Abstract
l-Methionine γ-lyase (EC 4.4.1.11) was purified to homogeneity from Brevibacterium linens BL2, a coryneform bacterium which has been used successfully as an adjunct bacterium to improve the flavor of Cheddar cheese. The enzyme catalyzes the α,γ elimination of methionine to produce methanethiol, α-ketobutyrate, and ammonia. It is a pyridoxal phosphate-dependent enzyme, with a native molecular mass of approximately 170 kDa, consisting of four identical subunits of 43 kDa each. The purified enzyme had optimum activity at pH 7.5 and was stable at pHs ranging from 6.0 to 8.0 for 24 h. The pure enzyme had its highest activity at 25°C but was active between 5 and 50°C. Activity was inhibited by carbonyl reagents, completely inactivated by dl-propargylglycine, and unaffected by metal-chelating agents. The pure enzyme had catalytic properties similar to those of l-methionine γ-lyase from Pseudomonas putida. Its Km for the catalysis of methionine was 6.12 mM, and its maximum rate of catalysis was 7.0 μmol min−1 mg−1. The enzyme was active under salt and pH conditions found in ripening Cheddar cheese but susceptible to degradation by intracellular proteases.
Methanethiol is associated with desirable Cheddar-type sulfur notes in good-quality Cheddar cheese (2, 27). The mechanism for the production of methanethiol in cheese is unknown, but it is linked to the catabolism of methionine (1, 15). l-Methionine γ-lyase (EC 4.4.1.11; MGL), also known as methionase, l-methionine γ-demethiolase, and l-methionine methanethiollyase (deaminating), is a pyridoxal phosphate (PLP)-dependent enzyme that catalyzes the direct conversion of l-methionine to α-ketobutyrate, methanethiol, and ammonia by an α,γ-elimination reaction (26). It does not catalyze the conversion of d enantiomers (24–26). MGL in Pseudomonas putida is a multifunctional enzyme system since it catalyzes the α,γ- and α,β-elimination reactions of methionine and its derivatives (24). In addition, the enzyme also catalyzes the β-replacement reactions of sulfur amino acids (24). Since its discovery in Escherichia coli and Proteus vulgaris by Onitake (19), this enzyme has been found in various bacteria and is regarded as a key enzyme in the bacterial metabolism of methionine. However, this enzyme has not been purified to homogeneity from any food-grade microorganisms.
MGL is widely distributed in bacteria, especially in pseudomonads, and is induced by the addition of l-methionine to the culture medium (9, 28). The enzyme has been purified from Pseudomonas putida (25), Aeromonas sp. (26), Clostridium sporogenes (11), and Trichomonas vaginalis (16) and partially purified from and characterized for Brevibacterium linens NCDO 739 (4).
B. linens is a nonmotile, non-spore-forming, non-acid-fast, gram-positive coryneform bacterium normally found on the surfaces of Limburger and other Trappist-type cheeses. This organism tolerates salt concentrations ranging between 8 and 20% and is capable of growing in a broad pH range from 5.5 to 9.5, with an optimum pH of 7.0 (20). In Trappist-type cheeses, brevibacteria depend on Saccharomyces cerevisiae to metabolize lactate, which increases the pH of the curd, as well as to produce growth factors that are important for their growth (20). Interest in B. linens has focused around its ability to produce an extracellular protease, which has recently been isolated (21), and its ability to produce high levels of methanethiol (3, 9, 10, 22).
B. linens produces various sulfur compounds, including methanethiol, that are thought to be important in Cheddar-like flavor and aroma (3, 9, 10, 22). Ferchichi et al. (9) suggested that MGL is responsible for the methanethiol-producing capability of B. linens but did not provide definitive evidence. Weimer et al. (28) proposed that B. linens BL2 is responsible for Cheddar-type flavor development in low-fat cheese, but again conclusive evidence was lacking. In this study, MGL was purified to homogeneity from B. linens BL2 and its physical and chemical properties were examined.
MATERIALS AND METHODS
Chemicals.
l-Ethionine, l-methionine sulfone, l-methionine sulfoxide, l-cysteine, S-methyl-l-cysteine, O-actetyl-l-serine, dl-selenomethionine, dl-selenocysteine, S-adenosylhomocysteine, and S-adenosylmethionine were purchased from Fluka (Ronkonkoma, N.Y.). PLP was obtained from Boehringer Mannheim GmbH (Mannheim, Germany); all other chemicals were obtained from Sigma Chemical Co. (St. Louis, Mo.). Size exclusion standards for the calibration of the gel filtration column were obtained from Pharmacia Biotechnology (Uppsala, Sweden). Unless otherwise mentioned, all reagents used in this study were of analytical grade.
