) OR634953, and (TUB) OR634948.
Note: Apiospora dematiacea morphologically resembles A. hydei, characterized by conidiogenous cells aggregated in clusters on hyphae and globose conidia in surface view, with a pale equatorial slit when viewed from the side [8]. However, A. dematiacea is distinguishable from A. hydei due to its hyphae-like conidiogenous cells and more varied conidial shapes that include sterile cells. From a phylogenetic perspective, while Apiospora dematiacea shares a sister relationship with A. hydei, it constitutes a distinct lineage. A comparison of nucleotide base pairs between the ex-type strain of A. hydei (CBS 114990) and our newly isolated strain of Apiospora dematiacea (KUNCC 23-14202) reveals differences of 1183/1185 bp (99%), 578/579 (99%, including 1 gap), 411/432 bp (95%, including 12 bp gaps), and 778/794 bp (98%, including 2 bp gaps) in the LSU, ITS, TEF1-α, and TUB2 sequences, respectively. This confirms that they are distinct species.
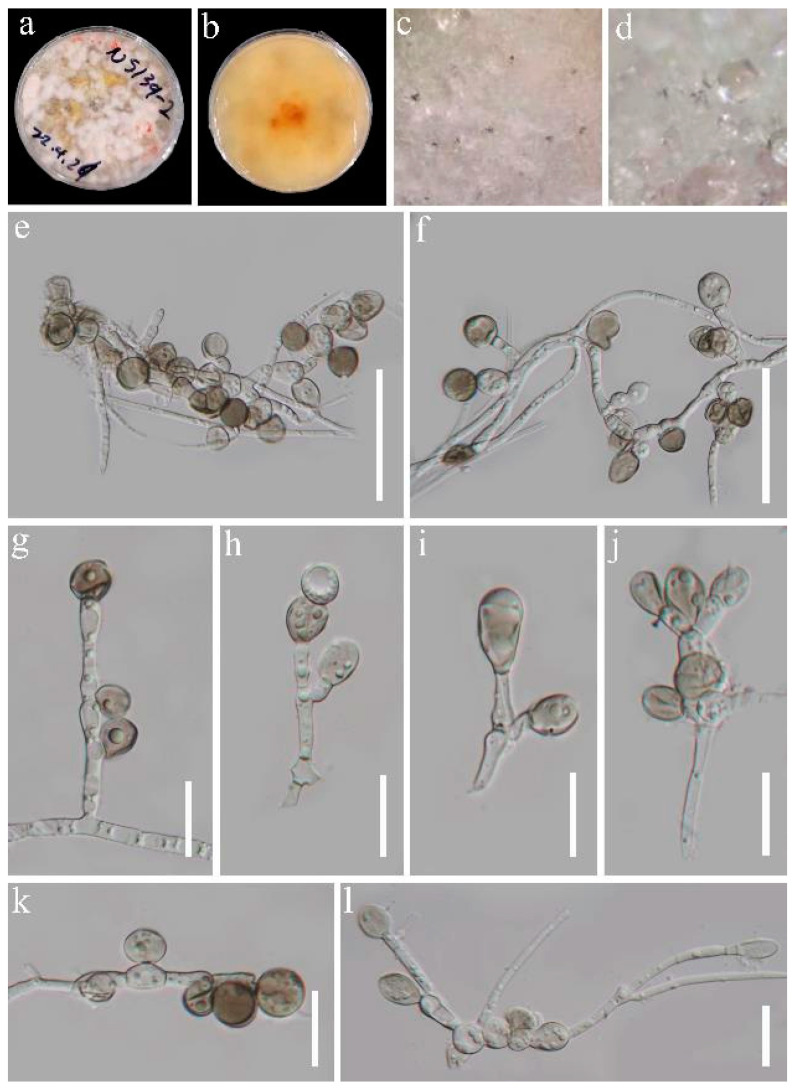
Apiospora dicranopteridis J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 4)
Figure 4.
