Abstract
Defined microbial communities were developed by combining selective enrichment with molecular monitoring of total community genes coding for 16S rRNAs (16S rDNAs) to identify potential polychlorinated biphenyl (PCB)-dechlorinating anaerobes that ortho dechlorinate 2,3,5,6-tetrachlorobiphenyl. In enrichment cultures that contained a defined estuarine medium, three fatty acids, and sterile sediment, a Clostridium sp. was predominant in the absence of added PCB, but undescribed species in the δ subgroup of the class Proteobacteria, the low-G+C gram-positive subgroup, the Thermotogales subgroup, and a single species with sequence similarity to the deeply branching species Dehalococcoides ethenogenes were more predominant during active dechlorination of the PCB. Species with high sequence similarities to Methanomicrobiales and Methanosarcinales archaeal subgroups were predominant in both dechlorinating and nondechlorinating enrichment cultures. Deletion of sediment from PCB-dechlorinating enrichment cultures reduced the rate of dechlorination and the diversity of the community. Substitution of sodium acetate for the mixture of three fatty acids increased the rate of dechlorination, further reduced the community diversity, and caused a shift in the predominant species that included restriction fragment length polymorphism patterns not previously detected. Although PCB-dechlorinating cultures were methanogenic, inhibition of methanogenesis and elimination of the archaeal community by addition of bromoethanesulfonic acid only slightly inhibited dechlorination, indicating that the archaea were not required for ortho dechlorination of the congener. Deletion of Clostridium spp. from the community profile by addition of vancomycin only slightly reduced dechlorination. However, addition of sodium molybdate, an inhibitor of sulfate reduction, inhibited dechlorination and deleted selected species from the community profiles of the class Bacteria. With the exception of one 16S rDNA sequence that had the highest sequence similarity to the obligate perchloroethylene-dechlorinating Dehalococcoides, the 16S rDNA sequences associated with PCB ortho dechlorination had high sequence similarities to the δ, low-G+C gram-positive, and Thermotogales subgroups, which all include sulfur-, sulfate-, and/or iron(III)-respiring bacterial species.
The extensive industrial use of polychlorinated biphenyls (PCBs) during the 20th century has resulted in the release of an estimated several million pounds of PCBs into the environment (2). Due to the hydrophobicity and chemical stability of these compounds, PCBs ultimately accumulate in subsurface anaerobic sediments, where reductive dechlorination by anaerobic microorganisms is proposed to be an essential step in PCB degradation and detoxification (6). Although anaerobic reductive dechlorination has been documented in the environment and in the laboratory, attempts to identify and isolate anaerobic PCB-dechlorinating microbes by classical enrichment and isolation techniques have been unsuccessful (for a review, see reference 2). Isolation of anaerobic PCB-dechlorinating microbes has been hindered in part by the inability to maintain and sequentially transfer dechlorinating consortia in defined medium. May et al. (24) were the first to demonstrate that single colonies could be obtained by plating highly enriched PCB-dechlorinating enrichment cultures on agar-solidified media. Although two of the colonies exhibited para dechlorination activity when transferred back to liquid enrichment medium, the colonies contained a mixed community of microorganisms and dechlorination required the addition of sediment to the medium. More recently, highly enriched PCB-ortho-dechlorinating enrichment cultures were developed from Baltimore Harbor sediments in minimal media that contained sediments and a single congener (3) or Aroclor 1260 (37). These were the first confirmed reports of sustained ortho dechlorination of PCBs throughout sequential transfers in medium with estuarine sediments. Finally, Cutter et al. demonstrated that a consortium of PCB-ortho-dechlorinating anaerobes from Baltimore Harbor could be sequentially transferred and maintained in minimal medium without the addition of sterile sediment (9). With the ability to maintain PCB dechlorination in a completely defined medium, highly enriched PCB-dechlorinating consortia could be developed by sequential transfers in medium that contained the minimal growth requirements for dechlorinating species.
The current study identifies putative PCB-dechlorinating anaerobes in ortho-dechlorinating enrichment cultures by a comprehensive approach that combines traditional selective enrichment techniques with molecular monitoring (SEMM). Microbial consortia enriched for PCB ortho dechlorination in minimal medium were analyzed by comparative sequence analysis of genes coding for 16S rRNA (16S rDNA) amplified from total community DNAs. Protocols were developed for chromosomal DNA extraction from sediment, 16S rDNA amplification by PCR, cloning of partial 16S rDNA PCR fragments, screening by restriction fragment length polymorphism (RFLP) analysis, and DNA sequencing for comparative sequence analysis. By utilizing these techniques, shifts in the microbial community were monitored as the cultures were further enriched for PCB-dechlorinating anaerobes by elimination of undefined medium components (i.e., sediment), changes in carbon source, and addition of selective physiological inhibitors. The results presented herein demonstrate the applicability of the SEMM approach for the selection and monitoring of highly defined PCB-dechlorinating microbial consortia.
MATERIALS AND METHODS
Enrichment cultures.
Enrichment cultures were initiated as described previously (9). Briefly, sediment samples collected from the Northwest Branch of Baltimore Harbor, Baltimore, Md. (39°16.8′N, 76°36.1′W), were used to inoculate sterile, anaerobic estuarine salts medium that did not contain added sulfate to a final concentration of 5% (dry wt/vol). Where indicated, sodium acetate, alone or with sodium propionate and butyrate, was added to a final concentration of 2.5 mM (each). The congener 2,3,5,6-tetrachlorobiphenyl (2,3,5,6-CB; AccuStandard, Inc., New Haven, Conn.) was solubilized in acetone and added to a final concentration of 173 μM. For the inhibitor studies, bromoethanesulfonic acid (BES), vancomycin, and sodium molybdate were dissolved in deionized water, filter sterilized, and added to final concentrations of 3 mM, 100 μg/ml, and 20 mM, respectively. All cultures were incubated in the dark at 30°C. PCBs were extracted and analyzed by gas chromatography coupled with an electron capture detector using a 16-point standard curve for each congener as described previously (3).
Extraction of genomic DNA.
