Abstract
Y chromosomal ampliconic genes (YAGs) are important for male fertility, as they encode proteins functioning in spermatogenesis. The variation in copy number and expression levels of these multicopy gene families has been studied in great apes; however, the diversity of splicing variants remains unexplored. Here, we deciphered the sequences of polyadenylated transcripts of all nine YAG families (BPY2, CDY, DAZ, HSFY, PRY, RBMY, TSPY, VCY, and XKRY) from testis samples of six great ape species (human, chimpanzee, bonobo, gorilla, Bornean orangutan, and Sumatran orangutan). To achieve this, we enriched YAG transcripts with capture probe hybridization and sequenced them with long (Pacific Biosciences) reads. Our analysis of this data set resulted in several findings. First, we observed evolutionarily conserved alternative splicing patterns for most YAG families except for BPY2 and PRY. Second, our results suggest that BPY2 transcripts and proteins originate from separate genomic regions in bonobo versus human, which is possibly facilitated by acquiring new promoters. Third, our analysis indicates that the PRY gene family, having the highest representation of noncoding transcripts, has been undergoing pseudogenization. Fourth, we have not detected signatures of selection in the five YAG families shared among great apes, even though we identified many species-specific protein-coding transcripts. Fifth, we predicted consensus disorder regions across most gene families and species, which could be used for future investigations of male infertility. Overall, our work illuminates the YAG isoform landscape and provides a genomic resource for future functional studies focusing on infertility phenotypes in humans and critically endangered great apes.
Keywords: transcript isoform, diversity, ampliconic gene, Y chromosome, great apes
Significance.
Ampliconic genes on the Y chromosomes in great apes encode proteins functioning in spermatogenesis and play important, yet not fully understood, roles in male reproductive health. Copy number variation of ampliconic genes within and across great ape species has been recently investigated, yet transcript diversity of ampliconic genes remains largely unknown. To address this critical knowledge gap, we used a combination of capture-probe hybridization and PacBio long-read sequencing technology, providing the first comprehensive understanding of isoform landscape of each of the nine multicopy Y chromosome ampliconic gene families in great apes. Here, we demonstrate high isoform diversity across great apes, which recapitulates previously observed high levels of copy number variations. This variation at the gene copy and transcript number is thought to reflect differences in mating patterns and sperm competition levels across species. By providing a freely available repository of ampliconic gene transcript isoforms, this study offers a crucial tool for researchers seeking to understand the genetic basis of male reproductive biology and potential targets for therapeutic interventions in humans and critically endangered great apes.
Introduction
Y chromosomal ampliconic genes (YAGs) are expressed exclusively in testis (Skaletsky et al. 2003), encode proteins functioning in spermatogenesis, and are important for male fertility. In humans, YAGs belong to nine multicopy gene families—BPY2, CDY, DAZ, HSFY, PRY, RBMY, TSPY, VCY, and XKRY. Most of these gene families are shared among primates (Cortez et al. 2014); however, some were pseudogenized or completely deleted in certain great ape lineages, which is potentially linked to interspecies differences in mating patterns (Hughes et al. 2010; Cechova et al. 2020). Although the variability in copy number (Oetjens et al. 2016; Tomaszkiewicz et al. 2016; Ye et al. 2018; Vegesna et al. 2020) and in gene expression levels (Fagerberg et al. 2014; Vegesna et al. 2019, 2020) of YAGs has been recently explored in great apes, we still do not have a full understanding of whether this variability is reflected in transcript diversity. In fact, sequences of full-length transcripts for YAG families across great apes are lacking. Moreover, their precise cellular functions in the male germline are still not completely deciphered (Lahn and Page 1997; Zou et al. 2003; Stouffs 2004; Wong et al. 2004; Yen 2004; Navarro-Costa 2012). Investigating the transcriptional landscape of YAGs is expected to inform the diversity in sperm characteristics and male fertility phenotypes observed across great apes.
Recently, great ape YAG transcripts were assembled from short-read RNA-seq data (Vegesna et al. 2020). However, only one consensus transcript per gene family was generated for most families and, in some cases, even that representative sequence was incomplete. These limitations arise from the fact that many YAG families are expressed at low levels (Fagerberg et al. 2014; Vegesna et al. 2019, 2020), leading to a small number of sequencing reads and problematic transcript assemblies. In another study (Cortez et al. 2014), gorilla YAG transcript sequences were reconstructed from short-read RNA-seq data originating from testis of one individual, which also resulted in only one transcript per gene family. In the same study, orangutan transcript sequences for YAG families were predicted from whole-genome DNA sequencing data and were not based on the analysis of transcripts expressed in testis. The annotation of the published chimpanzee Y chromosome is incomplete, missing full-length coding sequences of all six YAG families present in this species (Hughes et al. 2010). Therefore, the complete repertoire of transcript sequences of great ape YAGs resolved at a nucleotide level is still unavailable.
To fill this critical gap, we captured (with hybridization) polyadenylated YAG transcripts from testis samples of six great ape species: human, chimpanzee, bonobo, gorilla, Sumatran orangutan, and Bornean orangutan, and sequenced them using Pacific Biosciences (PacBio) Iso-Seq protocol. Obtaining this unique data set allowed us to compare YAG transcripts across great ape species that diverged from each other between ∼0.4 and ∼13 million years ago (Glazko 2003; Locke et al. 2011). Specifically, we asked the following questions: 1) How diverse are YAG transcript isoforms across great apes? 2) Do nonhuman YAG transcripts share splicing patterns with the well-annotated YAGs on the human Y chromosome? 3) Have any transcripts accumulated nonsense mutations, leading to pseudogenization? 4) Do any of the YAGs conserved across great ape species evolve under purifying selection? 5) Which YAG isoforms are lineage-specific, and which exhibit signatures of positive selection? By addressing these questions, we deciphered the isoform landscape of YAG sequences in great apes. The resulting data and the analyses described herein will be critical for designing evidence-based strategies for preserving reproductive success of these species, all of which (except for humans) are endangered.
Results
Sequencing of Polyadenylated Y Chromosomal Ampliconic Gene Transcripts in Six Great Ape Species
To study the evolution of YAG transcripts across great apes, we isolated total RNA from testis samples of six great ape species and generated two cDNA technical replicates for each sample (supplementary fig. S1 and table S1, Supplementary Material online). We used gene family–specific hybridization capture probes (supplementary table S2, Supplementary Material online) validated with male-specific primers (supplementary table S3, Supplementary Material online) to pull down YAG cDNAs and generate two PacBio Iso-Seq libraries, which were sequenced. We identified full-length nonchimeric (FLNC) transcripts (supplementary table S4, Supplementary Material online) and clustered them with isONclust according to the following workflow (supplementary fig. S2, Supplementary Material online). We assigned the clusters to their respective gene families and performed error correction of the clustered and gene family–assigned FLNCs with IsoCon (supplementary fig. S3, Supplementary Material online). We then identified replicate-supported transcripts and mapped them to one human genomic copy with the largest number of exons per each YAG family and to the databases of human and nonhuman transcripts. Finally, we identified identical and species-specific transcripts and tested for selection acting on protein-coding transcripts (supplementary fig. S4, Supplementary Material online).
