Abstract
Microsporidia, as a group, cause a wide range of infections, though two species of microsporidia in particular, Enterocytozoon bieneusi and Encephalitozoon intestinalis, are associated with gastrointestinal disease in humans. To date, the mode of transmission and environmental occurrence of microsporidia have not been elucidated due to lack of sensitive and specific screening methods. The present study was undertaken with recently developed methods to screen several significant water sources. Water concentrates were subjected to community DNA extraction followed by microsporidium-specific PCR amplification, PCR sequencing, and database homology comparison. A total of 14 water concentrates were screened; 7 of these contained human-pathogenic microsporidia. The presence of Encephalitozoon intestinalis was confirmed in tertiary sewage effluent, surface water, and groundwater; the presence of Enterocytozoon bieneusi was confirmed in surface water; and the presence of Vittaforma corneae was confirmed in tertiary effluent. Thus, this study represents the first confirmation, to the species level, of human-pathogenic microsporidia in water, indicating that these human-pathogenic microsporidia may be waterborne pathogens.
Waterborne infections are a leading cause of human morbidity and mortality worldwide, but over 50% of these infections are caused by unknown agents. The reservoirs of these waterborne pathogens can be animals, humans, or the environment itself, and such agents can cause a variety of diseases, including giardiasis, cryptosporidiosis, typhoid fever, and hepatitis. New agents of waterborne disease are recognized on a regular basis, and, in general, any human pathogen excreted in feces or urine has the potential to be waterborne. One such group of microorganisms, which has only recently been recognized as an etiologic agent of disease in humans, is the microsporidia.
“Microsporidia” is a nontaxonomic name used to describe protozoan parasites belonging to the phylum Microspora. Microsporidia have recently been listed by the Centers for Disease Control and Prevention as important emerging pathogens. This is because seven species of microsporidia have been shown to be etiologic agents of disease in humans, especially immunocompromised or immunosuppressed individuals. Two species in particular, Enterocytozoon bieneusi and Encephalitozoon intestinalis, have been shown to be responsible for intestinal disease manifestations in AIDS patients (1, 15), and Encephalitozoon cuniculi has been detected in feces (7). A recent study showed that up to 40% of patients with AIDS-associated diarrheas were shedding microsporidia (8). Recent studies have also identified these pathogens in relation to immunocompetent individuals (11, 13). Thus, it can logically be assumed that there may be the potential for waterborne transmission of these agents. A recent epidemiological study has shown direct correlation of Encephalitozoon intestinalis infection with the use of well water and groundwater (3a). This could implicate groundwater as a potential source of infection not only for enteroviruses and enteric bacteria but also for microsporidia. Up until this point, however, the presence of human-pathogenic microsporidia has not been confirmed in water.
The major obstacle hindering the detection of microsporidia in the environment has been the small size of the human-pathogenic microsporidia (<2 μm in diameter), which has made it virtually impossible to positively identify the species of isolates visually without the use of transmission electron microscopy (12). Because of the difficulty of using transmission electron microscopy on environmental samples, there has been no confirmed documentation of human-pathogenic microsporidia associated with water samples. Another important factor that makes it difficult to correctly identify microsporidia in the environment is the presence of other genera of microsporidia that are not associated with pathogenicity in humans but which do infect nonhuman targets, including fish and insects. These other members of the Microspora have been detected in water and have been shown to cross-react with antibodies developed against the human pathogens (4), thus prohibiting the use of immunofluorescence assay (IFA) analysis.
In order to provide the initial link to the hypothesis that microsporidia are waterborne pathogens, we have used molecular methods developed in our laboratory to screen a variety of water sources, including raw sewage, tertiary effluent, surface water, and groundwater, for the presence of human-pathogenic microsporidia. Water concentrates from these sources were purified by density gradient flotation; subsequently, community DNA was extracted. Following this, human-pathogenic microsporidium-specific PCR screening was performed, resulting in the generation of PCR amplicons; these were then sequenced and database homology searches were performed in order to determine the species of the microsporidia.
MATERIALS AND METHODS
Water sampling.
