Abstract
The mortality rates of invasive fungal infections remain high because of the limited number of antifungal drugs available and antifungal drug resistance, which can rapidly evolve during treatment. Mutations in key resistance genes such as ERG11 were postulated to be the predominant cause of antifungal drug resistance in the clinic. However, recent advances in whole genome sequencing have revealed that there are multiple mechanisms leading to the microevolution of resistance. In many fungal species, resistance can emerge through ERG11-independent mechanisms and through the accumulation of mutations in many genes to generate a polygenic resistance phenotype. In addition, genome sequencing has revealed that full or partial aneuploidy commonly occurs in clinical or microevolved in vitro isolates to confer antifungal resistance. This review will provide an overview of the mutations known to be selected during the adaptive microevolution of antifungal drug resistance and focus on how recent advances in genome sequencing technology have enhanced our understanding of this process.
Keywords: fungi, pathogen, antifungal drugs, resistance, microevolution, adaptation, mutator, ploidy, aneuploidy, phenotypic diversity
1. Introduction
Fungal infections pose an escalating health problem; however, their contribution to the global burden of disease remains under-recognized. It has been estimated that 1.7 billion people are infected with fungi each year, resulting in 1.5 million deaths annually from invasive infections [1,2]. Although there are more than 600 species of fungi that can cause disease in humans, more than 90% of all reported deaths result from invasive infections with the opportunistic pathogens Cryptococcus neoformans, Candida albicans, Aspergillus fumigatus and Pneumocystis jirovecii [1,3]. Other species within the Cryptococcus species complex (C. deneoformans and C. gattii), Candida genera (C. glabrata, C. tropicalis, C. parapsilosis, C. krusei and C. auris) and Aspergillus genera (A. flavus, A. terreus, A. niger and A. nidulans) also cause human invasive infections [1,2,4,5,6]. In addition, moulds in the Fusarium, Scedosporium, Mucorales and Lomentospora genera can cause life-threatening invasive infections, which, although rarer, have high mortality, because of high resistance rates or inherent resistance [7]. Other significant invasive fungal infections are restricted to endemic regions and are caused by infections with thermally dimorphic pathogenic species including Coccidioides immitis, Coccidioides posadasii, Blastomyces dermatitidis and Histoplasma capsulatum (USA); Paracoccidioides brasiliensis and Paracoccidioides lutzii (Brazil); and Talaromyces marneffei (Southeast Asia) [8,9,10]. The World Health Organisation (WHO) has identified C. neoformans, C. albicans, A. fumigatus and C. auris as a critical group of species requiring priority research development and public health action to improve responses and prevent the development of antifungal drug resistance [7]. C. glabrata, Histoplasma spp., Mucorales, Fusarium spp., C. tropicalis and C. parapsilosis are classified as high priority by the WHO, and Scedosporium spp., Lomentospora prolificans, Coccidioides spp., C. krusei, C. gattii, T. marneffei, P. jirovecii and Paracoccidioides spp. are classified as medium priority [7].
Invasive fungal infections are difficult to treat and result in a high mortality rate, often surpassing 50% and increasing to up to 90% for some species if treatment is delayed [1,7,11]. The major contributing factors to mortality are the limited number of antifungal drugs available and antifungal drug resistance, which results in ineffectual treatment or relapse [12,13,14,15,16]. Resistance has been described for every class of antifungal drugs, is common in some drug classes such as azoles and evolves rapidly during treatment [13].
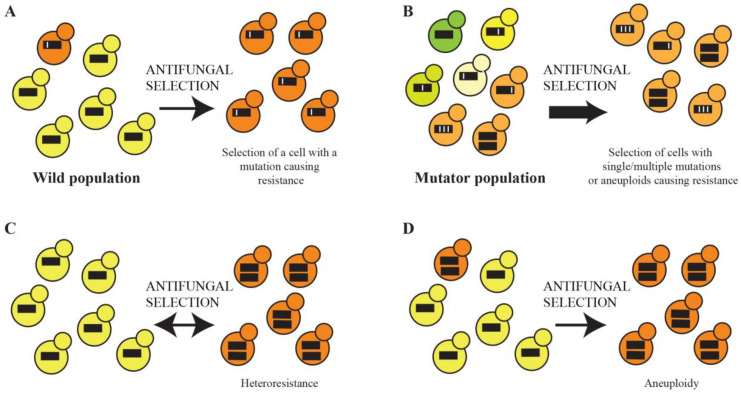
Fungal cells possessing mutations causing antifungal resistance are selected for in the clinic and become predominant in the population in a short timeframe in a process termed adaptive microevolution (Figure 1A). Adaptive microevolution is enhanced by an increased mutation rate, which provides higher genetic diversity within a population on which selection can act (Figure 1B). Isolates with elevated mutation rates, termed mutators, are associated with the enhanced evolution of antifungal resistance [17]. Recent advances in next-generation sequencing technology have revealed that resistance can emerge through single mutations in key genes or via the accumulation of mutations in many genes (polygenic), changes to the transcriptome and aneuploidy (chromosome duplications). In the presence of an antifungal, fungi can undergo a process termed heteroresistance, where transient aneuploidy occurs to confer resistance (Figure 1C). If the aneuploidy becomes permanent, these stable aneuploids are selected for in a clinical population (Figure 1D). This review will provide an overview of the mutations known to be selected during the adaptive microevolution of antifungal drug resistance and focus on how recent advances in technology have enhanced our understanding of this process.
Figure 1.
Adaptive evolution of antifungal resistance. (A). Cells in the population that possess a mutation resulting in antifungal drug resistance are selected and become predominant in the population. (B). An elevated mutation rate provides higher genetic diversity within a population on which selection for antifungal-resistant cells can occur. (C). Transient aneuploidy (heteroresistance) occurs in the presence of an antifungal to confer resistance. (D). Permanent aneuploidy-conferring antifungal resistance is selected for in a clinical population. The thick black lines represent schematic chromosomes, with white lines representing mutations.
2. Antifungal Drugs
The limited numbers of antifungal drugs and the tapered pipeline for the development of novel antifungals are widely recognised challenges for clinical mycology [1]. Many antifungal drugs can cause anaphylactic reactions or other life-threatening side effects, including renal or liver damage, and there has only been a single new class of antifungal, the echinocandins, released in the last few decades [18,19]. In addition, the use of antifungal drugs is limited by the type of administration, unfavourable drug interactions, bioavailability in target tissues and restricted activity [1,19]. There are four main classes of antifungals used to treat invasive fungal infections: azoles, polyenes, pyrimidine analogues and echinocandins. A fifth class of antifungals, allylamines, is only used for treating superficial infections [19] (Figure 2).
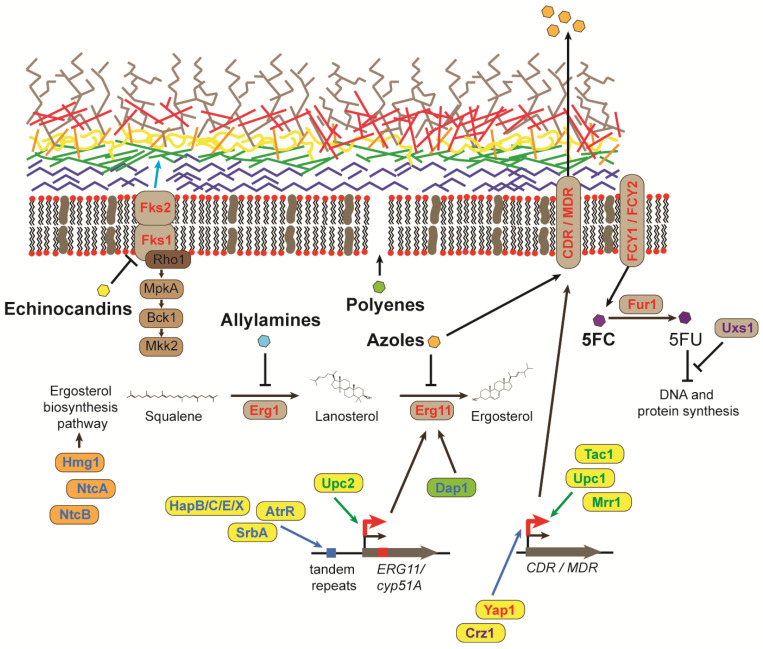
Figure 2.
Molecular mechanisms of resistance to antifungal drugs. Schematic of the fungal cell membrane and wall showing the mechanism of action of the five antifungals and the mechanisms of resistance (5FC: 5-fluorocytosine and 5FU: 5-fluorouracil). Mechanisms common to several different fungal species are indicated in red (text, arrows or boxes), those specific to A. fumigatus in blue (text and boxes), to C. neoformans in purple text and to Candida species in green text. The fungal cell wall comprises chitin (blue), ß-1,3-glucan (light green), ß1,6- glucan (orange), proteins (yellow), α-1,3-glucan (red) and galactomannans (brown). Transcription factors and proteins that regulate ergosterol biosynthesis are shown in yellow and orange, respectively. Damage Resistance Protein 1 (Dap1) complex is shown in green.