Replicate-Supported Transcripts
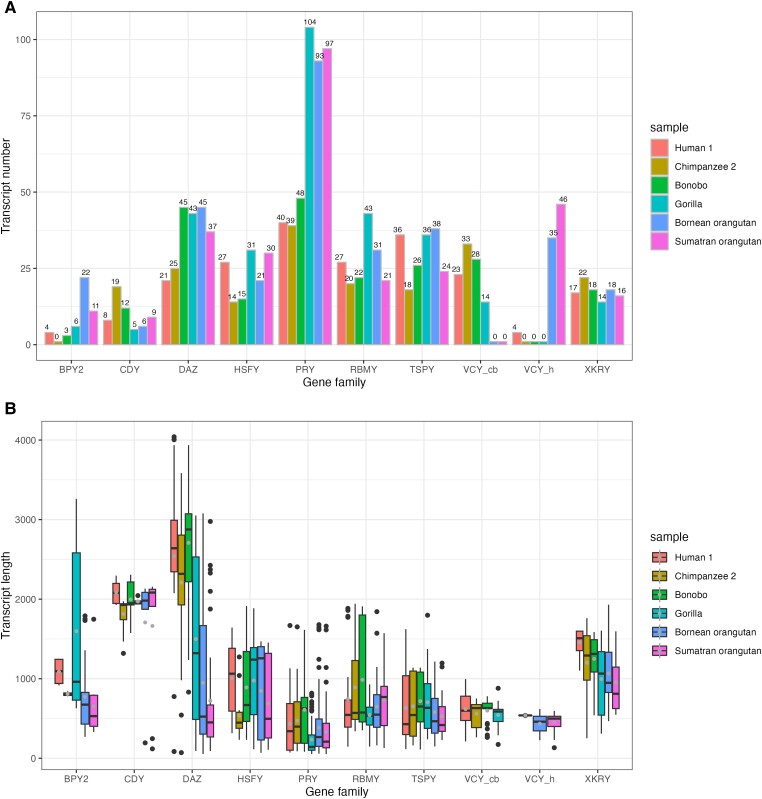
We identified 1,510 replicate-supported transcripts, that is, isoforms shared between the two technical replicates (supplementary table S7, Supplementary Material online), in six samples. Each replicate-supported transcript was supported by at least two reads (in each technical replicate) with an average of 92 reads (min = 2, max = 3,245, and med = 44; all but 11 replicate-supported transcripts were supported by more than two reads). The total number of replicate-supported transcripts per sample ranged from 190 (for chimpanzee) to 315 (for Bornean orangutan), with an average of 256 (supplementary table S6, Supplementary Material online). The number of replicate-supported transcripts per gene family differed substantially among gene families and among species (fig. 1A, supplementary table S6, Supplementary Material online). The longest transcripts belonged to the CDY and DAZ gene families with mean lengths of 1,868 and 1,687 bp, respectively (fig. 1B). The longest transcript was a human DAZ transcript (4,041 bp) followed by a chimpanzee DAZ transcript (3,584 bp; fig. 1B, supplementary tables S8 and S9, Supplementary Material online). The shortest transcripts belonged to the PRY gene family with a mean length of 374 bp (fig. 1B, supplementary table S8, Supplementary Material online).
Fig. 1.
(A) Number of replicate-supported transcripts presented separately for each great ape species and gene family. (B) Distribution of lengths of replicate-supported transcripts per gene family per species. Dark dots outside bars represent outliers, light dots inside bars represent mean lengths, and horizontal lines represent median lengths. VCY_h, captured with the human-specific probe; VCY_cb, captured with the chimpanzee- and bonobo-specific probe.
Analysis of Transcripts with Complete Open Reading Frames
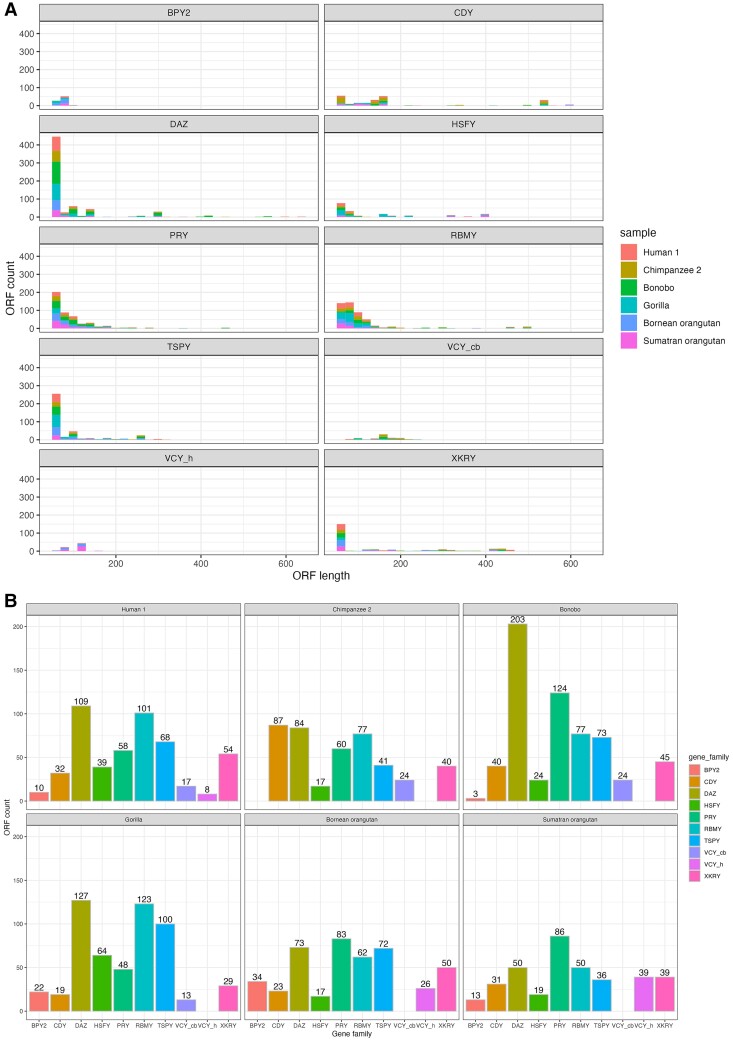
We predicted complete open reading frames (cORFs), that is, sequences that begin with a start codon and end with a stop codon, in the replicate-supported transcripts. Because most cORFs we found were <50 amino acids (aa) long (supplementary dataset S2, Supplementary material online), we suspect them to be artifacts. However, an abundance of short proteins was found in mammalian genomes (Frith et al. 2006); therefore, we have chosen an ORF length threshold of >50 aa for the downstream analysis of transcripts with cORFs (supplementary datasets S3 and S16, Supplementary material online; fig. 2A). In total, we identified 7,966 cORFs longer than 50 aa in our replicate-supported transcripts.
Fig. 2.
(A) Distribution of predicted cORF lengths (in amino acids) per gene family per species. VCY_h, captured with the human-specific probe; VCY_cb, captured with the chimpanzee- and bonobo-specific probe). (B) Number of cORFs per gene family per species.
We could detect cORFs in replicate-supported transcripts in all YAG families (fig. 2B, supplementary dataset S16, Supplementary material online) in all the species analyzed with one exception. In chimpanzee, cORF for BPY2 was missing among replicate-supported transcripts; however, a BPY2 cORF was present in one of the technical replicates. In general, the lengths of proteins predicted from these cORFs in human and nonhuman apes were similar to the lengths of the corresponding human proteins reported in the literature. For example, a previous study (Lahn and Page 1997) predicted human BPY2 proteins to be 106 aa long. We found cORFs corresponding to BPY2 proteins of this length in our human sample and of a similar length in our gorilla sample (fig. 2A). The same study showed human VCY protein to be 125 aa long (Lahn and Page 2000), and we found predicted homologs of the same length in our human sample (fig. 2A). Some gene families in certain great ape lineages have more cORFs than other species, which could indicate more proteins and thus more pronounced functional relevance in those species, such as DAZ in bonobo and chimpanzee for increased sperm functions.
Homology to Human YAG Protein-Coding Transcripts
First, to study homology to human YAG protein-coding transcripts, we used BLASTN (Camacho et al. 2009) to align all replicate-supported transcripts against publicly available Y transcript data sets for human, as available in ENSEMBL (supplementary dataset S4, Supplementary material online), chimpanzee, as available in ENSEMBL (supplementary dataset S5, Supplementary material online), gorilla Y, as published by Cortez et al. (2014) (supplementary dataset S6, Supplementary material online), and the predicted orangutan Y chromosome transcripts generated by Cortez et al. (2014) (supplementary dataset S7, Supplementary material online). Because we could not find any replicate-supported transcripts that fully covered known transcripts from the above data sets (supplementary datasets S4–S7, Supplementary material online) at 100% sequence identity, we limited our subsequent analyses to protein-coding sequences.
Second, we aligned our nonhuman replicate-supported transcripts to the annotated human reference YAG protein-coding sequences (supplementary dataset S8, Supplementary material online) using BLASTN (Camacho et al. 2009) (see Materials and Methods for details; supplementary table S10, Supplementary Material online). For several great ape species, we found replicate-supported transcripts homologous to human reference transcripts to be either completely absent or of reduced length in some YAG families (supplementary table S10, Supplementary Material online), suggesting deletion or pseudogenization. No homologous BPY2 transcripts were found in chimpanzee and bonobo; however, as mentioned above, such sequences were present in one chimpanzee technical replicate. BPY2 homologs differed substantially in length between human and gorilla (covering only 60–71% of the human reference with 98–99% of sequence identity) and even more so between each orangutan species and human (covering 34–37% of the human reference with 93–96% of sequence identity). HSFY, PRY, and XKRY homologs were short or absent in chimpanzee and bonobo, confirming previous findings (Bellott et al. 2014; Cortez et al. 2014; Cechova et al. 2020). VCY homologs were found in bonobo and in both species of orangutan, which contradicts previous results (Cortez et al. 2014; Cechova et al. 2020) and requires further investigation. XKRY homologs were present in gorilla (covering 80% of the human reference with 86.8% of sequence identity) and both species of orangutan (64–100% coverage or % of aligned sequence with 84–93.3% of sequence identity) but were absent from both bonobo and chimpanzee (supplementary table S10, Supplementary Material online), which confirms previous studies (Hughes et al. 2010; Cechova et al. 2020).
