Abstract
We examined the ability of a soil bacterium, Agrobacterium radiobacter J14a, to degrade the herbicide atrazine under a variety of cultural conditions, and we used this bacterium to increase the biodegradation of atrazine in soils from agricultural chemical distribution sites. J14a cells grown in nitrogen-free medium with citrate and sucrose as carbon sources mineralized 94% of 50 μg of [14C-U-ring]atrazine ml−1 in 72 h with a concurrent increase in the population size from 7.9 × 105 to 5.0 × 107 cells ml−1. Under these conditions cells mineralized the [ethyl-14C]atrazine and incorporated approximately 30% of the 14C into the J14a biomass. Cells grown in medium without additional carbon and nitrogen sources degraded atrazine, but the cell numbers did not increase. Metabolites produced by J14a during atrazine degradation include hydroxyatrazine, deethylatrazine, and deethyl-hydroxyatrazine. The addition of 105 J14a cells g−1 into soil with a low indigenous population of atrazine degraders treated with 50 and 200 μg of atrazine g−1 soil resulted in two to five times higher mineralization than in the noninoculated soil. Sucrose addition did not result in significantly faster mineralization rates or shorten degradation lag times. However, J14a introduction (105 cells g−1) into another soil with a larger indigenous atrazine-mineralizing population reduced the atrazine degradation lag times below those in noninoculated treatments but did not generally increase total atrazine mineralization.
Use of atrazine (2-chloro-4-ethylamino-6-isopropylamino-s-triazine) in the United States was estimated in 1991 to be 34 to 41 million kilograms annually (30). Contamination of soil from pesticide mixing, loading, storage, and rinsing at agricultural chemical dealerships in the Midwest is a concern due to potential contamination of surface water and groundwater. The U.S. Environmental Protection Agency (EPA) reported that atrazine has been found in the groundwater of approximately 25 states due to both point and nonpoint sources. In a 1987 study of Iowa public water systems, 16 of the 18 wells with detections of pesticides, including atrazine, were located within 1,000 feet of an agricultural chemical dealership (12). High levels of pesticides and nitrate in soils, surface water, and groundwater were found at 28 dealerships in Iowa, with maximum atrazine concentrations of 1,100 μg g−1 in soil, 16 μg liter−1 in surface water, and 1,500 μg liter−1 in groundwater (14), which are well above the U.S. EPA maximum contaminant level of 3 μg liter−1 for drinking water. Low-cost strategies for bioremediation of atrazine and other pesticides in soil and groundwater at agrichemical dealership sites are needed.
The degradation of atrazine occurs predominantly by biological processes, including N-dealkylation, dechlorination, and ring cleavage. Atrazine biodegradation can be initiated by N-dealkylation of the ethyl or isopropyl side chains to produce deethylatrazine (DEA) or deisopropylatrazine (DIA) (3, 6, 22–24). Dechlorination has been reported as an early step in atrazine metabolism (6, 20), and two different s-triazine hydrolase enzymes (11, 25) have been characterized. In some microorganisms, complete biodegradation of atrazine to ammonia and CO2 has been obtained (19, 28, 32). These previous studies show a wide variation in the kinetics and extent of atrazine degradation and the ability of bacteria to grow on atrazine. In addition, the stimulatory or inhibitory effects of additional carbon or nitrogen sources on atrazine degradation varies among the bacteria described in the literature. An understanding of these factors is necessary in order to improve the effectiveness of these bacteria in bioremediation applications.
Previous studies have shown that atrazine-degrading bacteria applied as single strains or as consortia can increase degradation of atrazine in soil, but these treatments vary in their effectiveness. Soil contaminated with 1,500 μg of atrazine g−1 was inoculated with the Pseudomonas strain ADP, resulting in the degradation of approximately 17% of the atrazine (19). Addition of citrate to the soil increased the degradation to 70% of the aged atrazine. Addition of a Pseudomonas strain resulted in mineralization of over 60% of 10 μg of atrazine g−1 soil in 49 days and an atrazine half-life of 1 day (33). However, the addition of the same strain to another low-pH soil and a soil with a high organic-matter content resulted in much slower atrazine degradation, with half-lives of 19 and 22 days, respectively. Mixed cultures of microorganisms have also increased the biodegradation of atrazine when added to soils containing relatively small amounts of atrazine (3, 4).
The objectives of the present study were to determine the factors that govern the atrazine biodegradation by a newly isolated bacterium and to determine its potential effectiveness in degrading atrazine in soil. We describe here the isolation and characterization of Agrobacterium radiobacter J14a, a strain which is capable of utilizing atrazine as a sole nitrogen source. Experiments were also conducted to determine the effects of secondary carbon and nitrogen substrates on atrazine degradation in culture and in soils contaminated with atrazine.
MATERIALS AND METHODS
Chemicals.
Atrazine, DEA, DIA, deethyldeisopropylatrazine (DEDIA), simazine, and cyanazine were purchased from ChemService, West Chester, Pa. Propazine was obtained from Riedel-de Haen, Seelze, Germany. Ametryne and prometon were obtained from the EPA, Research Triangle Park, N.C. All chemicals exceeded 96% purity and were used without further purification. Hydroxyatrazine (HA), deethyl-hydroxyatrazine (DEHA), and deisopropyl-hydroxyatrazine (DIHA) were obtained from Robert Lerch (USDA-ARS Cropping Systems and Water Quality Research Unit, University of Missouri, Columbia, Mo.). The [14C-U-ring]atrazine (18.96 mCi mmol−1, 98% radiochemical purity) and [N-ethyl-14C]atrazine (13.18 mCi mmol−1, 98% radiochemical purity) were purchased from Sigma Chemical Co., St. Louis, Mo.
Enrichment and isolation.
Surface soil collected from an atrazine-treated corn field near Sheldon, Nebraska, served as the inoculum for nitrogen-limited enrichment cultures. This enrichment medium, designated BMA, contains a basal minimal salts medium supplemented with 1 g each of sodium citrate and sucrose, 20 ml of vitamin solution (5 mg of thiamine, 2 mg of d-biotin, 2 mg of folic acid, 10 mg of nicotinamide, and 10 mg of pyridoxine per liter), 20 ml of trace elements solution (21), and 50 mg of atrazine in 1 liter of deionized water. After several transfers into fresh BMA liquid medium, a mixed culture was obtained. Due to production of large amounts of polysaccharide on BMA and 0.5 strength tryptic soy agar (TSA) (Difco Laboratories, Detroit, Mich.) plates, it was difficult to obtain a pure culture by streaking for isolation. We used the Percoll (Pharmacia Fine Chemicals, Piscataway, N.J.) density centrifugation method (27) to separate the organisms in the culture. Isolates from each of three bands were screened for the ability to degrade atrazine, and the atrazine-degrading isolate J14a was used in further experiments. A second atrazine-degrading bacterium was also isolated, which we do not describe in this report.
