Abstract
A study of the presence of human viruses (adenoviruses, enteroviruses, and hepatitis A viruses [HAVs]) in environmental and shellfish samples was carried out by applying DNA and cDNA amplification techniques by PCR. The detection of human adenoviruses by PCR was also examined as a potential molecular test to monitor viral pollution. The samples studied were urban and slaughterhouse sewage, river water, seawater, and shellfish. Enteroviruses were quantified by PFU in Buffalo green monkey kidney cells and fecal coliforms and phages of Bacteroides fragilis HSP40 were also evaluated in some of the samples. The amplification of viral DNA and cDNA has shown a high prevalence of human viruses that would not be detected by the use of classical techniques, such as the quantification of PFU in cell lines. The results of the analysis of slaughterhouse sewage samples together with the test of farm animal feces indicate that the adenoviruses and the HAVs detected in the environment are mostly of human origin. A significative correlation between the detection of human viruses by PCR and the values of bacteriophages of B. fragilis HSP40 in urban raw sewage was observed. Human adenoviruses were the viruses most frequently detected throughout the year, and all the samples that were positive for enteroviruses or HAVs were also positive for human adenoviruses. The results suggest that the detection of adenoviruses by PCR could be used as an index of the presence of human viruses in the environment where a molecular index is acceptable.
Human viruses are present in water contaminated by sewage. Large numbers of viruses are excreted in human feces and urine, and even at low concentrations they can cause illness when ingested. These diseases include paralysis, meningitis, respiratory disease, epidemic vomiting and diarrhea, myocarditis, congenital heart anomalies, infectious hepatitis, and eye infections. Epidemiological studies are difficult, since those infected by the virus may act as carriers but show no symptoms. The disease may become apparent only after another person has become infected, which may occur at a distance from the original source (27).
Shortcomings in microbiological quality standards have been revealed on several occasions, when viruses were isolated from drinking water supplies, freshwater, seawater, or shellfish that met the current standards of bacterial indices (14). Furthermore, there does not appear to be a correlation between the numbers of viruses present in the water and the number of fecal coliforms, which is the parameter most frequently used to establish the quality of water.
The only viral parameter that is included in European regulations governing the quality of water resources is the presence of enteroviruses, since many of them are vaccine-related polioviruses and can be isolated and quantified as PFU in cell culture (13).
The development of nucleic acid-based methods has facilitated the detection of viruses that replicate poorly or not at all in cell cultures. Amplification of viral nucleic acid by PCR is the current method of choice. PCR or reverse transcriptase PCR (RT-PCR) (to detect RNA viral genomes) is sensitive, specific, and rapid (16).
We used PCR to study the distribution of human viruses, enteroviruses, adenoviruses, and hepatitis A viruses (HAVs), in seawater, river water, sewage, and shellfish. We compared the results with the other classical parameters, the number of enterovirus PFU in cell cultures and the number of fecal coliforms, and also correlated the human viruses detected by PCR with bacteriophages of Bacteroides fragilis HSP40. The last parameter has been suggested as an indicator of viral pollution in sewage. Our data strongly suggest that the human adenovirus PCR test described here could be a molecular index for the presence of human viruses in the environment in those situations in which the verification of the infectiousness of the viruses is not necessary.
MATERIALS AND METHODS
Viruses and cells.
Adenovirus types 2 (prototype) and 12 (prototype-like) were grown on A549 cells, and poliovirus type 1 (strain LSc 2ab) was propagated in Buffalo green monkey kidney (BGM) cells growing in Eagle minimal essential medium (MEM) (Auto-pow; ICN Biomedicals Inc.) containing 5% fetal bovine serum. HAV strain HM175 was propagated in FRhK-4 cells growing in MEM supplemented with 15% fetal bovine serum. The viruses were partially purified, stored frozen at −80°C, and used in this study as positive controls.
Enterovirus and bacteriophage plaque assays.
Infectious enteroviruses from samples were grown and assayed as PFU in monolayers of BGM cells, the standard cell line used to assay environmental samples for enteroviruses (4). Briefly, cells were grown to confluent monolayers in 90- by 15-mm tissue culture dishes. Before cells were exposed to the sample, the growing medium was poured off and 1 ml of sample that had been previously decontaminated with 30% chloroform (this process was repeated if the sample presented toxicity for the cells) was inoculated onto each plastic dish. The cells were overlaid with MEM without fetal bovine serum, and 0.7% agar. After 5 days of incubation at 37°C in 5% CO2 in air, the cells were fixed with formaldehyde and stained with crystal violet, and plaques were counted.
B. fragilis HSP40, grown on Bacteroides phage recovery medium (26), was used in the quantification of B. fragilis phages. The phages were quantified by the double-agar-layer (PFU) method. Water samples were decontaminated by filtration through polyvinylidene difluoride membranes (pore size, 0.22 μm; Millipore) before quantification of phages infecting B. fragilis (26).
Bacteriological analysis.
Fecal coliforms were enumerated by standard methods (3).
Samples.
Fifteen raw domestic sewage samples were collected from the sewerage network of Barcelona, Spain, and three samples were taken after settling in a sewage purification plant (100% positive for fecal coliforms, with a mean concentration of 1.7 × 106 CFU/100 ml). Each sample was collected in a sterile 500-ml polyethylene container, kept at 4°C for less than 8 h until the viral particles were quantified by PFU or concentrated in phosphate-buffered saline (PBS), and stored frozen at −80°C.
