Abstract
Leuconostoc carnosum was shown to be the specific spoilage organism in vacuum-packaged, sliced, cooked ham showing spoilage during 3 weeks of shelf life. Identification of the specific spoilage organism was done by use of phenotypic data and ClaI, EcoRI, and HindIII reference strain ribopatterns. One hundred L. carnosum isolates associated with the production and spoilage of the ham were further characterized by pulsed-field gel electrophoresis (PFGE), together with some meat-associated Leuconostoc species: L. citreum, L. gelidum, L. mesenteroides subsp. dextranicum, and L. mesenteroides subsp. mesenteroides. ApaI and SmaI digests divided the industrial L. carnosum strains into 25 different PFGE types, ApaI and SmaI types being consistent. Only one specific PFGE type was associated with the spoiled packages. This type also was detected in air and raw-meat mass samples. The spoilage strain did not produce bacteriocins. Only seven isolates belonging to three different PFGE types produced bacteriocins. Similarity analysis of the industrial L. carnosum strains revealed a homogeneous cluster which could be divided into eight subclusters consisting of strains having at most three-fragment differences. The L. carnosum cluster was clearly distinguished from the other meat-associated leuconostoc clusters, with the exception of the L. carnosum type strain. Ribotyping can be very helpful in the identification of L. carnosum, but its discriminatory power is too weak for strain characterization. PFGE provides good discrimination for studies dealing with the properties of homogeneous L. carnosum strains.
Lactic acid bacteria (LAB) are the major spoilage bacteria in vacuum-packaged, cooked meat products (1, 2, 10, 13, 25, 27, 31, 38, 44, 47, 56). Lactobacillus and Leuconostoc have been the main genera associated with the spoilage of these products, Lactobacillus sake and Lactobacillus curvatus being isolated commonly (12, 16, 18, 19, 24, 27, 30, 35, 39, 43–46). Compared to aerobic spoilage bacteria, spoilage LAB produce their typical sensory changes, such as souring, gas formation, and/or slime formation, later, at the stationary phase (29, 44), and a vacuum-packaged product is usually expected to maintain good sensory quality for at least 3 to 4 weeks. However, due to an increased level of LAB contamination or particularly active spoilage strains, spoilage may occur during the shelf-life period, subjecting the producer to recalls (30, 31, 33, 46).
In an LAB contamination study of vacuum-packaged, sliced, cooked ham, 982 LAB isolates from the spoiled product and production line were characterized in order to determine the underlying reasons for fluctuations in product quality (4, 6). Many lots had been showing spoilage changes, i.e., sour odor and taste, before the sell-by date. In that study, ribotyping (21) was used as a tool for contamination analysis. Based on EcoRI and HindIII ribopatterns, two major spoilage LAB types, types G and A, were detected. Contamination with these spoilage LAB was shown to have occurred postcooking, and a probable site of air-mediated contamination from the macerated raw-meat mass to the cooked product was revealed. Because type G showed the typical EcoRI and HindIII ribopatterns of L. sake (5), no further identification or characterization studies were warranted. However, the most important specific spoilage organism, type A, was not identified to the species level. Type A had been detected as the dominant type in the macerated raw-meat mass and in the spoiled packages with the strongest changes in sensory characteristics (6). It had also persisted in the plant during the 1-year study period, consisting of two separate large-scale contamination experiments (4, 6).
In this study, we set out to identify type A LAB to the species level and characterize in more detail the 100 isolates possessing the type A EcoRI and HindIII ribopatterns. Since phenotypic characteristics alone are seldom sufficient for species identification of LAB (15), a reference strain library was created by ribotyping and was used with phenotypic data. Pulsed-field gel electrophoresis (PFGE) was applied in order to provide further strain-level characterization. Production of bacteriocins was determined for evaluation of the impact of this characteristic in a population associated with process contamination and product spoilage.
MATERIALS AND METHODS
Bacterial strains.
One hundred type A LAB possessing the same EcoRI and HindIII ribopatterns had been isolated during a contamination study of a meat plant (6). All isolates were gram-positive, oval cocci isolated from a macerated raw-meat mass, air in the macerating room, surfaces and air in the cooking room, worker’s gloves, surfaces of the ham prior to slicing, and vacuum-packaged, sliced, cooked ham cultured on the sell-by date. Isolates originating from different sources are listed in Table 1.
TABLE 1.
Division of the isolates into different types and certain phenotypic properties
| Typea | Isolatesb | Restriction enzyme profile
|
Production ofc:
|
||
|---|---|---|---|---|---|
| ApaI | SmaI | Slime from sucrose | Bacteriocin | ||
| A I-a | M2n, M2h | A1 | S1 | − | − |
| A I-b | M3h, M6f, M6h | A2 | S2 | − | − |
| A I-c | P31a, M41, M4m | A3 | S3 | − | − |
| A I-d | I27e | A4 | S4 | + | − |
| A I-e | M5i, M5j | A5 | S5 | − | − |
| A I-f | P36b | A6 | S6 | − | − |
| A I-g | M6j | A7 | S7 | + | − |
| A I-h | V8a–o, V9a–o, V11a–m, V13a–o, I2b, I27a, M5o, M6a | A8 | S8 | − | − |
| A I-i | M6o | A9 | S9 | − | − |
| A I-j | M2e, M2l, M2o, M3f, M3l | A10 | S10 | − | + |
| A I-k | M6g | A11 | S11 | + | − |
| A II-a | I1b | A12 | S12 | − | − |
| A II-b | I1c, I1f | A13 | S13 | − | − |
| A II-c | I26b, I28b | A14 | S14 | + | − |
| A II-d | I2a | A15 | S15 | − | − |
| A III-a | M2j, M3e, M3m | A16 | S16 | + | − |
| A III-b | M1e | A17 | S17 | + | − |
| A III-c | M5k | A18 | S18 | + | − |
| A IV | M2d | A19 | S19 | − | + |
| A V-a | M3o, M6i | A20 | S20 | + | − |
| A V-b | M1f | A21 | S21 | + | + |
| A VI | M1j | A22 | S22 | + | − |
| A VII-a | I27f | A23 | S23 | + | − |
| A VII-b | M1i | A24 | S24 | − | − |
| A VIII | M1c | A25 | S25 | − | − |
Types sharing the same roman numeral differ by at most three bands in the restriction enzyme profiles.