Bacterial strain and growth conditions.
B. linens BL2, obtained from the Utah State University culture collection, was frozen (−70°C) in Trypticase soy broth (TSB) containing 30% glycerol and stored at −70°C until further use. Before each use, a frozen stock culture was thawed and grown in 5 ml of TSB at 25°C with aeration (250 rpm) for two transfers prior to inoculation for further studies.
Enzyme assays.
Amounts of free thiol groups were determined by the method of Laakso and Nurmikko (12). The assay mixture contained 50 mM potassium phosphate (KP; pH 7.2), 10 mM l-methionine, 0.02 mM PLP, 0.25 mM 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB), and the enzyme in cell extracts (CEs) or in pure form in a final volume of 1.0 ml. The reaction mixture was incubated quiescently at 25°C for 1 h and observed at 412 nm in a double-beam model UV2100U spectrophotometer (Shimadzu Scientific Instruments, Inc., Pleasanton, Calif.). The concentration of thiols produced was determined from a standard curve obtained with solutions of known concentrations of ethanethiol.
α-Ketobutyrate produced by the α,γ elimination of methionine was measured by derivatizing the reaction mix with 3-methyl-2-benzothiazolone hydrazone (23). The assay mixture (18) contained 50 mM KP (pH 7.2), 10 mM l-methionine, 0.02 mM PLP, and >0.015 U of the enzyme in a final volume of 0.5 ml. The reaction mixture was incubated at 25°C for 1 h, and the reaction was stopped by the addition of trichloroacetic acid to a final concentration of 5%. After centrifugation at 16,000 × g for 2 min, the α-ketobutyrate formed in the supernatant solution was determined with 3-methyl-2-benzothiazolone hydrazone (23).
Isolation and purification of MGL.
Purification steps were carried out at 0 to 5°C unless otherwise mentioned. B. linens BL2 was grown in TSB with 0.1% l-methionine at 25°C for 36 h with shaking at 250 rpm. Cells from 10 liters, grown in 10 2-liter baffle flasks, were harvested by centrifugation (6,000 × g for 10 min at 4°C) and washed twice with 50 mM KP (pH 7.2). Washed cells were resuspended in 50 mM KP (pH 7.2) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM EDTA, 0.02 mM PLP, 2% ethanol, and 1 mg of lysozyme per ml and incubated at 37°C for 1 h to lyse the cells. DNase I (0.25 μg/ml; Sigma Chemical Co.) was then added to the cell lysate, stirred at room temperature for 1 h, and centrifuged (10,000 × g for 1 h at 4°C), and the supernatant was decanted and treated as the CE.
The CE was fractionated with ammonium sulfate (AS). Crystalline AS was added gradually over a period of 15 min to a beaker containing the CE in an ice bath. Proteins precipitated by 55% saturation with AS and by 65% saturation with AS were collected by centrifugation (10,000 × g for 15 min at 4°C) and dissolved in 50 mM KP buffer (pH 7.2) containing 1 mM PMSF, 1 mM EDTA, 0.02 mM PLP, and 2% ethanol. This was then desalted with a 30-kDa-Ultrafree centrifugal ultrafiltration device (Millipore Corporation, Bedford, Mass.) by centrifugation at 4,000 × g at 4°C.
The retentate with MGL activity was filtered though a 0.22-μm-pore-size low-level-protein binding filter (Millipore Corporation) before being loaded on an anion-exchange column (DEAE-Fractogel [650 M]; EM Separations, Gibbstown, N.J.) equilibrated with 20 mM sodium phosphate (NaP; pH 7.2). The column was washed with the same buffer containing 0.15 M NaCl until the absorbance at 280 nm of the eluate decreased to less than 0.05 absorbance units. The enzyme was then eluted with the same buffer containing 0.41 M NaCl.
The fraction containing the enzyme was desalted with a 30-kDa-Ultrafree centrifugal filter unit (4,000 × g for 1 h at 4°C; Millipore Corporation). Solid AS was added to the concentrated fraction to a final concentration of 1.5 M. The sample was centrifuged (10,000 × g for 30 min at 4°C) and then filtered with a 0.22-μm-pore-size low-level-protein binding filter; Gelman Sciences, Ann Arbor, Mich.) to remove all precipitated proteins before being injected on a Porous PH hydrophobic-interaction chromatography column (PerSeptive Biosystems, Framingham, Mass.). The column was equilibrated with 1.5 M AS dissolved in 20 mM NaP (pH 7.2). Proteins were eluted at 5 ml/min by linearly decreasing the AS concentration from 100 to 0%. Absorbance was monitored at 280 and 480 nm with a diode array detector (Beckman Instruments, Fullerton, Calif.) connected to a high-performance liquid chromatograph (System Gold; Beckman Instruments).