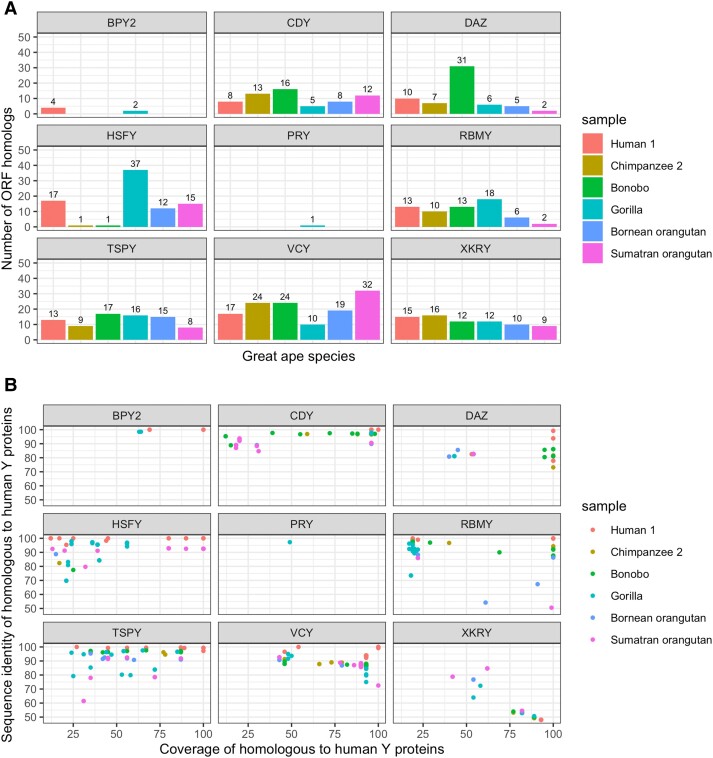
We also aligned the predicted cORFs in replicate-supported transcripts to human reference Y chromosome proteins (supplementary dataset S9, Supplementary material online) using BLASTP (see Materials and Methods for details). The resulting high-confidence cORF homologs per gene family per species are presented in figure 3 and supplementary dataset S9, Supplementary Material online. CDY, DAZ, RBMY, TSPY, and VCY ORF homologs were abundant across all great ape species (fig. 3A). Among YAG families, CDY ORF homologs aligned to human proteins with the highest coverage. The largest numbers of CDY and DAZ ORF homologs were observed in bonobo. Only single HSFY ORF homolog with low coverage (17–25%) but relatively high identity (77–82%) was found in each of the chimpanzee and bonobo samples (fig. 3A, supplementary dataset S9, Supplementary Material online). BPY2 ORF homologs were present only in human and gorilla samples. PRY homologs were missing in all species but gorilla (fig. 3A), indicating that these sequences are either not expressed or expressed below our detection level.
Fig. 3.
Analysis of high-confidence cORFs homologous to human Y chromosome ampliconic transcripts per gene family per species. (A) Numbers of cORFs. (B) Coverage and sequence identity in alignments to homologous human Y proteins.
Proteins that were predicted from cORFs CDY, DAZ, RBMY, and TSPY in chimpanzee and bonobo had high coverage and sequence identity when aligned to human Y proteins (fig. 3B). Across great apes, the highest protein alignment length was observed for the CDY cORF homologs, followed by RBMY and DAZ cORF homologs (fig. 3B). HSFY cORF homologs in Sumatran orangutan had high sequence identity to, and covered most of, the corresponding human Y proteins (fig. 3B). Short HSFY cORF homologs of 50–250 aa were found in gorilla, and longer ones (300–400 aa) were observed in both species of orangutan. Most VCY and XKRY cORF homologs from nonhuman great apes covered ∼100–150 aa of the corresponding human Y proteins (fig. 3B).
Transcripts with Identical Protein-Coding Sequences
To gain insights into evolutionary conservation or divergence of YAG transcripts across great ape species that diverged from each other between ∼0.4 and ∼13 million years ago (supplementary fig. S12, Supplementary Material online), we identified replicate-supported transcripts with identical cORFs (but potentially differing at 5′ and/or 3′ untranslated regions [UTRs]) within (between the two samples for human or chimpanzee) or between great ape species analyzed (supplementary table S13 and dataset S14, Supplementary material online). We found that all gene families had shared cORFs between the two closely related orangutan species (supplementary table S13, Supplementary Material online). In contrast, some gene families (BPY2 and VCY) consistently did not have shared cORFs even between closely related Pan species, chimpanzee and bonobo (supplementary table S13, Supplementary Material online). In the whole data set, most shared cORFs were present in DAZ, PRY, RBMY, and TSPY gene families (supplementary table S13, Supplementary Material online). DAZ and PRY cORFs were shared among human, chimpanzee, bonobo, and gorilla but not between any one of them and each of the two orangutans, suggesting that human–gorilla is the largest evolutionary distance at which they are conserved. Identical TSPY cORFs were present within the same species (human or chimpanzee) and between some species (human and chimpanzee, human and bonobo, chimpanzee and bonobo, and the two species of orangutan; supplementary table S13, Supplementary Material online).
Species-Specific Protein-Coding Transcript Sequences
Subsequently, we were able to identify species-specific transcripts with cORFs (supplementary table S16, Supplementary Material online) and species-specific protein-coding sequences (supplementary table S14, Supplementary Material online). Across most gene families, at least one protein-coding transcript per gene family was species specific. The highest number of species-specific protein-coding transcripts was present in the PRY gene family: it ranged from 16 to 57 per species (supplementary table S14, Supplementary Material online). BPY2 had the lowest number of species-specific protein-coding transcripts, including only three, three, and two transcripts specific to gorilla, Bornean orangutan, and Sumatran orangutan, respectively.
Species-Specific Protein Prediction and Identification of Protein Domains
Next, we predicted species-specific proteins and identified their domains, because these domains might be tailored for specialized functions unique to each species (supplementary dataset S17, Supplementary material online). Most interestingly, we detected consensus disorder regions of at least 20 amino acids across all gene families (but BPY2 and XKRY) and most great ape species except for CDY in both species of orangutan and human 1, DAZ in human 2 and gorilla, HSFY in bonobo, PRY in Bornean orangutan, RBMY in Sumatran orangutan, and TSPY in human 2, chimp 1, and Sumatran orangutan. These identified regions could be used for future investigations of the relationship between protein disorder and fertility-related diseases, such as male infertility, as many disease-associated proteins contain intrinsically disordered regions (supplementary dataset S17, Supplementary material online).
Among the ten species-specific predicted BPY2 proteins, one protein in Sumatran orangutan featured a signature of a >20-aa region of a membrane-bound protein predicted to be embedded in the membrane. When investigating species-specific predicted CDY proteins, we encountered a wide array of chromodomain or chromodomain Y-like signatures characteristic of this protein family. Moreover, many CDY proteins exhibited lengthy (>100-amino acid) signatures associated with clpP/crotonase, involved in protein degradation (Mabanglo and Houry 2022), or enoyl-CoA hydratase/isomerase, implicated in fatty acid metabolism (Mills et al. 2022). Notably, CDY proteins in both orangutan species bore 30-amino acid-long signatures within signal peptide regions, which could be potentially linked to protein transport or secretion during spermatogenesis. Most species-specific predicted DAZ proteins contained DAZ domains, DAZ repeats, and RNA recognition motifs, consistent with the characteristics of this protein family. Interestingly, we identified a specific domain, NADH-ubiquinone oxidoreductase chain 4L, a core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (complex I), in one predicted DAZ protein in Bornean orangutan, suggesting its potential involvement in the regulation of mitochondrial function during spermatogenesis. Additionally, a signal peptide was found in one predicted DAZ protein in gorilla.