Azoles are the largest and most widely used class of antifungal agents because of their broad-spectrum activity and oral administration, which is useful in resource-limited settings [1,19]. Azole antifungals inhibit the biosynthesis of ergosterol, a crucial component of the cell membrane, which disrupts fungal growth and replication. Azoles bind the iron in the active site of the enzyme lanosterol 14 alpha-demethylase, causing a block in the ergosterol biosynthesis pathway and the accumulation of toxic sterols [19]. Similar to azoles, allylamine antifungals target ergosterol biosynthesis by inhibiting an essential enzyme, in this case, squalene epoxidase, which leads to the accumulation of squalene and increasing membrane permeability [19,20]. Polyenes are broad-spectrum antifungals that bind ergosterol in the cell membrane, inducing an extramembranous sterol sponge that destabilizes the membrane and generates membrane pores, which causes the leakage of cellular content and death [19,21,22]. However, use is limited by the need for intravenous administration and severe side effects [19]. 5-fluorocytosine (5-FC) is a pyrimidine analogue used in synergistic combination with polyene amphotericin B. 5-FC enters the fungal cell via the cytosine permease enzyme, where it is converted by cytosine deaminase into the active form, 5-fluorouracil (5-FU). 5-FU can compete with uracil to disrupt RNA and subsequent protein synthesis and can also inhibit DNA synthesis through the inhibition of thymidylate synthase. The newest class of broad-spectrum antifungals, echinocandins, inhibit the synthesis of ß-1,3-glucan in the fungal cell wall, which results in osmotic instability and cell death [23]. Use is limited because of poor absorption, a short half-life and the requirement for daily intravenous administration [19].
3. Mutations in Genes Involved in Ergosterol Biosynthesis Confer Antifungal Resistance
Resistance to azole antifungals is commonly attributed to the selection of mutants that over-express or alter the ERG11/cyp51A gene encoding the target enzyme of the ergosterol biosynthesis pathway, lanosterol 14 alpha-demethylase [24,25,26,27,28,29] (Figure 2). Single-nucleotide mutations in ERG11 have been shown to lead to resistance in C. albicans, C. glabrata, C. tropicalis, C. krusei, C. parapsilosis and C. albicans clinical isolates, which typically over-express ERG11 [24,27,28,30]. C. auris clinical isolates possess ERG11 sequence variants that account for their intrinsic multi-drug resistance to azoles [28,31]. ERG11 mutations have also been associated with fluconazole resistance in C. neoformans clinical isolates [32,33,34,35]. Mutations in other genes of the ergosterol pathway can also lead to azole resistance but are less commonly observed. In C. glabrata, the resistance of microevolved strains can occur via mutations in ERG3 and ERG4 [30]. In C. albicans, mutations in ERG3, ERG2 or ERG6 confer resistance by preventing the generation of toxic sterol intermediates; however, ERG3A and ERG3B mutants in A. fumigatus do not [36,37].
In aspergillosis patients where an azole-resistant isolate is obtained through environmental exposure, the most common mechanism resulting in resistance is the duplication of a 34 bp tandem repeat in the cyp51A (ERG11 homologue) promoter, in combination with a specific substitution (TR34/L98H) [14,38,39]. This mutation was found to be correlated with exposure to agricultural azoles and confers pan-azole resistance [39,40,41]. Agricultural use of the triazoles tebuconazole and propiconazole to control fungal diseases in plant crops has increased and results in the persistent contamination of soil, sewage and wastewater in the environment [40,41,42]. A recent surveillance study in Vietnam, where the use of fungicides is widespread and poorly regulated, showed that azole resistance occurs predominantly in isolates from cultivated soils, and 95.2% of A. fumigatus environmental isolates were resistant to at least one azole [42].
More pan-azole resistance alleles arising from various alterations to the cyp51A promoter tandem repeat have since been discovered in both the clinic and the environment in various locations around the world [39,40,43]. A recent study analysing 1190 azole-resistant A. fumigatus isolates, obtained from the environment from regions all over the globe, predominately carried the cyp51A TR34/L98H (60.7%) or TR46/Y121F/T289A (15.0%) alleles [39]. cyp51A mutations that occur during human infection are commonly missense mutations that prevent the azole from binding to the 14 α-demethylase enzyme azole target [6,25,40,44].
Genes regulating the transcription of ergosterol biosynthesis genes and ergosterol production also play a role in azole susceptibility (Figure 2). The sterol regulatory element binding protein (SREBP) pathway is required for adaptation to hypoxia and sterol homeostasis in fungi [45]. Under low oxygen, the SREBP transcription factor in C. neoformans (Sre1) and A. fumigatus (SrbA) activates genes required for ergosterol biosynthesis, and deletion results in increased susceptibility to azole antifungals [46,47,48]. SrbA has been shown to bind directly to the tandem repeats in the cyp51A promoter [6,48]. An additional transcription factor, encoded by atrR, also binds the tandem repeats in the cyp51A promoter and is required for normal tolerance to azoles [49]. The homologous gene to SRE1/srbA in C. albicans, CPH2, is not necessary for ergosterol biosynthesis [50]. Rather, in C. albicans, a different transcription factor, Upc2, regulates the expression of ergosterol biosynthesis genes [51]. The disruption of UPC2 in C. albicans increases azole susceptibility and overexpression or activating mutations cause azole resistance in vitro [51,52]. In C. albicans clinical isolates, gain-of-function UPC2 mutants contribute to the increased expression of ERG11 and fluconazole resistance [53,54,55,56]. The deletion of UPC2 in C. glabrata decreases the expression of ergosterol biosynthesis genes [57].
cyp51A expression in A. fumigatus is also regulated by a transcriptional complex containing HapB, HapC and HapE and an additional factor, HapX [38]. A mutation in hapE was initially found in an azole-resistant clinical isolate via whole genome sequencing [44]. The deletion of HapB, HapC, HapE and HapX and the expression of HapEP88L result in increased resistance to azoles [38]. HMG CoA reductase encoded by hmg1 is a sterol-sensing protein bound to the endoplasmic reticulum that initiates ergosterol biosynthesis. hmg1 mutations generated in the sterol-sensing domain or identified in clinical isolates result in altered sterol levels and azole resistance [58,59]. Mutations in the NtcA and NtcB subunits of the Negative co-factor two (Ntc2) complex, which regulates ergosterol biosynthesis, also result in pan-azole resistance [60]. Mutations in subunits of the Damage Resistance Protein 1 (Dap1) complex, which regulates cyp51a and Erg5 function, also result in resistance [61].
Similar to azoles, allylamine antifungals target ergosterol biosynthesis [20] (Figure 2). Terbinafine is an allylamine antifungal commonly used to treat dermatophyte infections. Mutations in the gene encoding the target enzyme (ERG1) confer resistance to terbinafine in clinical isolates of Trichophyton interdigitale and Trichophyton rubrum [62,63]. The introduction of the equivalent mutation in A. fumigatus and C. albicans also confers terbinafine resistance [62,64].
4. The Overexpression of Genes Encoding Efflux Pumps Confers Azole Resistance
The overexpression of MDR1 genes encoding efflux pumps of the major facilitator superfamily (MFS) or CDR genes encoding efflux pumps of the ATP-binding cassette (ABC) superfamily occurs in azole-resistant clinical isolates of C. albicans, C. parapsilosis, C. krusei, C. auris and C. glabrata [16,65,66] (Figure 2). Long-term therapy of oropharyngeal candidiasis in AIDS patients results in the constitutive expression of the CDR1, CDR2 and MDR1 genes [67,68]. The transcription factors that control the expression of efflux pumps in C. albicans and C. parapsilosis are Tac1, Mrr1 and Upc1 [69,70,71]. Another transcription factor, Cap1, cooperates with Mrr1 in C. albicans [69]. Cph1 and Mcm1 are additional negative and positive regulators of MDR1 expression, respectively [72,73]. C. albicans and C. parapsilosis gain-of-function mutations in TAC1 result in the constitutive expression of CDR1 and CDR2 in azole-resistant clinical isolates and in vitro [29,69,70,71,74]. Likewise, MRR1, UPC1 and CAP1 gain-of-function mutations result in the overexpression of MDR1 in azole-resistant clinical isolates (MRR1 and UPC1) or in vitro (CAP1) [29,53,69,70,71,75]. Mutations in TAC1 and UPC1 and the overexpression of CDR1 are also responsible for azole resistance in C. auris [76,77].
Almost all azole-resistant C. glabrata clinical isolates and those from in vitro evolution experiments possess activating mutations in the PDR1 gene, which encodes a transcription factor that induces the expression of CDR1 [27,30,78,79,80,81]. Pdr1 is regulated in part by the Hst1 deacetylase, which regulates gene expression by interacting with the mediator complex [81]. The deletion of HST1 and components of the mediator complex result in fluconazole resistance [82,83].