Sampling was described previously (6). Sources were as follows: M, raw-meat mass; P, surface; I, air; V, spoiled product. Groups of lowercase letters indicate a series of isolates; e.g., a–o indicates 15 isolates from V8a to V8o.
+, production; −, no production.
In order to obtain a library for species identification, the following reference strains were ribotyped with ClaI, EcoRI, and HindIII: Leuconostoc carnosum NCFB (National Collection of Food Bacteria) 2776T, Leuconostoc citreum (Leuconostoc amelibiosum) D1 (35), Leuconostoc fallax CCUG (Culture Collection of University of Gothenburg) 30061T, Leuconostoc gelidum NCFB 2775T, Leuconostoc lactis CCUG 30064T, Leuconostoc mesenteroides subsp. mesenteroides DSM (Deutsche Sammlung von Mikroorganismen) 20343T, Leuconostoc mesenteroides subsp. cremoris CCUG 21965T, Leuconostoc mesenteroides subsp. dextranicum DSM 20484T, Leuconostoc pseudomesenteroides DSM 20193T, Weissella halotolerans ATCC (American Type Culture Collection) 35410T, Weissella viridescens ATCC 12706T, and Weissella paramesenteroides DSM 20288T. In addition, the previously established (5, 7) ClaI, EcoRI, and HindIII Lactobacillus ribotypes were compared with the Leuconostoc and Weissella ribotypes characterized in this study.
The meat-associated reference strains L. carnosum NCFB 2776T, L. citreum (L. amelibiosum) D1 (35), L. gelidum NCFB 2775T, L. mesenteroides subsp. dextranicum DSM 20484T, L. mesenteroides subsp. mesenteroides DSM 20343T, L. pseudomesenteroides DSM 20193T, and W. paramesenteroides DSM 20288T were characterized by PFGE along with the industrial isolates.
All strains were maintained in MRS broth (Difco, Detroit, Mich.) at −70°C and cultured with MRS broth or MRS agar (Oxoid, Basingstoke, United Kingdom) as previously described (28).
Phenotypic characterization.
The anaerobic growth of all industrial isolates on Rogosa selective Lactobacillus agar (Orion Diagnostica, Espoo, Finland) was determined, and the scheme of Villiani et al. (55) was used for the presumptive identification of Leuconostoc spp. Gas production from glucose was tested with modified MRS broth in Durham tubes (51). Production of ammonia from arginine was observed by the method of Briggs (14), and dextran formation was studied with 5% sucrose-containing agar (22). Fermentation of carbohydrates was determined by use of the API 50 CH Lactobacillus identification system (Biomerieux, Marcy l’Etoile, France) for five randomly selected isolates (I27a, M1f, V8a, M6f, and P31a), which were also tested for the ability to produce different lactic acid isomers by an enzymatic method (57) with d- and l-lactate dehydrogenases (Boehringer GmbH, Mannheim, Federal Republic of Germany). The five randomly selected isolates were also tested for growth in MRS broth at 8, 10, 15, and 37°C.
Bacteriocin determination.
The agar spot test method modified by Schillinger and Lücke (48) was used for screening bacteriocin activity. Based on existing literature, L. mesenteroides subsp. mesenteroides DSM 20343T was selected as the indicator bacterium (3, 26, 40, 54, 58).
In vitro isolation of DNA and ribotyping for species identification.
Reference strains and the five randomly selected industrial isolates, already known to possess similar EcoRI and HindIII ribotypes, were characterized with ClaI, EcoRI, and HindIII (New England BioLabs, Beverly, Mass.). These enzymes were selected because they characterize LAB well (4–6). DNA was isolated by the guanidium thiocyanate method of Pitcher et al. (42) as modified by Björkroth and Korkeala (4) by combined lysozyme and mutanolysin treatments. Restriction endonuclease treatment of 3 μg of DNA was done as specified by the manufacturer (New England BioLabs). Genomic blotting was done by vacuum blotting (Vacugene; Pharmacia, Uppsala, Sweden), and the ribosomal DNA probe for ribotyping was labeled by reverse transcription (avian myeloblastosis virus reverse transcriptase [Promega, Madison, Wis.]; Dig DNA labeling kit [Boehringer]) as previously described by Blumberg et al. (11). Membranes were hybridized at 68°C as described by Björkroth and Korkeala (5). Similarity between all ribopatterns was determined visually.
In situ DNA isolation and PFGE.
