SUMMARY
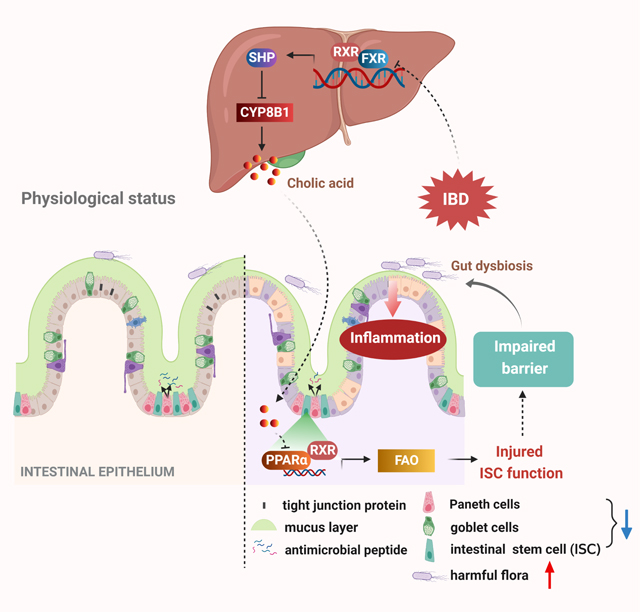
Although disrupted bile acid (BA) homeostasis is implicated in inflammatory bowel disease (IBD), the role of hepatic BA metabolism in the pathogenesis of colitis is poorly understood. Here, we found that cholic acid (CA) levels were increased in patients and mice. CYP8B1, which synthesizes CA, was induced in livers of colitic mice. CA-treated or liver Cyp8b1-overexpressing mice developed more severe colitis with compromised repair of the mucosal barrier, whereas Cyp8b1-knockout mice were resistant to colitis. Mechanistically, CA inhibited peroxisome proliferator-activated receptor (PPAR) α, resulting in impeded fatty acid oxidation (FAO) and impaired Lgr5+ intestinal stem cell (ISC) renewal. A PPARα agonist restored FAO and improved Lgr5+ ISC function. Activation of the farnesoid X receptor (FXR) suppressed liver CYP8B1 expression and ameliorated colitis in mice. This study reveals a connection between the hepatic CYP8B1-CA axis and colitis via regulating intestinal epithelial regeneration, suggesting BA-based strategies might be beneficial in IBD treatment.
Keywords: bile acids, CYP8B1, intestinal stem cell, epithelial mucosal barrier, liver-gut axis
eTOC
Xie and colleagues propose a regulatory mechanism of hepatic bile acid metabolism in colitis pathogenesis. They find that the hepatic CYP8B1-cholic acid metabolic axis impairs Lgr5+ intestinal stem cell renewal by repressing PPARα, thus exacerbating intestinal injury. Hepatic FXR activation or CYP8B1 knockout restore damaged epithelial barrier and alleviate colitis.
Graphical Abstract
INTRODUCTION
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is an idiopathic and relapsing disorder. It is characterized by the chronic mucosal inflammation that causes severe damage to the epithelial layer. While the triggers for IBD are still not fully understood, genetic and environment factors, as well as gut dysbiosis are functionally integrated to promote overactive mucosal immune responses (Ramos and Papadakis, 2019). The recent two decades witnessed remarkable progress in the development of immune-targeting therapies for IBD treatment, including anti-inflammatory agents, immunosuppressants, and the burgeoning biologics (Hindryckx et al., 2018). However, their clinical applications are limited due to unstable efficacy, severe side effects, and drug resistance. In addition to the suppression of inflammation, the healing of injured mucosa and restoration of the epithelial barrier are associated with long-term remission of IBD (Bernstein, 2015). Therefore, it is necessary to further understand the etiology of IBD, especially the regulatory mechanism of epithelial function in its pathogenesis.
Studies have shown that 1.4%–7.5% of IBD patients are accompanied by primary sclerosing cholangitis (PSC), and 70%–80% of PSC patients spontaneously develop IBD (Annese, 2019), suggesting that IBD and hepatobiliary diseases may be interrelated via liver-gut communication. Bile acids (BAs) are the final products of cholesterol catabolism, acting as amphiphilic emulsifiers facilitating the absorption of lipids from intestine and biliary excretion of cholesterol and phospholipids. They mainly modulate farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 (GPBAR1, TGR5) to regulate BA enterohepatic circulation, energy metabolism, and immune function (Schaap et al., 2014). BAs also regulate intestinal epithelial barrier, cell death and proliferation, and mucus secretion (Hegyi et al., 2018). The primary BAs are synthesized in liver via the classical and the alternative pathways. In the classical pathway, cholesterol is metabolized to 7α-hydroxycholesterol by the rate-limiting enzyme cholesterol 7α-hydroxylase (cytochrome P450 7A1, CYP7A1) and then to cholic acid (CA) by sterol 12α-hydroxylase (CYP8B1), while in the alternative pathway, sterol 27-hydroxylase (CYP27A1) and oxysterol 7α-hydroxylase (CYP7B1) convert cholesterol into chenodeoxycholic acid (CDCA). Based on their chemical structures, BAs can be divided to 12α-hydroxylated BAs (12α-OH BAs, e.g. CA and its derived deoxycholic acid (DCA), and their conjugates), and non-12α-OH BAs (e.g. CDCA, and its derived muricholic acid (MCA), ursodeoxycholic acid (UDCA), lithocholic acid (LCA), and their conjugates). It is important to note that CYP8B1 controls the content of CA and CA/CDCA or 12α-OH/non-12α-OH BAs (Jiao et al., 2022). Clinical studies have shown that serum CA was markedly increased in active UC teenage patients (Ejderhamn and Strandvik, 1991), and PSC patients with UC had higher plasma CA fraction and CA/CDCA ratios when compared with the PSC patients without IBD (Mousa et al., 2020), implying that hepatic CYP8B1-CA metabolic axis plays an important role in IBD development via the liver-gut regulation. However, the mechanism of this relationship remains unknown.
Intestinal epithelial barriers, that include both physical and chemical barriers, segregate gut commensal microbes and the host immunity to maintain intestinal homeostasis. Epithelial cell damage and subsequent barrier dysfunction are the prominent features of IBD (Atreya and Neurath, 2015). Under physiological conditions, Lgr5+ intestinal stem cells (ISCs) have the ability to quickly self-renew every 3 to 5 days to maintain barrier function and adapt to different stress stimuli (Peterson and Artis, 2014). When injury occurs, Lgr5+ ISCs replenish damaged epithelial cells, and generate specific types of cells such as secretory goblet cells and Paneth cells to maintain the intestinal mucus layer (Liu et al., 2021). Several reports have shown that BA metabolism has a potential effect on self-renew function of Lgr5+ ISCs (Huang et al., 2020; Mroz et al., 2018; Sorrentino et al., 2020). Especially, BAs physiologically activate Lgr5+ ISCs via TGR5 (Sorrentino et al., 2020). These observations prompted us to explore whether hepatic BA metabolism coordinates epithelial regeneration under injury.
Here, we demonstrated that hepatic CYP8B1-CA pathway dampened Lgr5+ ISC regeneration by repression of peroxisome proliferator-activated receptor alpha (PPARα)-dependent fatty acid oxidation (FAO), and ultimately promoted colitis, thus uncovering the functional importance of the BA-based liver-gut axis during colitis pathogenesis.
RESULTS
Bile acid metabolism is dysregulated in active UC patients and experimental colitic mice
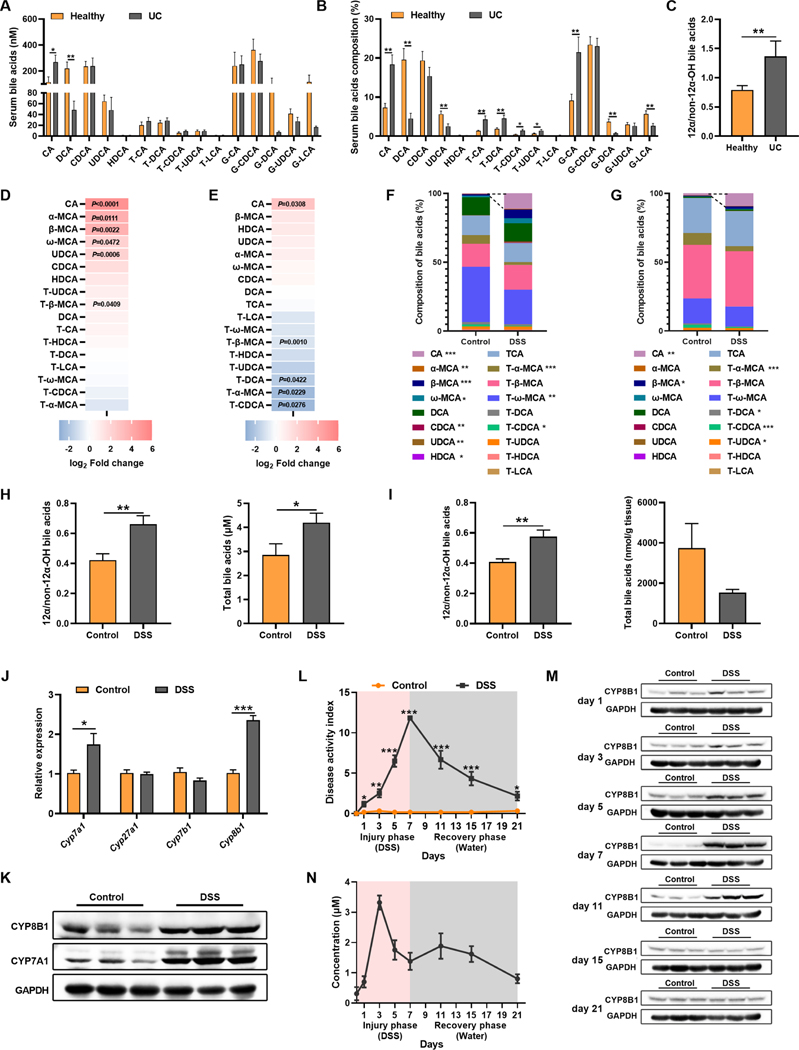
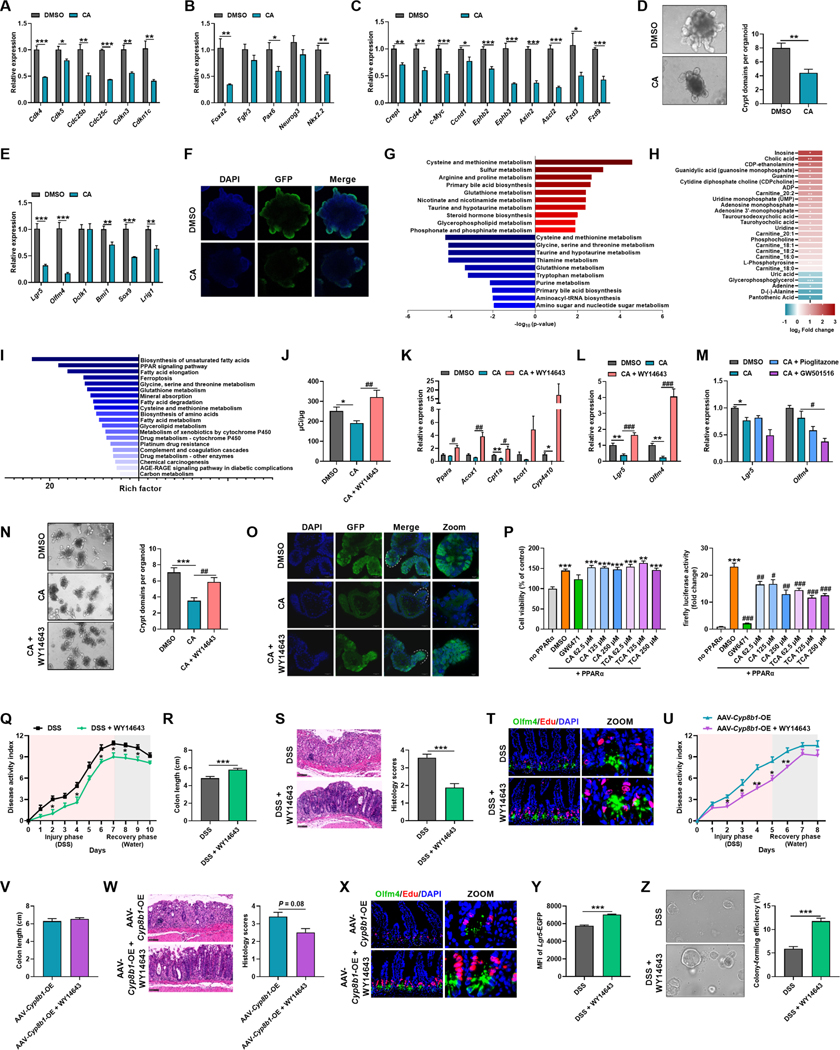
To investigate the association between BA metabolism and colitis, we determined BA levels and found that both absolute concentration and proportion of CA together with 12α-OH/non-12α-OH BAs were markedly higher in active UC patients relative to healthy controls (Figure 1A–1C). Consistently, CA and 12α-OH/non-12α-OH BAs were also increased in DSS-induced colitic mice when compared to controls (Figure 1D–1I). These data indicated the potential upregulation of CYP8B1 in liver, which was confirmed both at the mRNA and protein levels (Figure 1J and 1K). Dynamic analysis of CYP8B1 and CA during and after DSS treatment revealed that the levels of CYP8B1 and CA started to rise as early as day 1 upon DSS challenge, prior to when the obvious intestinal injury occurred; they returned to normal levels after DSS removal (Figure 1L–1N), suggesting that CA accumulates in intestine due to upregulation of CYP8B1 in liver during colitis development.
Figure 1. CYP8B1-cholic acid metabolic pathway is activated in UC patients and experimental colitic mice.
(A-C) Serum BAs concentration (A), proportion (B), and 12α-OH/non-12α-OH BA (C).
(D-K) Mice were treated with DSS for 7 days (n = 6 mice/group). Fold change of individual BA in serum (D) and intestine (E), BAs proportions in serum (F) and intestine (G), 12α-OH/non-12α-OH BA and total BAs in serum (H) and intestine (I), mRNAs of indicated genes (J) and representative immunoblots of indicated proteins (K) in liver. (L-N) Mice were treated with DSS for 7 days followed by 14 days of water (n = 4–6 mice/group). DAI (L), representative immunoblots of CYP8B1 in liver (M), serum CA level (N). (A-J, L) Mean ± SEM. Unpaired Student’s t-test. * P < 0.05; ** P < 0.01; *** P < 0.001.
CA supplementation exacerbates intestinal injury and dysregulates the mucosal barrier
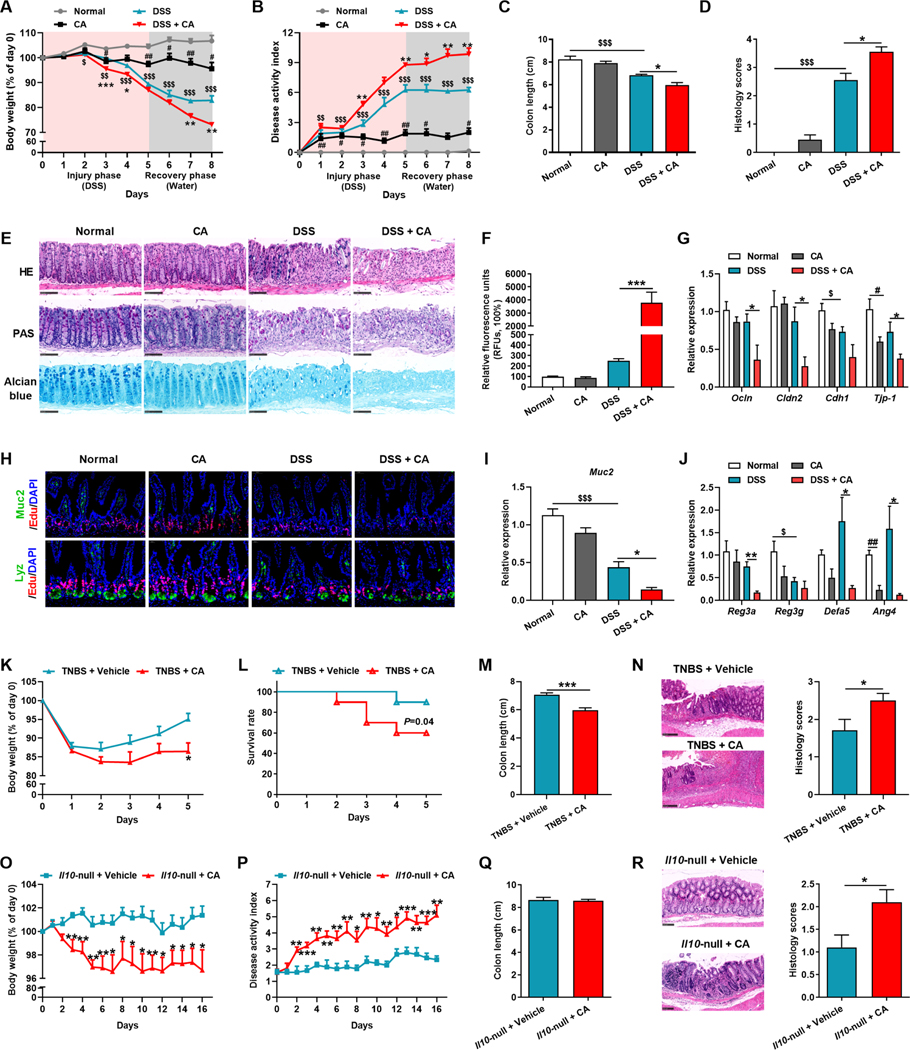
Since we found no direct evidence indicating that CA exacerbates colitis, CA supplementation was performed in colitic mice to mimic CA elevation (Figure S1A). CA treatment accelerated body weight loss, increased the disease activity index (DAI), and retarded the recovery from DSS-induced colitis (Figure 2A–2C). Histopathological evaluation further revealed that intestinal injury and epithelial mucosal barrier impairment were more evident after CA treatment (Figure 2D and 2E).