The methods described herein for the phylogenetic analysis of the enrichment cultures are slightly modified from those described previously (13). Depending upon the culture turbidity, between 1 and 10 ml of culture was anaerobically withdrawn and utilized for extraction of bulk genomic DNA (final yield, greater than 100 ng as estimated by visualization on an agarose gel stained with ethidium bromide). The culture sample was centrifuged, and the cell and sediment pellet was resuspended in 250 μl of sterile TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). The resuspended pellet was added to a 2.2-ml screw-cap conical tube that contained 2.5 g of autoclaved zirconia-silica beads (0.1 mm), and 250 μl each of sodium phosphate buffer (0.1 M, pH 8.0) and TS-SDS buffer (0.1 M NaCl, 0.5 M Tris [pH 8.0], 10% [wt/vol] sodium dodecyl sulfate). The sample was cooled on ice for 10 min and then homogenized for 5 min with a Mini-Bead Beater (Biospec, Bartlesville, Okla.) at 4°C to lyse cells. Debris was removed by centrifugation for 5 min at 14,000 × g. Crude DNA in the supernatant was purified twice with equal volumes of trissaturated phenol and chloroform-isoamyl alcohol (24:1), followed by extraction with an equal volume of chloroform. Approximately 200 μl of Phase-Lock gel (5 Prime-3 Prime, Inc., Boulder, Colo.) was utilized to promote separation of the phases and allow easier visualization of the interface. The decanted aqueous phase was diluted to 1 ml with sterile deionized water. Humic acids, which inhibit PCR (32, 34), were extracted from nucleic acids by addition of 0.125 g of insoluble polyvinylpolypyrrolidone (Sigma, St. Louis, Mo.) to the 1 ml of diluted crude DNA extract (17, 30). The polyvinylpolypyrrolidone was removed by centrifugation for 5 min at 14,000 × g, and the chromosomal DNA was recovered by precipitation with an equal volume of isopropanol at −20°C. The DNA was pelleted by centrifugation, and then the pellet was washed with 70% ethanol and centrifuged again at high speed. The supernatant was discarded, and the DNA was dried under vacuum for 5 min. Further removal of humic acids was achieved by electrophoresis of the DNA extract in a 1.3% low-melting-point agarose gel (Fisher Scientific, Fairlawn, N.J.) containing 2% soluble polyvinylpyrrolidone (40). The chromosomal DNA band was excised from the gel and recovered with a Promega Wizard PCR Prep Kit (Promega, Madison, Wis.) in accordance with the manufacturer’s instructions.
PCR amplification and cloning.
PCR was utilized to amplify bacterial and archaeal 16S rDNAs from the mixed community of genomic DNAs. Universal primers 519F (5′-CAG CA/CG CCG CGG TAA TA/TC-3′) and 1406R (5′-ACG GGC GGT GTG TA/GC-3′) were utilized for the amplification of bacterial 16S rDNAs (21). Archaeal 16S rDNAs were amplified with specific archaeal primers 21F (5′-TTC CGG TTG ATC CYG CCG GA-3′) and 958R (5′-TCC GGC GTT GAM TCC AAT T-3′) (11). All PCR amplifications were performed by using the GeneAmp PCR kit with Taq DNA polymerase (Perkin Elmer, Inc.) in a PTC200 thermal cycler (MJ Research, Watertown, Mass.). Conditions for PCR were as follows: an initial denaturation step of 1.5 min at 94°C; 30 amplification cycles of denaturation (30 s at 94°C), annealing (30 s at 55°C), and elongation (30 s at 72°C); and a final extension step of 5 min at 72°C. The PCR products were purified by utilizing the QIAquick PCR purification kit (Qiagen, Inc., Chatsworth, Calif.). Plasmid libraries for both domains were generated by ligating 2 μl of purified PCR fragments into the pCRII vector (Invitrogen, Carlsbad, Calif.) in accordance with the manufacturer’s instructions. The ligation reactions were transformed into the Escherichia coli INVαF′ competent cells supplied with the Invitrogen Original TA Cloning Kit.
Library screening.
Ninety-six randomly chosen clones were selected from colonies and grown overnight in Luria broth with kanamycin (100 μg/ml). The partial 16S rDNA fragments were amplified directly from 2 μl of an overnight-grown Luria broth culture added to 48 μl of PCR mixture using the following PCR conditions: 1 cycle of 3 min at 95°C; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final extension step of 72°C for 5 min. Subsequently, the PCR products were digested separately with the restriction endonucleases HaeIII and HhaI (New England Biolabs, Inc., Beverly, Mass.). The restriction digests were electrophoresed in a 3% Trevi-Gel (TreviGen, Gaithersburg, Md.) and visualized with SYBR Green I nucleic acid gel stain (Molecular Bio-Probes, Eugene, Oreg.) by using a Fluoroimager (Molecular Dynamics, Sunnyvale, Calif.). Clones were categorized according to their distinct RFLPs.
Sequencing and analysis.
At least two representative clones for each unique RFLP were sequenced for comparative phylogenetic analysis. Plasmid DNA was purified with the Qiagen Plasmid Mini Kit (Qiagen, Inc.), and the sequence was determined after dye terminator cycle sequencing on an ABI 373 Automated Sequencer (Applied Biosystems, Foster City, Calif.). Initially, the clones were sequenced from the flanking 5′ end with a T7 sequencing primer and from the flanking 3′ end with an M13 reverse sequencing primer, both located on the pCRII vector, to obtain the complete fragment sequence.
Sequences were analyzed with the National Center for Biotechnology Information basic local alignment search tool via the BLASTN program (1) and the SIM_RANK program of the Ribosomal Database Project (28).
Chimeric sequence evaluation.
Screening methods similar to those described previously by Snaidr et al. (29) were utilized for chimera screening. First, the sequences were manually aligned and then analyzed by using a software package that takes into account misalignments in secondary structure that could result from chimeras (7). Second, short sequences (∼300 bp) of both the 16S rDNA 5′ and 3′ flanking regions were then submitted to both the BLASTN and SIM_RANK programs for comparative phylogenetic analysis of whole and partial gene sequences. Third, partial sequences were evaluated with the Check_Chimera program of the Ribosomal Database Project. To further minimize chimera formation, high-molecular-weight genomic DNA and PCR products were size fractionated in agarose gels prior to library construction. In addition, both bacterial and archaeal clone libraries were generated and screened from three replicate PCRs.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences used to generate a phylogenetic tree are as follows: Clostridium litorale, X77845; Dehalobacter restrictus, U84497; Dehalococcoides ethenogenes, AF004928; Desulfitobacterium dehalogenans, L28946; Desulfitobacterium frappieri, U40078; Desulfobacter postgatei, M26633; Desulfomonile tiedjei, M26635; Desulfonema ishimotoei, U45992; Desulfosarcina variabilis, M34407; Desulfothiovibrio peptidovorans, U52817; Desulfotomaculum orientis, M34417; Desulfovibrio desulfuricans, M34113; Desulfuromonas acetexigens, U23140; Desulfuromusa succinoxidans, X79415; Fervidobacterium nodosum, M59177; Geobacter metallireducens, L07834; Geotoga petraea, L10658; Pelobacter propionicus, X70954; Petrotoga miotherma, L10657; Syntrophospora bryantii, M26491; Syntrophus gentianae, X85132; Thermoanaerobacter brockii, L09165; Thermosipho africanus, M83140; Thermotoga maritima, M21774.