Sampling of surface water and groundwater was performed by the USEPA Information Collection Rule (ICR) method for the concentration and detection of protozoan parasites (10). Surface water was collected by using a Homelite AP-125 portable water pump connected by a two-way valve (ACE Hardware) to a hose with a Snapi-tite quick connect (Melnor). The hose was connected to a plastic filter holder (Ametek, Plymouth, Mich.) that in turn was connected to a flow meter (1/2 by 1/2 in.) (Kent, Ocala, Fla.). The two-way valve was used to slow water flow to 1 gal per min, and the end of a screened six-foot intake hose was submerged in the water source. Groundwater samples were collected with the same filter cartridge apparatus, though submersible pumps at the bottoms of the wells provided sampling pressure. For surface water and groundwater samples, a 1DPPY cartridge filter (Cuno, Meriden, Conn.) in a plastic filter holder was used to trap the parasites.
Wastewater samples were collected with dipping rods and sterile 1-liter polypropylene bottles which had been previously coated with elution solution (10× phosphate-buffered saline, 1.0% Tween 80, 1.0% sodium dodecyl sulfate, and 1% Antifoam A [Sigma Chemical Co., St. Louis, Mo.]). Two liters of raw and 6 liters of treated effluent were collected. Raw sewage was collected from the treatment facilities’ inlet canal, while effluent was obtained from the chute used to transfer the secondary treated water to the tertiary treatment plant, which passed the effluent by pressure filtration through mixed-medium filters. Bottles were placed immediately on ice and processed within 24 h.
Modified ICR purification.
Filters were processed within 96 h of collection by dividing 3 liters of elution solution (10) into two 4-liter beakers. The filters were placed on a large piece of aluminum foil. Residual water contained in the bag was poured into one of the beakers, and the bag was rinsed with elution solution. Filters were cut open and hand washed in the elution solution for a total of 30 min to release particulates, including microsporidia, trapped within the filter. After washing, the elution solution was concentrated by centrifugation in 750-ml plastic centrifuge bottles at 3,600 rpm (2,000 × g) for 10 min in a Beckman GS-6 swinging bucket rotor centrifuge; to help ensure that spores remained intact, no brake was applied. Sample supernatant was aspirated and discarded, and the pellets were resuspended in an appropriate volume of elution solution (10) and pooled into one sample that was again centrifuged, at 2,800 rpm (1,050 × g) for 10 min. Supernatants were again removed, and the final pellet was resuspended in an equal volume of 20% formalin and stored at 4°C until further processing. Raw and effluent water samples were processed, as described, by centrifugation, pooling, and resuspension in 20% formalin.
Flotation purification.
Flotation purification of pelleted water concentrates was performed in accordance with the USEPA protocol (10). Percoll-sucrose with a specific gravity of 1.10 was employed to separate spores from denser particulate matter. Pellets resuspended in 20 ml of 1× phosphate-buffered saline were placed in 50-ml conical centrifuge tubes. This sample was carefully underlaid with Percoll-sucrose and centrifuged at 2,800 rpm (1,050 × g) for 10 min. The upper aqueous layer (20 ml), containing the spores, was then aspirated off with an additional 5 ml from the interface. This sample was transferred to a new 50-ml conical centrifuge tube, and the volume was adjusted to 50 ml. This was then centrifuged at 2,800 rpm (1,050 × g) for 10 min and the supernatant was removed, leaving a 1-ml volume in addition to the pelleted sample.
Community DNA extraction.
Total DNA was extracted directly from the samples by using a QIAamp Tissue Kit (Qiagen, Inc., Santa Clarita, Calif.) and a modified protocol. The protocol, which is described below, uses the manufacturer’s reagents, which are designated as they appear in the manufacturer’s protocol. Up to 100 μl of pelleted water concentrate was resuspended in 180 μl of buffer ATL by vortexing. To this, 20 μl of proteinase K (100 mg/ml) was added and the sample was once again vortexed. The sample was then incubated at 55°C for 4 h in a shaking water bath. Following this incubation, 200 μl of buffer AL was added and the sample was vortexed thoroughly and incubated in a 70°C water bath for 10 min, followed by a 10 min incubation in a 98°C water bath. Following these incubation steps, the samples were once again vortexed and then centrifuged at 14,000 rpm (16,000 × g) for 2 min to pellet any solids. The supernatant was transferred to a clean microcentrifuge tube and the pellet was discarded. Following this, 210 μl of 100% ethanol was added to the sample, which was then thoroughly vortexed for 1 min. The sample was then added carefully to QIAamp spin columns, preventing any contamination of the outside of the tube, and centrifuged at 6,000 × g for 1 min. The membrane-captured DNA was subsequently washed twice, according to the manufacturer’s instructions, and eluted. Elution was performed by adding 100 μl of molecular-grade water which had been preheated to 70°C to the column and incubating the columns in a hybridization incubator for 10 min at 70°C. The samples were then centrifuged for 1 min at 14,000 rpm (16,000 × g). In order to increase the quantity of DNA recovered, the eluted water was reapplied to the top of the column and the column was once again incubated in the hybridization incubator for 10 min at 70°C. Finally, the column was centrifuged at 14,000 rpm, and up to 80 μl of the flow-through was used for 100-μl PCRs.