The majority of HSFY proteins exhibited heat shock factor DNA-binding domains, whereas certain HSFY proteins in gorilla, human 2, and Sumatran orangutan also featured winged helix DNA-binding domains, potentially contributing to protein–DNA and protein–protein interactions. Furthermore, we discovered a specific domain exclusive to the HSFY predicted protein in the human 1 sample, comprising a >20-amino acid-long region of a membrane-bound protein predicted to be embedded in the membrane. Among the protein families studied, PRY proteins displayed the most diverse representation of protein domains, encompassing signatures of ATP synthase and cytochrome subunits, as well as tubulin domains, suggesting their potential involvement in energy production during spermatogenesis. Predicted RBMY proteins were found to contain RNA recognition motifs such as RRM, RBD, or RNP domains and signal peptides. Notably, specific signatures related to transformer-2 sex-determining proteins, associated with the regulation of alternative splicing of pre-mRNA, were identified in predicted RBMY proteins in human 1, bonobo, chimp 2, and Bornean orangutan, indicating their potential role in regulating alternative splicing during spermatogenesis. Moreover, five RBMY proteins in both human samples exhibited signal peptides within transmembrane regions (SignalP-TM), potentially related to protein transport or secretion during spermatogenesis.
Most of the species-specific predicted TSPY proteins across all great apes demonstrated signatures of nucleosome assembly proteins, suggesting their potential involvement in chromatin remodeling, gene expression regulation, and epigenetic modifications during spermatogenesis. Regarding VCY proteins, most of the species-specific predicted members displayed signatures of either the variable charge X/Y family or testis-specific basic protein Y 1-related signatures, consistent with the characteristics of this protein family. When investigating species-specific XKRY proteins, extensive XK-related protein regions, often exceeding 200 amino acids, were identified in most species-specific predicted proteins, featuring small internal segments of a membrane-bound protein predicted to be embedded in the membrane. Intriguingly, one XKRY-predicted protein in human 2 exhibited a signature of a region of a membrane-bound protein predicted to be outside the membrane, either in the cytoplasm or in the extracellular region.
Noncoding Transcripts
Additionally, we identified many YAG transcripts without any cORFs, that is, noncoding transcripts (supplementary table S15, Supplementary Material online). Most of them were present in the PRY gene family (min = 9, max = 85, and med = 37 across species). The lowest number of noncoding transcripts was observed in the XKRY, BPY2, and CDY gene families (med = 0 and maximum of 1, 2, and 2, respectively, across species).
Alternative Splicing Patterns
To study splicing patterns of YAG transcripts across great ape species with divergence time between ∼0.4 and ∼13 million years ago (supplementary fig. S12, Supplementary Material online), we mapped cORF-containing replicate-supported transcripts to one ampliconic gene copy (picked up at random) per gene family on the human Y chromosome (figs. 4 and 5; supplementary figs. S4–S11, Supplementary Material online showing only transcripts with cORFs; supplementary datasets S15 and S16, Supplementary material online).
Fig. 4.
Conserved splicing patterns of DAZ transcripts with complete ORFs captured with gene-specific hybridization probes and mapped to one human gene copy ENSG00000188120. The human DAZ consensus coding sequence DAZ_CCDS48209.1 was also mapped to show the protein-coding exons (top row). Transcripts are grouped by species. Each colored block represents a location of an exon, and each arrow indicates the forward “>” or reverse “<” direction as mapping to the human genomic copy. The lines between exons represent introns.
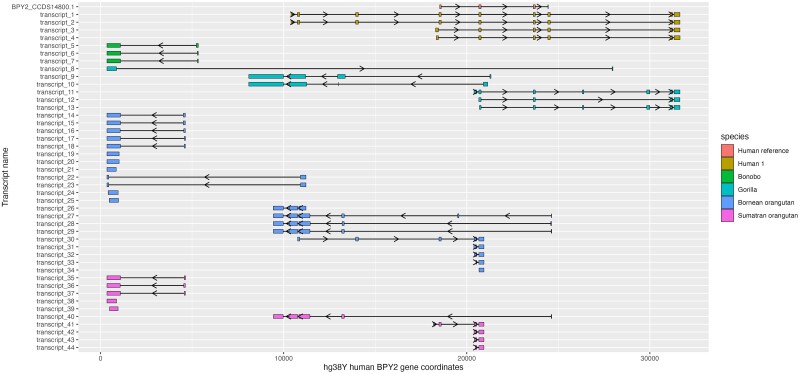
Fig. 5.
Divergent splicing patterns of BPY2 transcripts with complete ORFs captured with gene-specific hybridization probes and mapped to one human gene copy ENSG00000183753. The human BPY2 consensus coding sequence BPY2_CCDS14800.1 was also mapped to show the protein-coding exons (top row). Transcripts are grouped by species. Each colored block represents a location of an exon, and each arrow indicates the forward “>” or reverse “<” direction as mapping to the human genomic copy. The lines between exons represent introns.
For a number of YAG families, splicing patterns of nonhuman ape transcripts closely resembled that of human transcripts we recovered. Conserved splicing patterns are best exemplified by the longest DAZ nonhuman transcripts, which, when mapped to the human genomic copy, had the exons’ locations similar to that of human transcripts (fig. 4). Many nonhuman CDY transcripts recapitulated the splicing patterns of human transcripts (supplementary fig. S4, Supplementary Material online). Most nonhuman HSFY transcripts mapped to the same exon locations as did human transcripts (supplementary fig. S5, Supplementary Material online). Most nonhuman RBMY transcripts followed the locations of exons of human transcripts, with a few diverse transcripts found in gorilla and in both species of orangutan (supplementary fig. S7, Supplementary Material online). Most nonhuman TSPY transcripts followed the splicing patterns of human transcripts (supplementary fig. S8, Supplementary Material online). VCY transcripts from bonobo and gorilla, which were captured with the human-specific probe, mapped to the exon locations of human transcripts (supplementary fig. S9, Supplementary Material online). VCY transcripts from chimpanzee and bonobo, captured with the Pan-specific probe, recapitulated the exon locations of chimpanzee transcripts (supplementary fig. S10, Supplementary Material online).
Other gene families had more divergent patterns of exon–intron structure among great apes. None of the exon locations for the BPY2 transcripts from nonhuman samples recapitulated those found in the human sample (fig. 5). For example, many BPY2 transcripts from bonobo and both species of orangutan mapped upstream from the human transcripts (fig. 5). Additionally, all analyzed species but human and bonobo had BPY2 transcripts originating from both strands of this gene (fig. 5). All PRY transcripts from nonhuman samples mapped to only one exon of the human consensus coding sequence (supplementary fig. S6, Supplementary Material online). Most nonhuman XKRY transcripts mapped to the same two exons of human transcripts (supplementary fig. S11, Supplementary Material online); however, all XKRY transcripts from both species of orangutan were missing the noncoding exon found in all the other species, and two XKRY transcripts from Bornean orangutan had longer protein-coding exonic sequences (supplementary fig. S11, Supplementary Material online).
Selection Tests
To test for selection, we limited our analysis to gene families that were present in five or more ape species as well as in the outgroup species (macaque): CDY, DAZ, HSFY, RBMY, and TSPY. We did not detect any evidence of positive selection in these gene families; however, we did identify lower ratios of nonsynonymous over synonymous substitution rates in CDY in macaque and DAZ in human (supplementary table S9, Supplementary Material online). Nevertheless, the values were not significantly lower than one, rejecting the hypothesis that these sequences evolved under purifying selection (supplementary table S9, Supplementary Material online).