Sequences of the partial 16S rDNA clones exhibiting RFLP types 1, 4, 5, 11, 15, 17, 24, 25, 40, 105, 108, 109, and 144 were submitted to GenBank under accession no. AF058000 to AF058012, respectively.
RESULTS
Effects of PCB on community profiles.
Selective enrichment techniques were used to establish ortho-dechlorinating enrichment cultures. Concomitantly, the cultures were monitored by screening the 16S rDNA community for putative PCB-ortho-dechlorinating microorganisms within these enrichment cultures. The diversity of the microbial community was minimized from the outset by the use of a minimal estuarine medium that contained sterilized Baltimore Harbor sediments. Further, the enrichment cultures were incubated with a single PCB congener, 2,3,5,6-CB, to facilitate monitoring of the rate and extent of dechlorination and to select for congener-specific dechlorinating organisms that were capable of reductively dechlorinating the parent congener and its trichlorinated intermediate (3).
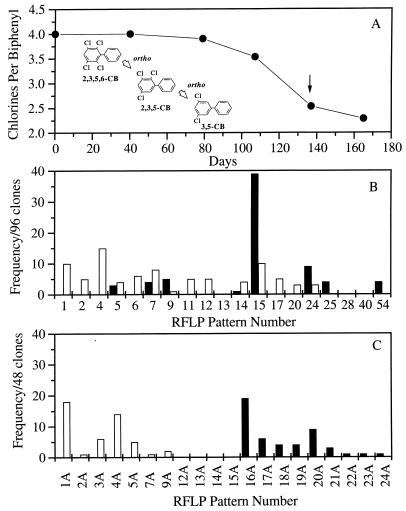
Enrichment cultures that exhibited ortho dechlorination of 2,3,5,6-CB were generated by three sequential transfers (10% inoculum) of Baltimore Harbor sediments in estuarine medium supplemented with a mixture of three fatty acids: propionate, butyrate, and acetate (3, 9). Following the third sequential transfer, the only dechlorination pathway observed for these cultures, ortho dechlorination of 2,3,5,6-CB (Fig. 1A, inset), was observed in the PCB-containing culture after 79 days and achieved a maximum rate after 107 days (Fig. 1A). Approximately 75% of the parent congener was converted to 3,5-CB after 160 days. Duplicate enrichment cultures that did not contain a PCB were maintained and sequentially transferred concurrently with the PCB-dechlorinating enrichment cultures. Both dechlorinating and nondechlorinating enrichment cultures were methanogenic.
FIG. 1.
(A) Rate of chlorine removal from 2,3,5,6-CB by enrichment cultures containing 0.1% Baltimore Harbor sediment. The dechlorination pathway of 2,3,5,6-CB by ortho-dechlorinating enrichment cultures is shown in the inset. (B) Community profiles of bacterial 16S rDNA clones from Baltimore Harbor enrichment cultures incubated with (□) and without (■) 2,3,5,6-CB. Samples for phylogenetic analysis were taken at day 137, as indicated for panel A. Both enrichment cultures were amended with a mixture of three fatty acids as carbon sources. (C) Community profiles of archaeal 16S rDNA clones from Baltimore Harbor enrichment cultures incubated with (□) and without (■) 2,3,5,6-CB.
Community profiles analyzed at 137 days after the third sequential transfer of dechlorinating and nondechlorinating enrichment cultures are shown in Fig. 1B. Sixteen predominant RFLP types were identified in the cultures, and 16S rDNA fragments from two representative clones for each pattern were subjected to comparative sequence analysis. Eight RFLP types, 1, 2, 4, 6, 11, 12, 17, and 20, were detected exclusively in cultures that contained the PCB congeners. RFLP type 4, the most predominant clone, accounting for 30% of the selected clones, showed the highest sequence similarity to the δ subgroup (Table 1). RFLP type 1, the second most predominant clone, accounted for 20% of the selected clones and showed the highest sequence similarity to the Thermotogales subgroup. Of the remaining clones, RFLP types 11 and 12 had the highest sequence similarity to the low-G+C gram-positive subgroup, RFLP types 4, 6, and 20 had the highest sequence homology to members of the δ subgroup, and RFLP type 17 exhibited the highest sequence similarity to the deeply branching species Dehalococcoides ethenogenes (25). Only one representative clone with RFLP type 6 was identified because the partial 16S rDNA insert was unstable and often lost from the vector prior to sequencing.
TABLE 1.
Phylogenetic affiliations of predominant RFLP types from PCB-ortho-dechlorinating enrichment cultures based on bacterial 16S rRNA gene sequences
| RFLP type | Closest phylogenetic relative | % Similarity to closest relative |
|---|---|---|
| 1 | Thermotoga maritima | 85 |
| 2 | Bacteroides eggerthii | 89 |
| 4 | Desulfosarcina variabilis | 93 |
| 5 | Desulfothiovibrio peptidovorans | 87 |
| 6 | Desulfuromonas thiophila | 94 |
| 7 | Clostridium litorale | 91 |
| 9 | Desulfonema magnum | 82 |
| 11 | Syntrophospora bryantii | 94 |
| 12 | Unidentified oil field bacterium | 75 |
| 15 | Clostridium litorale | 99 |
| 17 | Dehalococcoides ethenogenes | 89 |
| 20 | Pelobacter acidigallici | 86 |
| 24 | Acholeplasma laidlawii | 84 |
| 25 | Desulfonema magnum | 94 |
| 28 | Desulfovibrio caledoniensis | 95 |
| 40 | Syntrophus gentianae | 94 |
| 54 | Clostridium litorale | 84 |
| 105 | Desulfuromonas thiophila | 96 |
| 108 | Desulfuromonas acetexigens | 99 |
| 109 | Desulfovibrio sp. | 92 |
| 116 | Desulfovibrio sp. | 86 |
| 130 | Uncultured eubacterium | 89 |
| 138 | Unidentified low-G+C gram-positive sp. | 96 |
| 144 | Desulfovibrio sp. strain B650 | 98 |
| 146 | Desulfovibrio sp. | 91 |
RFLP types 7 and 14 showed the highest sequence similarity to the low-G+C gram-positive subgroup. Both patterns were detected in the presence and absence of a PCB but increased significantly (≥50%) in medium that contained a PCB. The remaining clones, which had high sequence similarity to members of the δ subgroup (RFLP type 25) and the low-G+C gram-positive subgroup (RFLP types 5, 9, 15, 24, and 54), were either detected at similar frequencies in both cultures, increased in the frequency of detection relative to one another, or detected only in the PCB-free culture. The results suggest that species represented by the latter clones do not have a significant role in PCB ortho dechlorination.