To identify the human or nonhuman origin of the detected viruses, 17 slaughterhouse sewage samples (containing urine, feces, and intestinal contents of animals, all diluted in water) taken from different sampling points near Barcelona (mean concentration of fecal coliforms, 108 CFU/100 ml) were analyzed. Twelve samples of farm animal feces (from cows, hens, chickens, ducks, pigs, or sheep) were also analyzed after being suspended (20 g of sample) in 10% (wt/vol) phage buffer (20 mM Na2HPO4, 22 mM KH2PO4, 85 mM NaCl, 1 mM MgSO4, 0.1 mM CaCl). Each sample was collected from a different location and each was a mixture of the samples taken in a section of the farm with 5 to 8 feces samples mixed for cows, 10 to 12 for pigs or sheep, and more than 20 for hens, chickens, and ducks. All the samples were kept at 4°C for less than 8 h until their treatment and frozen storage.
Twenty 50-liter river water samples were collected from the Llobregat River at two locations and concentrated by the positively charged filters (Zeta-plus; CUNO) according to the method described by Sobsey and Jones (22) (volumes analyzed were equivalent to 2 liters of river water for human adenovirus detection and 1 liter of river water for enterovirus and HAV detection).
Twenty-three seawater samples were taken at different sites along the Barcelona coastline. Fourteen 50-liter samples were concentrated in the same way as the river water samples, and nine 500-ml samples were concentrated by a modification of the membrane-filtration swirling-elution method described by Sinton et al. (5a, 21). The previously described procedure was adapted to seawater analysis, and the treatment used with other environmental samples was also applied. Briefly, 500 ml of seawater was filtered through a nitrate and acetate cellulose membrane (0.22-μm pore size; Millipore) without any previous treatment of the sample, and the viruses were retained by electrostatic forces. Viruses were eluted by adding 2 ml of glycine buffer (0.25 N, pH 9.5), and the eluate was concentrated by the method described below. The equivalent volume of seawater analyzed by PCR was 50 ml for human adenovirus detection and 25 ml for the analysis of enterovirus and HAV. Viral concentrates of seawater were also artificially contaminated with poliovirus and analyzed for the presence of inhibitors of the RT-PCR amplification.
Six mussel samples (Mytillus galloprovincialis) and one clam sample (Mercenaria mercenaria) were obtained from shellfish growing in areas with different levels of fecal pollution. Each mussel or clam sample was a mixture of approximately 1,000 g of mussel or clam and was kept at 4°C for less than 8 h and cleaned, and 100 g of the homogenized material, corresponding to 10 to 15 mussels or 30 to 40 clams, was treated for the analysis of the different parameters. Mussels artificially seeded with poliovirus were analyzed for the presence of inhibitors of the RT-PCR amplification. The commercial mussel samples used as a negative control were obtained from the market and showed negative results in our laboratory for the studied viruses. Commercial mussels are those that theoretically comply with the bacterial standards of the European Union regulation either with or without a previous depuration treatment.
Recovery of viral particles and nucleic acid extraction.
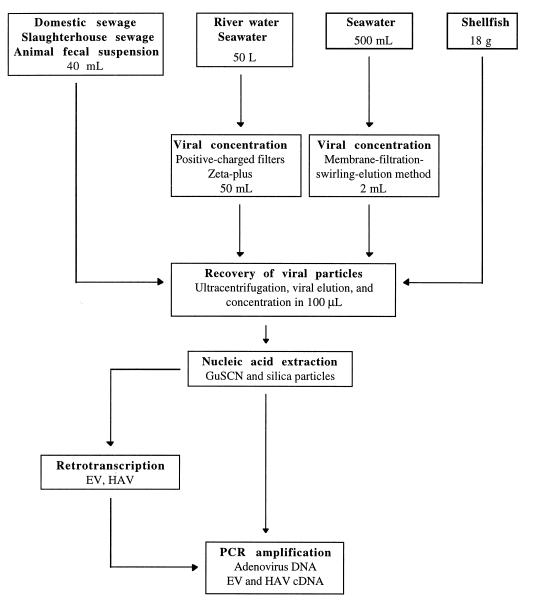
The methods used for the recovery of viral particles and the nucleic acid extraction (Fig. 1) were chosen on the basis of previous studies (8, 20). Briefly, 40 ml of sample was ultracentrifuged (229,600 × g for 1 h at 4°C) to pellet all the viral particles together with any suspended material. The sediment was eluted by mixing it with 5 ml of 0.25 N glycine buffer (pH 9.5) on ice for 30 min, and the suspended solids were separated by centrifugation (12,000 × g for 15 min) after the addition of 5 ml of 2× PBS. The viruses were then pelleted by ultracentrifugation (229,600 × g for 1 h at 4°C), resuspended in 0.1 ml of 1× PBS, and stored at −80°C (Fig. 1). Consistent with previous studies, the yield was 70%.
FIG. 1.
Procedure for the detection of adenovirus, enterovirus, and HAV in environmental samples. GuSCN, guanidinium thiocyanate; EV, enterovirus.