Fractions were desalted with 30-kDa-Ultrafree centrifugal filter units (Millipore Corporation), after which the fraction containing activity was applied to a Mono Q H 5/5 anion-exchange column (Pharmacia Biotechnology) equilibrated with 20 mM NaP buffer (pH 8.0). Proteins were eluted at 1 ml/min with a linear gradient from 0.3 to 0.4 M NaCl.
The peak with MGL activity was then injected on a Superose 12 HR 10/30 (Pharmacia Biotechnology) gel filtration column equilibrated with 0.02 M KP buffer (pH 7.2). The protein was eluted with the same buffer, and the peak with activity was concentrated with a 30-kDa-Ultrafree centrifugal filter unit. The enzyme was either assayed immediately or stored at −70°C in 0.1 M KP buffer (pH 7.2).
Protein assay.
The total protein content in each CE was determined by the bicinchoninic acid assay (Pierce Chemical Co., Rockford, Ill.) according to the manufacturer’s instructions. Bovine serum albumin was used to obtain a standard curve.
PAGE.
Discontinuous denaturing polyacrylamide gel electrophoresis (PAGE) was performed with a 12% resolving and a 4% stacking gel as described by Laemmli (13) in a Protean II Xi Cell (Bio-Rad Laboratories, Hercules, Calif.) at 30 mA for 6 h. The proteins were visualized by silver staining (Phastgel silver kit; Pharmacia Biotechnology). Broad-range molecular mass marker proteins (Bio-Rad Laboratories) were used as references.
Estimation of molecular mass of the enzyme.
The molecular mass of the purified enzyme was estimated by size exclusion chromatography with a Superose 12 HR 10/30 gel filtration column (Pharmacia Biotechnology) equilibrated with 20 mM KP buffer (pH 7.2) and calibrated with ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (68 kDa), ovalbumin (45 kDa), β-lactoglobulin (36 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa).
Substrate specificity of the purified enzyme.
The ability of the purified enzyme to catalyze elimination reactions against amino acids and substituted amino acids was tested separately with l-methionine, d-methionine, l-ethionine, l-methionine sulfone, l-methionine sulfoxide, α-keto γ-methyl thiobutyrate (KMTB), dl-homocysteine, l-cystathionine, l-cysteine, d-cysteine, l-cystine, S-methyl-l-cysteine, O-actetyl-l-serine, dl-selenomethionine, dl-selenocysteine, S-adenosylhomocysteine, S-adenosylmethionine, dl-homocysteic acid, and l-djenkolic acid. Activity was measured by incubating the enzyme in 0.05 M KP buffer (pH 7.5) containing 0.02 mM PLP at 25°C for 1 h with 10 mM each substrate. We measured α-keto acids produced from each substrate, with the exception of KMTB, for which thiol production was determined.
Influence of temperature and pH.
Optimum temperature for 1-h assays was determined by assaying activity over temperatures ranging from 4 to 50°C in 0.05 M KP buffer (pH 7.5), with each buffer being made at each of the tested temperatures. Temperature stability of the enzyme was determined by incubating the enzyme in 0.05 M KP buffer (pH 7.5) for up to 1 h at temperatures ranging from 4 to 50°C. Aliquots were removed at various times, and residual activity was measured at 25°C in 0.05 M KP buffer (pH 7.5) by determining the amount of α-ketobutyrate produced with 10 mM Met as the substrate. Thermal inactivation parameters were obtained from a plot of ln k versus 1/T, where k is the rate constant for denaturation of the enzyme and T is the absolute temperature. The slope of the line represents Ea/R, where Ea is the energy of activation for the denaturation reaction and R is the universal gas constant expressed in joules per mole per degree.
The stability and optimum pH of the enzyme were determined at 25°C with 50 mM potassium citrate (pH 4.0 to 6.5), 50 mM KP (pH 6.5 to 8.0), and 50 mM potassium-glycine-NaOH (pH 8.0 to 10.0) buffers. The pH stability of the enzyme was determined by incubating the enzyme at each pH with 0.02 mM PLP for 24 h at 4°C. Residual activity was measured by incubating the enzyme in 0.05 M KP buffer (pH 7.5) at 25°C for 1 h and by determining the amount of α-ketobutyrate produced with 10 mM Met as the substrate.
Influence of inhibitors.