The community profiles of methanogenic archaea enriched in the presence and absence of a PCB differed significantly (Fig. 1C). Seven predominant RFLP types were detected in the actively dechlorinating culture. RFLP types 1A, 4A, and 5A had the highest sequence similarity to the Methanosarcinales subgroup, whereas RFLP types 2A, 3A, 7A, and 9A had the highest sequence similarity to the Methanomicrobiales subgroup (Table 2). Conversely, none of the clones detected in the presence of a PCB were detected in the PCB-free enrichment culture. RFLP types 16A, 19A, 20A, 21A, 22A, and 24A had the highest sequence similarity to the Methanosarcinales subgroup, and the remaining clones, with RFLP types 17A, 18A, and 23A, had the highest similarity to the Methanomicrobiales subgroup. Although the community profiles differed in the absence and presence of a PCB congener, both cultures exhibited similar distributions of species belonging to the autotrophic, hydrogen-utilizing order Methanomicrobiales and the aceticlastic and methylotrophic order Methanosarcinales. This preliminary characterization represented a baseline community profile for the PCB-dechlorinating and nondechlorinating enrichment cultures.
TABLE 2.
Phylogenetic affiliations of predominant RFLP types from PCB-ortho-dechlorinating enrichment cultures based on archaeal 16S rRNA gene sequences
| RFLP type | Closest phylogenetic relative | % Similarity to closest relative |
|---|---|---|
| 1A | Methanosaeta concilii | 91 |
| 2A | Methanoculleus marisnigri | 90 |
| 3A | Methanoplanus limicola | 90 |
| 4A | Methanohalophilus mahii | 87 |
| 5A | Methanohalobium evestigatum | 81 |
| 7A | Methanogenium organophilum | 96 |
| 9A | Methanospirillum hungatei | 87 |
| 16A | Methanosaeta concilii | 99 |
| 17A | Methanoplanus petrolearius | 94 |
| 18A | Methanogenium organophilim | 96 |
| 19A | Methanosaeta concilii | 96 |
| 20A | Methanohalophilus mahii | 86 |
| 21A | Methanosaeta concilii | 96 |
| 22A | Methanosaeta concilii | 99 |
| 23A | Methanoplanus limicola | 92 |
| 24A | Methanosaeta concilii | 99 |
Effects of Baltimore Harbor sediment on ortho-dechlorinating consortia.
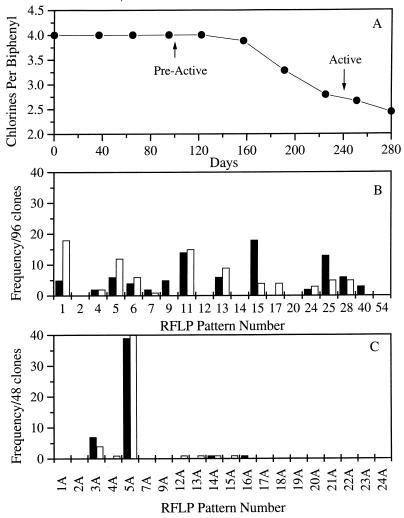
To eliminate the effects of putative alternative electron acceptors (e.g., humic acids, SO42−, Fe2+) and undefined nutrients that may be present in Baltimore Harbor sediments, PCB-dechlorinating enrichment cultures were sequentially transferred in completely defined estuarine medium that contained 2,3,5,6-CB and three fatty acids as carbon sources without the addition of sterile sediments (9). After four sequential transfers in the absence of sediments, dechlorination of 2,3,5,6-CB was detected after an extensive lag period (>100 days) and the congener was completely transformed to 3,5-CB after 240 days (Fig. 2A). Methane production was observed in the sediment-free enrichment cultures.
FIG. 2.
(A) Reductive dechlorination of 2,3,5,6-CB in sediment-free Baltimore Harbor enrichment cultures with a mixture of three fatty acids as carbon sources. Sediment was removed by dilution after four sequential transfers. The enrichment culture was sampled for phylogenetic analysis prior to the onset of dechlorination (preactive, day 102) and during ortho dechlorination (active, day 240). (B) Community profiles of bacterial 16S rDNA clones from sediment-free Baltimore Harbor enrichment cultures prior to (■) and following (□) the onset of ortho dechlorination. (C) Community profiles of archaeal 16S rDNA clones from sediment-free Baltimore Harbor enrichment cultures prior to (■) and following (□) the onset of ortho dechlorination.
Community profiles were compared before and after the onset of dechlorination in the fourth sequential enrichment culture transfer in defined medium (Fig. 2B). Of the 14 predominant RFLP types previously detected in PCB-dechlorinating cultures with sediment, 10 were detected in the sediment-free cultures. As observed in the previous cultures, RFLP type 1 was the predominant species, accounting for 36% of the clones detected. Of the seven remaining RFLP types that appeared exclusively in the PCB-dechlorinating enrichment culture with sediment, only four were detected in the absence of sediment (RFLP types 4, 6, 11, and 17) and only the relative detection frequencies of RFLP type 5 increased significantly with the onset of dechlorination. The absence of RFLP types 2, 9, 12, 14, 20, and 54 indicated that these species were diluted out to undetectable levels after sediment was deleted. Although this observation suggests that the latter species are not required for ortho dechlorination of 2,3,5,6-CB, it does not rule out the possibility that they are capable of dechlorination but lacked specific growth factors provided by the sediments. The three remaining clones, RFLP types 28, 40 (δ subgroup), and 13 (low-G+C gram-positive subgroup), were not observed previously in medium that contained sediment but were selectively enriched in the absence of sediment.