The method used for the recovery of viral particles from shellfish samples was a modification of the protocol for environmental water samples described above. Briefly, the outer surface was cleaned with water and ethanol (96%), shucked, and drained of excess fluid. One hundred grams of shellfish tissue was homogenized in a blender and eluted with 200 ml of glycine buffer (0.25 N, pH 10). After orbital shaking for 15 min, the solids were pelleted by centrifugation at 2,500 × g for 15 min, and the supernatant was adjusted to pH 7. The viruses were pelleted by ultracentrifugation of 8 ml of supernatant at 229,600 × g for 1 h at 4°C, resuspended in 200 μl of 1× PBS, and stored at −80°C.
Nucleic acid was extracted by the method described by Boom et al. (5), using guanidinium thiocyanate and adsorption of the nucleic acids to silica particles. This method showed the best recovery yield compared to extraction methods tested in previous studies (7, 8, 20).
Oligonucleotide primers.
The sequences (Table 1), specificities, and sensitivities of the primers used were described in previous studies (1, 2, 7, 8, 20). The specificities of the enterovirus primers was previously evaluated against 24 enterovirus strains, and the primers were shown to be able to recognize all of them. The adenovirus hexon primers used in the two-step amplification were able to detect the 47 human adenovirus serotypes. The sensitivity was checked by a limiting dilution experiment described by Puig et al. (20) and was found to be in the range of 1 to 10 viral particles after nested PCR amplification.
TABLE 1.
Oligonucleotide primers used for PCR amplification of human adenovirus, enterovirus, and HAV
| Virus type (region)a | Position | Amplification reaction | Primers | Product size (bp) | Sequence |
|---|---|---|---|---|---|
| Ad2 (hexon) | |||||
| Ad40 (hexon) | 18858–18883b | First | hexAA1885 | 301 | 5′-GCCGCAGTGGTCTTACATGCACATC-3′ |
| Ad41 (hexon) | 19136–19158b | First | hexAA1913 | 5′-CAGCACGCCGCGGATGTCAAAGT-3′ | |
| Ad2 (hexon) | 18937–18960b | Nested | nehexAA1893 | 143 | 5′-GCCACCGAGACGTACTTCAGCCTG-3′ |
| Ad2 (hexon) | 19051–19079b | Nested | nehexAA1905 | 5′-TTGTACGAGTACGCGGTATCCTCGCGGTC-3′ | |
| Polio 1 (5′NTR) | |||||
| CV B4 (5′NTR) | 64–83c | First | Ent 1d | 540 | 5′-CGGTACCTTTGTACGCCTGT-3′ |
| Polio 1 (5′NTR) | 578–597c | First | Ent 2 | 5′-ATTGTCACCATAAGCAGCCA-3′ | |
| Polio 1 (5′NTR) | 430–450c | Nested | neEnt 1 | 123 | 5′-TCCGGCCCCTGAATGCGGCTA-3′ |
| CV B4 (5′NTR) | 547–567c | Nested | neEnt 2 | 5′-GAAACACGGACACCCAAAGTA-3′ | |
| HAV (5′NTR) | 332–352 | First | HAV1 | 368 | 5′-TTGGAACGTCACCTTGCAGTG-3′ |
| HAV (5′NTR) | 680–700 | First | HAV2 | 5′-CTGAGTACCTCAGAGGCAAAC-3′ | |
| HAV (5′NTR) | 371–391 | Nested | neHAV1 | 290 | 5′-ATCTCTTTGATCTTCCACAAG-3′ |
| HAV (5′NTR) | 641–661 | Nested | neHAV2 | 5′-GAACAGTCCAGCTGTCAATGG-3′ |
Ad, adenovirus; CV, coxsackievirus.
Sequence position refers to the Ad2 hexon region.
Sequence position refers to the coxsackievirus B4 5′NTR.
Modified from Gow et al. (9).
Enzymatic amplification.
The reaction mixture for reverse transcription contained 5 μl of the nucleic acids extracted (25 μl of RNA solution for analysis of enterovirus or HAV in shellfish) plus 1.5 mM MgCl2, 1× PCR buffer II (Perkin-Elmer Roche, Inc.) containing 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 200 μM concentrations of each deoxynucleotide triphosphate and the corresponding external primer for enterovirus or HAV (0.35 μM Ent 2 or HAV2, respectively) in a total volume of 10 μl (50 μl for shellfish). The reaction mixture was incubated at 95°C for 5 min before the addition of 50 U (250 U for shellfish) of Moloney murine leukemia virus reverse transcriptase (Perkin-Elmer Roche, Inc.) and 10 U (50 U for shellfish) of RNase inhibitor (Perkin-Elmer Roche, Inc.). Temperature was cycled as follows: 30 min at 42°C and 5 min at 95°C.
For a typical one-step reaction, 10 μl of extracted viral DNA was used for adenovirus (corresponding to 4 ml of sewage sample, 2 liters of river or seawater, or 50 ml of seawater when viruses were concentrated by the filtration-swirling method), and 10 μl of the cDNA solution (corresponding to 2 ml of sewage sample, 1 liter of river or seawater, or 25 ml of seawater when viruses were concentrated by the filtration-swirling method) was used for enterovirus and HAV detection. For shellfish analysis, we used 25 μl of the DNA extracted for adenovirus or 50 μl of cDNA for the detection of enterovirus and HAV, having tested the equivalent of 3.6 g of tissue for each virus. Amplification was carried out in a 50-μl reaction mixture (100 μl of reaction mixture for shellfish) containing 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleotide triphosphate, 0.08 μM each adenovirus primer or 0.15 μM the enterovirus or HAV primer, and 2 U of Ampli Taq DNA polymerase (Perkin-Elmer Cetus). Thermal cycling of the amplification mixture was performed in a programmable heat block (Gene Amp PCR System 2400; Perkin-Elmer). In all PCR assays, the first cycle of denaturation was carried out for 4 min at 94°C. The conditions for the amplification were denaturing at 92°C for 90 s, annealing at 55°C for 90 s, and extension at 72°C for 120 s.