Cells were harvested from 2 ml of MRS broth cultures grown overnight at 30°C. DNA isolation in situ from agarose blocks was performed as described by Maslow et al. (37) with the modifications described by Björkroth et al. (9). Initially, 11 rare-cutting restriction enzymes, ApaI, AscI, EagI, MluI, NotI, NruI, RsrII, SacII, SmaI, XbaI, and XhoI, were tested for the cleavage of DNA of three strains (NCFB 2776T, M6f, and I27a). ApaI and SmaI, which produced convenient numbers of fragments with discriminatory patterns, were chosen for the cleavage of all strains. The samples were electrophoresed through a 1.2% (wt/vol) agarose gel (SeaKem Gold; FMC BioProducts, Rockland, Maine) in 0.5× TBE (45 mM Tris, 4.5 mM boric acid [pH 8.3], 1 mM sodium EDTA) at 14°C by use of a Gene Navigator system with the hexagonal electrode (Pharmacia). Interpolation ramping from 0.5 to 15 s for 20 h at 200 V was used for both enzyme digests.
PFGE data management.
Photographs of the PFGE banding patterns were scanned with a ScanJet 4c/T scanner (Hewlett-Packard Co., Boise, Idaho). Numerical analysis of macrorestriction patterns was performed with a GelCompar system (version 4.0; Applied Maths, Kortijk, Belgium). The similarity between all pairs was expressed by Dice coefficient correlation, and clustering by the unweighted pair-group method with arithmetic averages was used for the construction of the dendrogram. Types were considered closely related (53) in the presence of at most a three-band difference (one genetic event). This relationship was indicated in the type nomination by a shared roman numeral.
RESULTS
The 100 isolates did not grow on Rogosa selective Lactobacillus agar; all produced gas from glucose but did not produce ammonia from arginine. Fifteen isolates (11 different PFGE types) produced slime from sucrose, and bacteriocins were produced by 7 isolates (Table 1). The five isolates tested produced only d-lactic acid and had similar fermentation patterns for the utilization of ribose, d-glucose, d-fructose, α-methyl-d-glucoside, N-acetylglucosamine, cellobiose, saccharose, trehalose, β-gentiobiose, d-turanose, and gluconate. Growth occurred at 8, 10, and 15°C but not at 37°C.
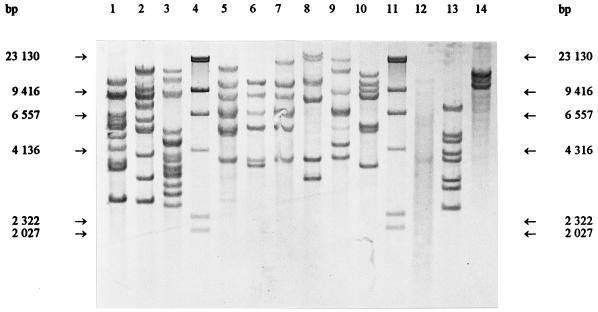
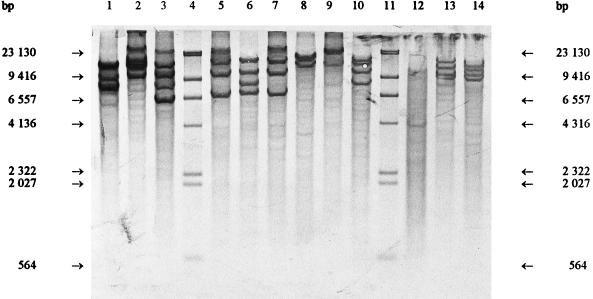
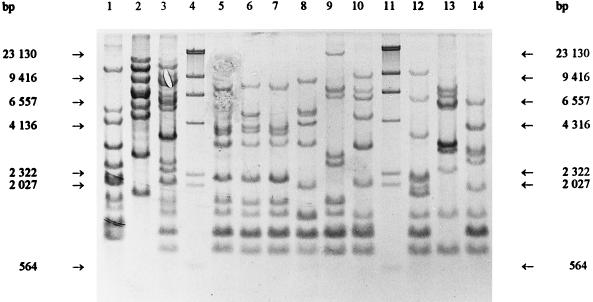
Previously determined oval cell morphology and the phenotypic characteristics typical of leuconostocs led to the comparison with the Leuconostoc and Weissella type strains. Figures 1, 2, and 3 show that the ClaI, EcoRI, and HindIII ribotypes, respectively, of the reference strains differed clearly from the Lactobacillus ribotypes obtained previously (5, 7). The ClaI, EcoRI, and HindIII ribopatterns of the industrial isolates were found to be identical to those of L. carnosum NCFB 2776T (Fig. 1, 2, and 3, lanes 10). All of the other type strains were distinct from L. carnosum NCFB 2776T. Based on the phenotypic data and the identical ribopatterns, the industrial isolates were classified as L. carnosum. HindIII and EcoRI generated the least distinguishing ribotypes for the Leuconostoc and Weissella species. ClaI was the only enzyme distinguishing L. mesenteroides subsp. mesenteroides from L. mesenteroides subsp. dextranicum (Fig. 1, lanes 5 and 7).
FIG. 1.
ClaI ribopatterns. Lanes 4 and 11, phage lambda DNA cleaved with HindIII as a fragment size marker; lane 1, Weissella viridescens ATCC 12706T; lane 2, Weissella halotolerans ATCC 35410T; lane 3, Weissella paramesenteroides DSM 20288T; lane 5, Leuconostoc mesenteroides subsp. mesenteroides DSM 20343T; lane 6, Leuconostoc mesenteroides subsp. cremoris CCUG 21965T; lane 7, Leuconostoc mesenteroides subsp. dextranicum DSM 20484T; lane 8, Leuconostoc pseudomesenteroides DSM 20193T; lane 9, Leuconostoc carnosum NCFB 2776T; lane 10, Leuconostoc gelidum NCFB 2775T; lane 12, Leuconostoc lactis CCUG 30064T; lane 13, Leuconostoc fallax CCUG 30061T; lane 14, Leuconostoc citreum (Leuconostoc amelibiosum) D1.