Compounds tested for their inhibitory effects included dl-propargylglycine, hydroxylamine, l-penicillamine, N-ethylmaleimide, iodoacetate, EDTA, Tris, and glycine. The enzyme was incubated with 1 and 10 mM concentrations of the compound for 10 min before addition of substrate (10 mM l-Met). The amount of α-ketobutyrate produced after incubation at 25°C for 1 h was then determined.
Kinetic studies.
Enzyme kinetics for the α,γ-elimination reaction was determined with methionine as the substrate and by measuring the amount of methanethiol produced with DTNB (12). The enzyme was incubated with 0.05 M KP (pH 7.5), 0.02 mM PLP, 0.28 mM DTNB, and 0.1 to 40 mM Met. The reaction was started by the addition of substrate, and product formation was monitored continuously at 412 nm with a model UV2100U double-beam spectrophotometer (Shimadzu Scientific Instruments, Inc.). The Kms and maximum rates of metabolism for the enzyme reactions were estimated from Eadie-Hofstee plots.
Absorption spectrum of the enzyme.
The absorption spectrum of the enzyme was measured with a diode array detector attached to a high-performance liquid chromatography unit (Beckman Instruments) by gel filtration chromatography on a Superose 12 HR 10/30 column during the final purification stage of the enzyme.
RESULTS
Purification of MGL.
Purification of MGL to homogeneity was accomplished in five successive steps (Table 1). Eighty percent of the enzyme activity precipitated between 55 and 65% AS. In the second step, the enzyme eluted between 0.21 and 0.41 M NaCl during anion-exchange chromatography on DEAE-Fractogel. In the third step, we used hydrophobic-interaction chromatography on a column and the enzyme was eluted with 0.75 M AS. During the fourth step, done by anion-exchange chromatography with a Mono Q Porous HP column, the enzyme eluted with 0.37 M NaCl. The enzyme was finally separated from other proteins by gel filtration chromatography on Superose 12. This scheme resulted in a homogeneous enzyme (Fig. 1), which was purified 82-fold, with an activity yield of 0.2%.
TABLE 1.
Purification of MGL from B. linens BL2
| Purification step | Total protein (mg) | Total activity (Ua) | Sp act (U/mg) | Purification factor | Yield (%) |
|---|---|---|---|---|---|
| CE | 495 | 1,389 | 2.8 | 1.0 | 100 |
| AS fractionation | 182 | 1,100 | 6.0 | 2.2 | 79.2 |
| DEAE-Fractogel anion-exchange chromatography | 75 | 522 | 6.8 | 2.4 | 37.6 |
| Porous PH hydrophobic-interaction chromatography | 1.2 | 86 | 71.5 | 25.5 | 6.2 |
| Mono Q anion-exchange chromatography | 0.08 | 14 | 177.3 | 63.3 | 1.0 |
| Superose 12 gel filtration | 0.01 | 2 | 228.9 | 81.7 | 0.2 |
One unit is defined as 1 nmol of thiol generated per min.
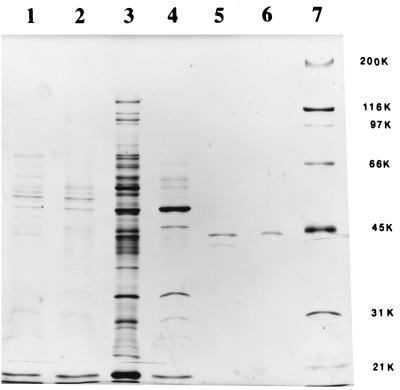
FIG. 1.
Sodium dodecyl sulfate-PAGE analysis of different stages during the purification of MGL from B. linens BL2 of fractions containing enzyme activity. Lanes: 1, CE; 2, 55 to 65% ammonium sulfate fraction; 3, DEAE-Fractogel column; 4, hydrophobic-interaction column; 5, Mono Q; 6, Superose 12; 7, molecular mass marker proteins. Approximate molecular masses in kilodaltons (K) are indicated on the right.
Enzyme size and absorbance characteristics.
The molecular mass of the native enzyme was estimated to be approximately 170 kDa and was determined during the final stage of purification with a Superose 12 gel filtration column. When the gel was electrophoresed under denaturing conditions by sodium dodecyl sulfate-PAGE (Fig. 1), a single band with an approximate molecular mass of 43 kDa was observed. Analysis of the absorption spectrum of the purified enzyme demonstrated a peak at 420 nm in addition to a peak at 280 nm.
Substrate specificities.