Apiospora dicranopteridis (HKAS 129877, holotype). (a,b) Cultures on PDA from above and below; (c,d) colonies on PDA; (d–l) conidiophores, conidiogenous cells with conidia. Scale bars: (e,f) 50 μm; (g–l) 20 μm.
Index Fungorum number: IF901117; Facesoffungi number: FoF14875
Etymology: referring to the fungal host genus, Dicranopteris.
Holotype: HKAS 129877
Culture characteristics: Colonies on PDA medium circular, edge entire, floccose at the surface with dense, white aerial mycelia, growly fast, reaching 55 mm diam after 10 d at 25 °C, cottony, velvety, loose, white from above, yellow to yellowish whites in reverse. Vegetative hypha: septate, branched, sometimes coiled, guttulate, hyaline to pale brown.
Description: Endophytic in the stems of Dicranopteris pedata. Sexual morph: Undetermined. Asexual morph: Conidiophores are cylindrical, septate, branched, smooth-walled, and often reduced to conidiogenous cells. Conidiogenous cells 6–15 × 3.5–10 µm ( = 10 × 6 µm, n = 25) µm, solitary to aggregated in clusters arising from dense aerial hyphae, mono- to polybasic, sympodial, sub-globose to doliiform to cylindrical, smooth, subhyaline. Conidia amerospores, aseptate, globose or sub globose, 10.5–13 µm diam. ( = 11.5 µm, n = 15), cylindrical to broadly clavate 14–17(–22) × (6–)8–10.5 µm ( = 16 × 9 µm, n = 8), with rounded at the apex and a slightly narrower and truncate base, smooth to finely roughened guttules, without an equatorial germ slit, hyaline to pale brown.
Material examined: China, Guizhou Province, Qianxinan Buyi and Miao Autonomous Prefecture, Ceheng County (24°59′44″ N 105°50′16″ E), isolated from the healthy stems of Dicranopteris pedata nearby the roadside, 16 March 2022, J.Y. Zhang, 139-2 (HKAS 129877, holotype), ex-type living cultures, KUNCC23-14171; Ibid., isolated from the root of D. pedate, 16 March 2022, J.Y. Zhang, 170-4 (GZAAS 23-0780, paratype), living cultures, KUNCC 23-14177; Ibid, Anlong County, Jia Jia Ya Kou (24°59′23″ N; 105°35′20″ E), isolated from the healthy leaf of D. pedate, 16 March 2022, J.Y. Zhang, 223-4 (HKAS 129895, paratype), living cultures, GZCC 23-0712; Ibid, isolated from the healthy rhizome of D. ampla, 16 March 2022, J.Y. Zhang, 225-1 (HKAS 129898, paratype), living cultures, GZCC 23-0708.
GenBank accession numbers: KUNCC23-14171: (LSU) OR590336, (ITS) OR590342, (TEF1-a) OR634950, (TUB2) OR634944; KUNCC 23-14177: (LSU) OR590337, (ITS) OR590343, (TEF1-a) OR634951, (TUB2) OR634945; GZCC 23-0712: (LSU) OR590338, (ITS) OR590345, (TEF1-a) OR634952, (TUB2) OR634947; GZCC 23-0708: (ITS) OR590344, (TUB2) OR634946.
Notes: Apiospora dicranopteridis is morphologically distinct from other Apiospora species by its mono- or polyblastic, elongated cylindrical conidiogenous cells and globose to cylindrical to broadly clavate, hyaline to pale brown conidia. Phylogenetically, four strains (GZCC 23-0708, GZCC 23-0712, KUNCC23-14177, and KUNCC23-14171) representing Apiospora dicranopteridis sp. nov. formed a distinct clade. They share a sister relationship with A. koreana (KUC21332) and A. qinlingensis (CFCC 52303), reinforcing the notion that they are separate species.
Apiospora globosa J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 5)
Figure 5.
Apiospora globosa (HKAS 129921, holotype). (a,b) Cultures on WA from above and below; (c) colonies on WA; (d–f) conidiophores with conidia; (g–i) conidia. Scale bars: (d–f) 20 μm; (g–i) 10 μm.