Overall, the most predominant members of the methanogenic archaeal community did not change significantly with the onset of dechlorination in the sediment-free enrichment cultures, as indicated in Fig. 2C, and all were observed in previous cultures with sediment and the PCB congener. RFLP types 4A, 12A, and 14A were detected only after dechlorination was observed in the enrichment. RFLP types 3A, 5A, and 13A were detected both in the preactive and active cultures. RFLP type 15A was detected only in the absence of dechlorination. RFLP type 5A, the most predominant clone, had the highest sequence homology to members of the order Methanosarcinales, whereas the second most predominant clone, RFLP 3A, had the highest homology to members of the order Methanomicrobiales.
Effects of carbon source on ortho-dechlorinating consortia.
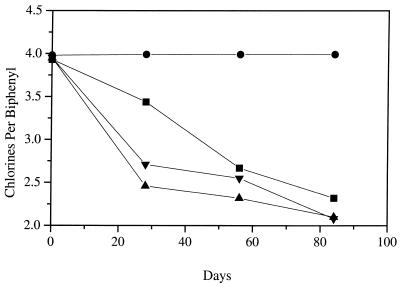
PCB-dechlorinating enrichment cultures grown with three fatty acids were sequentially transferred into defined estuarine medium that contained 2,3,5,6-CB and sediment with sodium acetate as the sole electron donor to minimize community diversity further. After three sequential transfers, dechlorination was detected within 28 days and the congener was completely transformed to 3,5-CB after 85 days (Fig. 3). Growth rates were not measured in cultures that contained sediment due to turbidity caused by the particles. However, enrichment cultures that contained sodium acetate had higher dechlorination rates than cultures that contained a mixture of three fatty acids. Cultures were methanogenic with sodium acetate.
FIG. 3.
Dechlorination rates of Baltimore Harbor cultures treated with physiological inhibitors. Symbols: ▴, no inhibitor; ■, 3 mM BES; •, 20 mM sodium molybdate; ▾, 100-μg/ml vancomycin.
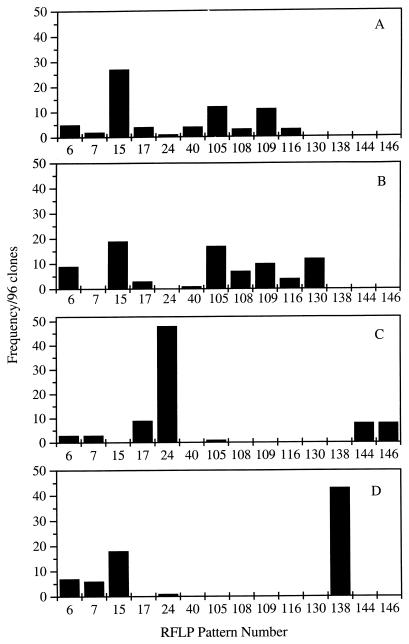
Community profiles were determined after three sequential transfers of the enrichment cultures with 2,3,5,6-CB and sodium acetate (Fig. 4A). Only 6 RFLP types, 6, 7, 15, 17, 24, and 40, of the 19 predominant RFLP types detected in the previous cultures that contained fatty acids were detected in cultures that contained only acetate as an electron donor. Interestingly, RFLP types 4, 5, 11, 13, 25, and 28, which were predominant in cultures that contained a mixture of fatty acids that included sodium acetate, were not detected in dechlorinating enrichment cultures grown with sodium acetate alone. These results suggest that growth of the latter species was linked to butyrate or propionate catabolism. The shift to acetate resulted in a significant overall change in the community. The most predominant RFLP types (105, 108, 109, and 116; frequency, ≥2/96 clones) detected in enrichment cultures containing sodium acetate were not detected previously, indicating that their growth may be linked specifically to acetate. All of the predominant RFLP types belonged to the δ subgroup.
FIG. 4.
Effects of physiological inhibitors on community profiles of Baltimore Harbor enrichment cultures enriched with 2,3,5,6-CB, acetate, and 0.1% Baltimore Harbor sediment. Panels: A, no inhibitor; B, 3 mM BES; C, 100-μg/ml vancomycin; D, 20 mM sodium molybdate.
Effects of selective inhibitors on ortho-dechlorinating consortia.
To further reduce community diversity and select for microbial species linked to ortho dechlorination of 2,3,5,6-CB with sodium acetate as the growth substrate, enrichment cultures were transferred into medium that contained physiological inhibitors. The inhibitors included BES, which selectively inhibits the methanogenic archaea (16); sodium molybdate, an analogue of sulfate, which selectively inhibits sulfate-reducing bacteria (31); and vancomycin, which selectively inhibits gram-positive bacteria by inhibiting biosynthesis of the cell wall peptidoglycan (27). Active cultures were transferred to medium that contained the selected physiological inhibitor and then sampled for analysis of the 16S rDNA community profile after the onset of dechlorination.
The addition of BES only slightly inhibited the rate of dechlorination, and nearly complete dechlorination of 2,3,5,6-CB to 3,5-CB occurred within 85 days (Fig. 3). The bacterial diversity and relative numbers of bacterial species in the BES-treated culture closely resembled those in untreated control cultures (Fig. 4A and B). Seven previously undescribed RFLP types were detected, but only RFLP type 130 (low-G+C gram-positive subgroup) was predominant at frequencies of ≥2/96 clones sampled. However, methanogenesis did not occur and archaeal rDNA was not detected by PCR, indicating that the methanogenic archaea were not required for ortho dechlorination of 2,3,5,6-CB to 2,3,5-CB and 3,5-CB with sodium acetate.
As expected, vancomycin caused a more significant shift in the bacterial community than BES (Fig. 4C). Interestingly, vancomycin, like BES, also inhibited methanogenesis and precluded detection of archaeal rDNA by PCR, confirming that the methanogenic archaea were not required for ortho dechlorination of 2,3,5,6-CB with sodium acetate. Five RFLP types, 6, 7, 17, 24, and 105, were detected previously in PCB-dechlorinating cultures that did not contain an inhibitor. Of the 10 RFLP types not detected previously, the two most predominant (frequency, ≥2/96 clones), 144 and 146, were most closely related to the δ subgroup.
The addition of sodium molybdate (final concentrations of 2 and 20 mM) completely inhibited dechlorination and inhibited methanogenesis of 2,3,5,6-CB (Fig. 3). Furthermore, the genomic yield of this culture was approximately 10-fold lower than that of the previous cultures, and the bacterial diversity was significantly reduced (Fig. 4D). As expected, RFLP types 40, 105, 108, 109, and 116, which had sequence similarity to the δ subgroup, were not detected in the molybdate culture. However, the relative detection frequency of RFLP type 6, which is also phylogenetically related to the δ subgroup, was similar to that of the positive control, along with low-G+C gram-positive RFLP types 7, 15, and 24. RFLP type 138 (low-G+C gram-positive subgroup) was detected only in this culture and, therefore, was unlikely to represent an ortho-dechlorinating species.