In C. neoformans and C. gattii, the expression of the ABC and MFS transporter genes is induced upon treatment with fluconazole [84,85]. S. cerevisiae-expressing C. gattii AFR1, AFR2 and MDR1 lead to higher resistance to fluconazole [84]. AFR1 is overexpressed in a fluconazole-resistant C. neoformans clinical isolate, and mice infected with an AFR1 mutant respond better to treatment with fluconazole [86,87]. AFR1 overexpression in a susceptible strain in vitro or gene deletion results in increased fluconazole resistance or susceptibility, respectively [85,87]. The expression of AFR1 is regulated by the CRZ1 and yap1 transcription factors [85].
The overexpression of efflux pumps encoded by atrI, cdr1B and mdr1 in A. fumigatus, occurs in azole-resistant clinical isolates and the overexpression of atrF, Afumdr1, Afumdr3 and Afumdr4 in vitro results in azole resistance [6]. Mutations in genes encoding transcription factors atrR and yap1, which regulate the expression of cdr1B and atrF, respectively, also confer resistance [88,89].
5. Mutations in Genes Encoding Glucan Synthases Result in Resistance to Echinocandin Antifungals
Resistance to echinocandins arises from mutations in the genes encoding the catalytic subunits of the target enzyme, 1,3-ß-D-glucan synthase complex, encoded by FKS genes (Figure 2). In C. albicans, C. krusei, C. auris and C. tropicalis, mutations specifically in FKS1 cause resistance [14,28,29,31,90,91]. Single-residue substitutions are commonly located in two FKS1 hot spots in C. albicans at amino acids 641–649 and 1357–1364 [92]. However, additional resistance mechanisms, yet to be identified, must also exist, as most echinocandin-resistant Candida isolates lack mutations within FKS1 [93]. In C. glabrata, resistance can be conferred by mutations in either FKS1 or FKS2 [14,30]. C. parapsilosis and Candida guilliermondii are intrinsically resistant to echinocandins because of a single nucleotide polymorphism that occurs in the FKS1 hotspot that confers resistance in other Candida species [94].
Echinocandin resistance in A. fumigatus clinical isolates is rare. One mutation in FKS1 has been found in an A. fumigatus echinocandin-resistant clinical isolate after micafungin treatment failure [95].
6. Resistance to Polyenes and the Pyrimidine Analogue 5-FC
Amphotericin B resistance is associated with reduced fitness, so, as a consequence, clinical resistance is rare despite over 50 years of use as a monotherapy to treat invasive infections [96,97]. Similar to azoles, resistance can arise through mutations in the ERG genes of the ergosterol biosynthesis pathway. Missense mutations in ERG3 and ERG6 can confer amphotericin B resistance in C. glabrata and C. auris [98,99,100]. However, most Candida amphotericin B-resistant strains have not been characterised at the gene level but rather by detecting changes in the sterol composition of membranes. The only amphotericin B-resistant C. neoformans isolate carries a mutation in ERG2 [101].
5-FC is used only in combination with amphotericin B, as resistance to 5-FC emerges frequently. Mutations in FCY1 and FCY2, permeases required for 5-FC transport, and in FUR1, a gene that encodes a uracil phosphoribosyltransferase that converts 5-FC into toxic 5-FU, confer resistance to 5-FC in C. albicans, C. glabrata, C. auris and C. neoformans [31,102]. Mutations in a gene encoding an enzyme that converts UDP-glucuronic acid into UDP-xylose (UXS1), which results in altered nucleotide metabolism, also confers resistance in C. neoformans by suppressing the toxicity of 5-FC and its derivative, 5-FU [103] (Figure 2).
7. Mutation Rate Enhances the Microevolution of Drug Resistance
The microevolution of antifungal resistance is significantly increased by an elevated mutation rate, which bestows the fungal population with higher genetic diversity upon which selection can act [17]. Strains that exhibit an elevated mutation rate, termed mutators, exhibit the rapid emergence of azole resistance in vitro in C. neoformans, Cryptococcus deuterogattii, C. glabrata and A. fumigatus [104,105,106,107]. In clinical populations, the most frequently mutated gene giving rise to a mutator phenotype is the MSH2 gene of the DNA mismatch repair (MMR) pathway, although an MLH1 variant (also in the MMR pathway) has been found in C. auris [108]. Non-synonymous variation in MSH2 has been discovered in clinical populations of C. deuterogattii, C. neoformans, C. glabrata and A. fumigatus [12,104,105,106,107,109,110,111,112,113,114]. However, the exact prevalence of MSH2 mutators in clinical populations and their clinical relevance remains controversial, as a correlation with antifungal resistance is often not found. Challenges in measuring mutation rates have led to many studies reporting sequence variance without confirming an increased mutation rate, and some MSH2 alleles in C. glabrata previously called mutators have subsequently been shown not to result in a mutator phenotype [105,111,112,113,114,115]. Nevertheless, mutations in MSH2 are strongly associated with the emergence of resistance to azoles, polyenes, pyrimidine analogues and echinocandins in vitro [104,105,106,107]. In C. glabrata, resistance to azoles, amphotericin B and echinocandins in msh2Δ mutants can arise from mutations in PDR1 (azoles), ERG6 (amphotericin B), FKS1 and FKS2 (echinocandins) [105]. Whole genome sequencing of 5-FC-resistant msh2Δ mutants in C. deuterogattii revealed mutations in FUR1, FCY2 and UXS1 [103]. Whole genome sequencing of azole and amphotericin B-resistant msh2Δ mutants in C. neoformans revealed polygenic resistance, where mutations accumulate in genes that alter stress signalling, cellular efflux, membrane trafficking and epigenetic modification [116].
8. Whole Genome Sequencing Reveals That the Microevolution of Drug Resistance Can Be Polygenic
Although mutations in single key genes such as ERG11 and PDR1 appear to be the predominant mode of azole resistance in C. albicans and C. glabrata, respectively, this is likely not the case in other pathogenic fungi, where resistance is also driven by ERG11-independent mechanisms [27,117]. Between 50 and 70% of fluconazole-resistant C. neoformans clinical isolates lack any mutations in ERG11 [26,32,33,34,35]. A recent study of the C. gattii Pacific Northwest outbreak also concluded that neither the overexpression of ERG11 nor mutations within the gene were responsible for the resistance to fluconazole in these isolates [118]. In addition, greater than 50% of A. fumigatus azole-resistant clinical isolates do not possess mutations in the regulatory or coding regions of cyp51A [6,39,65,119,120]. Recent advances in next-generation sequencing technology have enabled more studies utilizing mutational profiling to follow the emergence of antifungal drug resistance. Genome sequencing of clinical isolates over the course of infection has been performed, as well as of resistant isolates generated from in vitro microevolution experiments, where isolates are passaged in a laboratory in low concentrations of antifungals. These types of studies have revealed that resistance likely emerges through the accumulation of mutations in many genes (e.g., a polygenic phenotype).
An analysis of the mutational profiles of C. albicans clinical isolates from oral candidiasis patients revealed mutations in genes required for filamentous growth, cell adhesion, biofilm formation, cell cycle and stress, drug responses and carbohydrate binding, as well as changes in ploidy [121]. Transcriptomic analysis of the evolution of a C. glabrata clinical isolate over time from azole susceptibility to posaconazole resistance and clotrimazole resistance to fluconazole/voriconazole resistance showed that only the population with resistance to all azoles had a gain-of-function PDR1 mutation, whereas intermediate strains possessed alternative resistance mechanisms [122]. In C. auris, the mutational spectrum, coupled with an analysis of the transcriptome of fluconazole-resistant in vitro microevolved strains, suggests mutations commonly accumulate in genes encoding transcription factors (TAC1B, UPC2, ZCF18 and ZCF22), but there are a large number of different mechanisms that promote drug resistance, including changes in ploidy and multiple pathways leading to resistance, including efflux transporter upregulation and transcriptional changes in ribosome biogenesis, RNA metabolism and sugar transport [77,108,123].
Whole genome sequencing of in vitro microevolved azole-resistant msh2 (mutator) isolates in C. neoformans has shown aneuploidy and mutations accumulating in the genes of some of the same biological processes shown by transcriptomic studies to be differentially expressed in response to azole exposure, such as stress signalling, transmembrane transport, epigenetic modification, translation, transcription and carbohydrate metabolism [116,124]. Azole-resistant microevolved strains accumulated mutations in genes that encode the components required for membrane trafficking (KES1 and ALP3) and epigenetic modification (RLF2, EAF1, EAF6, YAF9 and SWC4), and the deletion of these genes resulted in fluconazole resistance [116].
In vitro microevolution experiments on voriconazole resistance in A. fumigatus showed resistant strains did not possess mutations in cyp51A, hmg1 or hapE, but transcriptomic analysis of these strains showed resistance was likely due to the overexpression of transcription factor asg1, which has been shown to regulate the expression of several ABC and MFS transporter genes and genes of the ergosterol biosynthesis pathway [125].