The external primers were used in the first 30 cycles of amplification. Then, 1 μl was added to a new batch of 50 μl of PCR mixture containing 0.16 μM each nested primer (nEnt1/nEnt2 for enterovirus, nHAV1/nHAV2 for HAV, and nehexAA1893/nehexAA1905 for adenovirus detection) in a new 30-cycle amplification. The results were analyzed by gel electrophoresis on a 2% NuSieve GTG plus 1% Seakem ME agarose gel (FMC Bioproducts, Rockland, Maine) that was stained with ethidium bromide.
Quality control of the amplification method.
To reduce the probability of sample contamination by amplified DNA molecules, standard precautions were applied in all the manipulations. Separate areas were used for reagents, treatment of samples, and manipulation of amplified samples. All the samples were analyzed twice in independent experiments, and a negative control was added every two samples (a negative control is an amplification reaction mixture with the same reagents as those in the test tubes of the samples but without the inoculum of viral nucleic acids). Treatment with restriction enzyme AluI for the amplified enteroviral cDNA and uracil-DNA-glycosylase treatments for the adenoviral tests were performed in a previous study (20), but they were not considered necessary for the routine analysis in this study. Undiluted samples and 1/10 dilutions of the nucleic acid extracts were analyzed on highly polluted samples in order to avoid false negatives because of inhibition of the reactions. This could occur in a minority of the samples, according to the positive results observed in samples supplemented with viruses.
Sequencing of the nested HAV products.
To evaluate the variability of the detected HAV strains and also as an extra control for testing the specificity of the applied technique and for testing for the absence of cross-contamination, nested HAV amplicons from half of the positive samples were sequenced. We expected to detect sequence variability compared to the HAV strain used as control. Briefly, nested product was purified by electroelution, followed by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and precipitation with ethanol (100%) and 1:10 (vol/vol) sodium acetate (3 M, pH 5.5). Both strands of the purified DNA were sequenced with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems Inc.) with Ampli Taq DNA polymerase FS according to the manufacturer’s instructions. The results were checked with the ABI PRISM 377 automated sequencer (Applied Biosystems Inc.). The sequences were compared with all the sequences of the GenBank and the EMBL with the FASTA program (version 9.0; University of Wisconsin Genetics Computer Group, Madison, Wis.).
RESULTS
Domestic sewage water samples.
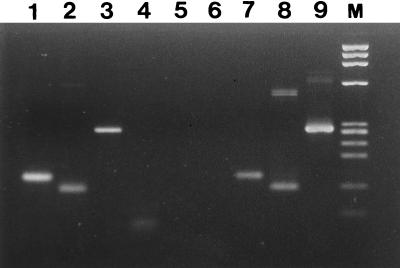
In this study, we analyzed 15 raw domestic sewage samples collected monthly in 1994 and 1995. Three effluent samples collected after settling had occurred in a primary treatment of the sewage were also analyzed (Table 2). The results obtained by nested PCR amplification showed that the human viruses most often detected were adenoviruses which were present in 14 (93%) of the raw sewage samples and in 2 of the 3 effluent samples. Enteroviruses were present in six (40%) of the influent sewage samples as indicated by nested PCR; in one of these, PFUs were detected in 20 ml of sample. The results of infectious enterovirus were probably underestimated because of the high toxicity levels of the samples. Many of the samples were toxic for the cells. None of the effluent samples was positive for enterovirus either by nested PCR or by cell culture. In terms of the other human viruses, four raw sewage samples were positive for HAV (27%), three were also positive for enterovirus and adenovirus (Fig. 2), and one was positive for adenovirus. None of the effluent samples was positive for HAV. Adenovirus type 2 was added to the unique sewage sample that showed negative results for human adenovirus by PCR, and amplified adenoviral DNA was observed. The results showed the expected sensitivity of the test (10 to 100 viral particles) to be as in a previous study (20), proving the absence of PCR inhibitors and therefore the absence of a false-negative result.
TABLE 2.
Viral parameters of sewage water samples
| Sample | PCR resulta
|
Enterovirus on cell cultureb | B. fragilis bacteriophagesc | ||
|---|---|---|---|---|---|
| HAd | HAV | EV | |||
| January | + | − | − | T | 34 |
| February | + | − | − | T | 41 |
| March | − | − | − | T | 25 |
| April | + | − | + | T | 49 |
| May | + | − | + | 14 | 209 |
| June | + | + | + | T | 34 |
| July | + | + | − | 0 | 85 |
| August | + | − | + | 0 | 160 |
| September | + | − | − | 0 | 44 |
| October | + | + | + | 0 | 33 |
| November | + | − | − | 0 | 62 |
| December | + | − | − | 0 | 74 |
| February | |||||
| Influent | + | + | + | 0 | 110 |
| Effluent | + | − | − | 0 | 63 |
| April | |||||
| Influent | + | − | − | 0 | 48 |
| Effluent | + | − | − | 0 | 139 |
| September | |||||
| Influent | + | − | − | 0 | 32 |
| Effluent | − | − | − | 0 | 23 |
Results obtained by nested PCR analysis of 4 ml of sewage for the detection of adenovirus or 2 ml for the detection of enterovirus or HAV. HAd, human adenovirus; EV, enterovirus.