The activities of the purified enzyme on various substrates were determined by the production of α-keto acids. The production of thiols from KMTB was monitored. The purified enzyme was capable of catalyzing the α,γ elimination of a number of substrates. dl-Ethionine and dl-homocysteine produced more activity than with methionine (Table 2). Addition of oxygen atoms to the sulfur atom of methionine, as was observed by using l-methionine sulfone and l-methionine sulfoxide as substrates, resulted in over 50% decrease in activity. The enzyme activities against S-adenosylmethionine and S-adenosylhomocysteine were less than 7% of that compared to methionine. When the sulfur atom of methionine was replaced with a selenium atom, the enzyme was still able to catalyze the elimination reaction. l-Cysteine was degraded to 2% of the level of activity on methionine; however, addition of a methyl group to the sulfur atom of cysteine made it much more susceptible to degradation. The d enantiomers of methionine and cysteine did not serve as substrates for the enzyme.
TABLE 2.
Substrate specificities of purified MGL from B. linens BL2
| Substrate | Relative activity (%)a |
|---|---|
| dl-Homocysteine | 134 ± 22 |
| dl-Ethionine | 111 ± 8 |
| l-Methionine | 100 |
| dl-Selenomethionine | 80 ± 8 |
| l-Methionine sulfone | 45 ± 1 |
| l-Methionine sulfoxide | 42 ± 4 |
| S-Methyl l-cysteine | 36 ± 5 |
| O-Acetyl l-serine | 18 |
| S-Adenosylmethionine | 7 ± 1 |
| l-Cysteine | 2 ± 1 |
| S-Adenosylhomocysteine | 1 ± 1 |
| d-Methionine | 0 |
| dl-Methionine dl-sulfoximine | 0 |
| l-Cystathionine | 0 |
| O-Succinyl l-homoserine | 0 |
| l-Cystine | 0 |
| KMTB | 0b |
| l-Djenkolic acid | 0 |
Determined by α-keto acid production. The values are means of three determinations ± standard deviations.
Determined by thiol production.
Kinetic parameters.
The Km for the catalysis of methionine as determined from the rates of methanethiol and α-ketobutyrate production was found to be 6.12 mM, and the maximum rate of metabolism as determined from Eadie-Hofstee plots was found to be 7.0 μmol min−1 mg−1.
Influence of temperature and pH.
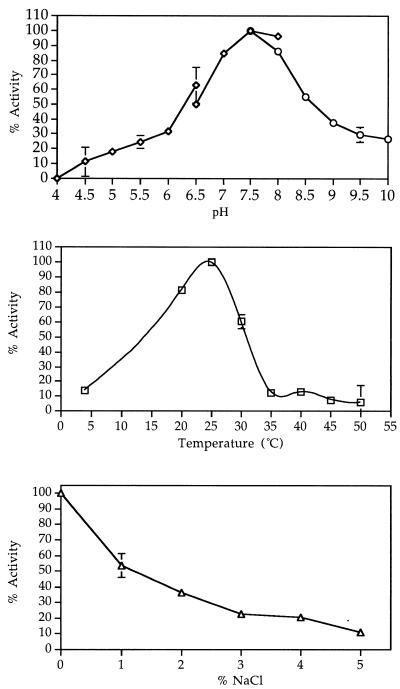
The pH optimum for the α,γ elimination of methionine was 7.5 to 8.0 (Fig. 2A). At pH 5.5 the enzyme retained over 20% of its activity, at pH 4.5 its activity decreased to 10%, and at pH 4.0 it became inactivated. At pH 7.5 the enzyme had highest activity at 25°C (Fig. 2B).
FIG. 2.
Effect of pH (A) (in K-citrate [diamonds], KP [triangles], and K-Gly-NaOH [circles]), temperature (B), and NaCl (C) on the activity of MGL from B. linens BL2.
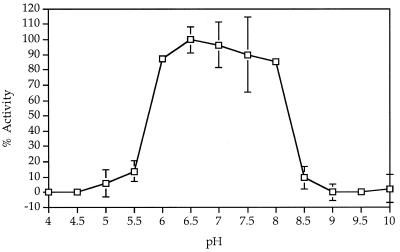
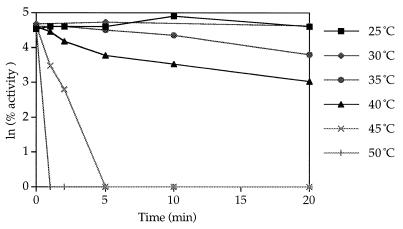
The enzyme was stable at pHs ranging from 6.0 to 8.0 for 24 h (Fig. 3). Partially purified as well as pure enzyme could be stored on ice at 4°C in 0.05 M KP (pH 7.5) with 0.02 mM PLP without significant loss of activity for over 2 weeks. Freezing and thawing the enzyme solution resulted in a loss of over 60% of the enzyme activity, and the enzyme was labile at temperatures greater than 30°C (Fig. 4). The denaturation reaction demonstrated first-order kinetics and had a standard free energy of activation of 186 kJ mol−1.