9. Heteroresistance Caused by Transient Aneuploidy and Permanent Aneuploidies in Clinical Isolates
Recent advances in whole genome sequencing and mutational profiling have also revealed that large-scale alterations to the genome, such as changes in ploidy (the number of chromosome sets), are a common mechanism utilized by fungi to adapt to environmental stress and generate azole resistance [121,126,127,128]. Exposure to azole antifungals has been shown to result in transient aneuploidy in C. neoformans in a process called heteroresistance [129,130]. Upon exposure to azoles, one or more aneuploidies (chromosome duplications) rapidly develop; however, normal ploidy is re-established when the azole is removed because of reduced fitness [127,129,130]. In response to increasing fluconazole concentrations in C. neoformans, chromosome 1 containing ERG11 and AFR1 is duplicated, followed by the subsequent duplication of chromosomes 4, 10 and 14 [127]. The transient aneuploidy of chromosome 1 is concomitant with increased fluconazole MIC and clinical relapse in cryptococcal meningitis patients [131].
In addition, whole genome sequencing has revealed many antifungal-resistant clinical isolates possess permanent aneuploidies. C. neoformans clinical isolates exposed to azoles commonly have disomy of chromosome 1, which results in azole resistance [127,131,132,133]. Exposure to fluconazole in vitro rapidly leads to entire or segmental disomy of chromosome 1 (92% of isolates) and chromosome 4 (36%) in combination with other disomies [134]. Aneuploidy of other chromosomes (2, 4, 6, 8–10, 12–14) has also been observed [12,131,135,136,137,138,139]. One study showed that 8.5% of clinical isolates contain a duplicated chromosome (commonly 1, 9, 12 or 14), but only 4% of these aneuploid isolates displayed azole resistance [139]. Partial and full chromosomal duplications in clinical populations reduce fitness in vivo [139]. In total, 43.75% (7/16) of azole-resistant A. fumigatus chronic pulmonary aspergillosis clinical isolates, which do not possess a mutation in cyp51A, display aneuploidy of chromosomal regions containing genes associated with azole resistance, cyp51A, cyp51B or cyp51ec, as well as those encoding MFS and ABC transporters and transcription factors [120].
In C. albicans clinical isolates, azole resistance can arise from large genome rearrangements, including the translocations of chromosomal arms; the duplication of the chromosomal region of the left arm of chr5 containing ERG11 and TAC1 to produce an isochromosome (i(5L)); trisomies of chr3, chr4, chr5 or chr7; and loss-of-heterozygosity in the chromosomal regions containing TAC1 (chr5) and MRR1 (chr3); and the formation of new chromosomes via the duplication of segments containing a centromere and the addition of telomeric ends [27,75,121,126,140,141,142,143,144,145,146,147,148]. Long repeat sequences drive the plasticity of the C. albicans genome; for example, the recombination of a long-inverted repeat sequence at the centromere of chr5 is required for the formation of i(5L) [146,149]. Resistance can be attributed to the increased gene dosage of CDR1, CDR2, CRZ1 (transcription factor) and MRR1 on chr3 and TAC1 or ERG11 on chr5 [146]. One study predicted that at least 50% of fluconazole-resistant isolates are aneuploid [140]. Recently, a study by [150] showed that different concentrations of fluconazole can select for different genotypic outcomes. Lineages of C. albicans that evolved in fluconazole concentrations close to the MIC50 of their ancestor acquired aneuploidies and copy number variations, whereas lineages evolved above the ancestral MIC50 acquired mutational changes [150]. In C. albicans, resistance to posaconazole generated through in vitro experimental evolution also results in aneuploidy and cross-tolerance to fluconazole [145]. Loss of chr5 or combined trisomy of the right arm and monosomy of the left arm of chr5 also leads to caspofungin resistance in C. albicans [151,152].
Azole-resistant strains selected directly in vitro or during microevolution experiments also gain permanent aneuploidies [153]. A. flavus strains, selected for voriconazole resistance, contain duplications of chromosome 8 or a segmental duplication of chromosome 3, which contains atrA but no cyp51A mutations [154]. Numerous recent studies on C. auris that sequenced and compared the genomes of parental fluconazole-susceptible strains and experimentally evolved fluconazole-resistant strains showed the rapid generation of the aneuploidies of chromosome 5 (which contains the regulator TAC1B) or 3, segmental aneuploidy of chromosome 1 (contains ERG11), loss of subtelomeric regions, karyotype alterations and the generation of supernumerary chromosomes (centromere-inclusive chromosomal duplications of segments of chromosome 5) [77,108,123,155]. Aneuploids are also commonly found in microevoution experiments on fluconazole resistance in C. glabrata [30]. Fluconazole-resistant C. neoformans MSH2 strains possess permanent aneuploidies of chromosomes 1 and 4 [116].
10. Conclusions
There is a critical need for the development of novel antifungals given the restricted number available, limitations on use and the emergence of resistance. Resistance is rapidly becoming an important issue that will worsen without the introduction of new antifungals for use in the clinic. Mutations in key resistance genes such as ERG11 were once thought to be the predominant cause of antifungal drug resistance. However, the recent use of whole genome sequencing has shown that the microevolution of resistance is far more complicated, and there is still a long way to go to understand this process. Although mutations in single genes such as ERG11 and PDR1 are the predominant cause of resistance in Candida species, a large proportion of clinical isolates of other fungal species lack ERG11-dependent resistance mechanisms and instead possess accumulated mutations in many genes in order to generate a polygenic resistance phenotype. Currently, it is impossible to determine the precise contribution, if any, of every mutational change observed in the genomes of resistant strains. Sexual outcrossing is not possible for most pathogenic fungi, meaning that the association between mutations and resistance phenotypes is difficult to analyse. Their phenotypic contribution to resistance could be confirmed by regenerating the mutation in an antifungal-susceptible strain using gene editing technology; however, this process would be unrealistic to perform for such a large number of mutations. In addition, these complex mutational profiles are coupled with highly plastic genomes—where aneuploidy is rapidly generated either transiently or permanently—and transcriptional changes, which must be separated from adaptive responses. Understanding the many factors contributing to the emergence of resistance is crucial for the development of effective future treatment strategies.
Data Availability Statement
Not applicable. All articles included in the review are openly available in the NIH National Library of Medicine (https://pubmed.ncbi.nlm.nih.gov/).
Conflicts of Interest
The author declares no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Park B.J., Wannemuehler K.A., Marston B.J., Govender N., Pappas P.G., Chiller T.M. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 3.Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi. 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slavin M.A., Chakrabarti A. Opportunistic fungal infections in the Asia-Pacific region. Med. Mycol. 2012;50:18–25. doi: 10.3109/13693786.2011.602989. [DOI] [PubMed] [Google Scholar]
- 5.Wisplinghoff H., Bischoff T., Tallent S.M., Seifert H., Wenzel R.P., Edmond M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 6.Arastehfar A., Carvalho A., Houbraken J., Lombardi L., Garcia-Rubio R., Jenks J.D., Rivero-Menendez O., Aljohani R., Jacobsen I.D., Berman J., et al. Aspergillus fumigatus and aspergillosis: From basics to clinics. Stud. Mycol. 2021;100:100115. doi: 10.1016/j.simyco.2021.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organisation . WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. World Health Organization; Geneva, Switzerland: 2022. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 8.Knox K.S., Hage C.A. Histoplasmosis. Proc. Am. Thorac. Soc. 2010;7:169–172. doi: 10.1513/pats.200907-069AL. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Martinez R., Mendez-Tovar L.J. Blastomycosis. Clin. Dermatol. 2012;30:565–572. doi: 10.1016/j.clindermatol.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen C., Barker B.M., Hoover S., Nix D.E., Ampel N.M., Frelinger J.A., Orbach M.J., Galgiani J.N. Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clin. Microbiol. Rev. 2013;26:505–525. doi: 10.1128/CMR.00005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam H.S., Jeon K., Um S.W., Suh G.Y., Chung M.P., Kim H., Kwon O.J., Koh W.J. Clinical characteristics and treatment outcomes of chronic necrotizing pulmonary aspergillosis: A review of 43 cases. Int. J. Infect. Dis. 2010;14:e479–e482. doi: 10.1016/j.ijid.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Rhodes J., Beale M.A., Vanhove M., Jarvis J.N., Kannambath S., Simpson J.A., Ryan A., Meintjes G., Harrison T.S., Fisher M.C., et al. A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection. G3. 2017;7:1165–1176. doi: 10.1534/g3.116.037499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coelho C., Casadevall A. Cryptococcal therapies and drug targets: The old, the new and the promising. Cell. Microbiol. 2016;18:792–799. doi: 10.1111/cmi.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamaletsou M.N., Walsh T.J., Sipsas N.V. Invasive fungal infections in patients with hematological malignancies: Emergence of resistant pathogens and new antifungal therapies. Turk. J. Haematol. 2018;35:1–11. doi: 10.4274/tjh.2018.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lestrade P.P., Bentvelsen R.G., Schauwvlieghe A., Schalekamp S., van der Velden W., Kuiper E.J., van Paassen J., van der Hoven B., van der Lee H.A., Melchers W.J.G., et al. Voriconazole resistance and mortality in invasive aspergillosis: A multicenter retrospective cohort study. Clin. Infect. Dis. 2019;68:1463–1471. doi: 10.1093/cid/ciy859. [DOI] [PubMed] [Google Scholar]