Enterovirus PFU obtained from 20 ml of sewage; T, toxic effect.
Bacteriophage PFU obtained from 1 ml of sample.
FIG. 2.
Agarose gel electrophoresis showing amplified DNA after nested PCR of one sewage sample that was positive for human adenovirus, enterovirus, and HAV (lanes 1, 2, and 3, respectively). Lanes 4, 5, and 6 are the corresponding negative controls. Lanes 7, 8, and 9 are positive controls. Lane M, molecular weight standard φX174 HaeIII digest.
The results from the three samples taken at the entry and the effluent of the sewage treatment plant showed that, as expected (10), treatment does not completely remove the viruses; human adenoviruses and bacteriophages of B. fragilis HSP40 were detected in the effluent. These samples were collected in February, April, and September the year after the other sewage samples were collected.
Slaughterhouse sewage and farm animal feces samples.
The results of the seventeen slaughterhouse sewage samples analyzed are shown in Table 3. Three of the samples (numbers 9, 10, and 11) that were not expected to contain human contamination were positive for enterovirus, probably of animal origin, and none of them was positive for adenoviruses or HAV. Samples 1, 4, 5, and 6, which included some human fecal contamination (sewage from toilets is mixed with the sewage of animal origin), showed positive results for human adenoviruses, and three of them were also positive for enterovirus. None of the samples was positive for HAV.
TABLE 3.
Microbial parameters of slaughterhouse sewage samples
| Sample no. | Origin | Human fecal cross-contamination | PCR resulta
|
||
|---|---|---|---|---|---|
| HAd | HAV | EV | |||
| 1 | Pigs | Yes | + | − | + |
| 2 | Kids and calves | Yes | − | − | − |
| 3 | Sheep and calves | Yes | − | − | − |
| 4 | Chickens | Yes | + | − | − |
| 5 | Pigs | Yes | + | − | + |
| 6 | Pigs | Yes | + | − | + |
| 7 | Pigs | Yes | − | − | + |
| 8 | Pigs | Yes | − | − | + |
| 9 | Sheep and calves | No | − | − | + |
| 10 | Sheep and calves | No | − | − | + |
| 11 | Pigs, sheep, and calves | No | − | − | + |
| 12 | Pigs, sheep, and calves | No | − | − | − |
| 13 | Pigs, sheep, and calves | No | − | − | − |
| 14 | Pigs, sheep, and calves | No | − | − | − |
| 15 | Pigs, sheep, and calves | No | − | − | − |
| 16 | Pigs, sheep, and calves | No | − | − | − |
| 17 | Pigs, sheep, and calves | No | − | − | − |
Results obtained by nested PCR analysis of 4 ml of sample for the detection of adenovirus or 2 ml of sample for the detection of enterovirus or HAV. HAd, human adenovirus; EV, enterovirus.
None of the 12 fecal samples from cows, hens, chickens, ducks, pigs, or sheep was positive for any of the viruses analyzed by nested PCR, but they were taken from a small group of animals. To control the presence of inhibitors of the PCR that could be found in fecal samples from the animals, we added low levels (about 104 viral particles) of adenovirus type 2 and poliovirus type 1 to 1 aliquot of two samples of feces from pigs and one sample of feces from hens. Poliovirus type 1 was also added to one sample of chicken feces, two samples from cows, and one sample of sheep feces. The samples always showed positive results in the undiluted sample or in some cases only after dilution to 1:10.
Correlation between the presence of human viruses and bacteriophages of B. fragilis HSP40.
We compared the numbers of phages infecting B. fragilis HSP40 with the numbers of human viruses, as detected by PCR, by applying Kendall’s nonparametric test. There was a significant positive correlation (r = 0.439, P = 0.025) between the log B. fragilis phages and the results for human viruses by PCR in sewage samples. The detection of human viruses was compared with the values of bacteriophages by a quantification in a semiquantitative way. Samples were scored 0 if they were negative for viruses by PCR, 1 if they were positive for one viral parameter (adenovirus), and 2 if they were positive for adenoviruses plus enteroviruses or HAV.
River water samples.
We analyzed 23 river water samples from two sites with different levels of fecal pollution according to official data of the contamination level of the river from the Community Surveillance Program (three samples taken from site 2 in 1996 were less polluted). As in the sewage samples, the human virus most often detected was adenovirus, present in 15 of the 23 samples (65%) (Table 4). Enterovirus was detected in five samples (22%), one of which was also positive by cell culture (2 PFU in 20 ml of the viral concentrate), and we detected HAV in 10 samples (43%). More samples were found to be positive in 1996 than in 1995. All the samples positive for enterovirus or HAV were also positive for human adenoviruses.
TABLE 4.