FIG. 2.
EcoRI ribopatterns. Lanes 4 and 11, phage lambda DNA cleaved with HindIII as a fragment size marker; lane 1, Weissella viridescens ATCC 12706T; lane 2, Weissella halotolerans ATCC 35410T; lane 3, Weissella paramesenteroides DSM 20288T; lane 5, Leuconostoc mesenteroides subsp. mesenteroides DSM 20343T; lane 6, Leuconostoc mesenteroides subsp. cremoris CCUG 21965T; lane 7, Leuconostoc mesenteroides subsp. dextranicum DSM 20484T; lane 8, Leuconostoc pseudomesenteroides DSM 20193T; lane 9, Leuconostoc carnosum NCFB 2776T; lane 10, Leuconostoc gelidum NCFB 2775T; lane 12, Leuconostoc lactis CCUG 30064T; lane 13, Leuconostoc fallax CCUG 30061T; lane 14, Leuconostoc citreum (Leuconostoc amelibiosum) D1.
FIG. 3.
HindIII ribopatterns. Lanes 4 and 11, phage lambda DNA cleaved with HindIII as a fragment size marker; lane 1, Weissella viridescens ATCC 12706T; lane 2, Weissella halotolerans ATCC 35410T; lane 3, Weissella paramesenteroides DSM 20288T; lane 5, Leuconostoc mesenteroides subsp. mesenteroides DSM 20343T; lane 6, Leuconostoc mesenteroides subsp. cremoris CCUG 21965T; lane 7, Leuconostoc mesenteroides subsp. dextranicum DSM 20484T; lane 8, Leuconostoc pseudomesenteroides DSM 20193T; lane 9, Leuconostoc carnosum NCFB 2776T; lane 10, Leuconostoc gelidum NCFB 2775T; lane 12, Leuconostoc lactis CCUG 30064T; lane 13, Leuconostoc fallax CCUG 30061T; lane 14, Leuconostoc citreum (Leuconostoc amelibiosum) D1.
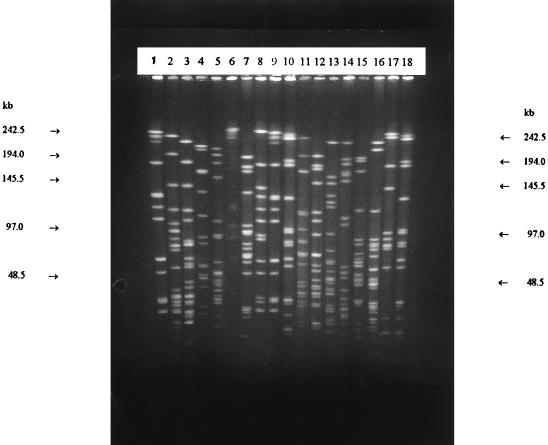
Both ApaI and SmaI generated 25 different patterns for the meat plant isolates when one-band differences are noted. The ApaI types were consistent with the SmaI types (Table 1). All meat-associated reference strains, with the exception of L. carnosum NCFB 2776T, were clearly distinguished from the industrial isolates (Fig. 4 and 5). Both ApaI and SmaI resulted in convenient numbers of fragments for macrorestriction analysis (Fig. 4). However, SmaI cleaved the DNA efficiently, whereas some partial digestion was occasionally noted with ApaI. Because of the better reproducibility, SmaI patterns were chosen for the numerical analysis.
FIG. 4.
SmaI (lanes 1 to 9) and ApaI (lanes 10 to 18) ribopatterns. Lanes 1, 9, 10, and 18, Leuconostoc carnosum NCFB 2776T; lanes 2 and 11, Leuconostoc mesenteroides subsp. mesenteroides DSM 20343T; lanes 3 and 12, Leuconostoc mesenteroides subsp. dextranicum DSM 20484T; lanes 4 and 13, Leuconostoc pseudomesenteroides DSM 20193T; lanes 5 and 14, Weissella paramesenteroides DSM 20288T; lanes 6 and 15, Leuconostoc gelidum NCFB 2775T; lanes 7 and 16, Leuconostoc citreum (Leuconostoc amelibiosum) D1; lanes 8 and 17, Leuconostoc carnosum V-8a.
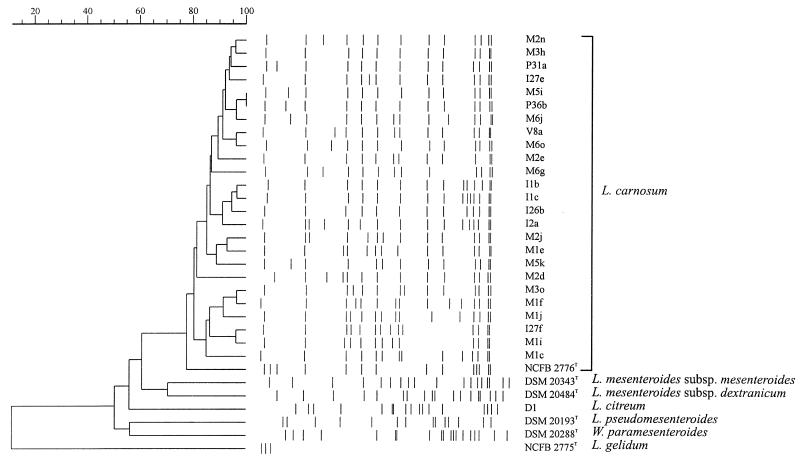
FIG. 5.