Figure 2. Administration of CA in colitic mice exacerbates intestinal injury.
(A-J) Mice were treated with/without DSS for 5 days followed by 3 days of water together with vehicle or CA. Body-weight loss (A), DAI (B), colon length (C), histology score (D), representative H&E, Periodic Acid-Schiff (PAS) and Alcian blue staining of intestine sections (E), relative fluorescence intensity of FITC-dextran in serum (F), mRNAs of indicated genes in intestine (G), representative Muc2 or Lyz with Edu staining of intestine sections (H), Muc2 (I) and AMPs (J) mRNAs in intestine. (A-E) n = 8–10 mice/group. (F-J) n = 5 mice/group.
(K-R) TNBS-induced colitic mice (K-N, n = 6–10 mice/group) and Il10-null mice (O-R, n = 9–11 mice/group) were gavaged with vehicle or CA. Body-weight loss (K and O), survival rate (L), DAI (P), colon length (M and Q), representative H&E staining and histology score of intestine sections (N and R).
(E, H, N, R) Scale bars, 100 μm. (A-D, F-G, I-R) Mean ± SEM. (A-D, F, G, I, J) One-way ANOVA with Dunnett’s post hoc test; *, DSS + CA vs DSS; #, CA vs Normal; $, DSS vs Normal. (K-R) Unpaired Student’s t-test. */#/$ P < 0.05; **/##/$ $ P < 0.01; ***/$ $ $ P < 0.001. See also Figure S1–S3.
Recent evidence indicates that mucosal barrier dysfunction contributes to IBD development (Camara-Lemarroy et al., 2018). To further explore the effects of CA on mucosal barrier, intestinal permeability was assessed by FITC-dextran assay. Serum of CA-treated colitic mice exhibited higher absorption of fluorescence than control mice, indicative of a leaky gut (Figure 2F). Meanwhile, CA significantly damaged the intestinal barrier as indicated by the diminished mRNA expression of tight junction-related proteins and Mucin2 (Muc2), which is produced by goblet cells and forms the skeleton of the intestinal mucus (Figure 2G–2I). Furthermore, CA treatment also decreased the expression of lysozyme (Lyz, a Paneth cell marker) and antimicrobial peptides (AMPs) (Figure 2H and 2J). Exogenous CA supplementation also accelerated disease progression and exacerbated intestinal injury in the trinitron-benzene-sulfonic acid (TNBS)-induced CD model (Figure 2K–2N), Il10-null spontaneous colitis model (Figure 2O–2R), and T cell-transfer model (Figure S2A–S2E).
However, CA exerted little influence on the proportion of different immune cells in mesenteric lymph nodes (MLNs) and gut lamina propria mononuclear cells (LPMCs) (Figure S3A and S3B). Correspondingly, CA did not have a direct effect on the activation of bone marrow-derived macrophages (BMDMs) or T cells in vitro (Figure S3C and S3D). In another cohort with 15 UC patients, correlation analysis also revealed that CA level was not positively correlated with inflammatory indices (Figure S3E), suggesting that CA had limited relevance on the immunological pathogenesis.
Without DSS treatment, CA itself reduced the body weight, but has no impact on soft stool, occult blood, morphological changes in the intestinal epithelium, epithelial permeability and did not change or only mildly decreased mucin and the expression of tight junction proteins, Muc2, and AMPs (Figure 2A–2J). These findings suggested that CA could not significantly disrupt barrier function to induce colitis.
Collectively, CA at pathological levels potentiates colitis synergistically with DSS-induced injury, and resultant epithelial damage, mainly by inducing the impairment of epithelial mucosal barrier, rather than directly regulating immune cells, indicating that CA functions as a pivotal enterohepatic metabolite in IBD pathogenesis.
Hepatic CYP8B1 overexpression accelerates while disturbance decelerates colitis
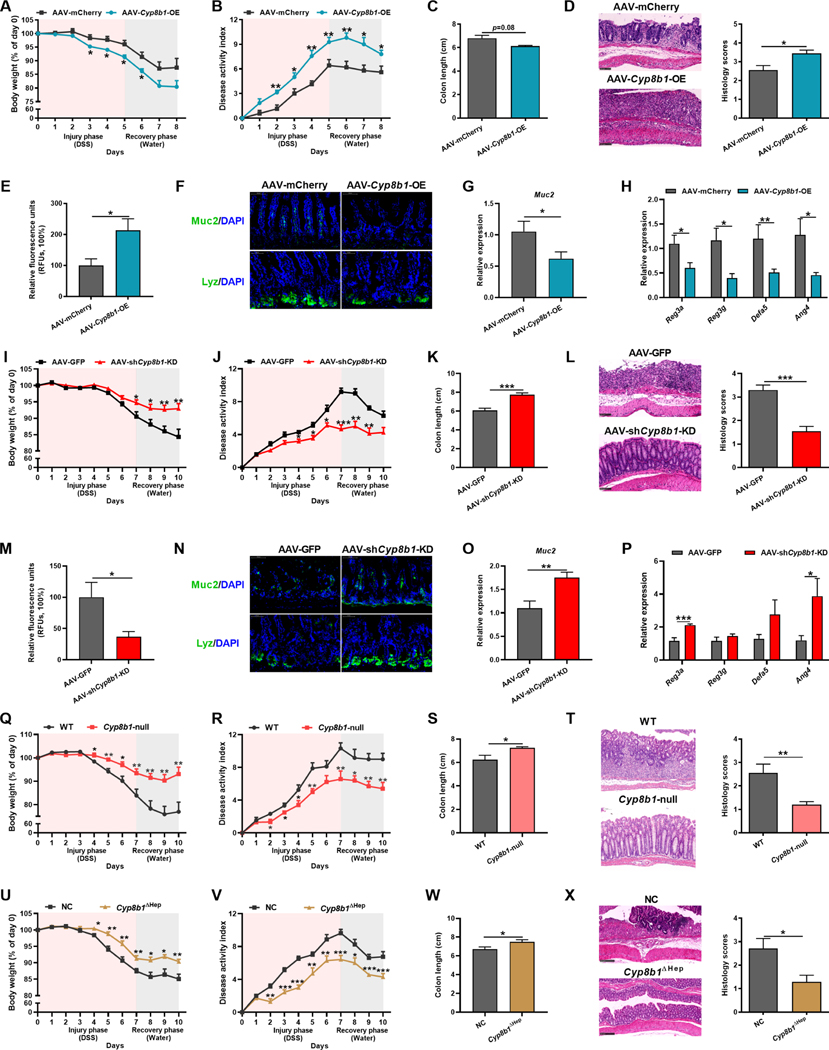
Next, to investigate the role of hepatic CYP8B1 in intestinal injury, we overexpressed CYP8B1 in liver by using adeno-associated virus (AAV-Cyp8b1-OE), which successfully elevated the CA levels without affecting remarkably other genes involved in BA metabolism (Figure S1B, S2F–S2J). AAV-Cyp8b1-OE mice phenocopied the CA supplemented mice when compared with scramble controls (AAV-mCherry) (Figure 3A–3H). Similarly, immune responses were not directly influenced (Figure S3F and S3G). These data revealed that hyper-activated CYP8B1 in liver could drive epithelial mucosal barrier dysfunction.
Figure 3. CYP8B1 overexpression potentiates colitis in mice, whereas CYP8B1 ablation ameliorates colitis.
(A-H) AAV-Cyp8b1-OE and AAV-mCherry mice were treated with DSS for 5 days followed by 3 days of water (n = 5–8 mice/group). Body-weight loss (A), DAI (B), colon length (C), representative H&E staining and histology score of intestine sections (D), relative fluorescence intensity of FITC-dextran in serum (E), representative Muc2 or Lyz staining of intestine sections (F), Muc2 (G) and AMPs (H) mRNAs in intestine.
(I-P) AAV-shCyp8b1-KD and AAV-GFP mice were treated with DSS for 7 days followed by 3 days of water (n = 8–10 mice/group). Body-weight loss (I), DAI (J), colon length (K), representative H&E staining and histology score of intestine sections (L), relative fluorescence intensity of FITC-dextran in serum (M), representative Muc2 or Lyz staining of intestine sections (N), Muc2 (O) and AMPs (P) mRNAs in intestine.
(Q-X) Cyp8b1-null mice (Q-T, n = 9–10 mice/group) and Cyp8b1ΔHep mice (U-X, n = 7–8 mice/group) were treated with DSS for 7 days followed by 3 days of water. Body-weight loss (Q and U), DAI (R and V), colon length (S and W), representative H&E staining and histology score of intestine sections (T and X).
(D, F, L, N, T, X) Scale bars, 100 μm. (A-E, G-M, O-X) Mean ± SEM. Unpaired Student’s t-test. * P < 0.05; ** P < 0.01; *** P < 0.001. See also Figure S1–S3 and S5.
In contrast to mice overexpressing CYP8B1, knockdown of CYP8B1 by AAV-shCyp8b1 in DSS model and Il10-null model featured significantly attenuated colitis symptoms (Figure 3I–3P, S1C, S2K–S2S). The whole-body and hepatic-specific CYP8B1 knockout mice also exhibited restrained CA synthesis in company with improvement of colitis (Figure 3Q–3X, S1D and S1E, S2T–S2Z, S4A–S4F). 16S rRNA sequencing data showed that hepatic CYP8B1 overexpression or knockdown did not obviously alter the structure of gut microbiota (Figure S5A–S5N), while CA could still potentiate DSS-induced colitis in microbiota-depleted mice (Figure S5O–S5S).
These observations collectively support the notion that the hepatic CYP8B1 plays an essential role in colitis pathogenesis by increasing intestinal injury and impairing mucosal barrier. The repression of liver CYP8B1 activity ameliorates DSS-induced intestinal injury, which is independent of immune response and gut microbiota.
CA accumulation triggers Lgr5+ intestinal stem cell loss and hinders epithelial regeneration
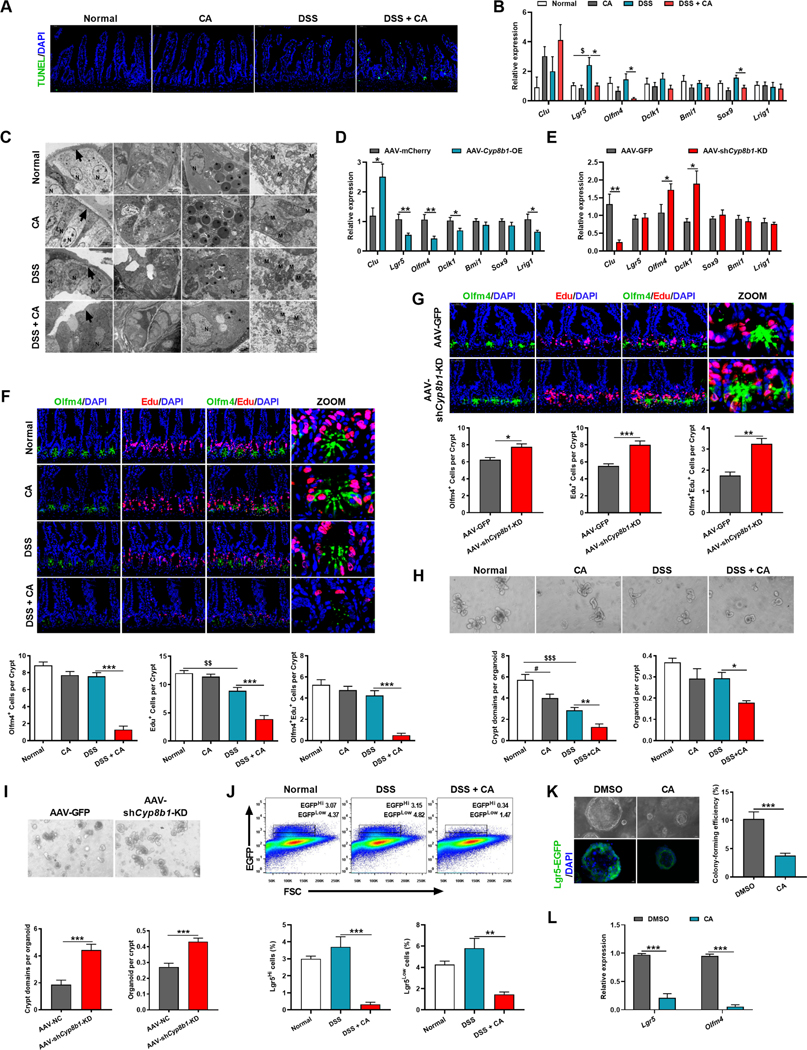
Mechanistic exploration was carried out to elucidate how CA influences integrity of the intestinal barrier. Neither CA nor DSS alone noticeably caused cell death in the epithelium, while CA supplementation in DSS-treated mice decreased proliferating cells and remarkably increased epithelial cell damage encompassing the entire crypt-villus epithelium, from the base of the crypt to the top of villi, consistent with elevated expression of the DNA damage marker Clu (Figure 4A and 4B). Similar patterns were observed in AAV-Cyp8b1-OE colitic mice, while the opposite trends were noted after CYP8B1 ablation (Figure 4C, 4D, S4G and S4H).Transmission electron microscopy (TEM) visualized significant morphological changes in the intestinal epithelium of CA-treated colitic, including disordered intestinal epithelial cells, loss of goblet cells, damaged Paneth cells, decreased mucus and eosinophil cytoplasmic granules, swelled mitochondria, as well as destruction of crypt structure (Figure 4E).
Figure 4. CA treatment reduces Lgr5+ intestinal stem cells and dampens epithelial regeneration.
(A) Representative TUNEL staining of intestinal sections of CA-treated colitic mice.
(B-D) mRNAs of indicated genes in intestine of CA-treated colitic mice (B, n = 5 mice/group), AAV-Cyp8b1-OE colitic mice (C, n = 5–6 mice/group) and AAV-shCyp8b1-KD colitic mice (D, n = 8 mice/group).
(E) Representative TEM of intestinal epithelial layer (arrows: epithelium, N: nucleus, asterisks: mucus and eosinophil cytoplasmic granules, M: mitochondria) in CA-treated colitic mice. (F and G) Representative Edu and Olfm4 staining and quantitation in intestine of CA-treated colitic mice (F) and AAV-shCyp8b1-KD colitic mice (G). n = 5 mice/group.
(H and I) Representative images and quantitation of the crypts isolated from CA-treated colitic mice (H) and AAV-shCyp8b1-KD colitic mice (I) in primary cultures (n = 5 mice/group).
(J) Representative FACS plots of Lgr5-EGFP+ cells in the crypts of CA-treated colitic mice (n = 3 mice/group).
(K and L) Representative images and colon-forming efficiency (K) and Lgr5 and Olfm4 mRNA expression (L) of isolated Lgr5+ ISCs cultured for 6 days (n = 6 biological replicates/group).
Scale bars, 50 μm (A, F, G), 100 μm (H, I, K). (B-D, F-L) Mean ± SEM. (B, F, H, J) One-way ANOVA with Dunnett’s post hoc test. (C, D, G, I, K, L) Unpaired Student’s t-test. */$ P < 0.05; **/$ $ P < 0.01; *** P < 0.001. See also Figure S4.