Isolate identification.
Fatty acid profile analysis of the isolate was performed by Microcheck, Inc., Northfield, Vt. Microcheck subcultured the isolate onto TSA, incubated the plates overnight at 28°C, harvested and extracted the cells, and analyzed the cellular fatty acids of the isolate by high-resolution gas chromatography. The isolate’s profile was compared for similarity to the profiles of their 1,700-strain database. The substrate utilization of J14a was examined by using Biolog plates (Biolog, Inc., Hayward, Calif.). The inoculum for the Biolog plates was grown on TSA overnight and, after inoculation, the plates were incubated for 24 h in the dark at 30°C. The substrate utilization patterns were determined by absorbance measurements with an automated microplate reader and compared with the Biolog strain database. The results obtained from the fatty acid and Biolog identification system analysis were confirmed by tests for 3-ketolactose production, carrot tumorigenesis, Kovac’s oxidase, catalase, and urease. Flagellum numbers and position were observed under a transmission electron microscope by the phosphotungstic acid negative-staining technique.
Inoculum preparation.
Unless otherwise stated, the inoculum for all of the experiments was prepared by growing bacteria in 50 ml of either BMA or 0.5-strength tryptic soy broth (TSB) for 3 days at 28°C on a rotary shaker at 120 rpm. Cultures were pelleted by centrifugation at room temperature at 7,000 × g for 10 min. Cells were rinsed twice with 20-ml aliquots of sterilized 0.0125 M phosphate buffer (pH 7.2) and quantified by plate count techniques.
Mineralization and degradation.
Mineralization of [14C-U-ring]atrazine was used to confirm the atrazine-degrading ability of the isolate. We prepared triplicate biometer flasks (Bellco, Vineland, N.J.) containing 50-μg ml−1 and 1,500 Bq of [14C-U-ring]atrazine in 50 ml of BMA. Three uninoculated controls were also monitored for 14CO2 production. At each sampling time the entire 10-ml volume of NaOH in the biometer sidearm was removed and replaced with fresh NaOH. A 3-ml aliquot of the NaOH was transferred to a scintillation vial with 15 ml of Ultima Gold scintillation cocktail (Packard Instrument Company, Meriden, Conn.) and analyzed with a Packard 1600 liquid scintillation counter (LSC). The radioactivity counted by the LSC was corrected for quenching and background radioactivity. The final degrader population in each flask was determined by plating. Additional flasks of BMA containing 50 μg of unlabeled atrazine ml−1 were monitored for atrazine concentration and metabolite production by high-pressure liquid chromatography (HPLC) and J14a cell counts. The Monod and logistic growth models (29) were used to describe atrazine mineralization:
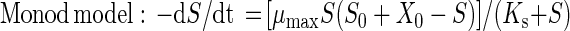 |
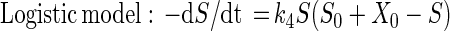 |
where S0 is the initial substrate concentration (in micrograms per milliliter), X0 is the amount of substrate (in micrograms per milliliter) required to produce the initial population density equal to B0 (the initial biomass), and X0 = B0/Y (yield). The rate constant k4 is equivalent to μmax/Ks, where μmax is the maximum specific growth rate (per hour) and Ks is the half-saturation constant (i.e., the substrate concentration [in micrograms per milliliter] when μ is half of μmax). An additional parameter, ζ, is needed to estimate S from measures of 14CO2 respired and 14C in the medium; ζ is the constant fraction of radiolabeled substrate incorporated into the cells. Parameters for these models were estimated by nonlinear regression analysis (29).
The incorporation of atrazine into J14a cells was determined with [ethyl-14C]atrazine. BMA medium was prepared with or without C sources (vitamins, sucrose, and citrate) containing 50 μg and 3,750 Bq of [ethyl-14C]atrazine ml−1 and inoculated as described previously. Three uninoculated controls were also included. The production of 14CO2 was monitored, and the 14C in the cells was sampled at 24-h intervals by removing 1 ml of medium from each inoculated flask and filtering it through a 0.22-μm (pore size) nylon filter. The filter was rinsed with 10 ml of phosphate buffer. The filter was placed in a scintillation vial with 15 ml of cocktail and counted with the LSC to determine the cell-associated radioactivity. An additional 200 μl of the medium was also removed to determine the total 14C in the medium. At the end of the experiment, approximately 5% of the radioactivity remained in the medium. This cell-free, filtered medium was passed through a cyclohexyl, 1,000-mg solid-phase extraction cartridge (United Chemical Technologies, Inc., Horsham, Pa.) to concentrate the analytes. The columns were eluted with 2 ml of methanol, and the atrazine and metabolite concentrations were determined by HPLC.
In order to determine the metabolites produced during atrazine degradation, J14a cells (approximately 1010 CFU ml−1) were incubated with atrazine, and the medium was analyzed by HPLC for metabolic products. The inoculum was grown in BMA or 0.5 strength TSB (400 ml) in a rotary shaker at 120 rpm and 28°C for 72 h. The cells were centrifuged, washed with phosphate buffer, and resuspended in BMA medium. TSB- and BMA-grown cells (2.5 ml) containing approximately 1010 CFU ml−1 each were dispensed into sterilized 25-ml glass centrifuge tubes, which were then treated with 50 μg of atrazine ml−1 (with or without [14C-U-ring]atrazine) and incubated on a rotary shaker at 120 rpm and 28°C. Four tubes for each cell type were sacrificed at 4 and 24 h. Samples were diluted to 100 ml with water, and the cells were lysed by sonication for 6 min at a 50% duty cycle. Cellular debris was pelleted by centrifugation at 5,000 rpm for 20 min. The supernatant was diluted to 250 ml with water, and 1 M KH2PO4 (pH 2.5) was added to obtain a final solution concentration of 0.05 M KH2PO4. Samples were passed through cyclohexyl (C8), 1,000-mg solid-phase extraction cartridges (United Chemical) and eluted with 3 ml of methanol to quantify the atrazine, DEA, DIA, and DEDIA. The aqueous phase was then passed through an SCX 3-ml Bond Elut cation-exchange extraction cartridge (Varian, Harbor City, Calif.) and eluted with 3 ml of 75% 0.5 M KH2PO4 (pH 7.5)–25% acetonitrile to analyze for the presence of HA, DEHA, and DIHA. Each sample was filtered through a 0.22-μm-pore-size filter into an HPLC vial. Basal minimal salts medium was spiked with 1-μg ml−1 solutions of HA, DEHA, and DIHA, to determine the extraction efficiency. Samples were sent to Robert Lerch for HPLC analysis for hydroxylated metabolite production (18). The radiolabeled samples were analyzed by Waters HPLC with a Radiomatic radioactivity detector (Packard Instruments) for atrazine and the chlorinated metabolites DEA, DIA, and DEDIA.