Dendrogram based on SmaI ribopatterns. The similarity between all pairs was expressed by Dice coefficient correlation, and the unweighted pair-group method with arithmetic averages was used for the construction of the dendrogram.
Figure 5 shows the dendrogram of the industrial isolates and the reference strains. L. carnosum formed a homogeneous cluster, within which eight subclusters consisted of strains having at most three-band differences. Reference strains, with the exception of the L. carnosum type strain, clustered separately from the industrial isolates. Isolates associated with the sensorially spoiled products all showed the type A I-h pattern (Fig. 4, lanes 8 and 17) and belonged to the largest cluster, consisting of A I types (Fig. 5 and Table 1). Type A I-h was also detected in two raw-meat mass samples and two air samples, one from the macerating room and one from the postcooking form removal area.
DISCUSSION
L. carnosum was identified as the specific spoilage organism in the vacuum-packaged, cooked ham studied here. This species was described along with L. gelidum by Shaw and Harding in 1989 (50). It belongs to the main Leuconostoc cluster designated Leuconostoc sensu stricto and shares 97 to 99% rRNA homology with the other sensu stricto species: L. citreum, L. gelidum, L. lactis, L. mesenteroides, and L. pseudomesenteroides (15). Characterization studies of L. carnosum have been sparse and have been done with a limited number of strains (50, 58). Studies associated with L. carnosum have mainly focused on the production and purification of bacteriocins produced by this species (3, 20, 23, 26, 40, 41, 49, 52, 54).
L. carnosum seems to be strongly associated with ham products. In an earlier meat production plant contamination study (6), type A was found to dominate in the microflora of the raw-pork mass macerated overnight. In this plant, we noted that L. carnosum contamination occurred mainly in ham, whereas L. sake and L. curvatus have been detected in a variety of products (4, 6). Approximately 36% of the spoilage flora in Vienna sausages has been reported to consist of leuconostocs (17). When these Leuconostoc species were identified (18), the absence of L. carnosum was emphasized. In another characterization study of the LAB causing spoilage in vacuum-packaged, processed meats, a high prevalence of bacteriocin-producing psychrotropic leuconostocs was revealed (58). In that study, nine isolates were identified as L. carnosum; eight of these nine originated from different types of ham and one originated from sliced turkey. The strains forming the L. carnosum cluster (III) in the work describing this species (50) were from cold-stored, vacuum-packaged beef, pork, bacon, cooked ham, and luncheon meat. Compared with ham and other whole-meat products, emulsion sausages have more variable raw materials, such as different meat mixtures, pork skin emulsion, and spices, and undergo a different type of processing. The process and ingredients used for ham manufacturing may favor the survival and/or growth of L. carnosum. However, an adequate cooking process, considered to be the most important factor destroying LAB on products prior to packaging (1, 33, 34, 36), and the use of nitrite are similar in the production of emulsion sausages and whole-meat products.
PFGE characterization of L. carnosum confirmed the assumption that the raw-meat mass was the major source of contamination. The type of LAB contamination in a product has been considered to reflect the type of contamination in the processing facility (25, 38). Various LAB types were shown to contaminate the environment associated with the ham processing line studied here (4, 6). The greatest diversity in the different types of LAB was found in the environmental surface samples (4, 6). However, the majority of these LAB types have never been isolated from packaged products (4, 6). Only type A I-h isolates associated with the spoiled packages (V isolates), raw material (M5o and M6a isolates), and air of the macerating room (isolate I2b) and postcooking form removal area (isolate I27a) contaminated the products before they were transferred to the slicing-packaging department. The products were contaminated with a spoilage organism from the raw-meat mass before they entered the slicing line. In this case, the slicing line and the slicing room were not the main site and source of contamination, as is so often thought (25, 38). This route of contamination may be more common than is generally considered, also explaining the link between raw-meat mass and cooked ham.
Identification of species of the genus Leuconostoc is difficult (15, 55), which apparently is the main reason for the sparse population characterizations published. Leuconostoc spp. are phenotypically related to Weissella spp., heterofermentative lactobacilli, and pediococci and form a natural phylogenetic group with Weissella confusa, W. halotolerans, Weissella kandleri, Weissella minor, and W. viridescens (15). Due to the variable results obtained, sugar fermentation patterns are of little value in the species identification and could lead to misclassification (15). For presumptive identification, the scheme proposed by Villiani et al. (55) was found practical. However, in this scheme L. carnosum is supposed to form dextran. Only 15 of the 100 isolates tested here (11 of the established 25 PFGE types) formed slime from sucrose, lessening the value of this characteristic in L. carnosum identification.
It has been stated that reliable differentiation between L. carnosum and L. gelidum is impossible without DNA-DNA hybridization (15). Our results indicate that ribotyping can be used to distinguish L. carnosum from the other phenotypically related leuconostocs. However, care must be taken when enzymes are selected for species identification by ribotyping. Using HindIII-based ribopatterns, Villiani et al. (55) could not distinguish L. mesenteroides subsp. mesenteroides from L. mesenteroides subsp. dextranicum and L. lactis. We found HindIII and EcoRI to be the least distinguishing enzymes and ClaI to be the only enzyme generating a clear one-band shift in the patterns of these two subspecies (Fig. 1, lanes 5 and 7). ClaI may thus provide better results for the discrimination of L. mesenteroides subspecies. However, the HindIII pattern of the L. lactis type strain was clearly distinguished from the patterns of the L. mesenteroides subspecies (Fig. 3, lanes 5, 7, and 12). Despite its value in species identification and LAB contamination studies dealing with a diversity of species, ribotyping cannot be used for strain characterization when such a homogeneous population, such as the population of L. carnosum isolated from the meat production plant studied here, is assessed.