The intestine epithelium self-renews rapidly which is dictated by ISC to counteract intestinal damage and mucosal barrier dysfunction (Peterson and Artis, 2014). By analyzing the transcripts associated with three ISC subsets: crypt base columnar cell (CBC), also known as Lgr5+ ISC (Lgr5 and Olfm4), reserve stem cell (Dclk1 and Bmi1), and a mixed-type of stem cell (Sox9 and Lrig1), both CA supplementation and CYP8B1 overexpression dramatically reduced Lgr5+ ISCs while CYP8B1 disruption promoted the expression of ISC markers in both DSS and Il10-null models (Figure 4B, 4D, 4E, S4G–S4J). Immunofluorescent staining of Olfm4 (downstream of Lgr5) further indicated a loss of Lgr5+ ISCs in the crypt compartments of CA-treated colitic mice and AAV-Cyp8b1-OE mice. Edu+Olfm4+ cells are reported indices of proliferative Lgr5+ ISCs (Chen et al., 2022; Nusse et al., 2018; Thorne et al., 2018). CA supplementation and CYP8B1 overexpression remarkably decreased both Edu+ cells and Edu+Olfm4+ cells in crypts of colitic mice with less cells emanating from the crypt (Figure 4F and S4K), whereas CYP8B1 knockdown reserved competent Lgr5+ ISCs upon DSS treatment (Figure 4G).
Since self-renew by dividing and differentiating into the specialized cells are the principal features of ISCs (Andersson-Rolf et al., 2017), changes in ISCs function by CA were then examined. Intestinal crypts derived from the intestines of CA-treated or AAV-Cyp8b1-OE colitic mice produced fewer outgrowths when cultured ex vivo and they failed to self-expand into organoids and exhibited less budding, while crypts derived from AAV-shCyp8b1-KD colitic mice had superior budding and organoid-forming ability (Figure 4H, 4I and S4L). Consistently, organoids from CA and AAV-Cyp8b1-OE colitic mice failed to re-form after disaggregation, while organoids from AAV-shCyp8b1-KD colitic mice re-formed secondary organoids (Figure S4M–S4O). CA treatment directly dampened the compensation of both Lgr5Hi and Lgr5Low cells in the crypt after a 3-day recovery (Figure 4J), indicating that the Lgr5+ cell pool was disturbed by CA in colitic mice. Furthermore, the addition of CA at pathological concentrations (50 μM) resulted in the failure of spherical organoid formation and reduced expression of Lgr5+ ISC markers (Figure 4K and 4L). These findings suggested that the regenerative response of intestinal epithelium to injury is restrained when the CYP8B1-CA axis is activated under colitis.
CA impairs Lgr5+ intestinal stem cell renewal by repressing fatty acid oxidation
Without DSS, CA alone had no obvious influence on the number and proliferating status of ISC. However, more Edu+Olfm4+ cells located in the crypt base in CA-treated mice, whereas these cells readily emanated from the base of the crypts in control mice, indicated that CA did perturb the regenerative response of ISCs (Figure 4F). In vitro, CA addition decreased levels of mRNAs related to cell cycle, cell differentiation and the Wnt pathway, and thus decreased the number of budding and the mRNAs of ISC markers, especially for the Lgr5+ ISCs (Figure 5A–5F). These findings suggested that CA slows down the regeneration of ISCs, but has no apparent influence on the physiological function of intestinal epithelium.
Figure 5. CA treatment inhibits PPARα signaling to impede fatty acid oxidation of Lgr5+ intestinal stem cells.
(A-F) Crypts isolated from Lgr5-EGFP-IRES-creERT2 mice were treated with or without CA for 6 days (n = 4 biological replicates/group). mRNAs of genes related to cell cycle (A), cell differentiation (B) and Wnt pathway (C), representative images of organoids and quantitation of organoid buddings (D), mRNAs of ISC markers (E), representative staining of Lgr5+ cells (F).
(G-I) Isolated crypts from CA-treated colitic mice (n = 4–6 mice/group). Top 10 upregulated (red) and downregulated (blue) KEGG pathway enrichments of differential metabolites (G), the heatmap of top altered metabolites (H), and top 20 downregulated gene set pathway enrichments (I). (J-O) Crypts isolated from Lgr5-EGFP-IRES-creERT2 mice were cultured with CA together with/without WY14643/Pioglitazone/GW501516 for 6 days (n = 3–4 biological replicates/group). β-oxidation activity (J), mRNAs of indicated genes (K-M), representative images and quantitation of organoid buddings (N), representative staining of Lgr5+ cells (O). (P) Cytotoxicity and PPARα reporter assay (n = 4 biological replicates/group).
(Q-X) Colitic WT (Q-T, n = 7–8 mice/group) or AAV-Cyp8b1-OE (U-X, n = 5 mice/group) mice were administered with vehicle or WY14643. DAI (Q and U), colon length (R and V), representative H&E staining and histology score of intestine sections (S and W), representative intestinal Edu and Olfm4 staining (T and X).
(Y and Z) The statistics of Lgr5-EGFP MFI (Y) and representative images and colon-forming efficiency (Z) of Lgr5Hi cells in the crypts from colitic Lgr5-EGFP-IRES-creERT2 mice treated with vehicle or WY14643 (n = 5 mice/group).
Scale bars, 100 μm (D, F, N, O, S, W, Z), 50 μm (T, X). (A-E, J-N, P-S, U-W, Y, Z) Mean ± SEM. Unpaired Student’s t-test (A-E, Q-S, U-W, Y, Z). One-way ANOVA with Dunnett’s post hoc test (J-N, P). */# P < 0.05; **/## P < 0.01; ***/### P < 0.001. See also Figure S6.
FXR and TGR5 are the most well-known BA-activated receptors and regulate many physiological and pathological processes (Vallim and Edwards, 2009). CA exerts agonistic effects on FXR and TGR5. However, the influence of CA on Lgr5+ ISC function was independent of FXR or TGR5, since CA treatment still suppressed the ISC function and worsened colitis in Fxr-null and Tgr5-null mice (Figure S6).
On the other hand, accumulating evidence indicate cellular metabolism as a key regulator of stem cell activation and differentiation (Folmes et al., 2012; Ito and Suda, 2014). KEGG pathway enrichment of metabolomics data showed that CA mainly perturbed amino acid and nucleic acid metabolism in crypts from colitic mice, coincident with the disrupted cell cycle and limited proliferation capability (Figure 5G). Typically, metabolomics analysis showed that CA markedly augmented acylcarnitines with different chain lengths (Figure 5H), indicative of suppressed FAO in crypts. Consistently, RNA-seq showed that CA inhibited multiple metabolic pathways, especially the PPAR signaling pathway in crypts (Figure 5I), which plays a central role in the control of fatty acid metabolism (Poulsen et al., 2012). Indeed, FAO capability was lower in CA-treated organoids and the mRNAs associated with FAO were also downregulated by CA (Figure 5J and 5K). These findings proved that CA inhibited PPAR-mediated FAO in crypts.
The retardment of intestinal wound healing by CA is dependent on PPARα
The PPAR family consists of three members, PPARα, PPARγ, and PPARδ. Only pharmacologic activation of PPARα by WY14643 rescued Lgr5+ ISC loss after CA treatment (Figure 5J–5M). WY14643 increased the budding of organoids and the number of Lgr5+ ISCs in crypt cultures (Figure 5N and 5O). CA was reported to antagonize PPARα activity (Sinal et al., 2001), which was further verified by reporter assays (Figure 5P). PPARα agonism attenuated colitis symptoms in DSS-treated mice and even in AAV-Cyp8b1-OE colitic mice (Figure 5Q–5X). Of note, WY14643 enhanced the EGFP mean fluorescence intensity (MFI) of Lgr5Hi population and the stemness of Lgr5Hi ISCs on colony formation (Figure 5Y and 5Z).
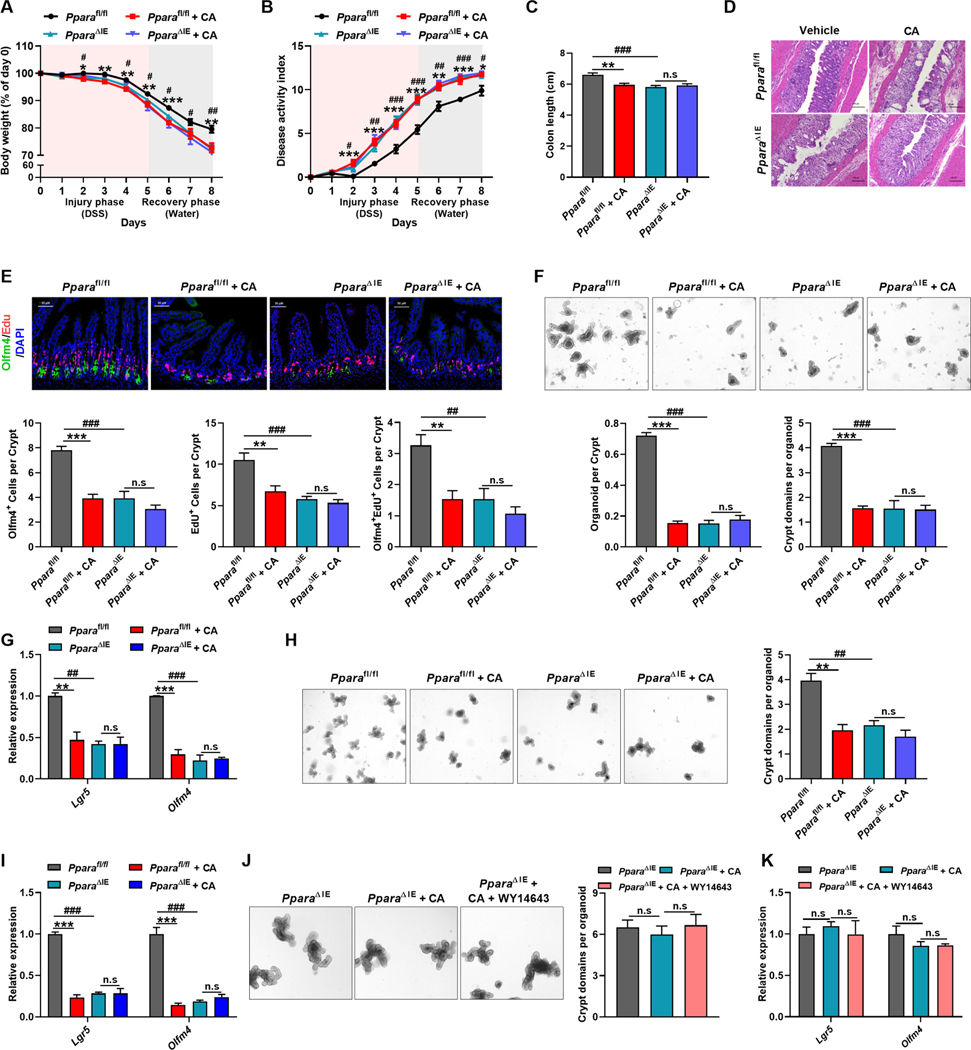
To verify the intrinsic correlation between Lgr5+ ISCs damage and intestinal PPARα signaling, intestinal-specific PPARα knockout (PparaΔIE) mice were used. Intestinal knockout of PPARα aggravated colitic symptoms to a similar extent in CA-treated Pparafl/fl mice (Figure 6A–6C). Histological analysis further demonstrated that intestinal injury was aggravated and Edu+Olfm4+ ISCs were decreased in colitic PparaΔIE mice. However, exogenous CA treatment neither worsened colitis and intestinal injury nor decreased Edu+Olfm4+ ISCs in PparaΔIE mice (Figure 6D and 6E). Consistently, crypts derived from colitic PparaΔIE mice had defective budding, organoid-forming and passaging ability, and reduced expression of Lgr5+ ISCs markers, while CA supplement exerted no additional effects on the crypts of colitic PparaΔIE mice (Figure 6F–6I). As expected, the impact of WY14643 on organoids were offset in crypts isolated from PparaΔIE mice (Figure 6J and 6K). These data demonstrated that CA impairs the regenerative function of Lgr5+ ISCs via repressing PPARα-mediated FAO, thus exacerbating intestinal injury during colitis.
Figure 6. The impairment of CA on intestinal barrier function is dependent on PPARα.
(A-I) Pparafl/fl and PparaΔIE mice were treated with DSS for 5 days followed by 3 days of water together with vehicle or CA. Body-weight loss (A), DAI (B), colon length (C), representative H&E staining of intestine sections (D), representative Edu and Olfm4 staining and quantitation in intestine (E), representative images and quantitation of the intestinal crypts and mRNAs of indicated genes of crypts in primary (F and G) and secondary (H and I) culture. (A-C) n = 8–9 mice/group. (E) n = 5 mice/group. (F-J) n = 3 mice/group.
(J and K) Representative images and quantitation (J, n = 3 biological replicates/group) and mRNAs of indicated genes (K, n = 4 biological replicates/group) of crypts from PparaΔIE mice cultured with indicated treatments for 6 days.
Scale bars, 50 μm (E), 100 μm (D, F, H, J). (A-C, E-K) Mean ± SEM. One-way ANOVA with Dunnett’s post hoc test. *, Pparafl/fl + CA vs Pparafl/fl; #, PparaΔIE vs Pparafl/fl. */# P < 0.05; **/## P < 0.01; ***/### P < 0.001.
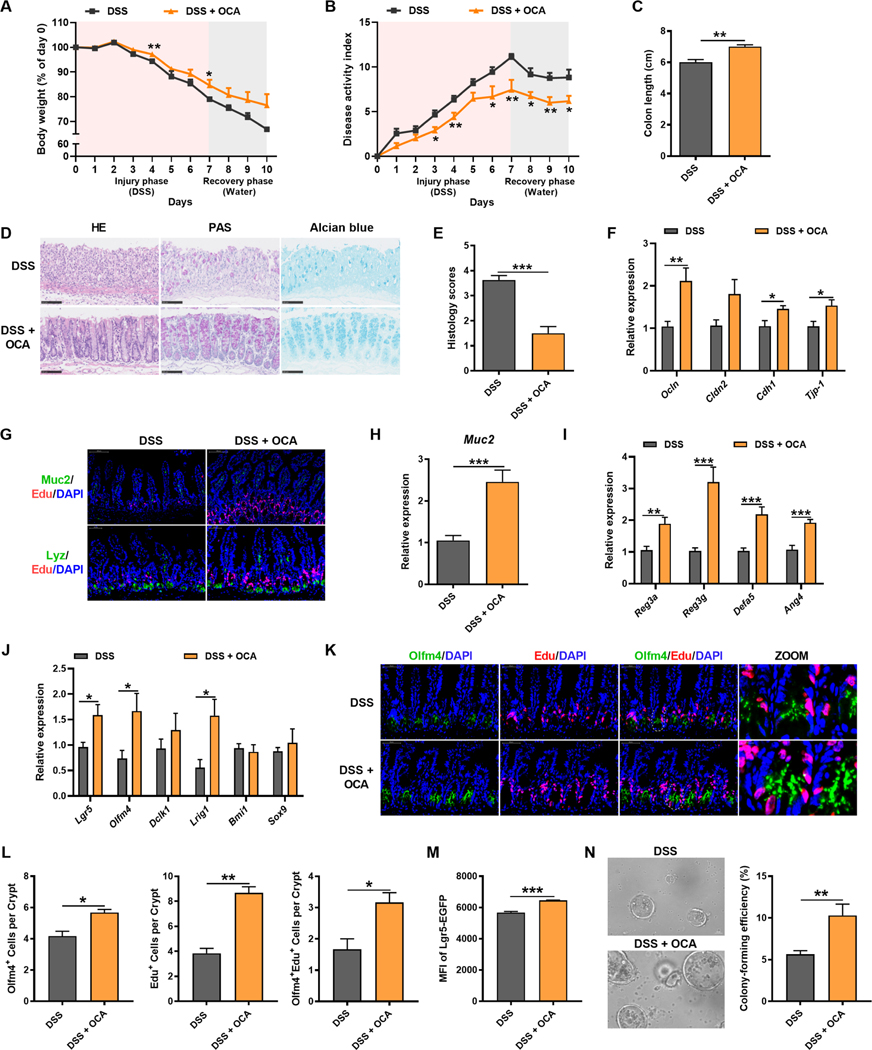
OCA treatment restores damaged epithelial mucosal barrier and alleviates colitis by targeting hepatic FXR-CYP8B1 signaling
The above findings suggested CYP8B1 as a potential target for IBD treatment. However, no specific CYP8B1 inhibitor has been described. Either hepatic or intestinal FXR activation could downregulate CYP8B1 expression (Kong et al., 2012). Notably, both hepatic and intestinal FXR signalings were suppressed in colitic mice (Figure S7A–S7C). OCA, an clinically used FXR agonist, has been reported to inhibit intestinal inflammation and preserve the barrier in experimental IBD models (Gadaleta et al., 2011). However, it remains unclear whether the therapeutic action of OCA on IBD involves altering BA metabolism and epithelial regeneration.