BMA medium was modified to assess the requirements for J14a growth and atrazine metabolism. Treatments were as follows: (i) minimal salts medium amended with the carbon sources, trace elements, and vitamins, but without any atrazine; (ii) basal minimal salts, carbon sources, trace elements, and atrazine as the sole N source (no vitamins); (iii) carbon- and nitrogen-limited medium containing only the basal minimal salts, trace elements, vitamins, and atrazine; (iv) complete BMA medium with 5 g of NH4NO3 liter−1; or (v) basal minimal salts medium, atrazine, vitamins, and trace elements, without the carbon sources. Treatments were prepared in triplicate and were incubated for 5 days on the rotary shaker at 120 rpm and 28°C in a completely randomized design. Initial and final atrazine concentrations were determined by HPLC. Initial and final populations in each flask were determined by plating.
Experiments were conducted to determine if organic nitrogen sources inhibit atrazine degradation. Mineralization of atrazine in BMA and TSB media was compared with J14a cells grown in 0.5-strength TSB medium through three successive 24-h transfers prior to inoculation. In both media, the atrazine concentration was 50 μg ml−1, including 1,500 Bq of [14C-U-ring]atrazine. Sampling occurred as described previously. The initial and final J14a populations were determined by the drop plate technique.
In order to determine if the atrazine-degrading enzymes were retained with the cells or released into the growth medium, a cell extract of J14a was prepared by growing 50 ml of J14a in BMA medium, centrifuging the culture at 7,000 × g for 15 min to pellet the intact cells, and filtering the supernatant through a 0.22-μm-pore-size disposable sterile bottle-top filtering apparatus (Corning Glass Works, Corning, N.Y.). A 5-ml portion of the cell extract was added to triplicate flasks containing 50 ml of BMA medium supplemented with 50 μg of atrazine ml−1 and 100 μg of chloramphenicol ml−1 to inhibit the growth of any cells that passed through the filter. The flasks were incubated on a rotary shaker at 120 rpm and 28°C for 120 h. Initial and final atrazine concentrations were determined by HPLC.
Another experiment was performed to determine if the atrazine-degrading enzymes are produced constitutively or are induced. BMA medium amended with 50 μg of atrazine ml−1 and 100 μg of chloramphenicol ml−1 was inoculated with J14a cells grown in 0.5-strength TSB through three successive 24-h transfers and incubated for 120 h. Concentrations of herbicide were determined by HPLC. The J14a population sizes were determined by plating.
Substrate range.
Triplicate flasks containing minimal salts medium were treated with vitamins, carbon sources, trace elements, and one of the following herbicides: atrazine, 50 μg ml−1; ametryne, 5 μg ml−1; cyanazine, 50 μg ml−1; prometon, 5 μg ml−1; propazine, 5 μg ml−1; or simazine, 1 μg ml−1. Each flask was inoculated with approximately 5 × 107 cells ml−1 of J14a and incubated on the rotary shaker for 120 h. Initial and final herbicide concentrations were determined by HPLC.
HPLC analysis.
HPLC was performed with a Waters HPLC system with a model 490E UV detector operated at 220 nm and a Nova-Pak C18, 10-cm, radially compressed column. Deionized water and acetonitrile (ACN) were used to separate atrazine, DEDIA, DIA, and DEA starting at 25% ACN, followed by a linear gradient to 75% ACN at 10 min, then returning to 25% ACN by 13 min, and holding those conditions until 15 min at 1.8 ml min−1. Calibration standards contained DEDIA, DIA, DEA, and atrazine with retention times of 1.4, 2.5, 4.1, and 9.3 min, respectively. A modified gradient was used to determine the concentrations of the additional s-triazines used in the substrate range experiment. The mobile-phase flow rate was increased to 2 ml min−1. The gradient started at 25% ACN, reached 75% ACN at 8 min, held at those conditions for 4 min, ramped down to 25% ACN–75% water by 14 min, and held at those conditions for 5 min. Retention times for simazine, cyanazine, atrazine, prometon, ametryne, and propazine were 7.0, 7.1, 7.6, 9.5, 9.9, and 10.0 min, respectively. The [14C]atrazine HPLC method starts with 25% ACN–75% water for 3 min, changing to 75% ACN by 11 min and back to 25% ACN by 16 min, and holds those conditions until 20 min at a flow rate of 1.0 ml min−1. Atrazine and metabolites were quantified by using a Packard RAM detector. Counting efficiency and background were determined with standards prepared from [14C]atrazine solutions.
Inoculation and degradation in soil.
Soil was collected at two agricultural chemical dealerships in Iowa by removing the top 5 cm of soil from locations which appeared to be impacted by the dealerships’ pesticide spills and runoff. These dealerships are code-named Alpha and Bravo, and the soils are referenced by the same names. The soil from Alpha had the following characteristics: a sandy loam texture with 75% sand, 17% silt, and 8% clay; 3.2% organic C and 0.07% total N; and a pH of 7.9 (2:1 slurry). The soil from Bravo was a loamy sand with 78% sand, 18% silt, and 4% clay; 2.4% organic C and 0.05% total N; and a pH of 6.5 (2:1 slurry). Soil at both sites consisted of mixtures of soil, sand, and limestone that had been used to maintain a surface at the site. Soils were passed through a 4-mm sieve and 50-g (dry weight basis) portions of soil were adjusted to 10% moisture in Bellco biometer flasks. Atrazine-mineralizing populations were determined with [14C-U-ring]atrazine as an N source in a most-probable-number (MPN) technique (17) prior to the start of the experiment. Each biometer was treated with atrazine solutions providing 50 or 200 μg of atrazine g−1 of soil and 3,337 Bq of [14C-U-ring]atrazine. After a thorough mixing, flasks were incubated at 25°C in the dark. After 3 days of incubation to allow for herbicide sorption to the soil, three flasks at each herbicide concentration were treated with 1% (wt/wt) sucrose, 105 J14a cells g−1 of soil, or both sucrose and J14a and incubated for 60 more days. The 14CO2 produced was measured by LSC analysis of the NaOH traps. Additional flasks treated with atrazine (but no 14C), sucrose, and J14a were prepared as described and incubated in a similar way to monitor the atrazine-mineralizing populations by the MPN technique.
Also at day 63, a 10-g subsample from each biometer flask was extracted twice with 30 ml of methanol. Methanol was added to the soil; the samples were then shaken at 190 rpm on a reciprocating shaker for 1 h, allowed to sit for 24 h, and shaken again for 30 min. The soil was removed from the methanol by centrifugation and then reextracted. Total 14C in combined methanol extracts was determined by LSC. Extracts were analyzed by HPLC to determine atrazine, DEA, DIA, and DEDIA concentrations. Duplicate 1-g aliquots of dried, ground soil were combusted with an OX500 biological oxidizer (R. J. Harvey Instrument Corporation, Hillsdale, N.J.) to determine residual 14C in the soil after extraction.