Microbial parameters of river water samples
| Sample description (year collected) | PCR resulta
|
Enterovirus on cell cultureb | Concn of fecal coliformsc | ||
|---|---|---|---|---|---|
| HAd | HAV | EV | |||
| Site 1 (1995) | |||||
| January | + | − | − | 0 | 6,800 |
| February | + | − | − | 0 | 1,000 |
| March | + | + | − | 0 | 200 |
| May | − | − | − | 0 | 4,200 |
| June | − | − | − | 0 | 39,000 |
| August | − | − | − | 0 | 43,000 |
| September | + | + | − | 0 | 3,800 |
| October | − | − | − | 0 | 6,000 |
| November | + | − | − | 0 | 2,400 |
| December | + | + | − | 0 | 3,200 |
| Site 1 (1996) | |||||
| February | + | − | − | 0 | 4,400 |
| March | + | + | + | 0 | 10,000 |
| April | + | + | + | 2 | 1,000 |
| May | + | − | + | 0 | 2,950 |
| June | − | − | − | 0 | 2,800 |
| July | + | + | + | 0 | 16,000 |
| September | − | − | − | 0 | NTd |
| October | + | + | − | 0 | 900 |
| November | + | + | + | 0 | NT |
| December | + | + | − | 0 | NT |
| Site 2 (1996) | |||||
| January | + | + | − | 0 | <10 |
| April | − | − | − | 0 | <10 |
| July | − | − | − | 0 | <10 |
Results obtained by nested PCR analysis of 2 ml of concentrate (corresponding to 2 liters of river water) for the detection of adenovirus, and 1 ml of concentrate (corresponding to 1 liter of river water) for the detection of enterovirus or HAV. HAd, human adenovirus; EV, enterovirus.
Enterovirus PFU obtained from 20 ml of concentrate (equivalent to 20 liters of river water) on BGM cells.
Results expressed as CFU/100 ml.
NT, not tested.
Although fecal coliform concentrations of <10 CFU/100 ml were found in the three samples taken from site 2, adenovirus and HAV were detected in one of them when nested PCR amplification was applied.
Seawater samples.
None of the 14 seawater samples concentrated by positively charged filters showed positive results for any of the human viruses tested for by nested PCR or cell culture, although enteroviruses were detected by nested PCR when they were added to the concentrated sample. These samples were taken during the summer months from different points along the Catalan coast with low levels of fecal pollution (fecal coliform levels between 1 × 101 and 3.4 × 103 CFU/100 ml).
The data for the samples concentrated by the modified membrane-filtration swirling-elution method are shown in Table 5. We detected adenovirus in seven of nine samples analyzed, of which two were also positive for enterovirus and HAV, two were positive for enterovirus, and one was positive for HAV.
TABLE 5.
Microbial parameters of seawater samples
| Sample no. | PCR resulta
|
Concn of fecal coliformsb | ||
|---|---|---|---|---|
| HAd | HAV | EV | ||
| 1 | + | + | + | 16,875 |
| 2 | + | + | − | 13,800 |
| 3 | + | + | + | 11,400 |
| 4 | + | − | − | 24,540 |
| 5 | − | − | − | 430 |
| 6 | + | − | − | 220 |
| 7 | + | − | + | 330 |
| 8 | + | − | + | 160 |
| 9 | − | − | − | 100 |
Results obtained by nested PCR analysis of 50 ml of seawater for the analysis of adenovirus and 25 ml of sample for the analysis of enterovirus or HAV by the modified concentration method described by Sinton et al. (21). HAd, human adenovirus; EV, enterovirus.
Results expressed as CFU/100 ml.
Shellfish samples.
Samples were taken from three sites with different levels of fecal pollution. Sites 2 and 3 were more polluted according to the information supplied by the National Center of Aquaculture of Catalonia, which had carried out a survey of the microbiological quality of the area. None of the samples from site 1 was positive for any human virus after nested PCR amplification of 3.6 g of tissue extract. Samples from sites 2 and 3 were positive for adenovirus and enterovirus by PCR (Table 6). All the samples were toxic for BGM cells.
TABLE 6.
Microbial parameters of shellfish samples
| Sampling site and sample no. | Type of sample | PCR resulta
|
Concn of fecal coliformsb | ||
|---|---|---|---|---|---|
| HAd | HAV | EV | |||
| Site 1 | |||||
| 1 | Mussel | − | − | − | 275 |
| 2 | Mussel | − | − | − | 800 |
| 3 | Mussel | − | − | − | 55 |
| 4 | Mussel | − | − | − | 2 |
| Site 2 | |||||
| 1 | Mussel | + | − | + | 220 |
| 2 | Mussel | + | − | + | 300 |
| Site 3 | |||||
| 1 | Clam | + | − | + | >480 |
Results obtained by nested PCR analysis of 3.6 g of tissue for each virus. HAd, human adenovirus; EV, enterovirus.
CFU/1 g of tissue.
Three mussel sample extracts were artificially contaminated with polioviruses that were successfully detected, demonstrating the absence of RT-PCR inhibitors in these samples.
Seasonal distribution.
The results obtained by nested PCR analysis showed that enteric viruses were found in sewage and river water samples year-round. Adenoviruses and HAV were detected throughout the year, whereas no sewage or river water sample was positive for enteroviruses in the cooler months from November to February during the period studied (1995 and 1996).
Analysis of the sequences of the HAVs detected.
The nested amplicons of 8 of 17 positive samples for the HAV (about 50%), were sequenced. The most frequent isolates (present in 7 of 8 samples) were closely related to HAV strain HM175, showing 2 nucleotide differences in the 250 nucleotides analyzed (A-569 and A-610 in environmental strain instead of G-569 and G-610 in the positive control HAV HM175). One seawater sample showed seven nucleotide differences with the control strain; the HAV MBB strain was the most closely related, with only four nucleotide differences.