OCA activated both hepatic and intestinal FXR signaling, and thus depleted the expression of hepatic CYP8B1 and decreased CA synthesis (Figure S7D–S7J). In line with previous studies, OCA held therapeutic actions on DSS-induced colitis (Figure 7A–7J). Immunofluorescent staining of Olfm4 and Edu further confirmed that OCA promoted Lgr5+ ISC proliferation (Figure 7K and 7L). In addition, crypts derived from OCA-treated colitic mice had superior budding, organoid-forming, and passaging ability (Figure S7K and S7L). Similar to PPARα agonism, OCA also preserved the Lgr5Hi population and maintained the potential of ISCs on colony formation. Moreover, Lgr5Hi ISCs isolated from OCA-treated colitic mice produced larger spherical organoids (Figure 7M and 7N). These observations indicated that FXR agonism restored the intestinal barrier function by promoting the ISC renewal and epithelial regeneration. Following epithelial barrier restoration, OCA treatment obviously decreased the population of different immune cells in MLNs and LPMCs, along with reduced colonic proinflammatory cytokines, which implied a resolution of intestinal inflammation (Figure S7M and S7N). These data identified FXR as a promising therapeutic target in IBD via suppression of liver CYP8B1.
Figure 7. OCA treatment alleviates colitis in mice by improving epithelial mucosal barrier function.
Mice were treated with DSS for 7 days followed by 3 days of water together with vehicle or OCA.
(A) Body-weight loss. (B) DAI. (C) Colon length. (D) Representative H&E, PAS and Alcian blue staining of intestine sections. (E) Histology score. (F, H-J) mRNAs of indicated genes in intestine. n = 7–9 mice/group. (G) Representative Muc2 or Lyz with Edu staining of intestine sections. (K and L) Representative Edu and Olfm4 staining and quantitation in intestine (n = 5 mice/group). (M) The statistics of Lgr5-EGFP MFI (M, n = 3 mice/group) and representative images and colon-forming efficiency of Lgr5Hi cells in the crypts (N, n = 5 mice/group) from colitic Lgr5-EGFP-IRES-creERT2 mice treated with vehicle or OCA.
Scale bars, 100 μm (D, G, N), 50 μm (K). (A-C, E, F, H-J, L-N) Mean ± SEM. Unpaired Student’s t-test. * P < 0.05; ** P < 0.01; *** P < 0.001. See also Figure S7.
FXR is mainly expressed in the liver and intestine, with considerably lower expression in immune tissues (Figure S7O). To investigate whether the action of OCA depends on liver or intestinal FXR, FxrΔHep and FxrΔIE mice were used. OCA was still able to ameliorate the colitis symptoms in FxrΔIE mice (Figure S7P–S7S), while the therapeutic effects almost disappeared in FxrΔHep mice (Figure S7T–S7W), demonstrating that the protection of OCA is hepatic FXR-dependent and thereby the contribution of hepatic FXR-CYP8B1 pathway in liver-gut interaction is confirmed during colitis development.
DISCUSSION
BA enterohepatic circulation are important components of the “liver-gut communication”. IBD may cause disorders in BA homeostasis, while these changes in-turn promote IBD through BA receptor-mediated signal pathways and other mechanisms (Franzosa et al., 2019). Herein, we establish an inter-organ regulatory mechanism of hepatic BA metabolism in IBD pathogenesis, which delineates the functional importance of “liver-gut axis” theory in IBD. During colitis, the BA pool size was not changed in active UC patients compared to healthy controls, implying that CYP7A1 might not be the key enzyme regulating BA metabolism under IBD although induction of CYP7A1 was observed in colitic mice. Instead, CYP8B1 is activated and produces excessive CA that is released into the intestinal lumen and leashes the epithelial regenerative program by damaging PPARα-mediated Lgr5+ ISC renewal function, which eventually decelerates the repair of injured epithelial mucosal barrier and worsens colitis. Although resolution of inflammation is a major goal of IBD therapy, growing evidence suggest that mucosal healing is positively related to a better outcome (Yamamoto-Furusho and Parra-Holguín, 2021). Our findings provide a rationale for the therapeutic options of IBD via liver FXR agonism to counter the deleterious effects of the CYP8B1-CA axis on intestinal barrier function and thus colitis.
CYP8B1 is the main enzyme regulating CA synthesis. Thus, changes in CA levels after DSS insult should correlate with CYP8B1 levels. However, the dynamic study showed that CYP8B1 expression and serum CA levels after DSS-induced intestinal injury were not correlated, since serum CA levels peaked earlier than CYP8B1 protein expression. This lack of correlation could be due to a preferential uptake of intestinal CA to the serum soon after DSS administration followed by loss of BAs through DSS-induced diarrhea at later times. Serum CA levels decreased from day 3 after DSS, while hepatic CYP8B1 expression continued to rise, possibly as a result of the lower serum and probably liver CA. More importantly, supplementation with CA and overexpression of CYP8B1, exacerbated the colitis phenotype, whereas disruption of CYP8B1 ameliorated colitis, supporting the view that the hepatic CYP8B1 plays an important role in IBD development via liver-gut communication. Additional experiments are needed to further explore the connection between hepatic CYP8B1 expression and serum CA levels.
Studies have reported that BAs such as CA were increased in colitis, inducing gut dysbiosis with a reduced gut microbial diversity but proliferation of several aggressive pathogens to induce inflammation (Wohlgemuth et al., 2011; Zhou et al., 2020). However, hepatic CYP8B1 overexpression or knockdown only slightly altered the gut microbiome and CA increased pathogens such as E. coli at late colitis stage, suggesting that microbiota is changed as a consequence to CA accumulation in gut. Gut resident-immune cells are also recognized as crucial drivers of tissue destruction in IBD (Neurath, 2019). Nevertheless, CA did not affect the function of immune cells. It is difficult to determine the relative contribution of each event (mucosal healing, innate immunity, and gut microbiome). Our results suggest that the aggravation of colitis by CA is independent of its direct regulation of gut microbiota and immune cells, but might be attributed to its ability to increase the epithelial permeability after injury and decrease the regeneration capability, leading to a reduction of antibacterial mucins, AMPs and invasion of harmful flora that eventually trigger inflammation. It remains to be clarified the consequences of CA-impaired epithelial barrier.
Dysregulation of epithelial barrier function may directly disrupt mucosal homeostasis and lead to the IBD development (Hegyi et al., 2018). At the active stage of UC, inflammation-induced tissue damage leads to multiple ulcers. Generally, after inflammation is alleviated, epithelial cells can be repaired quickly through the organized recovery, proliferation, and differentiation from ISCs. Once the function of ISCs impaired, epithelial repair is disturbed and persistent ulceration would commence (Okamoto and Watanabe, 2005). In recent years, growing evidence suggest that mucosal healing is positively related to a better outcome (Lichtenstein and Rutgeerts, 2010). Thus, therapeutic approaches to achieve mucosal healing might be beneficial for IBD patients. BAs are among the most abundant metabolites in gut and are receiving more and more interests as critical regulators of epithelial function in health and disease (Hegyi et al., 2018). Under physiological conditions, BAs represent an intrinsic stimulus that dictates Lgr5+ ISC proliferation to sustain daily epithelial regeneration (Sorrentino et al., 2020). Herein, CA accumulation under the pathological situation of colitis was observed to induce DNA fragmentation and mitochondria dysfunction, thus triggering apoptosis in epithelial cells at the base of the crypt. Previous studies have addressed the importance of Lgr5+ ISCs in maintaining epithelial regeneration and barrier function (Metcalfe et al., 2014; Tan et al., 2021). It is possible that CA dampens Lgr5+ ISC renewal and intestinal regeneration after acute insults, which is proven by our results that both CA treatment and Cyp8b1-overexpression markedly defect the function of Lgr5+ ISCs. Thus, targeting liver BA metabolism to improve mucosal healing might provide a previously unappreciated perspective for UC treatment.
CA as a hydrophobic BA was reported to directly destroy egg phosphatidylcholine liposomes resulting in membrane lysis, and also modulated tight junction structure and barrier function (O’Connor et al., 1985; Raimondi et al., 2008), but the inhibitory mechanism of CA on Lgr5+ ISC renewal is still unclear. As an endogenous ligand, CA could activate FXR and TGR5 signalings to regulate the metabolic processes (Sarenac and Mikov, 2018). FXR restrained Lgr5+ cancer stem cell proliferation (Fu et al., 2019), while TGR5 activated Lgr5+ ISC renewal (Sorrentino et al., 2020). However, the present study proved that neither FXR nor TGR5 mediates the inhibition of Lgr5+ ISCs by CA.
Acylcarnitines transport the fatty acids into mitochondrion where they can be oxidized for energy. They can also be transported outside tissues, and an increase of plasma acylcarnitines has been linked to the progression of various diseases (Violante et al., 2013), suggestive of an obstructed FAO. PPARα, PPARγ and PPARδ regulate the transcription of multiple critical genes involved in FAO that are crucial for ISC survival and maintenance. Disruption of Cpt1a, encoding the rate-limiting enzyme in FAO, under the control of PPARα and PPARδ, was found to reduce the proliferation of ISCs, increase crypt apoptosis, and inhibit Lgr5+ ISC function under high-fat diet feeding, while short-term fasting activates PPARδ-CPT1α-mediated FAO to augment ISC function in mice (Mana et al., 2021; Mihaylova et al., 2018). Other than PPARγ and PPARδ, CA was reported to be an antagonist of PPARα (Sinal et al., 2001). Consistent with these findings, CA was found to decrease the expression of FAO-related genes including Cpt1a, and the number and budding of organoids derived from Lgr5+ ISC cultures. Similar results were obtained when organoids were treated with a PPARα antagonist (Stojanović et al., 2021). PPARα agonist largely reversed the loss of FAO activity by CA and boosted Lgr5+ ISC function. Previous studies have shown that intestinal PPARα and PPARδ double knockout mice have almost normal ISC numbers and function under normal physiologic conditions, which only pose mild to moderate stress to ISCs and do not significantly damage epithelial barrier function (Mana et al., 2021). However, we demonstrated that intestinal knockout of PPARα accelerates colitis in the case of severe intestinal epithelial injury, suggesting that PPARα might be an important subtype in intestinal epithelial repair function. Our data indicate an essential role for PPARα-FAO pathway in CA-induced ISC dysfunction.
At present, there are no agents targeting CYP8B1 on the market. To validate the translational significance of our finding, we first applied the AAV system, which is widely favored as a gene therapy vector, to deliver shCyp8b1 to the liver of colitic mice. Knockdown of hepatic Cyp8b1 ameliorated DSS-induced intestinal injury and improved mucosal barrier function, confirming CYP8B1 as a potential therapeutic target for IBD treatment. Next, OCA, a potent and selective FXR agonist was chosen to treat experimental colitis since it could indirectly suppress CYP8B1 expression (Kong et al., 2012). It was reported that OCA prevents chemical-induced intestinal inflammation and barrier damage via counteracting with NF-κB signaling in enterocytes and immune cells, and retention of dendritic cells in the spleen (Gadaleta et al., 2011; Massafra et al., 2016). However, the exact mechanism was not fully clarified because FXR is abundantly expressed in liver and intestine compared to immune tissues. In the current study, OCA significantly inhibited the expression of CYP8B1 in liver, as well as decreased the accumulation of CA in intestine, reprogrammed Lgr5+ ISC renewal, and thus relieved colitis. Although it is difficult to discriminate whether intestinal or hepatic FXR contributes more profoundly to the induction of CYP8B1 in colitis, the protective effect of OCA in colitis was found to be hepatic FXR-dependent rather than intestinal FXR-dependent, confirming a major contribution of the hepatic FXR-CYP8B1 pathway in IBD development.
In conclusion, our findings establish a cross-organ regulatory mechanism in IBD pathogenesis in which the CYP8B1-CA pathway acts as a central node linking liver BA metabolism and intestinal epithelial wound healing by coordinating the Lgr5+ ISC renewal program via the PPARα-FAO axis. From a therapeutic angle, direct disruption of CYP8B1 by the AAV-delivery system or alternatively, activation of FXR with selective and potent agonists in the liver could constitute a promising strategy for IBD treatment by enhancing the regenerative capacity of the intestinal epithelium.
LIMITATIONS OF THE STUDY
In a preliminary study, we found that complete BA clearance by BA sequester cholestyramine was not conducive to alleviate colitis as OCA did, indicating different BAs might have diverse effects on the ISC function. It is also known that humans and mice possess different BA profiles which might bring additional complexity in humans that cannot be elucidated in mice. Thus, it is necessary to further investigate the role of different BA metabolic pathways in the progression of IBD in humans. Moreover, the involvement of the directly regulatory effect by FXR agonists on immune cells could not be excluded during this process, since a reduced infiltration of immune cells was noted in OCA-treated mice. More clinical studies should be directed to validate FXR as a therapeutic target for IBD and combined therapies of FXR agonists with immune-targeting agents for the treatment of relapsing and refractory ulcerative colitis.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Cen Xie (xiecen@simm.ac.cn).
Materials availability
All unique/stable reagents and materials generated in this study will be made available upon completion of a Materials Transfer Agreement.