Only one type, A I-h (Fig. 4, lanes 8 and 17), from the largest lineage, was associated with the sensorially spoiled packages; however, even the production environment was not overwhelmingly contaminated by this specific organism. Strains of this type may possess characteristics that aid in growth niche occupation. Specific spoilage organisms have been considered to have better competitive ability, enabling them to prevail in the microflora present (8, 13, 32). Differences in the generation time, production of bacteriocins, strong ability to produce slime or volatile compounds causing sensorial spoilage, and better resistance to different stress factors, such as cold, heat, and disinfectants, are factors considered to be associated with specific spoilage organisms. For the L. carnosum population studied here, the production of bacteriocins was not found to be a common characteristic, as reported by Yang and Ray (58). The nine L. carnosum isolates studied by Yang and Ray (58) all inhibited L. mesenteroides. The true general impact of bacteriocin production in the development of spoilage flora is still not clear. Studies of bacteriocin production have mainly focused on the use of bacteriocins or cultures producing bacteriocins as biopreservatives. Biopreservatives are inoculated at a high initial concentration or a dense population in a freshly prepared product. This situation differs clearly from the situation in which some or one species in a contaminating flora gradually occupies a niche in a package and, finally, when reaching the stationary phase, spoils the product.
Molecular typing methods also provide valuable information for applied microbiology. They can contribute to knowledge of different bacterial populations associated with food processing and enable future research to be focused accurately on specific spoilage organisms and their specific characteristics. Such work will rely mainly on the reliable species identification and good strain characterization of specific spoilage organisms.
REFERENCES
- 1.Allen J R, Foster E M. Spoilage of vacuum-packed sliced processed meats during refrigerated storage. Food Res. 1960;25:19–25. [Google Scholar]
- 2.Alm F, Erichsen I, Molin N. The effect of vacuum packaging on some sliced processed meat products as judged by organoleptic and bacteriological analysis. Food Technol. 1961;15:199–203. [Google Scholar]
- 3.Becker B, Holzapfel W H, von Holy A. Effect of pH and the bacteriocin carnocin 54 on growth and cell morphology of two Leuconostoc strains. Lett Appl Microbiol. 1994;19:126–128. [Google Scholar]
- 4.Björkroth J, Korkeala H. Evaluation of Lactobacillus sake contamination in vacuum packaged sliced cooked meat products by ribotyping. J Food Prot. 1996;59:398–401. doi: 10.4315/0362-028X-59.4.398. [DOI] [PubMed] [Google Scholar]
- 5.Björkroth J, Korkeala H. rRNA gene restriction patterns as a characterization tool for Lactobacillus sake strains producing ropy slime. Int J Food Microbiol. 1996;30:293–302. doi: 10.1016/0168-1605(96)00955-5. [DOI] [PubMed] [Google Scholar]
- 6.Björkroth J, Korkeala H. Use of rRNA gene restriction patterns to evaluate lactic acid bacterium contamination of vacuum-packaged sliced cooked whole-meat products in a meat processing plant. Appl Environ Microbiol. 1997;63:448–453. doi: 10.1128/aem.63.2.448-453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Björkroth J, Korkeala H. Characterization of Lactobacillus fructivorans spoilage in ketchup. J Food Prot. 1997;60:505–509. doi: 10.4315/0362-028X-60.5.505. [DOI] [PubMed] [Google Scholar]
- 8.Björkroth J, Korkeala H. Ropy slime-producing Lactobacillus sake strains possess a strong competitive ability against a commercial biopreservate. Int J Food Microbiol. 1997;38:117–123. doi: 10.1016/s0168-1605(97)00097-4. [DOI] [PubMed] [Google Scholar]
- 9.Björkroth J, Ridell J, Korkeala H. Characterization of Lactobacillus sake strains associated with production of ropy slime by randomly amplified polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) patterns. Int J Food Microbiol. 1996;31:59–68. doi: 10.1016/0168-1605(96)00964-6. [DOI] [PubMed] [Google Scholar]
- 10.Blickstad E, Molin G. The microbial flora of smoked pork loin and frankfurter sausage stored in different gas atmospheres at 4°C. J Appl Bacteriol. 1983;54:45–56. doi: 10.1111/j.1365-2672.1983.tb01299.x. [DOI] [PubMed] [Google Scholar]
- 11.Blumberg H M, Kielbauch J A, Wachsmuth I K. Molecular epidemiology of Yersinia enterocolitica O:3 infections: use of chromosomal DNA restriction fragment length polymorphism of rRNA genes. J Clin Microbiol. 1991;29:2368–2374. doi: 10.1128/jcm.29.11.2368-2374.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borch E, Molin G. Numerical taxonomy of psychrotrophic lactic acid bacteria from prepacked meat and meat products. Antonie Leeuwenhoek. 1988;54:301–323. doi: 10.1007/BF00393522. [DOI] [PubMed] [Google Scholar]