- 16.Beardsley J., Halliday C.L., Chen S.C., Sorrell T.C. Responding to the emergence of antifungal drug resistance: Perspectives from the bench and the bedside. Future Microbiol. 2018;13:1175–1191. doi: 10.2217/fmb-2018-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyce K.J. Mutators enhance adaptive micro-evolution in pathogenic microbes. Microorganisms. 2022;10:442. doi: 10.3390/microorganisms10020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyriakidis I., Tragiannidis A., Munchen S., Groll A.H. Clinical hepatotoxicity associated with antifungal agents. Expert Opin. Drug Saf. 2017;16:149–165. doi: 10.1080/14740338.2017.1270264. [DOI] [PubMed] [Google Scholar]
- 19.Campoy S., Adrio J.L. Antifungals. Biochem. Pharmacol. 2017;133:86–96. doi: 10.1016/j.bcp.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Shafiei M., Peyton L., Hashemzadeh M., Foroumadi A. History of the development of antifungal azoles: A review on structures, SAR, and mechanism of action. Bioorg. Chem. 2020;104:104240. doi: 10.1016/j.bioorg.2020.104240. [DOI] [PubMed] [Google Scholar]
- 21.Anderson T.M., Clay M.C., Cioffi A.G., Diaz K.A., Hisao G.S., Tuttle M.D., Nieuwkoop A.J., Comellas G., Maryum N., Wang S., et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol. 2014;10:400–406. doi: 10.1038/nchembio.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcon-Gonzalez J.M., Jimenez-Dominguez G., Ortega-Blake I., Carrillo-Tripp M. Multi-phase solvation model for biological membranes: Molecular action mechanism of amphotericin B. J. Chem. Theory Comput. 2017;13:3388–3397. doi: 10.1021/acs.jctc.7b00337. [DOI] [PubMed] [Google Scholar]
- 23.Maligie M.A., Selitrennikoff C.P. Cryptococcus neoformans resistance to echinocandins: (1,3)beta-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 2005;49:2851–2856. doi: 10.1128/AAC.49.7.2851-2856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morio F., Loge C., Besse B., Hennequin C., Le Pape P. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: New substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 2010;66:373–384. doi: 10.1016/j.diagmicrobio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Meis J.F., Chowdhary A., Rhodes J.L., Fisher M.C., Verweij P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150460. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodero L., Mellado E., Rodriguez A.C., Salve A., Guelfand L., Cahn P., Cuenca-Estrella M., Davel G., Rodriguez-Tudela J.L. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 2003;47:3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whaley S.G., Rogers P.D. Azole Resistance in Candida glabrata. Curr. Infect. Dis. Rep. 2016;18:41. doi: 10.1007/s11908-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 28.Bidaud A.L., Chowdhary A., Dannaoui E. Candida auris: An emerging drug resistant yeast—A mini-review. J. Mycol. Med. 2018;28:568–573. doi: 10.1016/j.mycmed.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Sitterle E., Coste A.T., Obadia T., Maufrais C., Chauvel M., Sertour N., Sanglard D., Puel A., D’Enfert C., Bougnoux M.E. Large-scale genome mining allows identification of neutral polymorphisms and novel resistance mutations in genes involved in Candida albicans resistance to azoles and echinocandins. J. Antimicrob. Chemother. 2020;75:835–848. doi: 10.1093/jac/dkz537. [DOI] [PubMed] [Google Scholar]
- 30.Ksiezopolska E., Schikora-Tamarit M.A., Beyer R., Nunez-Rodriguez J.C., Schuller C., Gabaldon T. Narrow mutational signatures drive acquisition of multidrug resistance in the fungal pathogen Candida glabrata. Curr. Biol. 2021;31:5314–5326.e10. doi: 10.1016/j.cub.2021.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes J., Abdolrasouli A., Farrer R.A., Cuomo C.A., Aanensen D.M., Armstrong-James D., Fisher M.C., Schelenz S. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 2018;7:43. doi: 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sionov E., Chang Y.C., Garraffo H.M., Dolan M.A., Ghannoum M.A., Kwon-Chung K.J. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14alpha-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 2012;56:1162–1169. doi: 10.1128/AAC.05502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selb R., Fuchs V., Graf B., Hamprecht A., Hogardt M., Sedlacek L., Schwarz R., Idelevich E.A., Becker S.L., Held J., et al. Molecular typing and in vitro resistance of Cryptococcus neoformans clinical isolates obtained in Germany between 2011 and 2017. Int. J. Med. Microbiol. 2019;309:151336. doi: 10.1016/j.ijmm.2019.151336. [DOI] [PubMed] [Google Scholar]
- 34.Bosco-Borgeat M.E., Mazza M., Taverna C.G., Cordoba S., Murisengo O.A., Vivot W., Davel G. Amino acid substitution in Cryptococcus neoformans lanosterol 14-alpha-demethylase involved in fluconazole resistance in clinical isolates. Rev. Argent. Microbiol. 2016;48:137–142. doi: 10.1016/j.ram.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Gago S., Serrano C., Alastruey-Izquierdo A., Cuesta I., Martin-Mazuelos E., Aller A.I., Gomez-Lopez A., Mellado E. Molecular identification, antifungal resistance and virulence of Cryptococcus neoformans and Cryptococcus deneoformans isolated in Seville, Spain. Mycoses. 2017;60:40–50. doi: 10.1111/myc.12543. [DOI] [PubMed] [Google Scholar]
- 36.Kelly S.L., Lamb D.C., Baldwin B.C., Corran A.J., Kelly D.E. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 1997;272:9986–9988. doi: 10.1074/jbc.272.15.9986. [DOI] [PubMed] [Google Scholar]
- 37.Alcazar-Fuoli L., Mellado E., Garcia-Effron G., Buitrago M.J., Lopez J.F., Grimalt J.O., Cuenca-Estrella J.M., Rodriguez-Tudela J.L. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: Role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 2006;50:453–460. doi: 10.1128/AAC.50.2.453-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gsaller F., Hortschansky P., Furukawa T., Carr P.D., Rash B., Capilla J., Muller C., Bracher F., Bowyer P., Haas H., et al. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog. 2016;12:e1005775. doi: 10.1371/journal.ppat.1005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burks C., Darby A., Gomez Londono L., Momany M., Brewer M.T. Azole-resistant Aspergillus fumigatus in the environment: Identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog. 2021;17:e1009711. doi: 10.1371/journal.ppat.1009711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chowdhary A., Sharma C., Kathuria S., Hagen F., Meis J.F. Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. J. Antimicrob. Chemother. 2014;69:555–557. doi: 10.1093/jac/dkt397. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhary A., Kathuria S., Xu J., Meis J.F. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013;9:e1003633. doi: 10.1371/annotation/4ffcf1da-b180-4149-834c-9c723c5dbf9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duong T.N., Le T.V., Tran K.H., Nguyen P.T., Nguyen B.T., Nguyen T.A., Nguyen H.P., Nguyen B.T., Fisher M.C., Rhodes J., et al. Azole-resistant Aspergillus fumigatus is highly prevalent in the environment of Vietnam, with marked variability by land use type. Environ. Microbiol. 2021;23:7632–7642. doi: 10.1111/1462-2920.15660. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M., Feng C.L., Chen F., He Q., Su X., Shi Y. Triazole resistance in Aspergillus fumigatus clinical isolates obtained in Nanjing, China. Chin. Med. J. (Engl.) 2017;130:665–668. doi: 10.4103/0366-6999.201609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camps S.M., Dutilh B.E., Arendrup M.C., Rijs A.J., Snelders E., Huynen M.A., Verweij P.E., Melchers W.J. Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS ONE. 2012;7:e50034. doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bien C.M., Espenshade P.J. Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell. 2010;9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y.C., Bien C.M., Lee H., Espenshade P.J., Kwon-Chung K.J. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 47.Chun C.D., Liu O.W., Madhani H.D. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willger S.D., Puttikamonkul S., Kim K.H., Burritt J.B., Grahl N., Metzler L.J., Barbuch R., Bard M., Lawrence C.B., Cramer R.A., Jr. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul S., Stamnes M., Thomas G.H., Liu H., Hagiwara D., Gomi K., Filler S.G., Moye-Rowley W.S. AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. mBio. 2019;10:e02563-18. doi: 10.1128/mBio.02563-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lane S., Di Lena P., Tormanen K., Baldi P., Liu H. Function and regulation of Cph2 in Candida albicans. Eukaryot. Cell. 2015;14:1114–1126. doi: 10.1128/EC.