RESULTS
Identification.
J14a is a gram-negative, motile, rod-shaped bacterium with several peritrichous flagella visible under transmission electron microscopy. Fatty-acid analysis of J14a showed an excellent match (similarity index = 0.629) with A. radiobacter and a good match (similarity index = 0.450) with A. rubi. J14a also matched the Biolog profile for A. radiobacter. When grown on TSA medium, J14a produced generally round, beige, mucoid, opaque, smooth-edged colonies that became rough edged with age. J14a showed positive reactions for catalase, 3-ketolactose production, urease, and Kovac’s oxidase test. The 3-ketolactose production is unique to two biovars in the genus Agrobacterium, and this positive reaction by J14a is fairly strong proof that the isolate belongs to the genus Agrobacterium. J14a was negative for the carrot tumorigenesis test.
Metabolism and growth.
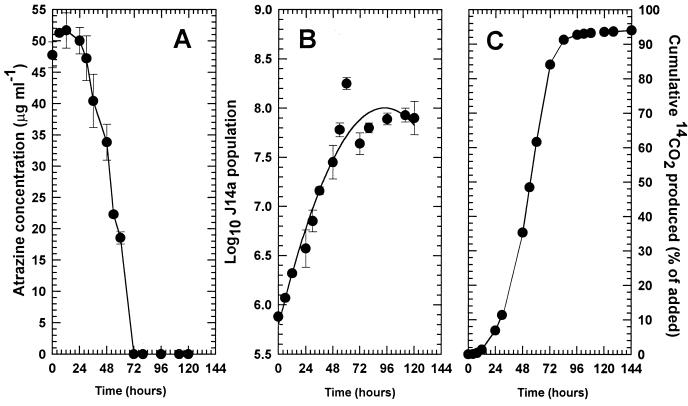
J14a mineralized approximately 94% of the [14C-U-ring]atrazine in BMA medium after a lag time of approximately 12 h (Fig. 1). Atrazine disappearance and cell growth (Fig. 1) coincide with 14CO2 appearance. Cell growth levels off at approximately 72 h, which coincides with atrazine concentration decreasing below the HPLC detection limit of 25 ng ml−1. Mineralization of [14C-U-ring]atrazine in an experiment similar to that reported in Fig. 1 (data not shown) was described slightly better by the Monod model than by the logistic model, based on residual sums of squares of 1.87 and 21.1, respectively. Parameter estimates for both models are presented in Table 1. Ring mineralization was nearly complete (94%) with little carbon incorporation into microbial biomass (estimated to be 6% at maximum), while the model value of 0.03 μg of C μg−1 substrate for ζ is equivalent to 1.5% C incorporated. Differences between the observed and predicted estimates of 14C in the biomass may be due to incomplete metabolism of atrazine or experimental error (measurement error or incomplete mass balance), because incorporation of the triazine ring C would not be expected. Other microorganisms metabolize the triazine ring to urea, then to CO2 and NH4+ (10, 28). The Monod parameter Ks was estimated to be 38 μg ml−1, but there is significant uncertainty in this estimate. This may be due to the use of supersaturated solutions of atrazine (50 μg ml−1), which exceed the water solubility of 33 μg ml−1. Above this concentration atrazine forms microprecipitates and resolubilization may limit biodegradation. However, the initial concentration estimates from the models (S0) were near the measured value of 50 μg ml−1, which suggests that the entire amount of atrazine is available to the organisms.
FIG. 1.
Degradation of 50 μg of [14C-U-ring]atrazine ml−1 in N-limited medium by strain J14a. Atrazine remaining in the medium (A) is shown in relation to cell density (B) and 14CO2 production (C).
TABLE 1.
Parameters describing kinetics of [14C-U-ring]atrazine mineralization by A. radiobacter J14a
| Model | Kinetics parameter (SD)a
|
|||||
|---|---|---|---|---|---|---|
| S0 | X0 | k4 | μmax | Ks | ζ | |
| Logistic model | 49.7 (0.5) | 0.3 (0.05) | 0.00098 (0.0003) | NA | NA | 0.03 (0.002) |
| Monod model | 50.3 (0.1) | 1.1 (0.1) | NA | 0.113 (0.008) | 37.5 (7.8) | 0.03 (0.0006) |
Parameter estimates and standard deviations are given. NA, not applicable (the model does not estimate this parameter).
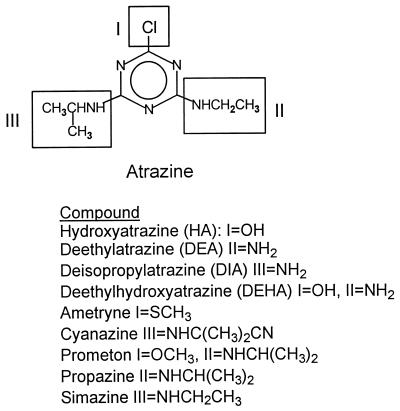
The metabolites produced by J14a during atrazine degradation were DEA, DEHA, and HA (Fig. 2, Table 2), based on retention times with detection by UV and 14C measurement. An unknown 14C-labeled compound consistently eluted after atrazine during the HPLC analysis. The compounds DIA, DEDIA, and DIHA were not detected. No apparent difference in the amount of atrazine degraded or metabolites produced was found between samples taken at 4 h versus those taken at 24 h. Cells grown in TSB produced more HA than cells grown in BMA, whereas the concentrations of DEHA and DEA produced were essentially the same for cells grown in either medium.
FIG. 2.
Structure of atrazine, atrazine metabolites, and related triazines degraded by strain J14a.
TABLE 2.
Atrazine and metabolites recovered 4 h after the addition of [14C-U-ring]atrazine to cultures of A. radiobacter J14a
| Mediuma | Amt recovered (% initial 14C [SD])b of:
|
||||
|---|---|---|---|---|---|
| Atrazine | HA | DEHA | DEA | Unknown | |
| TSB | 30.0 (0.1) | 16.2 (0.8) | 2.6 (0.1) | 3.0 (0.3) | 0.8 (1.2) |
| BMA | 25.8 (2.1) | 1.8 (0.1) | 2.1 (0.1) | 4.1 (0.2) | 1.8 (0.1) |
TSB and BMA were inoculated with ca. 1010 cells of strain J14a.
Data are means and standard deviations from triplicate flasks, expressed as the percentage of the initial 14C. Recoveries of 14C do not include losses of 14CO2.