PCR.
The PCR primers used in this analysis have been described previously (3, 6). The forward primer (5′-CAC CAG GTT GAT TCT GCC TGA C-3′) and the reverse primer (5′-CCT CTC CGG AAC CAA ACC CTG-3′) amplify the small-subunit ribosomal DNA (SSU-rDNA) of microsporidia, resulting in amplicon sizes of 250 bp for Enterocytozoon bieneusi, 268 bp for Encephalitozoon cuniculi, 270 bp for Encephalitozoon intestinalis, and 279 bp for Encephalitozoon hellem. PCR conditions were as follows: Taq Gold (Perkin-Elmer Corp., Norwalk, Conn.)-induced hot start cycling conditions consisting of 10 min of denaturation at 95°C, followed by 40 cycles of denaturation at 94°C for 45 s, annealing at 58°C for 20 s, and extension at 72°C for 40 s. A final extension step consisting of 5 min at 72°C was also included.
Sequencing.
PCR products were purified by using a QIAquick PCR purification kit (Qiagen) and were resuspended in sterile H2O. The forward PCR primer was then used for dye termination PCR sequencing, which was performed at the University of Arizona’s Laboratory of Molecular Systematics and Evolution sequencing facility.
SSU-rDNA sequence analysis.
Database searching was performed with BLAST 2.0 on the National Center for Biotechnology Information’s World Wide Web site (http://www.ncbi.nlm.nih.gov).
RESULTS AND DISCUSSION
Enterocytozoon bieneusi and Encephalitozoon intestinalis are etiologic agents of gastrointestinal disease in humans, and their spores are shed in feces. For this reason, we feel that they may be waterborne. In order to provide a link between water and microsporidia, we developed a PCR method which was subsequently used to screen a variety of water sources, including raw sewage (Fig. 1), water treatment effluents, surface water, and groundwater, for the presence of human-pathogenic microsporidia. Of the 14 water samples screened by the PCR method, 7 showed amplification of microsporidial SSU-rDNA. The PCR amplicons from positive samples were purified and sequenced with the forward PCR primer described by Dowd et al. (3). Sequences were then entered into the National Center for Biotechnology Information’s BLAST 2.0 search engine and subjected to database homology testing. The results of the sequence analyses are shown in Table 1. The species identified most often was Encephalitozoon intestinalis. One sample showed the presence of Enterocytozoon bieneusi, and one sample showed the presence of Vittaforma corneae.
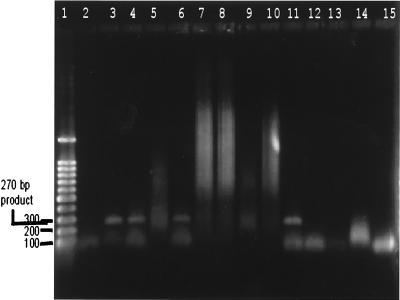
FIG. 1.
Typical PCR screening of water concentrates showing several positive results after community DNA extraction. After such screening, PCR products are purified and sequenced and the species of the microsporidia are identified by computer database homology comparison. Lane 1, 100-bp DNA ladder; lanes 2 to 14, replicate PCRs of community DNA extracted from raw sewage samples; lane 15, negative control PCR derived from sterile double-distilled water and used to ensure that no cross-reactivity occurred during community DNA extractions and PCR setup. Several of the lanes show PCR products of the size predicted for microsporidia. Lanes 3, 4, 6, 9, and 11 are all presumptively positive, showing products between 250 and 270 bp. Lane 9 has a very faint band, of the size predicted for Enterocytozoon bieneusi, while the other lane have bands of the size predicted for Encephalitozoon intestinalis.