DISCUSSION
Molecular screening of the 16S rDNAs from the total community of genomic DNAs was used to characterize microbial consortia in PCB-ortho-dechlorinating enrichment cultures without isolation of heretofore unculturable dechlorinating species. Bias can be introduced at various stages in the protocol, particularly during cell lysis and PCR amplification. Therefore, to minimize screening bias, a physical cell lysis method, bead mill homogenization, was used to effectively lyse all cell types, including those most recalcitrant to physical and enzymatic treatments (22, 26). To minimize PCR bias, separate primers were used for bacterial and archaeal phylogenetic domains. The primers were tested with Baltimore Harbor enrichment cultures and determined empirically to yield greater community diversity than other “universal” primers previously described (data not shown). In addition, PCR parameters, including use of a denaturant (formamide), temperature, and ion concentration, were optimized to yield maximum diversity in the community profiles of Baltimore Harbor enrichment cultures. Other factors, such as species-specific 16S rDNA copy number and PCR bias for a low-G+C template, also affect the quantitative assessment of microbial communities (14), and as a result, this approach can provide only an estimate of the actual abundance of microorganisms in each enrichment. In the current study, all enrichment cultures were sequentially transferred from the same inoculum source and grown under similar conditions. Throughout the study, community profile comparisons of duplicate cultures and of sequential transfers of identical treatments were reproducible (data not shown). Therefore, it was possible to determine whether an individual species was associated with PCB dechlorination by assaying for the coexistence or mutual exclusion of its RFLP type with dechlorination after treatment with physiological inhibitors. By monitoring the rates of dechlorination and relative frequencies of detection of specific RFLP types associated with PCB dechlorination, this approach was used to establish a highly defined PCB-ortho-dechlorinating community and to monitor the effects of sequential culture transfers and treatments on specific community members.
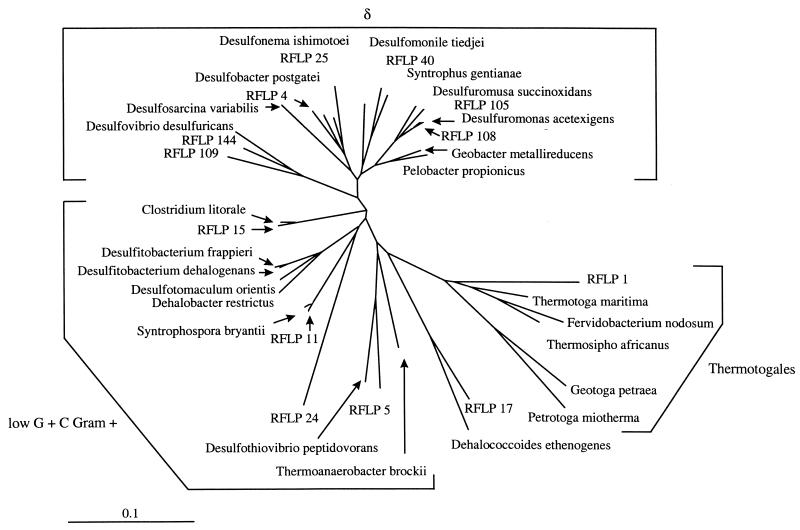
Previous attempts to identify and isolate anaerobic PCB dechlorinators by selective enrichment and isolation techniques have been unsuccessful (2). The failure to identify these species is likely due to the development of previous enrichment cultures in complex, undefined medium, which resulted in selection for faster-growing, non-PCB-dechlorinating microorganisms that likely outcompete PCB dechlorinators. By using the SEMM approach, conditions were developed that would maintain cultures of PCB-dechlorinating consortia indefinitely in a defined minimal medium. While other molecular approaches have been described for the isolation of bacteria from the environment (19, 23, 33), this is the first reported application of a molecular approach for the development of a defined PCB-dechlorinating consortium in a minimal medium. By reducing the medium complexity, the community diversity in a PCB-dechlorinating consortium was systematically reduced with the addition of medium components and physiological inhibitors that selectively promoted the growth of species involved in ortho dechlorination of 2,3,5,6-CB. Screening of the microbial communities by RFLP of PCR-amplified 16S rDNA as the cultures were selectively enriched provided a means for effectively monitoring the effects of treatments on individual species and, by a process of elimination, enabled us to identify species that are most likely to catalyze PCB dechlorination. In addition, the phylogeny of individual RFLP types was determined by comparative sequence analysis of the PCR-amplified 16S rDNA fragments (Fig. 5).
FIG. 5.
Phylogenetic tree inferred from comparative sequence analyses of partial 16S rDNA sequences from several predominant clones obtained from PCB-ortho-dechlorinating enrichment cultures. For construction of a phylogenetic tree, approximately 890-bp segments of selected sequences were aligned manually with a collection of known bacterial 16S rRDAs (for nucleotide sequence accession numbers, see Materials and Methods) obtained from the GenBank database by using software described by Chun (7). Evolutionary distances, expressed as estimated changes per 100 nucleotides, were calculated from the percentages of similarity by using the correction of Jukes and Cantor (18). A dendogram was constructed with PHYLIP based on the unweighted pair group method with arithmetic averages (15). The bar represents 0.1 U of evolutionary distance.
By sequentially transferring cultures in both the presence and the absence of 2,3,5,6-CB, species that had a selective growth advantage with the congener were enriched, as indicated by differences in the community profiles. However, several RFLP types were present under both culture conditions, indicating that these species utilized alternative electron acceptors to PCB for growth. Possible mechanisms included (i) methanogenic carbon dioxide reduction by hydrogen-utilizing methanogens via interspecies hydrogen exchange with propionate- and butyrate-utilizing acetogens or acetate-dismutating species, which include low-G+C gram-positive species such as clostridia and members of the δ subgroup; (ii) dismutation of acetate by aceticlastic methanogens; (iii) fatty acid oxidation with unknown dissimilatory electron acceptors in sediment; and (iv) fatty acid oxidation with PCB as a dissimilatory electron acceptor.