Index Fungorum number: IF901402; Facesoffungi number: FoF14658.
Etymology: referring to the globose to subglobose conidia
Holotype: HKAS 129921
Culture characteristics: Colonies on WA medium irregulate, with several dark spots, flat with undulate edge, reaching 33 mm diam after 15 d at 25 °C, hyaline to light brown. Vegetative hyphae are thin, sparse, septate, branched, guttulate, and hyaline, some curled in a ring structure.
Description: Endophytic in the stems of Dicranopteris linearis. Sexual morph: Undetermined. Asexual morph: Conidiophores undistinguishable, hyphae-like. Conidiogenous cells are undistinguishable and hyphae-like. Conidia produced directly from vegetative hypha inside the WA culture, 4.5–8.5 µm diam ( = 6 µm, n = 20), aseptate, globose to subglobose, smooth to finely roughened, light yellow to gold to black.
Material examined: China, Guizhou Province, Anshun City, Ziyun Miao Buyi Autonomous County, Getu River Scenic Spot (25°48′26″ N 106°4′24″ E), isolated from the healthy stem of Dicranopteris linearis in a disturbed forest, 2 August 2022, J.Y. Zhang, S4-1 (dry WA culture, HKAS 129921, holotype; dry culture of WA-carrot mixture, GZAAS 23-0790), ex-type living cultures, KUNCC 23-14210.
GenBank accession numbers: (LSU) OR590340, (ITS) OR590347, and (TEF1-α) OR634954.
Notes: Phylogenetically, Apiospora globosa forms a distinct clade that is sister to the species Apiospora neosubglobosa (KUMCC 16-0203 and JHB 006). While Apiospora neosubglobosa has been described with only a sexual morph [9,22], our new species produces an asexual morph in culture. Morphologically, Apiospora globosa resembles A. xenocordella in conidial shape but has notably different conidiogenous cells and conidial size (4.5–8.5 µm diam. vs. 9–10 µm diam.). Apiospora globosa possesses indistinct, hyphae-like conidiogenous cells, whereas A. xenocordella features globose to clavate to doliiform conidiogenous cells [8].
4. Discussion
The genus Apiospora is relatively well studied, with species distributed across tropical, subtropical, temperate, and cold climates globally [8,12,14,41,51]. Members of the Apiospora species can function as endophytes [16,19,59,60], pathogens [8,19,61], or saprobes [5,10,13,23], found on various hosts, including various plants, air, water, soil debris, home dust, food, and the gut of insects [8,11,12,14,59,61]. They do not exhibit a clear lifestyle preference or pronounced sensitivity to environmental change. A fungus-host distribution of Arthrinium species (most of which have been synonymized under Apiospora) was provided by Wang et al. [11]. The data showed that Poaceae and Cyperaceae are the dominant host plant families, especially the former [10,12,14,17,20,22,42,62,63]. There have been no previous reports of Apiospora species from ferns, likely due to the neglect of fungi on ferns [64,65]. In this study, four new species (Apiospora aseptata, A. dematiacea, A. dicranopteridis, and A. globosa) were reported as endophytes isolated from three medicinal ferns—Dicranopteris ampla, D. linearis, and D. pedate—based on evidence from morphology and phylogenetic analyses of a concatenated dataset of LSU, ITS, TEF1-α, and TUB2 sequences. This study represents the first report of Apiospora species from Dicranopteris species, expanding the host diversity knowledge of Apiospora species.