TABLE 1.
Analysis of water for microsporidia by IFA, PCR, and sequencinga
| Sample type | Result of:
|
Speciesb | Accession no.c | Scored | % Similaritye | Pf | |
|---|---|---|---|---|---|---|---|
| IFA | PCR | ||||||
| Raw sewage | + | + | Encephalitozoon (Septata) intestinalis | L39113 | 391 | 97 | 1e-107 |
| Tertiary effluentg | + | + | Encephalitozoon (Septata) intestinalis | L39113 | 418 | 98 | 1e-115 |
| Tertiary effluentg | + | + | V. corneae | gb U11046 | 376 | 98 | 1e-90 |
| Tertiary effluentg | + | − | nd | nd | nd | nd | nd |
| Surface waterh | + | − | nd | nd | nd | nd | nd |
| Surface waterh | + | + | Encephalitozoon (Septata) intestinalis | U09929 | 289 | 96 | 9e-77 |
| Surface waterh | nd | + | Encephalitozoon (Septata) intestinalis | U39113 | 391 | 97 | 1e-98 |
| Surface wateri | nd | + | Enterocytozoon bieneusi | L16868 | 363 | 98 | 6e-99 |
| Controlj | + | + | Encephalitozoon (Septata) intestinalis | U39297 | 307 | 98 | 2e-82 |
| Controlj | nd | + | nd | nd | nd | nd | nd |
| Groundwater | − | + | Encephalitozoon (Septata) intestinalis | U39297 | 365 | 96 | 1e-85 |
| Groundwaterk | + | − | nd | nd | nd | nd | nd |
| Groundwaterk | + | − | nd | nd | nd | nd | nd |
| Groundwaterk | + | − | nd | nd | nd | nd | nd |
| Groundwaterk | + | − | nd | nd | nd | nd | nd |
| Groundwaterk | + | − | nd | nd | nd | nd | nd |
Results of PCR sequencing and species determination of microsporidia by using computer database homology searching. This method matches the unknown sequence with its closest relative in sequence databases. Similar sequences are ranked in order of highest homology and given statistical scores based on this homology. nd, not determined.
Species with the highest homology to the unknown sequence.
Accession number of the sequence with highest homology to the unknown sequence.
Ranking score given to the sequence with highest homology.
Percent base-to-base similarity between unknown and homologous sequences. It should be noted that in almost every instance the bases missed were due to a call of N by the sequencing program.
P, probability (on a scale of 0 to 1) that the sequence identified occurred by chance.
Activated sludge followed by pressure filtration through mixed-medium filters and extended chlorination.
Impacted with tertiary wastewater effluent.
No impact with discharged domestic wastewater.
Control, groundwater samples seeded with Encephalitozoon intestinalis spores.
Potentially impacted by surface wastewater infiltration.
One of the questions raised by molecular analysis is related to the potential of PCR to detect other species of microsporidia in water, especially those species that infect fish, insects, and amphibians but not humans. However, the ability of PCR sequencing and database searching to distinguish between very closely related species has already been shown (3) and allows human and nonhuman microsporidia to be distinguished. Thus, we feel that because the PCR sequencing approach was able to differentiate among the three closely related Encephalitozoonidae and was able to detect and determine the species of Enterocytozoon bieneusi, its ability to confirm their presence in the environment is justified. In most cases, comparisons of amplicons showing less than 100% homology to the database sequences were due to the computer sequencing software’s inability to identify bases during sequencing, as when the sequencing software incorporated an “N” in place of an actual base due to a weak or covered signal from the actual base. Taking this and the fidelity of Taq polymerase into account, we felt that at least 96% homology was necessary for definitive species determination of microsporidia. Therefore, if the percent homology was 95% or below, then a result was listed only as presumptive for the presence of the identified organism. If the percent homology was 96% or greater, then the identification was considered confirmed.