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacPherson S., Akache B., Weber S., De Deken X., Raymond M., Turcotte B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 2005;49:1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasicek E.M., Berkow E.L., Flowers S.A., Barker K.S., Rogers P.D. UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot. Cell. 2014;13:933–946. doi: 10.1128/EC.00221-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunkel N., Liu T.T., Barker K.S., Homayouni R., Morschhauser J., Rogers P.D. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell. 2008;7:1180–1190. doi: 10.1128/EC.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heilmann C.J., Schneider S., Barker K.S., Rogers P.D., Morschhauser J. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 2010;54:353–359. doi: 10.1128/AAC.01102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoot S.J., Smith A.R., Brown R.P., White T.C. An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 2011;55:940–942. doi: 10.1128/AAC.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flowers S.A., Barker K.S., Berkow E.L., Toner G., Chadwick S.G., Gygax S.E., Morschhauser J., Rogers P.D. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell. 2012;11:1289–1299. doi: 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whaley S.G., Caudle K.E., Vermitsky J.P., Chadwick S.G., Toner G., Barker K.S., Gygax S.E., Rogers P.D. UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob. Agents Chemother. 2014;58:4543–4554. doi: 10.1128/AAC.02217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Losada L., Sugui J.A., Eckhaus M.A., Chang Y.C., Mounaud S., Figat A., Joardar V., Pakala S.B., Pakala S., Venepally P., et al. Genetic analysis using an isogenic mating pair of Aspergillus fumigatus identifies azole resistance genes and lack of MAT locus’s role in virulence. PLoS Pathog. 2015;11:e1004834. doi: 10.1371/journal.ppat.1004834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rybak J.M., Ge W., Wiederhold N.P., Parker J.E., Kelly S.L., Rogers P.D., Fortwendel J.R. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in Aspergillus fumigatus. mBio. 2019;10:e00437-19. doi: 10.1128/mBio.00437-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furukawa T., van Rhijn N., Fraczek M., Gsaller F., Davies E., Carr P., Gago S., Fortune-Grant R., Rahman S., Gilsenan J.M., et al. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat. Commun. 2020;11:427. doi: 10.1038/s41467-019-14191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song J., Zhai P., Zhang Y., Zhang C., Sang H., Han G., Keller N.P., Lu L. The Aspergillus fumigatus damage resistance protein family coordinately regulates ergosterol biosynthesis and azole susceptibility. mBio. 2016;7:e01919-15. doi: 10.1128/mBio.01919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osborne C.S., Leitner I., Favre B., Ryder N.S. Amino acid substitution in Trichophyton rubrum squalene epoxidase associated with resistance to terbinafine. Antimicrob. Agents Chemother. 2005;49:2840–2844. doi: 10.1128/AAC.49.7.2840-2844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh A., Masih A., Khurana A., Singh P.K., Gupta M., Hagen F., Meis J.F., Chowdhary A. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses. 2018;61:477–484. doi: 10.1111/myc.12772. [DOI] [PubMed] [Google Scholar]
- 64.Rocha E.M., Gardiner R.E., Park S., Martinez-Rossi N.M., Perlin D.S. A Phe389Leu substitution in ergA confers terbinafine resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2006;50:2533–2536. doi: 10.1128/AAC.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraczek M.G., Bromley M., Buied A., Moore C.B., Rajendran R., Rautemaa R., Ramage G., Denning D.W., Bowyer P. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013;68:1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- 66.Wasi M., Khandelwal N.K., Moorhouse A.J., Nair R., Vishwakarma P., Bravo Ruiz G., Ross Z.K., Lorenz A., Rudramurthy S.M., Chakrabarti A., et al. ABC transporter genes show upregulated expression in drug-resistant clinical isolates of Candida auris: A Genome-wide characterization of ATP-Binding Cassette (ABC) transporter genes. Front. Microbiol. 2019;10:1445. doi: 10.3389/fmicb.2019.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanglard D., Kuchler K., Ischer F., Pagani J.L., Monod M., Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 1995;39:2378–2386. doi: 10.1128/AAC.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez-Ribot J.L., McAtee R.K., Lee L.N., Kirkpatrick W.R., White T.C., Sanglard D., Patterson T.F. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 1998;42:2932–2937. doi: 10.1128/AAC.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schubert S., Barker K.S., Znaidi S., Schneider S., Dierolf F., Dunkel N., Aid M., Boucher G., Rogers P.D., Raymond M., et al. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob. Agents Chemother. 2011;55:2212–2223. doi: 10.1128/AAC.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berkow E.L., Manigaba K., Parker J.E., Barker K.S., Kelly S.L., Rogers P.D. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob. Agents Chemother. 2015;59:5942–5950. doi: 10.1128/AAC.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morio F., Pagniez F., Besse M., Gay-Andrieu F., Miegeville M., Le Pape P. Deciphering azole resistance mechanisms with a focus on transcription factor-encoding genes TAC1, MRR1 and UPC2 in a set of fluconazole-resistant clinical isolates of Candida albicans. Int. J. Antimicrob. Agents. 2013;42:410–415. doi: 10.1016/j.ijantimicag.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 72.Riggle P.J., Kumamoto C.A. Transcriptional regulation of MDR1, encoding a drug efflux determinant, in fluconazole-resistant Candida albicans strains through an Mcm1p binding site. Eukaryot. Cell. 2006;5:1957–1968. doi: 10.1128/EC.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo H.J., Tseng K.Y., Kao Y.Y., Tsao M.Y., Lo H.L., Yang Y.L. Cph1p negatively regulates MDR1 involved in drug resistance in Candida albicans. Int. J. Antimicrob. Agents. 2015;45:617–621. doi: 10.1016/j.ijantimicag.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 74.Coste A.T., Karababa M., Ischer F., Bille J., Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell. 2004;3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dunkel N., Blass J., Rogers P.D., Morschhauser J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 2008;69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rybak J.M., Munoz J.F., Barker K.S., Parker J.E., Esquivel B.D., Berkow E.L., Lockhart S.R., Gade L., Palmer G.E., White T.C., et al. Mutations in TAC1B: A novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio. 2020;11:e00365-20. doi: 10.1128/mBio.00365-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carolus H., Pierson S., Munoz J.F., Subotic A., Cruz R.B., Cuomo C.A., Van Dijck P. Genome-wide analysis of experimentally evolved Candida auris Reveals multiple novel mechanisms of multidrug resistance. mBio. 2021;12:e0333-20. doi: 10.1128/mBio.03333-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vermitsky J.P., Edlind T.D. Azole resistance in Candida glabrata: Coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 2004;48:3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai H.F., Krol A.A., Sarti K.E., Bennett J.E. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 2006;50:1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khalifa H.O., Arai T., Majima H., Watanabe A., Kamei K. Genetic basis of azole and echinocandin resistance in clinical Candida glabrata in Japan. Antimicrob. Agents Chemother. 2020;64:e00783-20. doi: 10.1128/AAC.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simonicova L., Moye-Rowley W.S. Functional information from clinically-derived drug resistant forms of the Candida glabrata Pdr1 transcription factor. PLoS Genet. 2020;16:e1009005. doi: 10.1371/journal.pgen.1009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orta-Zavalza E., Guerrero-Serrano G., Gutierrez-Escobedo G., Canas-Villamar I., Juarez-Cepeda J., Castano I., De Las Penas A. Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol. Microbiol. 2013;88:1135–1148. doi: 10.1111/mmi.12247. [DOI] [PubMed] [Google Scholar]
- 83.Borah S., Shivarathri R., Srivastava V.K., Ferrari S., Sanglard D., Kaur R. Pivotal role for a tail subunit of the RNA polymerase II mediator complex CgMed2 in azole tolerance and adherence in Candida glabrata. Antimicrob. Agents Chemother. 2014;58:5976–5986. doi: 10.1128/AAC.02786-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basso L.R., Jr., Gast C.E., Bruzual I., Wong B. Identification and properties of plasma membrane azole efflux pumps from the pathogenic fungi Cryptococcus gattii and Cryptococcus neoformans. J. Antimicrob. Chemother. 2015;70:1396–1407. doi: 10.1093/jac/dku554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang M., Sionov E., Khanal Lamichhane A., Kwon-Chung K.J., Chang Y.C. Roles of three Cryptococcus neoformans and Cryptococcus gattii efflux pump-coding genes in response to drug treatment. Antimicrob. Agents Chemother. 2018;62:e01751-17. doi: 10.1128/AAC.01751-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanguinetti M., Posteraro B., La Sorda M., Torelli R., Fiori B., Santangelo R., Delogu G., Fadda G. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect. Immun. 2006;74:1352–1359. doi: 10.1128/IAI.74.2.1352-1359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Posteraro B., Sanguinetti M., Sanglard D., La Sorda M., Boccia S., Romano L., Morace G., Fadda G. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol. Microbiol. 2003;47:357–371. doi: 10.1046/j.1365-2958.2003.03281.x. [DOI] [PubMed] [Google Scholar]