- 13.Borch E, Kant-Muermans M-L, Blixt Y. Bacterial spoilage of meat and cured meat products. Int J Food Microbiol. 1996;33:103–120. doi: 10.1016/0168-1605(96)01135-x. [DOI] [PubMed] [Google Scholar]
- 14.Briggs M. The classification of lactobacilli by means of physiological tests. J Appl Bacteriol. 1953;54:45–56. doi: 10.1099/00221287-9-2-234. [DOI] [PubMed] [Google Scholar]
- 15.Dellaglio F, Dicks L M T, Torriani S. The genus Leuconostoc. In: Wood B J B, Holzapfel W, editors. The genera of lactic acid bacteria. Glasgow, United Kingdom: Blackie Academic and Professional; 1995. pp. 235–278. [Google Scholar]
- 16.Dykes G A, Britz T J, von Holy A. Numerical taxonomy and identification of lactic acid bacteria from spoiled, vacuum-packaged Vienna sausages. J Appl Bacteriol. 1994;76:246–252. doi: 10.1111/j.1365-2672.1994.tb01623.x. [DOI] [PubMed] [Google Scholar]
- 17.Dykes G A, Cloete T E, von Holy A. Quantification of microbiological populations associated with the manufacture of vacuum-packaged, smoked Vienna sausages. Int J Food Microbiol. 1991;13:239–248. doi: 10.1016/0168-1605(91)90081-y. [DOI] [PubMed] [Google Scholar]
- 18.Dykes G A, Cloete T E, von Holy A. Identification of Leuconostoc species associated with the spoilage of vacuum-packaged Vienna sausages by DNA-DNA hybridization. Food Microbiol. 1994;11:271–274. [Google Scholar]
- 19.Dykes G A, Cloete T E, von Holy A. Taxonomy of lactic acid bacteria associated with vacuum-packaged processed meat spoilage by multivariate analysis of cellular fatty acids. Int J Food Microbiol. 1995;28:89–100. doi: 10.1016/0168-1605(94)00161-x. [DOI] [PubMed] [Google Scholar]
- 20.Felix J V, Papathanasopoulos M A, Smith A A, von Holy A, Hastings J W. Characterization of leucocin B-Ta11a: a bacteriocin from Leuconostoc carnosum Ta11a isolated from meat. Curr Microbiol. 1994;29:207–212. doi: 10.1007/BF01570155. [DOI] [PubMed] [Google Scholar]
- 21.Grimont F, Grimont P A D. Ribosomal ribonucleic acid gene restriction as potential taxonomic tools. Ann Inst Pasteur/Microbiol. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 22.Harrigan W F, McCance M E. Laboratory methods in food and dairy microbiology. New York, N.Y: Academic Press, Inc.; 1976. [Google Scholar]
- 23.Hastings J W, Stiles M E, von Holy A. Bacteriocins of leuconostocs isolated from meat. A review paper. Int J Food Microbiol. 1994;24:75–81. doi: 10.1016/0168-1605(94)90107-4. [DOI] [PubMed] [Google Scholar]
- 24.Holzapfel W H, Gerber E S. Abstracts of the 32nd European Meeting of Meat Research Workers. 1986. Predominance of Lactobacillus curvatus and Lactobacillus sake in the spoilage association of vacuum-packaged meat products; p. 26. [Google Scholar]
- 25.Kempton A G, Bobier S R. Bacterial growth in refrigerated, vacuum-packed luncheon meats. Can J Microbiol. 1970;16:287–297. doi: 10.1139/m70-053. [DOI] [PubMed] [Google Scholar]
- 26.Keppler K, Geisen R, Holzapfel W H. An α-amylase sensitive bacteriocin of Leuconostoc carnosum. Food Microbiol. 1994;11:39–45. [Google Scholar]
- 27.Korkeala H, Mäkelä P. Characterization of lactic acid bacteria isolated from vacuum-packed cooked ring sausages. Int J Food Microbiol. 1989;9:33–43. doi: 10.1016/0168-1605(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 28.Korkeala H, Lindroth S. Differences in microbial growth in the surface layer and at the center of vacuum-packaged cooked ring sausage. Int J Food Microbiol. 1987;4:105–110. [Google Scholar]
- 29.Korkeala H, Alanko T, Mäkelä P, Lindroth S. Shelf-life of vacuum-packed cooked ring sausages at different chill temperatures. Int J Food Microbiol. 1989;9:237–247. doi: 10.1016/0168-1605(89)90093-7. [DOI] [PubMed] [Google Scholar]
- 30.Korkeala H, Suortti T, Mäkelä P. Ropy slime formation in vacuum-packed cooked meat products caused by homofermentative lactobacilli and a Leuconostoc species. Int J Food Microbiol. 1988;7:339–347. doi: 10.1016/0168-1605(88)90060-8. [DOI] [PubMed] [Google Scholar]
- 31.Korkeala H J, Björkroth K J. Microbiological spoilage and contamination of vacuum-packaged cooked sausages: a review. J Food Prot. 1997;60:724–731. doi: 10.4315/0362-028X-60.6.724. [DOI] [PubMed] [Google Scholar]
- 32.Leisner J, Greer G G, Stiles M E. Control of beef spoilage by a sulfide-producing Lactobacillus sake strain with bacteriocinogenic Leuconostoc gelidum UAL187 during anaerobic storage at 2°C. Appl Environ Microbiol. 1996;62:2610–2614. doi: 10.1128/aem.62.7.2610-2614.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mäkelä P, Korkeala H. Lactobacillus contamination of cooked ring sausages at sausage processing plants. Int J Food Microbiol. 1987;5:323–330. [Google Scholar]
- 34.Mäkelä P, Korkeala H, Laine J. Raw materials of cooked ring sausages as a source of spoilage lactic acid bacteria. J Food Prot. 1990;53:965–968. doi: 10.4315/0362-028X-53.11.965. [DOI] [PubMed] [Google Scholar]