To further reduce selection to growth-linked or cometabolic PCB dechlorination, enrichment cultures were initiated and sequentially transferred into totally defined sediment-free medium. Although the medium complexity was reduced, the overall community diversity was reduced only slightly and the same phylogenetic groups (the δ, low-G+C gram-positive, Thermotogales, and Dehalococcoides subgroups) were detected, indicating that most species from the initial enrichment cultures adapted to growth without sediments. Past reports have indicated that sediments were required in order to maintain microbially mediated PCB-dechlorinating activity through sequential transfers, and several possible roles for sediment in the dechlorination process are discussed by Cutter et al. (9) and Boyle et al. (5). By developing a microbial community adapted to growth in defined medium, it was possible to further reduce the complexity of the ortho-dechlorinating community systematically by eliminating or substituting components.
The influence of the carbon source on the community of PCB-dechlorinating enrichment cultures was investigated. Changing the carbon source from a mixture of butyrate, propionate, and acetate to acetate as the sole electron donor caused a dramatic shift in the microbial community. Although the growth rates observed in enrichment cultures with the mixture of fatty acids were greater than rates observed in cultures with acetate alone, the dechlorination rate was greater in enrichment cultures that contained acetate alone. It is well documented that enrichment conditions, choice of PCB congener, and source of inoculum can influence dechlorinating activities (2). However, this is the first confirmed report of the influence of an electron donor on the community profile of a PCB-dechlorinating enrichment culture.
The overall results of this study show that the defined growth conditions supported the growth of only four phylogenetic subgroups among the bacteria, i.e., the δ, low-G+C gram-positive, and Thermotogales subgroups and a single species near the deeply branching species D. ethenogenes, and two phylogenetic subgroups among the archaea, i.e., the H2-CO2 utilizing Methanomicrobiales subgroup and the methylotrophic and aceticlastic Methanosarcinales subgroup (Fig. 5). The detection of the H2-CO2-utilizing methanogens indicates that hydrogen was likely generated by fatty acid-oxidizing acetogenic bacteria. This conclusion is supported by the observation that H2-CO2-utilizing Methanomicrobiales and methylotrophic and aceticlastic Methanosarcinales subgroup species are evenly distributed when enrichment cultures are grown on a mixture of fatty acids, but Methanosarcinales species become most predominant with acetate only. However, dechlorination was observed when methanogenesis and growth of all methanogenic archaea were inhibited by BES, indicating that methanogenic archaea are not required for acetate-mediated ortho dechlorination of 2,3,5,6-CB. The slight inhibition of dechlorination with BES treatment likely resulted from nonspecific inhibition of bacterial species that were involved in dechlorination. This conclusion is further supported by the observation that vancomycin treatment also inhibited methanogenesis and methanogen growth but had only a slight effect on the rate of dechlorination. A report by May et al. indicated that colonies of PCB-enriched consortia plated on solidified media para and/or meta dechlorinated 2,3,4-CB and 2,4,5-CB in the absence of methanogenesis (24). In contrast, the same cultures lost the ability to dechlorinate 2,5,3′,4′-CB and 3,4,2′-CB concurrently with the loss of methanogenic activity. Likewise, Ye et al. (38) reported that methanogenesis occurred concurrently with process H (meta, para) dechlorination of Aroclor 1242 but that process M (meta) dechlorination occurred in the absence of methanogenesis. Results of the current study show that ortho dechlorination of 2,3,5,6-CB is catalyzed in the absence of methanogenesis. These results, in conjunction with previous reports on para and meta dechlorination of individual congeners and Aroclors, support the hypothesis that different phylogenetic groups of bacteria and archaea dechlorinate selected PCB congeners.
RFLP type 15, which had high sequence similarity to Clostridium sp., was inhibited by the addition of vancomycin but not by molybdate. Reduction in the relative abundance of RFLP type 15 by the addition of vancomycin or by the removal of sediment did not affect the rate of removal of ortho chlorines from 2,3,5,6-CB, which suggests that RFLP type 15 does not have a role in dechlorination. Following pasteurization (80°C for 1 h) of cultures containing fatty acids and sediment, ortho dechlorination ceased, further supporting the conclusion that spore-forming microbes such as Clostridium spp. are not responsible for ortho dechlorination. In contrast, para and meta dechlorination of Aroclor 1242 by Hudson River sediments was shown to be resistant to pasteurization (36). Davenport et al. have reported that archaeal and clostridial 16S sequences are predominant in microcosms that meta and para dechlorinate 2′,3,4-CB (10). However, neither of the latter two studies reported ortho dechlorination, which further supports the hypothesis that different species exhibit congener specificity.
Species most frequently associated with ortho dechlorination of 2,3,5,6-CB in the Baltimore Harbor enrichment cultures had high sequence similarities to described species of dissimilatory sulfur- and sulfate/iron-reducing bacteria. In the presence of molybdate, ortho dechlorination of 2,3,5,6-CB was inhibited. Further, with the exception of one species, all of the 16S rDNA clones frequently associated with actively dechlorinating cultures cluster with the sulfate/iron-dissimilating δ subgroup or the elemental sulfur/thiosulfate/sulfite-dissimilating low-G+C gram-positive and Thermotogales subgroups. Ye proposed that spore-forming dissimilatory sulfate-reducing bacteria were responsible for process M (meta) dechlorination, since pasteurization and ethanol treatment did not inhibit dechlorinating activity in freshwater cultures but addition of molybdate did inhibit activity (39). In addition, described species that reductively dechlorinate aromatic or aliphatic compounds also cluster with sulfate or sulfur/iron reducers in the δ subgroup (e.g., Desulfomonile tiedjei, Pelobacter sp. TT4B strain 2CP1) and with the sulfur/thiosulfate/sulfite reducers in the low-G+C gram-positive subgroup (e.g., Desulfitobacterium dehalogenans and Desulfitobacterium frappieri) (4, 8, 12, 20, 35). Although species related to the Thermotogales subgroup have not been previously implicated in reductive dechlorination, several members of this phylum are capable of S0 reduction. Another species that was detected in ortho-dechlorinating enrichment cultures had the highest sequence similarity to the deeply branching species Dehalococcoides ethenogenes, which has been described as an obligate perchloroethylene-dechlorinating species (25). The consistent detection of this species in actively PCB-ortho-dechlorinating cultures and its absence from nondechlorinating cultures present the intriguing possibility that other obligate dehalogenating species exist.
In summary, SEMM has been shown to be an effective approach for developing community profiles associated with specific PCB-dechlorinating activities in a minimal defined medium. By using this approach, we have demonstrated that highly defined ortho-dechlorinating enrichment cultures have been developed and a stable microbial community has been maintained throughout sequential transfers in minimal growth conditions. Based on nutrient requirements of known species closely related to species identified in these ortho-dechlorinating enrichment cultures, efforts are currently under way to isolate and further characterize species from the enrichment community to confirm their role in catalysis of the dechlorination process.