Discussion
Alternative splicing is one of the major forces generating the diversity and driving evolution of phenotypes across the eukaryotic tree of life. In mammals, alternative splicing has been widely investigated across tissues (Merkin et al. 2012; Gueroussov et al. 2015; Zhang et al. 2017) and recently also across developmental stages (Mazin et al. 2021). With recent increases in accuracy and read length of sequencing technologies, transcriptomic studies are unraveling unprecedented levels of splicing variants’ diversity and complexity (Zhang et al. 2017; Bayega et al. 2018; Kuang and Canzar 2018; Oikonomopoulos et al. 2020; De Paoli-Iseppi et al. 2021). Deciphering transcript sequences of highly similar copies from multicopy gene families, however, still poses a significant challenge. Here, we focused on solving this problem by studying highly similar transcripts from multicopy YAG families and, for the first time, uncovered multiple transcripts for each of nine multicopy YAG families for all but one (Tapanuli orangutan) extant nonhuman great ape species. Earlier studies attempted to decipher transcript sequences of great ape YAGs using short-read RNA-seq data sets or to predict their full-length sequences using whole-genome DNA sequencing data (Cortez et al. 2014; Vegesna et al. 2020). However, such efforts resulted in reconstruction of only one consensus transcript per gene family and, in many cases, even that representative sequence was incomplete. A more specialized approach using reverse transcription-polymerase chain reaction (RT-PCR) and long-read sequencing was recently applied to capture multiple YAG transcripts per gene family from two human testis samples (Tomaszkiewicz and Makova 2018). Clustering YAG transcripts from long reads was achieved using a novel computational method (IsoCon, Sahlin et al. 2018) that is able to distinguish transcripts originating from separate copies of a YAG family, which can differ by just a few nucleotides or small insertions/deletions. Applying this method to two human testis samples, Sahlin et al. (2018) uncovered many novel YAG transcripts; however, because the primers were designed in the first and last protein-coding exons, full-length polyadenylated transcripts could not be obtained. To overcome this limitation, here we used gene family–specific hybridization capture probes and the Iso-Seq protocol to pull down UTR-containing YAG cDNAs for human and five other great ape species. Additionally, to gain higher confidence in the sequence accuracy of our transcripts, we generated and sequenced transcripts from two technical replicates per each sample and were able to identify transcripts supported by both replicates.
Diversity of YAG Transcripts across Great Apes
We recovered at least two transcript sequences per YAG family per species, except for BPY2 in chimpanzee (supplementary table S6, Supplementary Material online). Overall, we observed high variability in the number of transcripts per gene family per species. The high number of transcripts (303–315, supplementary table S6, Supplementary Material online) observed in both species of orangutan echoes the high copy number of YAGs reported previously in these species (supplementary table S12, Supplementary Material online) (Vegesna et al. 2020). Similarly, the low number of transcripts (190, supplementary table S6, Supplementary Material online) in chimpanzee is consistent with the low YAG copy number previously reported for this species (supplementary table S12, Supplementary Material online) (Vegesna et al. 2020). In gorilla, we found a high number of transcripts (299, supplementary table S6, Supplementary Material online) and also the highest numbers of species-specific YAG protein-coding sequences (supplementary table S12, Supplementary Material online) and noncoding transcripts (supplementary table S13, Supplementary Material online), despite its relatively low YAG copy number according to previous studies (Tomaszkiewicz et al. 2016; Vegesna et al. 2020) (supplementary table S15, Supplementary Material online).
Conserved and Divergent Patterns of Alternative Splicing of YAG Isoforms across Great Apes
Most gene families displayed conserved alternative splicing patterns across great apes, especially the longest DAZ, the single-exon CDY, the two-exon HSFY, and the multiexon TSPY and RBMY. Most of transcripts from these gene families were conserved (in terms of splicing patterns) in human, chimpanzee, and bonobo but were more divergent in gorilla and both species of orangutan. These divergent transcripts might encode proteins with different functions in spermatogenesis, a possibility that will have to be explored in future studies. Most divergent exon–intron structures were observed in the BPY2 and PRY gene families, whose peculiarities are discussed below. Notably, both BPY2, whose origin was only recently discovered (Cao et al. 2015), and PRY were excluded from previous evolutionary studies (e.g., Bhowmick et al. 2007) because of the lack of information on their detectable X-linked/autosomal copies and on their orthologs in other species.
Independent Evolutionary Origins of the BPY2 Gene Family Transcripts
Our study suggests that BPY2 splicing patterns are conserved in human and chimpanzee but are divergent among other great apes; a previous study showed that the BPY2 locus was acquired on the Y chromosome in the common ancestor of great apes (Cao et al. 2015). A full-length BPY2 transcript was present in one of the chimpanzee technical replicates, suggesting that low expression in that sample might have prevented us from capturing the transcript in the other replicate. Consistent with this, BPY2 was reported to have the second lowest expression levels among YAGs (after PRY) in a previous study focused on great ape ampliconic gene expression levels (Vegesna et al. 2020). Additionally, BPY2 is the only YAG family specifically expressed during postmeiotic sex chromosome repression (Lucotte et al. 2018), the stage that might not have been captured in this chimpanzee sample. Nevertheless, the BPY2 transcript with a cORF found in one chimpanzee technical replicate aligned to the human reference BPY2 transcript with high identity and over a large proportion of length (supplementary dataset S10, Supplementary material online). Similarly, BPY2 transcripts in orangutans mapped to human transcripts. In contrast, all BPY2 transcripts from bonobo mapped upstream of human transcripts, suggesting their distinct evolutionary origins. Approximately half of gorilla BPY2 transcripts mapped upstream of human transcripts. Interestingly, all species but human and bonobo had BPY2 transcripts originating from both strands of the gene.
Our analysis of protein-coding regions suggested that BPY2 cORFs in several species might produce proteins nonhomologous to human proteins. Protein alignments between the predicted cORFs in nonhuman great apes and human Y chromosome proteins produced no significant matches for BPY2 sequences in bonobo and both species of orangutan (fig. 3, supplementary table S11, Supplementary Material online). Gorilla BPY2 sequences had low identity and coverage when mapped to human Y BPY2 proteins. In contrast, the recovered from one technical replicate chimpanzee BPY2 cORF had high (98%) sequence similarity over the full length (106 aa) human BPY2 protein (supplementary dataset S12, Supplementary material online).
PRY Gene Family Is Undergoing Pseudogenization across Great Apes
Several lines of evidence in our results suggest that the PRY gene family is undergoing pseudogenization in most great ape species. First, most transcripts without cORFs we recovered map to the PRY gene family (supplementary table S15, Supplementary Material online). Second, all species but gorilla lack cORF-containing homologs to human PRY. Third, several PRY cORFs predicted for great apes are not homologous to human (supplementary table S11, Supplementary Material online) but are lineage specific (supplementary table S14, Supplementary Material online) and map upstream of the human PRY coding exons (supplementary fig. S10, Supplementary Material online). Consistent with these results, among YAGs, PRY has been previously reported to be expressed at the lowest level, if at all, in great apes (Vegesna et al. 2020).
Future Applications to Functional Genomics
Cellular functions of YAGs are poorly characterized and were investigated only at the gene, but not at the transcript, level. For example, previous studies demonstrated that deletions of Y chromosome regions containing certain YAGs, that is, azoospermia (AZF) regions, can lead to spermatogenic impairment—for example, spermatogenic arrest resulting in altered spermatozoa formation or a complete lack of sperm cells (azoospermia) (Kuroda-Kawaguchi et al. 2001; Repping et al. 2002). Some other studies attempted to correlate the copy number of specific YAG families with fertility levels; however, this led to inconclusive results (Giachini et al. 2009; Krausz et al. 2010; Nickkholgh et al. 2010). Here, we uncovered splicing variants of YAGs that in the future can be used to study differential sperm characteristics and male fertility phenotypes observed across great apes. Overall, our study provides an informative genomic resource of full-length YAG transcripts for future functional studies focusing on infertility phenotypes in humans and other great apes.
Limitations of the Study and Future Directions
Ampliconic genes have been previously reported to be expressed at low levels in testes (Fagerberg et al. 2014; Vegesna et al. 2019, 2020). Though our study captured the highest transcript diversity to date, some of the genes, such as BPY2 and PRY, were nevertheless not fully recovered in all the species. Thus, it would be recommended in the future to target these specific genes at a higher sequencing depth. Also, the hybridization capture protocol using only one short probe per gene family universal for all great ape species could have led to underrepresentation of recovered transcripts per gene family.
We have used one genomic copy per gene family from the human Y chromosome for tracking the splicing patterns of transcripts from human and nonhuman great ape samples. This allowed us to recapitulate the general conserved or divergent splicing patterns per gene family across great apes. In the future, a more detailed analysis with precise mapping to each well-annotated copy per gene family on the great ape Y chromosomes will alleviate the human-specific bias of this analysis (Chen et al. 2021). Also, novel computational tools need to be developed to cope with a well-known multimapping issue related to highly similar sequences. This is an ongoing work as the sequences of the Y chromosomes and their gene annotations from nonhuman great apes are being improved thanks to the efforts of the T2T Primate Consortium.