Data and code availability
16S rRNA gene sequencing and RNA-seq data were deposited at SRA and GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-rabbit IgG-HRP | Cell Signaling Technology | Cat# 7074S; RRID: AB_2099233 |
| Goat anti-mouse IgG-HRP | Cell Signaling Technology | Cat# 7076S; RRID: AB_330924 |
| Goat anti-rabbit IgG Alexa Fluro 488 conjugate | Cell Signaling Technology | Cat# 4412; RRID: AB_1904025 |
| Goat anti-rabbit IgG Alexa Fluor 488 conjugate | Abcam | Cat# ab150077; RRID: AB_2890897 |
| Goat anti-rabbit IgG horseradish peroxidase conjugate | Servicebio | Cat# G1215 |
| Goat anti-mouse IgG BioMag | Qiagen | Cat# 310007 |
| Hamster anti-mouse CD11c APC conjugate | BD Biosciences | Cat# 550261; RRID: AB_398460 |
| Hamster anti-mouse CD28 | BD Biosciences | Cat# 553295; RRID: AB_394764 |
| Hamster anti-mouse CD3e | BD Biosciences | Cat# 553057; RRID: AB_394590 |
| Hamster anti-mouse CD3 PerCP-Cy5.5 conjugate | BD Biosciences | Cat# 551163; RRID: AB_ AB_394082 |
| HRP-conjugated monoclonal mouse anti-GAPDH | KangChen | Cat# KC-5G5 |
| Mouse monoclonal anti-Muc2 | Santa Cruz Biotechnology | Cat# sc-59859; RRID: AB_784653 |
| Mouse monoclonal anti-FXR/NR1H4 | Santa Cruz Biotechnology | Cat# sc-25309; RRID: AB_628039 |
| Rabbit monoclonal anti-Lysozyme | Abcam | Cat# ab108508; RRID: AB_10861277 |
| Rabbit monoclonal anti-Olfm4 | Cell Signaling Technology | Cat# 39141; RRID: AB_2650511 |
| Rabbit polyclonal anti-GFP | Abcam | Cat# ab290; RRID: AB_303395 |
| Rabbit polyclonal anti-CYP8B1 | Abcam | Cat# ab191910; RRID: AB_2828000 |
| Rabbit polyclonal anti-CYP7A1 | Abcam | Cat# ab65596; RRID: AB_1566114 |
| Rat Anti-Mouse CD16/CD32 | BD Biosciences | Cat# 553142 |
| Rat anti-mouse Ly-6G PE conjugate | BD Biosciences | Cat# 551461; RRID: AB_394208 |
| Rat anti-mouse CD45 BV510 conjugate | BD Biosciences | Cat# 563891; RRID: AB_2734134 |
| Rat anti-mouse CD11b BUV395 conjugate | BD Biosciences | Cat# 563553; RRID: AB_2738276 |
| Rat anti-mouse F4/80 FITC conjugate | Thermo Fisher Scientific | Cat# 11-4801-85; RRID: AB_2637192 |
| Rat anti-mouse CD326 (Ep-CAM) PE conjugate | Biolegend | Cat# 118205; RRID: AB_1134176 |
| 7-AAD | BD Biosciences | Cat# 559925; RRID: AB_2869266 |
| Bacterial and virus strains | ||
| AAV2/8-TBG-mCherry | Hanbio | N/A |
| AAV2/8-TBG-m-Cyp8b1-3xflag-null | Hanbio | N/A |
| AAV2/9-CMV-eGFP | Genomeditech | N/A |
| AAV2/9-CMV-m-Cyp8b1-shRNA | Genomeditech | N/A |
| Biological samples | ||
| Human: UC patient serum | Shanghai Tenth People’s Hospital | N/A |
| Human: Healthy adult serum | Shanghai Tenth People’s Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Alcian blue | Sigma-Aldrich | Cat#: 109-09; CAS: 33864-99-2 |
| Acetonitrile | Thermo Fisher Scientific | Cat#: A955-4; CAS: 75-05-8 |
| Bacitracin | meilunbio | Cat#: MB1374; CAS: 1405-87-4 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat#: V900933; CAS: 9048-46-8 |
| B27 | Gibco | Cat#: 17504044 |
| CHIR99021 | MedChemExpress | Cat#: HY-10182; CAS: 252917-06-9 |
| Chloroform | Sinopharm | Cat#: 10006818; CAS: 67-66-3 |
| Cholic acid (CA) | Sigma-Aldrich | Cat#: C1129; CAS: 81-25-4 |
| DAPI | Sigma-Aldrich | Cat#: D1306; CAS: 28718-90-3 |
| DAPI | Abcam | Cat#: ab104139; CAS: 28718-90-3 |
| Dextran sodium sulfate (DSS) | MP Biomedicals | SKU: 02160110-CF; CAS: 9011-18-1 |
| Dispase II | Sigma-Aldrich | Cat#: D4693; CAS: 42613-33-2 |
| DMEM/F12 | Gibco | Cat#: C11330500BT |
| DNase I | Sigma-Aldrich | Cat#: 10104159001 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#: D4540; CAS: 67-68-5 |
| d5-TCA | Toronto research chemicals | Cat#: T008852; CAS: 81-24-3 |
| Epidermal growth factor (EGF) | Peprotech | Cat#: 315-09 |
| Fetal bovine serum (FBS) | Gibco | Cat#: AHW87326 |
| FITC-dextran 4KD | Sigma-Aldrich | Cat#: 68059 |
| Fixative for TEM | Servicebio | Cat#: G1102 |
| Formic acid | Tokyo Chemical Industry | Cat#: F0654; CAS: 64-18-6 |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | Cat#: 100-0485 |
| Glutamax | Gibco | Cat#: 35050061 |
| GW501516 | MedChemExpress | Cat#: HY-10838; CAS: 317318-70-0 |
| GW6471 | MedChemExpress | Cat#: HY-15372; CAS: 880635-03-0 |
| Hematoxylin and eosin (H&E) | Servicebio | Cat#: G1003 |
| HEPES | Gibco | Cat#: 15630080 |
| IntestiCult Organoid Growth Medium (Mouse) with Supplement 1 and 2 | STEMCELL Technologies | Cat#: 06005 |
| Isopropanol | Thermo Fisher Scientific | Cat#: A461-4; CAS: 67-63-0 |
| L-Carnitine Inner Salt | Sigma-Aldrich | Cat#: C0158; CAS: 541-15-1 |
| Lipopolysaccharide (LPS) | Sigma-Aldrich | Cat#: 2880; CAS: 297-473-0 |
| Matrigel Matrix for Organoid Culture, Phenol Red-free, LDEV-free | Corning | Cat#: 356255 |
| Metronidazole | Meilunbio | Cat#: MB2200; CAS: 443-48-1 |
| Mouse colony-stimulating factor | Peprotech | Cat#: AF-315-02 |
| Methanol | Sigma-Aldrich | Cat#: 34860; CAS: 67-56-1 |
| 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazoliumbromide (MTT) | Sigma-Aldrich | Cat#: 2128; CAS: 298-93-1 |
| Na-Palmitate | Sigma-Aldrich | Cat#: P9767; CAS: 408-35-5 |
| N-acetylcysteine | Sigma-Aldrich | Cat#: A0737; CAS: 38520-57-9 |
| N2 | Gibco | Cat#: 17502001 |
| Neomycin | Meilunbio | Cat#: MB1716; CAS: 1405-10-3 |
| Nicotinamide | MedChemExpress | Cat#: HY-B0150; CAS: 98-92-0 |
| Noggin | Peprotech | Cat#: 250-38 |
| Obeticholic acid (OCA) | MedChemExpress | Cat#: HY-12222; CAS: 459789-99-2 |
| OsO4 | Ted Pella Inc. | Cat#: 18456; CAS: 20816-12-0 |
| Paraformaldehyde | Servicebio | Cat#: G1101 |
| Penicillin-Streptomycin | Gibco | Cat#: 15140122 |
| Periodic acid Schiff (PAS) | Servicebio | Cat#: G1008 |
| Pioglitazone | MedChemExpress | Cat#: HY-13956; CAS: 111025-46-8 |
| Protease inhibitor cocktail | Roche Life Science | Cat#: 11836170001 |
| RPMI 1640 | Gibco | Cat#: 72400047 |
| R-spondin-1 | Peprotech | Cat#: 315-32 |
| Scintillation cocktail | China Isotope & Radiation Corp. | Cat#: 1200-4 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | Cat#: V900859; CAS: 151-21-3 |
| Streptomycin | Meilunbio | Cat#: MB1275; CAS: 3810-74-0 |
| Taurocholic acid (TCA) | Sigma-Aldrich | Cat#: T4009; CAS: 345909-26-4 |
| Trinitron-benzene-sulfonic acid (TNBS) | Sigma-Aldrich | Cat#: P2297; CAS: 2508-19-12 |
| Triton X-100 | Beyotime | Cat#: ST795 |
| TrypLE | Gibco | Cat#: 12604013 |
| Type IV collagenase | Sigma-Aldrich | Cat#: C4-28 |
| UltraPure 0.5 M EDTA | Invitrogen | Cat#: 1557520 |
| WY14643 | MedChemExpress | Cat#: HY-16995; CAS: 50892-23-4 |
| Y-27632 | MedChemExpress | Cat#: HY-10071; CAS: 146986-50-7 |
| 4% paraformaldehyde | Servicebio | Cat#: G1101 |
| 5-Ethynyl-2’-deoxyuridine (Edu) | MedChemExpress | Cat#: HY-118411; CAS: 61135-33-9 |
| [9,10-3H]- palmitic acid | PerkinElmer | Cat#: NET043001MC |
| Critical commercial assays | ||
| BCA protein assay kit | Thermo Fisher Scientific | Cat#: 23255 |
| EDU staining kit | RiboBio | Cat#: C10310 |
| FIREFLYGLO Luciferase Assay Kit | Meilunbio | Cat#: MA0519 |
| Hieff qPCR SYBR Green Master Mix (High Rox) | Yeasen | Cat#: 11203ES08 |
| Hifair II 1st Strand cDNA Synthesis SuperMix (gDNA digester plus) | Yeasen | Cat#: 11123ES60 |
| In-situ cell death detection kit | Roche | Cat#: 11684795910 |
| LANCE® Ultra cAMP Kit | PerkinElmer | Cat#: TRF0262 |
| Mouse TNF-α ELISA kit | BD Biosciences | Cat#: 560478 |
| Mouse IL-6 ELISA kit | BD Biosciences | Cat#: 555240 |
| Mouse IFN-γ ELISA kit | BD Biosciences | Cat#: 555138 |
| Mouse IL-17 ELISA kit | Thermo Fisher Scientific | Cat#: BMS6001 |
| Mouse IL-1β ELISA kit | Thermo Fisher Scientific | Cat#: 88-7013-88 |
| QIAquick PCR purification kit | Qiagen | Cat#: 28104 |
| RNeasy mini kit | Qiagen | Cat#: 74106 |
| RNA simple total RNA kit | Tiangen Biotech | Cat#: DP419 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Fisher Scientific | Cat#: 34578 |
| Tissue protein extraction reagent | Thermo Fisher Scientific | Cat#: 78510 |
| Urine fecal occult blood test kit | Nanjing Jiancheng Bioengineering Institute | Cat#: C027-1-1 |
| Deposited data | ||
| The raw data of RNA-seq generated | This paper | GEO: GSE210290 |
| The raw data of 16S rRNA gene sequencing | This paper | SRA: PRJNA864099 |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Beijing Huafukang Bioscience Co. Inc. | N/A |
| Mouse: Il10-null | Gempharmatech | N/A |
| Mouse: Cyp8b1-null | Gempharmatech | N/A |
| Mouse: SCID | Gempharmatech | N/A |
| Mouse: Cas9LSL+/+ | The Jackson Laboratory | N/A |
| Mouse: Pparafl/fl | Luo et al., 2019 | N/A |
| Mouse: PparaΔIE | Luo et al., 2019 | N/A |
| Mouse: Fxr-null | Sinal et al., 2000 | N/A |
| Mouse: FxrΔIE | Kim et al., 2007 | N/A |
| Mouse: FxrΔHep | Kim et al., 2007 | N/A |
| Mouse: Lgr5-EGFP-IRES-creERT2 | Shanghai Model Organisms Center | NM-KI-200154 |
| Mouse: Tgr5-null | Provided by Prof. Xin Xie at Shanghai Institute of Materia Medica | N/A |
| Oligonucleotides | ||
| See Table S3 for qRT-PCR | This paper | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| Endnote | Endnote | http://endnote.com; |
| Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| FlowJo_V10 | BD Biosciences | https://www.flowjo.com/solutions/flowjo; |
| Leica Application Suite X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/details/product/leica-las-x-ls/ |
| NDP.view 2 | Hamamatsu | https://www.hamamatsu.com/jp/en/product/type/U12388-01/index.html |
| Pannoramic Viewer | 3DHISTECH | http://www.3dhistech.com/pannoramic_viewer |
| Prism 8 | GraphPad software | https://www.graphpad.com/ |
| Progenesis QI | Waters Corp. | http://www.nonlinear.com/progenesis/qi-for-proteomics/ |
| SIMCA 16 | Umetrics | http://umetrics.com/products/simca |
| Xcalibur | Thermo Fisher Scientific | http://chemistry.unt.edu/~verbeck/LIMS/Manuals/XCAL_Quant.pdf |
| Other | ||
| Acquity UPLC/Synapt XS HDMS system | Waters Corp. | N/A |
| Acquity BEH C18 column | Waters Corp. | SKU: 186002352 |
| Applied Biosystems 7900HT B Fast Real-Time PCR System | Applied Biosystems | N/A |
| BD LSRFortessa Fortessa Flow Cytometer | BD Biosciences | N/A |
| BD FACSAriaII Flow Cytometer | BD Biosciences | N/A |
| ChemiDoc MP Imaging System | Bio-Rad | N/A |
| Hitachi HT-7800 TEM | Hitachi | N/A |
| Illumina NovaSeq 6000 | Illumina | N/A |
| InfinityLab Poroshell 120 Hilic-Z column | Agilent technologies | 675775-924 |
| Leica TCS SPS microscope | Leica Microsystems | N/A |
| Leica DM6B laser microdissection systems | Leica Microsystems | N/A |
| LTQ Orbitrap mass spectrometer | Thermo Fisher Scientific | N/A |
| Olympus CKX53 microscope | Olympus | N/A |
| Spectramax M5 microplate reader | Molecular Devices | N/A |
| UPLC CSH column | Waters Corp. | SKU: 186005297 |
| Vanquish™ UHPLC | Thermo Fisher Scientific | N/A |
| 2450 Microplate Counter | PerkinElmer | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human samples
Serum samples were taken from 35 active UC patients and 35 healthy controls. Clinical information is summarized in Table S1. For correlation analysis, serum samples were taken from 15 active UC patients and the clinical information is summarized in Table S2. This study was approved by the Research Ethics Committee of Shanghai Tenth People’s Hospital, and written informed consent was given to all individuals before participation in the study.
Mouse Models
Male C57BL/6J mice (10- to 12-week-old, 23–25 g) and BALB/c mice were obtained from Beijing Huafukang Bioscience Co. Inc. Il10 deficient (Il10-null) mice (B6/JGpt-I-l10em1Cd4885/Gpt), whole-body Cyp8b1 knockout (Cyp8b1-null) mice and SCID mice were purchased from Gempharmatech. Cre-dependent Cas9 knock in (Cas9LSL+/+) mice were purchased from Jackson Laboratory. Lgr5-EGFP-IRES-creERT2 mice were purchased from Shanghai Model Organisms Center. Pparafl/fl and PparaΔIE mice were described previously (Luo et al., 2019). Fxr-null, intestinal Fxr-null (FxrΔIE) and hepatic Fxr-null (FxrΔHep) mice were described previously (Kim et al., 2007; Sinal et al., 2000). Tgr5-null mice were kindly provided by Prof. Xin Xie (Shanghai Institute of Materia Medica). Mice were maintained under a standard 12-h light/12-h dark cycle with water and food provided ad libitum. All experiments were performed according to the institutional ethical guidelines on animal care and approved by the Institute Animal Care and Use Committees at Shanghai Institute of Materia Medica and the National Cancer Institute.
METHOD DETAILS
Mouse treatments
For the induction of DSS-induced experimental colitis, C57BL/6J mice were put on drinking water containing 2% (w/v) DSS (MP Biomedicals) for 7 days. For CA treatment, C57BL/6J mice were administered with 400 mg/kg CA (i.g., Sigma-Aldrich) as reported (González-Peña et al., 2017; Srivastava et al., 2000) or vehicle concurrently treated with 2% DSS in drinking water for 5 days, followed by a 3-day recovery period on regular drinking water. For WY14643 or OCA treatment, C57BL/6J mice were administered 1 mg/kg WY14643 (i.g., MedChemExpress) or 20 mg/kg OCA (i.g., MedChemExpress) or vehicle concurrently treated with 2% DSS in drinking water for 7 days, followed by a 3-day recovery period on regular drinking water.
To monitor the severity of DSS-induced intestinal injury and inflammation, the body-weight loss ratio, stool consistency and rectal bleeding were assessed daily according to the described criterion. Briefly, for body weight, the changes were calculated in relative to the weight at baseline (100%). Weight loss of 1–5%, 5–10%, 10–20%, and >20% was scored as 1, 2, 3, and 4, respectively. Stool consistency was scored 0 for normal-formed pellets, 1 for soft but still formed stools, 2 for soft stools, 3 for very soft and wet stools, and 4 for watery diarrhea. To assess the occult blood in the stool, urine fecal occult blood test kit (Nanjing Jiancheng Bioengineering Institute) was used. 0 was scored for negative hemoccult, 1 for weakly positive hemoccult, 2 for positive hemoccult, 3 for visible blood traces in stool, and 4 for gross bleeding. The disease activity index (DAI) was calculated as the total of these three scores ranging from 0 (no inflammation) to 12 (severe colitis). The serum, liver, intestine and cecal content were collected at the endpoints of the experiments.
For the induction of TNBS-induced experimental colitis, 5% (w/v in H2O) TNBS (Sigma-Aldrich) was diluted with an equal volume of absolute ethanol to acquire 2.5% (w/v in 50% ethanol) TNBS solution. After being anesthetized with pentobarbital sodium, C57BL/6J mice were intrarectally administered with 100 μL 2.5% (w/v in 50% ethanol) TNBS solution via a 3.5 F catheter. The mice were then treated with 400 mg/kg CA (i.g.) or vehicle daily from day 0 to day 5. The body-weight loss ratio and survival rate were monitored.
For the induction of T cell transfer experimental colitis, the isolated CD3+CD4+CD25- T cells from BALB/c mice were injected (i.p.) into recipient SCID mice at a density of 3 × 105 per animal. After transfer, the recipient mice were treated with 400 mg/kg CA (i.g.) or vehicle for 15 days from day 0. The body-weight loss ratio and DAI were monitored every two days.
For the establishment of pseudo-germfree mice, C57BL/6J mice were treated with the antibiotic cocktail (1 mg/mL bacitracin, 1 mg/mL neomycin, 1 mg/mL streptomycin and 1 mg/mL metronidazole, Meilunbio) for 3 days, and replaced with 2% DSS for 5 days, followed by a 3-day recovery period on regular drinking water. The body-weight loss ratio and DAI were monitored every day.
Due to the limited quantity of mouse intestine samples, the animal studies were repeated for phenotype analysis, histological staining, TEM, intestinal permeability assays, qPCR, immunoblotting, bile acid analysis, crypt or ISC isolation, FACS. In addition, the susceptivity of different batches of mice to DSS was not consistent. Mice that were clearly under duress were killed in order to reduce pain and suffering before the end of study, according to the laboratory animal welfare guidelines. Thus, the animal number at the end of the experiment was always less than at the beginning of the experiment. The ranges for the mouse numbers in each experiment are indicated in figure legends, and the exact mouse numbers are found in the scatter plots within each figure.