FIG. 3.
Effect of pH on the stability of MGL from B. linens BL2.
FIG. 4.
Effect of temperature on the stability of MGL from B. linens BL2.
Enzyme inhibitors.
The enzyme was completely inhibited by 1 mM concentrations of carbonyl reagents, such as hydroxylamine, and by a 10 mM concentration of dl-penicillamine. dl-Propargylglycine completely inhibited enzyme activity at 1 mM (Table 3). Thiol-reducing agents, such as iodoacetate, did not decrease activity at 1 mM but did so at 10 mM. Metal-chelating agents did not influence enzyme activity. Enzyme activity decreased with increasing concentrations of NaCl (Fig. 2C). At a 5% NaCl concentration the enzyme retained 10% of its activity.
TABLE 3.
Effects of inhibitors on MGL activity from B. linens BL2
| Inhibitor | Relative activity (%)a at inhibitor concn (mM):
|
|
|---|---|---|
| 1 | 10 | |
| Hydroxylamine | 0 | 0 |
| dl-Penicillamine | 91 | 0 |
| dl-Propargylglycine | 0 | 0 |
| Iodoacetate | 97 | 79 |
| EDTA | 99 | 106 |
| Glycine | 99 | 79 |
| Tris | 89 | 78 |
Determined by α-keto acid production. Enzyme activity on 10 mM Met with no inhibitor added was considered to be 100% activity.
DISCUSSION
In this study MGL from B. linens BL2 was purified to homogeneity and subsequently characterized. MGL converts methionine to methanethiol, a compound associated with desirable Cheddar cheese flavor (2, 27). Although Collin and Law (4) reported the partial purification of MGL from B. linens, this is the first report of the purification to homogeneity of MGL from a food-grade microorganism. Purification was completed in five steps, with a 0.2% yield. The purification scheme of Nakayama et al. (18) was initially used in an attempt to purify the enzyme; however, different elution patterns were observed, suggesting a difference in the physical properties of the enzymes from the two species. Failure to include PMSF and EDTA in the cell lysis buffers resulted in a rapid loss of enzyme activity due to proteolytic degradation. By anion-exchange chromatography on DEAE-Fractogel, a 41% reduction in the yield of the enzyme was observed without a significant increase in specific activity; however, this step was found to be essential to remove lipids from the sample and prevent subsequent columns in the purification procedure from being poisoned.
The total molecular mass of the purified enzyme was 170 kDa, with four subunits of 43 kDa (Fig. 2). In this respect, the enzyme is similar to other purified MGLs (16, 25); however, MGL from C. sporogenes has been reported to contain two sets of nonidentical subunits (11).
The Km for the catalysis of methionine, as determined from the rates of methanethiol and α-ketobutyrate production, was found to be 6.12 mM. The Km of the brevibacterial enzyme is approximately six times higher than that observed for the pseudomonad enzyme. Ferchichi et al. (8) determined the approximate Km in CE of B. linens CNRZ 918 for l-methionine to range from 14 to 46 mM depending on the growth phase of the cells. However, since a CE is a complex environment containing many enzymes, it is difficult to make any comparisons. Since methionine is required by this strain (5) for growth, this Km value for a methionine-degrading enzyme is reasonable.
The purified MGL was capable of the catalysis of l amino acids (Table 2). In addition to sulfur and selenium-containing amino acids, the enzyme had catalytic activity with O-acetyl l-serine. Cystathionine and KMTB, however, were not substrates. These results indicate that the enzyme is capable of acting on C—S, C—Se, and C—O bonds but not on C—C bonds. Replacement of the thioether group of methionine with a sulfoxide or sulfone group decreased degradation. Unlike the enzyme purified from P. putida (6), the enzyme purified from B. linens BL2 did not degrade O-succinyl l-homoserine.
The brevibacterial enzyme is similar to the pseudomonad enzyme (18) in its substrate specificity, but it is distinctly different from MGL purified from C. sporogenes (11) in its inability to catalyze l-cystine and its lower activity against l-cysteine. It differs from the enzyme purified from Aeromonas sp. in that it has no activity toward cystathionine (7). The enzyme’s catalytic properties also differ from the T. vaginalis enzyme (16) in that it has much lower relative activities towards homocysteine, cysteine, and O-acetyl serine. Despite these differences, all reports note higher relative activity for homocysteine than that against methionine. It is interesting to note the high activity of this enzyme on homocysteine, a precursor of methionine in the methionine-biosynthetic pathway, suggesting that this may be a substrate-level regulatory function.