DISCUSSION
The molecular detection by PCR of adenoviruses, HAVs, and enteroviruses, when correctly applied, provides reliable data about the presence of these viruses in the environment, thus overcoming the technical limitations of the isolation of these viruses in cell culture.
In a previous study of viral prevalence in raw sewage or urban river water, Irving and Smith detected a high level of reoviruses, followed by enteroviruses and adenoviruses, when the viruses were isolated by cell culture (10). However, it is difficult to isolate many adenovirus strains in cell culture, and the slow-growing viruses are always underestimated when fast-growing enteroviruses are present (10, 24).
On the basis of our data, the number of enteroviruses isolated or detected by PFU in the environment has decreased in the last years, possibly because in Spain, as in many other countries, the oral poliovirus vaccine is administered to children at the ages of 2, 4, and 15 months, with an additional dose at the age of 6 years. The administration of the vaccine at these ages limits the distribution of the viruses in the environment in developed countries, because most of the viruses are expected to remain in disposable diapers, which are treated as solid waste. The polioviruses of the last dose of the attenuated vaccine do not normally multiply in the intestine. The number of adenovirus-positive samples is higher in this study than in previous data, mainly because the method (PCR) is more sensitive than cell culture, as we have shown in previous studies (8, 20).
Several reasons support the hypothesis that adenovirus detection by PCR is a better index for human viral contamination than detection of the presence of enteroviruses. One of the reasons is the high number of samples that are positive for human viruses but negative for enteroviruses. Human adenoviruses were detected by PCR in 14 of 15 sewage samples, whereas only 1 of 15 was positive for enterovirus by PFU and 6 of 15 were positive by PCR. Bacteriophages infecting B. fragilis HSP40 that are considered of human origin (25) were detected in all 15 sewage samples. The numbers of samples that were positive for viruses of human origin but negative for enteroviruses were even higher in other environments. A second and related reason is that adenoviruses and HAVs are more stable in various environments (including wastewater, seawater, and tapwater) than enterovirus, and they are more resistant to UV irradiation and other treatments in water purification plants (6, 10, 15, 23).
The sewage samples analyzed in the urban area of Barcelona contain high concentrations of fecal material and do not need complicated concentration procedures for the detection of the viral parameters. For this reason, we compared the numbers of bacteriophages of B. fragilis HSP40 in these samples with the numbers of viruses detected by PCR, and the significant correlation found with the levels of bacteriophages of B. fragilis HSP40 is interesting and indicates the need for further studies with a higher number of samples. The correlation between the positive results for human viruses and the values for bacteriophages of B. fragilis HSP40 in the sewage samples, both of human origin, could be evaluated in future studies since these two parameters are complementary in terms of the control of the virological quality of the environment and shellfish. Such studies would use PCR for the detection of viral nucleic acids and viral pathogens and would use bacteriophages as model microorganisms to test infectious viruses of human origin.
The study of the river water samples confirms the lack of correlation between fecal coliforms and human viruses (as determined by PCR or PFU), and samples that show a fecal coliform value of <10 CFU/100 ml have been found in this study to be positive for HAV and adenovirus.
In the first part of the study of seawater, 14 samples were concentrated by positively charged filters, as described by Sobsey and Jones (22) and as applied in previous studies (17, 19). In the second part, nine samples were concentrated by a procedure that is highly efficient for bacteriophage recovery (5a), which allows the identification of adenovirus- and enterovirus-positive seawater samples from polluted areas. The PCR method we developed was also able to identify the presence of viruses in shellfish.
The results of the tests on samples of nonhuman origin suggest that some nonhuman enteroviruses can be detected by the PCR procedure, but the adenoviruses and the HAVs detected are mostly of human origin.
The detection of the RNA viruses in the environment by PCR may be due to the fact that these viruses were recently infectious. According to Limsawat and Ohgaki, liberated RNA in wastewater could disappear in few minutes (12), and at medium- and low-level temperatures (37°C and lower), an endoribonuclease activity with different velocities which is associated with animal RNA viruses could be responsible for the inactivation of the RNA of single-stranded viruses without affecting the capsids (11, 18).
The detection of viruses in the environment by PCR, however, has important limitations: we do not know whether the detected viruses are infectious, the laboratory rules for personnel and materials must be very strict, and stringent quality controls must be applied in order to avoid false-negative or false-positive results. On the other hand, it is also clear that this approach is likely to provide a method with higher levels of specificity, sensitivity, and speed compared with the isolation of viruses in cell culture.
Although more studies need to be done, we can conclude that human adenovirus detection by PCR, because of the prevalence and stability of human adenoviruses and their mostly human origins, has very attractive features that indicate its usefulness as a molecular index of the presence of human viruses in the environment and shellfish. It is thus more indicative than enteroviruses of the presence of pathogenic human viruses such as HAV.
ACKNOWLEDGMENTS
This work was supported by the GRQ 94-1073 Generalitat de Catalunya and the research grant Ajut a la recerca 22.02.441.01-6(95) Generalitat de Catalunya.
S. Pina and M. Puig are fellows of the Generalitat de Catalunya. We thank SGAB and ATLL for cooperation in obtaining the samples. We also thank Serveis Científic-Tècnics of the University of Barcelona for their help in the sequencing of the PCR products.