The Apiospora-Arthrinium group has made certain achievements in bioactive secondary metabolites, with a high interest in agriculture, food, and the pharmaceutical industry [66,67,68,69]. The evaluation of the biological activities of Apiospora-Arthrinium spp. revealed this group has relatively biological activities for antifungal, antioxidant, and cellulolytic activity, especially Apiospora saccharicola [66]. A quick guide to secondary metabolites from the Apiospora–Arthrinium group was provided by Overgaard et al. [69], including the knowledge of 269 secondary metabolites and emphasizing some of the known biological or toxic compounds. For example, several species, including Apiospora arundinis, A. aurea, A. phaeosperma, A. sacchari, A. saccharicola, A. serenensis, A. terminalis, and Apiospora/Arthrinium spp., can produce 3-nitropropionic acid, which is associated with the food safety problem of poisonings or even deaths [70,71,72]. The study results of the genome sequence provided by Sørensen et al. [21] revealed that the Apiospora-Arthrinium group holds a high number of secondary metabolite gene clusters, which has attracted more attention to the compounds of this group [21,69]. In the current research context, we newly obtained seven endophytic taxa from the medicinal ferns of Dicranopteris rich in bioactive compounds and identified them as four new species (A. aseptata, A. dematiacea, A. dicranopteridis, and A. globosa) in the genus Apiospora, a fungal group that is currently attracting attention. These isolates are valuable fungi that are expected to explore secondary metabolites, which will also be the future research direction of our research.
All strains were initially cultured on a PDA medium. Most Apiospora strains sporulate on PDA substrate naturally under conventional conditions (room temperature 25–28°, natural light), which is consistent with many studies [8,11,16,19,23,41]. However, Apiospora globosa (KUNCC 23-14210) failed to sporulate on the PDA medium. Sporulation was later induced on various media, including CMM (Corn Meal Medium), MEA (malt extract agar), OA (oatmeal agar), SNA (synthrtic nutrient-poor agar), and WA (water agar). Eventually, sporulation of Apiospora globosa (KUNCC 23-14210) was successfully induced on WA and a WA-carrot mixture. During investigations of endophytic fungal diversity isolated from Dicranopteris in Guizhou, China, hundreds of isolates were selected from nearly a thousand endophytic strains for sporulation induction across multiple media (CMM, MEA, OA, PDA, SNA, and WA). Results showed that the sporulation rate on the WA medium exceeded that of other media. Therefore, WA is prioritized for inducing sporulation in plant endophytic fungi when the sporulation mechanism for this fungal group is not documented in previous publications.
Acknowledgments
Jing-Yi Zhang would like to thank Shaun Pennycook (Manaaki Whenua Landcare Research, New Zealand) for advising on the fungal names and Ning-Guo Liu and Chuan-Gen Lin for the guidance. Jing-Yi Zhang would also like to thank Mae Fah Luang University for granting me a tuition scholarship for my Ph.D. studies.
Author Contributions
Conceptualization, J.-Y.Z.; data curation, J.-Y.Z. and M.-L.C.; formal analysis, J.-Y.Z.; investigation, J.-Y.Z., M.-L.C. and Y.-X.W.; methodology, J.-Y.Z.; project administration, Y.-Z.L.; software, J.-Y.Z.; supervision, S.B. and Y.-Z.L.; writing—original draft preparation, J.-Y.Z.; writing—review and editing, S.B. and Y.-Z.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (NSFC 32060013), the Youth Science and Technology Talent Development Project from the Guizhou Provincial Department of Education (QJHKYZ [2021]263) and Youth Science and Technology Talent Development Project from Guizhou Provincial Department of Education (QJHKYZ [2022]345).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hyde K., Fröhlich J., Taylor J. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia. 1998;50:21–80. [Google Scholar]