Construction of AAV-Cyp8b1-OE or AAV-shCyp8b1 mice
AAV2/8-TBG-mCherry (AAV-mCherry) and AAV2/8-TBG-m-Cyp8b1-3xflag-null (AAV-Cyp8b1-OE) were constructed by Hanbio Biotechnology. TBG promoter was used to achieve hepatocyte-specific overexpression. AAV2/9-CMV-eGFP (AAV-GFP), AAV2/9-CMV-m-Cyp8b1-shRNA (AAV-shCyp8b1-KD, target sequence: ggtgtgaagatggcctctttc) were constructed by Genomeditech. For AAV transduction, the viruses (1 × 1011 genomic copies/mouse) were delivered via tail vein injection to 5- to 6-week-old mice for 4 weeks, and then the transfected mice were subjected to DSS treatment.
Construction of Cas9 knockout mice
The colony of Cas9LSL+/+ mice was maintained by crossing with the wild-type C57BL/6J mice. Liver-specific depletion of CYP8B1 was achieved via the CRISPR technology as previously described (Chen and Ding, 2022; Wei et al., 2020). Briefly, 8-week-old male Cas9LSL+/+ animals were randomly divided into groups, and AAV8 vectors expressing Cre recombinase and Cyp8b1-sgRNA (GCCTATCCTTGGTGATGCTA) were administered by tail vein injection. A firefly luciferase expression cassette was also included in the AAV vector to assist in vivo evaluation of delivery efficiency. AAV vectors with Cre recombinase and luciferase cassettes and no sgRNA were used as control viruses. Viruses were dissolved in 250 μL PBS and were administered at a dose of 2 × 1012 vector genomes per mouse.
Isolation of crypts, culture of intestinal organoids and treatments
Crypts were isolated from mice as described with modifications (Sorrentino et al., 2020). Briefly, intestines were isolated, and then flushed with PBS using a 50 mL syringe. The intestines were opened longitudinally and washed with cold PBS gently to remove the luminal contents. The tissue was cut into 2 mm2 fragments and further washed 3–4 times with cold PBS until the supernatant was clear. Intestinal samples were then incubated with 5 mM EDTA-PBS at 4 °C for 40 min and then the lysis buffer was replaced with PBS. The fragments were shaken vigorously and the suspension was filtered through a 70-μm filter. Enriched crypts were washed once with cold PBS and resuspended with DMEM/F12 medium (Gibco). The crypts were counted, plated with Matrigel (Corning) and IntestiCult Organoid Growth Medium with Supplement 1 and 2 (STEMCELL Technologies). Organoids were exposed to 50 μM CA (Sigma-Aldrich), 100 μM WY14643 (MedChemExpress), 10 μM pioglitazone (MedChemExpress) and 1 μM GW501516 (MedChemExpress) for 6 days. Medium was replaced every other day. The images of organoids were captured and the number of buds were counted under an Olympus CKX53 microscope. Clonogenicity (colony-forming efficiency) was calculated by plating 150 crypts and assessing organoid formation 3–7 days or as specified after initiation of cultures. For secondary organoid assays, primary organoids were dissociated by Gentle Cell Dissociation Reagent (GCDR, STEMCELL Technologies), centrifuged and resuspended with cold DMEM/F12. Cells were seeded onto Matrigel as previously described. Secondary organoids were enumerated on day 3, unless otherwise specified.
Histological analysis
Tissue specimens were fixed in 10% formalin for 12–24h, dehydrated and paraffin embedded. The sections were stained with hematoxylin and eosin (H&E). Histological evaluations were graded as follows: 0, no signs of inflammation; 1, low level inflammation with scattered mononuclear cells; 2, moderate inflammation with multiple foci of mononuclear cells; 3, high level inflammation with increased vascular density, marked wall thickening, moderate goblet cell loss and focal loss of crypts; and 4, maximal inflammation with transmural leukocyte infiltration, massive loss of goblet cells, extensive fibrosis, and diffuse loss of crypts. To visualize goblet cells in epithelial layer, paraffin-embedded tissue samples were stained with PAS to characterize the neutral mucins or Alcian blue to demonstrate acidic mucins. Images were acquired using Leica DM6B laser microdissection systems.
Edu assay
To measure the proliferation of intestinal epithelial cells, 5-ethynyl-2’-deoxyuridine (Edu, MedChemExpress) at 100 mg/kg was intraperitoneally injected into mice 2 h prior to killing. Intestines were collected and Edu positive cells were detected by Edu staining kit (RiboBio).
Immunohistochemistry and immunofluorescence
For tissue samples, paraffin-embedded intestinal sections were dewaxed in xylene and rehydrated through gradient alcohols. The masked antigens were retrieved by 0.01 M citrate buffer solution. After being blocked with 5% bovine serum albumin (BSA, Sigma-Aldrich), the tissue sections were stained with anti-Muc2 (Santa Cruz Biotechnology), anti-Lysozyme (Abcam) and anti-Olfm4 (Cell Signaling Technology). Immunohistochemistry was analyzed by goat anti-rabbit IgG conjugated to horseradish peroxidase (Servicebio). For immunofluorescence, signals were detected by goat anti-rabbit Alexa Fluor 488 conjugate (Abcam), and then counterstained with DAPI (Abcam). For intestinal organoids, after being fixed by 4% paraformaldehyde (Servicebio) and permeated by 0.5% Triton X-100 (Beyotime), the organoids were blocked by 2% BSA, incubated with anti-GFP (Abcam) at 4 °C overnight, and stained by goat anti-rabbit IgG Alexa Fluro 488 conjugate (Cell Signaling Technology) for 1 h, then 1 mg/mL DAPI (Sigma-Aldrich) was used to stain the nucleus. The images were captured and analyzed by Leica TCS SPS microscope and Leica Application Suite X software.
Quantitative real-time PCR
Total RNA was extracted using RNA simple total RNA kit (Tiangen Biotech) according to the manufacturer’s protocol and reverse transcribed by Hifair Ⅱ 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus, Yeasen). Real-time PCR was performed with Hieff qPCR SYBR Green Master Mix (High Rox, Yeasen) on an Applied Biosystems 7900HT Fast Real-Time PCR System. Real-time PCR primer sequences are included in the Table S3.
Immunoblotting
Intestines were lysed with sodium dodecyl sulfate sample buffer containing protease inhibitor cocktail (Roche Life Science). Protein concentrations were determined by the BCA protein assay kit (Thermo Fisher Scientific). Equal protein amounts were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The membranes were blocked with 5% BSA and then incubated overnight at 4 °C with primary antibodies against FXR (Santa Cruz Biotechnology), CYP8B1 (Abcam) and CYP7A1 (Abcam). Then the blots were incubated with anti-rabbit IgG or anti-mouse IgG antibody (Cell Signaling Technology), and HRP-conjugated monoclonal mouse anti-GAPDH (KangChen) as control for normalization. Signals were further visualized by SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) under ChemiDoc MP Imaging System (Bio-Rad).
Single cell preparation and flow cytometry assay
The single cell suspensions of mesenteric lymph nodes were prepared by grinding and filtering through a 40-μm nylon mesh strainer.
The single cell suspensions from intestinal lamina propria were prepared as previously described with slight modification (Zhou et al., 2019). Briefly, intestinal pieces were incubated in RPMI-1640 containing 10% FBS and 5 mM EDTA for 15min and then digested using 0.5 mg/mL Type IV collagenase (Sigma-Aldrich), 3 mg/mL dispase II (Sigma-Aldrich), and 0.1 mg/mL DNase I (Sigma-Aldrich) in a 37 °C shaking incubator for 30 min. Single cells were filtered using a 70-μm filter and then washed twice with PBS. Then the single cells were blocked with purified anti-mouse CD16/CD32 mAb (2.4G2, BD Biosciences) and stained with BV510-conjugated anti-CD45 mAb (BD Biosciences), BUV395-conjugated anti-CD11b mAb (BD Biosciences), FITC-conjugated anti-F4/80 mAb (Thermo Fisher Scientific), APC-conjugated anti-CD11c mAb (BD Biosciences), PE-conjugated anti-Ly6G mAb (BD Biosciences) and PerCP-Cy5.5-conjugated anti-CD3 mAb (BD Biosciences) in dark.
For single cells derived from crypts of Lgr5-EGFP-IRES-creERT2 mice, fresh intestinal crypts were incubated in TrypLE (Invitrogen) containing 10 μM Rho inhibitor Y-27632 (MedChemExpress) for 30–40 min at 37 °C to obtain a single-cell suspension. After terminating the digestion, the dissociated cells were resuspended in cold FACS buffer (1 mM EDTA-PBS containing 1% BSA and 10 μM Y-27632) and pass through 30-μm cell strainer. The single cells were stained with PE-conjugated anti-CD326 mAb (Biolegend) in dark and then resuspended in cold FACS buffe with the viability dye 7-AAD (BD Biosciences). Flow cytometry was performed on a BD LSR Fortessa Flow Cytometer or BD FACSAriaII Flow Cytometer, and the data were analyzed with FlowJo software (BD Biosciences).
TdT-mediated dUTP nick-end labeling (TUNEL) assay
Cell death was detected with an in-situ cell death detection kit (Roche) following the manufacture’s instruction. Briefly, paraffin-embedded intestinal sections were dewaxed in xylene and rehydrated through gradient alcohols. The slides were then incubated with 0.1% Triton X-100 in 0.1% sodium citrate buffer for 8 minutes followed by PBS washing. Next, the samples were incubated with the TUNEL reaction mixture in a humidified chamber at 37 °C for 60 min protected from light. This was followed by PBS washes and incubation with 1 mg/mL DAPI (Sigma-Aldrich) for 10 min. Fluorescence microscopy was performed using a Leica TCS SPS microscope.
Transmission electron microscopy (TEM) analysis
Intestinal sections were fixed in fixative for TEM (Servicebio) at 4 °C and then post-fixed with 1% OsO4 (Ted Pella Inc.) for 2 h. After being embedded in resin, ultrathin sections (70 nm) were cut and stained. The structures of colonic sections were analyzed using a Hitachi HT-7800 TEM.
FITC–dextran intestinal permeability assay
Intestinal permeability was determined using a permeability probe FITC–dextran 4KD (Sigma-Aldrich). Mice were fasted overnight and orally administered FITC-dextran (600 mg/kg). After 4 h, the fluorescence intensity of serum FITC-dextran was obtained at 480 nm excitation wavelength and 520 nm emission wavelength using a Spectramax M5 microplate reader (Molecular Devices).
Cell cultures and in vitro stimulation
Single Lgr5+ cells were isolated from crypts of Lgr5-EGFP-IRES-creERT2 mice by flow cytometry as described above. Sorted cells were collected, pelleted and embedded in Matrigel (Corning) followed by seeding on a 24-well plate (50 μL/well) and culture medium (Advanced DMEM/F12 supplemented with penicillin-streptomycin, 10 mM HEPES, 10 mM Glutamax, 1×N2, 1×B27 (all from Gibco), 1 mM N-acetylcysteine (Sigma-Aldrich), 5 μM CHIR99021 (MedChemExpress) and 10 mM nicotinamide (MedChemExpress)) containing growth factors 50 ng/mL EGF, 100 ng/mL Noggin, 500 ng/mL R-spondin-1 (all from Peprotech)) was overlaid. 10 mM Y-27632 (MedChemExpress) was included for the first 2 days to avoid anoikis. The entire medium was changed every other day. Single cells were exposed to 50 μM CA (Sigma-Aldrich) at the indicated concentrations for 6 days.
Bone marrow-derived macrophages (BMDMs) were differentiated as described previously (Lu et al., 2020). Briefly, BMDMs were separated from the femur and tibia bones of male C57BL/6J mice, and then cultured for 7 days in RPMI-1640 medium containing 10% FBS (Gibco) and 10 ng/mL of mouse colony-stimulating factor (M-CSF, Peprotech). The purity of macrophages (CD11b+F4/80+) was consistently >98% by flow cytometry analysis. Differentiated BMDMs were incubated with CA at the indicated concentrations for 24 h and the cytotoxicity assay was performed according to the 5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazoliumbromide (MTT, Sigma-Aldrich) method in 96-well microplates. Moreover, cells were treated with CA at the indicated concentrations in the presence of lipopolysaccharide (LPS, Sigma-Aldrich) for 24 h. After incubation, the supernatants were collected and used to quantify the cytokine production.
Purified T cells from male C57BL/6J mice were prepared by using immunomagnetic negative selection to deplete B cells as described previously with slight modification (Hou et al., 2009). Lymphocytes were incubated with magnetic particles bound to BioMag goat anti-mouse IgG (Qiagen). The purity of the resulting T cell populations was examined by flow cytometry analysis and was consistently >90%. Purified T cells were incubated with CA at the indicated concentrations for 24 h and then 5 mg/mL MTT was added to determine the cell viability. Moreover, purified T cells were cultured with 1 μg/mL anti-CD28 mAb (BD Biosciences) in 96-well flat-bottom plates coated with 5 μg/mL anti-CD3 mAb (BD Biosciences) for 24 h. CA was added simultaneously. After incubation, the supernatants were used to quantify the cytokine production.
Enzyme-linked immunosorbent assay (ELISA)
Intestines from mice were homogenized with tissue protein extraction reagent (Thermo Fisher Scientific) to extract total protein. The concentration of total protein was determined with the BCA protein assay kit (Thermo Fisher Scientific). Cytokines levels in tissue homogenates and culture supernatants were measured by mouse TNF-α, IL-6 and IFN-γ ELISA kits (BD Biosciences) and mouse IL-17 and IL-1β ELISA kit (Thermo Fisher Scientific), according to the manufacturer’s instructions
FAO measurement
The FAO activity was determined by quantifying the radioactivity of final product generated from 3H-palmitate β-oxidation. Crypts were seeded in 24-well plates and treated with DMSO or 50 μM CA or 50 μM CA combined with 100 μM WY14643 for 6 days. Then the culture medium was changed to basic medium containing [9,10-3H]- palmitic acid (PerkinElmer, Inc.), L-carnitine inner salt (Sigma-Aldrich) and Na-palmitate (Sigma-Aldrich). After incubation for 4 h at 37 °C, 300 μL culture medium was collected and mixed with 1 mL activated charcoal to absorb remaining [9,10-3H]-palmitic acid. After shaking for 30 min, the mixture was centrifuged and 300 μL supernatant was collected and mixed with 1 mL of scintillation cocktail (China Isotope & Radiation Corp.). The radioactivity was then determined by scintillation counter (2450 Microplate Counter, PerkinElmer, Inc.).
Reporter assay
For measurements of PPARα ligand activation, HEK293T cells were seeded in 96-well plates, and transfected with pSV-hRXRα and pGL4.11-PPRE-Luc with or without pSG5-hPPARα, at a density of 3 × 104 cells per well and cultured overnight. CA or taurocholic acid (TCA) at various concentrations (62.5, 125 and 250 μM) and 10 μM GW6471 (positive control) were added to the cells and the incubation lasted for 24 h at 37 °C. For measurement of FXR lignad activation, HEK293T cells were seeded in 96-well plates and transfected with pCMV-Script-hFXR and pGL4.11-hSHP-luciferase constructs, at a density of 3 × 104 cells per well and cultured overnight. CA or TCA at various concentration (62.5, 125 and 250 μM) and 10 μM OCA (positive control) were added to the cells and the incubation continued for 24 h at 37 °C. Luciferase activities were detected by use of the FIREFLYGLO Luciferase Assay Kit (Meilunbio, MA0519) according to the manufacturer’s instructions.
TGR5 activation assay
For the detection of TGR5 activation, HEK293T cells were seeded in 6-well plates at a density of 9×105 cells per well, and transfected with pCMV-Sport 6/h_TGR5 and CREB-luciferase reporter plasmid for 24 h. Cells were dissociated and resuspended with 0.1% BSA and 0.5 mM IBMX to a final density of 4×105 cells/mL. The suspension was treated with positive control INT777 (1 and 10 μM), CA (62.5, 125 and 250 μM) and TCA (62.5, 125 and 250 μM) for 30 min at room temperature. cAMP levels were measured by LANCE® Ultra cAMP Kit (PerkinElmer) according to the manufacturer’s instructions. The TR-FRET signal was read on a TR-FRET microplate reader (SYNERGY H1, Biotek, Inc).
16S rRNA gene sequencing
Fresh cecum content samples were collected for 16S rRNA sequencing by Shanghai Majorbio Bio-Pharm Technology Co.Ltd.. Microbiota DNA was extracted by fecal DNA isolation kit (QIAGEN, Dusseldorf, Germany). DNA was amplified using universal bacterial primers targeting the 16S rRNA variable V3-V4 region (341F: 5′-ACTCCTACGGGAGGCAGCAG-3′; 806R: 5′-GGACTACHVGGGTWTCTAAT-3′). Amplification products were separated using a 2.0% agarose gel and purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Bacterial DNA was sequenced on the Illumina HiSeq PE 150 platform and adapter-ligated DNA fragments were further sequenced according to standard protocols. UCLUST clustered the operational taxonomic units with 97% similarity. 16S rRNA gene sequencing analysis was performed as previously described (Lozupone and Knight, 2005).