- 35.Mäkelä P, Schillinger U, Korkeala H, Holzapfel W H. Classification of ropy slime producing lactic acid bacteria based on DNA-DNA homology, and identification of Lactobacillus sake and Leuconostoc amelibiosum as dominant spoilage organisms in meat products. Int J Food Microbiol. 1992;16:167–172. doi: 10.1016/0168-1605(92)90011-q. [DOI] [PubMed] [Google Scholar]
- 36.Mäkelä P M, Korkeala H J, Laine J J. Survival of ropy slime producing lactic acid bacteria in heat processes used in the meat industry. Meat Sci. 1992;31:463–471. doi: 10.1016/0309-1740(92)90028-3. [DOI] [PubMed] [Google Scholar]
- 37.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed-field electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and application. Washington, D.C: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 38.Mol J H H, Hietbring J E A, Mollen H W M, van Tinteren J. Observations on the microflora of vacuum packaged sliced cooked meat products. J Appl Bacteriol. 1971;34:377–397. doi: 10.1111/j.1365-2672.1971.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 39.Morishita Y, Shiromizu K. Characterization of lactobacilli isolated from meat and meat products. Int J Food Microbiol. 1986;3:19–29. [Google Scholar]
- 40.Papathanosopoulos M A, Hastings J W, von Holy A. Antibacterial activity of three Leuconostoc strains isolated from vacuum-packaged processed meats. J Basic Microbiol. 1994;34:173–182. doi: 10.1002/jobm.3620340307. [DOI] [PubMed] [Google Scholar]
- 41.Parente E, Moles M, Riccardi A. Leucocin F10, a bacteriocin from Leuconostoc carnosum. Int J Food Microbiol. 1996;33:231–234. doi: 10.1016/0168-1605(96)01159-2. [DOI] [PubMed] [Google Scholar]
- 42.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 43.Reuter G. Laktobazillen und eng verwandte Mikroorganismen in Fleisch und Fleischerzeugnissen. 2. Mitteilung: die Charakterisierung der isolierten Laktobazillenstämme. Fleischwirtschaft. 1970;50:954–962. [Google Scholar]
- 44.Reuter G. Untersuchungen zur Mikroflora von verpackten, aufgeschnittenen Brüh- und Kochwürsten. Arch Lebensmittelhyg. 1970;21:257–264. [Google Scholar]
- 45.Reuter G. Laktobazillen und eng verwandte Mikroorganismen in Fleisch und Fleischwaren. 4. Mitteilung: die Ökologie von Laktobazillen, Leuconostoc-Species und Pediokokken. Fleischwirtschaft. 1970;50:1397–1399. [Google Scholar]
- 46.Reuter G. Classification problems, ecology and some biochemical activities of lactobacilli of meat products. In: Can J C, Cutting C V, Whiting G C, editors. Lactic acid bacteria in beverages and food. London, England: Academic Press Ltd.; 1975. pp. 221–229. [Google Scholar]
- 47.Schillinger U, Lücke F-K. Lactic acid bacteria on vacuum-packaged meat and their influence on shelf life. Fleischwirtschaft. 1987;67:1244–1249. [Google Scholar]
- 48.Schillinger U, Lücke F-K. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55:1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schillinger U, Becker B, Holzapfel W H. Antilisterial activity of carnocin 54, a bacteriocin from Leuconostoc carnosum. Food Microbiol. 1995;12:31–37. [Google Scholar]
- 50.Shaw B G, Harding C D. Leuconostoc gelidum sp. nov. and Leuconostoc carnosum sp. nov. from chill-stored meats. Int J Syst Bacteriol. 1989;39:217–223. [Google Scholar]
- 51.Smittle R B, Cirigcliano M C. Salad dressings. In: Vanderzandt C, Splittstoesser D F, editors. Compendium of methods for the microbiological examination of foods. Washington, D.C: American Public Health Association; 1992. pp. 975–983. [Google Scholar]
- 52.Stiles M E. Bacteriocins produced by Leuconostoc species. J Dairy Sci. 1994;77:2718–2724. doi: 10.3168/jds.S0022-0302(94)77214-3. [DOI] [PubMed] [Google Scholar]
- 53.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Laack R L J M, Schillinger U, Holzapfel W H. Characterization and partial purification of a bacteriocin produced by Leuconostoc carnosum LA44A. Int J Food Microbiol. 1992;16:183–195. doi: 10.1016/0168-1605(92)90079-i. [DOI] [PubMed] [Google Scholar]
- 55.Villiani F, Moschetti G, Blaiotta G, Coppola S. Characterization of Leuconostoc mesenteroides by analysis of soluble whole-cell protein pattern, DNA fingerprinting and restriction of ribosomal DNA. J Appl Microbiol. 1997;82:578–588. doi: 10.1111/j.1365-2672.1997.tb03588.x. [DOI] [PubMed] [Google Scholar]
- 56.von Holy A, Cloete T E, Holzapfel W H. Quantification and characterization of microbial populations associated with spoiled, vacuum-packed Vienna sausages. Food Microbiol. 1991;8:95–104. doi: 10.1016/0168-1605(91)90081-y. [DOI] [PubMed] [Google Scholar]
- 57.Von Krush U, Lompe A. Schnellest zum qualitativen Nachweiss von l (+) and d (−) Milchsaure für die Bestimmung von Milchsaurebakterien. Milchwissenshaft. 1982;37:65–68. [Google Scholar]
- 58.Yang R, Ray B. Prevalence and biological control of bacteriocin-producing psychrotrophic leuconostocs associated with spoilage of vacuum-packaged processed meats. J Food Prot. 1994;57:209–217. doi: 10.4315/0362-028X-57.3.209. [DOI] [PubMed] [Google Scholar]