In the medium with 50 μg of [14C-U-ring]atrazine ml−1 as the sole C and N source, only 11% of the atrazine was mineralized by J14a, populations declined slightly from 7.6 × 105 to 3.1 × 105 CFU ml−1, and 9.4 ± 0.9 μg of atrazine ml−1 remained in the medium after 120 h. Atrazine degradation and cell growth in BMA medium containing atrazine as the sole N source (no vitamins) were similar to that shown in Fig. 1. Only limited growth was obtained in medium without any N source, showing that J14a does not fix N2 under these conditions. The addition of KNO3, (NH4)2SO4, or NH4NO3 in amounts far above those required for cell growth to minimal salts medium with sucrose and 50 μg of [14C-U-ring]atrazine ml−1 resulted in growth of cells, complete loss of atrazine from the medium, and mineralization reaching approximately 60% in 90 h for all three inorganic N sources (data not shown).
Strain J14a was also able to degrade atrazine in the presence of exogenous-N-containing substrates supplied in TSB. Cells grown through three successive transfers in TSB without atrazine and then inoculated into BMA medium mineralized an average of 88% of the [14C-U-ring]atrazine and grew from 5.1 × 107 to 2.6 × 109 CFU ml−1 (data not shown), which is similar to the results shown in Fig. 1. Cells prepared in the same manner and inoculated into TSB mineralized only about 63% of the [14C]atrazine during 120 h of incubation, and 37% of the 14C remained in the medium. The J14a population grew from 1.7 × 108 to 1.9 × 109 CFU ml−1. When the medium was passed through a C8 solid-phase extraction column, only 7% of the 14C was retained, while 30% passed through the column with the aqueous phase. This indicates that polar metabolites such as HA and hydroxy N-dealkylated metabolites were produced, as these are not retained on C8 columns. This increase in hydroxylated metabolite production in TSB media is consistent with the data presented in Table 2. Total recovery of 14C was 98% of that added initially.
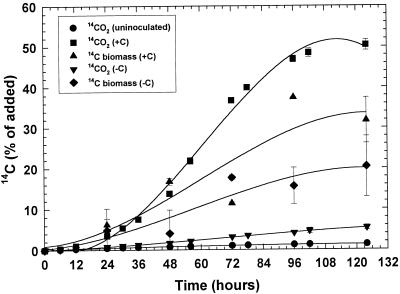
Mineralization and biomass incorporation of [ethyl-14C]atrazine was dependent upon the presence of an additional C source (sucrose) in the medium (Fig. 3). Uninoculated control flasks had an average of 1.3% of the 14C in NaOH traps, probably resulting from volatilization of atrazine. The ratio of biomass to 14CO2 incorporation was approximately 0.64 in medium with sucrose compared to 4.0 in C-limited medium without sucrose. After the experiment, samples of the media were passed through a C8 column and then through an SCX cation-exchange column to determine whether the remaining radioactivity was atrazine or metabolites. In C-amended medium the residual 14C (5% of added) was determined to be dissolved 14CO2 by release after acidification of the media. Nearly all of the 14C remaining in the C- and N-limited medium (60%) was determined to be atrazine. The average recovery of 14C as 14CO2, [14C]atrazine, metabolites remaining in the medium, and J14a-assimilated 14C was 98%, including 10% removal of 14C during sampling procedures. The J14a population in the C-amended medium grew from 7.3 × 106 to 2.8 × 108 CFU ml−1, while the population in the C- and N-limited medium remained unchanged throughout the experiment.
FIG. 3.
Mineralization and biomass incorporation of [ethyl-14C]atrazine by strain J14a in N-limited medium with or without additional carbon sources, expressed as the percentage of added 14C.
Atrazine-degrading enzymes appear to be constitutively produced by J14a and remain intracellular. TSB-grown J14a cells inoculated into BMA medium with 100 μg of chloramphenicol ml−1 decreased from 1.7 × 108 to 6.8 × 106 CFU ml−1 but still degraded 50 μg of atrazine ml−1. When TSB-grown cells were inoculated into BMA medium without chloramphenicol, complete degradation of the atrazine at 50 μg ml−1 occurred, with growth from 1.7 × 108 to 6.8 × 108 CFU ml−1. Culture supernatants did not degrade any atrazine, indicating that the atrazine-degrading enzymes are retained by the cells during degradation.
J14a degraded the herbicides ametryne (71% degraded), cyanazine (80%), prometon (62%), and simazine (100%) in N-limited medium (see Fig. 2). Metabolites of these compounds were not examined. Propazine was also degraded, but sampling problems caused by the low solubility of propazine make this result more qualitative than quantitative.
Inoculation and degradation in soil.
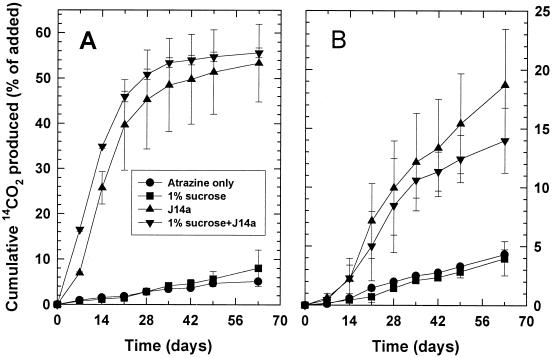
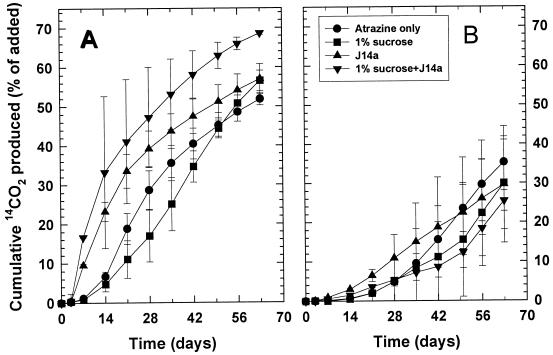
The addition of J14a to soils from the Alpha site resulted in two to five times more atrazine mineralization (significant at P ≤ 0.05) than that by the indigenous microbial community (Fig. 4). Sucrose addition had no effect on mineralization. The soils treated with 200 μg of atrazine g−1 and inoculated with J14a mineralized the atrazine continuously throughout the 63-day period, which is in contrast to the mineralization in inoculated soils treated with 50 μg of atrazine g−1 of soil, where mineralization is substantially slower after 35 days. Atrazine-mineralizing populations in the Alpha soil 63 days after atrazine addition were similar to those in the initial populations, despite the addition of 105 cells g of soil−1 at day 3 of the experiment (Table 3). Inoculation with J14a decreased both extractable atrazine and nonextractable bound residues remaining in the Alpha site soil treated with 50 μg g of atrazine−1, but it did not affect the distribution of 14C in soils treated with the higher concentration of atrazine (Table 3). Despite the differences in the fractional amount (%) of [14C]atrazine mineralized in the 50- and 200-μg g−1 treatments amended with J14a, the actual mass of atrazine mineralized was very similar in all four treatments, ranging from 27 to 38 μg g−1.