ACKNOWLEDGMENTS
We thank Lisa May for critical review of the manuscript.
This research was supported by Office of Naval Research Marine Environmental Quality Program grants N00014-96-1-0115 (K.S.) and N00014-96-1-0116 (H.M.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bedard D L, Quensen I J F. Microbial transformation and degradation of toxic organic chemicals. New York, N.Y: John Wiley & Sons, Inc.; 1995. Microbial reductive dechlorination of polychlorinated biphenyls; pp. 127–216. [Google Scholar]
- 3.Berkaw M, Sowers K R, May H D. Anaerobic ortho dechlorination of polychlorinated biphenyls by estuarine sediments from Baltimore Harbor. Appl Environ Microbiol. 1996;62:2534–2539. doi: 10.1128/aem.62.7.2534-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard B, Beaudet R, Villemur R, McSween G, Lépine F, Bisaillon J-G. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol. 1996;46:1010–1015. doi: 10.1099/00207713-46-4-1010. [DOI] [PubMed] [Google Scholar]
- 5.Boyle A W, Blake C K, Price W A I, May H D. Effects of polychlorinated biphenyl congener concentration and sediment supplementation on rates of methanogenesis and sediment supplementation on rates of methanogenesis and 2,3,6-trichlorobiphenyl dechlorination in an anaerobic enrichment. Appl Environ Microbiol. 1993;59:3027–3031. doi: 10.1128/aem.59.9.3027-3031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J F, Jr, Bedard D L, Brennan M J, Carnahan J C, Feng H, Wagner R E. Polychlorinated biphenyl dechlorination in aquatic sediments. Science. 1987;236:709–712. doi: 10.1126/science.236.4802.709. [DOI] [PubMed] [Google Scholar]
- 7.Chun J. Ph.D. thesis. Newcastle upon Tyne, United Kingdom: University of Newcastle upon Tyne; 1995. [Google Scholar]
- 8.Cole J R, Cascarelli A L, Mohn W W, Tiedje J M. Isolation and characterization of a novel bacterium growing via reductive dehalogenation of 2-chlorophenol. Appl Environ Microbiol. 1994;60:3536–3542. doi: 10.1128/aem.60.10.3536-3542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutter L A, Sowers K R, May H D. Microbial transformation of 2,3,5,6-tetrachlorobiphenyl under anaerobic conditions in the absence of soil or sediment. Appl Environ Microbiol. 1998;64:2966–2969. doi: 10.1128/aem.64.8.2966-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davenport G J, Champine J M, Dutta S K. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Assessment of in situ anaerobic PCB dechlorinators in a contaminated sediment consortium, abstr. Q-140; p. 479. [Google Scholar]
- 11.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWeerd K A, Mandelco L, Tanner R S, Woese C R, Suflita J M. Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium. Arch Microbiol. 1990;154:23–30. [Google Scholar]
- 13.Elberson M A. M.S. thesis. Baltimore: University of Maryland; 1996. [Google Scholar]
- 14.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Gunsalus R P, Romesser J A, Wolfe R S. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermotrophicum. Biochemistry. 1978;17:2374–2376. doi: 10.1021/bi00605a019. [DOI] [PubMed] [Google Scholar]
- 17.Holben W E, Jansson J K, Chelm B K, Tiedje J M. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 19.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krumholz L R, Sharp R, Fishbain S S. A freshwater anaerobe coupling acetate oxidation to tetrachloroethylene dehalogenation. Appl Environ Microbiol. 1996;62:4108–4113. doi: 10.1128/aem.62.11.4108-4113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leff L G, Dana J R, McArthur J V, Shimkets L J. Comparison of methods of DNA extraction from stream sediments. Appl Environ Microbiol. 1995;61:1141–1143. doi: 10.1128/aem.61.3.1141-1143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maltseva O, Oriel P. Monitoring of an alkaline 2,4,6-trichlorophenol-degrading enrichment culture by DNA fingerprinting methods and isolation of the responsible organism, haloalkaliphilic Nocardioides sp. strain M6. Appl Environ Microbiol. 1997;63:4145–4149. doi: 10.1128/aem.63.11.4145-4149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May H D, Boyle A W, Price W A I, Blake C K. Subculturing of a polychlorinated biphenyl-dechlorinating anaerobic enrichment on solid medium. Appl Environ Microbiol. 1992;58:4051–4054. doi: 10.1128/aem.58.12.4051-4054.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maymo-Gatell X, Chien Y, Gossett J M, Zinder S H. Isolation of a bacterium that reductively dechlorinated tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 26.Moré M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto M, Perkins H R. Physicochemical properties of vancomycin and iodovancomycin and their complexes with diacetyl-l-lysyl-d-alanyl-d-alanine. Biochem J. 1971;123:773–787. doi: 10.1042/bj1230773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen G J, Larsen N, Woese C R. The ribosomal database project. Nucleic Acids Res. 1991;19:2017–2021. doi: 10.1093/nar/19.suppl.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffan R J, Goksoyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor B F, Oremland R S. Depletion of adenosine triphosphate in Desulfovibrio by oxyanions of group VI elements. Curr Microbiol. 1979;3:101–103. [Google Scholar]
- 32.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl Environ Microbiol. 1996;62:4210–4215. doi: 10.1128/aem.62.11.4210-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai Y-L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 36.Williams W A. Stimulation and enrichment of two microbial polychlorinated biphenyl reductive dechlorination activities. Chemosphere. 1997;34:655–669. [Google Scholar]
- 37.Wu Q, Sowers K R, May H D. Microbial reductive dechlorination of Aroclor 1260 in anaerobic slurries of estuarine sediments. Appl Environ Microbiol. 1998;64:1052–1058. doi: 10.1128/aem.64.3.1052-1058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye D, Quensen III J F, Tiedje J M, Boyd S A. Anaerobic dechlorination of polychlorobiphenyls (Aroclor 1242) by pasteurized and ethanol-treated microorganisms from sediments. Appl Environ Microbiol. 1992;58:1110–1114. doi: 10.1128/aem.58.4.1110-1114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye D Y. Ph.D. thesis. East Lansing: Michigan State University; 1994. [Google Scholar]
- 40.Young C C, Burghoff R L, Keim L G, Minak-Bernero V, Lute J R, Hinton S M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl Environ Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]