- 2.Senanayake I.C., Maharachchikumbura S.S.N., Hyde K.D., Bhat J.D., Jones E.G., McKenzie E.H., Dai D.Q., Daranagama D.A., Dayarathne M.C., Goonasekara I.D. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes) Fungal Divers. 2015;73:73–144. doi: 10.1007/s13225-015-0340-y. [DOI] [Google Scholar]
- 3.Hyde K.D., Norphanphoun C., Maharachchikumbura S., Bhat D., Jones E., Bundhun D., Chen Y., Bao D., Boonmee S., Calabon M. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 4.Jiang N., Voglmayr H., Ma C.-Y., Xue H., Piao C.-G., Li Y. A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys. 2022;92:27. doi: 10.3897/mycokeys.92.86521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samarakoon M.C., Hyde K.D., Maharachchikumbura S.S., Stadler M., Gareth Jones E., Promputtha I., Suwannarach N., Camporesi E., Bulgakov T.S., Liu J.-K. Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes) Fungal Divers. 2022;112:1–88. doi: 10.1007/s13225-021-00495-5. [DOI] [Google Scholar]
- 6.Saccardo P.A. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti Soc. Veneziana-Trent. Istriana Sci. Nat. 1875;4:77–100. [Google Scholar]
- 7.Hawksworth D.L., Crous P.W., Redhead S.A., Reynolds D.R., Samson R.A., Seifert K.A., Taylor J.W., Wingfield M.J., Abaci Ö., Aime C. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2:105–111. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crous P.W., Groenewald J.Z. A phylogenetic re-evaluation of Arthrinium. IMA Fungus. 2013;4:133–154. doi: 10.5598/imafungus.2013.04.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai D.-Q., Jiang H.-B., Tang L.-Z., Bhat D.J. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 2016;7:1332–1345. doi: 10.5943/mycosphere/7/9/7. [DOI] [Google Scholar]
- 10.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 11.Wang M., Tan X.-M., Liu F., Cai L. Eight new Arthrinium species from China. MycoKeys. 2018;34:1–24. doi: 10.3897/mycokeys.34.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pintos Á., Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2021;7:197–221. doi: 10.3114/fuse.2021.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian X., Karunarathna S.C., Mapook A., Promputtha I., Xu J., Bao D., Tibpromma S. One new species and two new host records of Apiospora from bamboo and maize in Northern Thailand with thirteen new combinations. Life. 2021;11:1071. doi: 10.3390/life11101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhunjun C.S., Niskanen T., Suwannarach N., Wannathes N., Chen Y.-J., McKenzie E.H., Maharachchikumbura S.S., Buyck B., Zhao C.-L., Fan Y.-G. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022;114:387–462. doi: 10.1007/s13225-022-00501-4. [DOI] [Google Scholar]
- 15.Kwon S.L., Cho M., Lee Y.M., Kim C., Lee S.M., Ahn B.J., Lee H., Kim J.-J. Two unrecorded Apiospora species isolated from marine substrates in Korea with eight new combinations (A. piptatheri and A. rasikravindrae) Mycobiology. 2022;50:46–54. doi: 10.1080/12298093.2022.2038857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X., Chomnunti P., Doilom M., Daranagama D.A., Kang J. Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand. J. Fungi. 2022;8:248. doi: 10.3390/jof8030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phukhamsakda C., Nilsson R.H., Bhunjun C.S., de Farias A.R.G., Sun Y.-R., Wijesinghe S.N., Raza M., Bao D.-F., Lu L., Tibpromma S., et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022;114:327–386. doi: 10.1007/s13225-022-00502-3. [DOI] [Google Scholar]