FIG. 4.
Mineralization of 50 (A) or 200 (B) μg of [14C-U-ring]atrazine ml−1 added to soil from the Alpha site. Soils were amended with strain J14a (105 cells g of soil−1), sucrose, or both J14a and sucrose 3 days after atrazine treatment.
TABLE 3.
Effect of A. radiobacter J14a and sucrose on the population of atrazine-degrading bacteria and distribution of 14C from [14C-U-ring]atrazine after 63 days in soil
| Group (atrazine concn) | MPN (cells g of soil−1)a | % Extractable atrazine (SD)b | % Bound 14C (SD)b |
|---|---|---|---|
| Alpha soil (50 μg g−1) | |||
| Atrazine only | 20 | 65.7 (8.3) a | 16.7 (1.0) b |
| 1% sucrose | 23 | 64.7 (3.4) a | 19.4 (1.1) a |
| J14a | 490 | 20.0 (2.4) b | 14.4 (1.5) c |
| 1% sucrose+J14a | 460 | 12.7 (0.3) b | 10.8 (0.4) d |
| Alpha soil (200 μg g−1) | |||
| Atrazine only | 20 | 71.5 (4.9) a | 9.0 (1.4) a |
| 1% sucrose | 45 | 69.1 (2.9) a | 7.8 (0.7) a |
| J14a | <20 | 55.9 (12.7) a | 9.9 (2.2) a |
| 1% sucrose+J14a | 20 | 60.0 (2.0) a | 8.0 (0.2) a |
| Bravo soil (50 μg g−1) | |||
| Atrazine only | 11,000 | 11.6 (2.3) a | 21.0 (3.6) a |
| 1% sucrose | 4,600 | 13.3 (4.3) a | 19.0 (3.1) a |
| J14a | 7,900 | 11.2 (1.7) a | 19.4 (2.8) a |
| 1% sucrose+J14a | 17,000 | 5.9 (1.7) a | 12.1 (5.4) b |
| Bravo soil (200 μg g−1) | |||
| Atrazine only | 2,300 | 30.9 (10.7) a | 26.4 (0.8) a |
| 1% sucrose | 54,000 | 31.0 (12.3) a | 20.1 (3.1) a |
| J14a | 28,000 | 30.8 (11.2) a | 22.1 (5.9) a |
| 1% sucrose+J14a | 49,000 | 34.4 (3.9) a | 20.8 (0.6) a |
Initial populations were 420 and 1,400 cells g of soil−1 in Alpha and Bravo soils, respectively.
Means (percentage of added 14C) of three replications with the standard deviation given in parentheses. The different letters following the standard deviations indicate the means are significantly different (P ≤ 0.05) within each soil type and atrazine concentration group.
The Bravo site soil contained an active indigenous atrazine-mineralizing population of 1.4 × 103 degraders g of soil−1, (Table 3), which rapidly mineralized [14C-U-ring]atrazine (Fig. 5). In the soils treated with 50 μg of atrazine g−1, inoculation of J14a increased the initial atrazine mineralization rate over that in soils without J14a; however, only the treatment with both J14a and sucrose mineralized a significantly larger (P < 0.05) amount of atrazine at the end of the experiment. The soils treated with 200 μg of atrazine g−1 did not show statistically significant differences in the final amounts mineralized, but the two soils amended with J14a had a more rapid initial degradation rate than those without J14a. Greater amounts of bound 14C-labeled residues were formed in the soil from the Bravo site than in the soil from the Alpha site (Table 3). Recoveries of 14C from the Alpha and Bravo soils ranged from 79 to 93%, with an average recovery of 85%.
FIG. 5.
Mineralization of 50 (A) or 200 (B) μg of [14C-U-ring]atrazine ml−1 added to soil from the Bravo site. Soils were amended with strain J14a (105 cells g of soil−1), sucrose, or both J14a and sucrose 3 days after atrazine treatment.
DISCUSSION
In N-limited media with sucrose, J14a was capable of relatively rapid growth on atrazine, with a doubling time of 8.85 h at μmax. The μmax/Ks ratio is a general indicator of substrate use efficiency. For the Monod model this ratio is 3.0 × 10−3, compared to the 9.8 × 10−4 estimated by the logistic model (k4). For comparison, the analogous Vmax/Km ratios for purified s-triazine hydrolase enzymes from Rhodococcus corallinus NRRL B15444R and Pseudomonas strain ADP are 1.25 × 10−2 and 2.93 × 10−2, respectively (11, 25). In contrast, μmax/Ks ratios for 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria ranged from 8.89 × 10−3 to 0.1 due to generally smaller Ks values (calculated from data in reference 15). These comparisons indicate that J14a is slightly less competitive for atrazine than the purified bacterial enzymes and much less competitive than the 2,4-D-degrading bacteria are for 2,4-D. Furthermore, the value of 37.5 μg ml−1 for Ks indicates that atrazine concentrations to support μmax are unlikely to occur in soil systems due to solubility constraints.
Degradation of atrazine and simultaneous production of 14CO2 from [14C-U-ring]atrazine proceeded without significant accumulation of metabolites in the medium. The simultaneous production of small quantities of HA and DEA indicates that J14a produces enzymes that perform N-dealkylation and dechlorination concomitantly. DEHA is produced by the sequential action of these enzymes. The lack of DIA and DIHA accumulation indicates either that J14a preferentially removes the ethyl over the isopropyl side chain, which is consistent with other reports (5, 24) or that removal of the isopropyl side chain is the rate-limiting step of the J14a atrazine metabolic pathway and these metabolites are utilized immediately after production. Strain J14a is similar to strains M91-3 (28) and Pseudomonas strain ADP (19) in terms of dechlorination to produce HA followed by complete mineralization of the triazine ring, but DEA production was not reported for these other strains. The complete mineralization of the triazine ring by this bacterium is more extensive than with M91-3, Pseudomonas strain ADP, or the Pseudomonas strain YAYA6 (32), which mineralized between 40 and 80% of the atrazine. The identification of J14a as an Agrobacterium species is also different from most other triazine-degrading bacteria, which have been identified as Klebsiella (10, 16), Pseudomonas (6, 19, 32), and Rhodococcus (7, 10, 25, 31) species.