There are several potential technical reasons regarding why we were unable to find evidence of positive selection for any of the gene families. First, we did not test for positive selection in BPY2, PRY, VCY, and XKRY, which were either represented only in three species or missing from the outgroup, limiting the power of the analysis. Second, using codeml to test for positive selection in only six lineages may be underpowered, as using more species could yield more variation, thereby increasing the level of statistical power to detect positive selection. Third, even though the codeml software from the PAML package (Yang 2007) is usually used to estimate the nonsynonymous to synonymous substitution rate ratios of genes, it does not account for deletions and insertions in splicing variants. This methodological limitation calls for developing other selection test approaches that include gaps representing divergence states in the alignments.
Testis is composed of several cell types: mitotic spermatogonia, meiotic spermatocytes, and postmeiotic spermatids and somatic cell types, such as Sertoli and Leydig cells. Thus, capturing ampliconic gene transcripts from testis samples does not allow for cell-specific distinction among transcripts. Future studies focusing on identifying cell type–specific YAG transcripts using single-cell RNA-seq approaches should overcome this limitation. Several recent studies investigated the expression levels of X- versus Y-linked genes including ampliconic genes in separate cell types from human testis, but none of them focused on analyzing the expression at the isoform level (Sin et al. 2012; Lucotte et al. 2018). One of the most recent studies has focused on single-nucleus testis transcriptome data from 11 species including four great apes (Murat et al. 2022). However, except for one RBMY2 transcript from gorilla, no ampliconic transcripts were reported as a cell type marker for any of the analyzed species. Besides, genes on the Y chromosome in primates have undergone reduction in gene expression (Cortez et al. 2014), which required us to use this targeted (enrichment-based) approach, whereas it might not be needed for most genes on the other chromosomes. Additionally, it would be of interest to confirm all the transcripts we discovered in our study at the protein level, as was done in Ferrández-Peral et al. (2022).
Materials and Methods
To study the evolution of YAG transcripts across great apes, we isolated total RNA and synthesized oligo(dT)-primed cDNA from testis samples for six great ape species: human (two individuals), chimpanzee (two individuals), bonobo (one individual), gorilla (one individual), Bornean orangutan (one individual), and Sumatran orangutan (one individual). For each sample, we generated two technical replicates, which we started from separate aliquots of the same RNA stock and then each aliquot was processed separately (supplementary fig. S1, Supplementary Material online). After pulling down YAG cDNAs using gene family–specific hybridization capture probes (supplementary table S2, Supplementary Material online), two PacBio Iso-Seq libraries (one with cDNA centered around ∼2 kb and another one enriched for cDNA >3 kb) were pooled together and sequenced. We identified FLNCs (see Materials and Methods; supplementary table S4, Supplementary Material online) among circular consensus sequence (CCSs) generated from raw subreads. There were 15,529–87,639 of CCSs and 14,586–60,648 of FLNCs per technical replicate (supplementary table S4, Supplementary Material online). We next clustered FLNCs with isONclust (Sahlin and Medvedev 2020) and assigned the clusters to their respective gene families by aligning probe sequences to the FLNCs in each cluster. Finally, we performed error correction of the clustered and gene family–assigned FLNCs with IsoCon (Sahlin et al. 2018). This resulted in the total number of transcripts ranging from 526 to 2,302 per technical replicate (supplementary table S5, Supplementary Material online). This variation in part reflected differences in the sequencing yield per technical replicate (supplementary table S4, Supplementary Material online). To decrease differences in sequencing yield among samples, and also because biological replicates were unavailable for all the species, we removed human sample 2 and chimpanzee sample 1—the samples with the highest and the lowest average number of transcripts per technical replicate, respectively, in our data set (supplementary table S5, Supplementary Material online)—from subsequent analyses. We utilized these two samples to validate some of our findings from the other samples (see below). Briefly, our subsequent analyses consisted of 1) identifying replicate-supported transcripts; 2) mapping them to one human genomic copy per each YAG family and to the databases of human and nonhuman transcripts; 3) mapping predicted proteins to human Y proteins; 4) identifying identical (among samples or species) and species-specific transcripts; and 5) testing for selection acting on protein-coding transcripts (supplementary fig. S3, Supplementary Material online).
Samples, RNA Extraction, and Long-Read Sequencing
All human and nonhuman great ape samples were obtained and handled according to approved Institutional Review Board (IRB) and Institutional Biosafety Committee (IBC) protocols. Two human testis samples (IDs: A0119c and A014a) were provided by the Cooperative Human Tissue Network (CHTN) under Penn State IRB STUDY00005084. Two chimpanzee (Pan troglodytes) testis samples from individuals deceased from heart failure (IDs: 8720 and 9423) were provided by the University of Texas MD Anderson Cancer Center's Michale E. Keeling Center for Comparative Medicine and Research. One bonobo (Pan paniscus) (ID OR5013) and one Bornean orangutan (Pongo pygmaeus) (ID OR3405) testis samples were provided by San Diego Zoo Institute for Conservation Research. One western lowland gorilla (Gorilla gorilla gorilla) (ID 2006-0091) and one Sumatran orangutan (Pongo abelii) (ID 1991-0051) testis samples were provided by the Smithsonian Institution.
Total RNA was extracted from all eight testis samples (∼30 mg of tissue) using the RNeasy Mini Kit following the manufacturer-recommended protocol (Qiagen, United States). All samples had RIN value ≥6. Two technical replicates of cDNA were generated from each of the eight RNA samples using the SMARTer PCR cDNA Synthesis Kit (Clontech, United States). The universal 5′ cDNA primer and a 3′ barcoded technical replicate-specific oligo(dT) primer (Integrated DNA Technologies, United States) were used to prime the reactions (supplementary table S1, Supplementary Material online). The resulting cDNAs were used for a selective pulldown of YAG cDNAs using hybridization to in-house-designed biotinylated capture probes, synthesized by IDT. More detailed descriptions of the hybridization capture experiments are summarized in supplementary figure S1 and table S2, Supplementary Material online. To maximize enrichment, we performed the hybridization twice.
Enrichment and hybridization followed the PacBio protocol (https://www.pacb.com/wp-content/uploads/Procedure-Checklist-cDNA-Capture-Using-SeqCap-EZ-Libraries.pdf). Probes and primers (supplementary table S3, Supplementary Material online) used to validate male specificity were designed in house and synthesized by IDT. To design hybridization probes and primers, we generated consensus sequences using BWA-MEM (version 0.7.10) alignments (Li 2013). Illumina RNA-Seq, flow-sorted Y, and whole-genome sequencing data from chimpanzee, bonobo, gorilla, Bornean orangutan, and Sumatran orangutan (NCBI Sequence Read Archive under accession numbers SRR10392513–SRR10392518 and SRX7685072-SRX7685081) were aligned to the reference protein-coding sequences and visualized in Integrative Genomics Viewer (version 2.3.72) (Thorvaldsdóttir et al. 2013). To account for divergence among the great ape species, we have designed degenerate probes and primers provided in supplementary tables S2 and S3, Supplementary Material online. This was not done for VCY, which was found previously only in human and chimpanzee, and for which we used the nondegenerate probes and primers for these two species. Sixteen ampliconic cDNA samples (two per each of the eight samples) pooled in equimolar quantities were subsequently used for preparation of Iso-Seq libraries (one “standard” centered around cDNA ∼2 kb and one “longer” enriched for cDNA >3 kb) using the PacBio protocol (https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Iso-Seq-Express-Template-Preparation-for-Sequel-and-Sequel-II-Systems.pdf). The pooled samples were sequenced using the PacBio Sequel I instrument with four SMRT cells for the “standard” library and one SMRT cell for the “longer” library.
Analysis of Long Reads and Transcript Clustering
Long reads were analyzed according to the following workflow (supplementary fig. S2, Supplementary Material online). First, BAM files of CCSs were produced from the raw subreads using CCS (SMRTlink, version 6; --Polish --minPasses 3). Next, the BAM files from the five SMRT cells were merged and demultiplexed using lima (version 2.2.0, https://github.com/PacificBiosciences/barcoding; --isoseq --peek-guess), with 82% of CCSs passing quality filters. For each of the 16 resulting CCS files (two technical replicates for each of the eight samples), CCSs were classified as FLNC reads, based on the presence of poly(A) tails, which were subsequently removed using refine (Iso-Seq3, version 3.4.0, https://github.com/PacificBiosciences/IsoSeq; isoseq3 refine --require-polya). The resulting FLNC reads were clustered at the gene family level (separation by sequence similarity) with isONclust v0.0.6.1 (--t 1 --k 11 --w 15) (Sahlin and Medvedev 2020). Each read cluster produced by isONclust represented a gene family and was subsequently error corrected using IsoCon (--ignore_ends_len 15) (Sahlin et al. 2018). Finally, the error-corrected clusters were annotated to gene families by aligning probe sequences (supplementary table S2, Supplementary Material online) to the transcripts (Šošić and Šikić 2017) with annotate_clusters.py (script available at https://github.com/makovalab-psu/YAG_analysis). Specifically, each probe was forward and reverse complement and then aligned to all of the predicted transcripts in a cluster. The probe with the lowest edit distance E to a transcript in a cluster was voted with weight suggesting that the cluster belongs to the gene family for which the probe is designed. The cluster was then annotated to the gene family with the largest sum of transcript weights. We annotated a cluster rather than individual transcripts because some FLNC reads were partially missing the probe or had high edit distance to all of the gene families. However, these reads matched other regions of longer transcripts in the same cluster.