Finding human-pathogenic microsporidia in water could have been anticipated, but its importance should not be overlooked. The finding of microsporidia in raw sewage was the least surprising because several species of human-pathogenic microsporidia may be shed in feces and urine. However, their presence in tertiary effluent was more intriguing, since it may indicate that microsporidia can survive the wastewater treatment process, including mixed-medium filtration and chlorination. The structure of microsporidial spores indicates that they are potentially resistant to disinfection. This structure is characterized by electron-dense proteinaceous exospores and chitinous endospore layers (14). This structure is similar to other protozoan parasites, such as Giardia sp. cysts and Cryptosporidium sp. oocysts that have already been shown to be highly resistant to disinfection. Cryptosporidium parvum oocysts for instance, have been shown to require over 18 h of contact time for disinfection at the normal chlorine concentrations used in water treatment plants (5, 9). However, it should also be noted that PCR is able to detect both nonviable and viable organisms. Therefore, the microsporidia detected in the tertiary effluent may not be viable. Methods to evaluate the viability status of microsporidial water isolates are currently being developed in order to address this critical issue.
The presence of human-pathogenic microsporidia in surface water may suggest the presence of environmental reservoirs that include domestic and wild animals. Now, the detection of Encephalitozoon intestinalis and Enterocytozoon bieneusi in surface water may further lend credence to the potential for wild and/or domestic animal reservoirs. Like Giardia sp., which has environmental reservoirs including raccoons, and Cryptosporidium sp., which has cows as an accessory host, microsporidia may also have wild and/or domestic animals that are natural or accessory hosts. To lend further credence to this, Encephalitozoon intestinalis has recently been identified in a variety of animals, including cows, goats, pigs, and turkeys (1a). In any case, the role of water in the transmission of infection to humans could play a significant role.
Finally, the detection of microsporidia in groundwater is of the greatest interest and significance. Because of their small size, similar to most bacteria, it is possible that microsporidia could be transported in the subsurface, where they could eventually contaminate drinking wells. A recent epidemiological survey done in Mexico showed direct correlation between the incidence of diarrhea, in immunocompetent individuals infected with Encephalitozoon intestinalis, and the use of well water for drinking and food preparation (3a). It has been shown that enteroviruses and enteric bacteria can be transported in the subsurface over long distances, but few studies have looked at the transport of protozoan parasites in relation to groundwater (2). This is because most protozoa of clinical import are too large to be transported for any great distance in the terrestrial subsurface. Microsporidia, as mentioned, are similar in size to bacteria, so they may not be strained by soil matrices as readily as other protozoa. Thus, their small size increases their potential to be subsurface and groundwater contaminants.
Conclusions.
Microsporidia are a ubiquitous group of protozoan pathogens, many of which infect humans. Up until this point there has been no documentation of the environmental occurrence of microsporidia. This study screened a total of 14 water samples and 2 positive control water samples, which had been seeded with Encephalitozoon intestinalis spores, by PCR and found human-pathogenic microsporidia in 7 of these samples, not including the positive controls. Thus, half of the water samples screened during this study contained human-pathogenic microsporidia. Most of the PCR-positive water samples and the control samples were subsequently identified by sequencing and database homology analysis as Encephalitozoon intestinalis. However, two of these samples contained other species of microsporidia, namely, V. corneae and Enterocytozoon bieneusi. These results show that microsporidia may be waterborne pathogens. The data also suggest that there may be animal reservoirs for human-pathogenic microsporidia which have not yet been identified. Finally, the detection of microsporidia in groundwater samples indicates that there may be the potential for subsurface transport of these protozoan parasites.
Little is known epidemiologically about microsporidia, especially in immunocompetent humans, wild animals, and domestic animals. Though microsporidia have not been associated with any waterborne disease outbreaks to date, it may still be found in future studies, especially with the increasing awareness of microsporidia in the medical community and the advent of better methods for their detection, that they have this potential. Thus, this study has advanced the hypothesis that microsporidia are waterborne pathogens.
REFERENCES
- 1.Curry A, Canning E U. Human microsporidiosis. J Infect. 1993;27:229–236. doi: 10.1016/0163-4453(93)91923-d. [DOI] [PubMed] [Google Scholar]
- 1a.Dowd, S. E. Unpublished results.