RNA-Seq and data analysis
Total RNA from freshly sorted crypts was isolated using RNeasy mini kit (Qiagen, Germany). The library construction and sequencing were performed by Shanghai Biotechnology Corporation. Strand-specific libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation kit (Illumina, USA) following the manufacturer’s instructions. Purified libraries were quantified by Qubit 2.0 Fluorometer (Life Technologies, USA) and validated by Agilent 2100 bioanalyzer (Agilent Technologies, USA) to confirm the insert size and calculate the mole concentration. The samples were sequenced on the Illumina NovaSeq 6000 (Illumina, USA). After that, the clean reads were mapped to the mouse reference genome and the uniquely mapped fragments of each gene were counted by Stringtie (version 1.3.0). The expression levels from different samples were normalized by the Trimmed Mean of M values (TMM) method and the normalized expression levels of different samples were converted to FPKM (Fragments Per Kilobase of transcript per Million mapped fragments). Differentially expressed genes in RNA-seq data were determined by edgeR software (version 3.30.3). The pathway enrichment analysis was performed using the ClusterProfiler package (version 3.16.1) through R programming language (version 3.6.2).
Bile acid analysis
For serum samples, 50 μL serum was added to 100 μL cold acetonitrile solution containing 1 μM d5-TCA (Toronto Research Chemicals) as an internal standard. For tissue samples, 20 mg of tissue were homogenized with an acetonitrile solution containing 1 μM d5-TCA. After being precipitated, the upper supernatants were mixed with 0.1% formic acid solution. Then the concentrations of BAs were determined by Acquity UPLC/Synapt XS HDMS system (Waters Corp.). Chromatographic separation was achieved on an Acquity BEH C18 column (100 mm × 2.1 mm, 1.7 μm, Waters Corp.) at 45 °C with a gradient elution of mobile phase of 0.1% formic acid in water and 0.1% formic acid in acetonitrile (v/v) at a flow rate of 0.4 mL/min. Mass spectrometry was carried out under electrospray negative (ESI-) ion mode. A mass range of m/z 50 to 850 was acquired.
Metabolomics and lipidomics
Intestinal crypts were isolated as described above and extracted with methanol/water/chloroform mixtures. The organic phase and aqueous phase were subjected to evaporation to dryness under vacuum, respectively. The aqueous extracts were reconstituted with 65% acetonitrile for further metabolomics analysis. The organic extracts were suspended with chloroform: methanol (1:1, v/v) solution, diluted with isopropanol: acetonitrile: water (2:1:1, v/v/v) solution for further lipidomics analysis.
Metabolomics analysis was carried out on an Acquity UPLC/Synapt XS HDMS system via positive and negative ionization. Samples were separated by an InfinityLab Poroshell 120 Hilic-Z column (100 mm × 2.1 mm, 2.7 μm, Agilent technologies). Q-TOF mass analyzer was operated at 22, 000 mass resolution, and the scan range covered m/z 50–1500. The untargeted metabolomics data were processed with Progenesis QI software and the ultimate output termed ‘feature’ table with total ion ionization normalization was exported into SIMCA-P+15 software (Umetrics) for multivariate analysis. The databases, such as HMDB (http://www.hmdb.ca/), METLIN (https://metlin.scripps.edu/), KEGG (https://www.kegg.jp/), and as well as the in-house databases were applied for metabolite identification.
Lipidomics analysis was carried out by using a Vanquish UHPLC coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). Samples were eluted with gradient from a UPLC CSH column (100 mm × 2.1 mm, 1.7 μm, Waters Corp.). The MS analysis alternated between MS and data dependent MSn scans using dynamic exclusion. The scan range covered m/z 50–1500. For acylcarnitines analysis, data were proceeded with Xcalibur software to obtain the corresponding relative levels with internal standard normalization.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism 8.3.0 (GraphPad Software Inc., CA, USA). Experimental data are presented as mean ± SEM. Outliers were excluded from statistical analyses by the outlier analysis using the ROUT method (GraphPad Prism 8.3.0). Statistical significance was determined using unpaired two-tailed Student’s t-test between two groups and one-way ANOVA with Dunnett’s post hoc test for multi-group comparisons. Correlations were assessed by nonparametric Spearman’s test. The P values are noted in each figure legend and indicated as */$/# P < 0.05, **/$ $/## P < 0.01, ***/$ $ $/### P < 0.001. The statistical details for each experiment are indicated in the figure legend.
Supplementary Material
Highlights.
CA accumulates in the gut during colitis due to activation of hepatic CYP8B1
Exogenous CA or liver CYP8B1 overexpression potentiate intestinal injury
Excessive CA triggers Lgr5+ ISCs dysfunction by suppressing PPARα-mediated FAO
Activation of liver FXR and ablation of CYP8B1 expression alleviate colitis
ACKNOWLEDGMENTS
We acknowledge Dr. Hao Chen and Dr. Zhifu Xie from Shanghai Institute of Materia Medica for help with animal studies and FAO assays, as well as the valuable suggestions. We thank Prof. Shiyan Yu from Shanghai Jiao Tong University and Prof. Jun Qin from Shanghai Institute of Nutrition and Health for help with the ISC isolation. This study was supported by the National Key Research and Development Program of China (2021YFA1301200), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39020600), Shanghai Municipal Science and Technology Major Project, National Natural Science Foundation of China (91957116 and 82173873), the Shanghai Committee of Science and Technology (21S11907700) and the National Cancer Institute Intramural Research Program.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andersson-Rolf A, Zilbauer M, Koo BK, and Clevers H. (2017). Stem Cells in Repair of Gastrointestinal Epithelia. Physiology (Bethesda) 32, 278–289. 10.1152/physiol.00005.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annese V. (2019). A Review of Extraintestinal Manifestations and Complications of Inflammatory Bowel Disease. Saudi J Med Med Sci 7, 66–73. 10.4103/sjmms.sjmms_81_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya R, and Neurath MF (2015). IBD pathogenesis in 2014: Molecular pathways controlling barrier function in IBD. Nat Rev Gastroenterol Hepatol 12, 67–68. 10.1038/nrgastro.2014.201 [DOI] [PubMed] [Google Scholar]
- Bernstein CN (2015). Treatment of IBD: where we are and where we are going. Am. J. Gastroenterol 110, 114–126. 10.1038/ajg.2014.357 [DOI] [PubMed] [Google Scholar]
- Camara-Lemarroy CR, Metz L, Meddings JB, Sharkey KA, and Wee Yong V. (2018). The intestinal barrier in multiple sclerosis: implications for pathophysiology and therapeutics. Brain 141, 1900–1916. 10.1093/brain/awy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ME, Naeini SM, Srikrishnaraj A, Drucker DJ, Fesler Z, and Brubaker PL (2022). Glucagon-Like Peptide-2 Stimulates S-Phase Entry of Intestinal Lgr5+ Stem Cells. Cellular and molecular gastroenterology and hepatology. 10.1016/j.jcmgh.2022.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, and Ding Q. (2022). Optimized protocols for efficient gene editing in mouse hepatocytes in vivo using CRISPR-Cas9 technology. STAR Protoc 3, 101062. 10.1016/j.xpro.2021.101062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejderhamn J, and Strandvik B. (1991). Serum bile acids in relation to disease activity and intake of dietary fibers in juvenile ulcerative colitis. Digestion 50, 162–169. 10.1159/000200757 [DOI] [PubMed] [Google Scholar]
- Folmes CD, Dzeja PP, Nelson TJ, and Terzic A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11, 596–606. 10.1016/j.stem.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4, 293–305. 10.1038/s41564-018-0306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T, Coulter S, Yoshihara E, Oh TG, Fang S, Cayabyab F, Zhu Q, Zhang T, Leblanc M, Liu S, et al. (2019). FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 176, 1098–1112 e1018. 10.1016/j.cell.2019.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen ECL, Renooij W, Murzilli S, Klomp LWJ, Siersema PD, Schipper MEI, Danese S, et al. (2011). Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60, 463–472. 10.1136/gut.2010.212159 [DOI] [PubMed] [Google Scholar]
- González-Peña D, Giménez L, de Ancos B, and Sánchez-Moreno C. (2017). Role of dietary onion in modifying the faecal bile acid content in rats fed a high-cholesterol diet. Food & function 8, 2184–2192. 10.1039/c7fo00412e [DOI] [PubMed] [Google Scholar]
- Hegyi P, Maleth J, Walters JR, Hofmann AF, and Keely SJ (2018). Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol. Rev 98, 1983–2023. 10.1152/physrev.00054.2017 [DOI] [PubMed] [Google Scholar]
- Hindryckx P, Vande Casteele N, Novak G, Khanna R, D’Haens G, Sandborn WJ, Danese S, Jairath V, and Feagan BG (2018). The Expanding Therapeutic Armamentarium for Inflammatory Bowel Disease: How to Choose the Right Drug[s] for Our Patients? J Crohns Colitis 12, 105–119. 10.1093/ecco-jcc/jjx117 [DOI] [PubMed] [Google Scholar]
- Hou LF, He SJ, Wang JX, Yang Y, Zhu FH, Zhou Y, He PL, Zhang Y, Yang YF, Li Y, et al. (2009). SM934, a water-soluble derivative of arteminisin, exerts immunosuppressive functions in vitro and in vivo. Int Immunopharmacol 9, 1509–1517. 10.1016/j.intimp.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Huang D, Xiong M, Xu X, Wu X, Xu J, Cai X, Lu L, and Zhou H. (2020). Bile acids elevated by high-fat feeding induce endoplasmic reticulum stress in intestinal stem cells and contribute to mucosal barrier damage. Biochem Biophys Res Commun 529, 289–295. 10.1016/j.bbrc.2020.05.226 [DOI] [PubMed] [Google Scholar]
- Ito K, and Suda T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15, 243–256. 10.1038/nrm3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao TY, Ma YD, Guo XZ, Ye YF, and Xie C. (2022). Bile acid and receptors: biology and drug discovery for nonalcoholic fatty liver disease. Acta pharmacologica Sinica 43, 1103–1119. 10.1038/s41401-022-00880-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, and Gonzalez FJ (2007). Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res 48, 2664–2672. 10.1194/jlr.M700330-JLR200 [DOI] [PubMed] [Google Scholar]
- Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, and Guo GL (2012). Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology (Baltimore, Md.) 56, 1034–1043. 10.1002/hep.25740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein GR, and Rutgeerts P. (2010). Importance of mucosal healing in ulcerative colitis. Inflamm Bowel Dis 16, 338–346. 10.1002/ibd.20997 [DOI] [PubMed] [Google Scholar]
- Liu CY, Cham CM, and Chang EB (2021). Epithelial wound healing in inflammatory bowel diseases: the next therapeutic frontier. Translational research : the journal of laboratory and clinical medicine. 10.1016/j.trsl.2021.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, and Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology 71, 8228–8235. 10.1128/aem.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Li H, Fan C, Qi Q, Yan Y, Wu Y, Feng C, Wu B, Gao Y, Zuo J, et al. (2020). RIPK1 inhibitor ameliorates colitis by directly maintaining intestinal barrier homeostasis and regulating following IECs-immuno crosstalk. Biochem Pharmacol 172, 113751. 10.1016/j.bcp.2019.113751 [DOI] [PubMed] [Google Scholar]
- Luo Y, Xie C, Brocker CN, Fan J, Wu X, Feng L, Wang Q, Zhao J, Lu D, Tandon M, et al. (2019). Intestinal PPARα Protects Against Colon Carcinogenesis via Regulation of Methyltransferases DNMT1 and PRMT6. Gastroenterology 157, 744–759.e744. 10.1053/j.gastro.2019.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana MD, Hussey AM, Tzouanas CN, Imada S, Barrera Millan Y, Bahceci D, Saiz DR, Webb AT, Lewis CA, Carmeliet P, et al. (2021). High-fat diet-activated fatty acid oxidation mediates intestinal stemness and tumorigenicity. Cell Rep 35, 109212. 10.1016/j.celrep.2021.109212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massafra V, Ijssennagger N, Plantinga M, Milona A, Ramos Pittol JM, Boes M, and van Mil SW (2016). Splenic dendritic cell involvement in FXR-mediated amelioration of DSS colitis. Biochim Biophys Acta 1862, 166–173. 10.1016/j.bbadis.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Metcalfe C, Kljavin NM, Ybarra R, and de Sauvage FJ (2014). Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell stem cell 14, 149–159. 10.1016/j.stem.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, Abu-Remaileh M, Clavain L, Erdemir A, Lewis CA, et al. (2018). Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell stem cell 22, 769–778 e764. 10.1016/j.stem.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa OY, Juran BD, McCauley BM, Vesterhus MN, Folseraas T, Turgeon CT, Ali AH, Schlicht EM, Atkinson EJ, Hu C, et al. (2020). Bile Acid Profiles in Primary Sclerosing Cholangitis and Their Ability to Predict Hepatic Decompensation. Hepatology. 10.1002/hep.31652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz MS, Lajczak NK, Goggins BJ, Keely S, and Keely SJ (2018). The bile acids, deoxycholic acid and ursodeoxycholic acid, regulate colonic epithelial wound healing. Am J Physiol Gastrointest Liver Physiol 314, G378–G387. 10.1152/ajpgi.00435.2016 [DOI] [PubMed] [Google Scholar]
- Neurath MF (2019). Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol 20, 970–979. 10.1038/s41590-019-0415-0 [DOI] [PubMed] [Google Scholar]
- Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113. 10.1038/s41586-018-0257-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CJ, Wallace RG, Iwamoto K, Taguchi T, and Sunamoto J. (1985). Bile salt damage of egg phosphatidylcholine liposomes. Biochim Biophys Acta 817, 95–102. 10.1016/0005-2736(85)90072-0 [DOI] [PubMed] [Google Scholar]
- Okamoto R, and Watanabe M. (2005). Cellular and Molecular Mechanisms of the Epithelial Repair in IBD. Digest Dis Sci 50, S34–S38. 10.1007/s10620-005-2804-5 [DOI] [PubMed] [Google Scholar]
- Peterson LW, and Artis D. (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nature reviews. Immunology 14, 141–153. 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- Poulsen L, Siersbaek M, and Mandrup S. (2012). PPARs: fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol 23, 631–639. 10.1016/j.semcdb.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Raimondi F, Santoro P, Barone MV, Pappacoda S, Barretta ML, Nanayakkara M, Apicella C, Capasso L, and Paludetto R. (2008). Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am J Physiol Gastrointest Liver Physiol 294, G906–913. 10.1152/ajpgi.00043.2007 [DOI] [PubMed] [Google Scholar]