Identification of Isoforms Shared between Technical Replicates
The sequencing and analysis strategy described above produced two clusters of transcripts, corresponding to two cDNA technical replicates per sample for each combination of sample and gene family. We let X and Y denote the two sets of transcripts corresponding to the transcripts from each of the two replicates for a given gene family. In order to eliminate random errors introduced by the PCR amplification of cDNA and subsequent sequencing (supplementary fig. S1, Supplementary Material online), we identified transcripts that were supported by both replicates by running “isoform_similarity.py” (script available at https://github.com/makovalab-psu/YAG_analysis). The script consists of two steps.
First, we identified supported transcripts between X and Y, denoted by Z (supplementary fig. S2B, Supplementary Material online). We called a transcript in one of the replicates to be supported if (a) it was an exact substring of a transcript in the other replicate and (b) had a maximum 5′ difference of 100 bp and a maximum 3′ difference of 30 bp to . Note that a transcript could be supported by several transcripts in the other replicate under this definition, but it only needed at least one supporting transcript in that other replicate to be classified as supported. Intuitively, the supported transcripts Z were those that were consistently predicted between replicates.
The supported transcripts could still be redundant due to experimental variability at the ends, such as 3′-end bias caused by 3′-poly(A) tail initiation of the cDNA synthesis. Therefore, as a second step, we removed the redundancy in Z by merging any redundant transcripts into the longest representation using the same criteria as for determining support. Specifically, we merged transcript into another transcript if (1) was an exact substring of and (2) fulfilled the criteria of 100 and 30 bp maximal 5′ and 3′ offsets to . We processed the transcripts greedily from shortest to longest transcript and, for each transcript , identified if there was a longer transcript it could be merged into, according to (1) and (2). If so, then we removed from the set. Whatever remained in the set after this processing was the set of merged transcripts (supplementary fig. S2B, Supplementary Material online).
After identifying supported transcripts and removing redundancy, we constructed a final set of nonredundant replicate-supported transcripts (supplementary dataset S1, Supplementary material online), which we, for brevity, from now on refer to as replicate-supported transcripts. The end offsets of 100 and 30 bp were chosen to be consistent with the parameters used by the Iso-Seq3 pipeline (https://github.com/PacificBiosciences/IsoSeq) for merging transcripts, except that we did not follow their recommendation on merging transcripts differing by any internal gaps of less than 10 bp. This is because we expected transcripts within the same gene family to be highly similar and differ by only small internal gaps, and thus, we kept such transcripts as separate ones.
Prediction of Coding Potential
We predicted ORFs for all replicate-supported transcripts using getorf (Rice et al. 2000) (supplementary dataset S2, Supplementary material online) and retained only complete ORFs of at least 50 amino acids in length (supplementary dataset S3, Supplementary material online).
YAG Homolog Sequence Similarity Search across Great Apes
We inferred statistically significant homologs according to Pearson (2013). We allowed protein sequence identity to be <20%, but we required E values to be <10−6 and bit scores to be >50. First, we aligned replicate-supported transcripts against human and chimpanzee Y chromosome transcripts using BLASTN (Camacho et al. 2009) (supplementary datasets S4 and S5, Supplementary material online). Transcripts for gorilla and Sumatran orangutan were aligned to predicted or experimentally deciphered publicly available transcript sequences for these species (Camacho et al. 2009) using BLASTN (supplementary datasets S6 and S7, Supplementary material online, respectively). Subsequently, we narrowed down the analysis to Y chromosome protein-coding sequences (supplementary dataset S8, Supplementary material online). To ensure that more divergent sequences were not missed, we also aligned the predicted ORFs against human Y chromosome proteins using BLASTP (Camacho et al. 2009) (supplementary dataset S9, Supplementary material online).
In cases where species had missing transcripts shared between technical replicates that were homologous to human Y chromosome reference sequence, we rechecked to see whether these sequences were present in the two previous steps of the analysis: 1) before merging into replicate-supported transcripts or 2) before clustering transcripts (raw FLNC per species). Separately, we used transcripts clustered per each technical replicate and raw FLNC sequences from each species to run BLASTN analysis (Camacho et al. 2009) against the human Y chromosome coding sequences (supplementary datasets S10 and S11, Supplementary material online, respectively). Additionally, we predicted ORFs from transcripts clustered per each technical replicate and raw FLNC and ran BLASTP analysis (Camacho et al. 2009) against the human Y chromosome proteins (supplementary datasets S12 and S13, Supplementary material online, respectively).
We also counted the number of identical protein-coding sequences between samples (supplementary table S13 and dataset S14, Supplementary material online). To do this, we converted each fasta sequence into a signature and identified identical signatures that were shared using script “fasta_to_hash_4.py” (available on GitHub).
Alternative Splicing Patterns and Classification of Splice Variants
To study alternative splicing patterns of YAGs across great apes, all replicate-supported transcripts were aligned to one human genomic copy per gene family (supplementary dataset S15, Supplementary material online) using uLTRA (version v0.0.4) (Sahlin and Mäkinen 2021) with default parameters. We have chosen a human gene copy per each gene family with the highest number of exons (and in cases when more than one copy had the same highest number of exons, we have chosen one of them randomly). This allowed us to infer the evolutionary origins of replicate-supported transcripts by comparing their exon–intron structure to that of human Y-specific reference genes.
Protein Prediction and Identification of Protein Domains
To identify functionally important protein domains and conserved sites, we conducted a comprehensive screening of species-specific predicted proteins against the most advanced InterPro protein resource (Paysan-Lafosse et al. 2023), which integrates data from more than 13 protein signature databases, including the most recent tools such as MobiDB-lite for disordered regions (Piovesan et al. 2021). We focused on identifying domains that were only found in species-specific predicted proteins, as these specific protein domains might be tailored for specialized functions unique to each species (supplementary dataset S17, Supplementary material online).
Selection Tests
For selection tests, we aligned the homologous sequences per gene family from all great apes to the reference human and macaque (outgroup) Y chromosome coding sequences. We used the codeml module of PAML (version 4.8, Yang 2007) to estimate nonsynonymous-to-synonymous substitution rate ratio (dN/dS) for orthologous YAGs in human, chimpanzee, bonobo, gorilla, Sumatran orangutan, Bornean orangutan, and macaque (outgroup). Protein-coding sequences of all YAG replicate-supported transcripts were aligned using CLUSTALW (Larkin et al. 2007). The phylogenies were generated with the neighbor-joining method (with 1,000 bootstrap replicas) as implemented in MEGAX (Kumar et al. 2018). First, for each YAG family, we tested for the difference in the dN/dS ratio between a branch of interest and other branches, where the null model with a background omega estimate ωo is compared against the alternative model assuming the branch-specific omega ωs is different from the background omega ωo. P values were calculated and corrected for multiple testing with Bonferroni correction. In this step, the one-ratio model (assuming one average dN/dS ratio for the entire tree) was compared with the two-ratio model (assuming the branch-specific dN/dS ratio ωs is different from the background dN/dS ratio ωo). There was no case where the branch-specific omega was > 1 and statistically significant (after Bonferroni correction for multiple testing), but we found two cases where the branch-specific omega was < 1 with a significant P value. Thus, second, we tested for purifying selection where the likelihood of the free model with branch-specific ωs < 1 was compared with a model with a single omega equal to 1. Alignments and all the input and output files used for PAML are provided at GitHub under https://github.com/makovalab-psu/YAG_analysis/tree/master/Data_files/Selection_tests. All the hypotheses for each gene are provided in the additional file “Selection_test_details” as an extension file to supplementary table S14, Supplementary Material online.