The purified enzyme had an absorbance spectrum typical of other PLP-containing enzymes, with a characteristic absorption maximum at 420 nm due to the azomethine linkage between the formyl group of PLP and an amino group of the enzyme (17). Additionally, carbonyl reagents, known inhibitors of PLP-containing enzymes, strongly inhibited this enzyme (Table 3). These data indicate that, like other MGLs purified so far, the MGL from B. linens BL2 is also a PLP-dependent enzyme. Furthermore, metal ion chelators did not affect enzyme activity, indicating that the enzyme is not a metalloenzyme.
The enzyme had highest activity at 25°C. However, this enzyme was not as temperature stable as the partially purified enzyme from B. linens NCDO 739 (4). Other proteins in the partially purified preparation may contribute to the temperature stability of the B. linens NCDO 739 enzyme. Similar pH stability and pH optimum curves were obtained as compared to those of the partially purified enzyme from B. linens NCDO 739. However, a notable difference is that the enzyme from B. linens BL2 had 18% residual activity at pH 5.0 compared to only 2% for the enzyme from B. linens NCDO 739. This result indicates that differences in the properties of the enzymes produced by different strains within the same genera may exist. Alternatively, impurities present in the partially purified enzyme preparation may affect activity.
The enzyme was active at the pH (Fig. 2A), temperature (Fig. 2B), and salt concentration (Fig. 2C) conditions existing in aging Cheddar cheese, suggesting that this enzyme may be active in the cheese matrix during ripening. However, residual activity in this environment should not exceed 0.6% of the activity under optimum conditions. Additionally, considering the susceptibility of this enzyme to proteolysis, it is likely that activity would rapidly decrease with cell lysis, supporting this hypothesis. Weimer et al. (28) tested CEs for the capacity to produce methanethiol under cheese-ripening conditions (pH 5.2, 4% NaCl) but detected no activity. This outcome may be explained by the fact that other enzymes, like proteases in the CEs, may have inactivated the enzyme. Additionally, evidence to support this theory is the rapid loss of the capacity to produce methanethiol observed for CEs stored in the absence of protease inhibitors (data not shown). Alternatively, methanethiol produced may be converted to other compounds, like S-methyl thioesters, by enzymes in the CE (14) which might be active under cheese-ripening conditions.
An important consideration when evaluating the role of this enzyme in the development of cheese flavor is whether the cells lyse. If the cells are alive and metabolically active, as was suggested by Weimer et al. (28), then methionine can be transported into the cells, where it will be converted into methanethiol. Subsequently, methanethiol may either be converted to other compounds or diffuse into the cheese matrix. If the cells lyse or the membranes are permeablized, transport of methionine into the cell will rely on diffusion and the enzyme will be exposed to many proteolytic enzymes in the cheese matrix. Consequently, the ability of this enzyme to impact cheese may be linked to the ability of the cell to resist lysis, persist, and remain metabolically active during cheese ripening.
ACKNOWLEDGMENT
This research was supported by the Utah Agricultural Experiment Station.
Footnotes
This is contribution 7006 from the Utah Agricultural Experiment Station.
REFERENCES
- 1.Alting A C, Engels W J, Schalkwijk S, Exterkate F A. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl Environ Microbiol. 1995;61:4037–4042. doi: 10.1128/aem.61.11.4037-4042.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aston J W, Dulley J R. Cheddar cheese flavor. Aust J Dairy Technol. 1982;37:59–64. [Google Scholar]
- 3.Boyaval P, Desmazeaud M J. Le point des connaissances sur Brevibacterium linens. Lait. 1983;63:187–216. [Google Scholar]
- 4.Collin J C, Law B A. Isolation and characterization of the l-methionine-γ-demethiolase from Brevibacterium linens NCDO 739. Sci Aliments. 1989;9:805–812. [Google Scholar]
- 5.Dias B, Weimer B. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl Environ Microbiol. 1998;64:3320–3326. doi: 10.1128/aem.64.9.3320-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esaki N, Nakayama T, Sawada S, Tanaka H, Soda K. Reactions of O-substituted l-homoserine catalysed by l-methionine γ-lyase and their mechanisms. Agric Biol Chem. 1984;48:1991–1996. [Google Scholar]