Strain J14a grew in media with atrazine as a sole N source, a finding which is similar to previous reports describing s-triazine ring cleavage to NH4+ and CO2 (10, 28). J14a is also capable of degrading and utilizing atrazine under conditions of simultaneous C and N limitations. However, under the conditions of these experiments growth did not occur, despite the incorporation of the [ethyl-14C]atrazine into the cellular biomass. The Rhodococcus TE1 strain dealkylates atrazine without concurrent growth (7), but the M91-3 and Pseudomonas YAYA6 strains grow on atrazine under C-limited conditions (28, 32). Under C-limited conditions, strain J14a incorporated approximately 20% of the ethyl side chain, but very little was mineralized as 14CO2. The ability to use C in the N-ethyl and N-isopropyl groups of atrazine for population maintenance or growth would confer an advantage to atrazine-degrading bacteria under conditions where atrazine concentrations are high, such as contaminated agrichemical dealership sites.
We determined the effect of several factors on the expression of triazine-degrading activity in strain J14a. Long-term culture in the absence of atrazine and degradation in the presence of chloramphenicol show that the degradation enzymes are produced constitutively. Mandelbaum et al. (21) reported that NH4NO3 suppressed atrazine degradation by a mixed culture, and Gan et al. (13) reported that NH4 suppressed atrazine mineralization in soil. These studies suggested that exogenous N might affect degradation in J14a. However, strain J14a degrades atrazine in the presence of the inorganic nitrogen sources NH4NO3, KNO3, and (NH4)2SO4, indicating that the J14a atrazine-metabolizing enzymes are not regulated by inorganic N concentrations in the environment. Degradative activity is not completely lost upon exposure to organic N sources, such as the organic components of TSB. However, mineralization of atrazine in TSB was incomplete (only about 63%), and greater concentrations of HA were produced in TSB-grown cells relative to BMA-grown cells. We do not know the mechanisms for this effect, and other studies have not addressed the effect of secondary substrates containing organic N; however, these types of compounds would be present in soil and may have analogous effects on the biodegradation process.
J14a also demonstrated a wide substrate range among a variety of s-triazine substrates in N-limited medium, unlike the Pseudomonas sp. isolated by Yanze-Kontchou and Gschwind (32), which could not degrade cyanazine or ametryne. J14a was capable of degrading every s-triazine substrate tested, although each of the triazines tested shares either an N-ethyl, an N-isopropyl, or a Cl functional group with atrazine (Fig. 2). The broad substrate range, constitutive enzyme production, insensitivity to inorganic N, and rapid dealkylation and dechlorination capabilities of J14a indicate that this organism would be ideal for addition to a spill site with a mixture of s-triazine herbicides.
We added strain J14a to soils collected from agricultural chemical dealership sites treated with atrazine at concentrations which are representative of those found at a wide range of sites. Our studies were conducted under conditions that constitute a rigorous test of bioaugmentation with J14a. We used soils from sites nearly devoid of vegetation and thus likely to be low in easily decomposable C. Furthermore, the addition of atrazine and water 3 days prior to J14a inoculation allowed the reestablishment of competing indigenous microbial populations and the sorption of atrazine. Initial sorption reactions are complete within 24 h, although sorption increases slowly thereafter (26). Other researchers (3, 33) simultaneously applied atrazine and bacterial inoculum into air-dried agricultural soils, which may have increased atrazine availability and reduced competing microbial populations.
Strain J14a augmentation was successful in increasing biodegradation of the herbicide at both concentrations in soil from the Alpha site, but it increased biodegradation only slightly in the Bravo soil. The different responses to J14a inoculation in these two soils appear to be related to the presence of indigenous atrazine degraders and to the competitiveness of strain J14a. In soil from the Alpha site, the indigenous atrazine degrader population was low and J14a was able to increase atrazine mineralization. However, the J14a populations declined from 105 cells g of soil−1 at inoculation to below 102 cells g−1 by 60 days after inoculation. The addition of sucrose did not appear to consistently increase either biodegradation or inoculum survival. Other bioaugmentation studies used inoculum levels 20 to 10,000 times larger than the 105 cells g of soil−1 used here, and the fate of those inoculated cells was not determined (3, 19, 33). The enhanced degradation obtained from J14a in the Alpha soil suggests that effective bioaugmentation can be achieved at lower inoculum levels, particularly if survival of the inoculated strains can be promoted.
The Bravo soils had larger initial populations of atrazine-degrading microorganisms, which were effective in mineralizing atrazine, and J14a addition increased the total mineralization above these levels in only one treatment. J14a addition increased mineralization over noninoculated treatments during the 7- to 35-day period, which suggests an initial impact of inoculation. However, by the end of the experiment, the total atrazine-degrading populations (J14a and other atrazine degraders) were below the 105 J14a cells g of soil−1 added initially. We cannot distinguish between J14a populations and other atrazine degraders in the Bravo soils, but some decline in the J14a population occurred.
Irrespective of our experimental treatments, the atrazine residues remaining in soils indicate that bioremediation was incomplete at 63 days. Estimated concentrations of atrazine in the soil water are well below the Ks for J14a measured in culture. It is uncertain how much of the solvent-extractable atrazine remaining in the soil is available to the J14a cells or other atrazine degraders. As residence time in soil increases, the concentration of atrazine in the solution and water-desorbable phases will decrease, while the proportion of atrazine bound to soil organic matter or clays increases and becomes much less bioavailable (1, 2). Preincubation of atrazine in a soil with a high organic-matter content (36%) lowered the bioavailability of the atrazine and limited degradation by a Pseudomonas strain (33).
Greater amounts of bound residue were formed in the Bravo soil, especially in the 200-μg g−1 treatments, than in the soil from the Alpha site. One possible explanation of this finding is that the microorganisms indigenous to Bravo soil may produce greater extracellular concentrations of HA than the Alpha site microorganisms or J14a. Sorption of HA (log Kom = 2.0 to 2.8) to soil organic matter is greater than that of atrazine (log Kom = 1.6 to 2.0), which could result in an accumulation of unavailable radiolabeled material in the bound residue fraction (9).
Agrobacterium strain J14a was isolated from soil and is capable of rapid atrazine metabolism. The constitutive expression of degradative enzymes and activity on a range of triazine herbicides suggest that strain J14a should be an effective bioaugmentation agent. The addition of this bacterium into a soil with an indigenous population that was ineffective in degrading atrazine resulted in significant increases in biodegradation. However, the biodegradation process was not complete, and the poor survival of J14a was a contributing factor. Long-term competition and survival of the inoculum and bioavailability of the chemical are important factors affecting the effectiveness of introduced microorganisms.
ACKNOWLEDGMENTS
We thank Jeremy Long for technical support in the experimentation; Todd Anderson, Ellen Arthur, and Joel Coats for arranging site sampling; Blythe Hoyle for help with the kinetics analysis; and Robert Lerch for chemical analysis.
This work was supported by the U.S. Environmental Protection Agency.