REFERENCES
- 1.Allard A, Girones R, Juto P, Wadell G. Polymerase chain reaction for detection of adenovirus in stool samples. J Clin Microbiol. 1990;28:2659–2667. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard A, Albinsson B, Wadell G. Detection of adenovirus in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J Med Virol. 1992;37:149–157. doi: 10.1002/jmv.1890370214. [DOI] [PubMed] [Google Scholar]
- 3.American Public Health Association. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C: American Public Health Association; 1995. [Google Scholar]
- 4.Arribas A, Bosch A, Lucena F, Pares R. Survey of viral pollution in Duero river (Spain): occurrence of natural virucidal phenomena. Environ Int. 1988;14:37–41. [Google Scholar]
- 5.Boom R, Sol C J A, Salimans M M M, Jansen C J, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Contreras, N. Personal communication.
- 6.Enriquez C E, Hurst C J, Gerba C P. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and wastewater. Water Res. 1995;29:2548–2553. [Google Scholar]
- 7.Girones R, Allard A, Wadell G, Jofre J. Application of PCR to the detection of adenoviruses in polluted waters. Water Sci Technol. 1993;27:235–241. [Google Scholar]
- 8.Girones R, Puig M, Allard A, Lucena F, Wadell G, Jofre J. Detection of adenovirus and enterovirus by PCR amplification in polluted waters. Water Sci Technol. 1995;31:351–357. [Google Scholar]
- 9.Gow J W, Behan W M H, Clements G B, Woodall C, Riding M, Behan P O. Enteroviral RNA sequences detected by polymerase chain reaction in muscle of patients with postviral fatigue syndrome. Br Med J. 1991;302:692–696. doi: 10.1136/bmj.302.6778.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irving L G, Smith F A. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl Environ Microbiol. 1981;41:51–59. doi: 10.1128/aem.41.1.51-59.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolakofsky D, Altman S. Endoribonuclease activity associated with animal RNA viruses. J Virol. 1978;25:274–284. doi: 10.1128/jvi.25.1.274-284.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limsawat S, Ohgaki S. Fate of liberated viral RNA in wastewater determined by PCR. Appl Environ Microbiol. 1997;63:2932–2933. doi: 10.1128/aem.63.7.2932-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucena F, Schwartzbrod L, Bosch A. The effect of a mass poliomyelitis vaccination program on the occurrence of enterovirus in seawater. Zentbl Bakteriol Hyg B. 1986;183:67–69. [PubMed] [Google Scholar]
- 14.Melnick J L, Metcalf T G. Distribution of viruses in the water environment. In: Fields B, et al., editors. Genetically altered viruses and the environment. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1985. pp. 95–102. [Google Scholar]
- 15.Meng Q S, Gerba C P. Comparative inactivation of enteric adenovirus, poliovirus and coliphages by ultraviolet irradiation. Water Res. 1996;30:2665–2668. [Google Scholar]
- 16.Metcalf T G, Melnick J L, Estes M K. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology, a trip of over 50 years. Annu Rev Microbiol. 1995;49:461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- 17.Muscillo M, Aulicino F A, Patti A M, Orsini P, Volterra L, Fara G M. Molecular techniques for the identification of enteric viruses in marine waters. Water Res. 1994;28:1–7. [Google Scholar]
- 18.Newman J F E, Brown F. Foot-and-mouth disease virus and poliovirus particles contain proteins of the replication complex. J Virol. 1997;71:7657–7662. doi: 10.1128/jvi.71.10.7657-7662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patti A M, Aulicino F A, Santi A L, Muscillo M, Orsini P, Bellucci C, La Rosa G, Mastroeni I, Volterra L. Enteric virus pollution of Tyrrhenian areas. Water Air Soil Pollut. 1996;88:261–267. [Google Scholar]
- 20.Puig M, Jofre J, Lucena F, Allard A, Wadell G, Girones R. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl Environ Microbiol. 1994;60:2963–2970. doi: 10.1128/aem.60.8.2963-2970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinton L W, Finlay R K, Reid A J. A simple membrane filtration-elution method for the enumeration of F-RNA, F-DNA and somatic coliphages in 100-ml water samples. J Microbiol Methods. 1996;25:257–269. [Google Scholar]
- 22.Sobsey M D, Jones B L. Concentration of poliovirus from tap water using positively charged microporous filters. Appl Environ Microbiol. 1979;37:588–595. doi: 10.1128/aem.37.3.588-595.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobsey M D, Shields P A, Hauchman F S, Hazard R L, Caton L W. Survival and transport of hepatitis A virus in soils, groundwater and wastewater. Water Sci Technol. 1986;18:97–106. [Google Scholar]
- 24.Tani N, Dohi Y, Kurumatani N, Yonemasu K. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol Immunol. 1995;39:577–580. doi: 10.1111/j.1348-0421.1995.tb02245.x. [DOI] [PubMed] [Google Scholar]
- 25.Tartera T, Lucena F, Jofre J. Human origin of Bacteroides fragilis bacteriophages present in the environment. Appl Environ Microbiol. 1989;55:2696–2701. doi: 10.1128/aem.55.10.2696-2701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tartera C, Araujo R, Michel T, Jofre J. Culture and decontamination methods affecting enumeration of phages infecting Bacteroides fragilis in sewage. Appl Environ Microbiol. 1992;58:2670–2673. doi: 10.1128/aem.58.8.2670-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. Report of WHO Scientific Group of human viruses in water, wastewater and soil. Technical Report Series, no. 639. Geneva, Switzerland: World Health Organization; 1979. [PubMed] [Google Scholar]