- 7.Esaki N, Soda K. l-Methionine γ-lyase from Pseudomonas putida and Aeromonas. Methods Enzymol. 1987;143:459–465. doi: 10.1016/0076-6879(87)43081-4. [DOI] [PubMed] [Google Scholar]
- 8.Ferchichi M, Hemme D, Nardi M, Pamboukdjian N. Production of methanethiol from methionine by Brevibacterium linens CNRZ 918. J Gen Microbiol. 1985;131:715–723. doi: 10.1099/00221287-131-4-715. [DOI] [PubMed] [Google Scholar]
- 9.Ferchichi M, Hemme D, Nardi M. Induction of methanethiol production by Brevibacterium linens CNRZ 918. J Gen Microbiol. 1986;132:3075–3082. doi: 10.1099/00221287-131-4-715. [DOI] [PubMed] [Google Scholar]
- 10.Hemme D, Bouillanne C, Metro F, Desmazeard M J. Microbial catabolism of amino acids during cheese ripening. Sci Aliments. 1982;2:113–123. [Google Scholar]
- 11.Kreis W, Hession C. Isolation and purification of l-methionine-α-deamino-γ-mercaptomethane-lyase (l-methionase) from Clostridium sporogenes. Cancer Res. 1973;33:1862–1865. [PubMed] [Google Scholar]
- 12.Laasko S, Nurmikko V. A spectrophotometric assay for demethiolating activity. Anal Biochem. 1976;72:600–605. doi: 10.1016/0003-2697(76)90572-8. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lamberet G, Auberger B, Bergere J L. Aptitude of cheese bacteria for volatile S-methyl thioester synthesis 1. Effect of substrates and pH on their formation by Brevibacterium linens GC171. Appl Microbiol Biotechnol. 1997;47:279–283. [Google Scholar]
- 15.Lindsay C R, Rippe J K. Enzymic generation of methanethiol to assist in the flavor development of Cheddar cheese and other foods. In: Parliment T H, Croteau R, editors. Biogeneration of aromas. Washington, D.C: American Chemical Society; 1986. pp. 286–308. [Google Scholar]
- 16.Lockwood B C, Coombs G H. Purification and characterization of methionine γ-lyase from Trichomonas vaginalis. Biochem J. 1991;279:675–682. doi: 10.1042/bj2790675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Carrion M. Vitamin B6, pyridoxal phosphate: chemical, biochemical and medical aspects. In: Dolphin D, Poulson R, Avramovic O, editors. Coenzymes and cofactors. Vol. 1. 1986. pp. 1–22. , part B. John Wiley and Sons, New York, N.Y. [Google Scholar]
- 18.Nakayama T, Esaki N, Lee W J. Purification and properties of l-methionine γ-lyase from Aeromonas sp. Agric Biol Chem. 1984;48:2367–2369. [Google Scholar]
- 19.Onitake J. On the formation of methylmercaptan from l-cystine and l-methionine by bacteria. J Osaka Med Assoc. 1938;37:263–270. [Google Scholar]
- 20.Purko N O, Nelson N O, Wood W A. The associative action between certain yeasts and Bacterium linens. J Dairy Sci. 1951;34:699–705. [Google Scholar]
- 21.Rattray F P, Bockelmann W, Fox P F. Purification and characterization of an extracellular proteinase from Brevibacterium linens ATCC 9174. Appl Environ Microbiol. 1995;61:3454–3456. doi: 10.1128/aem.61.9.3454-3456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharpe M E, Law B A, Phillips B A, Pitcher D G. Methanethiol production by coryneform bacteria: strains from dairy and human skin sources and Brevibacterium linens. J Gen Microbiol. 1977;101:345–349. doi: 10.1099/00221287-101-2-345. [DOI] [PubMed] [Google Scholar]
- 23.Soda K. A spectrophotometric microdetermination of keto acids with 3-methyl 2-benzothiazolone hydrazone. Agric Biol Chem. 1967;31:1054–1060. [Google Scholar]
- 24.Soda K, Tanaka H, Esaki N. Multifunctional biocatalysis: methionine γ-lyase. Trends Biochem Sci. 1983;8:214–217. [Google Scholar]
- 25.Tanaka H, Esaki N, Tammamoto T, Soda K. Purification and properties of methionine γ-lyase from Pseudomonas ovalis. FEBS Lett. 1976;66:307–311. doi: 10.1016/0014-5793(76)80528-5. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka H, Esaki N, Soda K. A versatile bacterial enzyme: methionine γ-lyase. Enzyme Microb Technol. 1985;7:530–537. [Google Scholar]
- 27.Urbach G. Contribution of lactic acid bacteria to flavour compound formation in dairy products. Int Dairy J. 1995;5:877–903. [Google Scholar]
- 28.Weimer B, Dias B, Madhavi U, Broadbent J, Brennand C, Jaegi J, Johnson M, Milani F, Steele J, Sisson D V. Influence of NaCl and pH on intracellular enzymes that influence Cheddar cheese ripening. Lait. 1997;77:383–398. [Google Scholar]