- 2.Dowd S E, Faries F C, Mitchell F, Wickersham L, Kinney A, Pillai S D. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. The occurrence and fate of Cryptosporidium sp. and Giardia sp. around confined animal feeding operations (CAFOs), abstr. Q-28; p. 460. [Google Scholar]
- 3.Dowd S E, Gerba C P, Enriquez F J, Pepper I L. PCR amplification and species determination of microsporidia in formalin-fixed feces after immunomagnetic separation. Appl Environ Microbiol. 1998;64:333–336. doi: 10.1128/aem.64.1.333-336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Enriquez, F. J. (University of Arizona). Personal communication.
- 4.Enriquez F J, Ditrich O, Palting J D, Smith K. Simple diagnosis of Encephalitozoon sp. microsporidial infections by using a panspecific antiexospore monoclonal antibody. J Clin Microbiol. 1997;35:724–729. doi: 10.1128/jcm.35.3.724-729.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fayer R. Effect of sodium hypochlorite exposure on infectivity of Cryptosporidium parvum oocysts for neonatal BALB/c mice. Appl Environ Microbiol. 1995;61:844–846. doi: 10.1128/aem.61.2.844-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedorko D P, Nelson N A, Cartwright C P. Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J Clin Microbiol. 1995;33:1739–1741. doi: 10.1128/jcm.33.7.1739-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzen C, Schwartz D A, Visvesvara G S, et al. Disseminated antibody confirmed Encephalitozoon cuniculi with asymptomatic infection of the gastrointestinal tract in a patient with AIDS. Clin Infect Dis. 1995;21:1480–1484. doi: 10.1093/clinids/21.6.1480. [DOI] [PubMed] [Google Scholar]
- 8.Kotler D P. Gastrointestinal manifestations of immunodeficiency infection. Adv Intern Med. 1995;40:197–241. [PubMed] [Google Scholar]
- 9.Peeters J E, Mazas E A, Masschelein W J, Martinez de Maturana I V, Debacker E. Effect of disinfection of drinking water with ozone or chlorine dioxide on survival of Cryptosporidium parvum oocysts. Appl Environ Microbiol. 1989;55:1519–1522. doi: 10.1128/aem.55.6.1519-1522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.USEPA . Microbial laboratory manual. ICR: information collection rule. Cincinnati, Ohio: USEPA; 1996. [Google Scholar]
- 11.Van Gool T, Vetter J C M, Weinmayr B, Van Dam A, Derouin R, Dankert J. High seroprevalence of Encephalitozoon species in immunocompetent subjects. J Infect Dis. 1997;175:1020–1024. doi: 10.1086/513963. [DOI] [PubMed] [Google Scholar]
- 12.Visvesvara G, Leitch S G J, Pieniazek N J, Da Silva A J, Wallace S, Slemenda S B, Weber R, Schwartz D A, Gorelkin L, Wilcox C M, Bryan R T. Short-term in vitro culture and molecular analysis of the microsporidian Enterocytozoon bieneusi. J Eukaryot Microbiol. 1995;42:506–510. doi: 10.1111/j.1550-7408.1995.tb05896.x. [DOI] [PubMed] [Google Scholar]
- 13.Visvesvara G, Leitch G, Moura H, Wallace S, Weber R, Bryan R. Culture, electron microscopy, and immunoblot studies on a microsporidian parasite isolated from the urine of a patient with AIDS. J Protozool. 1991;38:105S–110S. [PubMed] [Google Scholar]
- 14.Weber R, Bryan R T. Microsporidial infections in immunodeficient and immunocompetent patients. Clin Infect Dis. 1994;19:517–521. doi: 10.1093/clinids/19.3.517. [DOI] [PubMed] [Google Scholar]
- 15.Weber R, Kuster H, Keller R, Bachi T, Spycher M A, Briner J, Russi E, Luthy R. Pulmonary and intestinal microsporidiosis in a patient with the acquired immunodeficiency syndrome. Am Rev Respir Dis. 1992;146:1603–1605. doi: 10.1164/ajrccm/146.6.1603. [DOI] [PubMed] [Google Scholar]
- 16.Weber R, Bryan R T, Schwartz D A, Owen R L. Human microsporidial infections. Clin Microbiol Rev. 1994;7:426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittner M, Tanowitz H B, Weiss L M. Parasitic infections in AIDS patients. Parasitic Dis. 1993;7:569–586. [PubMed] [Google Scholar]