- 18.Pintos Á., Alvarado P. New studies on Apiospora (Amphisphaeriales, Apiosporaceae): Epitypification of Sphaeria apiospora, proposal of Ap. marianiae sp. nov. and description of the asexual morph of Ap. sichuanensis. MycoKeys. 2022;92:63–78. doi: 10.3897/mycokeys.92.87593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R., Li D., Zhang Z., Liu S., Liu X., Wang Y., Zhao H., Liu X., Zhang X., Xia J. Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China. MycoKeys. 2023;95:27–45. doi: 10.3897/mycokeys.95.96400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H., Dong W., Shu Y., Mapook A., Manawasinghe I., Doilom M., Luo M. Bambusicolous fungi in Guangdong, China: Establishing Apiospora magnispora sp. nov. (Apiosporaceae, Amphisphaeriales) based on morphological and molecular evidence. J. Fungal Biol. 2023;13:1–15. doi: 10.5943/cream/13/1/1. [DOI] [Google Scholar]
- 21.Sørensen T., Petersen C., Fechete L.I., Nielsen K.L., Sondergaard T.E. A highly contiguous genome assembly of Arthrinium puccinoides. Genome Biol. Evol. 2022;14:evac010. doi: 10.1093/gbe/evac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Q., Lv Y.-C., Xu X.-L., Deng Y., Wang F.-H., Liu S.-Y., Liu L.-J., Yang C.-L., Liu Y.-G. Morpho-Molecular Characterization of Microfungi Associated with Phyllostachys (Poaceae) in Sichuan, China. J. Fungi. 2022;8:702. doi: 10.3390/jof8070702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y., Liu J.K., Lin C.G., Chen Y.Y., Xiang M.M., Liu Z.Y. Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Front. Microbiol. 2021;12:661281. doi: 10.3389/fmicb.2021.661281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crous P.W., Hernández-Restrepo M., Schumacher R., Cowan D.A., Maggs-Kölling G., Marais E., Wingfield M.J., Yilmaz N., Adan O., Akulov A. New and interesting fungi. 4. Fungal Syst. Evol. 2021;7:255. doi: 10.3114/fuse.2021.07.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusari S., Lamshöft M., Zühlke S., Spiteller M. An endophytic fungus from Hypericum perforatum that produces hypericin. J. Nat. Prod. 2008;71:159–162. doi: 10.1021/np070669k. [DOI] [PubMed] [Google Scholar]
- 26.Rashmi M., Kushveer J., Sarma V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere. 2019;10:798–1079. doi: 10.5943/mycosphere/10/1/19. [DOI] [Google Scholar]
- 27.Schulz B., Boyle C., Draeger S., Römmert A.-K., Krohn K. Endophytic fungi: A source of novel biologically active secondary metabolites. Mycol. Res. 2002;106:996–1004. doi: 10.1017/S0953756202006342. [DOI] [Google Scholar]
- 28.Rana K.L., Kour D., Kaur T., Devi R., Negi C., Yadav A.N., Yadav N., Singh K., Saxena A.K. Microbial Endophytes. Elsevier; Amsterdam, The Netherlands: 2020. Endophytic fungi from medicinal plants: Biodiversity and biotechnological applications; pp. 273–305. [Google Scholar]
- 29.Stierle A., Strobel G., Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;260:214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- 30.Stierle A., Strobel G., Stierle D., Grothaus P., Bignami G. The search for a taxol-producing microorganism among the endophytic fungi of the Pacific yew, Taxus brevifolia. J. Nat. Prod. 1995;58:1315–1324. doi: 10.1021/np50123a002. [DOI] [PubMed] [Google Scholar]
- 31.Strobel G.A. Endophytes as sources of bioactive products. Microbes Infect. 2003;5:535–544. doi: 10.1016/S1286-4579(03)00073-X. [DOI] [PubMed] [Google Scholar]
- 32.PPG I. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016;54:563–603. doi: 10.1111/jse.12229. [DOI] [Google Scholar]