- 88.Ukai Y., Kuroiwa M., Kurihara N., Naruse H., Homma T., Maki H., Naito A. Contributions of yap1 mutation and subsequent atrf upregulation to voriconazole resistance in Aspergillus flavus. Antimicrob. Agents Chemother. 2018;62:e01216-18. doi: 10.1128/AAC.01216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hagiwara D., Miura D., Shimizu K., Paul S., Ohba A., Gonoi T., Watanabe A., Kamei K., Shintani T., Moye-Rowley W.S., et al. A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by co-regulating cyp51A and cdr1B expressions. PLoS Pathog. 2017;13:e1006096. doi: 10.1371/journal.ppat.1006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park S., Kelly R., Kahn J.N., Robles J., Hsu M.J., Register E., Li W., Vyas V., Fan H., Abruzzo G., et al. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 2005;49:3264–3273. doi: 10.1128/AAC.49.8.3264-3273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Balashov S.V., Park S., Perlin D.S. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 2006;50:2058–2063. doi: 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lackner M., Tscherner M., Schaller M., Kuchler K., Mair C., Sartori B., Istel F., Arendrup M.C., Lass-Florl C. Positions and numbers of FKS mutations in Candida albicans selectively influence in vitro and in vivo susceptibilities to echinocandin treatment. Antimicrob. Agents Chemother. 2014;58:3626–3635. doi: 10.1128/AAC.00123-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Castanheira M., Messer S.A., Rhomberg P.R., Pfaller M.A. Antifungal susceptibility patterns of a global collection of fungal isolates: Results of the SENTRY Antifungal Surveillance Program (2013) Diagn. Microbiol. Infect. Dis. 2016;85:200–204. doi: 10.1016/j.diagmicrobio.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 94.Garcia-Effron G., Park S., Perlin D.S. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: Implications for interpretive breakpoints. Antimicrob. Agents Chemother. 2009;53:112–122. doi: 10.1128/AAC.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jimenez-Ortigosa C., Moore C., Denning D.W., Perlin D.S. Emergence of echinocandin resistance due to a point mutation in the fks1 gene of Aspergillus fumigatus in a patient with chronic pulmonary aspergillosis. Antimicrob. Agents Chemother. 2017;61:e01277-17. doi: 10.1128/AAC.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moosa M.Y., Alangaden G.J., Manavathu E., Chandrasekar P.H. Resistance to amphotericin B does not emerge during treatment for invasive aspergillosis. J. Antimicrob. Chemother. 2002;49:209–213. doi: 10.1093/jac/49.1.209. [DOI] [PubMed] [Google Scholar]
- 97.Vincent B.M., Lancaster A.K., Scherz-Shouval R., Whitesell L., Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vandeputte P., Tronchin G., Berges T., Hennequin C., Chabasse D., Bouchara J.P. Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrob. Agents Chemother. 2007;51:982–990. doi: 10.1128/AAC.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rybak J.M., Barker K.S., Munoz J.F., Parker J.E., Ahmad S., Mokaddas E., Abdullah A., Elhagracy R.S., Kelly S.L., Cuomo C.A., et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin. Microbiol. Infect. 2022;28:838–843. doi: 10.1016/j.cmi.2021.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahmad S., Joseph L., Parker J.E., Asadzadeh M., Kelly S.L., Meis J.F., Khan Z. ERG6 and ERG2 are major targets conferring reduced susceptibility to Amphotericin B in clinical Candida glabrata isolates in Kuwait. Antimicrob. Agents Chemother. 2019;63:e01900-18. doi: 10.1128/AAC.01900-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kelly S.L., Lamb D.C., Taylor M., Corran A.J., Baldwin B.C., Powderly W.G. Resistance to amphotericin B associated with defective sterol delta 8-->7 isomerase in a Cryptococcus neoformans strain from an AIDS patient. FEMS Microbiol. Lett. 1994;122:39–42. doi: 10.1111/j.1574-6968.1994.tb07140.x. [DOI] [PubMed] [Google Scholar]
- 102.Edlind T.D., Katiyar S.K. Mutational analysis of flucytosine resistance in Candida glabrata. Antimicrob. Agents Chemother. 2010;54:4733–4738. doi: 10.1128/AAC.00605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Billmyre R.B., Applen Clancey S., Li L.X., Doering T.L., Heitman J. 5-fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in Cryptococcus. Nat. Commun. 2020;11:127. doi: 10.1038/s41467-019-13890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boyce K.J., Wang Y., Verma S., Shakya V.P.S., Xue C., Idnurm A. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. MBio. 2017;8:e00595-17. doi: 10.1128/mBio.00595-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Healey K.R., Zhao Y., Perez W.B., Lockhart S.R., Sobel J.D., Farmakiotis D., Kontoyiannis D.P., Sanglard D., Taj-Aldeen S.J., Alexander B.D., et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016;7:11128. doi: 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Billmyre R.B., Clancey S.A., Heitman J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife. 2017;6:e28802. doi: 10.7554/eLife.28802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dos Reis T.F., Silva L.P., de Castro P.A., do Carmo R.A., Marini M.M., da Silveira J.F., Ferreira B.H., Rodrigues F., Lind A.L., Rokas A., et al. The Aspergillus fumigatus mismatch repair MSH2 homolog is important for virulence and azole resistance. mSphere. 2019;4:e00416-19. doi: 10.1128/mSphere.00416-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burrack L.S., Todd R.T., Soisangwan N., Wiederhold N.P., Selmecki A. Genomic diversity across Candida auris clinical isolates shapes rapid development of antifungal resistance in vitro and in vivo. mBio. 2022;13:e0084222. doi: 10.1128/mbio.00842-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Byun S.A., Won E.J., Kim M.N., Lee W.G., Lee K., Lee H.S., Uh Y., Healey K.R., Perlin D.S., Choi M.J., et al. Multilocus Sequence Typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: Association with antifungal resistance, mutations in mismatch repair gene (msh2), and clinical outcomes. Front. Microbiol. 2018;9:1523. doi: 10.3389/fmicb.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hou X., Xiao M., Wang H., Yu S.Y., Zhang G., Zhao Y., Xu Y.C. Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob. Agents Chemother. 2018;62:e00153-18. doi: 10.1128/AAC.00153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singh A., Healey K.R., Yadav P., Upadhyaya G., Sachdeva N., Sarma S., Kumar A., Tarai B., Perlin D.S., Chowdhary A. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob. Agents Chemother. 2018;62:e00195-18. doi: 10.1128/AAC.00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delliere S., Healey K., Gits-Muselli M., Carrara B., Barbaro A., Guigue N., Lecefel C., Touratier S., Desnos-Ollivier M., Perlin D.S., et al. Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a french cohort of patients harboring low rates of resistance. Front. Microbiol. 2016;7:2038. doi: 10.3389/fmicb.2016.02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bordallo-Cardona M.A., Agnelli C., Gomez-Nunez A., Sanchez-Carrillo C., Bouza E., Munoz P., Escribano P., Guinea J. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob. Agents Chemother. 2019;63:e01876-18. doi: 10.1128/AAC.01876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Biswas C., Marcelino V.R., Van Hal S., Halliday C., Martinez E., Wang Q., Kidd S., Kennedy K., Marriott D., Morrissey C.O., et al. Whole genome sequencing of Australian Candida glabrata Isolates reveals genetic diversity and novel sequence types. Front. Microbiol. 2018;9:2946. doi: 10.3389/fmicb.2018.02946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shor E., Schuyler J., Perlin D.S. A novel, drug resistance-independent, fluorescence-based approach to measure mutation rates in microbial pathogens. mBio. 2019;10:e00120-19. doi: 10.1128/mBio.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Albehaijani S.H.I., Macreadie I., Morrissey C.O., Boyce K.J. Molecular mechanisms underlying the emergence of polygenetic antifungal drug resistance in msh2 mismatch repair mutants of Cryptococcus. JAC Antimicrob. Resist. 2022;4:dlac033. doi: 10.1093/jacamr/dlac033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ballard E., Melchers W.J.G., Zoll J., Brown A.J.P., Verweij P.E., Warris A. In-host microevolution of Aspergillus fumigatus: A phenotypic and genotypic analysis. Fungal Genet. Biol. 2018;113:1–13. doi: 10.1016/j.fgb.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gast C.E., Basso L.R., Jr., Bruzual I., Wong B. Azole resistance in Cryptococcus gattii from the Pacific Northwest: Investigation of the role of ERG11. Antimicrob. Agents Chemother. 2013;57:5478–5485. doi: 10.1128/AAC.02287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Denning D.W., Park S., Lass-Florl C., Fraczek M.G., Kirwan M., Gore R., Smith J., Bueid A., Moore C.B., Bowyer P., et al. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin. Infect. Dis. 2011;52:1123–1129. doi: 10.1093/cid/cir179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Khateb A., Gago S., Bromley M., Richardson M., Bowyer P. Aneuploidy is associated with azole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2023;67:e0125322. doi: 10.1128/aac.01253-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ford C.B., Funt J.M., Abbey D., Issi L., Guiducci C., Martinez D.A., Delorey T., Li B.Y., White T.C., Cuomo C., et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife. 2015;4:e00662. doi: 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cavalheiro M., Costa C., Silva-Dias A., Miranda I.M., Wang C., Pais P., Pinto S.N., Mil-Homens D., Sato-Okamoto M., Takahashi-Nakaguchi A., et al. A transcriptomics approach to unveiling the mechanisms of in vitro evolution towards fluconazole resistance of a Candida glabrata clinical isolate. Antimicrob. Agents Chemother. 