Data Access
PacBio Iso-Seq data for each of the samples were submitted to the NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA911852. Code is available at GitHub: https://github.com/makovalab-psu/YAG_analysis. All data sets underlying this article are available at GitHub: https://github.com/makovalab-psu/YAG_analysis/tree/master/Data_files/ and in its online Supplementary Material.
Supplementary Material
Acknowledgments
We thank Craig Praul and Dan Hannon from the Penn State Genomics Core Facility for performing PacBio sequencing. We are grateful to Andrew Cartoceti from the Smithsonian Institution, Oliver Ryder from the San Diego Zoo Institute for Conservation Research, and Sarah Dysart from MD Anderson Cancer Center for providing samples for this study. We thank Barbara Arbeithuber for early discussions on hybridization capture experiments, Bob Harris for providing us with the script for counting identical fasta sequences, and Barb McGrath for her critical reading of the manuscript. This study was funded by NIH Grant R01GM130691 (to K.D.M.), NIH Grant R01GM146462 (to P.M.), and NSF Grant DBI-2138585 (to P.M.).
Contributor Information
Marta Tomaszkiewicz, Department of Biology, The Pennsylvania State University, University Park, PA 16802, USA.
Kristoffer Sahlin, Department of Mathematics, Science for Life Laboratory, Stockholm University, Stockholm, Sweden.
Paul Medvedev, Department of Computer Science and Engineering, The Pennsylvania State University, University Park, PA 16802, USA; Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA; Center for Medical Genomics, The Pennsylvania State University, University Park, PA 16802, USA; Center for Computational Biology and Bioinformatics, The Pennsylvania State University, University Park, PA 16802, USA.
Kateryna D Makova, Department of Biology, The Pennsylvania State University, University Park, PA 16802, USA; Center for Medical Genomics, The Pennsylvania State University, University Park, PA 16802, USA; Center for Computational Biology and Bioinformatics, The Pennsylvania State University, University Park, PA 16802, USA.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Literature Cited
- Bayega A, et al. 2018. Transcript profiling using long-read sequencing technologies. Methods Mol Biol. 1783:121–147. [DOI] [PubMed] [Google Scholar]
- Bellott DW, et al. 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508:494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick BK, Satta Y, Takahata N. 2007. The origin and evolution of human ampliconic gene families and ampliconic structure. Genome Res. 17:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P-R, et al. 2015. De novo origin of VCY2 from autosome to Y-transposed amplicon. PLoS One 10:e0119651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cechova M, et al. 2020. Dynamic evolution of great ape Y chromosomes. Proc Natl Acad Sci U S A. 117:26273–26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N-C, Solomon B, Mun T, Iyer S, Langmead B. 2021. Reference flow: reducing reference bias using multiple population genomes. Genome Biol. 22:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, et al. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508:488–493. [DOI] [PubMed] [Google Scholar]
- De Paoli-Iseppi R, Gleeson J, Clark MB. 2021. Isoform age—splice isoform profiling using long-read technologies. Front Mol Biosci. 8:711733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrández-Peral L, et al. 2022. Transcriptome innovations in primates revealed by single-molecule long-read sequencing. Genome Res. 32:1448–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, et al. 2006. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachini C, et al. 2009. TSPY1 copy number variation influences spermatogenesis and shows differences among Y lineages. J Clin Endocrinol Metab. 94:4016–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko GV. 2003. Estimation of divergence times for major lineages of primate species. Mol Biol Evol. 20:424–434. [DOI] [PubMed] [Google Scholar]
- Gueroussov S, et al. 2015. An alternative splicing event amplifies evolutionary differences between vertebrates. Science 349:868–873. [DOI] [PubMed] [Google Scholar]
- Hughes JF, et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463:536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Giachini C, Forti G. 2010. TSPY and male fertility. Genes (Basel). 1:308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Canzar S. 2018. Tracking alternatively spliced isoforms from long reads by SpliceHunter. Methods Mol Biol. 1751:73–88. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, et al. 2001. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 29:279–286. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1997. Functional coherence of the human Y chromosome. Science 278:675–680. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 2000. A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum Mol Genet. 9:311–319. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN]. doi: https://arxiv.org/abs/1303.3997.
- Locke DP, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucotte EA, et al. 2018. Dynamic copy number evolution of X- and Y-linked ampliconic genes in human populations. Genetics 209:907–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabanglo MF, Houry WA. 2022. Recent structural insights into the mechanism of ClpP protease regulation by AAA+ chaperones and small molecules. J Biol Chem. 298:101781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin PV, Khaitovich P, Cardoso-Moreira M, Kaessmann H. 2021. Alternative splicing during mammalian organ development. Nat Genet. 53:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills CL, et al. 2022. Functional characterization of structural genomics proteins in the crotonase superfamily. ACS Chem Biol. 17:395–403. [DOI] [PubMed] [Google Scholar]
- Murat F, et al. 2022. The molecular evolution of spermatogenesis across mammals. Nature 613:308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Costa P. 2012. Sex, rebellion and decadence: the scandalous evolutionary history of the human Y chromosome. Biochim Biophys Acta. 1822:1851–1863. [DOI] [PubMed] [Google Scholar]
- Nickkholgh B, et al. 2010. Y chromosome TSPY copy numbers and semen quality. Fertil Steril. 94:1744–1747. [DOI] [PubMed] [Google Scholar]
- Oetjens MT, Shen F, Emery SB, Zou Z, Kidd JM. 2016. Y-chromosome structural diversity in the bonobo and chimpanzee lineages. Genome Biol Evol. 8:2231–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomopoulos S, et al. 2020. Methodologies for transcript profiling using long-read technologies. Front Genet. 11:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paysan-Lafosse T, et al. 2023. Interpro in 2022. Nucleic Acids Res. 51:D418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. 2013. An introduction to sequence similarity (‘homology’) searching. Curr Protoc Bioinformatics. 42:3.1.1–3.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D, et al. 2021. MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res. 49:D361–D367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, et al. 2002. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 71:906–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Sahlin K, Mäkinen V. 2021. Accurate spliced alignment of long RNA sequencing reads. Bioinformatics 37:4643–4651 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin K, Medvedev P. 2020. De novo clustering of long-read transcriptome data using a greedy, quality value-based algorithm. J Comput Biol. 27:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin K, Tomaszkiewicz M, Makova KD, Medvedev P. 2018. Deciphering highly similar multigene family transcripts from Iso-Seq data with IsoCon. Nat Commun. 9:4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin H-S, Ichijima Y, Koh E, Namiki M, Namekawa SH. 2012. Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res. 22:827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837. [DOI] [PubMed] [Google Scholar]
- Šošić M, Šikić M. 2017. Edlib: a C/C ++ library for fast, exact sequence alignment using edit distance. Bioinformatics 33:1394–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffs K. 2004. Expression pattern of the Y-linked PRY gene suggests a function in apoptosis but not in spermatogenesis. Mol Hum Reprod. 10:15–21. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszkiewicz M, et al. 2016. A time- and cost-effective strategy to sequence mammalian Y chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 26:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszkiewicz M, Makova K. 2018. Targeted sequencing of ampliconic gene transcripts from total human male testis RNA. Protoc Exch. doi: 10.1038/protex.2018.109 [DOI] [Google Scholar]
- Vegesna R, et al. 2020. Ampliconic genes on the great ape Y chromosomes: rapid evolution of copy number but conservation of expression levels. Genome Biol Evol. 12:842–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegesna R, Tomaszkiewicz M, Medvedev P, Makova KD. 2019. Dosage regulation, and variation in gene expression and copy number of human Y chromosome ampliconic genes. PLoS Genet. 15:e1008369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EYM, et al. 2004. Identification and characterization of human VCY2-interacting protein: VCY2IP-1, a microtubule-associated protein-like protein. Biol Reprod. 70:775–784. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Ye D, et al. 2018. High levels of copy number variation of ampliconic genes across major human Y haplogroups. Genome Biol Evol. 10:1333–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen PH. 2004. Putative biological functions of the DAZ family. Int J Androl. 27:125–129. [DOI] [PubMed] [Google Scholar]
- Zhang S-J, et al. 2017. Isoform evolution in primates through independent combination of alternative RNA processing events. Mol Biol Evol. 34:2453–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou SW, et al. 2003. Expression and localization of VCX/Y proteins and their possible involvement in regulation of ribosome assembly during spermatogenesis. Cell Res. 13:171–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.