- 33.Yang L., Huang Y., Lima L.V., Sun Z., Liu M., Wang J., Liu N., Ren H. Rethinking the ecosystem functions of Dicranopteris, a widespread genus of ferns. Front. Plant Sci. 2021;11:581513. doi: 10.3389/fpls.2020.581513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X.-L., Cheng X., Yang L.-M., Wang R.-R., Zheng Y.-T., Xiao W.-L., Zhao Y., Xu G., Lu Y., Chang Y. Dichotomains A and B: Two New Highly Oxygenated Phenolic Derivatives from Dicranopteris d ichotoma. Org. Lett. 2006;8:1937–1940. doi: 10.1021/ol060535i. [DOI] [PubMed] [Google Scholar]
- 35.Zakaria Z.A., Ghani Z.D.F.A., Nor R.N.S.R.M., Gopalan H.K., Sulaiman M.R., Abdullah F.C. Antinociceptive and anti-inflammatory activities of Dicranopteris linearis leaves chloroform extract in experimental animals. Yakugaku Zasshi. 2006;126:1197–1203. doi: 10.1248/yakushi.126.1197. [DOI] [PubMed] [Google Scholar]
- 36.Li X.-L., Yang L.-M., Zhao Y., Wang R.-R., Xu G., Zheng Y.-T., Tu L., Peng L.-Y., Cheng X., Zhao Q.-S. Tetranorclerodanes and clerodane-type diterpene glycosides from Dicranopteris dichotoma. J. Nat. Prod. 2007;70:265–268. doi: 10.1021/np0603166. [DOI] [PubMed] [Google Scholar]
- 37.Zakaria Z., Mohamed A., Mohd Jamil N., Rofiee M., Fatimah C., Mat Jais A., Sulaiman M., Somchit M. In vitro anticancer activity of various extracts of the Malaysian, available but neglected, plants (Muntingia Calabura And Dicranopteris Linearis) against Mcf-7 And Ht-29 cancer cell lines. E–PharmaNexus. 2008;1:10–17. [Google Scholar]
- 38.Kale M. Analysis of bioactive components of Dicranopteris Linearis (Burm. F.) Underwood whole plant ethanol extract by GC-MS. Pure Appl. Sci. Botany. 2015;34:9–12. doi: 10.5958/2320-3196.2015.00002.6. [DOI] [Google Scholar]
- 39.Li X.L., Tu L., Zhao Y., Peng L.Y., Xu G., Cheng X., Zhao Q.S. Terpenoids from two Dicranopteris species. Helv. Chim. Acta. 2008;91:856–861. doi: 10.1002/hlca.200890089. [DOI] [Google Scholar]
- 40.Nontachaiyapoom S., Sasirat S., Manoch L. Isolation and identification of Rhizoctonia-like fungi from roots of three orchid genera, Paphiopedilum, Dendrobium, and Cymbidium, collected in Chiang Rai and Chiang Mai provinces of Thailand. Mycorrhiza. 2010;20:459–471. doi: 10.1007/s00572-010-0297-3. [DOI] [PubMed] [Google Scholar]
- 41.Jiang N., Li J., Tian C. Arthrinium species associated with bamboo and reed plants in China. Fungal Syst. Evol. 2018;2:1–9. doi: 10.3114/fuse.2018.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang H.B., Hyde K.D., Doilom M., Karunarathna S.C., Xu J.C., Phookamsak R. Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. AJOM. 2019;2:254–268. doi: 10.5943/ajom/2/1/16. [DOI] [Google Scholar]
- 43.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat J., Buyck B., Cai L., Dai Y.-C., Abd-Elsalam K.A., Ertz D., Hidayat I. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 44.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 46.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 48.O’Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungusfusariumare nonorthologous. Mol. Phylogenet. Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 49.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Proc. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 51.Sun L.K., Park M. The genus Arthrinium (Ascomycota, Sordariomycetes, Apiosporaceae) from marine habitats from Korea, with eight new species. IMA Fungus. 2021;12:13. doi: 10.1186/s43008-021-00065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 54.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Boil. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nylander J. MrModeltest v2. Program Distributed by the Author. Evolutionary Biology Centre. Uppsala University, Sweden. 2004. [(accessed on 25 August 2023)]. Available online: https://www.ebc.uu.se/systzoo/staff/nylander.html.