2019;63:e00995-18. doi: 10.1128/AAC.00995-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Narayanan A., Kumar P., Chauhan A., Kumar M., Yadav K., Banerjee A., Sharma R.D., Rudramurthy S.M., Chakrabarti A., Sanyal K., et al. Directed evolution detects supernumerary centric chromosomes conferring resistance to azoles in Candida auris. mBio. 2022;13:e0305222. doi: 10.1128/mbio.03052-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Florio A.R., Ferrari S., De Carolis E., Torelli R., Fadda G., Sanguinetti M., Sanglard D., Posteraro B. Genome-wide expression profiling of the response to short-term exposure to fluconazole in Cryptococcus neoformans serotype A. BMC Microbiol. 2011;11:97. doi: 10.1186/1471-2180-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aruanno M., Gozel S., Mouyna I., Parker J.E., Bachmann D., Flamant P., Coste A.T., Sanglard D., Lamoth F. Insights in the molecular mechanisms of an azole stress adapted laboratory-generated Aspergillus fumigatus strain. Med. Mycol. 2021;59:763–772. doi: 10.1093/mmy/myaa118. [DOI] [PubMed] [Google Scholar]
- 126.Selmecki A., Gerami-Nejad M., Paulson C., Forche A., Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 127.Sionov E., Lee H., Chang Y.C., Kwon-Chung K.J. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;6:e10000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Almeida A.M., Matsumoto M.T., Baeza L.C., de Oliveira E.S.R.B., Kleiner A.A., Melhem Mde S., Mendes Giannini M.J. Molecular typing and antifungal susceptibility of clinical sequential isolates of Cryptococcus neoformans from Sao Paulo State, Brazil. FEMS Yeast Res. 2007;7:152–164. doi: 10.1111/j.1567-1364.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 129.Morrow C.A., Fraser J.A. Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin. Cell Dev. Biol. 2013;24:339–346. doi: 10.1016/j.semcdb.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 130.Chang Y.C., Khanal Lamichhane A., Kwon-Chung K.J. Cryptococcus neoformans, Unlike Candida albicans, Forms Aneuploid Clones Directly from Uninucleated Cells under Fluconazole Stress. mBio. 2018;9:e01290-18. doi: 10.1128/mBio.01290-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stone N.R., Rhodes J., Fisher M.C., Mfinanga S., Kivuyo S., Rugemalila J., Segal E.S., Needleman L., Molloy S.F., Kwon-Chung J., et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Investig. 2019;129:999–1014. doi: 10.1172/JCI124516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Semighini C.P., Averette A.F., Perfect J.R., Heitman J. Deletion of Cryptococcus neoformans AIF ortholog promotes chromosome aneuploidy and fluconazole-resistance in a metacaspase-independent manner. PLoS Pathog. 2011;7:e1002364. doi: 10.1371/journal.ppat.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou Z., Zhu C., Ip M., Liu M., Zhu Z., Liu R., Li X., Zeng L., Wu W. Molecular epidemiology and antifungal resistance of Cryptococcus neoformans from human immunodeficiency virus-negative and human immunodeficiency virus-positive patients in Eastern China. Front. Microbiol. 2022;13:942940. doi: 10.3389/fmicb.2022.942940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang F., Gritsenko V., Lu H., Zhen C., Gao L., Berman J., Jiang Y.Y. Adaptation to fluconazole via aneuploidy enables cross-adaptation to amphotericin B and flucytosine in Cryptococcus neoformans. Microbiol. Spectr. 2021;9:e0072321. doi: 10.1128/Spectrum.00723-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fu M.S., Liporagi-Lopes L.C., Dos Santos S.R.J., Tenor J.L., Perfect J.R., Cuomo C.A., Casadevall A. Amoeba predation of Cryptococcus neoformans results in pleiotropic changes to traits associated with virulence. mBio. 2021;12:e00567-21. doi: 10.1128/mBio.00567-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu G., Wang J., Choi J., Jung W.H., Liu I., Litvintseva A.P., Bicanic T., Aurora R., Mitchell T.G., Perfect J.R., et al. Variation in chromosome copy number influences the virulence of Cryptococcus neoformans and occurs in isolates from AIDS patients. BMC Genom. 2011;12:526. doi: 10.1186/1471-2164-12-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ormerod K.L., Morrow C.A., Chow E.W., Lee I.R., Arras S.D., Schirra H.J., Cox G.M., Fries B.C., Fraser J.A. Comparative genomics of serial isolates of Cryptococcus neoformans reveals gene associated with carbon utilization and virulence. G3. 2013;3:675–686. doi: 10.1534/g3.113.005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rhodes J., Desjardins C.A., Sykes S.M., Beale M.A., Vanhove M., Sakthikumar S., Chen Y., Gujja S., Saif S., Chowdhary A., et al. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics. 2017;207:327–346. doi: 10.1534/genetics.117.203836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sephton-Clark P., Tenor J.L., Toffaletti D.L., Meyers N., Giamberardino C., Molloy S.F., Palmucci J.R., Chan A., Chikaonda T., Heyderman R., et al. Genomic variation across a clinical cryptococcus population linked to disease outcome. mBio. 2022;13:e0262622. doi: 10.1128/mbio.02626-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Selmecki A., Forche A., Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Selmecki A.M., Dulmage K., Cowen L.E., Anderson J.B., Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Coste A., Turner V., Ischer F., Morschhauser J., Forche A., Selmecki A., Berman J., Bille J., Sanglard D. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Polakova S., Blume C., Zarate J.A., Mentel M., Jorck-Ramberg D., Stenderup J., Piskur J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ahmad K.M., Ishchuk O.P., Hellborg L., Jorgensen G., Skvarc M., Stenderup J., Jorck-Ramberg D., Polakova S., Piskur J. Small chromosomes among Danish Candida glabrata isolates originated through different mechanisms. Antonie Van Leeuwenhoek. 2013;104:111–122. doi: 10.1007/s10482-013-9931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kukurudz R.J., Chapel M., Wonitowy Q., Adamu Bukari A.R., Sidney B., Sierhuis R., Gerstein A.C. Acquisition of cross-azole tolerance and aneuploidy in Candida albicans strains evolved to posaconazole. G3. 2022;12:jkac156. doi: 10.1093/g3journal/jkac156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Todd R.T., Wikoff T.D., Forche A., Selmecki A. Genome plasticity in Candida albicans is driven by long repeat sequences. eLife. 2019;8:e45954. doi: 10.7554/eLife.45954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Anderson M.Z., Saha A., Haseeb A., Bennett R.J. A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinical isolate of Candida albicans. Microbiology. 2017;163:856–865. doi: 10.1099/mic.0.000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mba I.E., Nweze E.I., Eze E.A., Anyaegbunam Z.K.G. Genome plasticity in Candida albicans: A cutting-edge strategy for evolution, adaptation, and survival. Infect. Genet. Evol. 2022;99:105256. doi: 10.1016/j.meegid.2022.105256. [DOI] [PubMed] [Google Scholar]
- 149.Todd R.T., Selmecki A. Expandable and reversible copy number amplification drives rapid adaptation to antifungal drugs. eLife. 2020;9:e58349. doi: 10.7554/eLife.58349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Todd R.T., Soisangwan N., Peters S., Kemp B., Crooks T., Gerstein A., Selmecki A. Antifungal drug concentration impacts the spectrum of adaptive mutations in Candida albicans. Mol. Biol. Evol. 2023;40:msad009. doi: 10.1093/molbev/msad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang F., Zhang L., Wakabayashi H., Myers J., Jiang Y., Cao Y., Jimenez-Ortigosa C., Perlin D.S., Rustchenko E. Tolerance to caspofungin in Candida albicans is associated with at least three distinctive mechanisms that govern expression of FKS genes and cell wall remodeling. Antimicrob. Agents Chemother. 2017;61:e00071-17. doi: 10.1128/AAC.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sah S.K., Hayes J.J., Rustchenko E. The role of aneuploidy in the emergence of echinocandin resistance in human fungal pathogen Candida albicans. PLoS Pathog. 2021;17:e1009564. doi: 10.1371/journal.ppat.1009564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsai H.J., Nelliat A. A double-edged sword: Aneuploidy is a prevalent strategy in fungal adaptation. Genes. 2019;10:787. doi: 10.3390/genes10100787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barda O., Sadhasivam S., Gong D., Doron-Faigenboim A., Zakin V., Drott M.T., Sionov E. Aneuploidy Formation in the filamentous fungus Aspergillus flavus in response to azole stress. Microbiol. Spectr. 2023;11:e0433922. doi: 10.1128/spectrum.04339-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bing J., Hu T., Zheng Q., Munoz J.F., Cuomo C.A., Huang G. Experimental evolution identifies adaptive aneuploidy as a mechanism of fluconazole resistance in Candida auris. Antimicrob. Agents Chemother. 2020;65:e01466-20. doi: 10.1128/AAC.01466-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable. All articles included in the review are openly available in the NIH National Library of Medicine (https://pubmed.ncbi.nlm.nih.gov/).