REFERENCES
- 1.Alexander M. Biodegradation and bioremediation. San Diego, Calif: Academic Press, Inc.; 1994. Growth-linked biodegradation; pp. 10–14. [Google Scholar]
- 2.Alexander M. How toxic are toxic chemicals in soil? Environ Sci Technol. 1995;29:2713–2717. doi: 10.1021/es00011a003. [DOI] [PubMed] [Google Scholar]
- 3.Alvey S, Crowley D E. Survival and activity of an atrazine-mineralizing bacterial consortium in rhizosphere soil. Environ Sci Technol. 1996;30:1596–1603. [Google Scholar]
- 4.Assaf N A, Turco R F. Accelerated biodegradation of atrazine by a microbial consortium is possible in culture and soil. Biodegradation. 1994;5:29–35. doi: 10.1007/BF00695211. [DOI] [PubMed] [Google Scholar]
- 5.Assaf N A, Turco R F. Influence of carbon and nitrogen application on the mineralization of atrazine and its metabolites in soil. Pestic Sci. 1994;41:41–47. [Google Scholar]
- 6.Behki R M, Kahn S U. Degradation of atrazine by Pseudomonas: N-dealkylation and dehalogenation of atrazine and its metabolites. J Agric Food Chem. 1986;34:746–749. [Google Scholar]
- 7.Behki R, Topp E, Dick W, Germon P. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl Environ Microbiol. 1993;59:1955–1959. doi: 10.1128/aem.59.6.1955-1959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouquard C, Ouzzani J, Promé J-C, Michel-Briand Y, Plésiat P. Dechlorination of atrazine by a Rhizobium isolate. Appl Environ Microbiol. 1997;63:882–866. doi: 10.1128/aem.63.3.862-866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouwer W W M, Boesten J J T I, Siegers W G. Adsorption of transformation products of atrazine by soil. Weed Res. 1990;30:123–128. [Google Scholar]
- 10.Cook A M. Biodegradation of s-triazine xenobiotics. FEMS Microbiol Rev. 1987;46:93–116. [Google Scholar]
- 11.de Souza M L, Sadowsky M J, Wackett L P. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fawcett R S. Pesticides in groundwater: solving the right problem. Ground Water Monit Rev. 1989;9:5–7. [Google Scholar]
- 13.Gan J, Becker R L, Koskinen W C, Buhler D D. Degradation of atrazine in two soils as a function of concentration. J Environ Qual. 1996;25:1064–1072. [Google Scholar]
- 14.Gannon E. Site remediation: environmental clean-up of fertilizer and agri-chemical dealer sites, 28 Iowa case studies. Des Moines, Iowa: Iowa Natural Heritage Foundation; 1992. [Google Scholar]
- 15.Greer L E, Robinson J A, Shelton D R. Kinetic comparison of seven strains of 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1992;58:1027–1030. doi: 10.1128/aem.58.3.1027-1030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hapeman C J, Karns J S, Shelton D R. Total mineralization of aqueous atrazine in the presence of ammonium nitrate using ozone and Klebsiella terragena (strain DRS-I): mechanistic considerations for pilot scale disposal. J Agric Food Chem. 1995;43:1383–1391. [Google Scholar]
- 17.Jayachandran K, Stolpe N B, Moorman T B, Shea P J. Application of 14C-most-probable-number technique to enumerate atrazine-degrading microorganisms in soil. Soil Biol Biochem. 1998;30:523–529. [Google Scholar]
- 18.Lerch R N, Donald W W. Analysis of hydroxylated atrazine degradation products in water using solid-phase extraction and high-performance liquid chromatography. J Agric Food Chem. 1994;42:922–927. [Google Scholar]
- 19.Mandelbaum R T, Allan D L, Wackett L P. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandelbaum R T, Wackett L P, Allan D L. Rapid hydrolysis of atrazine to hydroxyatrazine by soil bacteria. Environ Sci Technol. 1993;27:1943–1946. [Google Scholar]
- 21.Mandelbaum R T, Wackett L P, Allan D L. Mineralization of the s-triazine by stable bacterial mixed cultures. Appl Environ Microbiol. 1993;59:1695–1701. doi: 10.1128/aem.59.6.1695-1701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masaphy S, Levanon D, Vaya J, Henis Y. Isolation and characterization of a novel atrazine metabolite produced by the fungus Pleurotus pulmonarius, 2-chloro-4-ethylamino-6-(1-hydroxyisopropyl)amino-1,3,5-triazine. Appl Environ Microbiol. 1993;59:4342–4346. doi: 10.1128/aem.59.12.4342-4346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirgain I, Green G A, Monteil H. Degradation of atrazine in laboratory microcosms: isolation and identification of the biodegrading bacteria. Environ Toxicol Chem. 1993;12:1627–1634. [Google Scholar]
- 24.Mougin C, Laugero C, Asther M, Dubroca J, Frasse P, Asther M. Biotransformation of the herbicide atrazine by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1994;60:705–708. doi: 10.1128/aem.60.2.705-708.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulbry W W. Purification and characterization of an inducible s-triazine hydrolase from Rhodococcus corallinus NRRL B-15444R. Appl Environ Microbiol. 1994;60:613–618. doi: 10.1128/aem.60.2.613-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak J M, Moorman T B, Karlen D L. Influence of soil aggregate size on atrazine sorption kinetics. J Agric Food Chem. 1994;42:1809–1812. [Google Scholar]
- 27.Putzer K P, Buchholz L A, Lidstrom M E, Remsen C C. Separation of methanotrophic bacteria by using Percoll and its application to isolation of mixed and pure cultures. Appl Environ Microbiol. 1991;57:3656–3659. doi: 10.1128/aem.57.12.3656-3659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radosevich M, Traina S J, Hao Y-L, Tuovinen O H. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol. 1995;61:297–302. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simkins S, Alexander M. Nonlinear estimation of the parameters of Monod kinetics that best describe mineralization of several substrate concentrations by dissimilar bacterial densities. Appl Environ Microbiol. 1985;50:816–824. doi: 10.1128/aem.50.4.816-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.U.S. Environmental Protection Agency. EPA’s pesticide programs. Publication no. 21T-1005. U.S. Washington, D.C: Environmental Protection Agency; 1991. [Google Scholar]
- 31.Van Zwieten L, Kennedy I R. Rapid degradation of atrazine by Rhodococcus sp. NI86/21 and by an atrazine-perfused soil. J Agric Food Chem. 1995;43:1377–1382. [Google Scholar]
- 32.Yanze-Kontchou C, Gschwind N. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl Environ Microbiol. 1994;60:4297–4302. doi: 10.1128/aem.60.12.4297-4302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanze-Kontchou C, Gschwind N. Mineralization of the herbicide atrazine in soil inoculated with a Pseudomonas strain. J Agric Food Chem. 1995;43:2291–2294. [Google Scholar]