- 57.Larget B., Simon D.L. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol. Boil. Evol. 1999;16:750–759. doi: 10.1093/oxfordjournals.molbev.a026160. [DOI] [Google Scholar]
- 58.Rambaut A., Drummond A. FigTree: Tree Figure Drawing Tool, version 1.2.2. Institute of Evolutionary Biology, University of Edinburgh; Edinburgh, UK: 2008. [Google Scholar]
- 59.Sharma R., Kulkarni G., Sonawane M.S., Shouche Y.S. A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience. 2014;55:118–123. doi: 10.1016/j.myc.2013.06.004. [DOI] [Google Scholar]
- 60.Chen T.Z., Zhang Y., Ming X.B., Zhang Q., Long H., Hyde K.D., Li Y., Wang Y. Morphological and phylogenetic resolution of Arthrinium from medicinal plants in Yunnan, including A. cordylines and A. pseudomarii spp. nov. Mycotaxon. 2021;136:183–199. doi: 10.5248/136.183. [DOI] [Google Scholar]
- 61.Zhao Y., Deng C., Chen X. Arthrinium phaeospermum causing dermatomycosis, a new record of China. Acta Mycol. Sin. 1990;9:232–235. [Google Scholar]
- 62.Kwon S.L., Cho M., Lee Y.M., Lee H., Kim C., Kim G.-H., Kim J.-J. Diversity of the Bambusicolous Fungus Apiospora in Korea: Discovery of New Apiospora Species. Mycobiology. 2022;50:302–316. doi: 10.1080/12298093.2022.2133808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang X. Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in northern Thailand. Biodivers. Data J. 2020;8:e58755. doi: 10.3897/BDJ.8.e58755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Razikin M.Z.M., Nagao H., Zakaria R. First report of Pteridocolous discomycetes, Lachnum lanariceps and L. oncospermatum, on decayed tree fern in Bukit Bendera (Penang Hill), Pulau Pinang, Malaysia. Ann. R. Bot. Gard. Peradiniya. 2014;36:407–410. [Google Scholar]
- 65.Kirschner R., Lee P.H., Huang Y.-M. Diversity of fungi on Taiwanese fern plants: Review and new discoveries. Taiwania. 2019;64:163–175. doi: 10.6165/tai.2019.64.163. [DOI] [Google Scholar]
- 66.Hong J.-H., Jang S., Heo Y.M., Min M., Lee H., Lee Y.M., Lee H., Kim J.-J. Investigation of marine-derived Fungal Divers. and their exploitable biological activities. Mar. Drugs. 2015;13:4137–4155. doi: 10.3390/md13074137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elissawy A.M., Ebada S.S., Ashour M.L., Özkaya F.C., Ebrahim W., Singab A.B., Proksch P. Spiroarthrinols a and B, two novel meroterpenoids isolated from the sponge-derived fungus Arthrinium sp. Phytochem. Lett. 2017;20:246–251. doi: 10.1016/j.phytol.2017.05.008. [DOI] [Google Scholar]
- 68.Pansanit A., Pripdeevech P. Antibacterial secondary metabolites from an endophytic fungus, Arthrinium sp. MFLUCC16-1053 isolated from Zingiber cassumunar. Mycology. 2018;9:264–272. doi: 10.1080/21501203.2018.1481154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Overgaard M.L., Aalborg T., Zeuner E.J., Westphal K.R., Lau F.A., Nielsen V.S., Carstensen K.B., Hundebøll E.A., Westermann T.A., Rathsach G.G. Quick guide to secondary metabolites from Apiospora and Arthrinium. Fungal. Biol. Rev. 2023;43:100288. doi: 10.1016/j.fbr.2022.10.001. [DOI] [Google Scholar]
- 70.Liu X., Luo X., Hu W. Studies on the epidemiology and etiology of moldy sugarcane poisoning in China. Biomed. Environ. Sci. 1992;5:161–177. [PubMed] [Google Scholar]
- 71.Birkelund T., Johansen R.F., Illum D.G., Dyrskog S.E., Østergaard J.A., Falconer T.M., Andersen C., Fridholm H., Overballe-Petersen S., Jensen J.S. Fatal 3-nitropropionic acid poisoning after consuming coconut water. Emerg. Infect. Dis. 2021;27:278. doi: 10.3201/eid2701.202222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao J., Jiang W., Wu X., He J., Li H., Wang T., Cheng L., Chen W., Mo L. First Report of Apiospora Mold on Sugarcane in China Caused by Apiospora arundinis (Arthrinium arundinis) Plant Dis. 2022;106:1058. doi: 10.1094/PDIS-02-21-0386-PDN. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.