- Ramos GP, and Papadakis KA (2019). Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin Proc 94, 155–165. 10.1016/j.mayocp.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarenac TM, and Mikov M. (2018). Bile Acid Synthesis: From Nature to the Chemical Modification and Synthesis and Their Applications as Drugs and Nutrients. Front Pharmacol 9, 939. 10.3389/fphar.2018.00939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap FG, Trauner M, and Jansen PL (2014). Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol 11, 55–67. 10.1038/nrgastro.2013.151 [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, and Gonzalez FJ (2000). Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744. 10.1016/s0092-8674(00)00062-3 [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Yoon M, and Gonzalez FJ (2001). Antagonism of the actions of peroxisome proliferator-activated receptor-alpha by bile acids. J. Biol. Chem 276, 47154–47162. 10.1074/jbc.M107000200 [DOI] [PubMed] [Google Scholar]
- Sorrentino G, Perino A, Yildiz E, El Alam G, Bou Sleiman M, Gioiello A, Pellicciari R, and Schoonjans K. (2020). Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 159, 956–968 e958. 10.1053/j.gastro.2020.05.067 [DOI] [PubMed] [Google Scholar]
- Srivastava RA, Srivastava N, and Averna M. (2000). Dietary cholic acid lowers plasma levels of mouse and human apolipoprotein A-I primarily via a transcriptional mechanism. European journal of biochemistry 267, 4272–4280. 10.1046/j.1432-1033.2000.01473.x [DOI] [PubMed] [Google Scholar]
- Stojanović O, Altirriba J, Rigo D, Spiljar M, Evrard E, Roska B, Fabbiano S, Zamboni N, Maechler P, Rohner-Jeanrenaud F, et al. (2021). Dietary excess regulates absorption and surface of gut epithelium through intestinal PPARα. Nature communications 12, 7031. 10.1038/s41467-021-27133-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SH, Phuah P, Tan LT, Yada S, Goh J, Tomaz LB, Chua M, Wong E, Lee B, and Barker N. (2021). A constant pool of Lgr5(+) intestinal stem cells is required for intestinal homeostasis. Cell reports 34, 108633. 10.1016/j.celrep.2020.108633 [DOI] [PubMed] [Google Scholar]
- Thorne CA, Chen IW, Sanman LE, Cobb MH, Wu LF, and Altschuler SJ (2018). Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Developmental cell 44, 624–633.e624. 10.1016/j.devcel.2018.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallim TQ, and Edwards PA (2009). Bile acids have the gall to function as hormones. Cell metabolism 10, 162–164. 10.1016/j.cmet.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Violante S, Ijlst L, Te Brinke H, Koster J, Tavares de Almeida I, Wanders RJ, Ventura FV, and Houten SM (2013). Peroxisomes contribute to the acylcarnitine production when the carnitine shuttle is deficient. Biochim. Biophys. Acta 1831, 1467–1474. 10.1016/j.bbalip.2013.06.007 [DOI] [PubMed] [Google Scholar]
- Wei Y, Tian C, Zhao Y, Liu X, Liu F, Li S, Chen Y, Qiu Y, Feng Z, Chen L, et al. (2020). MRG15 orchestrates rhythmic epigenomic remodelling and controls hepatic lipid metabolism. Nat Metab 2, 447–460. 10.1038/s42255-020-0203-z [DOI] [PubMed] [Google Scholar]
- Wohlgemuth S, Keller S, Kertscher R, Stadion M, Haller D, Kisling S, Jahreis G, Blaut M, and Loh G. (2011). Intestinal steroid profiles and microbiota composition in colitic mice. Gut Microbes 2, 159–166. 10.4161/gmic.2.3.16104 [DOI] [PubMed] [Google Scholar]
- Yamamoto-Furusho JK, and Parra-Holguín NN (2021). Emerging therapeutic options in inflammatory bowel disease. World journal of gastroenterology 27, 8242–8261. 10.3748/wjg.v27.i48.8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhou S-Y, Gillilland M, Li J-Y, Lee A, Gao J, Zhang G, Xu X, and Owyang C. (2020). Bile acid toxicity in Paneth cells contributes to gut dysbiosis induced by high-fat feeding. JCI insight 5. 10.1172/jci.insight.138881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, Putzel GG, Kabata H, Kelsen JR, Baldassano RN, et al. (2019). Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409. 10.1038/s41586-019-1082-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene sequencing and RNA-seq data were deposited at SRA and GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-rabbit IgG-HRP | Cell Signaling Technology | Cat# 7074S; RRID: AB_2099233 |
| Goat anti-mouse IgG-HRP | Cell Signaling Technology | Cat# 7076S; RRID: AB_330924 |
| Goat anti-rabbit IgG Alexa Fluro 488 conjugate | Cell Signaling Technology | Cat# 4412; RRID: AB_1904025 |
| Goat anti-rabbit IgG Alexa Fluor 488 conjugate | Abcam | Cat# ab150077; RRID: AB_2890897 |
| Goat anti-rabbit IgG horseradish peroxidase conjugate | Servicebio | Cat# G1215 |
| Goat anti-mouse IgG BioMag | Qiagen | Cat# 310007 |
| Hamster anti-mouse CD11c APC conjugate | BD Biosciences | Cat# 550261; RRID: AB_398460 |
| Hamster anti-mouse CD28 | BD Biosciences | Cat# 553295; RRID: AB_394764 |
| Hamster anti-mouse CD3e | BD Biosciences | Cat# 553057; RRID: AB_394590 |
| Hamster anti-mouse CD3 PerCP-Cy5.5 conjugate | BD Biosciences | Cat# 551163; RRID: AB_ AB_394082 |
| HRP-conjugated monoclonal mouse anti-GAPDH | KangChen | Cat# KC-5G5 |
| Mouse monoclonal anti-Muc2 | Santa Cruz Biotechnology | Cat# sc-59859; RRID: AB_784653 |
| Mouse monoclonal anti-FXR/NR1H4 | Santa Cruz Biotechnology | Cat# sc-25309; RRID: AB_628039 |
| Rabbit monoclonal anti-Lysozyme | Abcam | Cat# ab108508; RRID: AB_10861277 |
| Rabbit monoclonal anti-Olfm4 | Cell Signaling Technology | Cat# 39141; RRID: AB_2650511 |
| Rabbit polyclonal anti-GFP | Abcam | Cat# ab290; RRID: AB_303395 |
| Rabbit polyclonal anti-CYP8B1 | Abcam | Cat# ab191910; RRID: AB_2828000 |
| Rabbit polyclonal anti-CYP7A1 | Abcam | Cat# ab65596; RRID: AB_1566114 |
| Rat Anti-Mouse CD16/CD32 | BD Biosciences | Cat# 553142 |
| Rat anti-mouse Ly-6G PE conjugate | BD Biosciences | Cat# 551461; RRID: AB_394208 |
| Rat anti-mouse CD45 BV510 conjugate | BD Biosciences | Cat# 563891; RRID: AB_2734134 |
| Rat anti-mouse CD11b BUV395 conjugate | BD Biosciences | Cat# 563553; RRID: AB_2738276 |
| Rat anti-mouse F4/80 FITC conjugate | Thermo Fisher Scientific | Cat# 11-4801-85; RRID: AB_2637192 |
| Rat anti-mouse CD326 (Ep-CAM) PE conjugate | Biolegend | Cat# 118205; RRID: AB_1134176 |
| 7-AAD | BD Biosciences | Cat# 559925; RRID: AB_2869266 |
| Bacterial and virus strains | ||
| AAV2/8-TBG-mCherry | Hanbio | N/A |
| AAV2/8-TBG-m-Cyp8b1-3xflag-null | Hanbio | N/A |
| AAV2/9-CMV-eGFP | Genomeditech | N/A |
| AAV2/9-CMV-m-Cyp8b1-shRNA | Genomeditech | N/A |
| Biological samples | ||
| Human: UC patient serum | Shanghai Tenth People’s Hospital | N/A |
| Human: Healthy adult serum | Shanghai Tenth People’s Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Alcian blue | Sigma-Aldrich | Cat#: 109-09; CAS: 33864-99-2 |
| Acetonitrile | Thermo Fisher Scientific | Cat#: A955-4; CAS: 75-05-8 |
| Bacitracin | meilunbio | Cat#: MB1374; CAS: 1405-87-4 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat#: V900933; CAS: 9048-46-8 |
| B27 | Gibco | Cat#: 17504044 |
| CHIR99021 | MedChemExpress | Cat#: HY-10182; CAS: 252917-06-9 |
| Chloroform | Sinopharm | Cat#: 10006818; CAS: 67-66-3 |
| Cholic acid (CA) | Sigma-Aldrich | Cat#: C1129; CAS: 81-25-4 |
| DAPI | Sigma-Aldrich | Cat#: D1306; CAS: 28718-90-3 |
| DAPI | Abcam | Cat#: ab104139; CAS: 28718-90-3 |
| Dextran sodium sulfate (DSS) | MP Biomedicals | SKU: 02160110-CF; CAS: 9011-18-1 |
| Dispase II | Sigma-Aldrich | Cat#: D4693; CAS: 42613-33-2 |
| DMEM/F12 | Gibco | Cat#: C11330500BT |
| DNase I | Sigma-Aldrich | Cat#: 10104159001 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#: D4540; CAS: 67-68-5 |
| d5-TCA | Toronto research chemicals | Cat#: T008852; CAS: 81-24-3 |
| Epidermal growth factor (EGF) | Peprotech | Cat#: 315-09 |
| Fetal bovine serum (FBS) | Gibco | Cat#: AHW87326 |
| FITC-dextran 4KD | Sigma-Aldrich | Cat#: 68059 |
| Fixative for TEM | Servicebio | Cat#: G1102 |
| Formic acid | Tokyo Chemical Industry | Cat#: F0654; CAS: 64-18-6 |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | Cat#: 100-0485 |
| Glutamax | Gibco | Cat#: 35050061 |
| GW501516 | MedChemExpress | Cat#: HY-10838; CAS: 317318-70-0 |
| GW6471 | MedChemExpress | Cat#: HY-15372; CAS: 880635-03-0 |
| Hematoxylin and eosin (H&E) | Servicebio | Cat#: G1003 |
| HEPES | Gibco | Cat#: 15630080 |
| IntestiCult Organoid Growth Medium (Mouse) with Supplement 1 and 2 | STEMCELL Technologies | Cat#: 06005 |
| Isopropanol | Thermo Fisher Scientific | Cat#: A461-4; CAS: 67-63-0 |
| L-Carnitine Inner Salt | Sigma-Aldrich | Cat#: C0158; CAS: 541-15-1 |
| Lipopolysaccharide (LPS) | Sigma-Aldrich | Cat#: 2880; CAS: 297-473-0 |
| Matrigel Matrix for Organoid Culture, Phenol Red-free, LDEV-free | Corning | Cat#: 356255 |
| Metronidazole | Meilunbio | Cat#: MB2200; CAS: 443-48-1 |
| Mouse colony-stimulating factor | Peprotech | Cat#: AF-315-02 |
| Methanol | Sigma-Aldrich | Cat#: 34860; CAS: 67-56-1 |
| 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazoliumbromide (MTT) | Sigma-Aldrich | Cat#: 2128; CAS: 298-93-1 |
| Na-Palmitate | Sigma-Aldrich | Cat#: P9767; CAS: 408-35-5 |
| N-acetylcysteine | Sigma-Aldrich | Cat#: A0737; CAS: 38520-57-9 |
| N2 | Gibco | Cat#: 17502001 |
| Neomycin | Meilunbio | Cat#: MB1716; CAS: 1405-10-3 |
| Nicotinamide | MedChemExpress | Cat#: HY-B0150; CAS: 98-92-0 |
| Noggin | Peprotech | Cat#: 250-38 |
| Obeticholic acid (OCA) | MedChemExpress | Cat#: HY-12222; CAS: 459789-99-2 |
| OsO4 | Ted Pella Inc. | Cat#: 18456; CAS: 20816-12-0 |
| Paraformaldehyde | Servicebio | Cat#: G1101 |
| Penicillin-Streptomycin | Gibco | Cat#: 15140122 |
| Periodic acid Schiff (PAS) | Servicebio | Cat#: G1008 |
| Pioglitazone | MedChemExpress | Cat#: HY-13956; CAS: 111025-46-8 |
| Protease inhibitor cocktail | Roche Life Science | Cat#: 11836170001 |
| RPMI 1640 | Gibco | Cat#: 72400047 |
| R-spondin-1 | Peprotech | Cat#: 315-32 |
| Scintillation cocktail | China Isotope & Radiation Corp. | Cat#: 1200-4 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | Cat#: V900859; CAS: 151-21-3 |
| Streptomycin | Meilunbio | Cat#: MB1275; CAS: 3810-74-0 |
| Taurocholic acid (TCA) | Sigma-Aldrich | Cat#: T4009; CAS: 345909-26-4 |
| Trinitron-benzene-sulfonic acid (TNBS) | Sigma-Aldrich | Cat#: P2297; CAS: 2508-19-12 |
| Triton X-100 | Beyotime | Cat#: ST795 |
| TrypLE | Gibco | Cat#: 12604013 |
| Type IV collagenase | Sigma-Aldrich | Cat#: C4-28 |
| UltraPure 0.5 M EDTA | Invitrogen | Cat#: 1557520 |
| WY14643 | MedChemExpress | Cat#: HY-16995; CAS: 50892-23-4 |
| Y-27632 | MedChemExpress | Cat#: HY-10071; CAS: 146986-50-7 |
| 4% paraformaldehyde | Servicebio | Cat#: G1101 |
| 5-Ethynyl-2’-deoxyuridine (Edu) | MedChemExpress | Cat#: HY-118411; CAS: 61135-33-9 |
| [9,10-3H]- palmitic acid | PerkinElmer | Cat#: NET043001MC |
| Critical commercial assays | ||
| BCA protein assay kit | Thermo Fisher Scientific | Cat#: 23255 |
| EDU staining kit | RiboBio | Cat#: C10310 |
| FIREFLYGLO Luciferase Assay Kit | Meilunbio | Cat#: MA0519 |
| Hieff qPCR SYBR Green Master Mix (High Rox) | Yeasen | Cat#: 11203ES08 |
| Hifair II 1st Strand cDNA Synthesis SuperMix (gDNA digester plus) | Yeasen | Cat#: 11123ES60 |
| In-situ cell death detection kit | Roche | Cat#: 11684795910 |
| LANCE® Ultra cAMP Kit | PerkinElmer | Cat#: TRF0262 |
| Mouse TNF-α ELISA kit | BD Biosciences | Cat#: 560478 |
| Mouse IL-6 ELISA kit | BD Biosciences | Cat#: 555240 |
| Mouse IFN-γ ELISA kit | BD Biosciences | Cat#: 555138 |
| Mouse IL-17 ELISA kit | Thermo Fisher Scientific | Cat#: BMS6001 |
| Mouse IL-1β ELISA kit | Thermo Fisher Scientific | Cat#: 88-7013-88 |
| QIAquick PCR purification kit | Qiagen | Cat#: 28104 |
| RNeasy mini kit | Qiagen | Cat#: 74106 |
| RNA simple total RNA kit | Tiangen Biotech | Cat#: DP419 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Fisher Scientific | Cat#: 34578 |
| Tissue protein extraction reagent | Thermo Fisher Scientific | Cat#: 78510 |
| Urine fecal occult blood test kit | Nanjing Jiancheng Bioengineering Institute | Cat#: C027-1-1 |
| Deposited data | ||
| The raw data of RNA-seq generated | This paper | GEO: GSE210290 |
| The raw data of 16S rRNA gene sequencing | This paper | SRA: PRJNA864099 |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Beijing Huafukang Bioscience Co. Inc. | N/A |
| Mouse: Il10-null | Gempharmatech | N/A |
| Mouse: Cyp8b1-null | Gempharmatech | N/A |
| Mouse: SCID | Gempharmatech | N/A |
| Mouse: Cas9LSL+/+ | The Jackson Laboratory | N/A |
| Mouse: Pparafl/fl | Luo et al., 2019 | N/A |
| Mouse: PparaΔIE | Luo et al., 2019 | N/A |
| Mouse: Fxr-null | Sinal et al., 2000 | N/A |
| Mouse: FxrΔIE | Kim et al., 2007 | N/A |
| Mouse: FxrΔHep | Kim et al., 2007 | N/A |
| Mouse: Lgr5-EGFP-IRES-creERT2 | Shanghai Model Organisms Center | NM-KI-200154 |
| Mouse: Tgr5-null | Provided by Prof. Xin Xie at Shanghai Institute of Materia Medica | N/A |
| Oligonucleotides | ||
| See Table S3 for qRT-PCR | This paper | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| Endnote | Endnote | http://endnote.com; |
| Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| FlowJo_V10 | BD Biosciences | https://www.flowjo.com/solutions/flowjo; |
| Leica Application Suite X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/details/product/leica-las-x-ls/ |
| NDP.view 2 | Hamamatsu | https://www.hamamatsu.com/jp/en/product/type/U12388-01/index.html |
| Pannoramic Viewer | 3DHISTECH | http://www.3dhistech.com/pannoramic_viewer |
| Prism 8 | GraphPad software | https://www.graphpad.com/ |
| Progenesis QI | Waters Corp. | http://www.nonlinear.com/progenesis/qi-for-proteomics/ |
| SIMCA 16 | Umetrics | http://umetrics.com/products/simca |
| Xcalibur | Thermo Fisher Scientific | http://chemistry.unt.edu/~verbeck/LIMS/Manuals/XCAL_Quant.pdf |
| Other | ||
| Acquity UPLC/Synapt XS HDMS system | Waters Corp. | N/A |
| Acquity BEH C18 column | Waters Corp. | SKU: 186002352 |
| Applied Biosystems 7900HT B Fast Real-Time PCR System | Applied Biosystems | N/A |
| BD LSRFortessa Fortessa Flow Cytometer | BD Biosciences | N/A |
| BD FACSAriaII Flow Cytometer | BD Biosciences | N/A |
| ChemiDoc MP Imaging System | Bio-Rad | N/A |
| Hitachi HT-7800 TEM | Hitachi | N/A |
| Illumina NovaSeq 6000 | Illumina | N/A |
| InfinityLab Poroshell 120 Hilic-Z column | Agilent technologies | 675775-924 |
| Leica TCS SPS microscope | Leica Microsystems | N/A |
| Leica DM6B laser microdissection systems | Leica Microsystems | N/A |
| LTQ Orbitrap mass spectrometer | Thermo Fisher Scientific | N/A |
| Olympus CKX53 microscope | Olympus | N/A |
| Spectramax M5 microplate reader | Molecular Devices | N/A |
| UPLC CSH column | Waters Corp. | SKU: 186005297 |
| Vanquish™ UHPLC | Thermo Fisher Scientific | N/A |
| 2450 Microplate Counter | PerkinElmer | N/A |